Polycomb group proteins play important roles in developmental and cell proliferation processes. Baumann et al. demonstrate that the PRC1 protein CBX2 is critical for heterochromatin homeostasis, chromosome stability, and the prevention of premature cellular senescence.
Abstract
The polycomb group protein CBX2 is an important epigenetic reader involved in cell proliferation and differentiation. While CBX2 overexpression occurs in a wide range of human tumors, targeted deletion results in homeotic transformation, proliferative defects, and premature senescence. However, its cellular function(s) and whether it plays a role in maintenance of genome stability remain to be determined. Here, we demonstrate that loss of CBX2 in mouse fibroblasts induces abnormal large-scale chromatin structure and chromosome instability. Integrative transcriptome analysis and ATAC-seq revealed a significant dysregulation of transcripts involved in DNA repair, chromocenter formation, and tumorigenesis in addition to changes in chromatin accessibility of genes involved in lateral sclerosis, basal transcription factors, and folate metabolism. Notably, Cbx2−/− cells exhibit prominent decondensation of satellite DNA sequences at metaphase and increased sister chromatid recombination events leading to rampant chromosome instability. The presence of extensive centromere and telomere defects suggests a prominent role for CBX2 in heterochromatin homeostasis and the regulation of nuclear architecture.
Introduction
Cellular senescence is a major contributor to tissue dysfunction and age-related decline in organismal health. Several lines of evidence support the notion that accumulating DNA damage causes a progressive deterioration of large-scale chromatin organization during cellular aging (Niedernhofer et al., 2018). Persistent double-strand breaks activate the DNA damage response (DDR), arresting cell cycle progression, and induce apoptosis or cellular senescence to prevent the accumulation of deleterious mutations and potential neoplastic transformation (Bastians, 2015; Rao et al., 2017; Tanaka and Hirota, 2016). Hence, genome instability may induce changes in cell fate decisions as an immediate adaptive mechanism to prevent cancer development (Heng et al., 2013).
The molecular mechanisms regulating changes in the chromatin landscape during cellular senescence are not fully understood. However, changes in chromatin structure and function induced by altered epigenetic profiles are important contributing factors to the onset of cellular aging (O’Sullivan and Karlseder, 2012). Polycomb group (PcG) proteins are major epigenetic regulators essential for establishing heritable gene expression patterns and lineage commitment. As such, PcG proteins play an important role in cell proliferation, maintenance of adult stem cell populations, and regulation of stem cell self-renewal (Margueron and Reinberg, 2011; Mas and Di Croce, 2016; Morey et al., 2012; O’Loghlen et al., 2012; Pereira et al., 2010; Richly et al., 2011). PcG proteins aggregate dynamically to form two major enzymatic complexes, polycomb repressive complex (PRC) 1 and 2, and exhibit a critical role in the organization of chromatin structure (Cao et al., 2002; Simon and Kingston, 2013) and nucleosome compaction (Francis et al., 2004). The chromobox homologue protein 2 (CBX2) is a critical component of the PRC1 complex involved in antiviral innate immunity (Sun et al., 2019), neuronal (Gu et al., 2018), and gonadal differentiation (Garcia-Moreno et al., 2018; Katoh-Fukui et al., 1998, 2012), axial patterning during embryonic development, cell proliferation, and senescence (Coré et al., 1997, 2004). As an active subunit of the mammalian PRC1 complex, CBX2 is capable of inducing chromatin compaction in vitro (Tatavosian et al., 2015, 2019). Live cell imaging in mouse embryonic stem cells (mESCs) revealed that recombinant CBX2 associates with mitotic chromosomes, where it is critical to recruit additional members of the PRC1 complex (Zhen et al., 2014). We have previously shown that CBX2 knockout mice exhibit reduced germ cell viability, defects in DNA repair, and widespread chromosome abnormalities during meiosis (Baumann and De La Fuente, 2011). However, the mechanisms leading to deleterious chromosomal rearrangements and whether CBX2 plays a role in large-scale chromatin organization or chromatin accessibility in somatic cells remained to be determined. Importantly, the effects of loss of CBX2 on heterochromatin formation and nuclear architecture in vivo remained unexplored.
Here, we demonstrate that CBX2 is essential for the prevention of senescence-associated chromosomal rearrangements in mouse embryonic fibroblasts (MEFs). Loss of CBX2 leads to disruptions of nuclear architecture and large-scale chromatin structure and results in transcriptional dysregulation of zinc finger and homeobox transcription factors (TFs) as well as genes involved in retinoic acid (RA) response, DNA repair, cell cycle regulation, and tumorigenesis. Moreover, assays for transposase-accessible chromatin using sequencing (ATAC-seq) revealed that chromatin accessibility at regulatory elements, such as CpG islands at transcription start sites (TSSs), of several key genomic loci differs in Cbx2−/− fibroblasts at early passage and thus precedes the onset of senescence and genome instability. Senescent Cbx2−/− cells exhibit increased illegitimate mitotic recombination events and rampant chromosome instability indicative of break-fusion-bridge cycles leading to translocations and complex chromosomal rearrangements. These results reveal a critical role in maintenance of chromosome stability during cell division and the prevention of senescence-associated chromosomal rearrangements, providing important novel insight into the mechanisms leading to the onset of premature senescence in CBX2-deficient cells.
Results
CBX2 is required for maintenance of chromosome stability
Targeted deletion of Cbx2 induces proliferative defects in lymphoid precursor cells as well as cellular senescence as detected by increased β-galactosidase activity and abnormal accumulation of the cyclin-dependent kinase inhibitor p16INK4a (Coré et al., 2004). However, the mechanisms leading to the early senescent phenotype remained unexplored. The widespread chromosome abnormalities observed during meiosis in Cbx2−/− male and female germ cells (Baumann and De La Fuente, 2011) suggest a potential function in maintenance of chromosome stability in mitotic cells. Thus, we tested whether replicative senescence in CBX2-deficient cells is associated with the presence of numerical and/or structural chromosome abnormalities. Comparison of wild-type and Cbx2−/− MEFs at passage 2 (P2) revealed no significant differences in the proportion of metaphase spreads exhibiting chromosome breaks (10.4% and 11.9%, respectively; Fig. 1, A and B). However, analysis at P5 revealed a significant increase (P < 0.005) in the proportion of Cbx2−/− cells (58%) that exhibit both chromosome breaks and large-scale chromosomal rearrangements compared with (10.1%) wild-type controls (Fig. 1, A and B).
Figure 1.
Chromosome instability in Cbx2−/− fibroblasts. (A–D) Metaphases obtained from wild-type (n = 58) or Cbx2−/− (n = 68) fibroblast cultures showing base levels of chromosomal instability at P2. Culture to P5 significantly increases the incidence of chromosomal breaks and fragments (arrows, insets), fusions (arrowheads, inset) and premature centromere separation (asterisks, inset) in Cbx2−/− fibroblasts (n = 76) compared with wild-type controls (n = 68). Experiments were conducted in triplicate, and differences were considered statistically significant when P < 0.05 (two-way ANOVA and paired t tests). (E–F) Chromosome instability results in micronuclei formation (arrows) in interphase nuclei at P5 in Cbx2−/− (n = 76) compared with wild-type (n = 66) cells. (G–H) Loss of CBX2 significantly increases the number of γH2AX foci (green, arrows) per nucleus as well as the proportion of nuclei with >10 γH2AX foci in Cbx2−/− nuclei (n = 97) at P5 compared with wild-type controls (n = 115). The arrowheads mark a micronucleus. Experiments were conducted in triplicate, and differences were considered statistically significant when P < 0.05 (paired t tests).
Wild-type chromosomes demonstrated intact DAPI-stained pericentric heterochromatin domains and no evidence of large-scale chromosomal abnormalities at P5 (Fig. 1 A, inset). However, loss of CBX2 led to the emergence of chromosome breaks proximal to centromeric domains as well as a significant increase in the proportion (16%; P < 0.05) of metaphase cells with striking defects resembling end-to-end chromosome fusions (Fig. 1, A and C, arrowhead and inset). In addition, severe cases demonstrated catastrophic karyotypic aberrations including chromosome fragmentation (Fig. 1 A, arrows and insets), loss of sister chromatid cohesion (asterisk), and premature centromere separation (Fig. 1, A and D; 15%). These findings indicate that loss of CBX2 predisposes cells to chromosome instability and the formation of large-scale chromosomal rearrangements at P5. Consistent with the presence of structural chromosome aberrations, Cbx2−/− fibroblasts also exhibit a significant increase in the proportion of interphase nuclei (37%; P < 0.05) that exhibit micronuclei formation (Fig. 1 E, arrows; and Fig. S1) compared with only 17% in wild-type cells at P5 (Fig. 1 F). Moreover, analysis of DNA double strand breaks in interphase nuclei at P5 revealed a significant increase (P < 0.05) in the proportion of Cbx2−/− fibroblasts (18%) that accumulate double strand breaks with 10 or more γH2AX foci per nucleus as compared with wild-type cells (7%; Fig. 1, G and H, arrows). These results provide the initial evidence for a critical role of CBX2 in maintaining chromosome stability during mitotic cell division, and the prevention of prominent DNA damage and micronuclei formation.
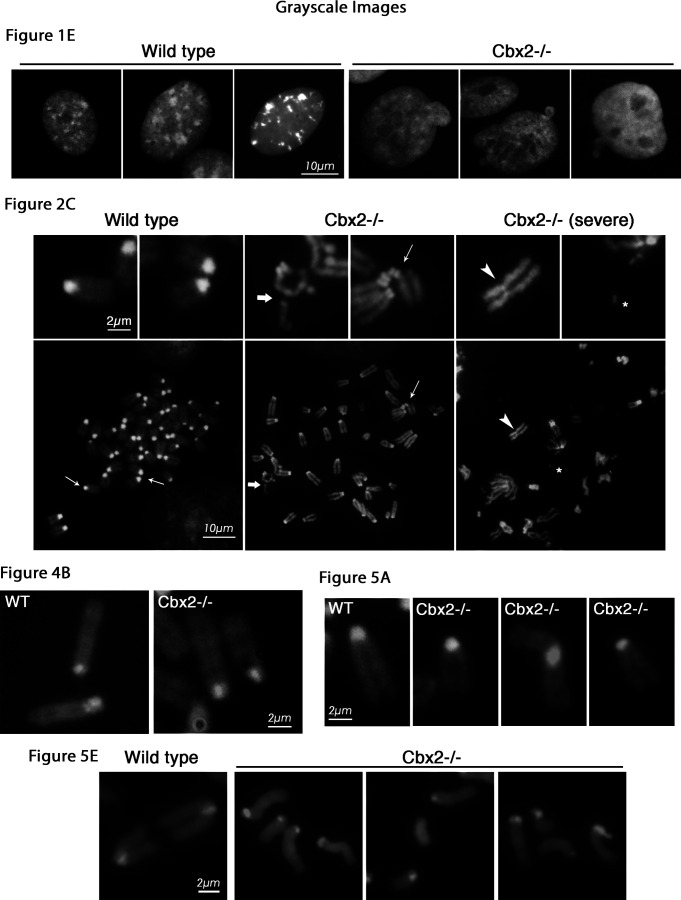
Figure S1.
Grayscale images. Additional example images (DAPI in grayscale) of wild-type and Cbx2−/− fibroblast nuclei to Fig. 1 E.Image compilation of DAPI grayscale-only images of chromosomes in Figs. 1, 2, 3, 4, and 5.
Loss of CBX2 induces abnormal centromere heterochromatin structure and nonrecurrent chromosome translocations
The high incidence of premature centromere separation defects in CBX2-deficient cells suggested that abnormal constitutive heterochromatin function might be an important mechanism leading to chromosome instability in this model. Thus, we conducted FISH with a major satellite DNA probe (Baumann et al., 2010). In wild-type fibroblasts at P5, major satellite DNA sequences are detected as a large domain of highly compact pericentric heterochromatin on each acrocentric metaphase chromosome (Fig. 2, A and C). In contrast, Cbx2−/− fibroblasts exhibit prominent signs of centromere instability associated with a spectrum of chromosome aberrations. For example, we found evidence for the presence of single chromatids lacking major satellite DNA (Fig. 2 C, bold arrow, inset) as well as recurrent breakpoints within or proximal to pericentric heterochromatin (Fig. 2, B and C, thin arrow, inset). Notably, in Cbx2−/− fibroblasts exhibiting severe chromosome defects, extreme centromere erosion resulted in the almost complete loss of satellite DNA and formation of chromosome fusions resembling a Robertsonian translocation (Fig. 2, B and C, arrowhead, inset). In addition, we found evidence for the presence of chromatin fragments containing major satellite DNA sequences that had become completely detached from metaphase chromosomes (Fig. 2, B and C, asterisk, inset). These results indicate that Cbx2−/− cells exhibit a major transition toward genome instability at P5. Importantly, our results also indicate that centromere erosion due to loss of satellite DNA and centromere breaks are important contributing factors to the onset of chromosome instability in CBX2-deficient cells.
Figure 2.
Centromeric heterochromatin aberrations in Cbx2−/− metaphases. (A) Illustration of major satellite DNA-FISH signals in normal chromosomes and appearance of defects such as double strand breaks within and erosion of centromeric sequences. (B and C) Major satellite FISH demonstrates chromosome breaks at centromeric and pericentric heterochromatin domains (red) in Cbx2−/− fibroblasts (arrow) as well as presence of single chromatids lacking major satellite DNA (bold arrow) and detached chromosome fragments consisting of major satellite sequences (asterisk) in Cbx2−/− (n = 48) compared with wild-type (n = 49) metaphases. Extreme centromere erosion leads to the formation of chromosome fusions (arrowhead). Data represent the mean values ± SD of three biological replicates. (D) Quantitative centromere size measurements following major satellite FISH (red) using threshold masks (green) reveal centromere decondensation leading to (E) an increased average EqDiameter and (F) reduced sphericity in Cbx2−/− metaphase chromosomes (n = 579) as compared with wild-type chromosomes (n = 1,259). Data represent the mean values ± SD of three biological replicates. KO, knockout.
CBX2 is the only PcG protein known to stably bind chromatin during mitosis (Zhen et al., 2014), suggesting a potentially important role in chromosome structure. Thus, we determined whether loss of CBX2 is associated with pericentric heterochromatin decondensation in mitotic chromosomes. Major satellite DNA domains (red) of wild-type metaphase chromosomes were analyzed using automated image analysis software (NIS elements; Nikon) and compared with data obtained from Cbx2−/− chromosomes using threshold masks (Fig. 2 D, green). The average equivalent diameter (EqDiameter) of pericentric heterochromatin domains in mitotic chromosomes was significantly larger (P < 0.001) in metaphase spreads of Cbx2−/− fibroblasts compared with wild-type cells (Fig. 2 E). In addition, chromatin domain sphericity, a measure of circularity based on area and perimeter computations, was significantly decreased (P < 0.001) in knockout chromosomes (Fig. 2 F), indicating that loss of CBX2 induces decondensation of pericentric heterochromatin, leading to poorly defined, irregularly shaped heterochromatin domains.
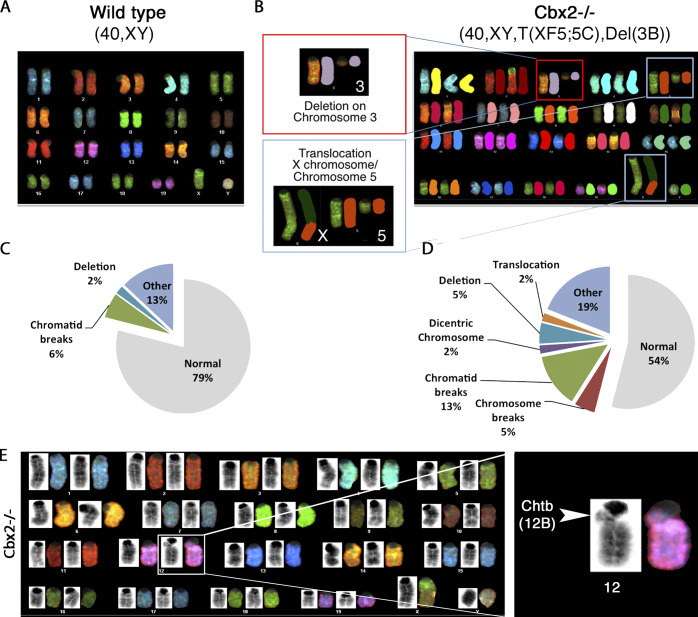
Next, we used spectral karyotyping (SKY) to conduct a genome-wide analysis of numerical and structural chromosome abnormalities and to determine the type of structural chromosome rearrangements associated with centromere instability in Cbx2−/− fibroblasts. Chromosome anomalies were classified according to the International Committee on Standardized Genetic Nomenclature for Mice, and the positions of chromosomal breakpoints were designated according to the standard G-banded karyotype of the mouse (Fig. 3). SKY analysis revealed a normal diploid (2n = 40, XY) chromosome complement in the majority (28 out of 30; 93.3%) of wild-type fibroblasts at P5 (Fig. 3 A). Notably, a similar proportion (26 out of 30; 86.6%) of Cbx2 mutant cells retained a diploid chromosome complement with only 13.3% of cells exhibiting a tetraploid (4n = 80; XXYY) chromosome number. However, we found evidence for the presence of more than one large-scale chromosomal rearrangement in the same cell in Cbx2−/− fibroblasts (Fig. 3 B). A representative example of the different types of large-scale rearrangements found in Cbx2−/− cells revealed the presence of a deletion in chromosome 3 resulting from a double strand break on band 3B, proximal to the centromere, resulting in the loss of most of the long arm in one copy of chromosome 3 (Del [3B]) albeit with retention of its centromere. Notably, the same cell also exhibited a double strand break in chromosome 5 (band 5C) resulting in an insertional translocation of a portion of chromosome 5 and its fusion to band F5 of the X chromosome leading to the formation of a derivative T(XF5;5C) chromosome (Fig. 3 B). These results indicate that chromosome or chromatid breaks, proximal to the centromere and/or pericentric heterochromatin domains in Cbx2−/− fibroblasts, result in the formation of complex chromosomal rearrangements, chromosome deletions, and translocations by P5. In contrast, no large-scale chromosomal translocations were detected in wild-type fibroblasts, although deletions of minute chromosome bands and chromatid breaks were present in a small proportion (2% and 6% of cells, respectively; Fig. 3 C). However, we found evidence for the presence of structural chromosome abnormalities in 46% of Cbx2−/− fibroblasts analyzed. Importantly, the type of nonrecurrent chromosome translocations detected by SKY analysis (Fig. 3 D) and the high proportion of chromosome (5%) and single chromatid breaks observed (13%; Fig. 3 E) suggest that large-scale chromosomal rearrangements are a result of centromere instability leading to break-fusion-bridge cycles in this model.
Figure 3.
Large-scale rearrangements in Cbx2−/− chromosome complements. (A and B) SKY analysis results from wild-type (A) and Cbx2−/− (B) fibroblast metaphase spreads at P5. Cbx2−/− metaphases present evidence for extensive, nonrecurrent chromosomal aberrations. The representative Cbx2−/− example shows a large deletion in chromosome 3 resulting from a double strand break in band 3B proximal to the centromere (Del[3B]). The same metaphase also exhibits a translocation of a portion of chromosome 5 (break at band 5C) onto band F5 on the long arm of the X chromosome, giving rise to the formation of a derivative T(XF5;5C) chromosome. (C and D) Proportion of metaphases with different types of chromosomal aberrations in wild-type and Cbx2−/− fibroblasts at P5 (n = 30 per sample). “Other”: acentric fragments, tri-radial figures, as well as chromosome and chromatids gaps. (E) G-banded karyotype obtained through SKY analysis of Cbx2−/− fibroblast metaphases at P5. The magnification shows a chromatid break within pericentric heterochromatin on the long arm of chromosome 12.
Increased centromere mitotic recombination in Cbx2−/− fibroblasts
To gain insight into the potential mechanisms leading to centromere instability, we used chromosome orientation FISH (CO-FISH) with strand-specific peptide nucleic acid (PNA) probes for centromeric minor satellite sequences (Bailey et al., 2010; De La Fuente et al., 2015) to detect the presence and quantify the frequency of centromere sister chromatid exchanges (C-SCE) resulting from illegitimate mitotic recombination. C-SCE were quantified following BrdU incorporation in wild-type and Cbx2−/− fibroblasts. After one cycle of DNA replication, the chromatid containing BrdU was digested by strand-selective exonuclease III cleavage, and a single stranded minor satellite probe against the leading strand was hybridized. Centromeres without occurrence of a chromatid exchange event exhibit a single minor satellite signal (Fig. 4, A and B). However, two minor satellite signals in the same chromosome can be detected following sister chromatid exchanges (Fig. 4, A and B). Comparison of centromeric mitotic recombination frequencies revealed that the majority of wild-type chromosomes (>90%) showed no evidence for the presence of C-SCE (Fig. 4 B), although basal levels of chromatid exchanges can be detected at low frequencies in wild-type chromosomes (Fig. 4 C). Notably, prominent double signals were detected at centromeric regions in nearly 40% of Cbx2−/− chromosomes, reflecting a significant (P < 0.05) increase in the frequency of C-SCE (Fig. 4, B and C). The number of minor satellite signals per chromosome also showed a significant increase (P < 0.05) in Cbx2−/− (1.38) compared with wild-type (1.09) chromosomes (Fig. 4 D). These results indicate that loss of CBX2 increases the frequency of illegitimate centromeric mitotic recombination events at minor satellite sequences and that impaired chromatin structure at tandem repeats leads to DNA double strand breaks and centromere instability. Notably, Cbx2−/− cells also exhibit a higher frequency (26%; P < 0.05) of chromosomes with abnormal recombination at euchromatin regions following BrdU incorporation as well as a significant increase in the average number of sister chromatid exchanges per chromosome (0.27; P < 0.05) compared with wild-type cells (19% and 0.2, respectively; Fig. 4 E, arrowheads).
Figure 4.
High incidence of centromere recombination in Cbx2−/− fibroblasts. (A) Schematic representation of CO-FISH (red) signal appearance in normal chromosomes (left) and following C-SCEs (two signals = recombined) within minor satellite sequences. (B and C) In Cbx2−/− chromosomes (n = 1,416), C-SCEs occurred with significantly increased frequency as indicated by double FISH signals (B, arrowheads; (C) and an increased number of minor satellite signals per chromosome (D) compared with wild-type controls (n = 1,440), in which the vast majority of chromosomes demonstrate single CO-FISH signals (arrows in B and C). Data represent the mean values ± SD of three biological replicates. (E) SCEs within euchromatic regions of chromosomes were analyzed using BrdU immunodetection (red) in metaphase spreads after pulse-incorporation of BrdU nucleotides. The proportion of chromosomes with SCE is significantly higher in Cbx2−/− chromosomes (n = 599) compared with wild-type controls (n = 688). Characteristic patterns for absence of SCE events (“no SCE”) with sister chromatids clearly distinguishable by bright or pale staining and examples of chromosomes displaying one or two SCE (arrowheads) are shown. Data represent the mean values ± SD of three biological replicates. KO, knockout.
Increased telomeric mitotic recombination leads to chromosome end-to-end fusions in Cbx2−− fibroblasts
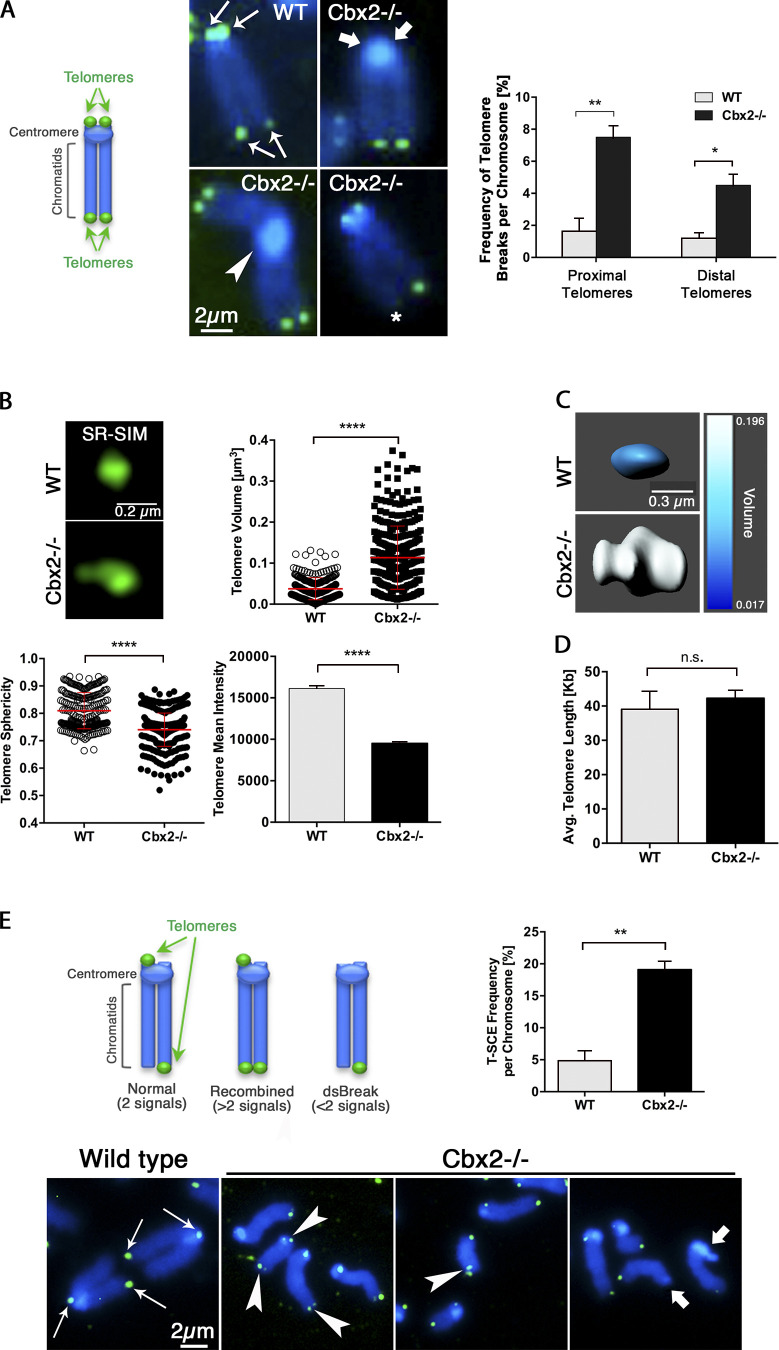
The type of chromosome fusions observed in Cbx2−/− fibroblasts suggests that telomeric sequences proximal to the centromere may also be affected. Thus, to assess potential defects in telomere structure, we used FISH with a PNA probe complementary to the telomeric DNA repeat sequence (TTAGGG). In wild-type metaphase spreads, intact chromosome ends were identified by the presence of four signals per chromosome (a single telomere signal proximal and distal to the centromere on each chromatid; Fig. 5 A, arrows). In contrast, we detected a spectrum of telomere defects in Cbx2−/− fibroblasts such as complete loss of proximal telomeric repeat sequences (bold arrows), distal telomere-free ends in one or both chromatids (identified by loss of telomere signal; Fig. 5 A, asterisk), as well as major chromosome rearrangements leading to the formation of centromere–centromere fusions resembling a Robertsonian translocation with large centromeres (Fig. 5 A, arrowhead). From the total number of individual chromosomes evaluated in wild-type (n = 1,729) and Cbx2−/− fibroblasts (n = 1,456), we found a similar low frequency (≤1.7%) of chromosome breaks at proximal and distal telomeres in wild-type cells. In contrast, 12% of the total number of chromosomes evaluated in Cbx2−/− fibroblasts exhibited evidence of breaks at proximal (7.5%) but also distal (4.5%) telomeres (Fig. 5 A). These results indicate that loss of CBX2 function is associated with a higher frequency of proximal telomere breaks. However, distal telomeres are also disrupted.
Figure 5.
Telomere dysfunction in Cbx2−/− fibroblasts. (A) Schematic representation of Telomere DNA-FISH signals (green) in mouse chromosomes. The position of n = 4 telomeres (arrows) per chromosome in wild-type chromosomes is indicated. Telomere-FISH analysis of P5 mitotic metaphase spreads indicates increased frequency of proximal (bold arrows) and distal (asterisk) telomere breaks in Cbx2−/− chromosomes compared with wild-type controls. The arrowhead indicates a Robertsonian-like centromere–centromere fusion in a Cbx2−/− metaphase. (B) SR-SIM imaging and quantitative measurement analysis of telomere volume, sphericity, and fluorescence intensity in wild-type (n = 239) and Cbx2−/− (n = 341) chromosomes. Data represent the mean values ± SD of three biological replicates. (C) Representative individual telomeres are shown following 3D surface rendering analysis using Imaris, including color scale–indicated volume differences. (D) Absolute telomere length quantification of wild-type and Cbx2−/− MEFs. Data represent the mean values ± SD of three independent biological replicates. (E) Schematic representation of telomere orientation-FISH signal appearance in normal and in chromosomes with occurrence of T-SCEs (more than two signals per chromosome = recombined) or double strand breaks (dsBreaks, less than two signals). Representative chromosomes of wild-type Cbx2 fibroblasts showing a single telomere signal (green) per chromatid (arrows). Significantly increased rates of T-SCEs (arrowheads) and signal-free chromatids as a result of chromosome breaks (bold arrows) in Cbx2−/− chromosomes (n = 968) compared with wild-type controls (n = 1,021). Data represent the mean values ± SD of three independent biological replicates.
Next, we used superresolution structured illumination (SR-SIM) and quantitative image analysis software (Imaris 9.3) to generate 3D renderings of telomere structure. Wild-type cells exhibit highly compact and spherical telomeres. In contrast, Cbx2−/− fibroblasts at P5 exhibit irregularly shaped, decondensed telomeres with areas of stretched chromatin (Fig. 5 B). Telomere volume, mean fluorescence intensity, and telomere sphericity (a measure of chromatin compaction) were compared between wild-type and Cbx2−/− metaphase chromosomes. Telomere mean volume was significantly increased in Cbx2−/− chromosomes (P < 0.001), while mean sphericity and fluorescence intensity showed decreased values (P < 0.001) compared with wild-type controls (Fig. 5 B). Importantly, these changes in telomere chromatin organization were also detected as a switch from blue to gray on the volume scale used to measure the 3D renderings of super-resolved telomeres (Fig. 5 C). To determine whether Cbx2−/− chromosomes also exhibit a global reduction of the average telomere length, we conducted absolute telomere length quantification assays on genomic DNA from wild-type and knockout fibroblasts at P5 (Fig. 5 D). Analysis of the average telomere length in kilobases using quantitative real-time PCR revealed no significant differences between wild-type and Cbx2−/− fibroblasts. These results indicate that loss of CBX2 induces decondensation of telomeric heterochromatin and formation of irregularly shaped telomeres by P5. Moreover, these results also indicate that large-scale structural defects might predispose to the loss of repetitive telomeric DNA sequences at both proximal and distal telomeres in a few chromosomes, leading to the formation of Robertsonian-like or end-to-end fusions. Notably, although telomere structural defects are detectable at P5, the average absolute telomere length remained unchanged, suggesting that telomere erosion in Cbx2−/− fibroblasts takes place progressively in a few chromosomes and before global changes in absolute telomere length.
Telomere repeat regions are subject to stringent regulatory mechanisms to prevent illegitimate recombination (Jaco et al., 2008). Thus, we used telomere CO-FISH to determine the mechanisms of progressive telomere erosion in Cbx2−/− chromosomes (Fig. 5 E). Telomere CO-FISH revealed that the frequency of telomere sister chromatid exchanges (T-SCEs) is only 4.9% per chromosome in wild-type fibroblasts (Fig. 5 E). However, T-SCE frequency increased significantly to 19.1% in Cbx2-deficient chromosomes (Fig. 5 E, arrowheads). Consistent with our conventional telomere FISH analysis, signal-free chromosome ends confirmed the presence of telomere DNA breaks (Fig. 5 E, bold arrows). These data indicate that loss of CBX2 interferes with proper telomeric heterochromatin compaction, leading to decondensed, fragile telomeres that exhibit increased levels of illegitimate mitotic recombination.
Loss of CBX2 disrupts nuclear architecture and induces senescence-associated distention of satellite DNA sequences (SADS)
Higher order chromatin organization is essential for the maintenance of chromosome stability in mammalian cells (O’Sullivan and Karlseder, 2012). However, senescent cells undergo dramatic changes in DNA organization and chromosome packaging (Freund et al., 2012; Swanson et al., 2013).
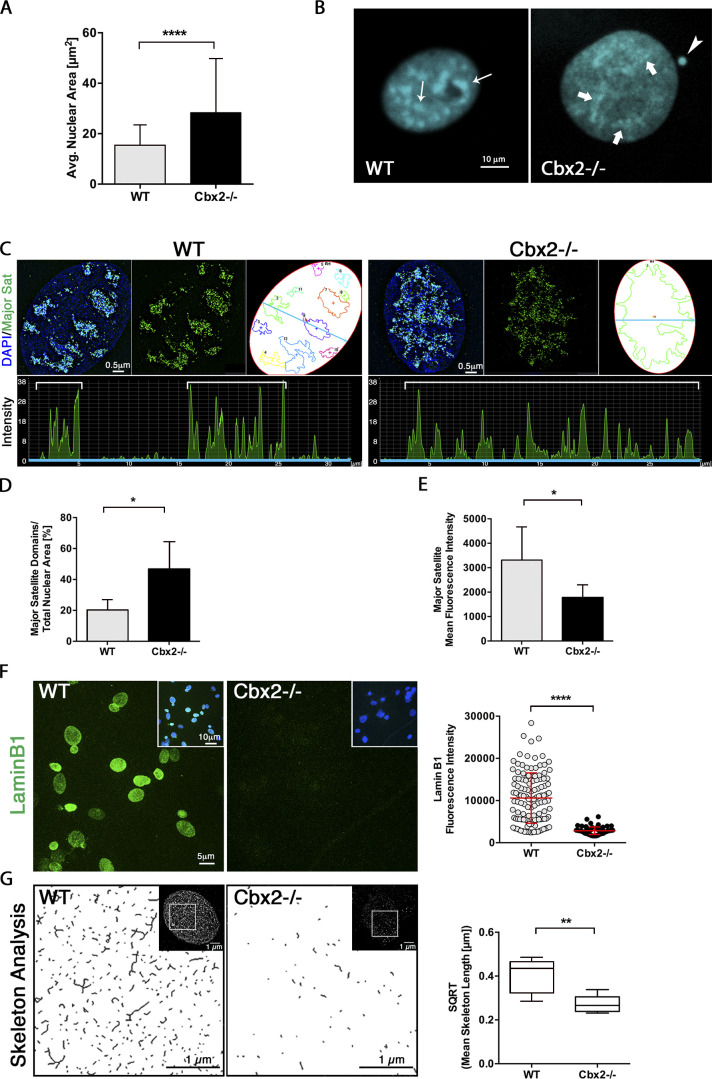
Confocal microscopy suggested that loss of CBX2 induced striking changes in the topological organization of pericentric heterochromatin domains at P5. To undertake a comprehensive analysis of nuclear architecture in single cells but with high throughput imaging, we used automated confocal microscopy in conjunction with multi-parameter image processing and high-content analysis (HCA). Notably, HCA revealed a significant increase (P < 0.001) in the nuclear area of Cbx2−/− fibroblasts (28.4 µm2) compared with wild-type cells (15.5 µm2; Fig. 6 A). Moreover, in wild-type cells at interphase, heterochromatin domains are organized as densely packed DAPI-stained chromocenters (Fig. 6 B, arrows). In contrast, Cbx2−/− fibroblasts exhibit striking changes in higher-order chromatin structure including loss of chromocenter organization and formation of highly decondensed pericentric heterochromatin (Fig. 6 B, bold arrows). Next, we determined whether the loss of chromocenter compaction in Cbx2−/− fibroblasts is associated with topological changes in the large-scale organization of major satellite sequences. SR-SIM provides a lateral resolution of approximately twice that of diffraction-limited microscopes and an axial resolution ranging between 150 and 200 nm, providing a powerful system for the analysis and reconstruction of topological chromatin organization and its interaction with the nuclear lamina (Schermelleh et al., 2008). Distention of SADS is a consistent and potentially ubiquitous marker of cellular senescence recently described in both human and mouse cells (Swanson et al., 2013). However, analysis of SADS higher-order chromatin unfolding beyond the diffraction limit of confocal microscopy remains unexplored. Using super-resolution analysis, networks of higher-order chromatin fibers can be resolved. Detection of major satellite sequences using DNA-FISH in combination with SR-SIM revealed that wild-type fibroblasts exhibit clearly delineated chromocenters that are formed by loops of tightly condensed chromatin fibers (Fig. 6 C). Consistent with the spatial separation of individual chromocenters, line-scan analysis revealed intermittent peaks of fluorescence intensity in the nucleoplasm. In contrast, major satellite sequences in Cbx2−/− fibroblasts were highly decondensed and, at times, fused into a large network of loosely connected chromatin fibers. Notably, line-scan analysis revealed the presence of continuous peaks of lower fluorescence intensity (Fig. 6 C). Consistent with a higher-order chromatin unfolding, we found evidence for a significant increase in the total nuclear area occupied by major satellite sequences in Cbx2−/− fibroblasts (46.8%, P < 0.05) compared with (20.5%) in wild-type fibroblasts, as well as a significant reduction in the average fluorescence intensity of major satellite sequences (Fig. 6, D and E) by both super-resolution and confocal microscopy. These findings are consistent with a phenotype of SADS, a hallmark of early nuclear chromatin changes during senescence (Swanson et al., 2013, 2015). Thus, loss of CBX2 induces striking changes in nuclear architecture and the large-scale organization of satellite DNA sequences by P5.
Figure 6.
Altered nuclear architecture and distention of satellite DNA sequences in Cbx2−/− cells. (A) Changes in nuclear architecture in Cbx2−/− cells (n = 63) are associated with an increased nuclear area compared with wild-type control cells (n = 120) as assessed by HCA. Data represent the mean values ± SD of three biological replicates and were analyzed using Mann–Whitney tests. (B) Confocal microscopy reveals large-scale decondensation of (DAPI-bright) heterochromatin blocks in Cbx2−/− nuclei (bold arrows) compared with wild-type nuclei presenting densely packed chromocenters (thin arrows). The arrowhead indicates a micronucleus in a Cbx2−/− fibroblast. (C) SR-SIM imaging of DNA-FISH–labeled pericentric heterochromatin (green) at DAPI-bright nuclear domains (blue). Threshold masks indicate areas detected by the major satellite FISH probe, and line scan graphs depict intermittent fluorescence intensity peaks corresponding to a cross-section of the nucleus (blue line). (D and E) The proportion of the nuclear area occupied by pericentric heterochromatin (D) and the mean fluorescence intensity of these domains (E) in wild-type (n = 5) and Cbx2−/− (n = 5) nuclei. (F) SR-SIM and HCA indicate a significant reduction in Lamin B1 (green) fluorescence intensity in Cbx2−/− fibroblasts at P5 (n = 68) compared with wild-type controls (n = 123). The insets show merged images of Lamin B1 (green) and DAPI (blue). (G) Skeleton analysis revealed significantly decreased mean Lamin B1 skeleton lengths in Cbx2−/− nuclei (n = 6) compared with wild-type controls (n = 9). Images represent magnified areas of the nuclei as indicated in the insets.
To test whether premature senescence in Cbx2−/− cells is also associated with abnormal structure and function of the nuclear lamina, we used high content confocal microscopy in conjunction with SR-SIM to quantify and structurally analyze Lamin B1 expression (Fišerová et al., 2019). HCA revealed a significant reduction (P < 0.001) in Lamin B1 fluorescence intensity in Cbx2−/− nuclei compared with wild-type controls (Fig. 6 F). SR-SIM revealed the presence of a dense network of Lamin B1 filaments distributed throughout the nuclear lamina in wild-type fibroblasts (Fig. 6 G). In contrast, and consistent with our HCA, Cbx2−/− fibroblasts exhibit a significant reduction in both the size and density of Lamin B1 filaments as detected by a Skeleton algorithm specifically designed for quantitative analysis of segmented, binary images of nuclear lamina filaments (Fišerová et al., 2019), confirming the presence of significantly shorter (P < 0.01) Lamin B1 branches (skeletons) in Cbx2−/− nuclei (Fig. 6 G). These findings indicate that loss of CBX2 is associated with several hallmarks of premature senescence such as altered nuclear architecture, SADS formation, and abnormal expression of key components of the nuclear lamina.
Altered expression levels of regulatory factors involved in DNA damage, chromosome instability, and tumorigenesis in Cbx2−/− fibroblasts
To determine the mechanisms leading to chromosome instability, we compared the transcriptomes of wild-type and Cbx2−/− fibroblasts at P2 using RNA sequencing (RNA-seq). This approach allowed for the comprehensive analysis of genome-wide transcriptional changes in mutant cells before the onset of chromosome instability and premature senescence. We identified 294 differentially expressed genes (DEGs) in Cbx2−/− fibroblasts (Fig. 7 A), of which 175 genes were significantly up-regulated (red, P < 0.05) and 119 genes were significantly down-regulated (blue, P < 0.05). Replicate samples clearly clustered by genotype as indicated by the heatmap and showed a high Pearson correlation coefficient (0.996) between replicates (Fig. 7 B and Fig. S2). Kyoto Encyclopedia of Genes and Genomes (Kegg) pathway analysis revealed an over-representation for papilloma viral infection, proteoglycans in cancer, and focal adhesion signaling pathways in Cbx2−/− MEFs (Fig. 7 C). Importantly, gene ontology (GO) analysis revealed an enrichment for biological processes, such as cell growth and death (17 genes), cancer (41 genes), DNA replication, repair, and aging, immunological factors, signal transduction, and amino acid metabolism (Fig. 7 D and Fig. S2), indicating that CBX2 plays a role in regulating the expression of factors associated with maintenance of genome stability and the prevention of neoplastic transformation. A total of 36 TFs were differentially expressed in Cbx2−/− MEFs compared with wild-type cells (Fig. 7 E). These TFs clustered preferentially into two main TF families: zinc finger zf-C2H2 and homeobox TFs (Fig. 7 F). Importantly, 15 (42%) of the differentially expressed TFs are known CBX2 targets in mESCs (Lau et al., 2017), suggesting a direct functional implication of CBX2 in the transcriptional control of these genes in MEFs.
Figure 7.
Loss of CBX2 alters the transcriptome profile of early passage fibroblasts. (A) MA plot of DEGs between wild-type (n = 2 samples) and Cbx2−/− (n = 2 samples) fibroblasts at P2 to visualize the differences between measurements taken in wild-type and Cbx2−/− samples. Red dots = up-regulated DEGs (n = 175), blue dots = down-regulated DEGs (n = 119), and gray dots representing non-DEGs (n = 17,147). (B) Heat map of DEGs indicating log10 transformed gene expression levels. The color scale indicates up- or down-regulated gene expression. (C) Kegg pathway enrichment for up- and down-regulated genes in Cbx2−/− cells. The gene number per pathway and the Q value are indicated by size and color scales, respectively. X axis represents the rich factor. (D) GO term analysis of biological processes for up- and down-regulated genes in Cbx2−/− fibroblasts with the x axis representing the number of genes detected per respective enriched pathway. (E) Gene expression heat map of DEGs encoding TFs. (F) DEG classification by TF families. The number of differentially expressed TFs per TF family in Cbx2−/− fibroblasts is indicated in black, while the number of DEG CBX2 target genes per TF family is indicated in red. (G) Significantly up- (red) or down- (blue) regulated key transcripts in Cbx2−/− fibroblasts. Asterisks indicate that correlative changes in chromatin accessibility were observed by ATAC-seq. Known CBX2 targets in mESCs are labeled in red.
Figure S2.
RNA-seq supplemental QC data and GO term classification. (A) Pearson correlation (corr) analysis between RNA-seq samples revealed strong correlation between the replicate samples of each genotype and led to the expected hierarchical clustering of the two wild-type samples and the two knockout samples, respectively. (B) A principal component analysis using an orthogonal transformation to convert a set of observations of variables into a set of values of linearly uncorrelated variables (principal components) also showed similar correlation between samples. (C) Box plot of the distribution of gene expression levels between samples, and (D) gene expression density map for all four samples indicating uniform gene expression levels. (E) The number of genes by different FPKM ranges across four samples. (F) GO classification of DEGs. Biological process (red), cellular component (blue), and molecular function (green). Coeff, coefficient; KO, knockout.
The top-most significantly down-regulated transcript in Cbx2−/− fibroblasts (−6.9-fold) was a processed thymine DNA glycosylase pseudogene (Tdg-ps2; Fig. 7 G), a paralog of the mismatch protein TDG, which is involved in DNA demethylation and epigenetic stability of several developmental genes known to be targets of the PRC (Cortázar et al., 2011). This was followed by Rpl26 (−5.29-fold), a component of the large ribosomal subunit implicated in DNA damage-induced cell senescence and regulation of p53 (Chen et al., 2017; Towers et al., 2015), and the RA early-inducible transcripts Raet1b (−4.7-fold) and Raet1c (−4.8-fold). Other important differentially expressed transcripts included Neuritin (Nrn1; −3.9-fold), commonly dysregulated in several human cancers (Dong et al., 2018; Kim et al., 2011), and LIM homeobox 8 (Lhx8), a polycomb-regulated developmental factor (Daino et al., 2018). In addition, Hmga1a, a high-mobility group protein associated with karyotypic abnormalities (Pierantoni et al., 2016), Sall1, a pericentric heterochromatin protein (Yamashita et al., 2007) known to interact with the telomere repeat binding factor TRF1 (Netzer et al., 2001) and Wilm’s tumor protein (WT1), a tumor suppressor implicated in genome stability (Shandilya and Roberts, 2015) showed altered expression levels. In contrast, significantly up-regulated transcripts included Rbm46, the ubiquitin-specific peptidase 2 (Usp2), the proto-oncogenes Spi1 and Fork head box D1, the morphogenic factors Wnt6 and Wnt10a, the Hmga1 splice variant Hmga1b, as well as the homeobox TF Nkx2.9 and the zinc finger TF Zic1. Notably, 23 known CBX2 target genes (Lau et al., 2017) showed differential expression levels in Cbx2−/− MEFs. However, due to limitations in the availability of validated CBX2 chromatin immunoprecipitation sequencing antibodies, direct genomic targets of CBX2 in fibroblasts remain to be elucidated. As is, we cannot formally establish whether individual expression changes in knockout fibroblasts are a direct consequence of loss of CBX2 or indirectly caused by changes in TF expression levels.
Changes in chromatin accessibility in Cbx2−/− fibroblasts
To determine whether the transcriptional dysregulation of several polycomb target genes observed in CBX2−/− cells is also associated with changes in chromatin accessibility, we conducted ATAC-seq analyses on parallel samples of P2 MEFs. ATAC-seq revealed a virtually identical peak distribution relative to genomic features in Cbx2−/− fibroblasts and wild-type cells (Fig. 8 A and Fig. S3). Heatmaps of tag distributions across TSSs (Fig. 8 B), merged regions, or genebodies (Fig. S3) revealed no major changes in tag distribution between Cbx2−/− and wild-type cells, indicating that loss of CBX2 does not induce global changes in chromatin accessibility. However, we found significantly differential ATAC-seq peaks (P < 0.01) in 520 genomic loci in Cbx2−/− fibroblasts compared with wild-type controls (Fig. 8 C and Fig. S3). Of these, 238 loci demonstrated loss of accessibility (red dots), while 282 loci showed increased accessibility (green dots). Interestingly, the top genomic loci with gain of chromatin accessibility involved three long noncoding RNAs (lncRNAs), Gm39154, Gm19299, and 9430037G07Rik, with unknown function (Fig. 8 D and Fig. S4). In several of the top 22 loci, changes of accessibility were also evident at several intergenic genomic regions (Fig. 8 D). The top coding transcripts showing gain in accessibility were the deubiquitinase (USP28; Fig. S4) involved in DDR during genotoxic stress (Wang et al., 2018) and the T cell lymphoma invasion and metastasis 2 gene (Fig. 8 D). On the other hand, the diaphanous-related formin 3 gene, which is involved in the spindle checkpoint (Damiani et al., 2016), the agouti signal protein (nonagouti), the kirre-like nephrin family adhesion molecule 3, and the sorting nexin 7 gene were the highest ranked protein coding loci showing a loss in chromatin accessibility.
Figure 8.
Loss of CBX2 induces changes in chromatin accessibility at key genomic loci in early passage MEFs. (A) The location of ATAC-seq peaks relative to genomic annotations is presented in pie charts for wild-type and Cbx2−/− fibroblasts at P2. (B) Heatmaps of tag distributions across TSS. (C) Volcano Plot of differential ATAC-seq peaks. Loci with significant loss (red, n = 238) and gain (green, n = 282) of chromatin accessibility in Cbx2−/− cells are highlighted. (D) Top-most regulated loci with gain (green bars) and loss (red bars) of accessibility in Cbx2−/− cells. (E) Comparative analysis relative to different genomic loci among all loci with gained versus all loci with lost accessibility. (F) UCSC browser view of ATAC-seq peak patterns at the Nrn1 locus. Cbx2−/− fibroblasts at P2 exhibit loss of chromatin accessibility at several CpG islands within the Nrn1 locus (red boxes). (G) Over-represented Kegg pathways identified in Cbx2−/− MEFs. (H) Top 10 over-represented GO terms.
Figure S3.
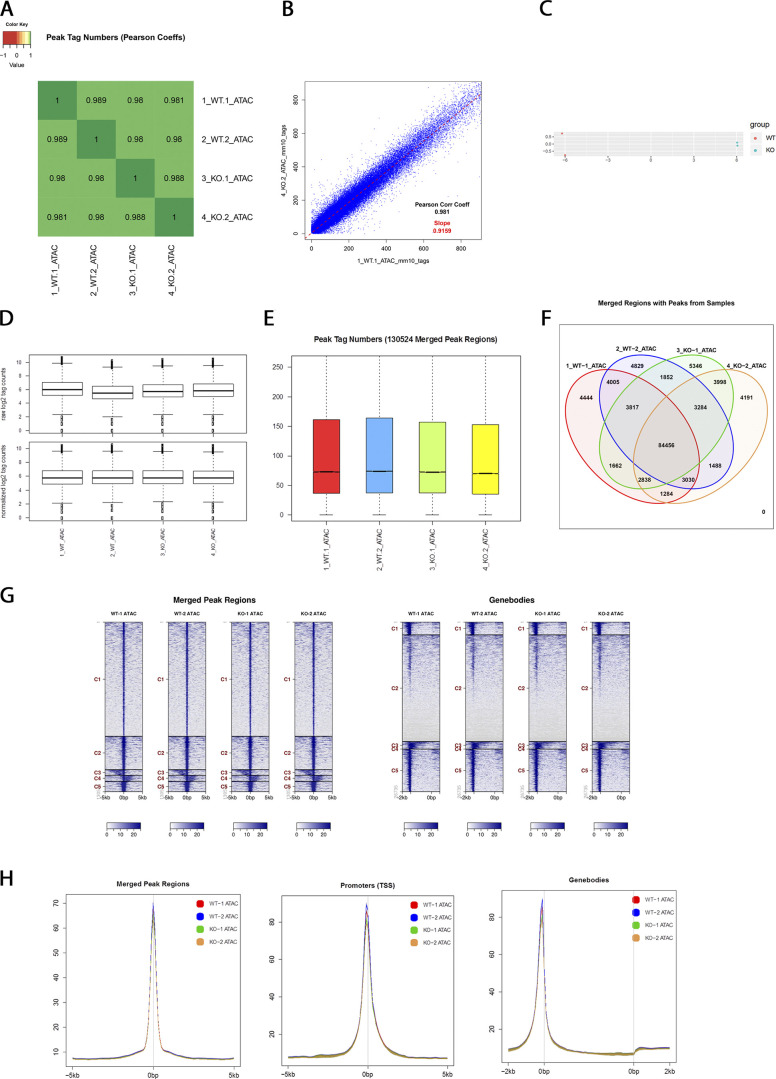
ATAC-seq supplemental QC data. (A and B) Pearson correlation of peak tag numbers. Correlation analysis between ATAC-seq samples revealed strong correlation between the two replicate samples of each genotype and led to the expected hierarchical clustering of the two wild-type samples and the two knockout samples, respectively. (C) A principal component analysis using an orthogonal transformation to convert a set of observations of variables into a set of values of linearly uncorrelated variables (principal components) also showed similar correlation between samples. (D) Box plots of raw and normalized peak counts from WT and Cbx2−/− samples. (E) Peak tag numbers of merged peak regions. (F) Venn diagram of merged regions with peaks from wild-type and Cbx2−/− samples. (G) Heatmaps of tag distributions in merged peak regions and genebodies. (H) Histograms showing the distance from peak centers at merged peak regions, TSSs, and genebodies.
Figure S4.
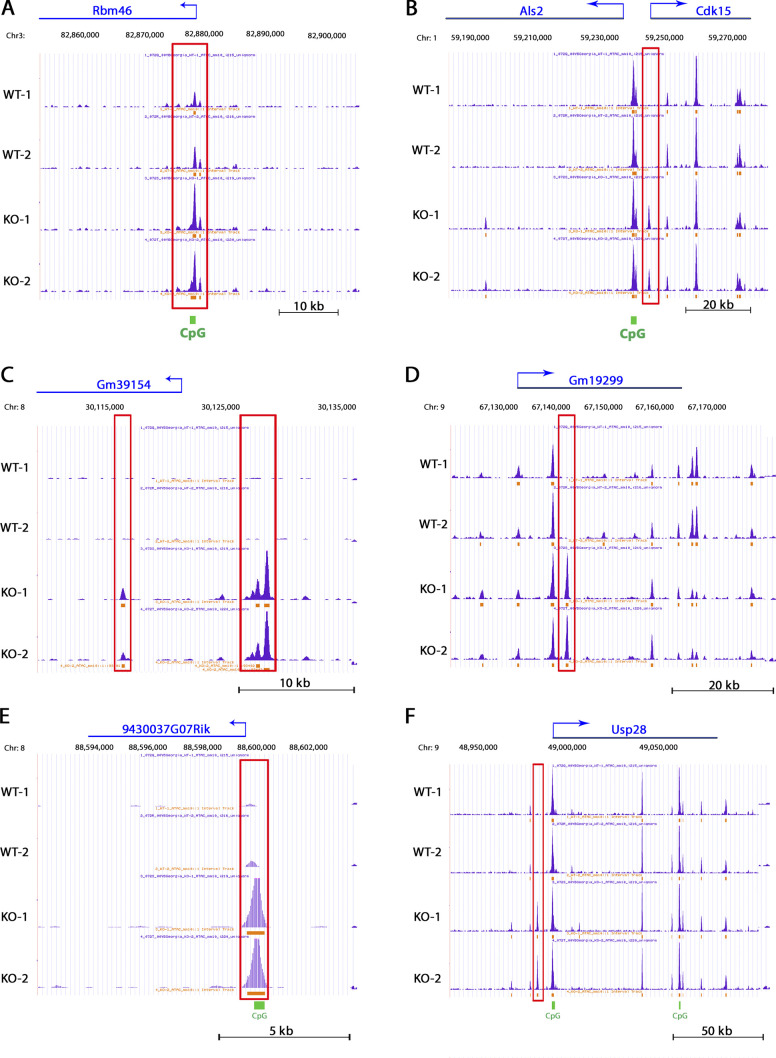
ATAC-seq genome browser views of key genomic loci. University of California Santa Cruz UCSC Genome Browser views of ATAC-seq peaks at the (A) Rbm46, (B) Als2, (C) Gm39154, (D) Gm19299, (E) 9430037G07Rik, and (F) Usp28 loci in wild-type and Cbx2−/− fibroblasts. Differential peaks are boxed in red. KO, knockout.
Of the 238 genomic regions with gains of chromatin accessibility, 46.3% corresponded to protein coding loci. This proportion was similar (46.9%) across the 282 regions with loss of accessibility (Fig. 8 E). In contrast, genomic regions coding for lncRNAs acquired chromatin accessibility at more than twice the rate than they lost peaks (19.4% and 7.7%, respectively) relative to the total number of changes per category. In addition, more intergenic regions lost accessibility (45.4%) than gained novel peaks (34.4%; Fig. 8 E). These findings support the notion that loss of CBX2 is associated with changes in chromatin accessibility not only at specific protein coding regions but also at intergenic and lncRNA loci. Importantly, loss of CBX2 may, depending on specific genomic loci, induce either increased or decreased chromatin accessibility.
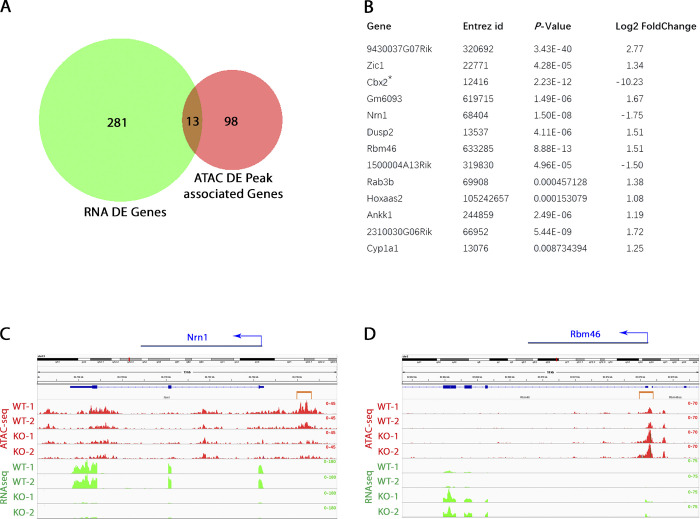
Due to the prominent role of PRC1 in the regulation of gene expression of many developmental genes (Cao et al., 2002; Di Croce and Helin, 2013; Isono et al., 2013), we sought to test whether loss of CBX2 alters chromatin accessibility at putative PRC1 target genes. ATAC-seq revealed that loss of CBX2 protein leads to a significant loss of chromatin accessibility at several CpG islands within the CBX2 target locus Nrn1 in MEFs at P2 (Fig. 8 F). The down-regulated CBX2 target gene and homeobox TF Nkx2.9 approached, but did not reach, statistical significance for chromatin accessibility changes, despite distinct changes in ATAC-seq peaks at CpG islands at this locus (Fig. S5). PRC1 components have previously been shown to alter chromatin accessibility at several enhancer sites in human cancer cells (Chan et al., 2018). We observed a gain of accessibility at a putative enhancer region upstream of the TSS of the Usp2 locus (Fig. S5). Testis-specific Usp2 isoforms localize to centrosomes in mouse spermatogenic cells (Lin et al., 2000), while suppression of Usp2 in triple negative human breast cancers improves chemo-responsiveness and cancer stem cell elimination (He et al., 2019). These results indicate that in addition to changes in pro-oncogenic and DDR loci, loss of CBX2 can also affect local chromatin accessibility at key TF genes and other critical target loci. Kegg pathway as well as GO term analyses revealed an enrichment of differential peaks in genes involved in immunological processes, amyotrophic lateral sclerosis (ALS), and folate and tRNA metabolism (Fig. 8, G and H). Importantly, changes in chromatin accessibility within 13 genes, including Nrn1, Rbm46, and Zic1, correlated with differential expression of their respective transcripts as determined by integrative ATAC-seq/RNA-seq analysis (Fig. 9, A–D).
Figure S5.
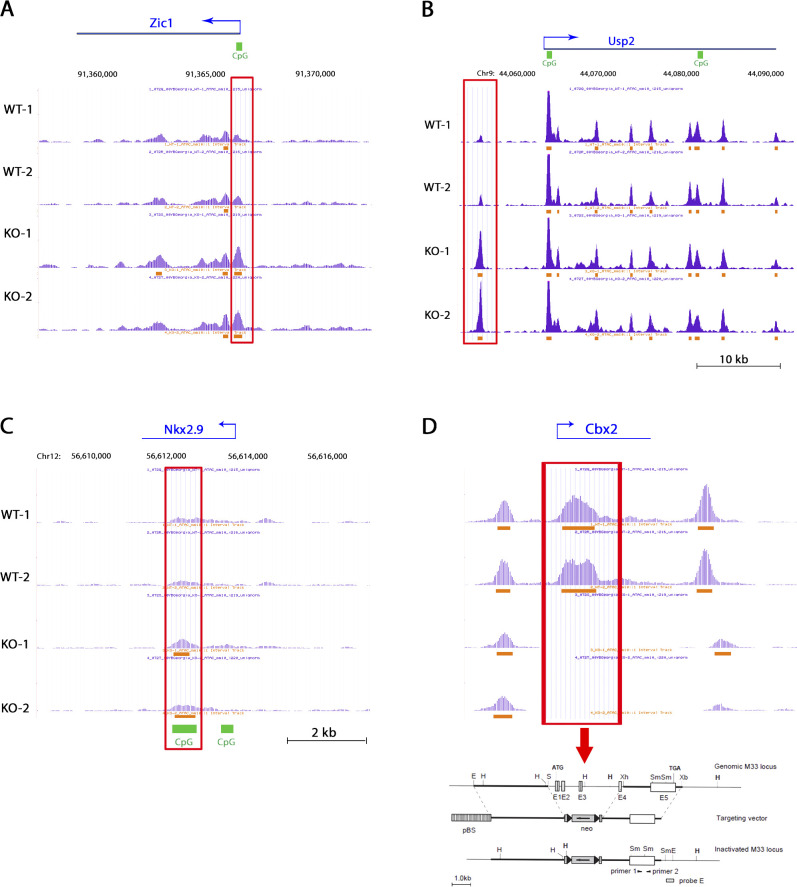
ATAC-seq genome browser views of additional key genomic loci. (A–C) University of California Santa Cruz UCSC Genome Browser views of ATAC-seq peaks at the (A) Zic1, (B) Usp2, and (C) Nkx2.9 locus in wild-type and Cbx2−/− fibroblasts. Differential peaks are boxed in red. (D) UCSC Genome Browser view of ATAC-seq peaks at the Cbx2 locus in wild-type and Cbx2−/− fibroblasts. Absence of peaks in Cbx2−/− cells at this locus is due to the targeting strategy used for generating the null allele. Cbx2 targeting strategy adapted from Coré et al. (1997). KO, knockout.
Figure 9.
Integrative ATAC-seq/RNA-seq analysis. (A) Venn diagram illustrating correlative changes in chromatin accessibility and gene expression in Cbx2−/− fibroblasts. DE, differentially expressed. (B) Loci with significant correlation in ATAC-seq and RNA-seq peaks. (C) Track alignment for ATAC-seq and RNA-seq tracks at the Nrn1 locus. (D) Track alignment for ATAC-seq and RNA-seq tracks at the TSS of the Rbm46 locus. The orange bracket indicates differential ATAC-seq peaks.
Together, our genome-wide analyses indicate that loss of CBX2 is associated with a significant reduction in transcripts for key enzymes required for heterochromatin function, cell cycle regulation, and activation of the DDR in early passage MEFs. Moreover, our results also revealed significant changes in chromatin accessibility at developmental and DDR loci in addition to key CBX2 target genes. Together, the transcriptional profiles of coding and noncoding RNAs in addition to changes in chromatin accessibility provide critical molecular insight into the mechanisms by which loss of CBX2 function leads to chromosome instability and cell senescence.
Discussion
CBX2 has recently emerged as a potential oncogene in different types of human cancers. A meta-analysis of >40,000 patient samples using the COSMIC and Oncomine databases revealed Cbx2 gene amplifications and increased mRNA expression in >30% of ovarian and breast tumors (Clermont et al., 2016). However, the cellular function(s) of CBX2 are not fully understood. Importantly, its potential role in maintenance of chromosome stability remained unexplored. Here, we provide new evidence indicating that CBX2 is required for the regulation of heterochromatin structure during interphase and chromosome stability during mitosis. Loss of CBX2 results in aberrant expression of genes involved in DDR, cell cycle regulation, cellular senescence, and cancer, providing novel mechanistic insight into its role in the regulation of chromosome stability. Importantly, by regulating the expression of both coding and noncoding RNAs associated with the DDR, CBX2 may be at the crossroads of DNA repair pathways, signaling the presence of chromosome instability during cell division and oncogene-induced neoplastic transformation. Centromere and telomere instability in Cbx2−/− fibroblasts results in increased mitotic recombination events at both minor satellite and telomeric DNA sequences, leading to chromosome breaks and formation of large-scale chromosomal rearrangements such as deletions, translocations, and end-to-end fusions. Our results also indicate that SADS during interphase, and decondensation of centromeric repeats during mitosis, increase the frequency of sister chromatid exchanges and lead to progressive centromere erosion. Notably, although accumulation of both proximal and distal telomere breaks occurs before any significant changes in absolute telomere length can be detected, the presence of both centromere and telomere breaks at a few chromosomes is sufficient to induce a major transition toward chromosome instability by P5. The onset of chromosome instability is also associated with emergence of major hallmarks of chromatin senescence such as changes in nuclear architecture, loss of Lamin B1 at the nuclear lamina, and micronuclei formation. Together, our results indicate that CBX2 plays a major role in the regulation of constitutive heterochromatin homeostasis and informs the mechanisms leading to premature senescence and large-scale chromosomal rearrangements induced by centromere instability.
Mechanisms of chromosome instability in Cbx2−/− fibroblasts
CBX2 is an important epigenetic reader that binds genomic regions marked by transcriptionally repressive histone modifications such as H3K27me3 and with lower affinity H3K9me3, and is capable of directly inducing nucleosome compaction in vitro through a stretch of positively charged amino acids (Grau et al., 2011; Kaustov et al., 2011). CBX2 exhibits an AT hook DNA binding domain and functions as an active subunit to recruit additional members of the PRC1 complex to the chromatin template during interphase and to mitotic chromosomes in mESCs (Kaustov et al., 2011; Zhen et al., 2014). However, its function on genome organization remains poorly understood. Transcriptome analyses indicate that loss of CBX2 is associated with altered expression of critical DNA damage signaling and repair proteins. Notably, the top down-regulated transcript in Cbx2−/− fibroblasts encodes a processed pseudogene of Tdg-ps2. Tdg-ps2 is a paralog of the gene coding for the mismatch protein TDG, which plays a critical role in the base-excision repair pathway by protecting CpG sites from spontaneous deamination of cytosine and 5mC and, thus, preventing G:T mismatches. TDG regulates the epigenetic stability of several Polycomb target genes and is involved in active DNA demethylation downstream of ten-eleven translocation dioxygenases (Cortázar et al., 2011; Cortellino et al., 2011; Dalton and Bellacosa, 2012; He et al., 2011). Down-regulation of TDG in melanoma cell lines causes cell cycle arrest, senescence, and genome instability (Mancuso et al., 2019). TDG has also been implicated in RA signaling processes by enhancing RXR/RAR binding to RA response elements in yeast (Um et al., 1998). Whether interference with transcriptional control through RA signaling plays a role in altered expression levels of the RA early-inducible proteins β and γ (Raet1b) and (Raet1c) observed in this study remains to be investigated.
The ribosomal protein L26 (RPL26) controls p53 translation in response to DNA damage in mouse and human cells (Chen and Kastan, 2010; Takagi et al., 2005) and stabilizes p53 protein through the formation of a ternary complex with HDM2 (Zhang et al., 2010). Importantly, RPL26 may function as a tumor suppressor in mouse tumor cell lines (Beck-Engeser et al., 2001; Takagi et al., 2005). Thus, down-regulation of Rpl26 in Cbx2−/− fibroblasts may attenuate p53 induction and limit the ability of these cells to respond to DNA damage. The high mobility group proteins Hmga1a and Hmga1b exhibit an AT-hook domain that allows for efficient binding to short AT-rich DNA regions and are important chromatin architectural factors with transcription regulatory activity (Munshi et al., 2001). High mobility group protein overexpression has major implications for breast cancer initiation and progression (Fusco and Fedele, 2007; Palmieri et al., 2012; Sgarra et al., 2018; Sumter et al., 2016). On the other hand, targeted deletion of Hmga1a induces down-regulation of spindle assembly checkpoint genes and micronuclei formation (Pierantoni et al., 2015; Pierantoni et al., 2016). Notably, transcripts for Hmga1a were significantly down-regulated in Cbx2−/− fibroblasts, potentially contributing to the onset of chromosome instability in mutant cells at P5. Together, these results indicate that loss of CBX2 function induces significant pro-oncogenic gene expression changes but also affects critical factors required for DDR and chromosome stability.
Consistent with previous studies using a double knockout model of the PRC1 components Ring1/Rnf2 in mESCs (Hodges et al., 2018; King et al., 2018), no global changes in chromatin accessibility at TSSs were detected in CBX2−/− fibroblasts. However, in contrast with other members of the PRC1 complex, targeted deletion of CBX2 induced changes in chromatin accessibility at regulatory regions of lncRNAs, the deubiquitinase (USP28), known to promote oncoprotein accumulation and p53 activation in response to chromosome instability (Meitinger et al., 2016; Wang et al., 2018), the T cell lymphoma invasion and metastasis 2 protein, which localizes to the nuclear envelope and has been implicated in regulation of nuclear morphology in MEFs (Woroniuk et al., 2018), as well as important neurodevelopmental genes such as ALS2. Alsin (Als2 and its C-terminal homologue Als2cl) play overlapping functions on endosome regulation and exhibit centrosome localization in human neuronal cells (Hadano et al., 2004; Millecamps et al., 2005). Importantly, an increase in accessibility upstream of the Als2 promoter was also associated with a significant up-regulation of Als2cl transcripts in CBX2−/− MEFs (Fig. S4).
It has been previously suggested that disruption of CBX2 may impair the repression of its target genes in a manner that may not require changes in chromatin accessibility (Illingworth, 2019; King et al., 2018). However, we found evidence for changes in chromatin accessibility in several CBX2 target genes, including Nrn1, Zic1, and Nkx2.9, that were associated with a corresponding change in transcription as identified by RNA-seq. In addition, we found CBX2 targets that showed altered transcriptional activity in mutant cells without changes in chromatin accessibility. Nkx2.9 encodes a homeodomain-containing TF known to be critical for embryogenesis (Jarrar et al., 2015). The Nkx2.9 promoter is bivalently marked and repressed but poised for transcription during early development, and PRC1 participates in its transcriptional repression (Mazzarella et al., 2011). Loss of Nkx2.9 leads to abnormal neuronal networks in the spinal cord (Holz et al., 2010; Pabst et al., 2003). In addition, overexpression of Zic1 in human cancers is associated with Survivin targeting-improved prognosis (Gu et al., 2019; Han et al., 2018). Notably, ZIC1 is also an important activator of Wnt signaling in Xenopus laevis (Merzdorf and Sive, 2006). Our results indicate that subtle albeit significant differences exist in the molecular phenotypes resulting from targeted deletion of different PRC1 components and that loss of CBX2 function affects chromatin accessibility at neural specifying Polycomb target genes in addition to nontarget genes and lncRNAs. Consistent with this notion, recent studies have also detected differences in chromatin accessibility at enhancer regions in Ring1a/Rnf2 knockout cells (Chan et al., 2018). PcG proteins are known to act in concert with an increasing number of lncRNAs to regulate gene expression and heterochromatin formation (Aguilo et al., 2011; Fatica and Bozzoni, 2014; Marchese et al., 2017). Importantly, emerging evidence indicates intricate regulatory interactions between CBX2 and lncRNAs (Kawaguchi et al., 2017; Sun et al., 2019; Yang et al., 2019) that might be critical for recruiting chromatin-remodeling proteins to specific loci or nuclear domains (Aguilo et al., 2011; Fatica and Bozzoni, 2014; Marchese et al., 2017).
Senescence-associated SADS and centromere erosion
SADS is an early senescence-associated phenomenon that indicates major changes in higher-order chromatin structure and function in both human and mouse cells (Swanson et al., 2013). The mechanisms of SADS formation and its consequences on centromere instability and genome organization are not fully understood. However, loss of CBX2 induced striking changes in the large-scale organization of satellite DNA and chromocenter formation. Notably, these changes in chromatin structure were also associated with a significant reduction in transcripts of factors essential for heterochromatin formation, including Sall1 and Hmga1a. HMGA1 binds pericentromeric heterochromatin in Drosophila melanogaster and mouse cells via its AT-hook domains and is strictly necessary for chromocenter organization, as loss of HMGA1 protein function induces abnormal chromocenter formation (Jagannathan et al., 2018). Importantly, with striking similarity to the phenotypes observed in Cbx2−/− fibroblasts, loss of HMGA1 leads to defective nuclear envelope integrity, micronuclei formation, and accumulation of DNA damage (Jagannathan et al., 2018). Our results indicate that loss of CBX2 results in SADS formation, abnormal chromocenter formation during interphase, and increased deleterious centromeric mitotic recombination events during mitosis. Thus, abnormal expression of factors involved in heterochromatin formation and subsequent distention of satellite sequences in CBX2−/− fibroblasts contribute to increased frequency of C-SCEs, centromere breaks, and progressive centromere erosion as a mechanism of chromosome instability. Mammalian centromeres are highly recombinogenic (Jaco et al., 2008). Therefore, any changes in protein complexes that bind pericentric and centromeric heterochromatin may predispose to increased recombination events at minor or major satellite DNA sequences. Previous studies indicate that the canonical PRC1 complex containing CBX2 also functions in the formation of atypical Polycomb chromatin domains capable of directly recognizing and binding pericentric heterochromatin in metaphase chromosomes of the early mouse embryo. Moreover, elegant biochemical studies have recently demonstrated that CBX2 undergoes a process of phase separation to form nuclear condensates that can concentrate DNA and nucleosomes in mESCs. Importantly, the phase separation function of CBX2 is coupled to its chromatin compaction ability (Plys et al., 2019; Tatavosian et al., 2015, 2019). Notably, phase separation of the heterochromatin protein HP1 has also been recently demonstrated as an important mechanism that contributes to the organization of constitutive heterochromatin in human somatic cells (Strom et al., 2017).
Increased centromere and telomere mitotic recombination and large-scale chromosome rearrangements
In mouse embryonic stem cells, centromeres are sixfold more recombinogenic than telomeres (Jaco et al., 2008). The presence of micronuclei, recurrent centromeric breaks in addition to the loss of entire chromosome arms and the type of segmental translocations detected through SKY analysis indicate that increased centromere recombination and centromere fission are the primary mechanism(s) of chromosome instability in CBX2−/− cells. However, both proximal and distal telomeres were affected. Because of the lack of evidence for any changes in absolute telomere length at P5, our results indicate that loss of CBX2 initially disrupts telomeric higher-order chromatin organization rather than telomere length. Consistent with this hypothesis, previous studies indicate that the presence of a few critically short telomeres is sufficient to induce chromosome fusions and translocations and that the shortest telomeres, rather than absolute telomere length, drive the onset of chromosome instability (Hemann et al., 2001). Aside from its role in chromocenter formation, HMGA1 has also been shown to be involved in telomere integrity in Arabidopsis thaliana (Charbonnel et al., 2018). Notably, Sall1, a pericentric heterochromatin protein, provides a functional link between centromeric and telomeric heterochromatin by establishing an interaction with the telomere repeat binding factor TRF1 (Netzer et al., 2001). Thus, altered expression levels of Hmga1a and Sall1 in Cbx2−/− fibroblasts may affect constitutive heterochromatin homeostasis at both centromeric and telomeric domains. The large-scale chromosome rearrangements observed here, together with changes in nuclear architecture, suggest that loss of CBX2 induces dramatic changes in meso-scale genome organization. In Drosophila cells, the PRC1 components Pc and Polyhomeotic proteins regulate nanoscale chromatin organization and gene expression by promoting the formation of protein clusters ranging from 30 nm to 700 nm. In turn, these protein clusters directly impact chromatin topology by establishing chromatin interactions at different scales of genome organization (Wani et al., 2016). Studies are in progress to determine whether loss of CBX2 function affects nanoscale chromatin organization in mammalian cells.
Together, our results provide novel molecular insight into the mechanisms leading to premature senescence in CBX2−/− cells and suggest that CBX2 functions at the crossroads of cellular senescence and oncogene-induced neoplastic transformation. Importantly, the frequent CBX2 gene amplification events and increased mRNA expression levels detected in a recent meta-analysis of a large sample of human tumors (Clermont et al., 2014) indicate that CBX2 is an important oncogenic factor with clear potential as a prognostic marker and therapeutic target in several types of human neoplasms. Our studies inform and contribute to the identification of specific target pathways required for heterochromatin formation, DNA repair, and cell cycle regulation that could be ideal candidates for novel drug design and the implementation of next-generation epigenetic therapy strategies in human oncology.
Materials and methods
Cell culture and sample preparation
All animal experiments were conducted in accordance with the recommendations and guidelines of the Institutional Animal Care and Use Committee and the Public Health Service Policy on Humane Care and Use of Laboratory Animals of the National Institutes of Health. Cbx2 null and wild-type MEFs were collected at 18.5 d post coitum from timed male embryos of Cbx2 heterozygote matings (C.129P2-Cbx2tm1Cim/J; Coré et al., 1997) according to standard procedures. Primary cell cultures were maintained in high-glucose DMEM (Gibco/Thermo Fisher Scientific) supplemented with 10% FBS (Hyclone, GE Healthcare), 1/100 (vol/vol) sodium pyruvate, 1/100 (vol/vol) L-glutamine, and 1/100 (vol/vol) penicillin/streptomycin (all Gibco/Thermo Fisher Scientific) before cytogenetic and molecular analysis of cultures at P2 and P5. Mitotic chromosome complements were spread onto glass slides following colchicine treatment (100 nM) for 2–5 h and hypotonic treatment with 75 mM KCl (Sigma-Aldrich) for 11 min before fixation with methanol/acetic acid (Sigma-Aldrich) or 2% PFA, 0.15% Triton X-100 for FISH and immunochemistry, respectively.
Immunochemistry
Immunochemical detection of subcellular protein localization was conducted on PFA-fixed interphase nuclei and metaphase spreads in blocking buffer containing 5% FBS and 0.05% Triton X-100 (Sigma-Aldrich) in PBS at 4°C overnight. A mouse γH2AX antibody (Abcam, 1:500) and a rabbit LaminB1 antibody (Abcam, 1:200) were both used by overnight incubation in blocking buffer at 4°C. Primary antibodies were detected using 488- and 555-coupled Alexa Fluor secondary antibodies (Molecular Probes/Thermo Fisher Scientific) at a dilution of 1:1,000 for 1 h at room temperature. Samples were counterstained and mounted using Vectashield containing DAPI (Vector Laboratories, Inc.) before image acquisition by conventional and confocal microscopy and SR-SIM as well as HCA as outlined below.
DNA-FISH
DNA-FISH analyses were performed using FITC-labeled telomere-specific PNA probes (TelC-FITC”-OO-TTAGGGTTAGGGTTAGGG; a generous gift from Dr. Maria Blasco [Centro Nacional de Investigaciones Oncológicas, Madrid, Spain] and Bio-Synthesis Inc., Lewisville, TX, respectively) as well as CY3-labeled major satellite-specific pericentromeric probes (Star-FISH Cambio). Surface spread chromosomes were dehydrated in serial ethanol washes (2 min each, 70%, 90%, 100% ethanol), air-dried, and denatured in 70% formamide (VWR International Ltd.) in 2 × saline sodium citrate (SSC) at 85°C for 10 min and subsequently chilled in ice-cold 70% ethanol for 5 min. 0.2 µg/ml telomeric repeat and major satellite probes were then denatured in PNA probe cocktail containing 70% formamide, 5 mM KCl, and 1 mM MgCl2 (Sigma-Aldrich), or manufacturer-supplied hybridization buffer (Cambio) for 10 min at 85°C and incubated at 30°C for 1 h, respectively. Overnight hybridization was performed at 30°C, and stringency washes were conducted in 50% formamide in 2 × SSC for 5 min each before mounting of coverslips in Vectashield mounting medium with DAPI and subsequent microscopic analysis.
SKY analysis
SKY analysis was conducted in collaboration with the Cytogenetic Laboratory at the Van Andel Research Institute (Grand Rapids, MI). Methanol/acetic acid–fixed surface spreads were analyzed by 24-color, whole-chromosome painting and simultaneous visualization of each chromosome pair in a metaphase spread. Images were recorded using a spectral interferogram (Applied Spectral Imaging). Classification of chromosome anomalies was conducted according to the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/anomalies), and the positions of chromosomal breakpoints were designated according to the standard G-banded karyotype of the mouse.
CO-FISH
CO-FISH analyses were conducted using a CY3-labeled minor satellite PNA probe complementary to the lagging strand (CY3-OO-AAAACACATTCGTTGGAA; a generous gift from Dr. Maria Blasco) as well as with a FITC-labeled PNA probe specific to the lagging strand of telomere repeat sequences (“TelC-FITC”; Bio-Synthesis Inc.) as previously described (Jaco et al., 2008) with minor modifications. Briefly, MEFs were cultured in medium containing 10 µM BrdU (Sigma-Aldrich) for one cell cycle to facilitate BrdU incorporation during S-phase. Metaphase chromosome complements were then surface spread as described above. Samples were stained with 500 µg/ml HOECHST 33258 (Molecular Probes/Thermo Fisher Scientific) for 15 min at room temperature, and BrdU-incorporated DNA was subsequently UV-nicked for 30 min in 2 × SSC and digested using 3 U/µl Exonuclease III (NEB Biolabs) for 30 min at 37°C. DNA was denatured at 75°C for 15 min in 70% formamide in 2 × SSC, and hybridization with the Cy3-labeled minor satellite or FITC-labeled telomere probes was performed at 37°C overnight. Stringency washes were conducted in 0.1% SSC for 5 min at room temperature before DNA counterstaining and mounting.
Analysis of sister chromatid exchanges by differential BrdU labeling
Sister chromatid exchanges were detected using pulse-incorporated BrdU (10 µM) that was added 17 h before supplementation of colchicine (100 nM) and metaphase chromosome spreading. Slides were treated with 2 N HCl for 20 min, extensively washed in PBS, and immunolabeled using a mouse anti-BrdU antibody (Roche, 1:200) overnight at 4°C in conjunction with an Alexa Fluor 555 anti-mouse secondary antibody. Detection of BrdU nucleotides by immunochemistry reveals strongly asymmetric fluorescent signals between homologous regions of sister chromatids.
Absolute telomere length quantification assay
Telomere length was directly quantified using the Absolute Mouse Telomere Length Quantification qPCR Assay Kit (ScienCell Research Laboratories) in conjunction with the FastStart Essential DNA Green Master mix (Roche Lifescience) on wild-type and knockout fibroblasts at P5. Results were normalized to an internal single copy reference amplicon derived from a sequence mapping to chromosome 10. In addition, a genomic DNA sample with known telomere length was used as a reference for calculating the absolute telomere length of the target samples (expressed as average telomere length [in kilobases] per telomere) in three biological replicates.
Transcriptome analysis by RNAseq
The transcriptomes of wild-type and Cbx2−/− fibroblasts at P2 were mapped and quantified by deep sequencing and quantifying the frequency of each gene represented in duplicate (RNA-seq; Mortazavi et al., 2008). 2 × 105 cells per replicate and genotype were used for RNA extraction and transcriptome analysis using a DNA nanoball sequencing platform, generating ∼4.62 GB per sample. The clean reads ratio was >99% for all four samples. The average mapping ratio with the reference genome using HISAT2 was 93.44%, with uniformity in the mapping result suggesting that the samples are comparable. Novel coding transcripts were merged with reference transcripts to obtain the complete reference. Bowtie2 was used to map clean reads to this reference (mapping ratio was 83.08%) before gene expression level computation for each sample with RNA-Seq by expectation-maximization. Differential expression was analyzed with DEseq2 (Love et al., 2014), and genes were classified as differentially expressed if the P value was <0.05 and the fold change was greater than two upward or downward.
ATAC-seq
ATAC-seq was used for mapping chromatin accessibility genome-wide by probing DNA accessibility with hyperactive Tn5 transposase and insertion of sequencing adapters into accessible regions of chromatin (Buenrostro et al., 2015). Sequencing reads were used to infer regions of increased accessibility. 100,000 cells per sample and replicate at P2 were used for ATAC-seq sequencing with Ilumina NextSeq 500. The paired-end 42 bp sequencing reads (PE42) were mapped to the genome using the BWA (Burrows-Wheeler aligner) algorithm with default settings. Only reads that passed Illumina’s purity filter, aligned with no more than two mismatches, and mapped uniquely to the genome were used in subsequent analyses. In addition, duplicate reads were removed. Genomic regions with high levels of transposition/tagging events were determined using the MACS2 peak calling algorithm (Zhang et al., 2008). Differentially enriched regions were identified using DESeq2 in GUAVA (Divate and Cheung, 2018) as described previously (Love et al., 2014), and regions with P value < 0.001 and fold change greater than four upward or downward are considered differentially enriched peaks. Peaks within 5,000 bp upstream and 3,000 bp downstream of the TSS were considered for the analysis. The peaks identified were associated and annotated with the nearest genes in GUAVA’s peak annotation algorithm, and these genes were then used to determine over-represented Kegg pathways and GO terms (Divate and Cheung, 2018).
Integrative analysis
To align the parameters of the two analyses, ATAC-seq data were annotated using GUAVA, and regions were considered differentially enriched peaks if the P value was <0.05 and the fold change greater than two upward or downward. Peaks within 5,000 bp upstream and 3,000 bp downstream of the TSS were considered for the analysis and annotated with the nearest genes by GUAVA (Divate and Cheung, 2018). These genes were compared with the DEGs found in RNAseq analysis, and genes present in both gene lists were analyzed and viewed by loading the sequence alignment data into the Integrative Genomics Viewer (Robinson et al., 2011).
Image acquisition, processing, and quantitative measurements
Epifluorescence images were acquired using a Leica DMRX/E fluorescence microscope (Leica Microsystems, Inc.) equipped with a PLAN APO 63×/1.20 water objective at room temperature. Fluorochromes used included Alexa Fluor 488– and Alexa Fluor 555–coupled secondary antibodies as well as CY3 and FITC-labeled fluorescent probes. Images were captured with a Leica DFC 350F camera using Openlab software (Improvision), and image processing was performed using Photoshop 8.0 (Adobe) for linear adjustments and cropping of fluorescent images. No gamma adjustments were made. SR-SIM was performed on an ELYRA S1 Superresolution microscope (Zeiss) on an Axio Observer Z1 inverted microscope stand equipped with three laser lines: 405 nm, 488 nm, and 561 nm. Images were acquired using a 100× oil objective and were processed with ZEN software (Zeiss) to generate super-resolved Z-stacks (0.01-µm steps), maximum projections, and colocalization line graphs.
Quantitative measurements were performed following establishment of identical fluorescence intensity threshold masks for all images before transformation into binary layers. Analyses were conducted using the NIS Elements 4.0 Automated Measurements Module (Advanced Research; Nikon Instruments Inc.) or Imaris 9.3 software (Bitplane AG) with multidimensional imaging software to measure the EqDiameter (the diameter of a sphere of equivalent volume of an irregularly shaped object), as well as volume and sphericity (the ratio of the surface area of a perfect sphere with the same volume as the measured object to the surface area of the object itself), and mean fluorescence intensity of the object. 3D renderings of individual telomeres were computed from 3D images by sequential preprocessing, segmentation, and connected component labeling steps. Surfaces were used to identify relevant entities within images for visualization and measurements of volume, fluorescence intensity, and sphericity. HCA of the images was conducted using an ImageXpress Micro Confocal imaging system (Molecular Devices, LLC) using DAPI and 488-nm filters. Data were analyzed using MetaXpress software (Molecular Devices, LLC) following detection of nuclei using DAPI fluorescence and measurement of nuclear surface areas and mean fluorescence intensity using the 488-nm filter. Skeleton analysis was conducted using the Skeletonize3D plugin of ImageJ software (National Institutes of Health) to perform skeletonization of binary images (8-bit) of Lamin B1 immunofluorescence staining as previously described (Fišerová et al., 2019). Briefly, following tagging of all pixels in a skeleton image, all junctions, triple and quadruple points, and branches were counted, and the mean branch length was measured.
Statistical analysis
All data presented were collected from three independent biological replicates (except for RNA-seq and ATAC-seq, which were conducted in duplicate experiments) and statistically analyzed using GraphPad Prism software. Comparison of all pairs was conducted using parametric and nonparametric tests (Mann–Whitney or t test) according to the sample distribution with GraphPad Prism software. Data are presented as means, and variation between individual replicates is indicated as the SD. Differences were considered significant when P < 0.05 and are indicated by different superscripts with *, P < 0.05; **, P < 0.01; ***, P < 0.005; and ****, P < 0.001.
Data availability
Next-generation RNA sequencing and ATAC-seq data is available on NCBI GEO (accession no. GSE156413).
Online supplemental material
Fig. S1 contains additional images of wild-type and Cbx2−/− fibroblast nuclei as well as grayscale images of all color metaphase spreads in Fig. 2, Fig. 4, and Fig. 5. Fig. S2 summarizes supporting information and quality control data pertaining to RNA-seq experiments. Fig. S3 contains supporting information and quality control data pertaining to ATAC-seq experiments. Fig. S4 shows University of California Santa Cruz UCSC Genome Browser tracks of the Rbm46, Als2/Cdk15, Gm39145, Gm19299, 9430037G07Rik, and Usp28 loci. Fig. S5 shows UCSC Genome Browser tracks of the Zic1, Usp2, Nkx2.9, and Cbx2 loci.
Acknowledgments
The authors thank Dr. M.K. Kandasamy for assistance with super-resolution structured illumination analysis at the Biomedical Microscopy Core facility, University of Georgia, and Julie Koeman at the Van Andel Research Institute, Grand Rapids, MI, for assistance with SKY analysis.
This work was supported by research grants to R. De La Fuente from the National Institutes of Health (HD093383), the National Science Foundation Center for Cell Manufacturing (CMaT; 164805), the Regenerative Engineering and Medicine Center at the University of Georgia, and the Georgia Cancer Coalition, and through a U.S. Department of Agriculture–National Institute of Food and Agriculture–National Institutes of Health dual-purpose grant (2020-67015-30882).
The authors declare no competing financial interests.
Author contributions: C. Baumann and R. De La Fuente conceived and designed the experiments. C. Baumann conducted the experiments and data analysis. X. Zhang conducted the GUAVA and Integrative RNA-seq/ATAC-seq analyses. C. Baumann and R. De La Fuente wrote the manuscript.
References
- Aguilo F., Zhou M.M., and Walsh M.J.. 2011. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 71:5365–5369. 10.1158/0008-5472.CAN-10-4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.M., Williams E.S., Cornforth M.N., and Goodwin E.H.. 2010. Chromosome Orientation fluorescence in situ hybridization or strand-specific FISH. Methods Mol. Biol. 659:173–183. 10.1007/978-1-60761-789-1_12 [DOI] [PubMed] [Google Scholar]
- Bastians H. 2015. Causes of Chromosomal Instability. Recent Results Cancer Res. 200:95–113. 10.1007/978-3-319-20291-4_5 [DOI] [PubMed] [Google Scholar]
- Baumann C., and De La Fuente R.. 2011. Role of polycomb group protein cbx2/m33 in meiosis onset and maintenance of chromosome stability in the Mammalian germline. Genes (Basel). 2:59–80. 10.3390/genes2010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C., Viveiros M.M., and De La Fuente R.. 2010. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet. 6 e1001137 10.1371/journal.pgen.1001137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck-Engeser G.B., Monach P.A., Mumberg D., Yang F., Wanderling S., Schreiber K., Espinosa R. III, Le Beau M.M., Meredith S.C., and Schreiber H.. 2001. Point mutation in essential genes with loss or mutation of the second allele: relevance to the retention of tumor-specific antigens. J. Exp. Med. 194:285–300. 10.1084/jem.194.3.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 109:21.29.1-21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., and Zhang Y.. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 298:1039–1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Chan H.L., Beckedorff F., Zhang Y., Garcia-Huidobro J., Jiang H., Colaprico A., Bilbao D., Figueroa M.E., LaCava J., Shiekhattar R., et al. 2018. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 9:3377 10.1038/s41467-018-05728-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C., Rymarenko O., Da Ines O., Benyahya F., White C.I., Butter F., and Amiard S.. 2018. The Linker Histone GH1-HMGA1 Is Involved in Telomere Stability and DNA Damage Repair. Plant Physiol. 177:311–327. 10.1104/pp.17.01789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., and Kastan M.B.. 2010. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 24:2146–2156. 10.1101/gad.1968910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Crutchley J., Zhang D., Owzar K., and Kastan M.B.. 2017. Identification of a DNA Damage-Induced Alternative Splicing Pathway That Regulates p53 and Cellular Senescence Markers. Cancer Discov. 7:766–781. 10.1158/2159-8290.CD-16-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont P.L., Sun L., Crea F., Thu K.L., Zhang A., Parolia A., Lam W.L., and Helgason C.D.. 2014. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. Br. J. Cancer. 111:1663–1672. 10.1038/bjc.2014.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont P.-L., Crea F., Chiang Y.T., Lin D., Zhang A., Wang J.Z.L., Parolia A., Wu R., Xue H., Wang Y., et al. 2016. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin. Epigenetics. 8:16 10.1186/s13148-016-0182-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coré N., Bel S., Gaunt S.J., Aurrand-Lions M., Pearce J., Fisher A., and Djabali M.. 1997. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 124:721–729. [DOI] [PubMed] [Google Scholar]
- Coré N., Joly F., Boned A., and Djabali M.. 2004. Disruption of E2F signaling suppresses the INK4a-induced proliferative defect in M33-deficient mice. Oncogene. 23:7660–7668. 10.1038/sj.onc.1207998 [DOI] [PubMed] [Google Scholar]
- Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A.L., Siegrist F., et al. 2011. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 470:419–423. 10.1038/nature09672 [DOI] [PubMed] [Google Scholar]
- Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 146:67–79. 10.1016/j.cell.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daino K., Nishimura M., Imaoka T., Takabatake M., Morioka T., Nishimura Y., Shimada Y., and Kakinuma S.. 2018. Epigenetic dysregulation of key developmental genes in radiation-induced rat mammary carcinomas. Int. J. Cancer. 143:343–354. 10.1002/ijc.31309 [DOI] [PubMed] [Google Scholar]
- Dalton S.R., and Bellacosa A.. 2012. DNA demethylation by TDG. Epigenomics. 4:459–467. 10.2217/epi.12.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani D., Goffinet A.M., Alberts A., and Tissir F.. 2016. Lack of Diaph3 relaxes the spindle checkpoint causing the loss of neural progenitors. Nat. Commun. 7:13509 10.1038/ncomms13509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R., Baumann C., and Viveiros M.M.. 2015. ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development. 142:1806–1817. 10.1242/dev.118927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L., and Helin K.. 2013. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20:1147–1155. 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- Divate M., and Cheung E.. 2018. GUAVA: A Graphical User Interface for the Analysis and Visualization of ATAC-seq Data. Front. Genet. 9:250 10.3389/fgene.2018.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Luo X., Niu Y., Yu N., Gao R., Wang H., Yang L., and Huang J.. 2018. Neuritin 1 expression in human normal tissues and its association with various human cancers. Int. J. Clin. Exp. Pathol. 11:1956–1964. [PMC free article] [PubMed] [Google Scholar]
- Fatica A., and Bozzoni I.. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15:7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Fišerová J., Maninová M., Sieger T., Uhlířová J., Šebestová L., Efenberková M., Čapek M., Fišer K., and Hozák P.. 2019. Nuclear pore protein TPR associates with lamin B1 and affects nuclear lamina organization and nuclear pore distribution. Cell. Mol. Life Sci. 76:2199–2216. 10.1007/s00018-019-03037-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N.J., Kingston R.E., and Woodcock C.L.. 2004. Chromatin compaction by a polycomb group protein complex. Science. 306:1574–1577. 10.1126/science.1100576 [DOI] [PubMed] [Google Scholar]
- Freund A., Laberge R.M., Demaria M., and Campisi J.. 2012. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 23:2066–2075. 10.1091/mbc.e11-10-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco A., and Fedele M.. 2007. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 7:899–910. 10.1038/nrc2271 [DOI] [PubMed] [Google Scholar]
- Garcia-Moreno S.A., Lin Y.-T., Futtner C.R., Salamone I.M., Capel B., and Maatouk D.M.. 2018. CBX2 is required during male sex determination to repress female fate at bivalent loci. bioRxiv. 10.1101/496984 (Preprint posted on December 14, 2018). [DOI] [Google Scholar]
- Grau D.J., Chapman B.A., Garlick J.D., Borowsky M., Francis N.J., and Kingston R.E.. 2011. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 25:2210–2221. 10.1101/gad.17288211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Wang X., Su D., Su X., Lin L., Li S., Wu Q., Liu S., Zhang P., Zhu X., et al. 2018. CBX2 Inhibits Neurite Development by Regulating Neuron-Specific Genes Expression. Front. Mol. Neurosci. 11:46 10.3389/fnmol.2018.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Guo X.K., Chen B.H., Gao X.J., Chen F., and Liu Q.. 2019. Prognostic and clinicopathological value of ZIC1 in patients with cervical squamous cell carcinoma. Oncol. Lett. 18:6621–6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadano S., Otomo A., Suzuki-Utsunomiya K., Kunita R., Yanagisawa Y., Showguchi-Miyata J., Mizumura H., and Ikeda J.E.. 2004. ALS2CL, the novel protein highly homologous to the carboxy-terminal half of ALS2, binds to Rab5 and modulates endosome dynamics. FEBS Lett. 575:64–70. 10.1016/j.febslet.2004.07.092 [DOI] [PubMed] [Google Scholar]
- Han W., Cao F., Gao X.J., Wang H.B., Chen F., Cai S.J., Zhang C., Hu Y.W., Ma J., Gu X., et al. 2018. ZIC1 acts a tumor suppressor in breast cancer by targeting survivin. Int. J. Oncol. 53:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 333:1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Lee H.J., Saha S., Ruan D., Guo H., and Chan C.H.. 2019. Inhibition of USP2 eliminates cancer stem cells and enhances TNBC responsiveness to chemotherapy. Cell Death Dis. 10:285 10.1038/s41419-019-1512-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Strong M.A., Hao L.Y., and Greider C.W.. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 107:67–77. 10.1016/S0092-8674(01)00504-9 [DOI] [PubMed] [Google Scholar]
- Heng H.H., Bremer S.W., Stevens J.B., Horne S.D., Liu G., Abdallah B.Y., Ye K.J., and Ye C.J.. 2013. Chromosomal instability (CIN): what it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 32:325–340. 10.1007/s10555-013-9427-7 [DOI] [PubMed] [Google Scholar]
- Hodges H.C., Stanton B.Z., Cermakova K., Chang C.Y., Miller E.L., Kirkland J.G., Ku W.L., Veverka V., Zhao K., and Crabtree G.R.. 2018. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 25:61–72. 10.1038/s41594-017-0007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz A., Kollmus H., Ryge J., Niederkofler V., Dias J., Ericson J., Stoeckli E.T., Kiehn O., and Arnold H.H.. 2010. The transcription factors Nkx2.2 and Nkx2.9 play a novel role in floor plate development and commissural axon guidance. Development. 137:4249–4260. 10.1242/dev.053819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R.S. 2019. Chromatin folding and nuclear architecture: PRC1 function in 3D. Curr. Opin. Genet. Dev. 55:82–90. 10.1016/j.gde.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K., Endo T.A., Ku M., Yamada D., Suzuki R., Sharif J., Ishikura T., Toyoda T., Bernstein B.E., and Koseki H.. 2013. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell. 26:565–577. 10.1016/j.devcel.2013.08.016 [DOI] [PubMed] [Google Scholar]
- Jaco I., Canela A., Vera E., and Blasco M.A.. 2008. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 181:885–892. 10.1083/jcb.200803042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M., Cummings R., and Yamashita Y.M.. 2018. A conserved function for pericentromeric satellite DNA. eLife. 7 e34122 10.7554/eLife.34122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrar W., Dias J.M., Ericson J., Arnold H.H., and Holz A.. 2015. Nkx2.2 and Nkx2.9 are the key regulators to determine cell fate of branchial and visceral motor neurons in caudal hindbrain. PLoS One. 10 e0124408 10.1371/journal.pone.0124408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh-Fukui Y., Tsuchiya R., Shiroishi T., Nakahara Y., Hashimoto N., Noguchi K., and Higashinakagawa T.. 1998. Male-to-female sex reversal in M33 mutant mice. Nature. 393:688–692. 10.1038/31482 [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y., Miyabayashi K., Komatsu T., Owaki A., Baba T., Shima Y., Kidokoro T., Kanai Y., Schedl A., Wilhelm D., et al. 2012. Cbx2, a polycomb group gene, is required for Sry gene expression in mice. Endocrinology. 153:913–924. 10.1210/en.2011-1055 [DOI] [PubMed] [Google Scholar]
- Kaustov L., Ouyang H., Amaya M., Lemak A., Nady N., Duan S., Wasney G.A., Li Z., Vedadi M., Schapira M., et al. 2011. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286:521–529. 10.1074/jbc.M110.191411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Machida S., Kurumizaka H., Tagami H., and Nakayama J.I.. 2017. Phosphorylation of CBX2 controls its nucleosome-binding specificity. J. Biochem. 162:343–355. 10.1093/jb/mvx040 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kang H.S., Kim T.W., and Kim S.J.. 2011. Differential methylation hybridization profiling identifies involvement of STAT1-mediated pathways in breast cancer. Int. J. Oncol. 39:955–963. [DOI] [PubMed] [Google Scholar]
- King H.W., Fursova N.A., Blackledge N.P., and Klose R.J.. 2018. Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res. 28:1494–1507. 10.1101/gr.237180.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau M.S., Schwartz M.G., Kundu S., Savol A.J., Wang P.I., Marr S.K., Grau D.J., Schorderet P., Sadreyev R.I., Tabin C.J., et al. 2017. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 355:1081–1084. 10.1126/science.aah5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Keriel A., Morales C.R., Bedard N., Zhao Q., Hingamp P., Lefrançois S., Combaret L., and Wing S.S.. 2000. Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures. Mol. Cell. Biol. 20:6568–6578. 10.1128/MCB.20.17.6568-6578.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso P., Tricarico R., Bhattacharjee V., Cosentino L., Kadariya Y., Jelinek J., Nicolas E., Einarson M., Beeharry N., Devarajan K., et al. 2019. Thymine DNA glycosylase as a novel target for melanoma. Oncogene. 38:3710–3728. 10.1038/s41388-018-0640-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese F.P., Raimondi I., and Huarte M.. 2017. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18:206 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., and Reinberg D.. 2011. The Polycomb complex PRC2 and its mark in life. Nature. 469:343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G., and Di Croce L.. 2016. The role of Polycomb in stem cell genome architecture. Curr. Opin. Cell Biol. 43:87–95. 10.1016/j.ceb.2016.09.006 [DOI] [PubMed] [Google Scholar]
- Mazzarella L., Jørgensen H.F., Soza-Ried J., Terry A.V., Pearson S., Lacaud G., Kouskoff V., Merkenschlager M., and Fisher A.G.. 2011. Embryonic stem cell-derived hemangioblasts remain epigenetically plastic and require PRC1 to prevent neural gene expression. Blood. 117:83–87. 10.1182/blood-2010-03-273128 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Anzola J.V., Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K., et al. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214:155–166. 10.1083/jcb.201604081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzdorf C.S., and Sive H.L.. 2006. The zic1 gene is an activator of Wnt signaling. Int. J. Dev. Biol. 50:611–617. 10.1387/ijdb.052110cm [DOI] [PubMed] [Google Scholar]
- Millecamps S., Gentil B.J., Gros-Louis F., Rouleau G., and Julien J.P.. 2005. Alsin is partially associated with centrosome in human cells. Biochim. Biophys. Acta. 1745:84–100. 10.1016/j.bbamcr.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S.A., and Di Croce L.. 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 10:47–62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., and Wold B.. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5:621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Munshi N., Agalioti T., Lomvardas S., Merika M., Chen G., and Thanos D.. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 293:1133–1136. 10.1126/science.293.5532.1133 [DOI] [PubMed] [Google Scholar]
- Netzer C., Rieger L., Brero A., Zhang C.D., Hinzke M., Kohlhase J., and Bohlander S.K.. 2001. SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/PIN2 and localizes to pericentromeric heterochromatin. Hum. Mol. Genet. 10:3017–3024. 10.1093/hmg/10.26.3017 [DOI] [PubMed] [Google Scholar]
- Niedernhofer L.J., Gurkar A.U., Wang Y., Vijg J., Hoeijmakers J.H.J., and Robbins P.D.. 2018. Nuclear Genomic Instability and Aging. Annu. Rev. Biochem. 87:295–322. 10.1146/annurev-biochem-062917-012239 [DOI] [PubMed] [Google Scholar]
- O’Loghlen A., Muñoz-Cabello A.M., Gaspar-Maia A., Wu H.-A., Banito A., Kunowska N., Racek T., Pemberton H.N., Beolchi P., Lavial F., et al. 2012. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell. 10:33–46. 10.1016/j.stem.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan R.J., and Karlseder J.. 2012. The great unravelling: chromatin as a modulator of the aging process. Trends Biochem. Sci. 37:466–476. 10.1016/j.tibs.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O., Rummelies J., Winter B., and Arnold H.H.. 2003. Targeted disruption of the homeobox gene Nkx2.9 reveals a role in development of the spinal accessory nerve. Development. 130:1193–1202. 10.1242/dev.00346 [DOI] [PubMed] [Google Scholar]
- Palmieri D., Valentino T., De Martino I., Esposito F., Cappabianca P., Wierinckx A., Vitiello M., Lombardi G., Colao A., Trouillas J., et al. 2012. PIT1 upregulation by HMGA proteins has a role in pituitary tumorigenesis. Endocr. Relat. Cancer. 19:123–135. 10.1530/ERC-11-0135 [DOI] [PubMed] [Google Scholar]
- Pereira C.F., Piccolo F.M., Tsubouchi T., Sauer S., Ryan N.K., Bruno L., Landeira D., Santos J., Banito A., Gil J., et al. 2010. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 6:547–556. 10.1016/j.stem.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Pierantoni G.M., Conte A., Rinaldo C., Tornincasa M., Gerlini R., Federico A., Valente D., Medico E., and Fusco A.. 2015. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget. 6:17342–17353. 10.18632/oncotarget.3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierantoni G.M., Conte A., Rinaldo C., Tornincasa M., Gerlini R., Valente D., Izzo A., and Fusco A.. 2016. Hmga1 null mouse embryonic fibroblasts display downregulation of spindle assembly checkpoint gene expression associated to nuclear and karyotypic abnormalities. Cell Cycle. 15:812–818. 10.1080/15384101.2016.1146835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plys A.J., Davis C.P., Kim J., Rizki G., Keenen M.M., Marr S.K., and Kingston R.E.. 2019. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33:799–813. 10.1101/gad.326488.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C.V., Asch A.S., and Yamada H.Y.. 2017. Emerging links among Chromosome Instability (CIN), cancer, and aging. Mol. Carcinog. 56:791–803. 10.1002/mc.22539 [DOI] [PubMed] [Google Scholar]
- Richly H., Aloia L., and Di Croce L.. 2011. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2 e204 10.1038/cddis.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarra R., Pegoraro S., Ros G., Penzo C., Chiefari E., Foti D., Brunetti A., and Manfioletti G.. 2018. High Mobility Group A (HMGA) proteins: Molecular instigators of breast cancer onset and progression. Biochim. Biophys. Acta Rev. Cancer. 1869:216–229. 10.1016/j.bbcan.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Shandilya J., and Roberts S.G.. 2015. A role of WT1 in cell division and genomic stability. Cell Cycle. 14:1358–1364. 10.1080/15384101.2015.1021525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., and Kingston R.E.. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 49:808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., and Karpen G.H.. 2017. Phase separation drives heterochromatin domain formation. Nature. 547:241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumter T.F., Xian L., Huso T., Koo M., Chang Y.T., Almasri T.N., Chia L., Inglis C., Reid D., and Resar L.M.. 2016. The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development. Curr. Mol. Med. 16:353–393. 10.2174/1566524016666160316152147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Cao X., and Wang C.. 2019. Polycomb chromobox Cbx2 enhances antiviral innate immunity by promoting Jmjd3-mediated demethylation of H3K27 at the Ifnb promoter. Protein Cell. 10:285–294. 10.1007/s13238-018-0581-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson E.C., Manning B., Zhang H., and Lawrence J.B.. 2013. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J. Cell Biol. 203:929–942. 10.1083/jcb.201306073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson E.C., Rapkin L.M., Bazett-Jones D.P., and Lawrence J.B.. 2015. Unfolding the story of chromatin organization in senescent cells. Nucleus. 6:254–260. 10.1080/19491034.2015.1057670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Absalon M.J., McLure K.G., and Kastan M.B.. 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 123:49–63. 10.1016/j.cell.2005.07.034 [DOI] [PubMed] [Google Scholar]
- Tanaka K., and Hirota T.. 2016. Chromosomal instability: A common feature and a therapeutic target of cancer. Biochim. Biophys. Acta. 1866:64–75. [DOI] [PubMed] [Google Scholar]
- Tatavosian R., Zhen C.Y., Duc H.N., Balas M.M., Johnson A.M., and Ren X.. 2015. Distinct Cellular Assembly Stoichiometry of Polycomb Complexes on Chromatin Revealed by Single-molecule Chromatin Immunoprecipitation Imaging. J. Biol. Chem. 290:28038–28054. 10.1074/jbc.M115.671115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavosian R., Kent S., Brown K., Yao T., Duc H.N., Huynh T.N., Zhen C.Y., Ma B., Wang H., and Ren X.. 2019. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 294:1451–1463. 10.1074/jbc.RA118.006620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers C.G., Guarnieri A.L., Micalizzi D.S., Harrell J.C., Gillen A.E., Kim J., Wang C.A., Oliphant M.U.J., Drasin D.J., Guney M.A., et al. 2015. The Six1 oncoprotein downregulates p53 via concomitant regulation of RPL26 and microRNA-27a-3p. Nat. Commun. 6:10077 10.1038/ncomms10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S., Harbers M., Benecke A., Pierrat B., Losson R., and Chambon P.. 1998. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 273:20728–20736. 10.1074/jbc.273.33.20728 [DOI] [PubMed] [Google Scholar]
- Wang X., Liu Z., Zhang L., Yang Z., Chen X., Luo J., Zhou Z., Mei X., Yu X., Shao Z., et al. 2018. Targeting deubiquitinase USP28 for cancer therapy. Cell Death Dis. 9:186 10.1038/s41419-017-0208-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani A.H., Boettiger A.N., Schorderet P., Ergun A., Münger C., Sadreyev R.I., Zhuang X., Kingston R.E., and Francis N.J.. 2016. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 7:10291 10.1038/ncomms10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woroniuk A., Porter A., White G., Newman D.T., Diamantopoulou Z., Waring T., Rooney C., Strathdee D., Marston D.J., Hahn K.M., et al. 2018. STEF/TIAM2-mediated Rac1 activity at the nuclear envelope regulates the perinuclear actin cap. Nat. Commun. 9:2124 10.1038/s41467-018-04404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K., Sato A., Asashima M., Wang P.C., and Nishinakamura R.. 2007. Mouse homolog of SALL1, a causative gene for Townes-Brocks syndrome, binds to A/T-rich sequences in pericentric heterochromatin via its C-terminal zinc finger domains. Genes Cells. 12:171–182. 10.1111/j.1365-2443.2007.01042.x [DOI] [PubMed] [Google Scholar]
- Yang F., Wen S., Zhang Y., Xu Y., Lv H., Zhu Y., Wang M., Su P., Huang C., and Tian Z.. 2019. Identifying potential metastasis-related long non-coding RNAs, microRNAs, and message RNAs in the esophageal squamous cell carcinoma. J. Cell. Biochem. 120:13202–13215. 10.1002/jcb.28594 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Yuan Y., Zhang W., Guan W., Wu Z., Jin C., Chen H., Zhang L., Yang X., et al. 2010. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 38:6544–6554. 10.1093/nar/gkq536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen C.Y., Duc H.N., Kokotovic M., Phiel C.J., and Ren X.. 2014. Cbx2 stably associates with mitotic chromosomes via a PRC2- or PRC1-independent mechanism and is needed for recruiting PRC1 complex to mitotic chromosomes. Mol. Biol. Cell. 25:3726–3739. 10.1091/mbc.e14-06-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Next-generation RNA sequencing and ATAC-seq data is available on NCBI GEO (accession no. GSE156413).