Shard et al. show that the emergence of neural stem cells from the Drosophila optic lobe neuroepithelium is an EMT-like process involving tissue-wide coordination. Neuralized acts downstream of NSC fate acquisition to downregulate the Crumbs complex to facilitate epithelium remodeling.
Abstract
Many tissues are produced by specialized progenitor cells emanating from epithelia via epithelial-to-mesenchymal transition (EMT). Most studies have so far focused on EMT involving single or isolated groups of cells. Here we describe an EMT-like process that requires tissue-level coordination. This EMT-like process occurs along a continuous front in the Drosophila optic lobe neuroepithelium to produce neural stem cells (NSCs). We find that emerging NSCs remain epithelial and apically constrict before dividing asymmetrically to produce neurons. Apical constriction is associated with contractile myosin pulses and involves RhoGEF3 and down-regulation of the Crumbs complex by the E3 ubiquitin ligase Neuralized. Anisotropy in Crumbs complex levels also results in accumulation of junctional myosin. Disrupting the regulation of Crumbs by Neuralized lowered junctional myosin and led to imprecision in the integration of emerging NSCs into the front. Thus, Neuralized promotes smooth progression of the differentiation front by coupling epithelium remodeling at the tissue level with NSC fate acquisition.
Introduction
During animal development, many complex organs are built by stem and progenitor cells originating from epithelial tissues (Norden, 2017). For instance, in vertebrate embryos, the anterior brain is produced by neuroepithelial cells that function as neural stem cells (NSCs), and several organs, such as the intestine and the pancreas, derive from endodermal epithelia. Similarly, acquisition of stem-like properties has been associated with epithelial-to-mesenchymal transition (EMT), at least in the context of carcinoma (Dongre and Weinberg, 2019). Thus, understanding how stem cells emerge from epithelia to build organs is an important issue. During Drosophila neurogenesis, a fixed number of NSCs, also termed neuroblasts, emerge from within the embryonic neuroectoderm via an EMT-like delamination process to build the larval brain (Hartenstein and Campos-Ortega, 1984; Hartenstein and Wodarz, 2013). As they become specified, NSCs undergo apical constriction driven by a pulsatile actomyosin meshwork (Simões et al., 2017; An et al., 2017; Sawyer et al., 2010; Martin and Goldstein, 2014) and delaminate as E-cadherin (E-cad) is removed from the apical-lateral membrane (Simões et al., 2017). Delaminating NSCs, however, maintain apical-basal polarity cues such that, upon division, cell fate determinants localize at the basal pole and the mitotic spindle lines up along this polarity axis (Schober et al., 1999; Knoblich, 2008).
A slightly different EMT-like process occurs in the larval brain for the formation of the optic lobe (OL) NSCs (Fig. 1 A). The outer proliferation center of the OL comprises a single layer of pseudostratified neuroepithelium (NE) cells that arise during embryogenesis and grow during larval development (Hofbauer and Campos-Ortega, 1990; Egger et al., 2007; Hakes et al., 2018). Then, in response to a systemic pulse of ecdysone, a self-perpetuating proneural wave sweeps the NE from medial to lateral during the third instar, eventually converting all NE cells into NSCs by the end of larval development (Dillard et al., 2018; Yasugi et al., 2008). These NSCs give rise to the medulla neurons that will make up part of the visual processing center of the adult brain. This proneural wave is proposed to travel through the NE via an excitable reaction-diffusion mechanism. Accordingly, an activator signal, the EGF Spitz, spreads in the NE by diffusion and self-induction and triggers negative feedback via the expression of Lethal of scute (L’sc), a proneural factor that promotes differentiation and thereby inhibits the production of active Spitz (Jörg et al., 2019). This creates a traveling front of EGF receptor (EGFR) signaling and proneural activity (Fig. 1 B). The traveling domain of proneural gene expression is called the transition zone (TZ). The TZ is therefore defined as a stripe of three to four epithelial cells expressing L’sc. L’sc induced by EGFR both self-activates and regulates the NE-to-NSC fate transition via Delta-Notch signaling (Wang et al., 2011; Weng et al., 2012; Yasugi et al., 2008, 2010; Sato et al., 2016; Jörg et al., 2019). NE cells therefore appear to follow a stereotyped fate trajectory: starting from a NotchON/proneuralOFF state in the NE, cells entering the TZ express increasing levels of L’sc to acquire a NotchON/proneuralON state before switching to a NotchOFF/proneuralON state as they adopt the NSC fate (Egger et al., 2010; Weng et al., 2012; Ngo et al., 2010; Wang et al., 2011; Orihara-Ono et al., 2011). The ON/OFF switch in Notch activity likely results from cis-inhibition by Delta (del Álamo et al., 2011; Contreras et al., 2018; Reddy et al., 2010). While the logic underlying progression of the TZ and cell fate switch within the TZ is partly understood, how epithelial NE cells undergo cell shape and polarity changes to become nonepithelial NSCs remains unexplored. Interestingly, while this epithelium-to-NSC transition exhibits similarities with NSC delamination in the embryo, there are also notable differences. First, instead of occurring in individual cells as seen in the embryo, this EMT-like transition involves a continuous stripe of cells undergoing epithelium remodeling in a coordinated manner at the tissue level. Second, newly specified NSCs do not delaminate from the NE to locate basally but rather remain in the same plane as NE cells (Yasugi et al., 2008; Ngo et al., 2010; Egger et al., 2007). Thus, the OL NE provides an opportunity to examine how an EMT-like process producing NSCs is regulated and coordinated at the tissue level.
Figure 1.
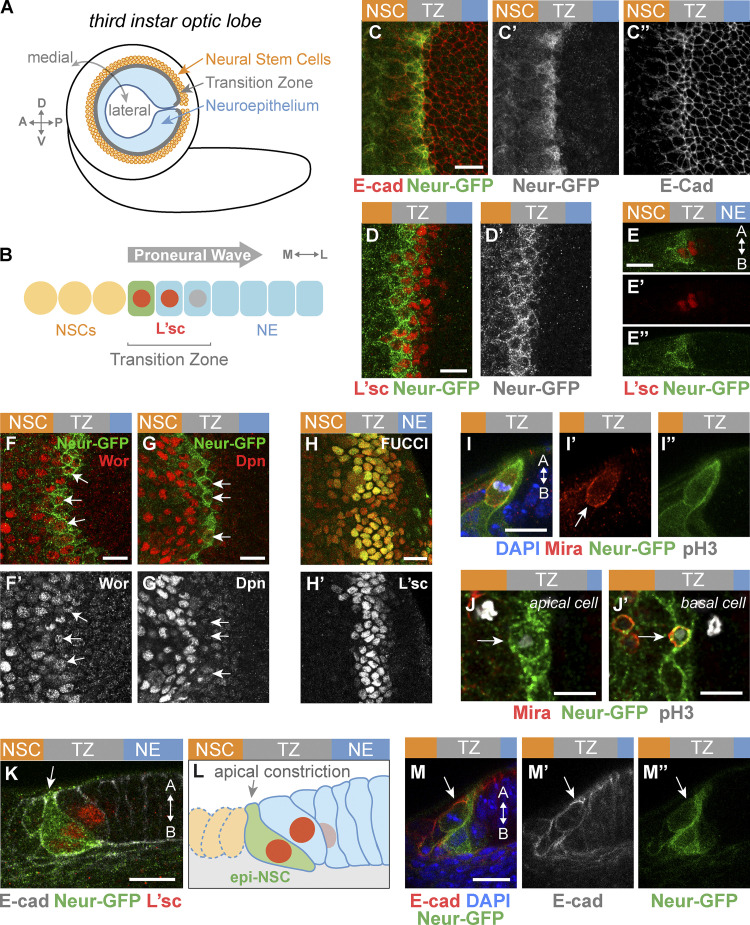
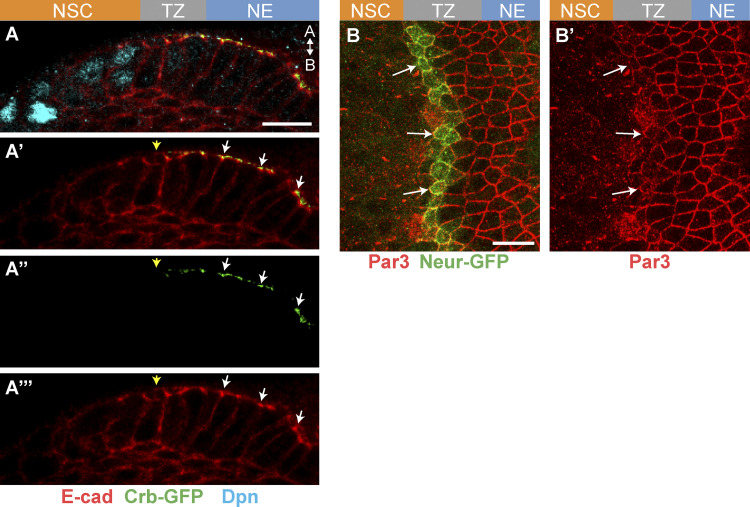
Neur marks emerging epithelial NSCs in the TZ. (A) Diagram of the third instar larval brain displaying the OL NE. P, posterior; A, anterior; D, dorsal; V, ventral. (B) The proneural wave sweeps the NE from medial to lateral, inducing NE–NSC transition in a region called the TZ defined by the graded expression of L’sc (red nuclei). The medial TZ cell appears in green. M, medial; L, lateral. (C–E″) Surface (C–D′) and cross section (E–E″; A, apical; B, basal) views showing that Neur-GFP (green) becomes expressed in the medial-most TZ cells (L’sc, red in D–E″). These cells showed increased E-cad levels (red in C). Lower levels of Neur-GFP were observed in NSCs (C′, D′, and E″). (F–G′) Most Neur-positive TZ cells (Neur-GFP, green) coexpressed Wor (red, F), while the NSC marker Dpn was up-regulated only in NSCs past the TZ (red, G). Arrows point to medial TZ cells. (H and H′) TZ cells (L’sc, H′) expressed CycB-RFP (red, H) and E2F1-GFP (green, H), indicating that they are in G2 phase. (I–J′) Medial TZ cells (Neur-GFP, green) appeared to undergo the first round of asymmetric cell division (pH3, phospho-histone H3 in white; DAPI, blue). This division was oriented along the apical-basal axis, and Mira localized asymmetrically (I–I″; Mira, red). The arrow in I′ indicates the basal crescent of Mira. A large apical cell (J) and a small basal cell (J′), likely corresponding to the first-born GMC, appeared to be produced by this asymmetric division. (K–M″) Cross-section views showing that the medial-most TZ cells marked by Neur-GFP (green) and L’sc (red in K) were apically constricted and had a characteristic teardrop shape while retaining apical AJs (K; E-cad, white). E-cad remained detectable as these cells divided (M–M″; E-cad, red; DAPI, blue). These apically constricted epithelial cells undergoing asymmetric cell division were named epi-NSCs (L). Scale bars = 10 µm.
The protein Crumbs (Crb) is a large transmembrane protein with a short intracellular tail that recruits the scaffolding protein Stardust (Sdt in flies, Pals1 in mammals). Sdt interacts with Pals1-associated tight junction (Patj) to form a Crb-Sdt-Patj complex (Bulgakova and Knust, 2009). The apical Crb complex is required to stabilize adherens junctions (AJs) and maintain epithelial integrity during tissue remodeling in Drosophila (Harris and Tepass, 2008; Bajur et al., 2019; Flores-Benitez and Knust, 2015; Campbell et al., 2009; Perez-Mockus et al., 2017b; Röper, 2012; Salis et al., 2017). Recently, the conserved E3 ubiquitin ligase Neuralized (Neur) was found to interact with specific isoforms of Sdt, and this interaction appeared to target the Crb complex proteins for endocytosis and degradation (Chanet and Schweisguth, 2012; Perez-Mockus et al., 2017b). This suggested that Neur expression in NSCs might promote their EMT-like delamination (Perez-Mockus and Schweisguth, 2017). Whether Neur regulates the epithelium-to-NSC transition in the OL is not known.
Here we found that emerging NSCs apically constrict one cell at a time and remain epithelial until they divide asymmetrically to produce the first-born differentiated cells. Neur is expressed in these newly specified NSCs and regulates apical constriction in part by down-regulating the Crb complex in a Sdt-dependent manner. The down-regulation of Crb by Neur in emerging NSCs also promotes the formation of supracellular actomyosin cables within the TZ. Live imaging of epithelium-to-NSC transition suggests a model whereby Neur acts downstream of NSC fate acquisition to coordinate epithelium remodeling at the tissue level in the TZ to produce a smooth differentiation front.
Results
Emerging NSCs retain epithelial properties
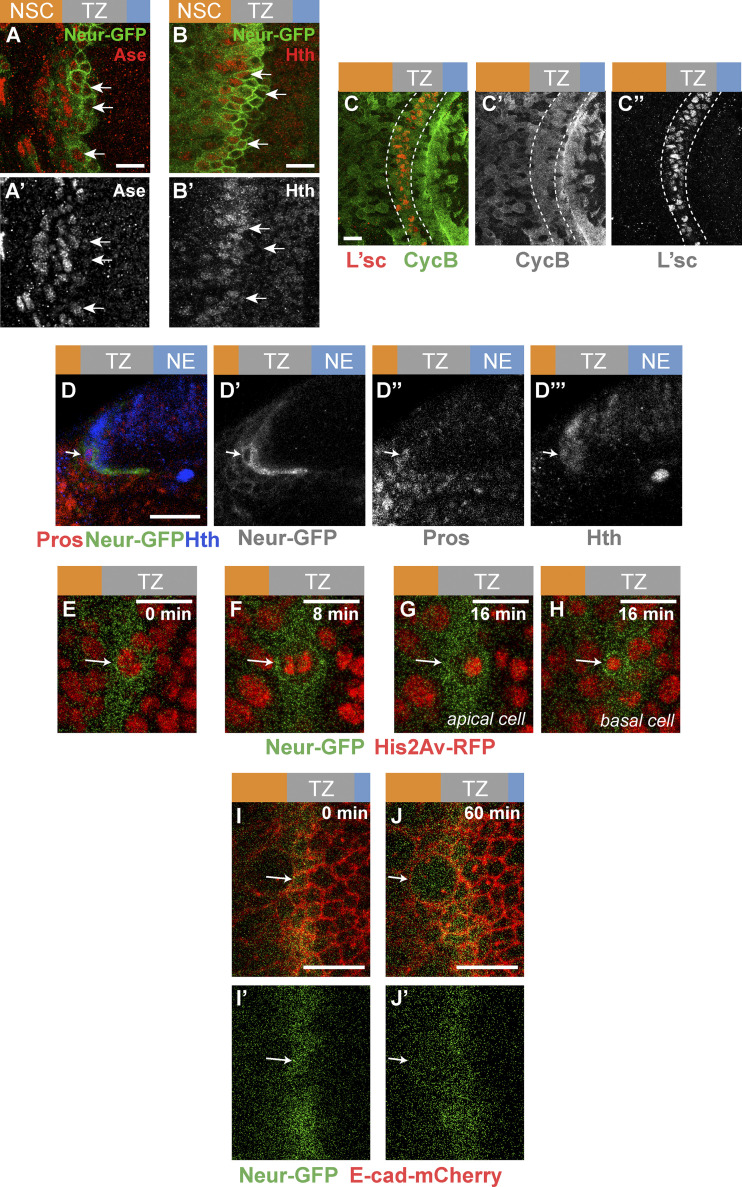
To study cell shape changes in emerging NSCs in the OL, we first identified an early NSC marker. Neur marks neural progenitors as they are singled out from proneural clusters in several epithelia (Huang et al., 1991; Castro et al., 2005; Rouault et al., 2010) and is expressed in medial TZ cells, based on a neur-lacZ transcriptional reporter (Contreras et al., 2018). Here, we further characterized the expression of Neur using a functional Neur-GFP bacterial artificial chromosome transgene (Perez-Mockus et al., 2017a). Our analysis showed that Neur is expressed in approximately one row of TZ cells at the medial edge of the NE (Fig. 1, C–E″). Neur was also detected at lower levels in NSCs. Moreover, most Neur-positive TZ cells expressed Worniu (Wor), a transcription factor associated with NSC identity (Ashraf and Ip, 2001; Cai et al., 2001; Fig. 1, F and F′), whereas the other NSC factors Asense (Ase) and the temporal identity factor Homothorax (Hth; Suzuki et al., 2013; Li et al., 2013) displayed only partial overlap with Neur-GFP (Fig. S1), and Deadpan (Dpn) was up-regulated only in NSCs past the TZ (Fig. 1, G and G′). Thus, Neur marks emerging NSCs in the TZ, and Wor is an early NSC identity factor during the NE-NSC transition.
Figure S1.
Molecular and cellular characterization of epi-NSCs. (A–B′) Neur expression (Neur-GFP, green) only partially overlapped with expression of the NSC markers Ase (red, A) and Hth (red, B). Arrows point to the nuclei of several medial TZ cells. (C–C″) TZ cells (L’sc, red) expressed CycB (green), supporting that they are in G2 phase. In contrast, heterogenous CycB expression is seen in the NE. (D–D″′) Cross-section view showing a small basal cell at the medial edge of the TZ (arrow) expressing Pros (red), Neur-GFP (green), and Hth (blue). This cell likely corresponds to a GMC derived from the asymmetric division of an epi-NSC. (E–H) Snapshots of a time-lapse video tracking an epi-NSC (Neur-GFP, green) entering mitosis (His2Av-RFP, red) and dividing asymmetrically to producing a large apical cell (E) and a small basal cell (F). Arrows point to the dividing epi-NSC and its progeny cells. (I–J′) Snapshots of a time-lapse video showing a surface view of the TZ (E-cad-mCherry, red). E-cad is retained apically as the epi-NSC (Neur-GFP, green; arrow) undergoes mitotic rounding. Scale bars = 10 µm.
We next studied the division mode of the Neur-positive TZ cells. As NE cells differentiate into NSCs, they gain the ability to divide asymmetrically to both self-renew and produce ganglion mother cells (GMCs; Hofbauer and Campos-Ortega, 1990; Egger et al., 2007). Given that Neur-positive TZ cells express some NSC identity genes, we wondered whether they divide asymmetrically like NSCs. While earlier observations suggested that TZ cells are in the G1 phase of the cell cycle (Weng et al., 2012; Orihara-Ono et al., 2011; Reddy et al., 2010), we first found using the FUCCI (fluorescent ubiquitination-based cell cycle indicator) system (Zielke et al., 2014) that all TZ cells are in G2 (Fig. 1, H and H′). Consistent with this, Cyclin B was detected in TZ cells (Fig. S1). Furthermore, we observed dividing Neur-positive TZ cells with asymmetric Miranda (Mira; Fig. 1, I–I″; Ikeshima-Kataoka et al., 1997). Analysis of these cells at telophase/cytokinesis indicated that two cells of different sizes were produced from the division (Fig. 1, J and J′). The small basal daughter cell appeared to be a GMC based on Prospero (Pros) staining (Spana and Doe, 1995; Figs. 1 J′ and Fig. S1). This GMC should be the first cell of the Hth lineage, since Neur-positive TZ cells begin to express Hth. Finally, we examined the division of these Neur-positive TZ cells using ex vivo live imaging of L3 larval brains expressing Neur-GFP and Histone2Av-RFP (Fig. S1). Tracking cells live, we observed that medial Neur-positive TZ cells divide asymmetrically to produce a larger apical cell and a small basal daughter cell inheriting Neur (Le Borgne and Schweisguth, 2003).
It was previously reported that the medial-most TZ cells have a characteristic teardrop shape suggestive of apical constriction (Orihara-Ono et al., 2011). We confirmed this observation by showing that the Neur-positive TZ cells apically constrict (Fig. 1, K–L). Importantly, these TZ cells remain epithelial with apical E-cad, and retain E-Cad as they undergo mitotic rounding and asymmetric division (Fig. 1, M–M″). Live imaging of larval brains expressing E-cad-mCherry and Neur-GFP further revealed that medial Neur-positive TZ cells exhibit a clear E-cad signal as they round up during mitosis, indicating that these TZ cells retain apical junctions when they enter mitosis (Fig. S1). In summary, our data strongly suggest that the medial TZ cells are epithelial cells in G2 phase that undergo a first round of asymmetric cell division to produce the earliest-born GMCs. Thus, the Neur-positive cells of the TZ have unique features: they express early NSC markers and undergo the first round of asymmetric cell division but differ from more mature NSCs by the presence of apical E-cad, a key feature of epithelial cells that is lost in NSCs (Fig. 1, M–M″; see also below). We refer here to this transitory epithelial-like NSC state as the epi-NSC state and to the Neur-positive TZ cells as the epi-NSCs.
Apical constriction by medial Myosin in epi-NSCs
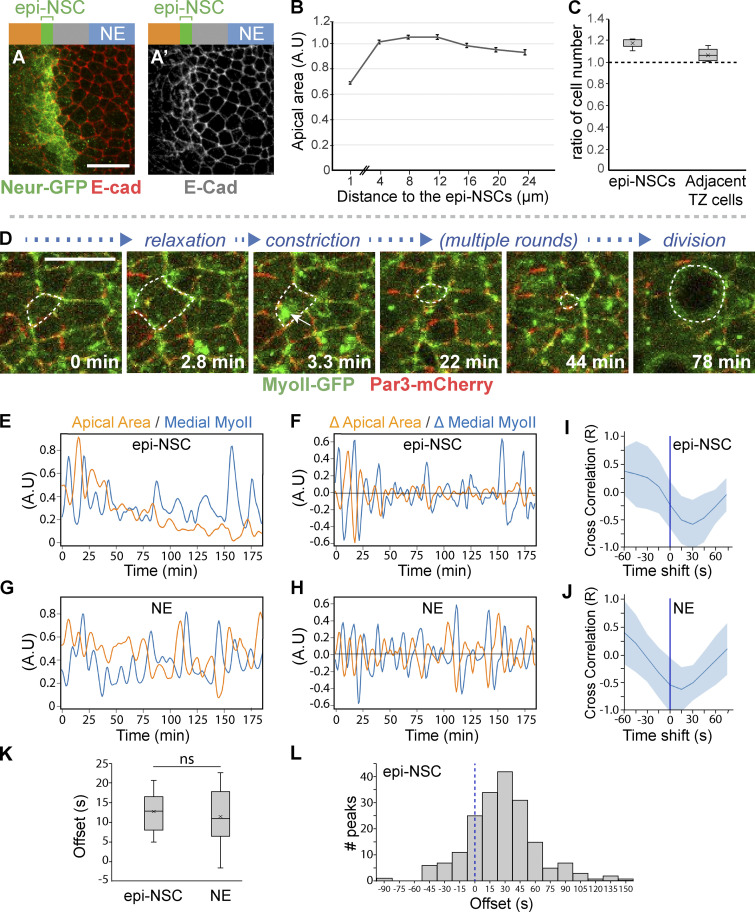
While epi-NSCs retain epithelial features until they divide, they appear to undergo apical constriction, which is suggestive of changes in epithelial organization. Using E-cad staining to quantify apical area (Fig. 2, A and B), we showed that medial TZ cells are indeed apically constricted. We further observed that apical constriction resulted in a slightly higher density of epi-NSCs compared with other TZ cells (Fig. 2 C), suggestive of cell–cell intercalation events within the TZ. In many epithelia, apical constriction can be induced by a contractile actomyosin meshwork (Martin et al., 2009; Martin and Goldstein, 2014). To monitor and correlate myosin dynamics with changes in apical area, we performed live imaging on ex vivo cultured brains from L3 larvae expressing a GFP-tagged allele of Myosin II (MyoII-GFP) and E-cad-mCherry (or partitioning defective 3 [Par3]-mCherry) to mark apical junctions. In these videos, epi-NSCs were detected as a row of apically constricted cells. Individual epi-NSCs undergoing a decrease of their apical area before division were selected for analysis. We observed that these epi-NSCs apically constricted through phases of apical area contraction and expansion (Fig. 2, D–F; and Video 1). Additionally, the levels of apical-medial MyoII appeared to pulse, suggesting phases of recruitment of MyoII at, and dissociation from, the apical cortex (Fig. 2, D–F). These MyoII pulses negatively correlated with apical area changes (Fig. 2 I). In addition, maximal accumulation of medial MyoII appeared to precede apical constriction by ∼13 ± 8 s (Fig. 2 K) with maximal accumulation of medial MyoII preceding apical reduction within 45 s in 46% of the constriction phases (Fig. 2 L). These data suggest that pulses of apical-medial MyoII drive apical constriction in epi-NSCs, as previously observed in several other epithelia (Martin et al., 2009; Martin and Goldstein, 2014) and in delaminating NSCs (Simões et al., 2017; An et al., 2017).
Figure 2.
Apical constriction of epi-NSCs correlates with pulses of medial-apical MyoII. (A) Surface view of wild-type NE stained for E-cad (red) and Neur-GFP (green) showing that epi-NSCs have smaller apical areas. Scale bars = 10 μm. (B) Quantification of apical area (normalized; A.U., arbitrary units) plotted against distance from epi-NSCs (in μm; x = 0 corresponds to a line joining the center of each epi-NSCs), showing that apical area decreases toward the medial edge of the TZ (n = 13 brains; means ± SEM of all pooled cells, 373–890 cells per binned distance). (C) Density along the D-V axis of epi-NSCs and other TZ cells relative to those of lateral NE cells (n = 8 brains; boxplots display the mean ratios, with the density of distal NE cells normalized to 1 for each brain). (D) Snapshots from a video of a brain expressing MyoII-GFP (green) and Par3-mCherry (red) showing an apically constricting TZ cell (dotted outline) undergoing phases of constriction and expansion. Constriction is associated with increased levels of medial-apical MyoII (arrow). Scale bars = 10 μm. (E–H) plots of apical area and MyoII intensity values over time for a representative epi-NSC and NE (G) cells, along with the rates of changes in apical area and MyoII levels (F and H). Increased MyoII levels were associated with reduced apical area in both epi-NSC and NE cells. (I and J) Cross-correlation values between area and myosin rates of change (mean ± SEM calculated for the different peaks) plotted as a function of the time shift in representative epi-NSC (10 contraction phases from one epi-NSC; n = 9; I) and NE cells (14 contraction phases from a single NE cell; n = 10; J). Anticorrelation was maximal with a 10–20-s time shift, i.e., offset, with maximal increase of MyoII preceding maximal rate of apical constriction. (K) Mean offset time between maximal rates of MyoII accumulation and area changes in epi-NSCs (n = 9) and NE cells (n = 10). MyoII accumulation similarly precedes constriction in both cell types (ns, no significant difference, two-tailed t test). (L) Offset times between maximal accumulation of MyoII and maximal constriction rates in epi-NSCs (n = 191 peaks, from 9 epi-NSCs).
Video 1.
Brain expressing MyoII-GFP (color-coded intensity) and Par3-mCherry (not depicted; used for segmentation) showing an apically constricting TZ cell (outline) undergoing phases of constriction and expansion. Changes in MyoII-GFP levels and apical area are shown on the right. Constriction is associated with increased levels of medial-apical MyoII. Frame rate is six frames per second.
Looking at MyoII dynamics in NE cells that are not undergoing apical constriction, we observed both medial MyoII pulses and fluctuations in apical area (Fig. 2, G–J; and Video 2). Maximal accumulation of MyoII preceded constriction by 12 ± 11 s (Fig. 2 K), indicating that pulses of apical-medial MyoII triggered constriction in NE cells. However, in contrast to epi-NSCs, NE cells did not apically constrict over time but rather displayed balanced phases of contraction and expansion. Apical constriction can result from medial MyoII pulses that persist long enough to let the energy stored dissipate in part via the remodeling of apical junctions and associated actin cortex (Martin and Wieschaus, 2010; Martin et al., 2009). Thus, differences in the cycle length of MyoII oscillations correlate with productive versus unproductive area fluctuations (Booth et al., 2014; Gorfinkiel and Blanchard, 2011; Blanchard et al., 2010). However, no significant difference was seen in the average duration of medial MyoII pulses between epi-NSCs and NE cells (not depicted). We therefore speculate that epi-NSCs might differ from NE cells in cortex remodeling such that remodeling is fast enough in epi-NSCs, but not in NE cells, to stabilize a loss of apical area upon constriction (Clément et al., 2017). In conclusion, contractile MyoII pulses appeared to direct sustained apical constriction in epi-NSCs but not in NE cells.
Video 2.
Brain expressing MyoII-GFP (color-coded intensity) and Par3-mCherry (not depicted; used for segmentation) showing an NE cell (outline) undergoing phases of constriction and expansion. Changes in MyoII-GFP levels and apical area are shown on the right. Constriction is associated with increased levels of medial-apical MyoII. Frame rate is six frames per second.
Junctional MyoII accumulation suggestive of supracellular cables within the TZ
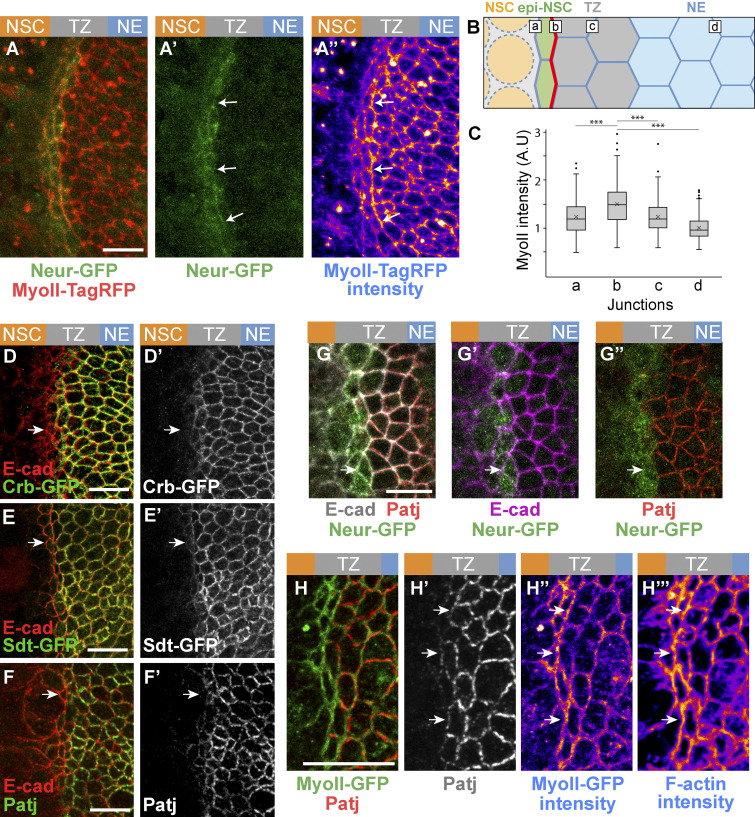
While studying MyoII dynamics, we observed high levels of junctional MyoII at cell–cell contacts between epi-NSCs and the more lateral TZ cells (Fig. 3, A–A″). Increased junctional MyoII was particularly noticeable where epi-NSCs line up to create a straight tissue interface with other TZ cells (Fig. 3 A″). We quantified the intensity of junctional MyoII along cell–cell contacts parallel to the differentiation front (Fig. 3 B) and showed that the epi-NSC/TZ junctions had significantly increased MyoII levels (Fig. 3 C). Additionally, increased junctional MyoII was accompanied by F-actin accumulation (Fig. 3, H–H″′). We therefore suggest that supracellular cables of actomyosin may form to produce enough tensile forces to line up epi-NSCs along this tissue interface (Röper, 2013). However, given the architecture of the brain, with the OL epithelium sandwiched between surface glia and neural tissues underneath, probing forces is technically challenging and was not performed.
Figure 3.
Low Crb in epi-NSCs correlates with polarized junctional MyoII. (A–A″) Snapshot of a brain expressing MyoII-TagRFP (red in A, intensity levels in A″) and Neur-GFP (green) showing an enrichment of junctional MyoII at the cell–cell contact between epi-NSC and more lateral TZ cells (arrows). Neur localized at the apical cortex (A′). (B) Diagram illustrating the different cell–cell contact used for the quantification of junctional MyoII, i.e., between NSCs and epi-NSCs (a), epi-NSCs and immediately adjacent TZ cells (b), between Neur-negative TZ cells (c) and lateral NE cells (d). (C) Box plots of junctional MyoII intensity at four distinct junctions as in B. MyoII is significantly enriched at b junctions relative to all others (n = 6, ***, P values <10−15, one-way ANOVA, Scheffé multiple comparison tests). (D–G″) Crb-GFP (green; D and D′), Sdt-GFP (green, all isoforms; E and E′) and Patj (green in F and F′; red in G and G″), but not E-cad (red in D, E, and F; magenta in G′′), were specifically down-regulated in apically constricted epi-NSCs (Neur-GFP, green, in G–G″). While E-Cad appeared to colocalize with Crb complex proteins in these apical z-projections, AJs are actually located just basal to Crb (see Fig. S2). (H–H″′) MyoII-GFP (green, H) was enriched along junctions lacking Patj (red; see arrows in H′ and H″). F-actin (phalloidin staining, H″′) was also enriched along junctions lacking Patj (arrows). Scale bars = 10 µm.
In other epithelia, supracellular MyoII cables have been observed along boundaries of cells with different levels of the apical protein Crb, and an anisotropic loss of Crb can cause their formation (Röper, 2012). To test whether Crb differentially accumulates in epi-NSCs relative to other TZ cells, we studied the pattern of Crb accumulation using GFP-tagged Crb. Crb-GFP localized apical to the AJs in NE and lateral TZ cells but was lost in epi-NSCs, identified as apically constricted medial TZ cells (Fig. 3 D and Fig. S2). The Crb complex proteins Patj and Sdt (observed using an allele tagging all isoforms of sdt with GFP) were similarly down-regulated in epi-NSCs (Fig. 3, E–G″). In contrast, the distribution of E-cad and Par3 remained largely unchanged (Fig. S2; note, however, that E-cad levels appeared to be slightly elevated, whereas Par3 was in part cytoplasmic in epi-NSCs, suggestive of junction remodeling). Additionally, we observed that MyoII was enriched at the apical membrane where Patj was down-regulated (Fig. 3, H–H″′). In summary, supracellular actomyosin cables appear to form at junctions juxtaposing TZ cells with high Crb and apically constricted epi-NSCs with low Crb, possibly creating a tension interface within the OL epithelium.
Figure S2.
Analysis of Crumbs and Par3 distribution. (A–A″′) Cross-section view showing that Crb-GFP (green) localizes just apical to AJs (E-cad, red) in NE cells (white arrows). Crb-GFP is down-regulated in medial-most TZ cells (yellow arrow) and was not detected in NSCs (Dpn, blue). (B and B′) epi-NSCs (Neur-GFP, green) retain Par3 (red) at junctions (see arrows), albeit at a slightly lower level consistent with junction remodeling in epi-NSCs. Scale bars = 10 µm.
Neur down-regulates Crb via Sdt in epi-NSCs
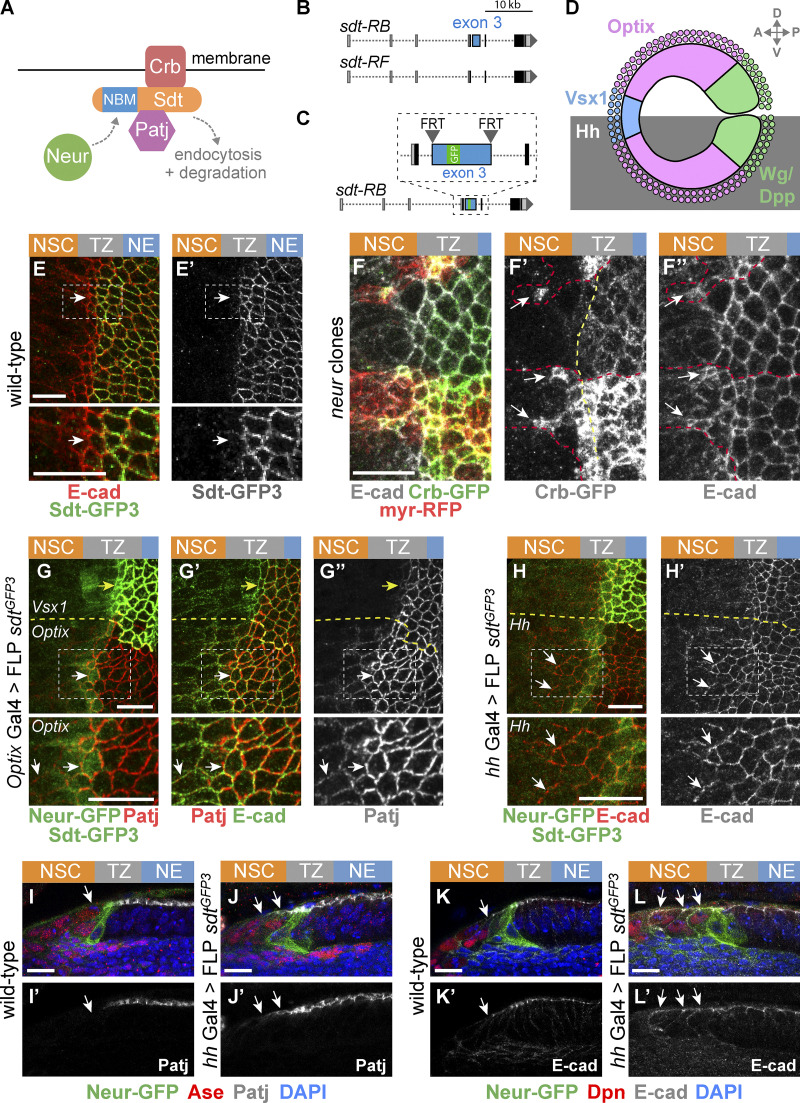
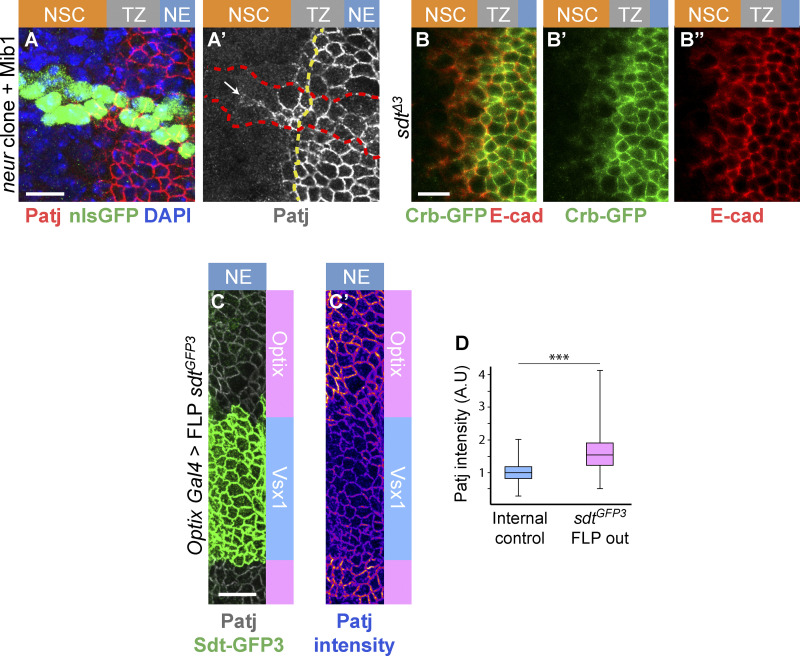
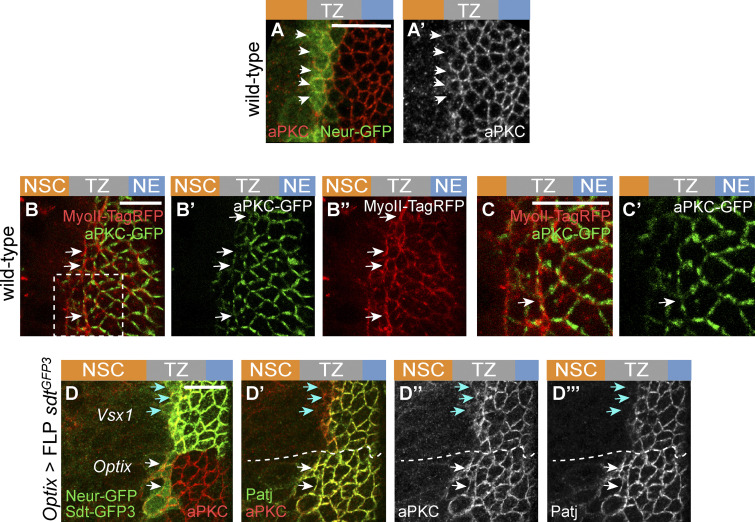
We next looked for candidate regulators of the Crb complex that might down-regulate Crb in epi-NSCs and first considered Neur. Indeed, while a key function of Neur is to target Delta for endocytosis (Lai et al., 2001; Le Borgne and Schweisguth, 2003; Weinmaster and Fischer, 2011), Neur can also down-regulate Crb, Std, and Patj, possibly via endocytosis and degradation of Crb complexes (Fig. 4 A; Perez-Mockus et al., 2017b). Neur directly interacts with a subset of Sdt isoforms that contain a Neur binding motif (NBM), encoded by exon 3, and targets Crb complexes that include these Sdt isoforms for degradation (Fig. 4 B). Using a knock-in allele of sdt that has GFP inserted into the sequence of exon 3 (Perez-Mockus et al., 2017b), we found that the NBM-containing isoforms of Sdt are expressed in the NE and TZ (Fig. 4, C and E). We therefore tested the role of Neur in the down-regulation of Crb. Crb-GFP was found to persist past the TZ boundary in neur mutant cells (Fig. 4, F–F″), indicating that Neur is required for the down-regulation of Crb in epi-NSCs. E-cad also appeared to persist in mutant NSCs close to the TZ (Fig. 4 F″), suggesting a delay in AJ disassembly in the absence of neur activity. Forced expression of the E3 ubiquitin ligase Mindbomb1 (Mib1), which can act redundantly with Neur in the regulation of Delta endocytosis (Lai et al., 2005; Le Borgne et al., 2005), did not rescue the delay in Patj down-regulation in neur mutant cells (Fig. S3). This indicates that Neur is unlikely to down-regulate Crb complex proteins in the OL via the regulated endocytosis of Delta. To test whether Neur down-regulates Crb via Sdt, we used a sdt allele that was engineered to flank the exon 3 with flippase recombinase target (FRT) sites such that only Neur-resistant isoforms of Sdt are expressed upon flippase (FLP)-mediated excision of this exon (Fig. 4 C). Using this tool, we compared the localization of Crb complex proteins in optix-Gal4 or hh-Gal4 cells expressing the Neur-resistant isoforms of Sdt with control cells in other domains of the NE (Fig. 4, G–G″). Patj was maintained at the apical surface of the Neur-positive epi-NSCs when exon 3 of sdt was excised. Moreover, both Patj and E-cad persisted in NSCs marked by Ase or Dpn, suggesting a delay in the EMT-like process (Fig. 4, H–L′). Of note, increased Patj accumulation was seen in NE cells upon deletion of exon 3 (Fig. S3), suggesting that the short Sdt isoforms promote the stability of the Crb complex even in cells that do not express Neur. Lastly, Crb-GFP was seen to persist in the most medial TZ cells (epi-NSCs) in sdtΔ3 mutant larvae that carry a deletion of exon 3 (Fig. S3). Together, these data demonstrate that Neur down-regulates the Crb complex via Sdt in epi-NSCs.
Figure 4.
Neur acts via Sdt to down-regulate Crb in epi-NSCs. (A) Working model of Crb regulation by Neur. Neur interacts directly with Sdt isoforms that include the NBM and targets the Crb complex for endocytosis and degradation. (B) Simplified view of the alternatively spliced sdt transcripts. A subset of the sdt mRNAs, such as RB but not RF, includes exon 3, which encodes the NBM of Sdt. (C) Molecular structure of the sdtGFP3 allele with GFP inserted into exon 3, itself flanked by FRT sites. (D) Diagram of the NE subdivided into spatial domains expressing different transcription factors including Optix and Vsx1, or signaling molecules including Dpp, Wg, and Hh, hence providing a toolbox of spatially restricted Gal4s. (E and E′) Sdt isoforms containing the sequence encoded by exon3 (Sdt-GFP3, green) were expressed in the NE and down-regulated in epi-NSCs (E-cad, red). (F–F″) Crb (Crb-GFP, green) was detected colocalizing with E-cad (white) in neur mutant cells past the TZ medial edge (arrows in F′ and F″). Mutant cells were marked by a membrane RFP (myrRFP, red) and had two copies of Crb-GFP, accounting for higher Crb-GFP levels. (G–G″) Expression of only Neur-resistant isoforms of Sdt, via the excision of exon 3 in the sdtGFP3 allele in the Optix domain (note the domain-specific loss of Std-GFP3, green in G), led to the detection of Patj (red in G) colocalizing with E-cad (green in G′) in mutant epi-NSCs (arrow; Neur-GFP, green in G) as well as in NSCs (arrowhead). In contrast, Patj was down-regulated in control epi-NSCs of the Vsx1 domain (yellow arrows). (H and H′) Excision of exon 3 of sdt in the Hh domain had a similar effect on E-Cad (red; see loss of Std-GFP3, green in G, in this domain; Neur-GFP, also green). (I–L′) Cross-section views showing that Patj (white in I–J′) was down-regulated in epi-NSCs (Neur-GFP, green) in the presence of Neur-sensitive Sdt in wild-type brains (I and I′) but not when only Neur-resistant Sdt was expressed upon excision of exon 3 in hh>FLP sdtGFP3 (J and J′). Note that Patj was observed at the apical cortex of NSCs (Ase, red) upon excision of exon 3 (J and J′). Likewise, E-cad (white in K–L′) was detected further within the NSC domain (Dpn, red) when Neur-resistant isoforms of Sdt were expressed (K–L′). DAPI (blue) marked all nuclei. Scale bars = 10 µm.
Figure S3.
Analysis of the down-regulation of Crb and Patj. (A and A′) Patj (red) was observed to persist at the apical cortex of neurIF65 mutant cells overexpressing Mib1 (using tub-Gal4 in MARCM clones marked by nuclear GFP, green, outlined in red in A′; DAPI, blue) past the TZ medial edge. The boundary between epi-NSCs and other TZ cells is shown as a yellow dotted line. (B–B″) Crb-GFP (green; E-cad, red) was observed to persist at the apical cortex of the most medial TZ cells (epi-NSCs) in the OL of larvae carrying a deletion of exon 3 of sdt (sdtΔ3). (C and D) Loss of the NBM-containing isoforms of Sdt in the Optix domains (marked by the loss of Sdt-GFP3, green, upon FLP-mediated excision of exon 3) resulted in slightly higher accumulation of Patj (white, B; intensity code, B′), as quantified in D (n = 4). Scale bars = 10 µm. ***, P < 10−3.
Neur regulates apical constriction in epi-NSCs in part via Sdt
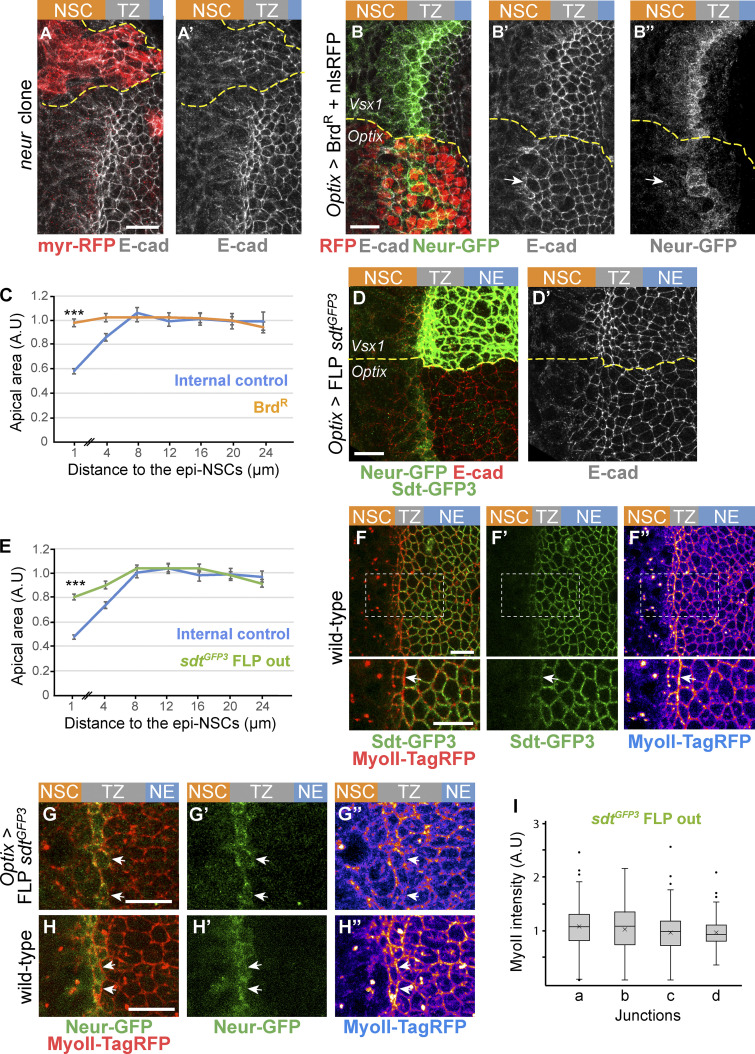
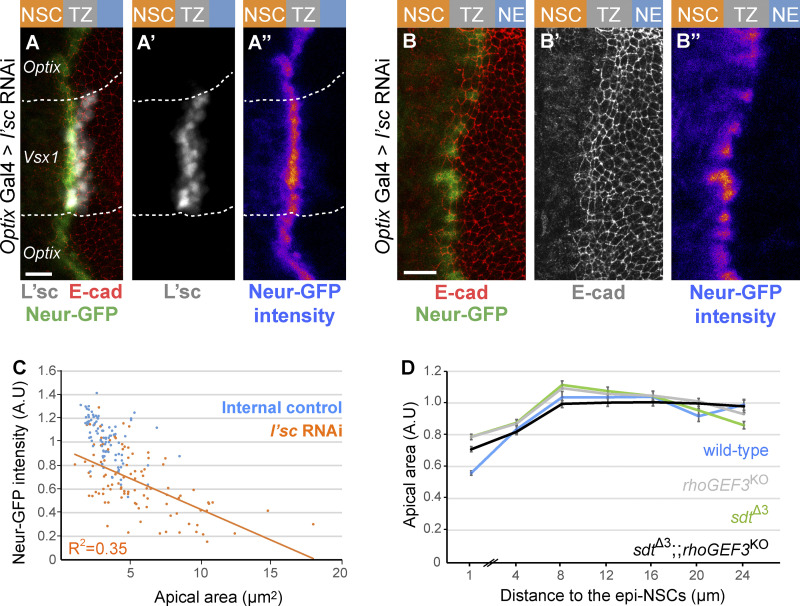
We next asked whether this regulation of Crb by Neur is required for the apical constriction of epi-NSCs. We found that medial TZ cells were no longer apically constricted in neur mutant clones (Fig. 5, A and A′). Additionally, ectopic expression of the Neur inhibitor BrdR, a stabilized version of Bearded (Brd; Bardin and Schweisguth, 2006; De Renzis et al., 2006; Perez-Mockus et al., 2017a), strongly reduced apical constriction and delayed the loss of E-cad in NSCs (Fig. 5, B and C). Finally, we used RNAi against the l’sc gene to perturb the expression of the neur gene, a direct target of the proneural factors (Miller et al., 2014; Rouault et al., 2010; Contreras et al., 2018), resulting in low and heterogeneous Neur-GFP expression (Fig. S4), and showed that Neur-GFP levels negatively correlated with apical area in epi-NSCs (Fig. S4). Together, these results show that Neur positively regulates the apical constriction of epi-NSCs.
Figure 5.
Neur is required for apical constriction and acts in part via Sdt. (A and A′) Loss of neur activity in neurIF65 MARCM clones (myr-RFP, red) led to defective apical constriction in cells located at the medial edge of the TZ as well as to the persistence of E-cad (white) in mutant NSCs. (B–B″) Inhibition of Neur by BrdR in the Optix domain (nuclear RFP, red) induced a loss of apical constriction of epi-NSCs (Neur-GFP, green) and persistence of apical E-cad (white) past the epi-NSCs (arrow). (C) Analysis of apical area in cells expressing BrdR and in control wild-type cells of the Vsx1 domain, plotted as in Fig. 2 B, showed that inhibition of Neur led to reduced apical constriction (n = 7 brains; means ± SEM were calculated using 34–210 cells per binned distance; ***, P = 8 × 10−29, two-tailed Student’s t test). (D and D′) Expression of only Neur-resistant isoforms of Sdt (loss of Sdt-GFP3), following excision of exon 3 of the sdtGFP3 allele, interfered with apical constriction of epi-NSCs (E-cad, white in D′5; compare apical area of cells at the medial edge of the TZ in the Optix vs. control Vsx1 domains). (E) Quantification of apical area in cells of the Optix domain expressing Neur-resistant isoforms of Sdt compared with control cells from the Vsx1 domain, plotted as in Fig. 2 B. The failure to down-regulate Sdt in the epi-NSCs (located at the 0–2-µm position along the x axis) resulted in intermediate level of apical constriction (n = 8 brains, means ± SEM were calculated using 82–319 cells per binned distance; ***, P = 5.9 × 10−26, two-tailed Student’s t test). (F–F″) In wild-type brains, MyoII (MyoII-TagRFP, red) is enriched at straight interfaces along the medial edge of the TZ where Neur-sensitive isoforms of Sdt (Sdt-GFP3, green) are down-regulated. The regions boxed in F–F″ are shown at higher magnification below. (G–H″) When only Neur-resistant isoforms of Sdt are expressed, upon FLP mediated excision of sdt exon 3 (G–G″), both increased MyoII levels (MyoII-TagRFP, red) and straight interfaces seen in wild-type brains (H–H″) were less clearly observed at the boundary between epi-NSCs (Neur-GFP, green) and adjacent TZ cells (see arrows). (I) Quantification of junctional MyoII along different cell–cell contacts (data plotted as in Fig. 2, B and C; see Fig. 2 C for the wild-type control; n = 9 brains; ns, nonsignificant difference, one-way ANOVA). Scale bars = 10 μm.
Figure S4.
Regulation of apical constriction by Neur and RhoGEF3. (A–B″) RNAi-mediated knockdown of l’sc expression in the Optix domain induced a strong loss of L’sc (white in A and A′) which was associated with a decrease in Neur-GFP (green in A; intensity in A″; compare epi-NSCs of the Optix and Vsx1 domains in A–A″). A high degree of cell-to-cell heterogeneity in Neur-GFP levels was seen upon l’sc knockdown (B–B″). (C) Quantification of apical area (normalized, A.U [arbitrary units]) in the NE of sdtΔ3 rhoGEF3 double mutants (black; n = 9 brains) plotted against the distance from the TZ medial edge (70–474 cells per binned distance). The phenotype of the double mutant appeared to be similar to those of rhoGEF3 (data from Fig. 6 D) and sdtΔ3 (data from Fig. 5 E; wild-type data from Fig. 6 D): double mutant cells located within 0–2 µm of the TZ medial edge do not have significantly different apical areas than either single mutant (ANOVA). (D) Neur-GFP intensity values (normalized to mean GFP values of control epi-NSCs from the same brain; maximal projection of apical Δz = 2 µm) were plotted against apical area for 102 epi-NSCs (n = 5 optix>l’sc RNAi brains). An inverse correlation between Neur-GFP intensity and apical area was observed, suggesting that Neur positively regulates apical constriction. Scale bars = 10 µm.
We next tested whether Neur regulates apical constriction via the regulation of Sdt. To do so, we quantified the apical area of TZ cells expressing only the Neur-resistant isoforms of Sdt upon excision of sdt exon 3. An increase in the apical area of epi-NSCs was observed (Fig. 5, D and E). Thus, Neur regulates apical constriction in epi-NSCs in part by inducing the down-regulation of the Crb complex. However, the effect seen upon excision of sdt exon 3 appeared to be milder than the one observed upon the inhibition of Neur by BrdR (Fig. 5, C and E). This suggests that Neur regulates apical constriction via additional targets. Consistent with this, Neur regulates apical constriction and actomyosin contractility independently of Sdt during ventral furrow formation in the embryo (Perez-Mockus et al., 2017a).
Loss of Crb down-regulation in epi-NSCs upon excision of exon 3 of sdt also resulted in a lack of MyoII enrichment along the epi-NSC/TZ junctions (Fig. 5, F–J), consistent with the notion that Crb anisotropy promotes the formation of supracellular MyoII cables. Crb may regulate MyoII levels via the regulated distribution of Rho kinase (Sidor et al., 2020) and possibly atypical PKC (aPKC; Röper, 2012). Consistent with this, low aPKC levels were detected at apical junctions of epi-NSCs where MyoII accumulated, and this distribution of apical aPKC appeared to depend on the down-regulation of Crb by Neur (Fig. S5). Thus, the down-regulation of Crb by Neur not only regulates apical constriction in epi-NSCs but also promotes the formation of supracellular MyoII cables.
Figure S5.
Regulation of apical constriction by Neur and RhoGEF3. (A and A′) Low levels of aPKC (red) were detected in epi-NSCs (Neur-GFP, green) in wild-type brains. (B–C′) Snapshot of a living brain showing that aPKC-GFP (green) localized uniformly at apical junctions of NE and TZ cells but appeared to localize in patches at the apical cortex of epi-NSCs. Note that MyoII-TagRFP (red) was observed to be enriched along junctions where aPKC were decreased (white arrows). The inset boxed in B is shown in C and CRNAis. (D–D″′) While low aPKC levels were observed at the apical cortex of control epi-NSCs in the Vsx1 domain (blue arrows), expression of the Neur-resistant isoforms of Sdt (upon excision of exon 3 of sdt in the Optix domain, hence the loss of Sdt-GFP3, green) led to the persistent accumulation of aPKC (red; white arrows in L–L″) and Patj (green; LRNAis and L″) in epi-NSCs (Neur-GFP, green in L). In the Vsx1 domain, Sdt-GFP3 is down-regulated in epi-NSCs (blue arrows). Scale bars = 10 µm.
RhoGEF3 regulates apical constriction independently of the regulation of Sdt by Neur
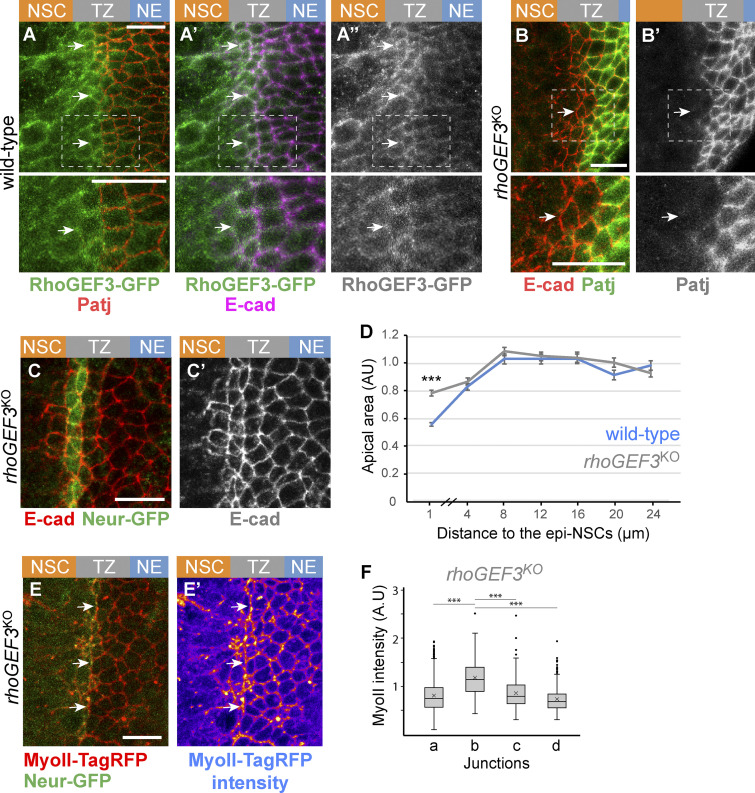
A second potential regulator of cortex remodeling in epi-NSCs is Rho guanine nucleotide exchange factor 3 (RhoGEF3), an exchange factor for Rac/Cdc42 (Couturier et al., 2017; Nakamura et al., 2017). Like Neur, RhoGEF3 is expressed in neural progenitor cells downstream of proneural factors (Couturier et al., 2017). Using a knock-in allele of RhoGEF3 tagged with GFP, we found that RhoGEF3 accumulated at the apical cortex of apically constricted TZ cells (Fig. 6, A–A″). To assay the role of RhoGEF3 in the TZ, we studied a rhoGEF3 knockout allele (Couturier et al., 2017). While the loss of rhoGEF3 had no effect on the down-regulation of Patj in epi-NSCs (Fig. 6, B and B′), it led to a partial loss of apical constriction (Fig. 6, C and D). Thus, RhoGEF3 regulates the apical constriction of epi-NSCs independently of Crb complex regulation. Analysis of apical constriction in sdtΔexon3, rhoGEF3KO double mutant brains showed that the double mutant had a phenotype similar to each single mutant (Fig. S4). Assuming that Neur-Sdt and RhoGEF3 act in a single linear pathway to regulate apical constriction, it is possible that RhoGEF3 acts downstream of Sdt. Alternatively, RhoGEF3 and Neur-Sdt may act in parallel pathways. Because the loss of rhoGEF3 had no effect on the down-regulation of the Crb complex, we wondered whether MyoII accumulated at the interface between epi-NSCs and other TZ cells in the absence of RhoGEF3. Using MyoII-TagRFP and Neur-GFP in cultured rhoGEF3 mutant brains, we found that junctional MyoII remained significantly enriched at the cell–cell contacts between epi-NSCs and their immediate TZ neighbors (Fig. 6, E and F; compare with Fig. 3 C). We suggest that low Crb, and not apical constriction, directs the formation of supracellular actomyosin cables (Röper, 2012).
Figure 6.
RhoGEF3 is required for apical constriction. (A–A″) RhoGEF3-GFP (green) was detected at low levels at apical junctions of the NE (Patj, red; E-cad, magenta) and appeared to be up-regulated in NSCs and epi-NSCs (white arrows); epi-NSCs were identified via the down-regulation of Patj. (B and B″) Down-regulation of Patj (green) appeared to occur normally in cells located at the medial edge of the NE in rhoGEF3 mutant brains (white arrows; E-cad, red). (C and D) Apical constriction of epi-NSCs (Neur-GFP, green in C) was reduced in rhoGEF3 mutant brains (C and C′; E-cad, red), as shown in the quantification (D) of the apical area in rhoGEF3 mutant (n = 9 brains) versus wild-type brains (n = 7; data presented as in Fig. 2 B, with 149–326 cells per binned distance; ***, P = 7 × 10−19, two-tailed Student’s t test). (E and E′) Junctional MyoII (MyoII-TagRFP, red) appeared to be enriched along at cell–cell contacts between epi-NSCs (Neur-GFP, green) and more lateral TZ cells (white arrows). (F) Box plots of MyoII intensity along junctions of the NE based on the four distinct junction identities described in Fig. 3 B, showing that b junctions had a significant enrichment of MyoII in rhoGEF3KO brains (n = 7 brains, b vs. a, b, or c, ***, P values <10−12, one-way ANOVA, Scheffé multiple comparison tests). Scale bars = 10 µm.
Coupling fate transition in individual cells with epithelium remodeling at the tissue scale
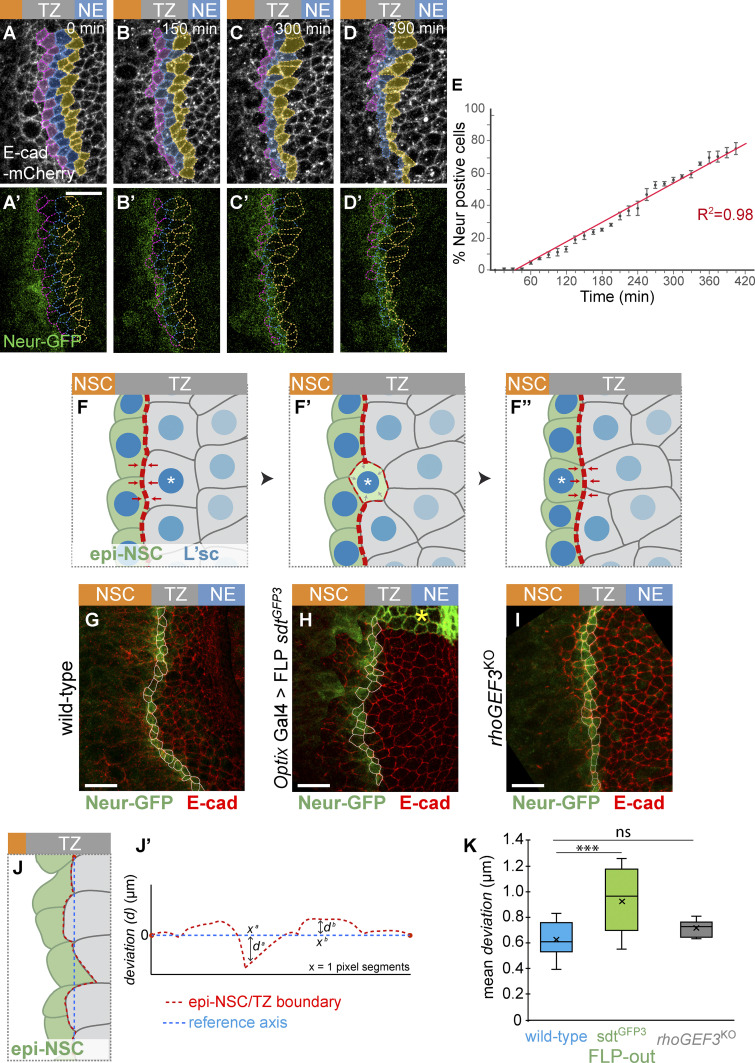
The dynamics of the NE-NSCs fate transition has so far mostly been considered with a particular focus on temporal dynamics (Egger et al., 2010; Weng et al., 2012; Ngo et al., 2010; Wang et al., 2011; Orihara-Ono et al., 2011). In contrast, how fate dynamics is coordinated in space along the differentiation front has not been examined. In particular, it is not clear whether all TZ cells juxtaposed to the epi-NSCs progress synchronously to form a new row of epi-NSCs, hence replacing the preexisting epi-NSCs in a quasi-simultaneous manner, leading to a saltatory progression of the epi-NSC front, or whether individual TZ cells become epi-NSCs in a more continuous manner, such that the epi-NSC front progresses gradually. To distinguish between these two possibilities, we followed the dynamics of TZ cell entry into the row of epi-NSCs live in cultured L3 brains. Neur-GFP and E-cad-mCherry were used to mark epi-NSCs and to outline epithelial cells, respectively. Tracking cells adjacent to the epi-NSCs, we monitored the timeline of TZ cells becoming epi-NSCs, i.e., express Neur-GFP, and found that the number of TZ cells becoming epi-NSCs increased at a constant rate over time (Fig. 7, A–E; a threshold value of Neur-GFP intensity was used to define when TZ cells become epi-NSCs). Thus, TZ cells do not adopt an epi-NSC state in a periodic manner, but rather individual TZ cells switch fate and integrate into the row of epi-NSCs one cell at a time in a gradual manner.
Figure 7.
Dynamics of of TZ-epi-NSC transition in space and time. (A–D′) Snapshots from a time-lapse video showing the dynamics of TZ-epi-NSC transition (E-cad-mCherry, white; Neur-GFP, green). TZ cells that have not yet become epi-NSCs were manually tracked (color code refers to the distance to epi-NSCs at t = 0). (E) Cumulative plot of TZ cells switching to the epi-NSC state, based on Neur-GFP expression over time (switch was defined by a threshold of 1 SD below the mean of Neur-GFP intensity in apically constricted epi-NSCs; n = 3 brains; 34–47 cells tracked per brain). Transition appeared to be a continuous process. (F–F″) Model of mechanical coupling by Neur. Acquisition of the epi-NSC state (green; L’sc levels in blue) is a probabilistic event occurring at the single-cell level (asterisk; F). It is associated with the expression of Neur that in turn induces apical constriction (F′). Neur down-regulates Crb in a cell-autonomous manner, hence moving the boundary between low and high Crb cells, hence the enrichment of junctional MyoII (red dotted line) resulting from the Crb anisotropy: MyoII accumulation increases at the lateral junction and decreases at the medial junction (F′). This is proposed to modify tension along this interface and promote cell–cell rearrangement to direct integration of the newly fated epi-NSCs into the one-cell row of epi-NSC at the medial edge of the TZ (F″). This mechanical coupling may ensure precise tissue architecture and regular spatial progression of the differentiation front. (G–I) Immunostainings showing the epi-NSCs (Neur-GFP, green; E-cad, red; cell outlines in white) located at the interface with the Neur-negative TZ cells which were used to evaluate precision of morphogenesis (G, wild-type; H, optix-Gal4 UAS-flp sdtGFP3: Neur-resistant cells do not express Sdt-GFP3, green; yellow star; I: rhoGEF3KO). (J and J′) The roughness of the epi-NSC/TZ junctional boundary (red dotted line in J and J′) was measured by calculating the mean of the deviation values (d in J′) along the x axis from a reference axis (blue dotted line in J and J′) linking the first junctional vertex to the last in groups of five epi-NSCs (green in J). See also Materials and methods. (K) Plot showing the mean values of deviation, or roughness, in wild-type (n = 12), optix-Gal4 UAS-flp sdtGFP3 (n = 10), and rhoGEF3KO (n = 6) brains (ns, not significant; ***, P = 0.00092, one-way ANOVA, Scheffé multiple comparison tests). Upon excision of exon 3 of sdt, epi-NSCs formed a less regular junctional interface. Scale bars = 10 μm.
Our observations therefore suggest a model whereby epithelial cortex remodeling in individual cells promotes smooth progression of the differentiation front at the tissue level by facilitating the organization of cells in space that are at a similar stage of fate transition (Fig. 7, F–F″). Specifically, we propose that as individual cells progress toward a epi-NSC fate via the up-regulation of L’sc, Neur is expressed, leading to the down-regulation of the Crb complex (Fig. 7 F′). This creates a new low-high Crb interface, with the previous low-high Crb cell–cell contact being converted into a low-low Crb interface. These changes impact on MyoII junctional accumulation, thereby repositioning the cell–cell contacts that are under tension and promoting the integration of the newly specified epi-NSC into the row of preexisting epi-NSCs (Fig. 7 F″). This model predicts that a failure to down-regulate Crb in cells progressing toward the epi-NSC state should result in defects in the integration of these cells into the row of epi-NSCs. To test this prediction, we evaluated how newly specified epi-NSCs integrate into the row of previously specified epi-NSCs by quantifying the roughness of the epi-NSC/TZ junctional boundary (Fig. 7, J and J′); as described in Rudolf et al. (2015), roughness measures the average variation of contour length between two points from a straight line). We found that the roughness of the epi-NSC/TZ boundary was significantly increased when the regulation of Crb by Neur is disrupted, i.e., upon excision of exon 3 of the sdtGFP3 allele (Fig. 7, G, H, and K). To test whether increased roughness is associated with loss of apical constriction or defective junctional MyoII enrichment, which are both perturbed when Sdt is no longer regulated by Neur, we examined rhoGEF3 mutants that exhibit defective apical constriction but detectable MyoII cables (Fig. 6, C–F). Similar roughness values were measured in wild-type and rhoGEF3 brains (Fig. 7, I and K). Thus, defects in junctional MyoII accumulation, and not defective apical constriction, might increase roughness of the epi-NSC/TZ boundary. We therefore suggest that the Crb-regulated accumulation of junctional MyoII downstream of Neur promotes the formation of a precisely lined-up single-cell row of epi-NSCs.
Discussion
The emergence of NSCs in the OL provides a good model to study how acquisition of a stem cell identity is associated with changes in epithelial tissue organization. Here we identify a transient cellular state during the NE-to-NSC transition that we named the epi-NSC state. It is characterized by the persistence of epithelial features, including apical E-cad, and up-regulation of NSC identity factors such as Wor, as well as other features including apical constriction, expression of Neur, and loss of the Crb complex. We find that TZ cells are in the G2 phase of the cell cycle and that epi-NSCs enter mitosis and divide asymmetrically to produce the first-born GMC. As individual TZ cells acquire this epi-NSC state and apically constrict, each emerging epi-NSC individually integrates the epi-NSC row. Neur in epi-NSCs regulates not only Delta signaling but also apical constriction, in part via the down-regulation of the Crb complex. Low Crb in epi-NSCs creates an anisotropy in Crb levels within the TZ. This anisotropy appears to result in the formation of supracellular MyoII cables. Down-regulation of the Crb complex by Neur is required for the proper integration of these emerging epi-NSCs into the row of previously established epi-NSCs. Thus, we propose that Neur links NSC fate specification with the formation of a continuous stripe of precisely lined-up epi-NSCs in the OL.
Neur regulates Notch signaling via the endocytosis of Delta and contributes to epithelium morphogenesis via its interaction with specific Sdt isoforms (Perez-Mockus et al., 2017b). These two activities of Neur coincide in the TZ, where Neur not only regulates Notch activity at the TZ (Contreras et al., 2018) but also down-regulates the Crb complex in epi-NSCs. Analysis of cells expressing only Neur-resistant Sdt isoforms showed that this activity of Neur is important for both apical constriction and formation of a smooth differentiation front. Neur is probably only one of several factors expressed in TZ cells downstream of the proneural activator L’sc that are involved in the regulation of apical constriction and cell–cell rearrangements. Indeed, our data indicate that RhoGEF3 also regulates apical constriction in epi-NSCs. In the ratchet model of apical constriction, first proposed in the context of ventral furrow formation (Martin et al., 2009), apical constriction is induced by the contractions of the medial actomyosin meshwork pulling onto apical junctions, combined with a rapid remodeling of the apical junctions, dissipating the energy stored (Mason et al., 2013; Martin et al., 2010; Clément et al., 2017). Because MyoII pulses were observed in both NE and epi-NSCs, our observation that only epi-NSCs, not NE cells, undergo apical constriction in response to repeated pulses of medial MyoII may suggest that the apical cortex is remodeled faster in epi-NSCs than in NE cells, hence contributing to stabilize a loss of apical area upon constriction (Clément et al., 2017). For instance, E-cad could be internalized at a faster rate in the absence of the Crb complex in epi-NSCs. This working hypothesis is consistent with the notion that the Crb complex stabilizes AJs and regulates MyoII at junctions to maintain epithelium integrity during morphogenesis (Harris and Tepass, 2008; Flores-Benitez and Knust, 2015; Campbell et al., 2009; Bajur et al., 2019; Perez-Mockus et al., 2017b; Röper, 2012; Salis et al., 2017; Sen et al., 2012). It is also consistent with our observation that loss of Crb regulation by Neur led to E-cad persisting in NSCs past the TZ. Future studies will address whether Neur promotes a faster turnover of E-cad via its regulatory activity on the Crb complex.
Both deletion of rhoGEF3 and loss of the regulation of Crb by Neur via the deletion of the sdt exon 3 reduced apical constriction of epi-NSCs without producing clear defects in NSC differentiation and OL development. This raises the question of the functional significance of apical constriction of epi-NSC. While it is possible that apical constriction facilitates EMT and cell–cell intercalation events at the TZ front in the OL, it may also be that apical constriction of epi-NSC merely reflects conservation of a morphogenetic program shared with other neural progenitors, e.g., delamination of NSCs from the neuroectoderm in the early fly embryo. Indeed, delaminating neuroblasts exhibit down-regulation of both Crb and E-cad preceding the first round of asymmetric division (Simões et al., 2017). In contrast, Crb but not E-cad is down-regulated in epi-NSCs in the OL before division. Whether this difference in E-cad distribution merely reflects a difference in geometry, e.g., delamination of individual cells surrounded by ectodermal cells, which reinforce their AJs in response to Notch versus progressive and collective changes along a front, or molecular differences in regulatory mechanisms remains to be explored. Interestingly, while Neur promotes EMT, Notch has been shown to prevent mesenchymal transition in fly imaginal disc epithelia (Boukhatmi et al., 2020) and human lung cancer (Ito et al., 2017). This raises the possibility that high Notch activity in the TZ blocks the NE-to-NSC transition in all TZ cells before the Notch ON/OFF switch. Thus, Neur in epi-NSCs could not only regulate an EMT-like process in epi-NSCs but also contribute to inhibit this same process in its TZ neighbors by promoting high Notch activity.
Reproducibility in morphogenesis is commonly observed. It relies in part on the precise decoding of positional information by gene regulatory networks (Petkova et al., 2019) to regulate cell shape changes at the single-cell level. In the OL, NE cells presumably read their distance to the NSC front through a combination of EGFR and Notch signaling to regulate L’sc levels. Reproducibility also often relies on tissue-scale mechanical coupling to organize cells in space (Eritano et al., 2020; Aliee et al., 2012; Yevick et al., 2019). For example, neural progenitors in the fish neural tube rearrange cell–cell contacts to establish sharp boundary through cell sorting. This sorting process corrects imprecision of spatial patterning through spatial averaging to establish sharp boundaries (Xiong et al., 2013). In the OL, the Neur-regulated Crb anisotropy within the TZ is proposed to result in the formation of supracellular MyoII cables that contribute to precisely line up Neur-expressing cells. In the absence of Crb regulation by Neur, the roughness of the epi-NSC/TZ boundary was increased, resulting in a less precise differentiation front. We therefore suggest that mechanical coupling of the Neur-expressing cells via supracellular MyoII cables contributes to define a spatially precise NSC front. While imprecision associated with the loss of the regulation of the Crb complex by Neur did not result in detectable defects in the expression of NSC markers, precision of this particular morphogenetic process might be relevant for fitness, for instance via the precise mapping to photoreceptor cells being simultaneously specified in the developing retina (Sato et al., 2013). Future studies will address whether Neur operates in other developmental contexts to regulate both Notch-dependent fate dynamics with changes in cell polarity and cortex contractility to mechanically couple at the tissue scale cells that are in a similar state and thereby ensure precision in morphogenesis.
Materials and methods
Flies
D. melanogaster flies were raised in food vials at 25°C, and larvae were allowed to develop until the desired stage. For all RNAi experiments, embryos and larvae were raised at 29°C instead of 25°C. Wild-type controls were w1118 flies. Otherwise, the following alleles and transgenes were used: E-cad-mcherry (FBal0247911) and crb-GFP-A (FBal0247913; Huang et al., 2009; both from Y. Hong); sdtGFP, tagging all Sdt isoforms (FBal0345001; Perez-Mockus et al., 2017b); sdtGFP3, a GFP-tagged and FRT-flanked knock-in allele of the sdt gene tagging all long isoforms of Sdt containing the NBM (FBal0345003; Perez-Mockus et al., 2017b); sdtΔ3, a mutant allele with a deletion of the GFP-tagged exon 3 from the sdtGFP3 allele (FBal0345002; Perez-Mockus et al., 2017b); rhoGEF3KO, a deletion of part of the RhoGEF3 locus generated by homologous recombination (FBal0338001; Couturier et al., 2017); MyoII-GFP (FBal0352225) and MyoII-TagRFP (FBal0352226), CRISPR knock-in alleles (Gracia et al., 2019; from M. Suzanne, CNRS, University of Toulouse, Toulouse, France) also noted sqheGFP and sqhTag-RFP; Neur-GFP (FBal0345008; Perez-Mockus et al., 2017a); RhoGEF3-GFP (FBal0337998; Couturier et al., 2017); aPKC-GFP (FBal0346728; Besson et al., 2015); ubi-Par3-mCherry (FBal0269882; Herszterg et al., 2013; from Y. Bellaïche, Institut Curie, Paris, France; also noted Baz-Cherry); optix-Gal4, also known as NP2631-Gal4 (FBti0034959; Gold and Brand, 2014; from A. Brand, Gurdon Institute, Cambridge, UK); hh-Gal4 (FBal0121730); FRT82B neurIF65 (FBal0012950); upstream activating sequence (UAS)-BrdR, a stabilized version of the protein Brd in which all K residues were mutated into R residues (FBal0355292; Perez-Mockus et al., 2017b); UAS-Mib1, a P element–based transgene carrying a mib1 cDNA under the control of UAS (Le Borgne et al., 2005); ubi-GFP-E2F1 combined with ubi-mRFP-CycB (FUCCI line, BDSC-55124); UAS-dsRNAi l’sc (BDSC-51443); Histone2Av-RFP (His2Av-RFP; BDSC-23651); and UAS-flp (FBti0040617). The following stocks were used for mosaic analysis with a repressible cell marker (MARCM) clone experiments: hs-flp UAS-nlsGFP tub-Gal4;; FRT82B tub-Gal80/TM6B (BDSC-86311) and hs-flp;; UAS-myrRFP tub-Gal4 FRT82B tub-Gal80/TM6B (from L. Cheng, University of Melbourne, Melbourne, Australia; same as above but with myrRFP, FBti0027896, as a clone marker). Larvae were heat-shocked for 1 h at 36.5°C to induce flp expression.
Immunostaining
Third-instar larval brains were dissected in PBS and fixed in 4% formaldehyde/PBS on shaker for 25 min. The brains were then washed 3 × 5 min with 0.3% Triton X-100 in PBS (PBST) and incubated on a shaker overnight at 4°C with primary antibodies. The brains were then washed 2 × 10 min with PBST and incubated with secondary antibodies for 3 h at room temperature or overnight at 4°C. After secondary antibody incubation and washes, brains were mounted in 4% N-propylgallate and 80% glycerol. The following antibodies were used: rabbit anti-aPKC (1:500; Santa Cruz Biotechnology), guinea pig anti-Patj (1:500; a gift from M. Krahn, University of Munster, Munster, Germany), rabbit anti-Par3 (1:500; a gift from A. Wodarz, Koeln University, Koeln, Germany), rat anti-DCAD2 E-Cad (1:200; DSHB), guinea pig anti-Dpn (1:5,000; a gift from J. Skeath, Washington University, St. Louis, MI), guinea pig anti-L’sc (1:1,200; a gift from M. Sato, University of Tokyo, Tokyo, Japan), mouse anti-Wor (1:1,000; a gift from Y. Cai, TLL, Singapore, Singapore), rabbit anti-Ase (1:5,000; a gift from Y.N. Jan, University of California, San Francisco, San Francisco, CA), rabbit anti-Hth (1:1,000; a gift from A. Salzberg, Technion, Haifa, Israel), mouse anti-Pros (1:50; MR1A from the DHSB), rabbit anti-Mira (1:1,000; a gift from F. Matsuzaki, RIKEN, Kobe, Japan), mouse anti-pH3 (1:400; Cell Signaling), rabbit anti-CycB (1:500; a gift from D. Glover, University of Cambridge, Cambridge, UK), rabbit anti-DsRed (1:200, Clontech), and goat anti-GFP (1:500; Abcam). All secondary antibodies were Alexa Fluor 488–, Cy3-, and Cy5-coupled antibodies from Jackson ImmunoResearch Laboratories and were diluted 1:500 in PBST. Brains were stained for actin using atto647N-phalloidin (1:1,000; Sigma-Aldrich). Brains were stained for DNA using DAPI (1 µg/ml, Sigma-Aldrich).
Confocal imaging and image data analysis
Images were acquired using a Zeiss LSM780 confocal microscope (acquisition software: Zen) with a 63× Plan Apochromat 1.4-NA differential interference contrast M27 objective (acquisition software: Zen). Samples were mounted in 4% N-propylgallate and 80% glycerol and imaged at room temperature. Cross-section views actually correspond to a single plane from a confocal z-stack taken as a lateral image of the NE. Images showing the nuclear layer of the NE were maximal projections of two to five planes from confocal z-stacks (Δz = 1 µm) using Fiji (Schindelin et al., 2012). Surface views of the NE were either maximum projections (three planes from a z-stack, Δz = 1 µm) or local z projections performed using Fiji. This facilitated constructing a flattened view of the curved epithelia for analysis. For local z projections, the following thresholds and postfilters were specified: neighborhood size = 30 pixels, binning = 1, Gaussian filter = 0, median postfilter size = 15 (to facilitate curvature smoothening), and Δz = 2–3. The E-cad signal was used as the reference plane for local z projections. Distances from the TZ were calculated by segmenting cells based on apical membrane signals (E-cad). E-cad signal was first smoothed by using the remove outliers tool and then by the median filtering 3D tool in Fiji. The E-cad signal was projected using the smooth manifold extraction plugin in Fiji. Cells were segmented (watershed segmentation) using the TissueAnalyzer plugin for Fiji and hand corrected. Cell distances to the TZ were calculated in Matlab by measuring the shortest distance of cell centroids to regions of interest drawn along apically constricted epi-NSCs expressing Neur. To estimate integration of Neur-positive TZ cells into the row of epi-NSC, we measured the roughness of the epi-NSC/TZ junctional boundary as described by Rudolf et al. (2015). The epi-NSC/TZ boundary was drawn following segmentation of local z-projected E-cad stainings using TissueAnalyzer. To avoid a bias in our analysis due to the intrinsic curvature of the tissue, we looked for local rather than global deviations. As such, for each OL, multiple groups of five epi-NSCs (4–12 groups per brain) were sampled to measure the mean orthogonal deviation (d) of the epi-NSC/TZ boundary relative to a reference axis linking the first vertex to the last in the group of five epi-NSCs. For each pixel (pixel size = 0.132 µm) that formed the boundary to the reference axis, a deviation value was computed as the orthogonal distance to the reference axis. Mean deviation values per brain were then plotted in Fig. 7 K.
Ex vivo brain culture
Third-instar larval brains were dissected in Schneider’s insect medium (Sigma-Aldrich) with 5% FBS (Gibco). Larval brains were then mounted in fibrinogen (MP Biomedicals; 5 µg diluted in 1 ml of Schneider’s insect medium solution), and clotting was induced using thrombin (MP Biomedicals) in 35-mm MatTek glass-bottom dishes. Brains were then cultured in 2 ml of Schneider’s insect medium containing 5% FBS, 0.5% penicillin (Gibco), Ecdysone (20 nM; Sigma-Aldrich), and insulin (200 µM; Sigma-Aldrich). Cultured brains were live imaged for 1–7 h.
Live imaging and image processing
To measure apical constriction and monitor MyoII dynamics, videos were acquired on a Zeiss LSM780 confocal microscope (acquisition software: Zen) with a 63× Plan Apochromat 1.4-NA differential interference contrast M27 objective using Δt = 15 s intervals and Δz = 1 µm (total of 20 z-sections). For segmentation, the videos were first locally z-projected using the E-cad-mCherry or Par3-mCherry signal as the reference plane. The following threshold and postfilters were specified: neighborhood size of 21 pixels, binning of 4, median postfilter size of 41, and Δz = 2 were maximum projected. Cells located at the medial edge of the NE that were observed to apically constrict during the course of the video as well as more lateral cells of the NE were hand-segmented at each time point using the TissueAnalyzer plugin for Fiji. We used custom-made programs for quantification and analysis. For each cell, apical area measurements were smoothed using a Gaussian filter (sigma value of 1.5× time step, i.e., ∼22 s). These values were then used to derive apical area rate of change: Δarea = area (tn+1) – area (tn). A contraction phase was defined as Δarea < 0. To identify contraction rate maxima, a threshold was set to exclude maxima that fell below the mean constriction rate for that cell to filter out noise.
To measure the medial–apical pool of MyoII, the segmentation mask was 1 pixel wide (1 pixel = 1 µm), and only signal that fell within the segmentation mask was included. MyoII intensity was quantified as the mean pixel intensity per cell per time point. For each cell, MyoII intensities were smoothed using a Gaussian filter (sigma value 1.5× time step, i.e., ∼22 s). These values were then used to derive medial myosin intensity rate of change: ΔMyoII intensity = MyoII intensity (tn+1) – MyoII intensity (tn). To identify peaks that would distinguish a phase of maximal accumulation of MyoII, a threshold was set such that peaks with amplitude values inferior to 25% of the mean of all peaks for each cell were discarded to filter out background noise.
Junctional MyoII was measured from snapshots of ex vivo cultured brains. The junctional MyoII signal was quantified in Fiji from maximum projections. Neur GFP was used to identify epi-NSCs, and junctional MyoII (MyoII-TagRFP) was measured by hand tracing regions of interest 2 pixels wide along junctions (1 pixel = 1 µm) and quantifying mean intensity.
Cross-correlation coefficients (R) between Δarea and ΔMyoII were determined by first identifying time points associated with maximal rates of constriction (thresholded as described above). We then considered an interval of 10 time points (∼150 s) and plotted the correlation coefficients between the two variables at each time point (for the plots, t0 s offset = time point of maximal peak of ΔMyoII). The correlation plots for each contraction phase were then merged to create an average cross-correlation plot per cell (8–40 contraction phases were identified per cell).
The average offset time between a maximum ΔMyoII and a maximum Δarea contraction was determined by first identifying the time points associated with maximal rates of area contraction (thresholded as described above). For each of these time points, the closest associated maximum ΔMyoII in time was identified (thresholded as described above), and the time offsets between the two maxima were calculated per contraction phase per cell. These offsets were then averaged per cell and pooled for all epi-NSCs and NE cells, respectively.
To monitor the dynamics of epi-NSC state acquisition, cultured brains were imaged using: Δt = 15 min intervals, Δz = 1 µm, and 20 z-sections. Images were first locally z-projected using the E-cad signal as the reference plane. The following threshold and postfilters were specified: neighborhood size = 30 pixels, binning = 3, Gaussian filter = 0, median postfilter size = 15 (to facilitate curvature smoothening), and Δz = 2 were used to locally maximum project. Neur-GFP and E-cad-mCherry were used to define epi-NSCs and NE membranes, respectively. Approximately three rows of NE cells adjacent to the Neur-positive epi-NSCs at t0 were tracked over the time lapse. At each time point, cells were segmented manually in Fiji, and the average Neur intensity was measured. Cells were defined as having acquired an epi-NSC state when their average Neur intensity signal surpassed a threshold of 1 SD below the mean Neur intensity of all apically constricted epi-NSCs at the medial edge.
Statistical analysis
For statistical calculations, R Studio was used to perform Student’s t tests (two tailed) to compare the means between two samples and one-way ANOVAs to compare the means of three or more samples; Scheffé multiple comparison tests were then used to perform post hoc analysis between the means.
Online supplemental material
Fig. S1 shows that epi-NSCs start to express Ase and Hth, and then divide to produce a basal GMC inheriting Neur while keeping its AJs. Fig. S2 shows that Crb is located just apical to AJs in the OL and that Par3 remains detectable at the apical cortex in epi-NSCs. Fig. S3 shows that the loss of neur activity or loss of the NBM-containing isoforms of Sdt delayed the down-regulation of Patj and Crb, and that the short Sdt isoforms stabilized Patj in the neur-negative cells of the NE. Fig. S4 shows that apical constriction negatively correlated with Neur levels. It also shows that sdtΔ3, rhoGEF3KO double mutant brains had a phenotype similar to each single mutant. Fig. S5 shows that aPKC and MyoII accumulate in a complementary manner along the epi-NSC/TZ boundary and that aPKC distribution in epi-NSCs depends on the regulation of Sdt by Neur. Videos 1 and 2 show that changes in apical area are associated with increased levels of medial-apical MyoII in both constricting epi-NSCs (Video 1) and nonconstricting NE cells (Video 2).
Supplementary Material
Acknowledgments
We thank Y. Bellaïche, A. Brand, Y. Cai, L. Cheng, D. Glover, Y. Hong, Y. N. Jan, M. Krahn, F. Matsuzaki, A. Salzberg, M. Sato, J. Skeath, M. Suzanne, A. Wodarz, the Developmental Studies Hybridoma Bank, Flybase, and Image Analysis Hub of the Institut Pasteur for reagents and resources. We thank S. Herbert (Image Analysis Hub), R. Levayer, J.-Y. Tinevez (Image Analysis Hub), T. Schweisguth, and L. Valon for help in segmentation, local z-projection, and image data analysis; M. Rujano for help in live imaging; and L. Couturier for technical help. We thank S. Chanet, E. Contreras, A. Hakes, and G. Perez-Mockus for critical reading.
This work was funded by the Agence Nationale de la Recherche (ANR-10-LABX-0073) and Fondation pour la Recherche Médicale (FRM-DEQ20180339219).
The authors declare no competing financial interests.
Author contributions: C. Shard and F. Schweisguth conceptualized this research. C. Shard did all experiments. J. Luna-Escalante performed programming for automatic data analysis. F. Schweisguth supervised the study and acquired funding. C. Shard and F. Schweisguth wrote the paper.
References
- Aliee M., Röper J.-C., Landsberg K.P., Pentzold C., Widmann T.J., Jülicher F., and Dahmann C.. 2012. Physical mechanisms shaping the Drosophila dorsoventral compartment boundary. Curr. Biol. 22:967–976. 10.1016/j.cub.2012.03.070 [DOI] [PubMed] [Google Scholar]
- An Y., Xue G., Shaobo Y., Mingxi D., Zhou X., Yu W., Ishibashi T., Zhang L., and Yan Y.. 2017. Apical constriction is driven by a pulsatile apical myosin network in delaminating Drosophila neuroblasts. Development. 144:2153–2164. 10.1242/dev.150763 [DOI] [PubMed] [Google Scholar]
- Ashraf S.I., and Ip Y.T.. 2001. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development. 128:4757–4767. [DOI] [PubMed] [Google Scholar]
- Bajur A.T., Iyer K.V., and Knust E.. 2019. Cytocortex-dependent dynamics of Drosophila Crumbs controls junctional stability and tension during germ band retraction. J. Cell Sci. 132 jcs228338 10.1242/jcs.228338 [DOI] [PubMed] [Google Scholar]
- Bardin A.J., and Schweisguth F.. 2006. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell. 10:245–255. 10.1016/j.devcel.2005.12.017 [DOI] [PubMed] [Google Scholar]
- Besson C., Bernard F., Corson F., Rouault H., Reynaud E., Keder A., Mazouni K., and Schweisguth F.. 2015. Planar cell polarity breaks the symmetry of PAR protein distribution prior to mitosis in Drosophila sensory organ precursor cells. Curr. Biol. 25:1104–1110. 10.1016/j.cub.2015.02.073 [DOI] [PubMed] [Google Scholar]
- Blanchard G.B., Murugesu S., Adams R.J., Martinez-Arias A., and Gorfinkiel N.. 2010. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 137:2743–2752. 10.1242/dev.045872 [DOI] [PubMed] [Google Scholar]
- Booth A.J.R., Blanchard G.B., Adams R.J., and Röper K.. 2014. A dynamic microtubule cytoskeleton directs medial actomyosin function during tube formation. Dev. Cell. 29:562–576. 10.1016/j.devcel.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhatmi H., Martins T., Pillidge Z., Kamenova T., and Bray S.. 2020. Notch Mediates Inter-tissue Communication to Promote Tumorigenesis. Curr. Biol. 30:1809–1820.e4. 10.1016/j.cub.2020.02.088 [DOI] [PubMed] [Google Scholar]
- Bulgakova N.A., and Knust E.. 2009. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 122:2587–2596. 10.1242/jcs.023648 [DOI] [PubMed] [Google Scholar]
- Cai Y., Chia W., and Yang X.. 2001. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J. 20:1704–1714. 10.1093/emboj/20.7.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K., Knust E., and Skaer H.. 2009. Crumbs stabilises epithelial polarity during tissue remodelling. J. Cell Sci. 122:2604–2612. 10.1242/jcs.047183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro B., Barolo S., Bailey A.M., and Posakony J.W.. 2005. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 132:3333–3344. 10.1242/dev.01920 [DOI] [PubMed] [Google Scholar]
- Chanet S., and Schweisguth F.. 2012. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nat. Cell Biol. 14:467–476. 10.1038/ncb2481 [DOI] [PubMed] [Google Scholar]
- Clément R., Dehapiot B., Collinet C., Lecuit T., and Lenne P.-F.. 2017. Viscoelastic Dissipation Stabilizes Cell Shape Changes during Tissue Morphogenesis. Curr. Biol. 27:3132–3142.e4. 10.1016/j.cub.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Contreras E.G., Egger B., Gold K.S., and Brand A.H.. 2018. Dynamic Notch signalling regulates neural stem cell state progression in the Drosophila optic lobe. Neural Dev. 13:25 10.1186/s13064-018-0123-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier L., Mazouni K., Bernard F., Besson C., Reynaud E., and Schweisguth F.. 2017. Regulation of cortical stability by RhoGEF3 in mitotic Sensory Organ Precursor cells in Drosophila. Biol. Open. 6:1851–1860. 10.1242/bio.026641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S., Yu J., Zinzen R., and Wieschaus E.. 2006. Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell. 10:257–264. 10.1016/j.devcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- del Álamo D., Rouault H., and Schweisguth F.. 2011. Mechanism and significance of cis-inhibition in Notch signalling. Curr. Biol. 21:R40–R47. 10.1016/j.cub.2010.10.034 [DOI] [PubMed] [Google Scholar]
- Dillard C., Narbonne-Reveau K., Foppolo S., Lanet E., and Maurange C.. 2018. Two distinct mechanisms silence chinmo in drosophila neuroblasts and neuroepithelial cells to limit their self-renewal. Development. 145 dev154534 10.1242/dev.154534 [DOI] [PubMed] [Google Scholar]
- Dongre A., and Weinberg R.A.. 2019. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20:69–84. 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- Egger B., Boone J.Q., Stevens N.R., Brand A.H., and Doe C.Q.. 2007. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2:1 10.1186/1749-8104-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Gold K.S., and Brand A.H.. 2010. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 137:2981–2987. 10.1242/dev.051250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eritano A.S., Bromley C.L., Bolea Albero A., Schütz L., Wen F.-L., Takeda M., Fukaya T., Sami M.M., Shibata T., Lemke S., et al. 2020. Tissue-Scale Mechanical Coupling Reduces Morphogenetic Noise to Ensure Precision during Epithelial Folding. Dev. Cell. 53:212–228.e12. 10.1016/j.devcel.2020.02.012 [DOI] [PubMed] [Google Scholar]
- Flores-Benitez D., and Knust E.. 2015. Crumbs is an essential regulator of cytoskeletal dynamics and cell-cell adhesion during dorsal closure in Drosophila. eLife. 4 e07398 10.7554/eLife.07398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold K.S., and Brand A.H.. 2014. Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev. 9:18 10.1186/1749-8104-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N., and Blanchard G.B.. 2011. Dynamics of actomyosin contractile activity during epithelial morphogenesis. Curr. Opin. Cell Biol. 23:531–539. 10.1016/j.ceb.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Gracia M., Theis S., Proag A., Gay G., Benassayag C., and Suzanne M.. 2019. Mechanical impact of epithelial-mesenchymal transition on epithelial morphogenesis in Drosophila. Nat. Commun. 10:2951 10.1038/s41467-019-10720-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes A.E., Otsuki L., and Brand A.H.. 2018. A newly discovered neural stem cell population is generated by the optic lobe neuroepithelium during embryogenesis in drosophila melanogaster. Development. 145 dev166207 10.1242/dev.166207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.P., and Tepass U.. 2008. Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183:1129–1143. 10.1083/jcb.200807020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V., and Campos-Ortega J.A.. 1984. Early neurogenesis in wild-type Drosophila melanogaster. Wihelm Roux Arch. Dev. Biol. 193:308–325. 10.1007/BF00848159 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., and Wodarz A.. 2013. Initial neurogenesis in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2:701–721. 10.1002/wdev.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszterg S., Leibfried A., Bosveld F., Martin C., and Bellaiche Y.. 2013. Interplay between the dividing cell and its neighbors regulates adherens junction formation during cytokinesis in epithelial tissue. Dev. Cell. 24:256–270. 10.1016/j.devcel.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Hofbauer A., and Campos-Ortega J.A.. 1990. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 198:264–274. 10.1007/BF00377393 [DOI] [PubMed] [Google Scholar]
- Huang F., Dambly-Chaudière C., and Ghysen A.. 1991. The emergence of sense organs in the wing disc of Drosophila. Development. 111:1087–1095. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A.M., and Hong Y.. 2009. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA. 106:8284–8289. 10.1073/pnas.0900641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J.B., Nabeshima Y., Doe C.Q., and Matsuzaki F.. 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 390:625–629. 10.1038/37641 [DOI] [PubMed] [Google Scholar]
- Ito T., Kudoh S., Ichimura T., Fujino K., Hassan W.A.M.A., and Udaka N.. 2017. Small cell lung cancer, an epithelial to mesenchymal transition (EMT)-like cancer: significance of inactive Notch signaling and expression of achaete-scute complex homologue 1. Hum. Cell. 30:1–10. 10.1007/s13577-016-0149-3 [DOI] [PubMed] [Google Scholar]
- Jörg D.J., Caygill E.E., Hakes A.E., Contreras E.G., Brand A.H., and Simons B.D.. 2019. The proneural wave in the Drosophila optic lobe is driven by an excitable reaction-diffusion mechanism. eLife. 8 e40919 10.7554/eLife.40919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Lai E.C., Deblandre G.A., Kintner C., and Rubin G.M.. 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell. 1:783–794. 10.1016/S1534-5807(01)00092-2 [DOI] [PubMed] [Google Scholar]
- Lai E.C., Roegiers F., Qin X., Jan Y.N., and Rubin G.M.. 2005. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 132:2319–2332. 10.1242/dev.01825 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., and Schweisguth F.. 2003. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell. 5:139–148. 10.1016/S1534-5807(03)00187-4 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Remaud S., Hamel S., and Schweisguth F.. 2005. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 3 e96 10.1371/journal.pbio.0030096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Erclik T., Bertet C., Chen Z., Voutev R., Venkatesh S., Morante J., Celik A., and Desplan C.. 2013. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 498:456–462. 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., and Goldstein B.. 2014. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development. 141:1987–1998. 10.1242/dev.102228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., and Wieschaus E.F.. 2010. Tensions divide. Nat. Cell Biol. 12:5–7. 10.1038/ncb0110-5 [DOI] [PubMed] [Google Scholar]
- Martin A.C., Gelbart M., Fernandez-Gonzalez R., Kaschube M., and Wieschaus E.F.. 2010. Integration of contractile forces during tissue invagination. J. Cell Biol. 188:735–749. 10.1083/jcb.200910099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., and Wieschaus E.F.. 2009. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 457:495–499. 10.1038/nature07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason F.M., Tworoger M., and Martin A.C.. 2013. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat. Cell Biol. 15:926–936. 10.1038/ncb2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.W., Rebeiz M., Atanasov J.E., and Posakony J.W.. 2014. Neural precursor-specific expression of multiple Drosophila genes is driven by dual enhancer modules with overlapping function. Proc. Natl. Acad. Sci. USA. 111:17194–17199. 10.1073/pnas.1415308111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Verboon J.M., and Parkhurst S.M.. 2017. Prepatterning by RhoGEFs governs Rho GTPase spatiotemporal dynamics during wound repair. J. Cell Biol. 216:3959–3969. 10.1083/jcb.201704145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo K.T., Wang J., Junker M., Kriz S., Vo G., Asem B., Olson J.M., Banerjee U., and Hartenstein V.. 2010. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev. Biol. 346:284–295. 10.1016/j.ydbio.2010.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C. 2017. Pseudostratified epithelia - cell biology, diversity and roles in organ formation at a glance. Journal of Cell Science. 130:1859–1863. 10.1242/jcs.192997 [DOI] [PubMed] [Google Scholar]
- Orihara-Ono M., Toriya M., Nakao K., and Okano H.. 2011. Downregulation of Notch mediates the seamless transition of individual Drosophila neuroepithelial progenitors into optic medullar neuroblasts during prolonged G1. Dev. Biol. 351:163–175. 10.1016/j.ydbio.2010.12.044 [DOI] [PubMed] [Google Scholar]
- Perez-Mockus G., and Schweisguth F.. 2017. Cell Polarity and Notch Signaling: Linked by the E3 Ubiquitin Ligase Neuralized? BioEssays. 39 1700128 10.1002/bies.201700128 [DOI] [PubMed] [Google Scholar]
- Perez-Mockus G., Mazouni K., Roca V., Corradi G., Conte V., and Schweisguth F.. 2017a Spatial regulation of contractility by Neuralized and Bearded during furrow invagination in Drosophila. Nat. Commun. 8:1594 10.1038/s41467-017-01482-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mockus G., Roca V., Mazouni K., and Schweisguth F.. 2017b Neuralized regulates Crumbs endocytosis and epithelium morphogenesis via specific Stardust isoforms. J. Cell Biol. 216:1405–1420. 10.1083/jcb.201611196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova M.D., Tkačik G., Bialek W., Wieschaus E.F., and Gregor T.. 2019. Optimal Decoding of Cellular Identities in a Genetic Network. Cell. 176:844–855.e15. 10.1016/j.cell.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B.V.V.G., Rauskolb C., and Irvine K.D.. 2010. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 137:2397–2408. 10.1242/dev.050013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K. 2012. Anisotropy of Crumbs and aPKC drives myosin cable assembly during tube formation. Dev. Cell. 23:939–953. 10.1016/j.devcel.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K. 2013. Supracellular actomyosin assemblies during development. Bioarchitecture. 3:45–49. 10.4161/bioa.25339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault H., Mazouni K., Couturier L., Hakim V., and Schweisguth F.. 2010. Genome-wide identification of cis-regulatory motifs and modules underlying gene coregulation using statistics and phylogeny. Proc. Natl. Acad. Sci. USA. 107:14615–14620. 10.1073/pnas.1002876107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf K., Umetsu D., Aliee M., Sui L., Jülicher F., and Dahmann C.. 2015. A local difference in Hedgehog signal transduction increases mechanical cell bond tension and biases cell intercalations along the Drosophila anteroposterior compartment boundary. Development. 142:3845–3858. doi:. 10.1242/dev.125542 [DOI] [PubMed] [Google Scholar]
- Salis P., Payre F., Valenti P., Bazellieres E., Le Bivic A., and Mottola G.. 2017. Crumbs, Moesin and Yurt regulate junctional stability and dynamics for a proper morphogenesis of the Drosophila pupal wing epithelium. Sci. Rep. 7:16778 10.1038/s41598-017-15272-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Suzuki T., and Nakai Y.. 2013. Waves of differentiation in the fly visual system. Dev. Biol. 380:1–11. 10.1016/j.ydbio.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Sato M., Yasugi T., Minami Y., Miura T., and Nagayama M.. 2016. Notch-mediated lateral inhibition regulates proneural wave propagation when combined with EGF-mediated reaction diffusion. Proc. Natl. Acad. Sci. USA. 113:E5153–E5162. 10.1073/pnas.1602739113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J.M., Harrell J.R., Shemer G., Sullivan-Brown J., Roh-Johnson M., and Goldstein B.. 2010. Apical constriction: a cell shape change that can drive morphogenesis. Dev. Biol. 341:5–19. 10.1016/j.ydbio.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M., Schaefer M., and Knoblich J.A.. 1999. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 402:548–551. 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Sen A., Nagy-Zsvér-Vadas Z., and Krahn M.P.. 2012. Drosophila PATJ supports adherens junction stability by modulating Myosin light chain activity. J. Cell Biol. 199:685–698. 10.1083/jcb.201206064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidor C., Stevens T.J., Jin L., Boulanger J., and Röper K.. 2020. Rho-Kinase Planar Polarization at Tissue Boundaries Depends on Phospho-regulation of Membrane Residence Time. Dev. Cell. 52:364–378.e7. 10.1016/j.devcel.2019.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S., Oh Y., Wang M.F.Z., Fernandez-Gonzalez R., and Tepass U.. 2017. Myosin II promotes the anisotropic loss of the apical domain during Drosophila neuroblast ingression. J. Cell Biol. 216:1387–1404. 10.1083/jcb.201608038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana E.P., and Doe C.Q.. 1995. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 121:3187–3195. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Kaido M., Takayama R., and Sato M.. 2013. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev. Biol. 380:12–24. 10.1016/j.ydbio.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Wang W., Liu W., Wang Y., Zhou L., Tang X., and Luo H.. 2011. Notch signaling regulates neuroepithelial stem cell maintenance and neuroblast formation in Drosophila optic lobe development. Dev. Biol. 350:414–428. 10.1016/j.ydbio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Weinmaster G., and Fischer J.A.. 2011. Notch ligand ubiquitylation: what is it good for? Dev. Cell. 21:134–144. 10.1016/j.devcel.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Haenfler J.M., and Lee C.Y.. 2012. Changes in Notch signaling coordinates maintenance and differentiation of the Drosophila larval optic lobe neuroepithelia. Dev. Neurobiol. 72:1376–1390. 10.1002/dneu.20995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Tentner A.R., Huang P., Gelas A., Mosaliganti K.R., Souhait L., Rannou N., Swinburne I.A., Obholzer N.D., Cowgill P.D., et al. 2013. Specified neural progenitors sort to form sharp domains after noisy Shh signaling. Cell. 153:550–561. 10.1016/j.cell.2013.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T., Sugie A., Umetsu D., and Tabata T.. 2010. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 137:3193–3203. 10.1242/dev.048058 [DOI] [PubMed] [Google Scholar]
- Yasugi T., Umetsu D., Murakami S., Sato M., and Tabata T.. 2008. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 135:1471–1480. 10.1242/dev.019117 [DOI] [PubMed] [Google Scholar]
- Yevick H.G., Miller P.W., Dunkel J., and Martin A.C.. 2019. Structural Redundancy in Supracellular Actomyosin Networks Enables Robust Tissue Folding. Dev. Cell. 50:586–598.e3. 10.1016/j.devcel.2019.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Korzelius J., van Straaten M., Bender K., Schuhknecht G.F.P., Dutta D., Xiang J., and Edgar B.A.. 2014. Fly-FUCCI: A versatile tool for studying cell proliferation in complex tissues. Cell Rep. 7:588–598. 10.1016/j.celrep.2014.03.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.