Abstract
Background
Glioblastoma (GBM) consists of devastating neoplasms with high invasive capacity, which have been difficult to study in vitro in a human-derived model system. Therapeutic progress is also limited by cellular heterogeneity within and between tumors, among other factors such as therapy resistance. To address these challenges, we present an experimental model using human cerebral organoids as a scaffold for patient-derived GBM cell invasion.
Methods
This study combined tissue clearing and confocal microscopy with single-cell RNA sequencing of GBM cells before and after co-culture with organoid cells.
Results
We show that tumor cells within organoids extend a network of long microtubes, recapitulating the in vivo behavior of GBM. Transcriptional changes implicated in the invasion process are coherent across patient samples, indicating that GBM cells reactively upregulate genes required for their dispersion. Potential interactions between GBM and organoid cells identified by an in silico receptor–ligand pairing screen suggest functional therapeutic targets.
Conclusions
Taken together, our model has proven useful for studying GBM invasion and transcriptional heterogeneity in vitro, with applications for both pharmacological screens and patient-specific treatment selection on a time scale amenable to clinical practice.
Keywords: glioblastoma, organoids, single-cell RNA sequencing, tumor cell heterogeneity, tumor invasion
Key Points.
Organoid technology and single-cell transcriptomics reveal cellular interactions of invasive GBM cells.
Transcriptional changes implicated during invasion suggest novel therapeutic targets for GBM.
Time scales are amenable to clinical practice and high-content drug screens.
Importance of the Study.
Human induced pluripotent stem cell‒derived organoids have recently emerged as biologically relevant in vitro models, while single-cell transcriptomics provides a powerful new approach for resolving gene expression at the cellular level. Here, we have combined both techniques to study cellular interactions of invasive GBM cells in human cerebral organoids. Our GBM co-culture assay successfully uncovers transcriptional changes implicated in the invasion process as well as potential ligand–receptor interactions between GBM and organoid cells, suggesting novel therapeutic targets for GBM. Our approach enables efficient quantitative studies of GBM and other invasive tumors in vitro, on time scales amenable to clinical practice and high-content drug screens.
Glioblastoma (GBM) is the most frequent and most aggressive primary brain tumor.1,2 Despite decades of intensive research, average survival time remains at 12–15 months from diagnosis.3 Surgical resection of GBM tumors is rarely complete because the tumor aggressively infiltrates the brain, with cells interconnecting via long membrane protrusions (microtubes).4 The resulting network enables multicellular communication through microtube-associated gap junctions, and increases tumor resistance to cell ablation and radiotherapy.5 Moreover, GBM cells interact with normal brain cells via soluble factors or direct cell-cell contacts to promote tumor proliferation and invasion.6
Two major challenges have impeded progress in the development of new GBM therapies. Firstly, there is increasing evidence for substantial genetic,7,8 epigenetic,9,10 and transcriptional heterogeneity11 between and within human tumors. Recent advances in single-cell RNA sequencing (scRNA-seq) technology have enabled the transcriptomic analysis of numerous tumor entities at the level of individual cells. However, in the case of GBM, resection of primary samples has resulted in limited insight into interactions between infiltrating tumor and normal brain cells, as isolation of neoplastic cells from the tumor periphery has proven challenging.12 How cellular heterogeneity of GBM cells and their interactions with normal brain cells relate to differences in proliferation or invasive capacity, which ultimately determine patient outcome, thus remains unknown.
A second challenge in the advancement of GBM therapies is the current lack of model systems to study defining properties of human GBM, especially invasion into the surrounding brain tissue. Previous in vitro models have suffered from limited physiological relevance or have been incompatible with the time scales for clinical decision making.6 Recent studies have shown that human cerebral organoids can be used as a platform for tumor cell transplantation or genetic engineering of tumors, enabling microscopic observation of tumor development.13–16 However, tumor cell interactions with normal brain cells have not been addressed yet.
Here, we developed an experimental approach to study the interaction of GBM and normal brain cells of the neuronal lineage in vitro, on clinically relevant time scales of less than 4 weeks. We used induced pluripotent stem cell (iPSC)–derived human cerebral organoids as a 3D scaffold for the invasion of patient-derived GBM cells and analyzed tumor microtube development by tissue clearing, confocal microscopy, and semi-automated quantification. In addition, we performed scRNA-seq of GBM cells before and after co-culture with organoid cells and identified a transcriptional program induced by the interactions between tumor and normal brain cells, suggesting potential therapeutic targets.
Materials and Methods
GBM Cell Culture
Primary tumor samples were received from the Edinger Institute of Frankfurt University Hospital. Informed consent was obtained prior to surgery. Experiments involving human patient material were performed in accordance with the Declaration of Helsinki and were approved by the ethics committee of the University Cancer Center Frankfurt (project #SNO-01-13). Patient-derived GBM cell cultures were established as described.17 Cells were cultured in suspension culture in 75 cm2 ultra-low attachment flasks in Neurobasal medium (Gibco) supplemented with 2 mM L-glutamine (Life Technologies), 1x B27 (Gibco), 2 µg/mL heparin, 20 ng/mL epidermal growth factor and 20 ng/mL basic fibroblast growth factor (R&D Systems). For passaging, spheroid cultures were dissociated using Accutase (StemCell Technologies) when they had reached diameters of ~100 µm, every 1–3 weeks. To confirm the invasive capacity of GBM cells in hydrogel matrix, spheroids were embedded in Matrigel (Corning).
Lentiviral Labeling and FACS
Second-generation replication-incompetent lentivirus was produced by FuGENE transfection (Promega) of HEK293T cells with the expression plasmid LeGO-G2, complemented with the packaging plasmids psPAX2 and pMD2.G (all from Addgene). GBM cells were infected on 3 consecutive days by spinoculation at 800g for 30–60 minutes, and green fluorescent protein (GFP)–expressing cells were isolated by fluorescence activated cell sorting (FACS). Lentivirally labeled GBM cells were maintained in neural maintenance medium and passaged every 1–3 weeks as described above.
Organoid Culture
The iPSC line 409b2 was obtained from the Riken Institute in Japan. For routine culture, iPSCs were maintained in mTeSR1 medium (StemCell Technologies) on tissue culture plates coated with Matrigel (Corning) and passaged every 4–5 days using Gentle Cell Dissociation Reagent (StemCell Technologies). For organoid seeding, iPSCs were dissociated with Accutase (StemCell Technologies) and transferred into AggreWell plates in Neural Induction Medium (StemCell Technologies) with 10 µM Y-27632 (StemCell Technologies) at a density of 1000 cells per cavity, following manufacturers’ instructions. Spheroid formation was confirmed visually after 24 hours, and spheroids were maintained in Neural Induction Medium (StemCell Technologies) with daily medium changes. After 5 days, spheroids were harvested from the AggreWell plates and embedded in Matrigel. Medium was changed to neural maintenance medium, a 1:1 mixture of N2- and B27-containing media (N2 medium: DMEM/F12 GlutaMAX, 1 × N2, 5 μg/mL insulin, 1 mM L-glutamine, 1 × non-essential amino acids, 100 μM 2-mercaptoethanol; B27 medium: Neurobasal, 1 × B27, 200 mM L-glutamine) supplemented with 50 U/mL penicillin and 50 mg/mL streptomycin, and exchanged every 2 days. Neural induction in 2D was performed after plating dissociated iPSCs onto Matrigel-coated plates, using the same culture media.
Organoid Invasion Assays
On day 24 of culture, organoids were removed from Matrigel by incubation with Dispase (Sigma) at 37°C and transferred to individual wells of a GravityTRAP ULA Plate (PerkinElmer). Labeled GBM cells were dissociated with Accutase and added to the organoid plate in neural maintenance medium at a concentration of 1000 cells per well. Plates were centrifuged at 100g for 3 min before returning to the incubator. After 2 days, Organoids were live stained with 100 nM SiR-actin (Spirochrome). The following day, organoids were harvested, fixed in 2% paraformaldehyde for 30 min, and embedded in Matrigel for immobilization. Tissue clearing was performed following the fructose/urea/α-thioglycerol (FRUIT) protocol as described.18 Immunohistochemistry and control invasion assays using MCF10AT spheroids grown in Matrigel or SH-SY5Y spheroids were performed as described in the Supplementary Methods. Confocal images were acquired on an LSM780 Axio Observer confocal laser scanning microscope (Zeiss) and analyzed using custom scripts in ImageJ and MatLab, as detailed in the Supplementary Methods.
Single-Cell RNA Sequencing
For scRNA-seq of GBM cell lines, cells were cultured in neural maintenance medium for 1–3 weeks after passaging, until the largest spheroids were ~100 µm in diameter. Cells were then dissociated with Accutase (StemCell Technologies), washed twice in phosphate buffered saline (PBS), and passed through a 20 µm cell strainer (PluriSelect).
For scRNA-seq of co-cultured GBM and organoid cells, neural progenitor cell (NPC) spheroids were generated by iPSC AggreWell plates as described above. After 7 days, NPC spheroids and lentivirally labeled GBM cells from separate cultures were dissociated using Accutase, mixed in a 1:1 ratio, and replated onto AggreWell plates at 1000 cells per cavity in neural maintenance medium. After 3 days, mixed spheroids were dissociated using Accutase, washed twice in PBS, and passed through a 20 µm cell strainer (PluriSelect). Cell isolation, library preparation, and analysis of scRNA-seq data were performed as described in detail in the Supplementary Methods. Briefly, RNA-seq libraries that contained at least 150 detected genes and at most 15% mitochondrial reads were selected for downstream processing. Adapting a previously published approach,19 aggregate expression for each gene across all cells was calculated as Ea = log(mean[Ej,1…n] + 1), where Ej is the counts-per-million expression value of the gene in cell j. Retained for analysis were 8533 genes with Ea > 2. Clustering and differential expression analysis were performed using the Seurat package as implemented in R.20 Gene set enrichment analysis21 was performed by computing overlaps between identified gene signatures and Gene Ontology (GO_C5) gene sets derived from the Molecular Signature Database (MSigDB, https://software.broadinstitute.org/gsea/msigdb). Potential receptor–ligand pairings were analyzed based on a list of 2557 previously published receptor–ligand pairs,22 by summing for each pair of cells the number of ligand–receptor pairs potentially connecting the pair.23
Data Availability
Raw sequencing data have been deposited at the European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/home) under accession number EGAS00001003852. Scripts used for analyzing transcriptome data and image data (in R, Fiji, and MatLab) are available from the authors upon request.
Results
Induced PSC–Derived Cerebral Organoids Provide a Scaffold for Glioblastoma Invasion
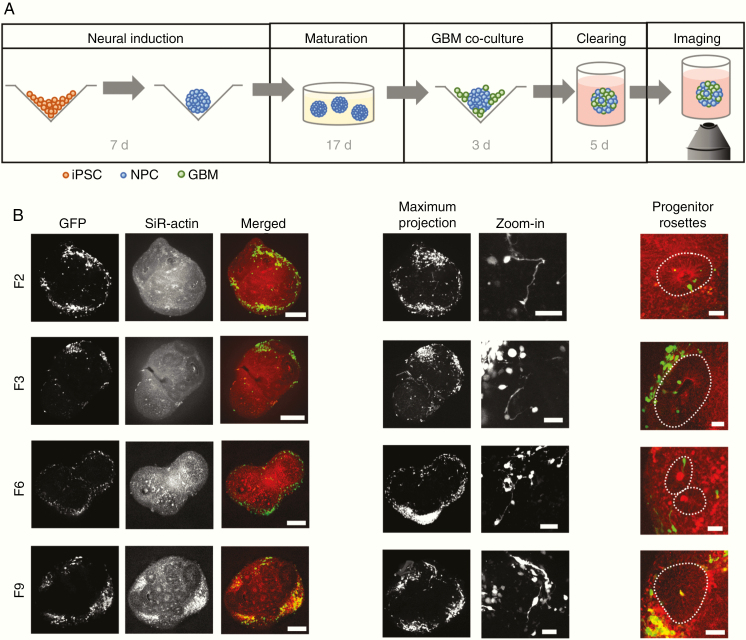
To study GBM invasion in a physiologically relevant 3D context, we adapted an established protocol for human iPSC-derived cerebral organoid development24 to achieve streamlined and reproducible production of organoids (Supplementary Methods and Supplementary Figure 1). From 24 days of age, cerebral organoids were co-cultured with fluorescently labeled GBM cells from 4 patient-derived cell lines (Figure 1A, Supplementary Table 1, and Supplementary Figure 1). Samples were fixed after 3 days and subjected to tissue clearing using the FRUIT protocol,18 enabling the visualization of tumor invasion by confocal microscopy. We found that tumor cells from all 4 GBM patients readily attached to and invaded into the organoids (Figure 1B). Tumor cells formed protrusions reaching to other cells over short and long distances (Figure 1B and Supplementary Figure 2A), consistent with tumor microtube formation observed in vivo in mice.4 GBM cells primarily invaded into the neuronal layers of the organoids, with little invasion into neural progenitor rosettes (Figure 1B and Supplementary Figure 2B). Conversely, we did not observe invasion of GBM cells into organoids grown from the breast cancer cell line MCF10AT25 or the neuroblastoma cell line SH-SY5Y (Supplementary Figure 2C).
Figure 1.
GBM invasion assay. (A) Experimental protocol. Following 7 days of neural induction, organoids were transferred to Matrigel and matured for 17 days. Organoids were then enzymatically released and co-cultured with GFP-labeled GBM cells for 3 days. Samples were embedded in Matrigel again for fixation, tissue clearing and confocal imaging. (B) GFP-labeled tumor cells from all 4 GBM patients invade into cerebral organoids (left; scale bars, 250 µm) where they form short-range and long-range connections (middle, maximum intensity projections over ~200–250 µm depth; scale bars, 50 µm). Invasion is largely restricted to neuronal layers, outside of neural progenitor rosettes indicated by dotted lines (right; scale bars, 50 µm).
Tumor Microtube Formation Recapitulates In Vivo Behavior of GBM Cells
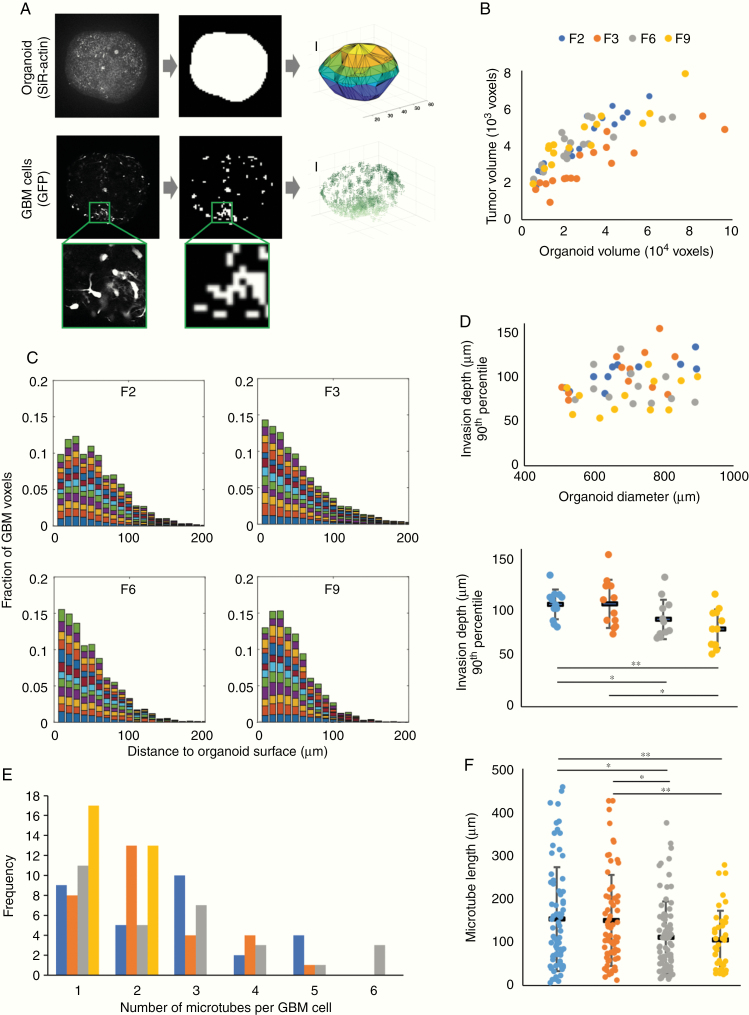
We developed a semi-automated image processing workflow to analyze the invasion process quantitatively (Figure 2A and Supplementary Methods), which we applied to a total of 66 organoids (n = 15–19 for each of the 4 patient GBM cell lines). We found that, for organoids of comparable sizes, the fraction of organoid volume taken up by tumor cells was similar across the 4 patient-derived cell lines (Figure 2B and Supplementary Figure 3A). The distribution of GBM cells within organoids was assessed by calculating the distances between GFP+ voxels across the same set of organoids. Tumor cells spread widely in all cases (Supplementary Figure 3B). To quantify invasion depth, we compared the distribution of distances of GFP+ voxels from the organoid surface across 12 similarly sized organoids for each patient-derived cell line. Invasion depths exceeded 100 µm in the majority of organoids (Figure 2C), with some cells detected at approximately 300 µm from the organoid surface. While migration depth of the most invasive cells (90th percentile of invasion depth) was uncorrelated with organoid size, we observed that cells from patients F6 and F9 were less invasive than cells from patients F2 and F3 (Figure 2D and Supplementary Videos); this suggests that the in vitro model may be able to reproduce intertumor heterogeneity in invasive behavior, although we cannot currently rule out that the observed differences in invasion depth stem from differences in 3D architecture between the scaffold organoids. By tracing membrane-bound cellular processes in images, we found that the number of microtubes per GBM cell ranged up to 6, with 2.2 ± 0.1 microtubes on average (Figure 2E). We quantified how many of these microtubes ended at other GBM cells, and identified between 0 and 4 such putative intratumoral connections per GBM cell, with an average of 1.2 ± 0.1 connections (Supplementary Figure 3C). Individual microtubes were up to 450 µm long (Figure 2F). Consistent with our earlier observation of intertumoral heterogeneity of invasive capacity, we found that microtube lengths differed between cell lines (Figure 2F). Interestingly, the cell lines with higher invasive capacity (F2 and F3) also showed longer microtubes; this observation suggests that GBM tumors extending longer microtubes may be able to colonize organoids more efficiently in vitro, consistent with recent in vivo reports that microtubes promote tumor dissemination by allowing GBM cells to exchange cytoplasmic molecules and even translocate nuclei over long distances.4,26
Figure 2.
Morphological features of patient-derived GBM cells invading organoids. (A) Image analysis workflow. To approximate the organoid surface, organoids were incubated with the live dye SiR-actin; following fixation and clearing, the actin signal was binarized and triangulated (top). Above-threshold GFP signal was used as a proxy for GBM cell location (bottom). Scale bars, 100 µm. (B) Total tumor cell volume as a function of organoid volume. (C) Distributions of distances of tumor voxels from the organoid surface of 12 organoids from each patient cell line with 500–900 µm diameter; each color represents one organoid. (D) Invasion depth of the most invasive cells from each cell line (90th percentile) compared with organoid size (top) and differences in invasion depth between patient cell lines (bottom; *P < 0.05, **P < 0.01, two-sided Student’s t-test). (E) Number of tumor microtubes per cell observed across 30 GBM cells from each patient. (F) Microtube lengths ranged up to almost 450 µm, with GBM cells from patients F2 and F3 developing longer microtubes than cells from patients F6 and F9 (*P < 0.05, **P < 0.01, two-sided Student’s t-test). In (D) and (F), black horizontal bars indicate mean values and error bars represent standard errors in the mean.
Single-Cell RNA Sequencing Reveals Transcriptional Heterogeneity Between Tumors and After Co-Culture with Organoid Cells
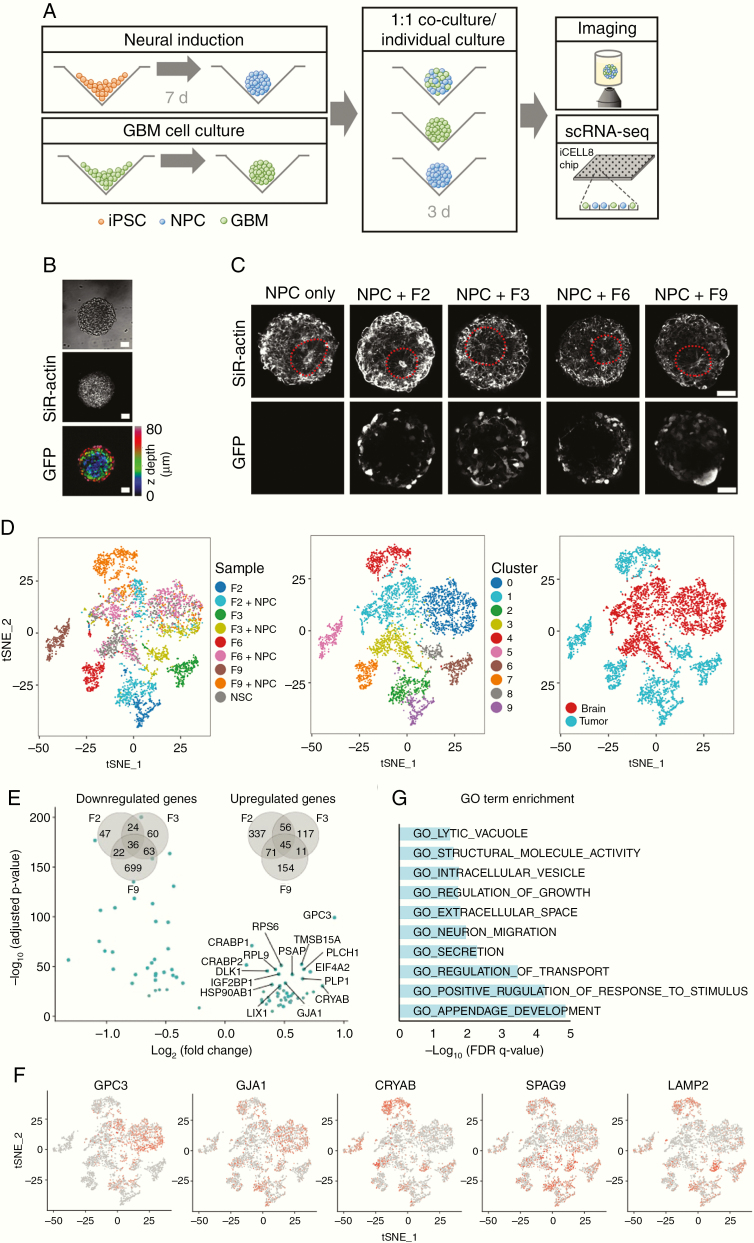
Our imaging results confirm that iPSC-derived cerebral organoids represent an effective model system for quantifying GBM invasion and tumor microtube formation in vitro. To further study heterogeneity of and interactions between GBM and organoid cells at the transcriptome level, we developed a more efficient workflow that could be applied on clinically relevant time scales and at higher throughput. In the modified assay, dissociated 7-day-old cerebral organoids were mixed with GBM cells from separate cultures at a 1:1 ratio and grown in co-culture for 3 days (Figure 3A). GBM cells from all 4 patient-derived cell lines readily mixed with dissociated organoid cells (Figure 3B and Supplementary Figure 4A). With or without addition of GBM cells, dissociated organoid cells efficiently reestablished the characteristic architecture of progenitor rosettes and neuronal layers observed in cerebral organoids, and membrane protrusions emanating from tumor cells were visible in all samples (Figure 3C). After 3 days of co-culture, mixed spheroids were dissociated and subjected to scRNA-seq. For comparison, we also dissociated and sequenced recomposed spheroids of organoid cells that had not been mixed with GBM cells (ie, NPCs) below, and GBM cells from all 4 patient-derived cell lines that had been grown separately as spheroids in the same culture medium (Figure 3A). Following preprocessing and quality control, we obtained 5083 single-cell transcriptional profiles with approximately 1400 genes detected per cell on average (Supplementary Figure 4B).
Figure 3.
Single-cell RNA-seq analysis of GBM cell interactions with cerebral organoid cells. (A) Protocol for the RNA-seq experiments. Following 7 days of neural induction, organoids and spheroids of lentivirally labeled GBM cells grown separately were enzymatically dissociated and mixed at a 1:1 ratio. After 3 days of co-culture, mixed spheroids were subjected to imaging or scRNA-seq using the iCell8 system. (B) Tumor cells mixed efficiently with organoid cells (see also Supplementary Figure 4). Scale bars, 50 µm. (C) With or without addition of GBM cells, dissociated organoid cells reestablished the characteristic 3D architecture of neural rosettes within 3 days. Scale bars, 50 µm. (D) t-SNE map showing all cells after quality control and PCA-based clustering, colored by sample origin (left), by cluster (middle), and by organoid or tumor cell identity (right). In addition to 3 clusters containing organoid cells, GBM cells clustered separately for each patient and before or after co-culture with organoid cells. (E) Volcano plot shows the 45 genes significantly up- or downregulated across all 3 patient-derived GBM cell lines upon co-culture with organoid cells (adjusted P < 0.05 for each patient separately). Venn diagrams quantify the overlap of differentially regulated genes detected from each patient. (F) Expression of differentially regulated genes visualized on a t-SNE map of all cells (t-SNE representation identical to panel D). (G) Gene Ontology–based gene set enrichment analysis of genes upregulated in all patient tumor cell lines upon co-culture with organoid cells.
Principal component analysis (PCA)–based clustering and 2D visualization by t-distributed stochastic neighbor embedding (t-SNE) maps revealed that GBM cells cultured alone clustered separately for each patient (clusters 5, 6, 7, and 9), confirming intertumoral heterogeneity (Figure 3D). This was also highlighted by differential expression of putative marker genes for GBM subtypes27 across patient samples (Supplementary Figure 4C). In agreement with previous scRNA-seq studies of GBM,11,28 we detected heterogeneous expression of gene signatures defining the classical, mesenchymal, neural, and proneural GBM subtypes29 within each patient-derived cell line, indicating that they comprise cells most representative of more than one subtype (Supplementary Figure 5). On average, cells from patients F2 and F3 mostly corresponded to the mesenchymal subtype, while cells from patients F6 and F9 most closely matched the neural subtype; as the mesenchymal subtype has been characterized as the most invasive,29,30 this is consistent with our earlier observation that F2 and F3 display higher invasive capacity in vitro.
We further identified 3 clusters (clusters 0, 1, and 3) containing cells from the unmixed organoids as well as cells from all 4 mixed samples, and concluded that the latter represent the organoid cells in the mixed samples (Figure 3D). The remaining clusters (2, 4, and 8) contain GBM cells from patients F2, F3, and F9 after co-culture with organoid cells. Note that as only 6 such cells were identified in the mixed sample from patient F6, they were excluded from further analyses and not displayed here.
Mixing GBM and Organoid Cells Leads to Upregulation of a Shared Set of Genes Across Patients
Differential gene expression testing between GBM cell clusters from mixed and unmixed samples revealed hundreds of genes that were significantly up- or downregulated upon co-culture with organoid cells (adjusted P < 0.05, log(fold change) > 0.15), and an overlap of 45 genes that were upregulated in all patients (Figure 3E). These included the homeobox transcription factor PAX6 (paired box 6), normally expressed in forebrain neural stem cells; the gap junction protein alpha 1 (GJA1) coding for connexin-43, which connects tumor microtubes in GBM4; glypican-3 (GPC3), a cell surface heparan sulfate proteoglycan and Wnt activator whose expression correlates with invasiveness of hepatocellular carcinoma31; collagen COL4A5, an extracellular matrix constituent; and several lysosomal, vesicular, and secretory proteins (Figure 3F and Supplementary Table 2). Gene set enrichment analysis of the 45 coherently upregulated genes confirmed that genes relating to growth regulation, neuronal migration, extracellular secretion, and stimulus response were enriched in this group (Figure 3G). Our results thus show that interactions between GBM and organoid cells increase expression of genes required for GBM network formation and invasion. Comparing our results with 4 distinct cell states identified in a recent scRNA-seq study of primary GBM,28 we noted a consistent shift in cell state composition toward a neural cell–like state in cells from patients F2 and F3 (Supplementary Figure 6).
Potential Ligand–Receptor Interactions Between Tumor Cells and Organoid Cells
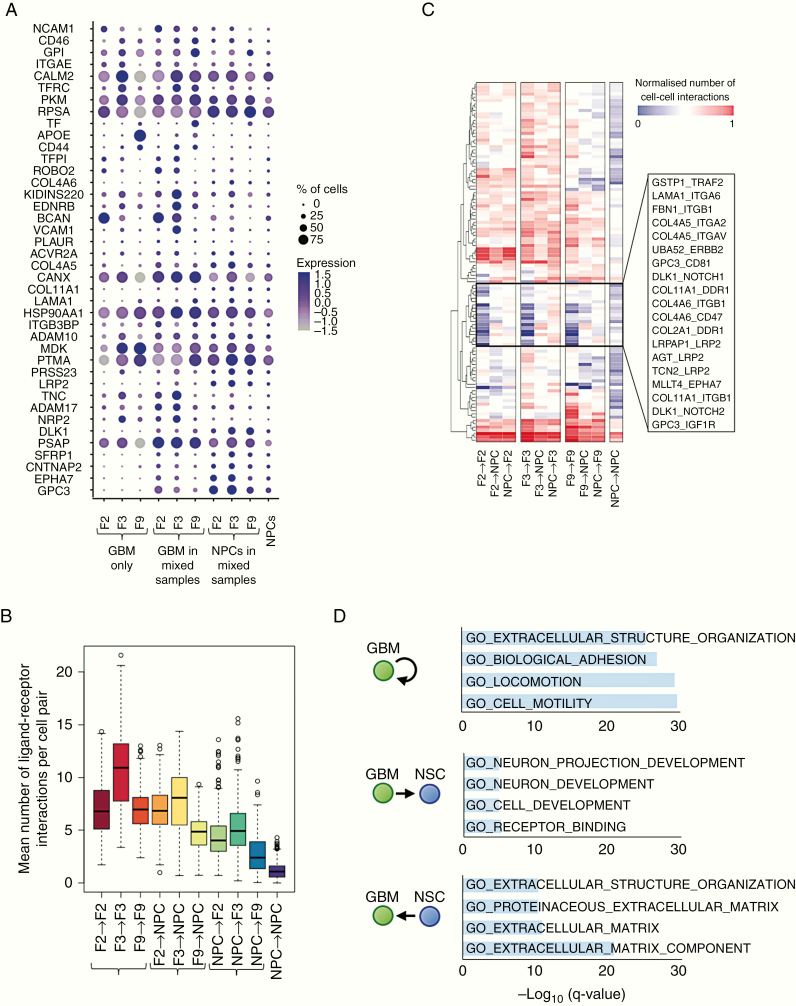
To investigate the nature of interactions between GBM and organoid cells, we considered the expression of 2,557 known ligand–receptor pairs22 across our samples, comprising a total of 1398 unique genes. Of these, 317 genes were expressed in our data, with approximately 13% expressed differentially between the same cell types in unmixed and mixed samples (Figure 4A).
Figure 4.
Potential ligand–receptor interactions between GBM and organoid cells. (A) Expression of ligand and receptor genes that are significantly upregulated in NPCs or in at least one GBM cell line upon co-culture (adjusted P < 0.05), averaged across GBM-only samples (left), GBM cells, or NPCs within mixed samples (middle), and the NPCs-only sample (right). Dot size corresponds to the fraction of cells expressing the gene in each group; dot color represents the average expression level. (B) The mean number of ligand–receptor pairs potentially connecting cells pairs of the given cell types, based on RNA expression levels across GBM-only samples (left), GBM cells, or NPCs within mixed samples (middle), and the NPCs-only sample (right). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. (C) Mean number of cell-cell interactions for different cell type combinations and selected ligand–receptor pairs. Hierarchical clustering reveals a group of ligand–receptor pairs (middle box) that are expressed at low levels in GBM cells or NPCs alone, but are upregulated upon co-culture. (D) Gene Ontology–based gene set enrichment analysis of ligand–receptor pairs preferentially expressed in the given cell type combinations.
Calculating the number of potential interactions between brain and tumor cells based on the expression of complementary receptors and ligands, we detected substantial crosstalk between cell types (Figure 4B). Hierarchical clustering of the number of cells potentially linked by each ligand–receptor pair revealed a group of ligand–receptor pairs that were expressed at low levels in the tumor-only and NPC-only cultures but presented many potential interactions between tumor cells and organoid cells in the mixed cultures (Figure 4C). These included several collagen-integrin interactions, GPC3 binding to insulin-like growth factor 1 receptor (IGF1R) or the cell cycle regulator CD81, and noncanonical Notch signaling (delta-like noncanonical notch ligand 1 [DLK1]/Notch 1, DLK1/Notch 2). Notably, despite the transcriptional heterogeneity we observed between patients, our approach detected consistently expressed potential interactions across all patient cell lines (Figure 4C and Supplementary Figure 7). Gene set enrichment analysis showed that the ligand–receptor pairs expressed at high levels in co-cultured samples are enriched for invasion-related genes (Figure 4D). Specifically, putative interactions in which GBM cells present the ligand and NPCs the receptor are enriched for genes involved in neuron projection development and receptor binding, whereas ligand–receptor pairs communicating in the opposite direction are enriched for extracellular matrix proteins.
Discussion
Despite its enormous therapeutic and prognostic significance, efficient methods to characterize the process of GBM invasion into human brain at a quantitative or transcriptional level are currently lacking. In this study, we present an in vitro model system in which lentivirally labeled patient-derived GBM cells invade into human cerebral organoids. By tissue clearing and confocal imaging, our approach shows that tumor cells extend up to 450 µm long membrane-bound processes after 3 days of invasion, recapitulating the development of GBM microtubes that has been observed in resected primary tumors and replicated in vivo in mice.4 Many of these processes terminate at distant tumor cells in our in vitro model, consistent with the development of an interconnected GBM network. By making GBM invasion experimentally accessible in vitro in a 3D tissue-like architecture, our experimental approach also enables the correlation of morphological phenotypes with transcriptional regulation by integration of imaging with single-cell sequencing. Here, scRNA-seq analysis of GBM and organoid cells separately or after co-culture revealed transcriptional changes induced by the interactions of tumor cells with their environment. Genes implicated in stimulus response, neuronal migration, secretion, and extracellular matrix were coherently upregulated across all tumor samples when mixed with NPCs, indicating that GBM cells sense the presence of neuronal cells and reactively amplify the transcription of genes supporting their dispersion. Among the upregulated genes was GJA1 (coding for connexin-43), known to enable multicellular communication via gap junctions in GBM networks in vivo.4
Heterogeneity between and within GBM tumors has impeded therapeutic progress for decades,32 with no targeted therapy available yet.33 Consistently, our imaging data suggest that GBM cells from different patients vary in their invasive capacity, although future studies should confirm differences in invasive capacity between GBM cell lines within the same organoids. While additional patient samples and more single-cell transcriptional profiles would be necessary to robustly link intertumoral differences in invasion behavior with specific transcriptional changes, our results corroborated the high degree of transcriptional heterogeneity between patients.11 However, we also detected a coherent element of transcriptional changes upon GBM and organoid cell co-culture indicating that targeting functional processes such as tumor microtube formation might improve therapeutic outcomes across patients.
Our analysis of ligand–receptor pair expression identified candidate pairs that may contribute to the invasion process, many of which have been linked with GBM progression by previous work. Glutathione S-transferase P has been shown to bind tumor necrosis factor receptor–associated factor 2 in vivo and in vitro, attenuating tumor necrosis factor signaling and thus enhancing resilience in tumor cells.34 Notch receptors 1 and 2 are known to associate with the transmembrane protein DLK1, but downstream effects in GBM remain unclear and may depend on heterogeneous cell states within the tumor.35 Binding of collagens to integrins, integrin-associated protein CD47, or discoidin domain receptor tyrosine kinase 1 all correlate with GBM proliferation and invasion.36–39 Integrin α6 as a receptor for the extracellular matrix protein laminin has also been detected in patient specimens of GBM and contributes to cancer stem cell proliferation in vitro.40
In addition to confirming these established signaling interactions, our in silico screen suggests putative interactions which may provide novel therapeutic targets for GBM. Binding of GPC3 to IGF1R or CD81 is known to contribute to hepatocellular carcinoma development and invasiveness41; immunotherapies targeting GPC3, which are showing promise in hepatocellular carcinoma,42,43 may also benefit GBM patients. Our results also suggest AFDN/MLLT4 (adherens junction formation factor/myeloid/lymphoid or mixed-lineage leukemia) as a ligand activating ephrin receptor A7, which has been linked to adverse outcome in primary and recurrent GBM.44 Finally, to our knowledge, the significance of fibrillin-integrin binding or of the low-density lipoprotein receptor related protein 2 interactions we detected have not been explored in GBM, but might present therapeutic opportunities. Further studies should explore the functional significance of these putative interactions.
Progress in organoid technology has led to the development of several organoid-based in vitro models for GBM within the past 2 years. Two studies have demonstrated that introducing oncogenic mutations in cerebral organoids initiates tumorigenesis,13,14 providing a model for studying the biological mechanisms underlying GBM formation and progression. Linkous et al recently showed that patient-derived GBM cells can invade, proliferate, and form microtubes within mature, 1–5 months old cerebral organoids,16 while da Silva et al demonstrated the feasibility of using younger organoids as a scaffold for GBM cell invasion.15 An earlier model used GBM invading into macroscopic human engineered neural tissue, with microarray data suggesting upregulation of extracellular matrix related transcription in one GBM cell line.45,46 Our study extends these findings to early-stage organoids generated by a highly reproducible and scalable protocol, thus enabling accelerated high-throughput screens. In contrast to previous studies, we also provide a transcriptomic characterization at the single-cell level of the GBM cell response to surrounding cerebral organoid cells.
Our results thus confirm the biological relevance of organoid-based model systems and show that interactions between GBM and organoid cells result in transcriptional changes detected by scRNA-seq. As our aim was to develop an experimental system suitable for high-throughput screens, we used organoids at 7 and 24 days of neural induction and performed scRNA-seq after just 3 days of co-culture; this potentially limits the accuracy of our results, since other cell types such as mature astrocytes are not represented in our organoids, and we cannot rule out that the transcriptional changes observed are at least partly transient reactions to being placed into co-culture.
In the future, we expect that our approach will further enable functional studies of GBM invasion that would be difficult or impossible to be conducted in vivo, including long-term imaging of network formation and multicellular communication. While we here used tissue clearing and confocal imaging for an endpoint quantification of tumor invasion, GBM-invaded organoids are similarly amenable to live imaging by two-photon or light-sheet microscopy. By combining imaging with recent single-cell RNA sequencing methodologies that provide transcriptome data for greater cell numbers,20,47 and with other single-cell sequencing modalities such as chromatin accessibility sequencing,48 our model could thus help resolve the functional, transcriptional, and epigenetic factors associated with different invasion behaviors of GBM or other tumors into human brain. It also provides the basis for high-content drug screens to assess patient-specific drug action on tumor and healthy brain cells, thus helping to identify the most effective drug at clinically relevant time scales.
Supplementary Material
Acknowledgments
The authors would like to thank Katharina Jechow for technical laboratory support, Jay Gopalakrishnan and Elke Gabriel for helpful discussions, and Monika Langlotz for FACS assistance.
Conflict of interest statement. The authors declare that they have no competing interests.
Authorship statement. Study design: TGK, CC, and RE. Protocol development for GBM invasion into cerebral organoids: TGK. Experimental work: TGK, SMT, and KJ. Data analysis: TGK and JP. Establishment of GBM cell cultures: TE, HP, and PA. Manuscript preparation: all authors.
Funding
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF; Federal Ministry of Education and Research); Heidelberg School of Oncology (postdoctoral fellowship to TGK); PhD stipends. SMT and TE were supported by the PhD program of the Helmholtz International Graduate School for Cancer Research (PhD fellowships to SMT, TE).
References
- 1. Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. [DOI] [PubMed] [Google Scholar]
- 2. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. [DOI] [PubMed] [Google Scholar]
- 3. Aldape K, Brindle KM, Chesler L, et al. . Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16(8):509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osswald M, Jung E, Sahm F, et al. . Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528(7580):93–98. [DOI] [PubMed] [Google Scholar]
- 5. Weil S, Osswald M, Solecki G, et al. . Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol. 2017;19(10):1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol. 2018;14(8):482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyer M, Reimand J, Lan X, et al. . Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A. 2015;112(3):851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sottoriva A, Spiteri I, Piccirillo SG, et al. . Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klughammer J, Kiesel B, Roetzer T, et al. . The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med. 2018;24(10):1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mazor T, Pankov A, Song JS, Costello JF. Intratumoral heterogeneity of the epigenome. Cancer Cell. 2016;29(4):440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel AP, Tirosh I, Trombetta JJ, et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darmanis S, Sloan SA, Croote D, et al. . Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ogawa J, Pao GM, Shokhirev MN, Verma IM. Glioblastoma model using human cerebral organoids. Cell Rep. 2018;23(4):1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bian S, Repic M, Guo Z, et al. . Genetically engineered cerebral organoids model brain tumor formation. Nat Methods. 2018;15(8):631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. da Silva B, Mathew RK, Polson ES, Williams J, Wurdak H. Spontaneous glioblastoma spheroid infiltration of early-stage cerebral organoids models brain tumor invasion. SLAS Discov. 2018;23(8):862–868. [DOI] [PubMed] [Google Scholar]
- 16. Linkous A, Balamatsias D, Snuderl M, et al. . Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26(12):3203–3211.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eisemann T, Costa B, Harter PN, et al. . Podoplanin expression is a prognostic biomarker but may be dispensable for the malignancy of glioblastoma. Neuro Oncol. 2019; 21(3):326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou B, Zhang D, Zhao S, et al. . Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat. 2015;9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puram SV, Tirosh I, Parikh AS, et al. . Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171(7):1611–1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macosko EZ, Basu A, Satija R, et al. . Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramilowski JA, Goldberg T, Harshbarger J, et al. . A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015;6:7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camp JG, Sekine K, Gerber T, et al. . Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546(7659):533–538. [DOI] [PubMed] [Google Scholar]
- 24. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148(1):313–319. [PMC free article] [PubMed] [Google Scholar]
- 26. Winkler F, Wick W. Harmful networks in the brain and beyond. Science. 2018;359(6380):1100–1101. [DOI] [PubMed] [Google Scholar]
- 27. Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134(3):505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neftel C, Laffy J, Filbin MG, et al. . An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carro MS, Lim WK, Alvarez MJ, et al. . The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng W, Tseng CJ, Lin TT, et al. . Glypican-3-mediated oncogenesis involves the Insulin-like growth factor-signaling pathway. Carcinogenesis. 2008;29(7):1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reardon DA, Wen PY. Glioma in 2014: unravelling tumour heterogeneity—implications for therapy. Nat Rev Clin Oncol. 2015;12(2):69–70. [DOI] [PubMed] [Google Scholar]
- 33. Touat M, Idbaih A, Sanson M, Ligon KL. Glioblastoma targeted therapy: updated approaches from recent biological insights. Ann Oncol. 2017;28(7):1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu Y, Fan Y, Xue B, et al. . Human glutathione S-transferase P1-1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene. 2006;25(42):5787–5800. [DOI] [PubMed] [Google Scholar]
- 35. Teodorczyk M, Schmidt MHH. Notching on cancer’s door: Notch signaling in brain tumors. Front Oncol. 2014;4:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamanaka R, Arao T, Yajima N, et al. . Identification of expressed genes characterizing long-term survival in malignant glioma patients. Oncogene. 2006;25(44):5994–6002. [DOI] [PubMed] [Google Scholar]
- 37. Gritsenko PG, Friedl P. Adaptive adhesion systems mediate glioma cell invasion in complex environments. J Cell Sci. 2018;131(15):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gritsenko PG, Ilina O, Friedl P. Interstitial guidance of cancer invasion. J Pathol. 2012;226(2):185–199. [DOI] [PubMed] [Google Scholar]
- 39. Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22(5):697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lathia JD, Gallagher J, Heddleston JM, et al. . Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6(5):421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Y, Liu H, Ding H. GPC-3 in hepatocellular carcinoma: current perspectives. J Hepatocell Carcinoma. 2016;3:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dargel C, Bassani-Sternberg M, Hasreiter J, et al. . T cells engineered to express a T-cell receptor specific for glypican-3 to recognize and kill hepatoma cells in vitro and in mice. Gastroenterology. 2015;149(4):1042–1052. [DOI] [PubMed] [Google Scholar]
- 43. Wu Q, Pi L, Le Trinh T, et al. . A novel vaccine targeting glypican-3 as a treatment for hepatocellular carcinoma. Mol Ther. 2017;25(10):2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang LF, Fokas E, Bieker M, et al. . Increased expression of EphA2 correlates with adverse outcome in primary and recurrent glioblastoma multiforme patients. Oncol Rep. 2008;19(1):151–156. [PubMed] [Google Scholar]
- 45. Nayernia Z, Turchi L, Cosset E, et al. . The relationship between brain tumor cell invasion of engineered neural tissues and in vivo features of glioblastoma. Biomaterials. 2013;34(33):8279–8290. [DOI] [PubMed] [Google Scholar]
- 46. Cosset E, Petty T, Dutoit V, et al. . Human tissue engineering allows the identification of active miRNA regulators of glioblastoma aggressiveness. Biomaterials. 2016;107:74–87. [DOI] [PubMed] [Google Scholar]
- 47. Zheng GX, Terry JM, Belgrader P, et al. . Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data have been deposited at the European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/home) under accession number EGAS00001003852. Scripts used for analyzing transcriptome data and image data (in R, Fiji, and MatLab) are available from the authors upon request.