Abstract
Background
The aim of this study was to predict isocitrate dehydrogenase (IDH) genotypes of gliomas using an interpretable deep learning application for dynamic susceptibility contrast (DSC) perfusion MRI.
Methods
Four hundred sixty-three patients with gliomas who underwent preoperative MRI were enrolled in the study. All the patients had immunohistopathologic diagnoses of either IDH-wildtype or IDH-mutant gliomas. Tumor subregions were segmented using a convolutional neural network followed by manual correction. DSC perfusion MRI was performed to obtain T2* susceptibility signal intensity-time curves from each subregion of the tumors: enhancing tumor, non-enhancing tumor, peritumoral edema, and whole tumor. These, with arterial input functions, were fed into a neural network as multidimensional inputs. A convolutional long short-term memory model with an attention mechanism was developed to predict IDH genotypes. Receiver operating characteristics analysis was performed to evaluate the model.
Results
The IDH genotype predictions had an accuracy, sensitivity, and specificity of 92.8%, 92.6%, and 93.1%, respectively, in the validation set (area under the curve [AUC], 0.98; 95% confidence interval [CI], 0.969–0.991) and 91.7%, 92.1%, and 91.5%, respectively, in the test set (AUC, 0.95; 95% CI, 0.898–0.982). In temporal feature analysis, T2* susceptibility signal intensity-time curves obtained from DSC perfusion MRI with attention weights demonstrated high attention on the combination of the end of the pre-contrast baseline, up/downslopes of signal drops, and/or post-bolus plateaus for the curves used to predict IDH genotype.
Conclusions
We developed an explainable recurrent neural network model based on DSC perfusion MRI to predict IDH genotypes in gliomas.
Keywords: angiogenesis, dynamic susceptibility contrast perfusion-weighted imaging, gliomas, isocitrate dehydrogenase mutations, recurrent neural network
Key Points.
1. The recurrent neural network model accurately predicted the IDH genotypes of gliomas using DSC perfusion MRI.
2. The model provides interpretable information from T2* susceptibility signal intensity-time curves for the prediction of IDH genotypes in gliomas.
Importance of the Study.
We developed an explainable recurrent neural network model with high diagnostic performance for the non-invasive prediction of IDH genotypes in gliomas using DSC perfusion MRI. Previous studies utilized relative cerebral blood volume (rCBV), which reflects tumor vascularity, to predict the IDH genotype of gliomas because IDH mutation is known to be associated with tumor angiogenesis in gliomas. However, there is a large overlap in rCBV values between IDH-wildtype and IDH-mutant groups, which leads to inaccurate predictions of IDH genotype. This study demonstrated that a recurrent neural network model, which learns sequential patterns, can distinguish these overlapped groups by utilizing raw multidimensional T2* susceptibility signal intensity-time curves obtained from DSC perfusion MRI, leading to improved and generalized diagnostic performance using an unseen test set. The model also provides interpretability by demonstrating which temporal features are crucial for the prediction of IDH genotypes based on molecular-biological backgrounds obtained using attention mechanisms.
Gliomas are the most frequent primary tumors of the central nervous system, exhibiting a devastating prognosis.1 According to the World Health Organization (WHO) classification system, gliomas are classified as grades I–IV based on histopathological and clinical criteria.2 The 5-year survival rate is only 5% for WHO grade IV gliomas or glioblastomas (GBM), with the median survival 14.6 months even after standard treatment including chemoradiotherapy with temozolomide.1,3 Moreover, the 5-year progression-free survival (PFS) rate is 50% in WHO grades II and III gliomas.4
Over the last decade, it has been shown that the presence of an isocitrate dehydrogenase (IDH) mutation is associated with overall survival as well as the diagnosis in gliomas.5,6 More than 80% of WHO grades II and III gliomas, or lower-grade gliomas (LGGs), and approximately 10% of secondary GBM have IDH mutations, the most common of which is the IDH1-R132H mutation.6 IDH mutation results in the loss of function of the enzyme that catalyzes the conversion of isocitrate to α-ketoglutarate as well as the gain of function of the enzyme to catalyze the conversion of α-ketoglutarate to (R)-2-hydroxyglutarate ((R)-2HG), an oncometabolite.7 However, IDH-mutant gliomas are less aggressive, easier to resect, and more sensitive to chemotherapy, particularly temozolomide, resulting in longer survival times than are found in IDH-wildtype gliomas.8 In contrast to IDH-mutant gliomas, in IDH-wildtype gliomas, aggressive surgical resection of the non-enhancing tumor does not provide any additional survival benefit.9 Moreover, IDH-wildtype LGGs have poor survival equal to that of GBM.10 Therefore, the preoperative prediction of the IDH genotype is crucial to treatment planning and prognosis prediction in patients with gliomas.
The conversion of α-ketoglutarate into (R)-2-hydroxyglutarate in IDH-mutant gliomas results in lower levels of hypoxia-inducible factor 1-alpha, a driver of hypoxia-initiated angiogenesis, in line with the less aggressive clinical course observed in IDH-mutant gliomas compared with IDH-wildtype gliomas.11–13 Recent studies have suggested that the IDH genotype is associated with and can be predicated using relative cerebral blood volume (rCBV) mapping obtained from dynamic susceptibility contrast (DSC) perfusion MRI,11,14 which has long been clinically used to investigate tumor angiogenesis. In particular, the IDH-mutant group had lower rCBV values than were found in the IDH-wildtype group,11,14 in line with the aforementioned studies.11–13 More recently, Zhang et al found that IDH-wildtype LGG vessels were molecularly distinct from the vasculature observed in IDH-mutated LGG.15 These previous results suggest that tumor angiogenesis differs according to the IDH genotype, and these differences may be distinguishable based on DSC perfusion MRI patterns.
Moreover, the long short-term memory (LSTM) model, a type of recurrent neural network (RNN) model, has demonstrated effective performance in various tasks, such as natural language processing, image captioning, genomic analysis, and medical diagnosis, recognizing patterns in sequential data.16–18 We hypothesized that an LSTM-based model could recognize specific patterns and classify multidimensional time-series data obtained from DSC perfusion MRI to distinguish IDH-wildtype gliomas from IDH-mutant gliomas. To the best of our knowledge, no deep learning–based methods that use raw T2* susceptibility signal intensity-time curves obtained from DSC perfusion MRI have previously been proposed for IDH genotype prediction in gliomas.
Finally, the purpose of this study was to develop an RNN model for the prediction of the IDH genotypes of gliomas with interpretability using preoperative multimodal MR imaging, including non-invasive DSC perfusion MRI.
Materials and Methods
Patients
The institutional review board of Seoul National University Hospital approved this retrospective study with a waiver of informed consent. From January 2013 to January 2018, enrolled in the study retrospectively were 603 patients who underwent treatment-naïve MR imaging. A total of 140 patients were excluded according to the exclusion criteria (Supplementary Fig. 1). Finally, a total of 463 patients who underwent all 4 conventional MRI and DSC perfusion MRI procedures were enrolled in the study. All of the enrolled patients had also undergone surgery or biopsy for tumors with immunohistopathological confirmation of the diagnosis. Detailed information regarding the tissue diagnosis and genetic analysis is provided in the Supplementary Material.
Imaging Acquisition
To obtain the T1-weighted imaging required for tumor segmentation, a T1-weighted 3D magnetization-prepared rapid acquisition gradient echo (MPRAGE) sequence was performed before and after the administration of gadobutrol (Gadovist, Bayer) at a dose of 0.1 mmol/kg of body weight in most of the enrolled patients. All of the DSC perfusion MRI protocols were included among the 3 dedicated protocols used in our institute. The MR scan parameters are provided in Supplementary Table 1.
Image Preprocessing
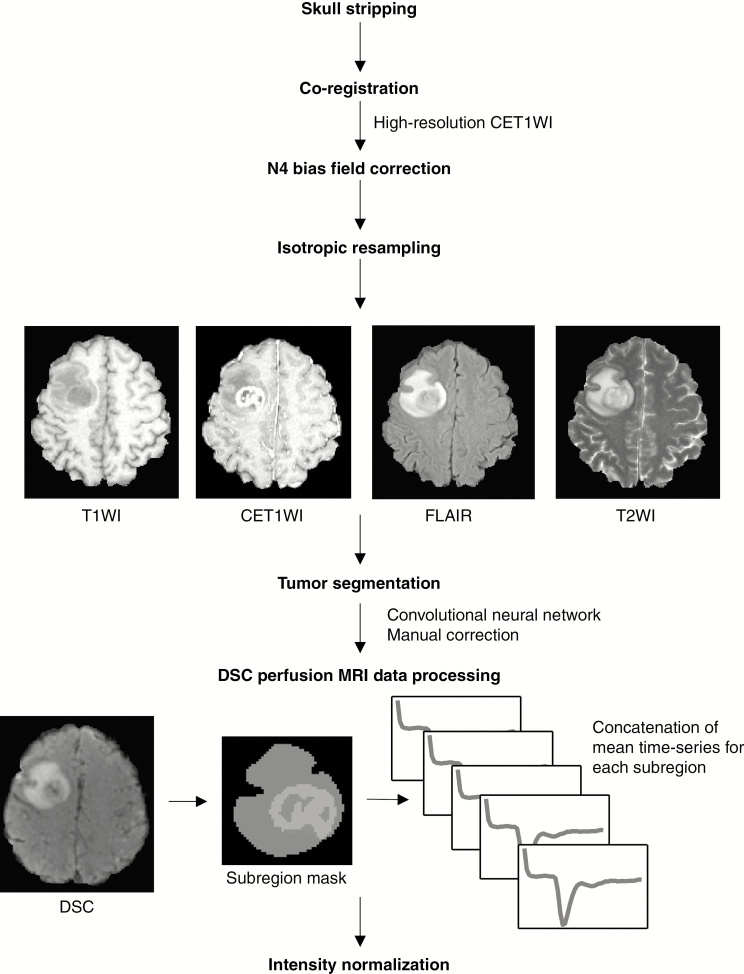
All patients underwent all 4 conventional MRI procedures, including T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), T2-weighted fluid attenuated inversion recovery (FLAIR) imaging, and contrast-enhanced T1-weighted imaging (CET1WI), which were required for tumor segmentation, as well as DSC perfusion MRI, which was required for the input of the neural network model. All MR images were co-registered to individual CET1W images that were mostly obtained using MPRAGE. The latter provided a submillimeter spatial resolution (0.7 mm), which was the highest resolution among the sequences. Intermodality co-registration as well as skull stripping were performed using NordicICE 4.1.3 (NordicNeuroLab). A mutual information-based algorithm was used to search for an optimal rigid transformation that aligned the 2 datasets with different modalities to achieve intermodality co-registration. N4 bias field correction was applied to remove all of the intensity non-uniformity with a low frequency.19 Next, all of the MR images were isotropically resampled to 1 mm with trilinear interpolation using FSL (FMRIB Software Library; http://www.fmrib.ox.ac.uk/fsl/).20 All procedures used during image preprocessing are summarized in Fig. 1.
Fig. 1.
MRI preprocessing pipeline. All the conventional MRI data were skull stripped, co-registered, and resampled. They were then submitted to tumor segmentation using convolutional neural networks with manual correction. The raw T2* signal intensity-time curve was obtained from DSC perfusion MRI using the subregion mask: peritumoral edema (blue), enhancing tumor core (green), and non-enhancing tumor core (red).
Tumor Segmentation
A fully automated segmentation tool that was the second best performing method in the international 2017 Brain Tumor Segmentation (BraTS) Challenge was utilized for segmentation. This tool used a cascade of fully convolutional neural network (CNN)21‒segmented whole tumors as subregions, as follows: the enhancing and non-enhancing tumor core and the peritumoral edema, according to the definition presented in the BraTS Challenge,22 using conventional MRI. More specifically, peritumoral edema, indicated by a peritumoral FLAIR high signal intensity lesion, was defined as a region clearly outside the enhancing and non-enhancing tumor core that presented as a non-enhanced T1-weighted as well as contrast-enhanced T1-weighted signal intensity abnormality (further details are provided in the Supplementary Material).21,22 Next, all tumor segmentations were manually corrected using 3D Slicer 4.8.1 (http://www.slicer.org/)23 by a neuroradiologist (K.S.C.) with 5 years of experience in neuroradiology.
DSC Perfusion MRI Data Processing and Normalization
Mean T2* susceptibility signal intensity-time curves were obtained for each of the tumor subregion and segmented using conventional MRI into the following categories: enhancing tumor, non-enhancing tumor, peritumoral edema, and whole tumor mask. The arterial input function (AIF, the fifth time course) was also obtained from DSC perfusion MRI. All of the time courses were normalized and concatenated to generate multidimensional time-series data. The detailed protocol used for the preprocessing of DSC perfusion MRI data is provided in the Supplementary Material.
Convolutional Long Short-Term Memory Network with Attention Mechanism
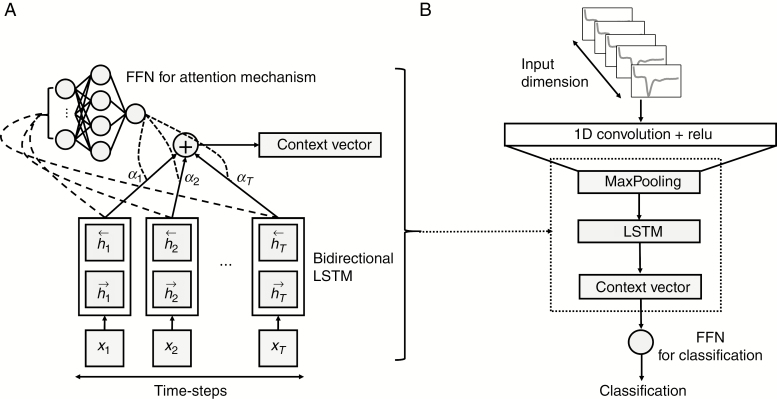
RNN is a deep learning model that learns sequential patterns or temporal dependencies within time-series data. In particular, LSTM, a type of RNN that effectively models sequences with varying lengths and captures long temporal dependencies and nonlinear dynamics, has achieved state-of-the-art results in tasks spanning natural language processing, image captioning, genomic analysis, and the analysis of medical data.16–18 In this study, bidirectional LSTM, an LSTM that allows the output units to be used to compute a representation that depends on both the past and the future, was used to reflect the context of inputs.24 First, 1-dimensional (1D) CNN was used as a powerful regional feature extractor in the sequential data. The LSTM then learned the temporal dependencies in the extracted features from the sequential data. In other words, the 1D CNN found a compact latent representation of the T2* susceptibility signal intensity-time curve, which was then fed into the bidirectional LSTM to learn the specific patterns that predicted the IDH genotypes of gliomas. This process generated 16 hidden states and 16 output states for every time step. Next, the weighted sum of the hidden states or the sequence outputs of the bidirectional LSTM network was used as a single condensed representation of the entire input sequence. More specifically, we used the bidirectional LSTM to produce a hidden state at each time step and then used a feed forward neural network (FFN) (Fig. 2A) attention function to assign importance (attention weights) , to each hidden state at time steps (Eq. 1).25 This enabled the model to predict a single target (ie, IDH genotype) per input sequence. Finally, the weighted sum of the hidden states, including the output weights of the attention function and a context vector , was fed into the single layer FFN for the classification, as illustrated in Fig. 2B. An overview of the model network structure is shown in Fig. 2B.
Fig. 2.
(A) Schematic of the feed-forward neural network attention mechanism and (B) overview of the convolutional LSTM with an attention model network structure.
| (1) |
Neural Network Model: Training, Validation, and Test Set
Among a total of 463 enrolled patients, 18 randomly chosen patients who contributed 144 samples (8 samples per patient using the sliding window technique, as described in the DSC perfusion MRI data processing and normalization section of the Supplementary Material) that were never seen by the model during training were used as a test set (this prevented these data from being mixed within the training and validation sets). The rest of the patients were divided at an approximately 8:1 ratio into generate training (n = 395; 3160 samples) and validation (n = 50; 400 samples) sets in a random manner. To report the more generalized performance of the model due to the small size of the test set, we performed 5-fold cross validation, in which 5 models were trained using each fold as a validation set and the rest of the fold as a training set. The average performance of the 5 models was used to evaluate the generalized performance of the model.
Neural Network Model: Evaluation
The model performance was evaluated by computing the accuracy, sensitivity, and specificity of the test set. Moreover, receiver operating characteristics (ROC) analysis was performed using sigmoid probabilities to obtain ROC curves and calculate the area under the curve (AUC). To calculate the 95% confidence intervals (CIs) of the AUC values, bootstrapping was performed and iterated 1000 times to create 1000 ROC AUCs. Although we used a small dataset to predict 1p/19q status (ie, IDH-mutant group; n = 1000 samples from 125 patients) to build a deep learning model, we also built an LSTM-based model for the prediction of 1p/19q status with hyperparameter modification.
Quantitative Analysis: Conventional Approach
Differences in age and the mean and 95th percentile values of the rCBV between the IDH genotype groups or among the WHO grade groups were analyzed using either Student’s t-test or one-way ANOVA (further details are provided in the statistical analysis section of the Supplementary Material). Using a conventional approach, we built a multivariate logistic regression model using age and mean rCBV values and compared its diagnostic performance with that of the LSTM-based model. Moreover, we built another logistic model using age and enhancement because they are known classifiers26 of IDH genotype based on disease prevalence.
Qualitative Analysis: Temporal Feature Interpretation
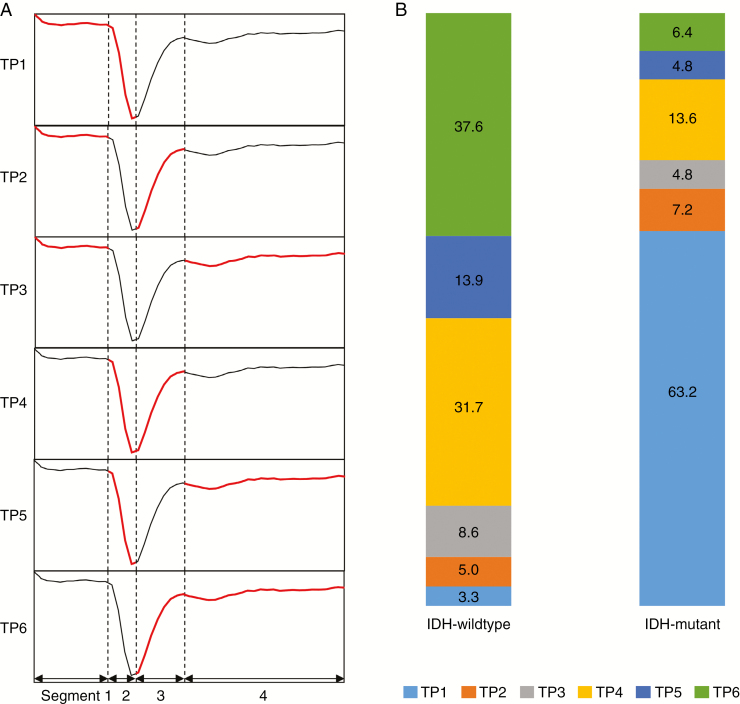
We used the attention layer, which consisted of a single FFN, to visualize and interpret temporal features. The attenuation mechanism enabled us to examine which time steps of the input sequences were critical for the model to achieve a classification by visualizing the weights of FFN as a heatmap.25 The multidimensional T2* susceptibility signal intensity-time curve was divided into 4 segments: the pre-contrast baseline, the downslope and upslope of the signal drop, and the post-bolus plateau. Among the 4 segments, 2 segments with the highest and second highest attentions were recorded in all 463 cases. Therefore, the 6 possible temporal patterns (TPs) (ie, if choosing 2 segments out of a total of 4 segments: patterns 1–6) for the heat map of attention weights and corresponding TPs were recorded in all 463 cases. A graphical definition of the TP is demonstrated in Fig. 4A.
Fig. 4.
Temporal patterns according to IDH genotype. (A) Graphical definition of the temporal patterns on the heat maps for attention weights overlaid upon T2* susceptibility intensity-time curves according to the segments with the highest and second highest attention weights: if the high attention weights were on segments 1 and 2; segments 1 and 3; segments 1 and 4; segments 2 and 3; segments 2 and 4; and segments 3 and 4 (red), the corresponding temporal patterns were defined as TP 1, TP 2, TP 3, TP 4, TP 5, and TP 6, respectively. (B) The frequencies of the temporal patterns of the heat maps of attention weights according to IDH genotype. The numbers represent percentages.Note: segment 1, pre-contrast baseline; segment 2, downslope of the signal drop; segment 3, upslope of the signal drop; and segment 4, post-bolus plateau.
Results
Patient Characteristics
A total of 3704 samples were generated from 463 patients with gliomas (272 males, 191 females; age, 52.2 ± 14.8 y; PFS, 21.1 ± 23.1 mo). There was no significant difference in age between male and female patients (P = 0.402). IDH mutations were significantly more frequent in LGG than in GBM (55.7% vs 9.7%; P < 0.0001). Moreover, the IDH-wildtype group was significantly older than the IDH-mutant group (56.1 vs 41.9 y, P < 0.0001). The incidence of 1p/19q codeletion was 27.6% (48 of 174) among LGGs. The detailed patient characteristics are summarized in Table 1.
Table 1.
Patient demographics and genetic information
| Total Patients, n = 463 | Age, mean ± SD, y | ||
|---|---|---|---|
| Sex | |||
| Male | 272 (58.7%) | 52.7 ± 15.3 | |
| Female | 191 (41.3%) | 51.6 ± 14.1 | |
| WHO grade and IDH | |||
| II | Wildtype | 17 (3.7%) | 48.7 ± 16.3 |
| Mutant | 15 (3.2%) | 44.3 ± 10.9 | |
| III | Wildtype | 60 (13.0%) | 52.2 ± 15.2 |
| Mutant | 82 (17.7%) | 40.9 ± 10.2 | |
| IV | Wildtype | 261 (56.4%) | 57.5 ± 13.5 |
| Mutant | 28 (6.0%) | 43.4 ± 13.9 | |
| WHO grade and 1p/19q | |||
| II | Codeleted | 7 (1.5%) | 41.4 ± 10.8 |
| Non-codeleted | 25 (5.4%) | 48.1 ± 14.6 | |
| III | Codeleted | 41 (8.9%) | 42.2 ± 9.3 |
| Non-codeleted | 101 (21.8%) | 47.1 ± 14.9 | |
| IV | Codeleted | 8 (1.7%) | 54.6 ± 16.7 |
| Non-codeleted | 281 (60.7%) | 56.1 ± 14.1 |
Model Performance
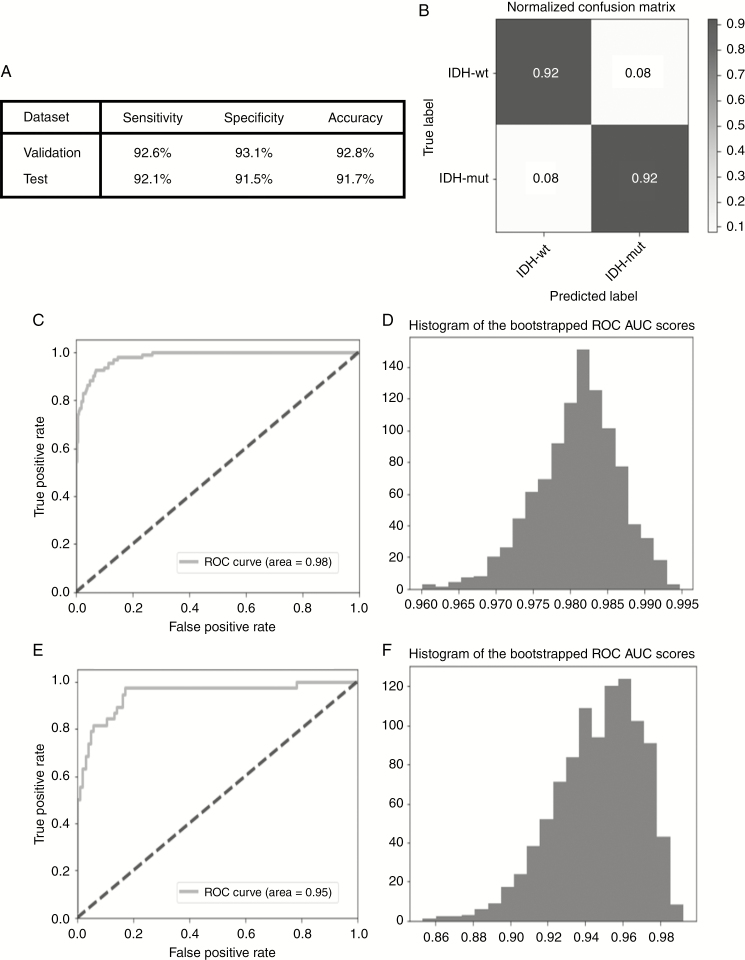
For the validation set, the IDH genotype predictions obtained using the optimized model had an accuracy, sensitivity, and specificity of 92.8%, 92.6%, and 93.1%, respectively. When the optimized model was applied to the unseen test set, its accuracy, sensitivity, and specificity were 91.7%, 92.1%, and 91.5%, respectively, for IDH genotype prediction. The diagnostic performance achieved in both the validation and test sets and the normalized confusion matrix for the test set are shown in Fig. 3A and B. The AUCs were 0.98 (0.969–0.991) and 0.95 (0.898–0.982) for the validation and test sets, respectively (Fig. 3C–F). In the 5-fold cross validation, the diagnostic performance of the model for each fold is shown in Supplementary Fig. 1 (mean AUC, 0.96 ± 0.02). In the subgroup analysis by WHO grade, the model had an accuracy, sensitivity, and specificity of 86.1%, 90.4%, and 83.8%, respectively, in LGG (AUC, 0.95; 0.943–0.963), and 91.0%, 88.4%, and 91.5%, respectively, in GBM (AUC, 0.96; 0.949–0.976) (Supplementary Fig. 2). There was no significant difference in the AUC between these groups (P = 0.331). The diagnostic performance of the LSTM-based model for predicting 1p/19q status is provided in the Supplementary Material.
Fig. 3.
Diagnostic performance and ROC analysis. (A) The diagnostic performance in both the validation and test sets and (B) the normalized confusion matrix for the test set. (C) ROC curve for the validation set (AUC, 0.98), (D) 95% CI AUC for the validation set using bootstrapping: 0.969–0.991, (E) ROC curve for the test set (AUC, 0.95), and (F) 95% CI AUC for the test set using bootstrapping: 0.898–0.982.
Quantitative Analysis: Conventional Approach
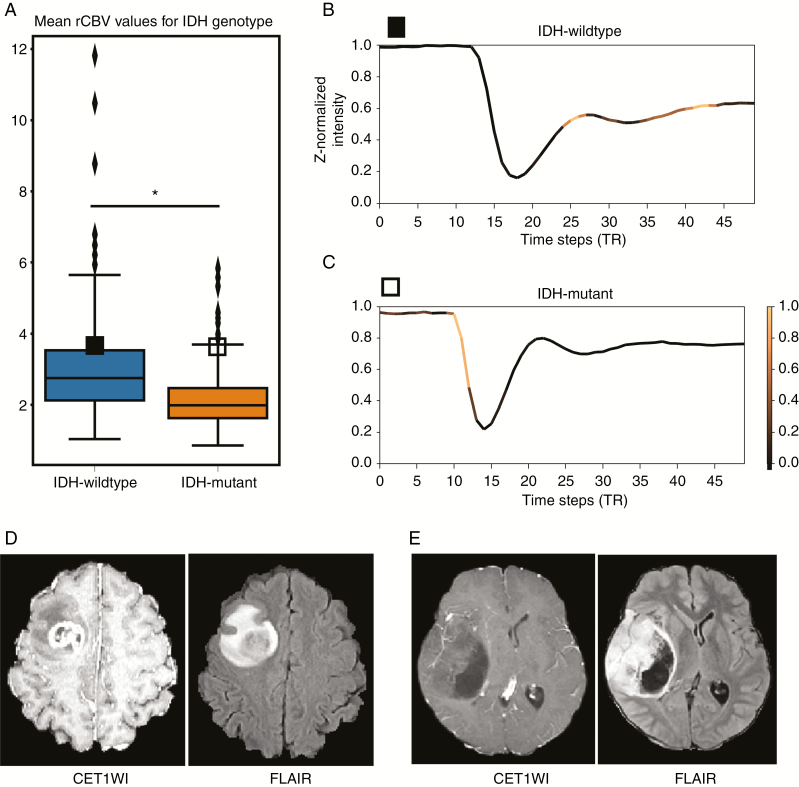
Both the mean and 95th percentile values of the rCBV were significantly higher in the IDH-wildtype group than in the IDH-mutant group (mean, 2.94 ± 1.25 vs 2.19 ± 0.90, P = 0.005; 95th percentile, 8.04 ± 3.55 vs 5.72 ± 2.52, P < 0.0001) (Supplementary Table 2). In the subgroup analysis by WHO grade, the mean rCBV values were significantly higher in the IDH-wildtype group than in the IDH-mutant group for both LGG and GBM (LGG, 2.53 ± 1.25 vs 2.11 ± 0.75, P < 0.0001; GBM, 3.07 ± 1.22 vs 2.49 ± 1.28, P < 0.0001) (Supplementary Fig. 3E). The min-max range of the mean rCBV showed that the ranges were largely overlapping (1.04–5.83) between the IDH-wildtype (1.04–11.81) and IDH-mutant groups (0.86–5.83) (Supplementary Table 2). The logistic regression model using age and enhancement achieved an accuracy, sensitivity, and specificity of 77.1%, 42.4%, and 89.9%, respectively (AUC, 0.80; 0.765–0.839), and therefore had a significantly poorer diagnostic performance than was achieved by the LSTM-based model (P < 0.0001). The diagnostic performance of the other logistic model using age and rCBV is provided in the Supplementary Material. The boxplots for the rCBV values that correspond to IDH genotypes and WHO grades are shown in Supplementary Fig. 3. Detailed results are provided in the Supplementary Material.
Qualitative Analysis: Temporal Feature Interpretation
Using the attention mechanism, we investigated which temporal features of the multidimensional T2* susceptibility signal intensity-time curve are crucial for the prediction of IDH mutation status. In other words, we sought to determine which time step or segment is important for classifying IDH genotypes in a given set of multidimensional time-series data. A single attention vector was obtained for each multidimensional input per patient. The specific patterns used to predict IDH genotype were recognized in the heatmaps of attention weights that were overlaid on the multidimensional T2* susceptibility signal intensity-time curves averaged from the whole tumor.
In the IDH-wildtype group, the most common TP was TP 6 (127 of 338; 37.6%), and the second most common pattern was TP 4 (107 of 338, 31.7%) (Fig. 4B). Conversely, in the IDH-mutant group, the most common TP was TP 1 (79 of 125; 63.2%), and the second most common pattern was TP 4 (17 of 125; 13.6%) (Fig. 4B). There were significant differences between the IDH-wildtype and IDH-mutant groups in the frequencies of the TPs obtained for T2* susceptibility signal intensity-time curves that were averaged from whole tumor (P < 0.0001) (Supplementary Table 3). Fig. 4B summarizes the profiles of the TPs of the heatmaps for attention weights overlaid on T2* susceptibility signal intensity-time curves that were averaged from whole tumor in both IDH-wildtype and IDH-mutant gliomas. In the subgroup analysis by WHO grade, there were also significant differences between the IDH-wildtype and IDH-mutant groups in the frequencies of TPs of T2* susceptibility signal intensity-time curves that were averaged from whole tumor (P < 0.0001) in both LGG and GBM (Supplementary Table 3). The most common TPs in the IDH-wildtype and IDH-mutant groups were TP 6 and TP 1, respectively, in both LGG and GBM. Moreover, among the correctly predicted cases of IDH-wildtype and IDH-mutant gliomas, some had nearly identical rCBV values (mean rCBV, 3.59 and 3.58, respectively, as illustrated in Fig. 5A), indicating that they could not be distinguished based on rCBV alone. In these two cases, the model produced patterns 6 and 1 in the IDH-wildtype and IDH-mutant glioma, respectively (Fig. 5B and C), with these pattern analyses resulting in a correct prediction (class probabilities of 0.849 and 0.956, respectively). Supplementary Fig. 4 also shows the different patterns observed in IDH-wildtype and IDH-mutant gliomas.
Fig. 5.
(A) Boxplots for the mean rCBV values corresponding to IDH genotypes. The vertical axis represents the mean rCBV values averaged from the whole tumor in arbitrary units. The asterisk (*) indicates that the mean rCBV values were significantly different between the groups (P = 0.005). Note that mean rCBV values according to IDH genotype largely overlap. Solid and hollow squares indicate the following cases of IDH-wildtype and IDH-mutant gliomas that had nearly the same rCBV values, respectively. (B), (C) Representative T2* susceptibility signal intensity-time curves from DSC perfusion MRI according to IDH genotype. The heatmap shows the normalized attention weights for each time step of the case belonging to the correct IDH genotype, as indicated by the color bar to the right of the graph. Brighter time steps indicate higher attention weights for IDH genotype prediction on the signal intensity-time curve. The vertical axis represents the z-normalized signal intensity in arbitrary units. The horizontal axis represents repetition time (the unit of time steps). Note that the T2* susceptibility signal intensity-time curve averaged from the whole tumor had a less steep upslope for the signal drop portion and an attenuated post-bolus plateau as well as a larger signal drop with a steeper downslope in the IDH-wildtype group than in the IDH-mutant group. The model therefore focused on the upslopes of the signal drop portion and post-bolus plateau in the IDH-wildtype group and on the pre-contrast baseline and the downslope of the signal drop in the IDH-mutant glioma. Corresponding CET1WI and FLAIR images for (D) IDH-wildtype and (E) IDH-mutant gliomas: note that the IDH-wildtype glioma shows thick, irregular enhancement with central necrosis, whereas the IDH-mutant glioma shows well-defined margins, reduced enhancement, and cyst-like necrosis.
Discussion
We developed an RNN model to predict IDH genotypes with a relatively large dataset (n = 463). To the best of our knowledge, this report describes the first “end-to-end” neural network model developed to predict IDH genotypes using raw T2* susceptibility signal intensity-time curves obtained from DSC perfusion MRI and bypassing postprocessing to generate rCBV maps.
This prediction model achieved high diagnostic performance with both interpretability and reproducibility for the test set. This was not only because the model was LSTM-based, allowing it to exhibit high performance in sequential learning,17 but also because of the following 3 features: (i) utilization of DSC perfusion MRI, which is better than conventional MRI at reflecting the specific tumor angiogenesis processes and vasculature of gliomas according to IDH genotype; (ii) we drastically reduced the number of parameters in the model with a relatively small dataset for deep learning to prevent overfitting and allow the model to be more effectively generalized than is possible with the CNN model when using the same numbers of 2D or 3D images; and (iii) we optimized the model by adding a convolutional layer, extracting semantic regional features and removing redundancy for highly correlated input sequential data, thus leading to better capture of temporal correlations as well as the attention layer17,25 for model interpretability.
Recently, CNN models developed to use conventional MRI (ie, T1WI, T2WI, FLAIR, and CET1WI) have been proposed as robust and non-invasive methods for obtaining preoperative IDH genotype predictions.27,28 However, previous studies suggested that IDH mutations are associated with tumor angiogenesis,11,14,15 best evaluated by DSC perfusion MRI, which provides a robust and clinically meaningful estimate of tumor angiogenesis. In particular, the (R)-2HG in the IDH-mutant group leads to a decrease in the level of hypoxia-inducible factor 1-alpha, a driver of hypoxia-initiated angiogenesis, consistent with the finding that the clinical course observed in the IDH-mutant group was more indolent than those of the IDH-wildtype group.11–13 Moreover, Chow et al revealed that GBM induces vascular dysregulation in peritumoral regions,29 the extent of which can be measured using blood oxygen level–dependent (BOLD) perfusion MRI, and that this area is larger in IDH-wildtype than in IDH-mutant gliomas. This finding could be used to differentiate IDH genotypes. Therefore, neural network models that utilize the raw DSC perfusion MRI signals obtained from not only the tumor core but also the peritumoral edema area will more plausibly predict IDH genotypes in gliomas, given the infiltrative nature of IDH-wildtype gliomas. More recently, Zhang et al found that IDH-wildtype LGG vessels are molecularly distinct from the vasculature of IDH-mutated LGG.15 More specifically, rCBV was lower in the IDH-mutant group than in the IDH-wildtype group,11,14,30,31 in line with the aforementioned studies.11–13 However, these studies showed that there was a large overlap in the ranges of rCBV between the IDH-wildtype and IDH-mutant groups,11,14 although these results may suggest that the tumor vasculature differs among IDH genotypes, and these differences may be distinguishable by DSC perfusion MRI patterns. In other words, no individual differences will be found when the rCBV of the glioma is within the overlapped range, and this will lead to the inaccurate prediction of the glioma’s IDH genotype. We hypothesized that we could develop a method to distinguish the IDH genotypes of gliomas in these overlapping rCBV groups by learning raw multidimensional DSC perfusion MRI signals using an LSTM-based model because these models can learn sequential patterns.
In addition, although leakage correction is supported by postprocessing software,32 rCBV assumes an intact blood–brain barrier, which is, however, disrupted in malignant gliomas and leads to incorrect estimations of rCBV. Moreover, previous studies reported that using different postprocessing software packages (required to convert the acquired images into rCBV maps) resulted in clinically significant differences in CBV images.33,34 Finally, permeability is one of the most potentially distinguishing features that can be derived from perfusion MRI data for the prediction of IDH genotypes.35 However, rCBV does not reflect the permeability of tumor vessels, which can instead be determined based on parameters such as the percentage of signal intensity recovery (PSR),36 which is derived from the ΔR2* curves obtained from DSC perfusion MRI.
Conversely, the idea of dividing voxels belonging to T2 high signal intensity lesions and contrast-enhanced lesions and then examining the raw DSC perfusion MRI time-intensity curves (ie, voxelwise T2* signal intensity-time curve analysis) is not a completely novel idea. Cha et al36 were able to differentiate GBM from cerebral metastasis using peak height (PH) and the PSR derived from the ΔR2* curve of DSC perfusion MRI, and these parameters are correlated with rCBV and Ktrans, respectively. This study extends the work of Cha et al36,37 by adding the use of deep learning algorithms. Moreover, these deep learning–based methods enabled the building of an end-to-end trainable model without the a priori selection of features (eg, volume, entropy, energy), and thereby might allow for greater clinical applicability and reproducibility compared with conventional radiomics approaches.27 Moreover, neural networks learn “representations,” and this leads to the discovery of novel significant features, whereas radiomics approaches require pre-engineered features with domain knowledge to be provided by human experts, thus reducing the probability of discovering new features.18 Considering this “end-to-end model” background, it is more appropriate to input raw DSC perfusion MRI data instead of postprocessed rCBV data when building a neural network model. In addition, the high performance achieved by the neural network model might be helpful for overcoming the limitations of current IDH genotyping. Currently, determining IDH genotype of gliomas requires a surgical biopsy, which requires general anesthesia and is therefore associated with possible risks. Moreover, immunohistochemistry to detect R132H misses approximately 15% of all IDH mutations,38 and sequencing might not be available, leading to delayed diagnosis.
Previous studies have developed many neural network models to provide advance predictions of fatal events, such as cardiac arrest, death, arrhythmia, and seizure, based on sequential medical data, including EEG, ECG, and other physiologic signals.18,39 Inspired by these previous studies, we utilized a convolutional bidirectional LSTM network with attention mechanisms, a neural network model for learning sequential data, to classify IDH genotypes from raw DSC perfusion MRI data. This approach also allowed the interpretation of time-series data.
Compared with a logistic regression model using age and rCBV, the LSTM-based model achieved significantly better diagnostic performance (AUC, 0.85 vs 0.95; P < 0.0001). In particular, the LSTM-based model had markedly improved sensitivity over that of the logistic regression model using the conventional approach (92.1% vs 52.0%).
In the subgroup analysis, the diagnostic performance of predictions of IDH genotypes between LGG and GBM showed that there was no significant difference between the ROC curves, suggesting that temporal features were not affected by WHO grade or by an imbalance in IDH status among the grades. Compared with the logistic regression model using known classifiers such as age and enhancement, the LSTM-based model achieved a higher diagnostic performance (P < 0.0001). There were significant differences in the TPs of the IDH-wildtype and IDH-mutant groups in both LGG and GBM, with TP 6 and TP 1 the most common TPs in IDH-wildtype and IDH-mutants, respectively (P < 0.0001). In the subgroup analysis performed using the conventional approach, mean rCBV values for the whole tumor were significantly higher in the IDH-wildtype group in both LGG and GBM (P < 0.0001).
To evaluate the generalizability of the performance of the model due to the small test set, we performed 5-fold cross validation of the LSTM-based model used to predict IDH genotypes. Two out of the 5 folds (folds 1 and 2) were slightly worse; however, the diagnostic performance (ie, sensitivity, specificity, accuracy, and AUC) was still similar to those of the validation and test sets (Supplementary Fig. 1A). The average AUC was also comparable to the AUCs for the test set (0.96 vs 0.95) (Supplementary Fig. 1B).
In the temporal feature analysis, we interpreted the multidimensional T2* susceptibility signal intensity-time curve from DSC perfusion MRI data (ie, signal intensity-time curve averaged from enhancing tumor, non-enhancing tumor, peritumoral edema, whole tumor, and AIF, respectively) that was overlaid on a heatmap of attention weights that was generated using a convolutional LSTM model. Attention mechanisms show where the convolutional LSTM model focuses for every time-step.17,25 In our model based on the T2* signal intensity-time curve, the upslope of the signal drop with the post-bolus plateau and the pre-contrast baseline with the downslope of the signal drop were useful in predicting IDH-wildtype and IDH-mutant glioma, respectively (Fig. 4). In other words, in terms of the prediction models, signal recovery was the most important feature of IDH-wildtype gliomas, in which it indicated that increased leaky/immature tumor vessels due to increased tumor angiogenesis were the crucial feature of IDH-wildtype gliomas,11,15 while tumor vascularity was the most significant feature of IDH-mutant glioma, in which it was correlated with PH as well as rCBV.36 In representative cases with nearly the same mean rCBVs in whole tumors, the model focused on the upslope of the signal drop and the post-bolus plateau of the T2* signal intensity-time curve obtained from the IDH-wildtype glioma (Fig. 5B). This parameter was correlated with signal recovery or vascular permeability, which was less steep and attenuated in the IDH-wildtype group than the IDH-mutant group. In addition, the model focused on the pre-contrast baseline and the downslope of the signal drop of the T2* signal intensity-time curve obtained from the IDH-mutant glioma (Fig. 5C), which was associated with tumor vascularity and was larger and steeper in the IDH-wildtype group than in the IDH-mutant group. Both of these results are consistent with previous studies11,15 that revealed that tumor angiogenesis is increased in the IDH-wildtype group compared with the IDH-mutant group, in which abnormal tumor vessels are leaky and immature.
There are several limitations to this study. First, there were differences in the magnetic field strengths of DSC perfusion MRI. Some were obtained using a 1.5 T scanner, and others were obtained using a 3 T scanner. Because T2* susceptibility signal intensity is strongly influenced by magnetic field strength, PH and PSR should be confounded by magnetic field strength, and this may lead to confusion in the neural network when predicting IDH genotypes. However, there was no significant association between magnetic field strengths for DSC perfusion MRI and IDH genotypes in the dataset (P = 0.053; Supplementary Table 4). This finding indicates that the magnetic field strength of the scanner did not affect the predicted IDH genotype, and the training, validation, and test sets were therefore divided after random shuffling of the total dataset. Second, although the model was designed to be explainable by radiologists, we used T2* susceptibility signal intensity-time curves obtained from subregions of tumors as the features of the input signals, and only a limited understanding of what exactly the model focuses on is possible given current attention mechanisms. Finally, because the proposed model utilizes only spatial information without incorporating the signal intensity, location, etc, of subregions of tumors from conventional MRI, incorporating the full set of conventional MRI data into the model may improve diagnostic performance.
In conclusion, we developed an LSTM-based model with high diagnostic performance to predict IDH genotypes in gliomas using DSC perfusion MRI. This approach is plausible because IDH mutation is associated with tumor angiogenesis. This non-invasive method might complement surgical biopsy, thereby improving treatment planning response evaluation when using anti-angiogenic therapies.
Funding
This research was supported by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2016M3C7A1914448 and NRF-2017M3C7A1031331), and by BK21 PLUS grant funded by the Ministry of Education (22A20151313464).
Supplementary Material
Acknowledgments
We would like to thank Byung-Hoon Kim, Yoseob Han, Yurim Kang, Moojin Yang, and Myungsu Chae for their invaluable assistance with the data collection and analysis.
Conflict of interest statement. The authors disclose no conflicts of interest related to this work.
Authorship statement. Study design: KSC, BJ, SHC. Data collection, analysis, interpretation: KSC. Figures: KSC, BJ. Manuscript writing: KSC, BJ, SHC. All authors revised and approved the final version of the manuscript.
References
- 1. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 4. Schomas DA, Laack NN, Rao RD, et al. Intracranial low-grade gliomas in adults: 30-year experience with long-term follow-up at Mayo Clinic. Neuro Oncol. 2009;11(4):437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. [DOI] [PubMed] [Google Scholar]
- 9. Beiko J, Suki D, Hess KR, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16(1):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Network CGAR. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kickingereder P, Sahm F, Radbruch A, et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep. 2015;5:16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye D, Ma S, Xiong Y, Guan KL. R-2-hydroxyglutarate as the key effector of IDH mutations promoting oncogenesis. Cancer Cell. 2013;23(3):274–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hong EK, Choi SH, Shin DJ, et al. Radiogenomics correlation between MR imaging features and major genetic profiles in glioblastoma. Eur Radiol. 2018;28(10):4350–4361. [DOI] [PubMed] [Google Scholar]
- 15. Zhang L, He L, Lugano R, et al. IDH mutation status is associated with distinct vascular gene expression signatures in lower-grade gliomas. Neuro Oncol. 2018;20(11):1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karpathy A, Fei-Fei L. Deep visual-semantic alignments for generating image descriptions. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):664–676. [DOI] [PubMed] [Google Scholar]
- 17. Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. In: Proc. International Conference on Learning Representations. 2015. http://arxiv.org/abs/1409.0473. Accessed April 26, 2019. [Google Scholar]
- 18. Lipton ZC, Kale DC, Elkan C, Wetzel R. Learning to diagnose with LSTM recurrent neural networks. In: Proc. International Conference on Learning Representations. 2016. http://arxiv.org/abs/1511.03677. Accessed April 26, 2019. [Google Scholar]
- 19. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. [DOI] [PubMed] [Google Scholar]
- 21. Wang G, Li W, Ourselin S, Vercauteren T. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Vol. 10670 Cham, Germany: Springer; 2017:178–190. [Google Scholar]
- 22. Menze BH, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30(9):1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE T Signal Proces. 1997;45(11):2673–2681. [Google Scholar]
- 25. Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33(21):3387–3395. [DOI] [PubMed] [Google Scholar]
- 26. Villanueva-Meyer JE, Wood MD, Choi BS, et al. MRI features and IDH mutational status of grade II diffuse gliomas: impact on diagnosis and prognosis. AJR Am J Roentgenol. 2018;210(3):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang K, Bai HX, Zhou H, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24(5):1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang P, Grinband J, Weinberg BD, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39(7):1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chow DS, Horenstein CI, Canoll P, et al. Glioblastoma induces vascular dysregulation in nonenhancing peritumoral regions in humans. AJR Am J Roentgenol. 2016;206(5):1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xing Z, Yang X, She D, Lin Y, Zhang Y, Cao D. Noninvasive assessment of IDH mutational status in World Health Organization grade II and III astrocytomas using DWI and DSC-PWI combined with conventional MR imaging. AJNR Am J Neuroradiol. 2017;38(6):1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bisdas S, Sanverdi E, Sudre C, Roettger D, Brandner S, Katsaros V.. The role of dynamic susceptibility contrast perfusion-weighted MRI in the estimation of IDH mutation in Gliomas [abstract]. J Clin Oncol. ; 2018;36(suppl 15):12063–12063. [Google Scholar]
- 32. Leu K, Boxerman JL, Cloughesy TF, et al. Improved leakage correction for single-echo dynamic susceptibility contrast perfusion MRI estimates of relative cerebral blood volume in high-grade gliomas by accounting for bidirectional contrast agent exchange. AJNR Am J Neuroradiol. 2016;37(8):1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelm ZS, Korfiatis PD, Lingineni RK, et al. Variability and accuracy of different software packages for dynamic susceptibility contrast magnetic resonance imaging for distinguishing glioblastoma progression from pseudoprogression. J Med Imaging (Bellingham). 2015;2(2):026001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korfiatis P, Kline TL, Kelm ZS, Carter RE, Hu LS, Erickson BJ. Dynamic susceptibility contrast-MRI quantification software tool: development and evaluation. Tomography. 2016;2(4):448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anzalone N, Castellano A, Cadioli M, et al. Brain gliomas: multicenter standardized assessment of dynamic contrast-enhanced and dynamic susceptibility contrast MR images. Radiology. 2018;287(3):933–943. [DOI] [PubMed] [Google Scholar]
- 36. Cha S, Lupo JM, Chen MH, et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. AJNR Am J Neuroradiol. 2007;28(6):1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lupo JM, Cha S, Chang SM, Nelson SJ. Dynamic susceptibility-weighted perfusion imaging of high-grade gliomas: characterization of spatial heterogeneity. AJNR Am J Neuroradiol. 2005;26(6):1446–1454. [PMC free article] [PubMed] [Google Scholar]
- 38. Andronesi OC, Rapalino O, Gerstner E, et al. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest. 2013;123(9):3659–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwon JM, Lee Y, Lee Y, Lee S, Park J. An algorithm based on deep learning for predicting in‐hospital cardiac arrest. J Am Heart Assoc. 2018;7(13):e008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.