Abstract
Glucocorticoids have multiple therapeutic benefits and are used both for immunosuppression and treatment purposes. Notwithstanding their benefits, glucocorticoid use often leads to hyperglycemia. Owing to the pathophysiologic overlap in glucocorticoid-induced hyperglycemia (GIH) and type 2 diabetes (T2D), we hypothesized that genetic variation in glucocorticoid pathways contributes to T2D risk. To determine the genetic contribution of glucocorticoid action on T2D risk, we conducted multiple genetic studies. First, we performed gene-set enrichment analyses on 3 collated glucocorticoid-related gene sets using publicly available genome-wide association and whole-exome data and demonstrated that genetic variants in glucocorticoid-related genes are associated with T2D and related glycemic traits. To identify which genes are driving this association, we performed gene burden tests using whole-exome sequence data. We identified 20 genes within the glucocorticoid-related gene sets that are nominally enriched for T2D-associated protein-coding variants. The most significant association was found in coding variants in coiled-coil α-helical rod protein 1 (CCHCR1) in the HLA region (P = .001). Further analyses revealed that noncoding variants near CCHCR1 are also associated with T2D at genome-wide significance (P = 7.70 × 10–14), independent of type 1 diabetes HLA risk. Finally, gene expression and colocalization analyses demonstrate that variants associated with increased T2D risk are also associated with decreased expression of CCHCR1 in multiple tissues, implicating this gene as a potential effector transcript at this locus. Our discovery of a genetic link between glucocorticoids and T2D findings support the hypothesis that T2D and GIH may have shared underlying mechanisms.
Keywords: corticosteroid, diabetes, genetics
Glucocorticoid hormones, when used at supraphysiologic concentrations, exert anti-inflammatory and immunosuppressive action by lowering inflammatory cytokines, reduced antibody receptor expression, and decreased T-cell function [1]. Owing to these therapeutic benefits, approximately 10 million Americans are prescribed glucocorticoids each year, with up to 0.9% of the population using these medications at any given time [2].
The use of steroids for therapeutic purposes is limited by their multiple side effects. Excessive glucocorticoid use causes decreased bone mass, glaucoma, increased risk for infections such as pneumocystis or reactivation of tuberculosis, and increased risk of hypertension. In addition, exogenous glucocorticoids can induce metabolic derangements such as central adiposity, hepatic steatosis, loss of skeletal muscle, insulin resistance, and hyperglycemia [3]. Glucocorticoid-induced hyperglycemia (GIH) has been reported in as many as 50% of glucocorticoid users [4, 5]. Patients with GIH can develop diabetic ketoacidosis or hyperosmolar state and are at increased risk of death [6, 7]. Although there is a positive correlation between the amount of steroids received and the risk of developing GIH, the patient-specific factors that predispose certain individuals are not known [8, 9].
The pathophysiology of GIH is similar to that of type 2 diabetes (T2D), including increased insulin resistance and gluconeogenesis, and decreased β-cell function. Glucocorticoids have a glycemic effect on the liver, pancreas, skeletal muscle, and adipose tissue. Through their actions on phosphoenolpyruvate carboxykinase and glucose-6-phosphatase, glucocorticoids increase gluconeogenesis in the liver [10]. Glucocorticoids inhibit translocation of the glucose transporter GLUT4 to the cell surface, thus reducing glucose uptake in response to insulin in adipose and skeletal muscle tissue [10-13]. Glucocorticoid-induced lipolysis and protein degradation additionally increases the substrates needed for gluconeogenesis [14]. In the pancreas, glucocorticoids inhibit the production and secretion of insulin acutely but lead to hyperinsulinemia after prolonged exposure. The shared pathophysiology between GIH and T2D suggests a potential overlap in the genetics and biology of these conditions.
The ongoing assembly of genome-wide association study (GWAS) data sets for T2D and related glycemic traits enables the comprehensive characterization of the genetic architecture of these traits. A GWAS resource does not exist for GIH; however, we can leverage our understanding of glucocorticoid biology to test whether glucocorticoid-related (GR) genes are enriched for variants robustly associated with T2D or other glycemic processes. We therefore analyzed curated gene lists to determine whether these genes were enriched for glycemic trait genetic variability. To identify which genes were driving this signal, we analyzed gene-specific whole-exome sequence and GWAS data. Coiled-coil α-helical rod protein 1 (CCHCR1), the gene with the strongest association, was further examined with independent validation in expression quantitative trait loci (eQTLs) data sets. Additionally, using colocalization [15] we evaluated the single-nucleotide variation (SNV, formerly single-nucleotide polymorphism) correlation with expression. We hereby identify CCHCR1 as a gene implicated in T2D.
1. Material and Methods
A. Enrichment Analyses
For enrichment analyses, we collated 3 GR gene sets: 1) “OMIM,” which includes genes from an Online Mendelian Inheritance in Man (OMIM) literature search for “glucocorticoid,” [16] 2) “glucocorticoid-responsive” genes [17], and 3) genes known to be involved in the glucocorticoid “biosynthesis” pathway [18] (Table1). The OMIM gene set was carefully curated with each individual gene evaluated for relevance. Glucocorticoid-responsive genes were identified from 3 separate gene expression data sets involving glucocorticoid treatment: lymphoblastic cell lines from the Childhood Asthma Management Program [19], human orthologs of mouse genes differentially expressed in C57BL/JGt mice treated with dexamethasone vs saline [20], and chromatin immunoprecipitation and RNA sequencing in A549 lung epithelia carcinoma cell lines treated with dexamethasone [21]. Genes were included in the glucocorticoid gene set if 2 out of 3 lists showed differential expression trending in the same direction for a given gene [17].
Table 1.
Curated gene lists
| Pathway | Genes |
|---|---|
| Glucocorticoid-responsive | DUSP1, NFKBIA, TFCP2L1, HLA-DRB1, MDK, SRGN, TNFAIP3, AGPAT2, SH2D4A, PASK, METTL7A, RHOU, CDKN2C, E2F7, PRDM1, ISG20, TNFSF10, ITGB1, CITED2, RABAC1, TXNIP, ABHD5, ERMAP, AURKA, ADAM19, CKAP4, SPDL1, KLF9, CEBPD, FKBP5, MAL, PDK4, ERRFI1, DDIT4, PERP, AHNAK, ADHFE1, TMEM56, CDK1, VCAN, DBN1, IGFBP4, KIF20B, DEPDC1, CENPK,BORA, CYSTM1, LYPD6B, HLA-DMA, ST8SIA4, C12orf75, ADORA2B, ALOX5AP, KLF5, TSPO,CAPG, CD9, COL4A3, FCER1G,GLRX, LMO7, LOX, RGS1, S100A6, SDC1,RASSF7, VAMP8, MAP3K6,CYTIP, SPRY1, TIPARP, HILPDA, FXYD5, PNPLA2,NCEH1, TMEM243, HOPX, PARD6B, CAMK4, CYPA1A,DDIT3, DNMT3A, GEM, GUCY1A3, ITGA1, LCK, PDE4B, PDGFRB, PLK1, SOX4, SPIB, ZEB1, ZNF207, FZD3, ENC1, BHLHE40,IER3,NREP, GDF15, IER2, ZEB2, PLK2, FNBP4, ANGPTL2, SLC39A6,PARM1, RND1, UBE2T, DACT1, ARRDC3, MARCKSL1, C1orf54, VASH2, NETO2, CDCA3, AFAP1L2,SGOL1, TUBB2B, BIRC3, GLUL, TCF7, SOCS1, TBC1D2, EMILIN2, BCL2, HES1, PRDX6, PHIP, ANG, PKIA, ERN1, OLR1, MMRN1, CGN, TIA1, GIT2, CCDC88A, CCND3, LTB4R,IL6ST, MT1X, MT2A, PER1, PYGB, IL1R2, HOMER2, CCHCR1, CHPT1, DEPTOR, SFXN5, CYP4V2,ABLIM1, SLC5A3, WIPI1, SEC14L1, SOD2, HRASLS2,TMEM62, ZC3H12A,SLC41A2, TMEM116,MAP3K8, SMARCC1,MAP3K14, INTS6, RHOB, ATP6V0A1, C5AR1, KLF6, GADD45A,FCGR2A, GCNT1, GCLC, HAGH,FOXN2, KCNK1, MYH6, POU5F1,MAPK13,PXN, MARCO,IL18RAP, PER2, P4HA2,LPIN2, HERPUD1, SPRY2, FSTL3, SERINC3, ELL2, ZNF281, PIK3R5, MKRN1, RASD1, SLC37A1, EPB41L4B, TRIB3, TMEM8A, PLEKHF2, SLC16A10, SLC25A29, RNF149, EPHB1, HOXB2, ID3,ITGA4, MEF2D,NAB2, CDK17,POLB, ST3GAL2,TNFSF9, SUCLA2, TRAF4, CLCF1,PPP1R15A,RASGRP3, MOXD1,SERTAD3, EVL, DPH5, ZFR, BCL11A, CXCR7, PELI1, PLEKHG1, TGIF2, DOCK7, L3HYPDH, MB21D1, RHOV, ARL5B |
| Biosynthesis | CYP11A1, CYP17A1, CYP21A2, CYP11B2, CYP11B1, HSD3B2, SULT2B1 |
| OMIM | NR3C1P1, MRAP, GLCCI1, SGK2,GLCCI1, SGK3, GMEB2, NR3C1, NNT, ARHGAP35, GMEB1, MC2R, CYP11B1, MCM4, TNFRSF18, AAAS, TSC22D3, SGK1, NCOA2, CYP3A4, MYOC, PTGS2, MC2R, CYP11B1, MCM4, DAX1, MIF, AR, STAR, PLN, FKBP5, BAG1, MTPN, DGKH, WBSCR22, ONECUT1, HSD11B1, PCK1, RWDD3, ADH6, PDCD2, TNFSF18, SLC30A2, ST13, TTLL5, CRY2, CYP17A1, CSN2, NR3C2, CALR, MT1A, CRY1, FPR2, ANXA1, CRH, NEDD4, EPHX1, CYP11B2, HDAC6,C YP19A1, GK,DRD1, SERPINE1, PHEX, MYC, TCF4, TLR7, HSD11B2, TLR9, MECP2, NOTCH1, NR5A1, ABCD1, AIP, ADCY9, ERVK-7, ERVK-4, ERVK-5, RSC1A1, DCAF6, ZFP36L2, CRHBP, ANXA11, TBX19, JMJD1C, UCN3, HSD17B8, RAB24, RBM14, ZNF395, DDX54, PPID, SRCAP, SLC9A3, HDAC2, NME3, UNC45A, SLC2A4RG, PDCD6, NME4, ABCA3, UMPS, RANBP9, RANBP10, TRIM27, KCNJ5,SCZD1, TRIM68, DMAP1, NME2, NFAT5, AMT, PDLIM7, PRPF6, RBFOX2, ANKRD11, SNW1, PDE11A, POMC, NCOA1, SCNN1G, HG6PD, SFTPA1, TXN, WNK4, CYP21A2, ARID1A, GIP,MVP, SS18, SMARCA4, IKZF1, HMGB1, DAXX, RARB, PPARA, SFTPB, PPARGC1B, TPH2, NME1, NAMPT, EGR1, CREB1, PPARGC1A, H6PD, HDAC1, CYP11A1, NPY, IL23A, NFKB1A, IL23R, AGT, RB1, DNMT1, THRB, CYP21A2, TGFB, PPARG1, APOE, GPR83, WIPF3, DEGS1, RASD1, CYP11B1, SMARCA2, STAT5A, IL10, TNF1 |
Abbreviation: OMIM, Online Mendelian Inheritance in Man.
We used the software MAGENTA [22] to test whether our 3 GR gene sets are enriched for glycemic variants using publicly available GWAS data, either associated with T2D from the DIAGRAM consortium [23-25] or quantitative glycemic traits from the MAGIC investigators [26-30]. For each phenotype tested, MAGENTA assigns each gene in the genome the P value of the most significant single-nucleotide variation (SNV) located −110 kilobases (kb) upstream and +40 kb downstream of the transcript. These values are corrected for gene size, SNV density, and linkage disequilibrium (LD). The genes within each set are then ranked according to likelihood of association with the given trait. MAGENTA then calculates a P value of enrichment for any given gene set based on at least 10 000 randomly permuted sets of the same size, to determine whether the defined gene set has overrepresentation of genes above an enrichment cutoff, set at either the 95th percentile or 75th of the associated P values. We tested if our 3 GR gene sets were enriched for variation in T2D and related glycemic phenotypes, including glycated hemoglobin (HbA1c), fasting glucose, fasting insulin (with and without adjustment for body mass index [BMI]), homeostasis model assessment for β-cell function (HOMA-β) and insulin resistance (HOMA-IR), 2-hour glucose and 2-hour insulin (with and without adjustment for BMI), and insulin secretion as measured by the corrected insulin response and the disposition index (DI).
B. Burden Tests
To identify the potential gene(s) driving the enrichment signal, gene burden tests were carried out using the 45K whole-exome sequence data set available in the Type 2 Diabetes Knowledge Portal (http://www.type2diabetesgenetics.org/) [31]. These exomes were obtained from more than 20 diabetes studies in multiple consortia and different countries. Five distinct ancestry groups were analyzed.
Variants were filtered based on potential deleterious effects on protein function, called masks. We used 7 different masks ranked by their levels of predicted deleterious effects in order of increasing deleteriousness. The strongest mask consisted of alleles predicted to cause loss of function by the LofTee algorithm (https://github.com/konradjk/loftee). The weaker masks included alleles predicted to be deleterious by bioinformatic algorithms, with the number of tools predicting deleteriousness correlating to the strength of the mask (the more tools predicting deleteriousness, the stronger the mask). These masks were then combined as previously described [31]. We employed 2 methods to collapse the results while accounting for the correlation among masks and multiple testing: 1) the “minimal P value” and 2) weighted tests. The “minimal P value” test takes the lowest P value across all masks and corrects for the effective number of tests performed on a gene. The weighted test collapses associations under a model whereby the phenotypic effects of alleles are directly proportional to their bioinformatically estimated deleteriousness. We assigned mask-specific allele weights according to their predicted deleteriousness, giving each variant a quantitative value estimating the fraction of loss-of-function variants. Full loss-of-function variants were given a value of 1, whereas synonymous variants were given a value of 0. In the “weighted burden” test, we used the sum of the weights of alleles carried by an individual as a predictor variable in place of the total number of alleles carried. Burden tests were performed using both SKAT [32] and Firth [33, 34] tests to determine if directionality of the effect affected the outcome.
C. Common and Rare Variant Analyses
To determine the candidacy of individual genes from gene burden analyses, we mined the comprehensive, publicly available GWAS data in the Type 2 Diabetes Knowledge Portal to examine whether the prioritized genes contained variants associated with T2D and/or related metabolic phenotypes. These phenotypes include T2D with and without adjustment for BMI, fasting glucose, fasting insulin, HbA1c, insulin sensitivity with and without adjustment for BMI, insulin at 30 minutes after an oral glucose tolerance test with and without adjustment for BMI, height, BMI, waist-hip ratio, waist circumference with and without adjustment for BMI, pericardial adipose tissue, triglycerides, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, coronary artery disease, chronic kidney disease, microalbuminuria, estimated glomerular filtration rate, diabetic kidney disease, end-stage renal disease, and palmitic acid levels [35]. The T2D portal performs association analyses on SNVs 250 kb upstream and downstream of the gene of interest. Independent replication of variants associated with T2D was obtained from the UK Biobank (UKBB) [36]. LD was calculated using LDlink [37].
D. Association Studies With Human Leukocyte Antigen Adjustment
The T2D candidate gene, CCHCR1, is located in the 6p21.3 region, the site of HLA gene loci whose alleles are strongly associated with type 1 diabetes (T1D). To determine whether the association observed in the UKBB was driven by the HLA-T1D association through CCHCR1 SNPs that might be in LD with T1D-associated SNVs, we tested the association of our SNVs both with T1D and T2D in the UKBB after adjusting for HLA SNVs associated with T1D. Starting with unrelated individuals of European ancestry in UKBB as described previously [38], we defined T1D and T2D as “probable” or “possible” cases based on previously described diabetes algorithms [39], expanded to include information from repeat assessment center visits. For controls, we used the “diabetes unlikely” subset, restricted to individuals older than or equal to age 55 years, and removed individuals with any indication of diabetes from repeat assessment center visit information, the touchscreen diabetes diagnosis question (field 2443), or diabetes International Classification of Diseases, ninth revision (ICD9) and tenth revision (ICD10) codes. In addition, controls with an HbA1c of 6.5% or greater were reclassified as “T2D cases” for the T2D case-control definition. T2D controls with an HbA1c of 5.7% or greater were removed.
To account for the association of T1D with HLA, we used 3 tagging SNVs to infer T1D-risk HLA-DR and HLA-DQ genotypes [40]. In brief, rs3104413, rs9273363, and rs2854275 predict high T1D-risk HLA-DR/DQ genotypes. We conditioned these HLA region SNVs as individual covariates as well as inferred HLA DR/DQ in our model. Logistic regression was performed and adjusted by covariates, including HLA SNVs and HLA-inferred genotypes (where appropriate), and the first 10 principal components to adjust for population stratification.
Because there are fewer than 1000 T1D cases in the UKBB, we examined data from a meta-analysis of T1D cases and controls genotyped on the ImmunoChip [41] to establish whether CCHCR1 SNVs are associated with T1D in models that adjust for the HLA haplotypes known to be associated with T1D. The complete meta-analysis includes more the 60 000 individuals of diverse ancestries, including participants previously studied as part of the Type 1 Diabetes Genetics Consortium (T1DGC [42]). A subset of 33 578 unrelated cases and controls of European ancestry were analyzed here to replicate and further assess CCHCR1 variant associations with T1D while controlling for T1D-associated HLA types. We used the first 10 principal components to control for population stratification. We conditioned the inferred HLA-DR genotype and on the 3 HLA DR/DQ SNVs [40].
E. Expression and Expression Quantitative Trait Loci Analyses
We used the Genotype-Tissue Expression (GTEx) portal (GTEx Analysis Release V8, dbGaP Accession phs000424.v8.p2) [43, 44] to examine the tissue expression profile of CCHCR1 and identify eQTLs for the top associated SNPs at the CCHCR1 locus from the common variant analysis. We additionally used LocusCompare [15] to evaluate for colocalization. Colocalization plots gene expression from eQTLs with association statistics from GWAS to infer the causal SNP. For LocusCompare, we used the T2D GWAS from Scott et al (2017) [45] and the GTEX data from v7 [43, 44].
2. Results
A. Glucocorticoid-Related Genes Are Enriched for Associations With Type 2 Diabetes and Glycemic Traits
We examined 3 complementary gene lists, an OMIM literature search set, a glucocorticoid responsive gene set, and the glucocorticoid biosynthesis pathway gene set to determine whether these genes were associated with T2D and glycemic traits (see Table 1). We tested for enrichment of T2D association in variants within these gene sets using MAGENTA [22]. MAGENTA revealed enrichment of all 3 GR gene sets with T2D or glycemic traits. At the 95% percentile of each score, the OMIM and the glucocorticoid biosynthesis gene sets were enriched for T2D genetic associations (P = .02 for both). The glucocorticoid biosynthesis gene set was also enriched for genetic associations with fasting insulin adjusted for BMI (P = .02). The glucocorticoid-responsive gene set demonstrated enrichment for genetic associations with fasting glucose (P = .03) [Table 2].
Table 2.
MAGENTA results showing an enrichment of glucocorticoid-related gene variant associations in type 2 diabetes and glycemic traits
| Phenotype | Gene list | 95% cutoff P | 75% cutoff P |
|---|---|---|---|
| T2D | OMIM | .025 | .340 |
| Expression | .807 | .252 | |
| Biosynthesis | .023 | .374 | |
| Fasting glucose | OMIM | .172 | .652 |
| Expression | .030 | .444 | |
| Biosynthesis | 1.000 | .821 | |
| Fasting insulin | OMIM | .091 | .011 |
| Expression | .972 | .425 | |
| Biosynthesis | 1.000 | 1.000 | |
| 2-h glucose | OMIM | .290 | .172 |
| Expression | .879 | .536 | |
| Biosynthesis | 1.000 | .467 | |
| 2-h insulin | OMIM | .931 | .779 |
| Expression | .557 | .652 | |
| Biosynthesis | 1.000 | .757 | |
| HOMA-β | OMIM | .561 | .332 |
| Expression | .795 | .272 | |
| Biosynthesis | 1.000 | .763 | |
| HOMA-IR | OMIM | .163 | .009 |
| Expression | .803 | .591 | |
| Biosynthesis | 1.000 | .766 | |
| 2-h glucose adj BMI | OMIM | .562 | .222 |
| Expression | .942 | .236 | |
| Biosynthesis | 1.000 | .467 | |
| HbA1c | OMIM | .977 | .777 |
| Expression | .693 | .857 | |
| Biosynthesis | 1.000 | .361 | |
| Fasting insulin main effect | OMIM | .558 | .150 |
| Expression | .696 | .464 | |
| Biosynthesis | 1.000 | .770 | |
| Fasting insulin adj BMI | OMIM | .721 | .039 |
| Expression | .797 | .411 | |
| Biosynthesis | 0.023 | 0.367 | |
| CIR insulin secretion | OMIM | 0.293 | 0.284 |
| Expression | 0.797 | 0.027 | |
| Biosynthesis | 1.000 | 0.823 | |
| DI insulin secretion | OMIM | 0.417 | 0.058 |
| Expression | 0.933 | 0.278 | |
| Biosynthesis | 1.000 | 1.000 |
Abbreviations: adj, adjusted; β, β cell; BMI, body mass index; CIR, corrected insulin response; DI, disposition index; HbA1c, glycated hemoglobin; HOMA, homeostatic model assessment; IR, insulin resistance; OMIM, Online Mendelian Inheritance in Man; T2D, type 2 diabetes.
B. Gene Burden Tests Highlight Candidate Genes as Potential Drivers of Enrichment Results
To determine if in aggregate, coding variants in any of the 399 genes from our GR gene sets suggested evidence for the involvement of individual genes in T2D, we performed gene burden tests using whole-exome sequence data from 20 791 T2D cases and 24 440 controls. After taking into account the number of filters and genes tested, no genes passed a Bonferroni-corrected significance threshold (P < 1.25 × 10–4). The most significant association was found in the CCHCR1 gene (P = .001, min-P Firth test), with provenance from the glucocorticoid-responsive expression gene set. Other genes approaching significance (defined by P value) include PKIA, ADCY9, ALOX5AP, SLC5A3, WIPI1, and NCEH1 (Table 3). The CCHCR1 association is driven by a common SNV (rs3130453, minor allele frequency = 0.48) that introduces a stop codon into specific isoforms (Trp78Ter in ENST00000396268/CCDS43445.1).
Table 3.
Top genes from gene burden test results using the minimum P test
| Gene | Test | β | P |
|---|---|---|---|
| CCHCR1 | Firth | .104 | .001 |
| PKIA | Firth | 1.65 | .002 |
| ADCY9 | Firth | .173 | .005 |
| SLC5A3 | Firth | 1.27 | .006 |
| WIPI1 | Firth | .707 | .006 |
| ALOX5AP | Firth | –1.18 | .006 |
| NCEH1 | SKAT | NA | .006 |
| NCEH1 | Firth | –.254 | .01 |
| TNFAIP3 | SKAT | NA | .01 |
| SS18 | SKAT | NA | .01 |
| DACT1 | Firth | –.24 | .02 |
| PASK | Firth | –.438 | .02 |
| ISG20 | SKAT | NA | .02 |
| DOCK7 | SKAT | NA | .02 |
| DEGS1 | SKAT | NA | .02 |
| PERP | Firth | –.333 | .02 |
| AR | Firth | .198 | .02 |
| ONECUT1 | SKAT | NA | .02 |
| ONECUT1 | Firth | –.603 | .03 |
| SLC30A2 | SKAT | NA | .03 |
| PARM1 | SKAT | NA | .03 |
| CRY2 | Firth | –.11 | .03 |
Abbreviation: CCHCR1, coiled-coil α-helical rod protein 1; NA, not applicable; SKAT, sequence kernel association test.
Because CCHCR1 has the strongest association with T2D risk among the GR genes, we sought to determine if variants in CCHCR1 itself, as opposed to other genes in the region, are driving the T2D association. We performed burden tests for genes located within 250 kb upstream and downstream of CCHCR1. Among the 18 genes in this region, CCHCR1 exhibited the most significant association, with GTF2H4 also approaching significance (P = 3.01 × 10–3) in the weighted Firth test; however, evaluation of common variation in GTF2H4 with T2D did not attain statistical significance.
C. Common Variation in CCHCR1 Is Associated With Type 2 Diabetes
To ascertain whether there is additional evidence for genetic association between the top 20 genes based on most significant P value from the burden-test analysis and T2D, we evaluated common variation at these loci in GWAS data sets with large numbers of cases and controls, because our gene burden testing had limited statistical power for rare variation [46]. Variants near the most significant gene from our burden-test analyses, CCHCR1, exhibited the strongest association with T2D, with multiple coding and noncoding variants reaching genome-wide significance (Table 4 and Fig. 1). As noted, CCHCR1 was previously identified in the largest meta-analysis of T2D GWAS [35]; however, the association was attributed to SNVs in the major histocompatibility complex (MHC; eg, rs601945) and was not investigated further. This association has also been observed in the Million Veteran Program T2D multiethnic analysis as a potential new T2D gene [47].
Table 4.
Genome-wide association studies analysis for type 2 diabetes using common variants ±/–250 kb CCHCR1
| dbSNP ID | Predicted impact | Major allele | Minor allele | P | OR | Gene |
|---|---|---|---|---|---|---|
| rs3131012 | Intron | T | C | 7.70 × 10–14 | 1.05 | CCHCR1 |
| rs2240063 | Intron | T | C | 9.80 × 10–14 | 1.05 | CCHCR1 |
| rs3130941 | Upstream gene variant | C | G | 2.20 × 10–13 | 1.07 | XXbac-BPG299F13 |
| rs9264024 | Upstream gene variant | A | G | 2.80 × 10–13 | 1.07 | XXbac-BPG299F13 |
| rs2240059 | Intron | C | T | 2.90 × 10–13 | 1.05 | CCHCR1 |
| rs3134782 | Upstream gene variant | G | A | 3.10 × 10–13 | 1.07 | XXbac-BPG299F13 |
| rs3130500 | Intron | T | A | 3.20 × 10–13 | 1.05 | CCHCR1 |
| rs3132535 | Intron | A | G | 3.60 × 10–13 | 1.05 | CCHCR1 |
| rs879882 | Intron | T | C | 4.00 × 10–13 | 1.05 | POU5F1 |
| rs3132520 | Intron | C | T | 4.00 × 10–13 | 1.05 | POU5F1 |
| rs2240064 | Intron | G | A | 4.70 × 10–13 | 1.05 | CCHCR1 |
| rs3130499 | Intron | T | C | 4.80 × 10–13 | 1.05 | CCHCR1 |
| rs3130450 | Intron | C | T | 4.80 × 10–13 | 1.05 | CCHCR1 |
| rs3130451 | Intron | A | G | 4.80 × 10–13 | 1.05 | CCHCR1 |
| rs3130520 | Upstream gene variant | T | C | 4.90 × 10–13 | 1.07 | XXbac-BPG299F13 |
| rs130078 | Synonymous | C | G | 5.30 × 10–13 | 1.05 | CCHCR1 |
| rs3130954 | Downstream variant | G | A | 5.40 × 10–13 | 1.07 | HCG27 |
| rs3130928 | Intron | C | A | 6.50 × 10–13 | 1.05 | POU5F1 |
| rs2022084 | Intron | A | G | 7.20 × 10–13 | 1.05 | CCHCR1 |
| rs3130498 | Intron | T | C | 7.20 × 10–13 | 1.05 | CCHCR1 |
| rs3132528 | 3’ UTR | C | T | 7.20 × 10–13 | 1.05 | TCF19 |
| rs3130929 | Intron | T | C | 7.20 × 10–13 | 1.05 | POU5F1 |
| rs3132523 | Intron | T | C | 7.20 × 10–13 | 1.05 | POU5F1 |
| rs2073723 | Intron | T | C | 8.00 × 10–13 | 1.05 | TCF19 |
| rs9263804 | Intron | C | T | 8.00 × 10–13 | 1.05 | POU5F1 |
| rs3132524 | Intron | T | C | 8.00 × 10–13 | 1.05 | POU5F1 |
| rs1065461 | 3’ UTR | T | C | 8.80 × 10–13 | 1.05 | TCF19 |
| rs3130501 | Intron | A | G | 8.80 × 10–13 | 1.05 | POU5F1 |
| rs3130502 | Intron | A | G | 9.70 × 10–13 | 1.05 | POU5F1 |
| rs3130504 | Intron | A | T | 1.10 × 10–12 | 1.05 | POU5F1 |
| rs3132522 | Intron | T | C | 1.20 × 10–12 | 1.05 | POU5F1 |
| rs3094193 | Intron | G | T | 1.20 × 10–12 | 1.05 | POU5F1 |
| rs3094192 | Intron | C | G | 1.30 × 10–12 | 1.05 | POU5F1 |
| rs3130931 | 5’ UTR | T | C | 1.40 × 10–12 | 1.05 | POU5F1 |
| rs3130456 | Upstream gene variant | C | A | 1.60 × 10–12 | 1.05 | CCHCR1 |
| rs3094189 | Intron | C | A | 1.70 × 10–12 | 1.07 | POU5F1 |
| rs2073721 | Missense | A | G | 1.80 × 10–12 | 1.05 | TCF19 |
| rs3130933 | Intron | T | C | 1.90 × 10–12 | 1.07 | TCF19 |
| rs3132533 | Intron | A | G | 2.00 × 10–12 | 1.05 | CCHCR1 |
| rs130073 | Synonymous | T | C | 2.30 × 10–12 | 1.05 | CCHCR1 |
| rs2073717 | Intron | G | C | 2.30 × 10–12 | 1.05 | CCHCR1 |
| rs3134748 | Regulatory region variant | C | T | 2.30 × 10–12 | 1.06 | |
| rs3130454 | Upstream gene variant | G | A | 2.60 × 10–12 | 1.05 | PSORS1C2 |
| rs3132537 | Intron | A | G | 2.90 × 10–12 | 1.05 | CCHCR1 |
| rs3131013 | Intron | T | C | 2.90 × 10–12 | 1.05 | CCHCR1 |
| rs3094663 | 5’ UTR | T | C | 3.20 × 10–12 | 1.05 | PSORS1C2 |
| rs743401 | Missense | C | T | 3.20 × 10–12 | 1.05 | CCHCR1 |
| rs3130532 | Intergenic variant | A | G | 3.30 × 10–12 | 1.06 | |
| rs9263787 | Intron | T | A | 3.50 × 10–12 | 1.05 | TCF19 |
Abbreviations: CCHCR1, coiled-coil α-helical rod protein 1; ID, identification; kb, kilobases; OR, odds ratio; UTR, untranslated region.
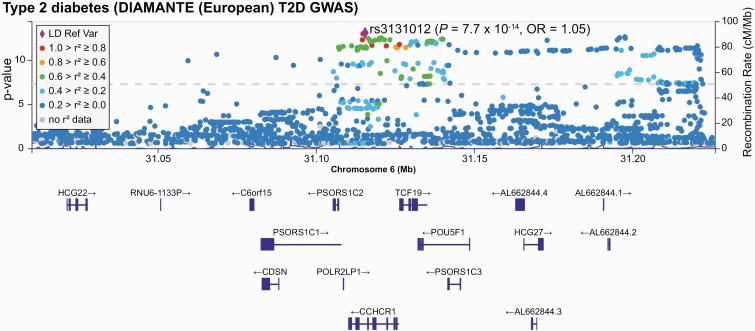
Figure 1.
Regional association plot of the type 2 diabetes association at chromosome 6p21.33 (location of coiled-coil α-helical rod protein 1 [CCHCR1]). Single-nucleotide variations (SNVs, formerly single-nucleotide polymorphisms [SNPs]) are plotted by position on chromosome 6 (x-axis) against association with type 2 diabetes from DIAMANTE (–log10 P value). The strongest signal at SNV rs3131012 is denoted by the purple diamond. Other SNVs are color-coded to reflect their linkage disequilibrium with the top SNV. The location and the direction of transcription for genes in the region are shown below the x-axis.
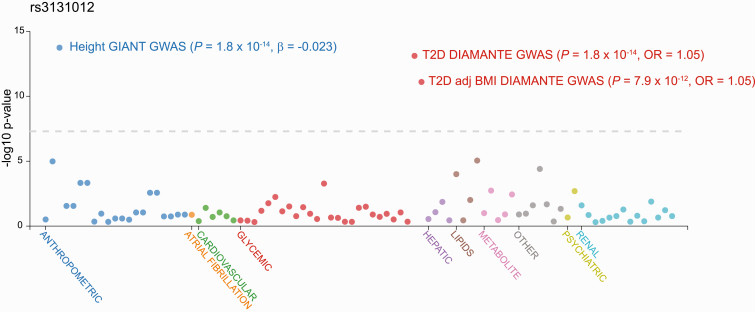
Our T2D-associated SNPs include a noncoding SNP (rs3131012) located in an intronic region of CCHCR1 (P = 7.7 × 10–14) and 2 missense SNPs (rs743401 E74K P = 3.2 × 10–12; rs2073721 M211V P = 3.2 × 10–12) predicted to be tolerated [48, 49]. Rs2073721, which lies in the TCF19 gene, is in modest LD (r2 = 0.3466 in European populations) with rs3131012, the most associated SNP. The CCHCR1 nonsense SNV (rs3130453) identified in our exome analysis, is also in modest LD (r2 = 0.3702) with rs3131012 and exhibits GWA with T2D (P = 1.5 × 10–9). Variants in CCHCR1 are associated with lipids (total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and triglycerides), height, and BMI (Fig. 2).
Figure 2.
Phenotype-wide association study (PheWAS) of rs3131012 in the AMP-T2D Knowledge Portal. We submitted a query for the top single-nucleotide variation (SNV, formerly single-nucleotide polymorphism [SNP]) rs3131012 to the multiple genome-wide association study (GWAS) databases contained in the AMP-T2D Knowledge Portal (www.type2diabetesgenetics.org). Phenotypes are listed along the x-axis including anthropometric, cardiovascular, glycemic, lipid, metabolite, renal, and other, plotted against association (–log10 P value). Multiple phenotypes are associated with this locus as revealed by the height GIANT GWAS and the type 2 diabetes DIAMANTE GWAS.
Given that CCHCR1 has not been previously confirmed as a gene of interest for T2D (though it was recently mentioned in a report by the Million Veteran Program during the course of this study available [47], we sought to further evaluate it in a large sample. We confirmed our finding in the UKBB, where SNVs at the CCHCR1 locus are associated with diabetes diagnosed by a doctor (rs3131012, P = 1.7 × 10–11; rs3130453, P = 3.2 × 10–8) as well as presence of insulin treatment (rs3131012, P = 2.4 × 10–9), malabsorption, lung function, height, thyroid disease, and psoriasis (Table 5). Initial UKBB summary results also show an association with T1D: This signal might be driven by LD between rs3131012 and rs3130453 with T1D-associated HLA genotypes/alleles. We examined the association of SNVs rs3131012 and rs3130453 with both T1D and T2D, with and without conditioning on HLA haplotypes. The association with T2D remained significant after conditioning on HLA, whereas the association with T1D was no longer significant. This result suggests the association of rs3131012 and rs3130453 with T1D was driven by HLA, whereas the association with T2D is distinct from the HLA and T1D risk (Table 6). In addition, analysis of the larger data set of 33 578 T1D cases and controls demonstrated that the T1D-CCHCR1 SNV association was attenuated by correction for HLA type, confirming that the T1D-CCHCR1 association is driven by HLA. Thus, the association of T2D with CCHCR1 is distinct from T1D (Table 7).
Table 5.
UK Biobank phenome-wide association study of phenotypes associated with rs3131012 and rs3130453 from a linear mixed model [33]
| Phenotype | rs3131012 | rs3130453 | ||
|---|---|---|---|---|
| P | β | P | β | |
| Noncancer illness code, self-reported: malabsorption/celiac disease | 1.2 × 10–78 | .003 | 2.6 × 10–99 | .0034 |
| Noncancer illness code, self-reported: psoriasis | 2.2 × 10–34 | .0032 | 1.0 × 10–53 | .004 |
| Diagnoses—main ICD10: K90 intestinal malabsorption | 1.9 × 10–24 | .0011 | 1.0 × 10–32 | .0012 |
| Noncancer illness code, self-reported: hyperthyroidism/thyrotoxicosis | 2.5 × 10–14 | .0016 | 3.7 × 10–11 | .0014 |
| Standing height | 1.7 × 10–13 | –.013 | 0.01 | –.0044 |
| FEV1 | 8.3 × 10–13 | –.015 | 3.4 × 10–10 | –.013 |
| Diabetes diagnosed by doctor | 1.7 × 10–11 | .0035 | 3.2 × 10–8 | .0029 |
| FEV1, best measure | 1.9 × 10–11 | –.015 | 4.7 × 10–10 | –.014 |
| FVC | 1.4 × 10–10 | –.013 | 2.3 × 10–5 | –.0083 |
| Treatment/medication code: insulin product | 2.4 × 10–9 | .0014 | 1.2 × 10–13 | .0018 |
Table 6.
Association of CCHCR1 single-nucleotide variations in UK Biobank with and without human leukocyte antigen adjustment
| SNV | UKBB T1D | UKBB T1D adj HLA type | UKBB T2D | UKBB T2D adj HLA type | ||||
|---|---|---|---|---|---|---|---|---|
| Estimate | P | Estimate | P | Estimate | P | Estimate | P | |
| rs3130453_T | 0.342 | 1.46 × 10–18 | 0.0244 | .563 | 0.0344 | .00331 | 0.0352 | .00431 |
| rs3131012_C | 0.282 | 9.45 × 10–13 | 0.0164 | .699 | 0.0575 | 1.09 × 10–06 | 0.0604 | 8.60 × 10–07 |
Abbreviations: adj, adjusted; CCHCR1, coiled-coil α-helical rod protein 1; HLA, human leukocyte antigen; SNV, single-nucleotide variation; T1D, type 1 diabetes; T2D, type 2 diabetes; UKBB, UK Biobank.
Table 7.
Association of CCHCR1 single-nucleotide variations in type 1 diabetes cohort with and without human leukocyte antigen adjustment
| SNV | T1D cohort | T1D cohort adj HLA type | ||
|---|---|---|---|---|
| Estimate | P | Estimate | P | |
| rs3130453_T | 0.318 | < 2 × 10–16 | 0.005 32 | .7896 |
| rs3131012_C | 0.231 | < 2 × 10–16 | –0.004 99 | .8024 |
Abbreviations: Abbreviations: adj, adjusted; CCHCR1, coiled-coil α-helical rod protein 1; HLA, human leukocyte antigen; SNV, single-nucleotide variation; T1D, type 1 diabetes.
D. Common Variation in CCHCR1 Is Associated With CCHCR1 Expression
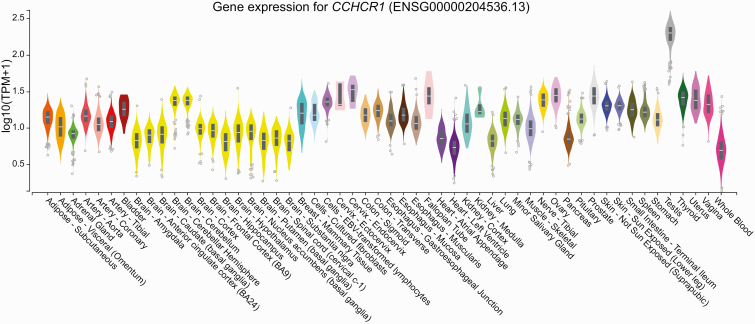
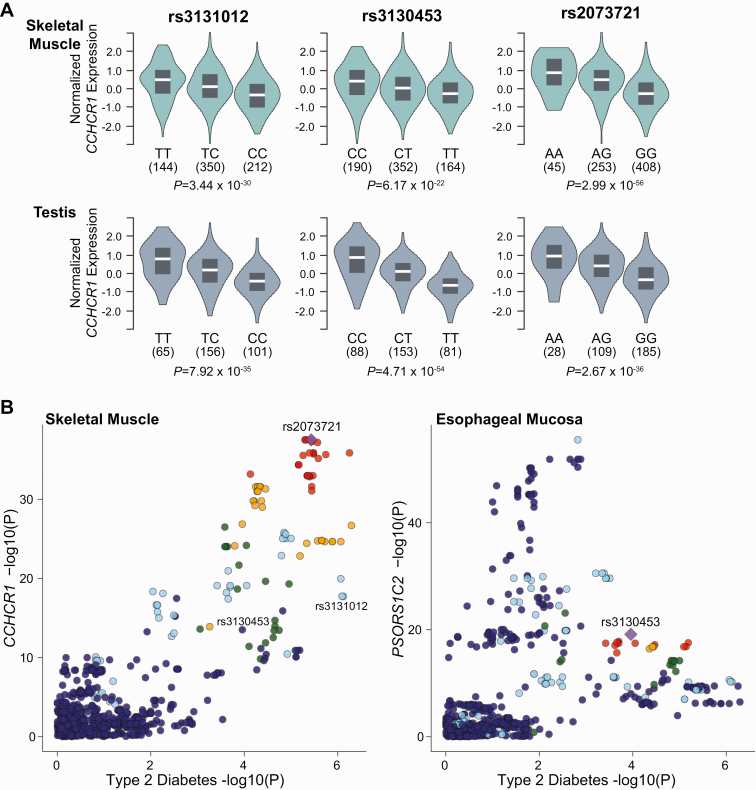
The coding variant associated with T2D through gene burden testing suggests CCHCR1 might be the effector transcript at this locus; however, we sought additional evidence supporting this hypothesis. We checked whether any of the most significant GWAS SNVs found in or near CCHCR1 affect gene expression levels in cis. CCHCR1 is broadly expressed across tissues, with the highest expression levels found in testes (Fig. 3) [43, 50]. We interrogated the GTEx database and found the most significant eQTL for the aforementioned SNVs is CCHCR1. We found that SNVs at CCHCR1 are eQTLs associated with lower CCHCR1 expression in testes (rs3131012, normalized effect size (NES) –0.54, P = 7.9 × 10–35; rs3130453, NES –0.64, P = 4.7 × 10–54, and rs2073721 NES –0.60 P = 2.7 × 10–36) and skeletal muscle (rs3131012, NES –0.39, P = 3.4 × 10–30; rs3130453, NES –0.33, P = 6.2 × 10–22, rs2073721 NES –0.60 P = 3.0 × 10– 56) (Table 8). Rs2073721, which lies in the TCF19 gene, colocalizes its T2D association with the CCHCR1 eQTL in skeletal muscle, whereas rs3130453 colocalizes with testis CCHCR1 expression, suggesting CCHCR1 is the T2D-relevant effector transcript (Fig. 4).
Figure 3.
Tissue-specific expression of coiled-coil α-helical rod protein 1 (CCHCR1) [43, 44].
Table 8.
Expression quantitative trait loci associated with CCHCR1 single-nucleotide variations in diabetes-related tissues
| SNV | Gene symbol | P | NES | Tissue |
|---|---|---|---|---|
| rs2073721 | CCHCR1 | 3.00 × 10–56 | –0.6 | Muscle—skeletal |
| HCG27 | 9.90 × 10–19 | 0.38 | Adipose—subcutaneous | |
| 7.90 × 10–10 | 0.3 | Adipose—visceral (Omentum) | ||
| 1.10 × 10–09 | 0.44 | Liver | ||
| 1.50 × 10–08 | 0.38 | Pancreas | ||
| 6.80 × 10–07 | 0.18 | Muscle—skeletal | ||
| PSORS1C2 | 2.10 × 10–17 | 0.5 | Adipose—subcutaneous | |
| 4.70 × 10–10 | 0.41 | Adipose—visceral (Omentum) | ||
| 1.80 × 10–07 | 0.49 | Liver | ||
| HLA-L | 2.30 × 10–14 | 0.38 | Adipose—subcutaneous | |
| 5.90 × 10–10 | 0.36 | Adipose—visceral (Omentum) | ||
| 2.70 × 10–08 | 0.28 | Muscle—skeletal | ||
| HCG22 | 1.40 × 10–13 | –0.39 | Adipose—subcutaneous | |
| 3.90 × 10–11 | –0.39 | Adipose—visceral (Omentum) | ||
| PSORS1C1 | 1.30 × 10–09 | 0.33 | Adipose—visceral (Omentum) | |
| 8.30 × 10–09 | 0.3 | Adipose—subcutaneous | ||
| 1.30 × 10–08 | 0.31 | Muscle—skeletal | ||
| DDAH2 | 9.70 × 10–07 | –0.13 | Adipose—visceral (Omentum) | |
| rs3131012 | CCHCR1 | 3.40 × 10–30 | –0.39 | Muscle—skeletal |
| 1.60 × 10–09 | 0.2 | Adipose—subcutaneous | ||
| HCG27 | 2.30 × 10–15 | 0.3 | Adipose—subcutaneous | |
| 6.90 × 10–10 | 0.26 | Adipose—visceral (Omentum) | ||
| 8.10 × 10–10 | 0.42 | Liver | ||
| 1.30 × 10–09 | 0.19 | Muscle—skeletal | ||
| XXbac-BPG299F13.17 | 1.20 × 10–07 | 0.16 | Muscle—skeletal | |
| rs3130453 | CCHCR1 | 6.20 × 10–22 | –0.33 | Muscle—skeletal |
| PSORS1C1 | 2.00 × 10–15 | 0.35 | Adipose—subcutaneous | |
| 1.90 × 10–11 | 0.33 | Adipose—visceral (Omentum) | ||
| 7.50 × 10–09 | 0.27 | Muscle—skeletal | ||
| PSORS1C2 | 9.20 × 10–15 | 0.4 | Adipose—subcutaneous | |
| 4.40 × 10–11 | 0.39 | Adipose—visceral (Omentum) | ||
| HLA-B | 8.30 × 10–09 | 0.26 | Pancreas | |
| 2.60 × 10–07 | 0.14 | Adipose—subcutaneous | ||
| 2.90 × 10–07 | 0.13 | Muscle—skeletal | ||
| 9.00 × 10–07 | 0.17 | Adipose—visceral (Omentum) | ||
| HLA-L | 2.20 × 10–09 | 0.26 | Adipose—subcutaneous | |
| 1.70 × 10–08 | 0.24 | Muscle—skeletal | ||
| 2.30 × 10–07 | 0.37 | Pancreas | ||
| 6.50 × 10–07 | 0.26 | Adipose—visceral (Omentum) | ||
| XXbac-BPG181B23.7 | 1.80 × 10–07 | –0.27 | Adipose—subcutaneous | |
| 1.40 × 10–07 | –0.23 | Adipose—visceral (Omentum) |
Abbreviations: CCHCR1, coiled-coil α-helical rod protein 1; HLA, human leukocyte antigen; NES, normalized effect size; SNV, single-nucleotide variation.
Figure 4.
A, The type 2 diabetes risk variants rs3131012, rs3130453, and rs 2073721 act as expression quantitative trait loci (eQTLs) for coiled-coil α-helical rod protein 1 (CCHCR1) gene expression in skeletal muscle, with additive allelic effects at each variant [43, 44]. Additionally, these variants, specifically rs2073721, colocalize in skeletal muscle tissue for CCHCR1 expression. Other genes, including B, HCG27, C, PSORS1C1, and D, PSORS1C2, act as eQTLs. However, they do not colocalize in diabetes-related tissues with their gene expression as illustrated by subcutaneous adipose tissue. Violin plots were created using GTEx v8. Colocalization plots were created using Scott et al (2017) [45] for genome-wide association study analysis and plotted against v7 GTEx expression data.
These SNVs are also eQTLs associated with increased expression of TCF19, HCG27, and PSORS1C1, PSORS1C2, and PSORS1C3 in other tissues. It is possible that these SNVs affect different genes in different directions (increased vs decreased), depending on tissue type. However, these eQTLs do not colocalize with the T2D association, suggesting these eQTLs are not part of the causative pathway in T2D (see Fig. 4). In summary, our data proposes CCHCR1 as a new T2D gene, and suggest decreased CCHCR1 expression based on loss of function and eQTL expression may be the mechanism of effect.
3. Discussion
Although T2D and GIH share pathophysiological manifestations, the extent to which the same molecular pathways contribute to their pathogenesis is not known. We leveraged the deep biological knowledge accrued about the glucocorticoid system and the extensive genomic association data recently generated for T2D and related glycemic traits to ascertain whether known GR genes are enriched for T2D-associated genetic variation. Using this approach, we detected a T2D association with SNVs in the CCHCR1 locus, supported by independent lines of genetic evidence (whole-exome sequence and GWAS) [51]. The additional identification of a cis-eQTL colocalizing with the T2D association signal further implicates CCHCR1 as the likely effector transcript. Through a systematic and comprehensive in silico analysis, we have therefore strengthened the prior probability that CCHCR1 is involved in T2D pathogenesis and open the door to functional and physiological validation experiments. Because glucocorticoids lead to alteration in glucose metabolism and insulin sensitivity, we propose that CCHCR1 provides one example of the potential genetic overlap between the 2 conditions.
Three major CCHCR1 transcripts encode different protein isoforms (consensus coding sequences [CCDS] 43445, 4695, and 47397). The common coding variant identified in our exome analyses (SNV rs3130453) results in an early stop codon (Trp78Ter) in the 2 longest isoforms (CCDS43445 and CCDS47397), but a tryptophan in the shortest protein isoform (CCDS4695). This T2D-associated nonsense/missense SNV (as well as a haplotype known as WWCC, referencing the amino acids in the risk haplotype) has previously been associated with an increased risk of psoriasis in multiple populations [52-54]. The connection between psoriasis and T2D, as well as between psoriasis treatment and hyperglycemia, and whether the various SNVs are acting through the same disease mechanisms, are key areas for future investigation.
Studies exploring the function of CCHCR1 SNPs have primarily been conducted in connection with its association to psoriasis. The CCHCR1 gene encodes a protein with 5 coiled-coil α-helical rod protein domains. The CCHCR1 protein has been linked to multiple, distinct biological processes, such as steroidogenesis, cytoskeleton regulation, and muscle differentiation [52, 55, 56]. CCHCR1 promotes steroidogenesis through its interaction with the mitochondrial steroidogenic acute regulatory protein (StAR), which regulates cholesterol transport to the inner mitochondrial membrane, a rate-limiting step in steroid biosynthesis [57, 58]. CCHCR1 localizes to either the centrosome or P-bodies affecting cytoskeleton-mediated processes, such as cell division, cell adhesion, and messenger RNA transport [52, 55, 59]. While the connection to steroidogenesis may suggest a physiological mechanism underlying our observed association between CCHCR1 SNVs and T2D, the cellular and physiological actions of CCHCR1 need further study to establish how genetic perturbation of CCHCR1 affects T2D risk. In addition, the many pleiotropic associations detected in the phenotype-wide association study of the UKBB suggest a fundamental biological role of this gene in general metabolism, consistent with the known multiple effects of glucocorticoids on several organ systems.
Though the CCHCR1 locus has been previously associated with T2D risk, the causative gene has remained unclear. This locus was first associated with T2D in 2014 through the lead SNV rs3130501, and has been replicated in other T2D studies, including the most extensive recent GWAS in Europeans [60, 61]. However, the causal gene was not identified, and the locus has been annotated based on the adjacent genes POU5F1 and TCF19 [25]. Our work highlights the inherent limitation of annotating associated SNVs with a given gene based on physical proximity alone. One reason for the lack of ascertainment at this locus is its proximity to the MHC, a region of high polymorphic burden with extensive and complex patterns of LD that hinder its analysis and the identification of causal genes. Though definitive confirmation requires functional validation through focused mechanistic studies outside the scope of this genetic exploration, our work provides evidence from exome sequence and expression data that CCHCR1 may be the causative gene at this T2D locus, and suggests variants, tissues, and the directionality of effect relevant to disease.
We recognize that demonstrating a genetic association is only the first step for understanding fully how a specific gene or pathway causes a clinical phenotype. Indeed, to elucidate a mechanism, investigators need to conduct functional experiments that reveal how a given genetic variant affects the regulation of a target gene, and how the effector transcript encoded by that gene alters cellular metabolism and organismal physiology. Unlike other observational association studies (eg, transcriptomics, metabolomics, epigenomics, epidemiology), genetic association studies have the singular advantage that genetic variation universally precedes the appearance of phenotype, as it is present at conception and is not subject to reverse causation. Thus, as long as potential confounders are controlled for, scientists may use genetic association and the unique unidirectional arrow of time to infer causality of the gene-phenotype association. Of course, for causality to be conclusively proven, experimental work must demonstrate that manipulating the gene variant rescues the phenotype. In the present in-depth in silico investigation, we have used rigorous statistical methods to raise the prior probability that a specific gene is causal, but our findings should be considered hypothesis-generating and are reported here for focused follow-up by the scientific community. Further genetic studies including a GWAS for GIH and a systematic test of the hypothesis that T2D and GIH share genetic determinants via LD score regression [62] could further quantify the shared genetics between these 2 phenotypes.
Acknowledgments
We wish to thank the AMP T2D program and the AMP T2D Knowledge Portal for providing the exome sequence data and analytic platform.
Financial Support: This work was supported by Harvard Catalyst, The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic health care centers; the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (Grants 1F32 DK115086–01A1 to L.B., K24 DK110550 to J.C.F., and 1DP3 DK111906 and 1R01 DK122586 to S.S.R.); and the Wagner Fellowship Fund (to C.C.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health.
Glossary
Abbreviations
- BMI
body mass index
- CCDS
consensus coding sequences
- CCHCR1
coiled-coil α-helical rod protein 1
- DI
disposition index
- eQTLs
expression quantitative trait loci
- GIH
glucocorticoid-induced hyperglycemia
- GR
glucocorticoid-related
- GWAS
genome-wide association study
- HbA1c
glycated hemoglobin
- HLA
human leukocyte antigen
- HOMA-β
homeostasis model assessment for β-cell function
- HOMA-IR
homeostasis model assessment for insulin resistance
- LD
linkage disequilibrium
- MHC
major histocompatibility complex
- OMIM
Online Mendelian Inheritance in Man
- SNV
single-nucleotide variation
- TD1
type 1 diabetes
- TD2
type 2 diabetes
- UKBB
UK Biobank
Additional Information
Disclosure Summary: L.N.B., J.M.M., C.C.R., S.S.R., L.C., and S.B.R.J. have nothing to disclose. J.C.F. has received a consulting honorarium from Janssen.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Liu XX, Zhu XM, Miao Q, Ye HY, Zhang ZY, Li YM. Hyperglycemia induced by glucocorticoids in nondiabetic patients: a meta-analysis. Ann Nutr Metab. 2014;65(4):324-332. [DOI] [PubMed] [Google Scholar]
- 2. Perez A, Jansen-Chaparro S, Saigi I, Bernal-Lopez MR, Minambres I, Gomez-Huelgas R. Glucocorticoid-induced hyperglycemia. J Diabetes. 2014;6(1):9-20. [DOI] [PubMed] [Google Scholar]
- 3. McDonough AK, Curtis JR, Saag KG. The epidemiology of glucocorticoid-associated adverse events. Curr Opin Rheumatol. 2008;20(2):131-137. [DOI] [PubMed] [Google Scholar]
- 4. Donihi AC, Raval D, Saul M, Korytkowski MT, DeVita MA. Prevalence and predictors of corticosteroid-related hyperglycemia in hospitalized patients. Endocr Pract. 2006;12(4):358-362. [DOI] [PubMed] [Google Scholar]
- 5. Hwang JL, Weiss RE. Steroid-induced diabetes: a clinical and molecular approach to understanding and treatment. Diabetes Metab Res Rev. 2014;30(2):96-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cagdas DN, Pac FA, Cakal E. Glucocorticoid-induced diabetic ketoacidosis in acute rheumatic fever. J Cardiovasc Pharmacol Ther. 2008;13(4):298-300. [DOI] [PubMed] [Google Scholar]
- 7. Fong AC, Cheung NW. The high incidence of steroid-induced hyperglycaemia in hospital. Diabetes Res Clin Pract. 2013;99(3):277-280. [DOI] [PubMed] [Google Scholar]
- 8. Kim SY, Yoo CG, Lee CT, et al. Incidence and risk factors of steroid-induced diabetes in patients with respiratory disease. J Korean Med Sci. 2011;26(2):264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albrechtsen A, Grarup N, Li Y, et al. D.E.S.I.R. Study Group; DIAGRAM Consortium . Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56(2):298-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Raalte DH, Ouwens DM, Diamant M. Novel insights into glucocorticoid-mediated diabetogenic effects: towards expansion of therapeutic options? Eur J Clin Invest. 2009;39(2):81-93. [DOI] [PubMed] [Google Scholar]
- 11. Weinstein SP, Wilson CM, Pritsker A, Cushman SW. Dexamethasone inhibits insulin-stimulated recruitment of GLUT4 to the cell surface in rat skeletal muscle. Metabolism. 1998;47(1):3-6. [DOI] [PubMed] [Google Scholar]
- 12. Dirlewanger M, Schneiter PH, Paquot N, Jequier E, Rey V, Tappy L. Effects of glucocorticoids on hepatic sensitivity to insulin and glucagon in man. Clin Nutr. 2000;19(1):29-34. [DOI] [PubMed] [Google Scholar]
- 13. Divertie GD, Jensen MD, Miles JM. Stimulation of lipolysis in humans by physiological hypercortisolemia. Diabetes. 1991;40(10):1228-1232. [DOI] [PubMed] [Google Scholar]
- 14. Tamez-Pérez HE, Quintanilla-Flores DL, Rodríguez-Gutiérrez R, González-González JG, Tamez-Peña AL. Steroid hyperglycemia: prevalence, early detection and therapeutic recommendations: a narrative review. World J Diabetes. 2015;6(8):1073-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boxiang Liu MG, Montgomery S. LocusCompare: a tool to visualize pairs of association. 2018.
- 16. Amberger JS, Bocchini CA, Scott AF, Hamosh A. . OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019 Jan 8;47(D1):D1038-D1043. doi: 10.1093/nar/gky1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sharma S, Kho AT, Chhabra D, et al. Glucocorticoid genes and the developmental origins of asthma susceptibility and treatment response. Am J Respir Cell Mol Biol. 2015;52(5):543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carroll TB, Aron DC, Findling JW, Tyrrell BChapter 9. Glucocorticoids and adrenal androgens. In: Gardner DG, Shoback D, eds. Greenspan’s Basic & Clinical Endocrinology. 9th ed. New York, NY: The McGraw-Hill Companies; 2011. [Google Scholar]
- 19. Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365(13):1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heine VM, Rowitch DH. Hedgehog signaling has a protective effect in glucocorticoid-induced mouse neonatal brain injury through an 11betaHSD2-dependent mechanism. J Clin Invest. 2009;119(2):267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reddy TE, Pauli F, Sprouse RO, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19(12):2163-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6(8):e1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scott RA, Lagou V, Welch RP, et al. DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moran A, Pekow P, Grover P, et al. Cystic Fibrosis Related Diabetes Therapy Study Group . Insulin therapy to improve BMI in cystic fibrosis-related diabetes without fasting hyperglycemia: results of the cystic fibrosis related diabetes therapy trial. Diabetes Care. 2009;32(10):1783-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahajan A, Go MJ, Zhang W, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu CT, Merino J, Rybin D, et al. Genome-wide Association Study of Change in Fasting Glucose over time in 13,807 non-diabetic European Ancestry Individuals . Sci Rep. 2019;9(1):9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walford GA, Gustafsson S, Rybin D, et al. Genome-Wide Association Study of the Modified Stumvoll Insulin Sensitivity Index Identifies BCL2 and FAM19A2 as Novel Insulin Sensitivity Loci . Diabetes. 2016;65(10):3200-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saxena R, Hivert MF, Langenberg C, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge . Nat Genet. 2010;42(2):142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance . Nat Genet. 2012;44(6):659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk . Nat Genet. 2010;42(2):105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flannick J, Mercader JM, Fuchsberger C, et al. Broad Genomics Platform; DiscovEHR Collaboration; CHARGE; LuCamp; ProDiGY; GoT2D; ESP; SIGMA-T2D; T2D-GENES; AMP-T2D-GENES . Exome sequencing of 20 791 cases of type 2 diabetes and 24 440 controls. Nature. 2019;570(7759):71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80(1):27-38. [Google Scholar]
- 34. Ma C, Blackwell T, Boehnke M, Scott LJ; GoT2D Investigators . Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37(6):539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elliott LT, Sharp K, Alfaro-Almagro F, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cole JB, Florez JC, Hirschhorn JN. Comprehensive genomic analysis of dietary habits in UK Biobank identifies hundreds of genetic associations. Nat Commun. 2020;11:1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eastwood SV, Mathur R, Atkinson M, et al. Algorithms for the capture and adjudication of prevalent and incident diabetes in UK Biobank. PloS One. 2016;11(9):e0162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen C, Varney MD, Harrison LC, Morahan G. Definition of high-risk type 1 diabetes HLA-DR and HLA-DQ types using only three single nucleotide polymorphisms. Diabetes. 2013;62(6):2135-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo Y, Kanai M, Choi W, et al. A High-Resolution HLA reference panel capturing global population diversity enables multi-ethnic fine-mapping in HIV host response. HIV/AIDS. medRxiv. 2020. doi: 10.1101/2020.07.16.20155606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Onengut-Gumuscu S, Chen WM, Burren O, et al. Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47(4):381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amberger JS, Bocchini CA, Schiettecatte FJM, Scott AF, Hamosh A. . OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Human Genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scott RA, Scott LJ, Mägi R, et al. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66(11):2888-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Flannick JA, Mercader JM, Fuchsberger C, et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vujkovic M, Keaton JM, Lynch JA, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52(7):680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lek M, Karczewski KJ, Minikel EV, et al. Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60 706 humans. Nature. 2016;536(7616):285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qi L, Cornelis MC, Kraft P, et al. Meta-Analysis of Glucose and Insulin-related traits Consortium (MAGIC); Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19(13):2706-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tervaniemi MH, Siitonen HA, Söderhäll C, et al. Centrosomal localization of the psoriasis candidate gene product, CCHCR1, supports a role in cytoskeletal organization. PloS One. 2012;7(11):e49920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Asumalahti K, Veal C, Laitinen T, et al. Psoriasis Consortium . Coding haplotype analysis supports HCR as the putative susceptibility gene for psoriasis at the MHC PSORS1 locus. Hum Mol Genet. 2002;11(5):589-597. [DOI] [PubMed] [Google Scholar]
- 54. Suomela S, Kainu K, Onkamo P, et al. Clinical associations of the risk alleles of HLA-Cw6 and CCHCR1*WWCC in psoriasis. Acta Derm Venereol. 2007;87(2):127-134. [DOI] [PubMed] [Google Scholar]
- 55. Tervaniemi MH, Katayama S, Skoog T, et al. Intracellular signalling pathways and cytoskeletal functions converge on the psoriasis candidate gene CCHCR1 expressed at P-bodies and centrosomes. BMC Genomics. 2018;19(1):432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Corbi N, Bruno T, De Angelis R, et al. RNA polymerase II subunit 3 is retained in the cytoplasm by its interaction with HCR, the psoriasis vulgaris candidate gene product. J Cell Sci. 2005;118(Pt 18):4253-4260. [DOI] [PubMed] [Google Scholar]
- 57. Tiala I, Suomela S, Huuhtanen J, et al. The CCHCR1 (HCR) gene is relevant for skin steroidogenesis and downregulated in cultured psoriatic keratinocytes. J Mol Med (Berl). 2007;85(6):589-601. [DOI] [PubMed] [Google Scholar]
- 58. Sugawara T, Shimizu H, Hoshi N, Nakajima A, Fujimoto S. Steroidogenic acute regulatory protein-binding protein cloned by a yeast two-hybrid system. J Biol Chem. 2003;278(43):42487-42494. [DOI] [PubMed] [Google Scholar]
- 59. Ling YH, Wong CC, Li KW, Chan KM, Boukamp P, Liu WK. CCHCR1 interacts with EDC4, suggesting its localization in P-bodies. Exp Cell Res. 2014;327(1):12-23. [DOI] [PubMed] [Google Scholar]
- 60. Bonàs-Guarch S, Guindo-Martínez M, Miguel-Escalada I, et al. Re-analysis of public genetic data reveals a rare X-chromosomal variant associated with type 2 diabetes. Nat Commun. 2018;9(1):321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mahajan A, Wessel J, Willems SM, et al. ExomeBP Consortium; MAGIC Consortium; GIANT Consortium . Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet. 2018;50(4):559-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bulik-Sullivan B, Finucane HK, Anttila V, et al. ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 . An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”