Abstract
Architectural DNA-binding proteins function to regulate diverse DNA reactions and have the defining property of significantly changing DNA conformation. Although the 1D movement along DNA by other types of DNA-binding proteins has been visualized, the mobility of architectural DNA-binding proteins on DNA remains unknown. Here, we applied single-molecule fluorescence imaging on arrays of extended DNA molecules to probe the binding dynamics of three structurally distinct architectural DNA-binding proteins: Nhp6A, HU, and Fis. Each of these proteins was observed to move along DNA, and the salt concentration independence of the 1D diffusion implies sliding with continuous contact to DNA. Nhp6A and HU exhibit a single sliding mode, whereas Fis exhibits two sliding modes. Based on comparison of the diffusion coefficients and sizes of many DNA binding proteins, the architectural proteins are categorized into a new group distinguished by an unusually high free-energy barrier for 1D diffusion. The higher free-energy barrier for 1D diffusion by architectural proteins can be attributed to the large DNA conformational changes that accompany binding and impede rotation-coupled movement along the DNA grooves.
Keywords: single-molecule fluorescence microscopy, protein–DNA sliding dynamics, bacterial nucleoid protein, HMGB chromatin protein, DNA conformational change
Introduction
Architectural DNA-binding proteins bind and bend DNA in a sequence-dependent or -independent manner, often assisting the function of other DNA-binding proteins in processes such as DNA recombination, transcription, replication, and DNA packaging [1-6]. For many of these proteins, the DNA sequence specificity of binding and DNA structural changes that accompany binding have been studied by a variety of molecular and structural methods [7-12]. Although they can avidly bind DNA in a qualitatively sequence-neutral manner, most structural studies have relied on specific DNA targets, sometimes employing DNA modifications or discontinuities, to obtain stable complexes [13-17]. Less is known about their non-specific binding properties and target search mechanisms. For other types of DNA-binding proteins, facilitated diffusion mechanisms, involving a combination of 3D diffusion and 1D diffusion along DNA, have been proposed to account for the rapid and efficient binding to rare targets [18-23], and indeed, many sequence-specific binding proteins have been directly observed to bind and diffuse along DNA in a non-specific manner [24-36]. Here, we examined the movement of several architectural DNA-binding proteins along DNA using single-molecule fluorescence microscopy.
Blainey et al. [27] proposed that 1D diffusion of DNA-binding proteins along DNA is controlled by their molecular size and free-energy barrier of sliding. When a protein moves from one position to a neighboring position on DNA, the protein “senses” the free-energy barrier. The free-energy barrier for a number of proteins that induce only modest structural changes to DNA upon binding have been determined to be relatively low [25,27,37]. On the other hand, DNA conformational changes stabilized by protein binding may increase the free-energy barrier and thereby slow 1D diffusion, in part by increasing the contacts between the protein and DNA. For example, nucleosomes, in which DNA is wrapped nearly twice around a histone octamers, do not significantly move along DNA in the absence of enhancing factors [38,39]. Molecular dynamics simulations are also consistent with the bending of DNA slowing 1D diffusion by increasing the contacts between the protein and DNA [40]. In addition, DNA containing kinks or flexure points has been shown to stabilize binding of architectural DNA binding proteins [41-45]. Accordingly, we hypothesize that DNA conformational changes captured or induced by protein binding can be an important factor in modulating the dynamic behavior of DNA-bound proteins.
To examine effects of DNA conformational changes on 1D diffusion along DNA, we focused on three abundant architectural DNA-binding proteins with distinct modes of DNA binding: Nhp6A, HU, and Fis (Fig. 1). Nhp6A is a monomer of 93 residues from Saccharomyces cerevisiae that possesses a high-mobility group B domain, which binds to the minor groove face, along with an N-terminal basic tail that wraps around the major groove on the opposite side [46-48]. The L-shape fold of Nhp6A introduces an overall DNA bend of about 70° in an NMR-based structure, severely unwinds the duplex, and widens the minor groove while compressing the major groove [15,49]. Nhp6A is associated predominantly within intergenic nucleosome-free promoter-regulatory regions of yeast chromatin [50]. Escherichia coli HU is a hetero-dimer of 90 residue subunits that bind DNA predominantly via long beta-ribbon arms inserted into the minor groove [9,51]. Like Nhp6A, HU binding induces flexible bends into DNA, which range in crystal structures from 105° to 139°, along with drastic changes in minor and major groove widths [16,45,52,53]. Fis is a homo-dimer of 98 residue subunits from E. coli, which contain a helix-turn-helix motif that inserts into the DNA major groove [17,54-57]. Crystal structures of Fis-DNA complexes reveal DNA bends of 65°–75° together with large changes in minor groove widths over the protein interface [17,57,58]. Greater bending angles have been estimated in solution depending on the DNA sequence flanking the primary interface [57,59-61]. Fis forms stable complexes to specific DNA segments whose shape can readily adapt to the protein interface [17,57,58]. Each of these proteins binds DNA at nanomolar affinities in a relatively sequence-neutral manner [46,53,62-65], enabling single-molecule tracking experiments to be performed.
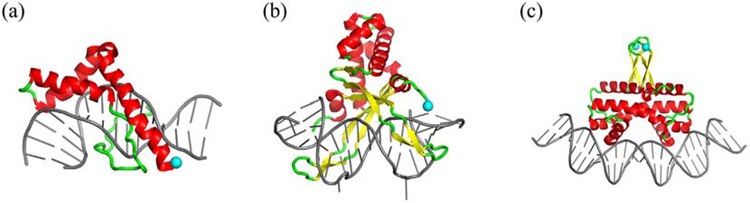
Fig. 1.
Structures of DNA complexes with Nhp6A (a), HU (b), and Fis (c). PDB codes used for Nhp6A, HU, and Fis are 1J5N, 1P71, and 3IV5, respectively. Protein α-helices, β-strands, and loops are red, yellow, and green, respectively. DNAs are gray. Light blue spheres denote fluorescent labeling sites.
In this report, we characterize the 1D diffusion properties for each of these three architectural DNA-binding proteins. The diffusion coefficients and sizes of these proteins reveal that they exhibit a higher free-energy barrier in 1D diffusion as compared with reported values for other DNA binding proteins. Our results suggest that 1D diffusion along DNA is determined not only by molecular size, as proposed previously [27], but also by DNA conformational changes associated with protein binding, and to a lesser degree, the magnitude of the interface surface area. To our knowledge, this is the first report to characterize the movement of architectural DNA-binding proteins along DNA.
Results
Labeling of proteins with fluorescent dye
For single-molecule fluorescence measurements, we labeled the three proteins with the fluorescent dye Atto488 using maleimide chemistry. For the labeling, we used otherwise native proteins containing single cysteines (Fig. 1): Nhp6A-A92C contains the cysteine at the C-terminus, HupB-cys91 contains the cysteine added to the C-terminus of the β chain of the α/β heterodimer, and Fis-Q21C contains the cysteine near the tip of the N-terminal β-hairpin arm. Each of the labeling positions is remote from the DNA binding surface, and fusions of GFP to the C-terminal end of Nhp6A and to the N-terminal end of Fis have been shown to have no detectable effects on DNA binding [66,67]. The labeling efficiencies were 90%, 11%, and 67% for Nhp6A, HU, and Fis subunits, respectively.
Dynamics of Nhp6A along DNA
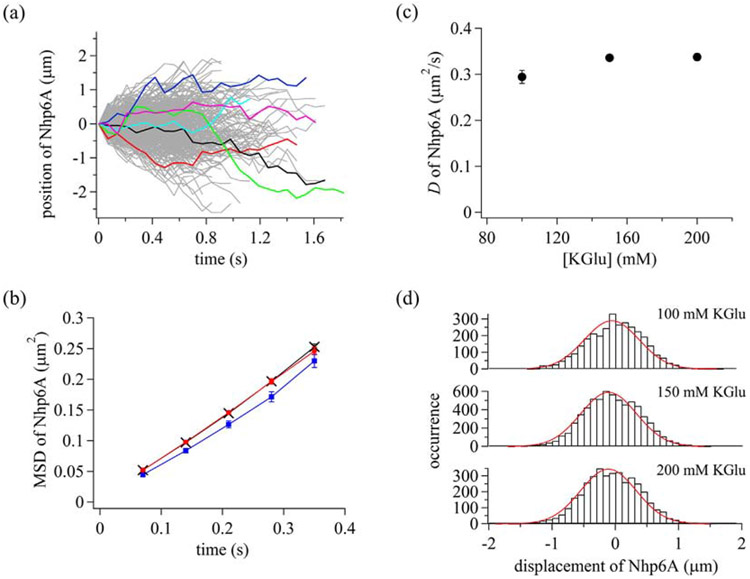
We flowed Atto488-Nhp6A into the cell containing an array of extended λ DNA molecules [68] and conducted single-molecule imaging (Supplementary Movie 1). DNA molecules were tethered at one end and stretched by flow pressure. To selectively illuminate protein molecules bound on DNA, we used highly inclined thin illumination [69]. A total of 1403 trajectories were obtained in the presence of 150 mM potassium glutamate (KGlu) (Fig. 2a). Nhp6A molecules moved toward or against the flow pressure and changed the movement direction at random timing. Mean square displacements (MSD) were linear with time over at least 280 ms, suggesting diffusional motion of Nhp6A along DNA. The average diffusion coefficient, D, obtained by fitting the MSD plot with a primary function having the slope of 2D, was 0.336 ± 0.005 μm2/s.
Fig. 2.
Dynamic properties of Nhp6A bound to DNA. (a) Single-molecule trajectories of Nhp6A in the presence of 150 mM KGlu. Some trajectories were colored to better visualize individual tracks. (b) Time courses of MSD of Nhp6A. Blue squares, black crosses, and red circles are data in the presence of 100, 150, and 200 mM KGlu, respectively. The errors are standard errors. The lines connecting symbols are to enhance visualization. (c) Diffusion coefficients of Nhp6A in different KGlu concentrations. The errors are fitting errors. (d) Displacement distributions of Nhp6A at time intervals of 280 ms. Red curves are the best-fitted curves based on a single Gaussian function.
To further characterize the diffusive behavior, we measured the salt dependence of 1D diffusion of Nhp6A along DNA. Increasing salt concentrations could promote repeated dissociation followed by reassociation with DNA (hopping), thereby increasing diffusion constants [70,71]. Alternatively, salt independence of 1D diffusion is indicative of movement of the protein with continuous contact with DNA (sliding) [25,72]. The MSD plots were linear within the 280-ms frames in 100–200 mM KGlu, suggesting 1D diffusion of Nhp6A in the different KGlu concentrations (Fig. 2b). The independence of D with KGlu concentrations suggests 1D sliding with continuous contact of Nhp6A with DNA (Fig. 2c). The D values in different KGlu concentrations are listed in Supplementary Table 1.
To examine the heterogeneity of 1D sliding dynamics, we next analyzed the displacement distribution. When a protein exhibits a single diffusion mode, the displacement distribution should be a single Gaussian function [37,73,74]. In contrast, the displacement distribution of a protein having multiple diffusion modes should be the sum of Gaussian functions with different diffusion coefficients [37,73,74]. The displacement distributions of Nhp6A in different KGlu concentrations fit well to a single Gaussian function (Eq. (2)), implying a single sliding mode of Nhp6A (Fig. 2d). D values and drift velocities obtained in the fitting are listed in Supplementary Table 1. Overall, our results demonstrate that Nhp6A exhibits a single sliding mode with continuous contact to DNA.
Dynamics of HU along DNA
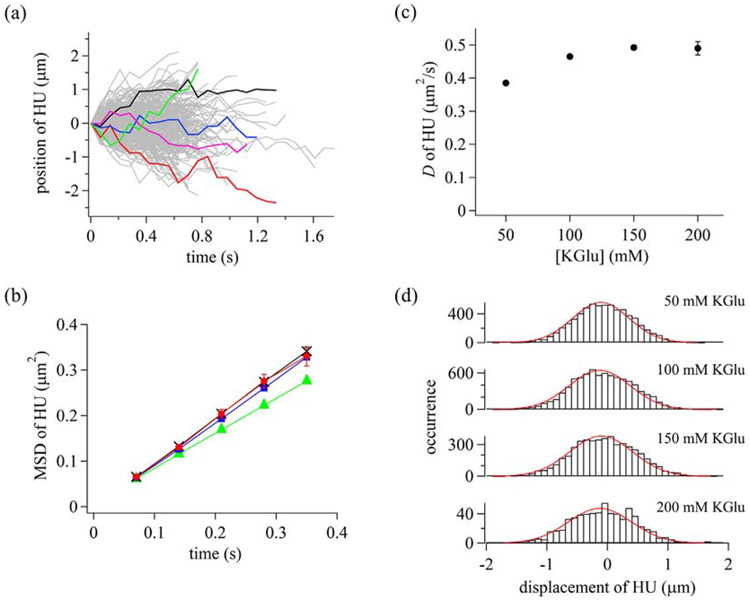
To examine dynamics of HU along DNA, we conducted single-molecule measurements of Atto488-HU in different KGlu concentrations. HU proteins were observed to travel along DNA in both directions with respect to buffer flow (Supplementary Movie 2). Single-molecule trajectories (100–1903) were obtained in 50–200 mM KGlu buffer (Fig. 3a). MSD plots were linear with time over 280 ms at various KGlu concentrations, suggesting diffusional motion of HU along DNA (Fig. 3b). The Dvalue of HU, obtained by fitting the slope of the MSD plot with 2D, was 0.492±0.007 μm2/s in the presence of 150 mM KGlu. The D value did not significantly change under the different KGlu concentrations tested, suggesting 1D sliding of HU with continuous contact to DNA (Fig. 3c and Supplementary Table 3). Furthermore, displacement distributions fit well to a single Gaussian function in different KGlu concentrations, suggesting a single sliding mode of HU (Fig. 3d). Diffusion coefficients and drift velocities are listed in Supplementary Table 2. We conclude that HU exhibits a single sliding mode with continuous contact to DNA, similar to Nhp6A.
Fig. 3.
Dynamic properties of HU bound to DNA. (a) Single-molecule trajectories of HU in the presence of 150 mM KGlu. Some trajectories were colored to better visualize individual tracks. (b) Time courses of MSD of HU. Green triangles, blue squares, black crosses, and red circles are data in the presence of 50, 100, 150, and 200 mM KGlu, respectively. The errors are standard errors. The lines connecting symbols are to enhance visualization. (c) Diffusion coefficients of HU in different KGlu concentrations. The errors are fitting errors. (d) Displacement distributions of HU at time intervals of 280 ms. Red curves are the best-fitted curves based on a single Gaussian function.
Dynamics of Fis along DNA
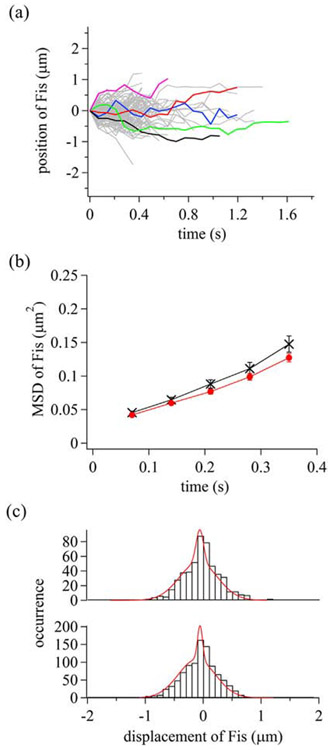
To examine dynamics of Fis binding along DNA, we conducted single-molecule measurements of Atto488-Fis in different KGlu concentrations (Supplementary Movie 3). In 100 mM KGlu, Fis covered the DNA shortly after the introduction of the labeled Fis at ~0.01 nM because of the slow dissociation of Fis from DNA [61,67,75], which prevented single-molecule measurements. In the presence of 150 and 200 mM KGlu, we obtained 118 and 224 trajectories, respectively, of molecules moving in either direction along DNA (Fig. 4a). A range of 88%–90% of the molecules were observed to be immobile, suggesting heterogeneous dynamics of Fis along DNA as described below. Here, we focus on the mobile molecules to understand the dynamics of Fis movement along DNA. MSD plots are linear with time over 350 ms, suggesting 1D diffusion of Fis along DNA (Fig. 4b). Average D values of Fis are 0.16 ± 0.01 and 0.14 ± 0.02 μm2/s in the presence of 150 and 200 mM KGlu, respectively, which are markedly lower than measured with Nhp6A and HU. The independence of the average D values with KGlu concentrations suggests 1D sliding of Fis with continuous contact to DNA.
Fig. 4.
Dynamic properties of Fis along DNA. (a) Single-molecule trajectories of mobile Fis molecules in the presence of 150 mM KGlu. Some trajectories were colored to better visualize individual tracks. (b) Time courses of MSD of Fis. Black crosses and red circles are data in the presence of 150 and 200 mM KGlu, respectively. The errors are standard errors. (c) Displacement distribution of Fis at time intervals of 280 ms. Red curves are the best-fitted curves based on the sum of two Gaussian functions. Top and bottom panels represent the data in the presence of 150 and 200 mM KGlu, respectively.
To examine the heterogeneity of sliding dynamics of Fis molecules displaying at least some mobility over the trajectory, we plotted displacement distributions of the mobile fraction (Fig. 4c). The displacement distributions in 150 and 200 mM KGlu fit well to the sum of two Gaussian functions, implying the presence of two sliding modes. The D values for the fast and slow modes are 0.19 ± 0.02 and 0.007 ± 0.006 μm2/s, respectively, in the presence of 150 mM KGlu. The D value obtained for the slow mode is within the average experimental accuracy of the position of a molecule on DNA (0.110 ± 0.009 μm), implying that Fis in the slow mode became stationary on DNA, or alternatively, a paused Fis became mobile during the evaluated time interval (Supplementary Fig. 1). The slow mode is consistent with the large fraction of immobile Fis. The fractions of Fis molecules exhibiting fast and slow modes were 86 ± 9% and 14 ± 5%, respectively, in the presence of 150 mM KGlu. D values, drift velocities, and fraction for each mode are listed in Supplementary Table 3. The presence of the two modes in the trajectories of the mobile Fis molecules demonstrates that Fis can convert from the fast mode to the slow mode and vice versa (Supplementary Fig. 1). The immobile or paused complexes presumably reflect Fis molecules associated with a DNA sequence that favorably adopts the optimal shape for stable complex formation.
Discussion
We characterized the movement of three architectural DNA-binding proteins along DNA using single-molecule imaging on DNA gardens. Nhp6A, HU, and Fis, which all generate large conformational changes into DNA upon binding, were observed to travel along stretched DNA molecules in either direction with respect to buffer flow. The salt concentration independence of diffusion by each of these proteins is consistent with sliding under continuous contact with DNA. Under physiological salt concentrations (150 mM KGlu), HU (D = 0.492 ± 0.007 μm2/s) moved on DNA at an average speed of 2.92 kb/s, Nhp6A (D = 0.336 ± 0.005 μm2/s) at 2.41 kb/s, and Fis (D = 0.16 ± 0.01 μm2/s) at 1.66 kb/s. Total distances traveled were measured to be up to 20 kb for Nhp6A and HU, and up to 12 kb for Fis before loss of the fluorescent signal by dissociation into solution or bleaching.
All of the Nhp6A and HU molecules exhibited a single sliding mode, whereas only a minority (about 10%) of the Fis proteins were observed moving on DNA, and some of these Fis complexes were observed to stop and start sliding during the course of imaging (Supplementary Fig. 1). We posit that the stationary Fis proteins are positioned at DNA segments that readily conform to the Fis binding interface and thus represent low energy complexes. The stationary Fis molecules were distributed throughout the λ DNA molecule, and their locations were not limited to binding peaks identified from in vivo chromatin immunoprecipitation experiments (Y. Bernatavichute and R.C.J., unpublished data). It has been shown that in order for Fis to form a stable DNA complex, the minor groove at the center of the binding site must narrow to about half its median width, then rapidly expand as the DNA extends outward to almost 50% wider than the median width, and then again become severely compressed at the binding site periphery. In addition, a minimum of about 55° of curvature over the protein interface is required to achieve critical contacts with the DNA backbone [17,57,58]. Although the DNA within complexes of Nhp6A and HU is also highly distorted, we do not detect pauses by these proteins within the temporal resolution of our experiments. We suspect, however, that they also transiently pause at DNA segments that more readily conform to low energy complexes.
The relatively slow diffusion coefficients exhibited by the architectural DNA-binding proteins, together with the random walk trajectories that are independent of the direction of buffer flow, are consistent with rotation-coupled sliding along the grooves of DNA [27,34,76,77]. As elaborated below, a comparison of the diffusion coefficients for these proteins to those reported for other DNA binding proteins believed to track the rotational pitch of DNA reveals that the architectural proteins exhibit an unusually high free-energy barrier for 1D sliding. We provide evidence that the higher free-energy barrier reflects the large DNA conformational changes that would be intrinsic to rotation-coupled sliding by these proteins.
High free-energy barriers for 1D diffusion are intrinsic to architectural DNA-binding proteins
To evaluate the diffusional behavior of architectural DNA-binding proteins, we plotted the diffusion coefficients against the protein radius for many DNA binding proteins (Fig. 5a). The diffusion coefficient for rotation-coupled motion is described by the equation:
| (1) |
where kB, T, η, BP, ROC, R, and ε denote the Boltzmann constant, temperature, solvent viscosity, the distance between two base pairs of DNA, the distance between DNA and a protein, the protein radius, and free-energy barrier, respectively [27,78]. The preexponential factor corresponds to the dependence of the protein size, while the exponential factor relates to the formation and dissociation of protein–DNA interactions. 6πηR and the rest in the denominator of the preexponential factor result from translational and rotational motions, respectively. Considering that the rotational motion should be much slower than the translational motion, we neglected the translational component [27]. Also, we assumed that ROC is the same as R [35]. D should correlate with R−3 [35,77]. For the three architectural proteins, we calculated Rg based on the protein structures within respective DNA complexes (1J5N for Nhp6A, 1P71 for HU, 3IV5 for Fis) and used Rg as R.
Fig. 5.
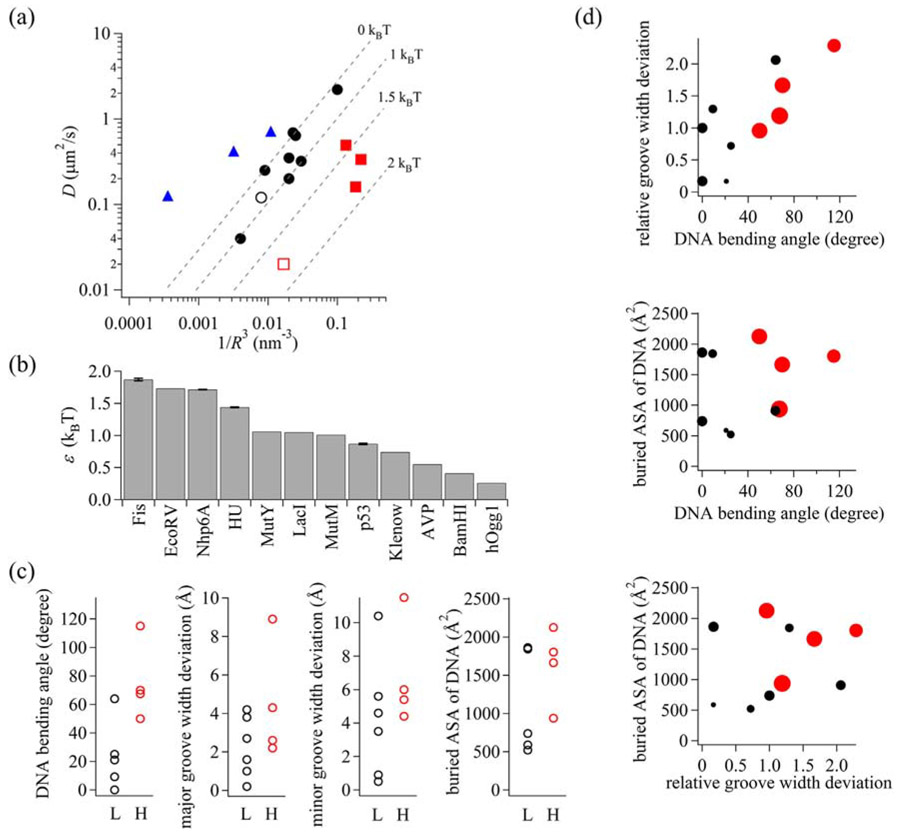
(a) Diffusion coefficient (D) versus radius (R) plot of DNA-binding proteins. Color coding is as follows: DNA-binding proteins with rotation-coupled motion exhibiting low free-energy barriers (black filled circles), architectural proteins (red filled squares), EcoRV (red open square), p53 (black open circle), and TALE proteins (blue triangles) with rotation-uncoupled motion. The dashed lines from top to bottom represent 0, 1, 1.5, 2 kBT, respectively, for the free-energy barrier that a protein feels moving along the helical pitch of DNA. (b) Free-energy barriers for proteins moving along the helical pitch of DNA. (c) Global DNA bending angles, maximum DNA groove-width deviations, and buried DNA accessible surface areas of protein–DNA complexes with high (H; > 1.1 kBT) and low (L; < 1.1 kBT) free-energy barriers. (d) Two-dimensional plots of DNA-binding proteins. Red and black circles represent DNA-binding proteins with high and low free-energy barriers, respectively. Size of the circle is proportional to the free-energy barrier.
The proteins in the D versus R plots are categorized into three groups: those possessing rotation-uncoupled motion and those possessing rotation-coupled motion with either low or high free-energy barriers. Most of the proteins traverse rotationally along DNA with a low free-energy barrier, as demonstrated by Xie and co-workers [27] (Fig. 5a, filled circles; Supplementary Table S4). Movement along DNA by these proteins is retarded by a free-energy barrier between 0 and 1.06 kBT (Fig. 5b and Supplementary Table S4). The free-energy barriers were estimated here to be slightly smaller than 1.1 ± 0.2 kBT obtained in the previous study [27] because of the assumption that ROC equals R. We include the free-energy barrier for p53 (0.87 ± 0.01 kBT), as determined from averaging two sliding modes [73]; p53 also falls within the low free-energy barrier group (Fig. 5a, open circle). In contrast, TALE proteins, analyzed by Schroeder and co-workers [35], are located to the left of the dashed line of the rotation-coupled motion with no free-energy barrier (Fig. 5a, triangles). TALE proteins belong to a class exhibiting primarily rotation-uncoupled motion.
The three architectural DNA-binding proteins analyzed in this study are positioned to the right of the dashed line representing the 1 kBT free-energy barrier (Fig. 5a, filled squares). The free-energy barriers of HU, Nhp6A, and Fis are 1.438 ± 0.005 kBT, 1.714 ± 0.004 kBT, and 1.87 ± 0.02 kBT, respectively (Fig. 5b and Supplementary Table S4). For Fis, we used the average diffusion coefficient; the free-energy barrier of the fast and slow modes of Fis is 1.8 ± 0.1 kBT and 2.6 ± 1.1 kBT, respectively. The slower diffusion observed by these three proteins fits with rotation-coupled diffusion, as opposed to the rotation-uncoupled diffusion identified for TALE proteins that traverse DNA extremely rapidly. The restriction enzyme EcoRV, which also exhibits rotation-coupled motion [77], is the only other protein in this set that displays a high free-energy barrier (1.73 kBT; Fig. 5a and b); by contrast, BamH1 is included within the low free-energy group. We conclude that the architectural DNA-binding proteins fall into a distinct group of rotation-coupled DNA binding proteins with a high free-energy barrier.
Mechanism for the high free-energy barrier observed for architectural DNA-binding proteins
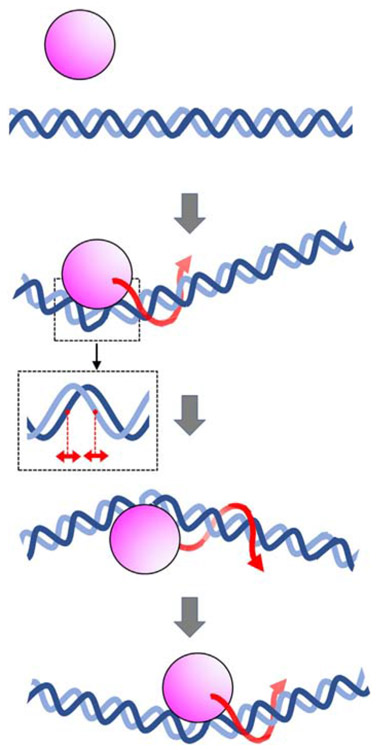
We considered whether the high free-energy barrier for sliding by the architectural DNA-binding proteins originates from the large DNA conformational changes that accompany binding. These include alterations in base pair twist, sometimes by intercalation of amino acid residues into the base stack, variations in major and minor groove widths, and bending of the DNA helical axis. These structural changes in DNA conformation are required for protein binding and would likely impede rotation-coupled sliding along DNA. To assess this, we compared global DNA bending angles and major and minor groove-width deviations within complexes of DNA binding proteins exhibiting high and low free-energy barriers (Fig. 5c). Data for other proteins were collated from examples where sliding kinetics and high-resolution structures, preferably of nonspecific DNA complexes, are available (Supplementary Table 4). In general, DNA-binding proteins with high free-energy barriers exhibit greater global DNA bending angles (P = 0.05 in a two-sample Kolmogorov–Smirnov test). There is a weaker correlation between the free-energy barrier and major and minor groove-width deviations. An additional potential determinant controlling the free-energy barrier may be the size of the protein–DNA interface. To evaluate this parameter, we extracted solvent-accessible DNA surface areas that are excluded within protein–DNA complexes that exhibit low and high free-energy barriers. As shown in Fig. 5c, there is a weak correlation between DNA surface areas contacted by the proteins and the free-energy barrier for diffusion. However, when combinations of the three parameters are considered (Fig. 5d), DNA-binding proteins with high free-energy barriers are distributed into distinct groups exhibiting higher values of each parameter tested (P = 0.05 in two-sample two-dimensional Kolmogorov–Smirnov tests for each comparison). This suggests that both groove-width deviations and the magnitude of the protein–DNA interface areas contribute to the high free-energy barrier. We note that the DNA parameters used in this analysis were extracted from crystal- or NMR-derived protein–DNA structures, which capture low-energy, stably bound, states that may not precisely mimic the sliding mode. In summary, we conclude that DNA bending has the greatest impact on rotation-coupled sliding along DNA, but DNA groove-width deviations and protein–DNA interface areas also contribute but to a lesser extent (Fig. 6).
Fig. 6.
Schematic diagram of an architectural DNA-binding protein traveling along DNA. Pink and blue represent the DNA-binding protein and DNA, respectively. The binding of architectural proteins to DNA is accompanied by large conformational changes in the DNA structure, including changes in helical twist, major and minor groove widths (shown in inset), and axis curvature. Red arrows represent rotation-coupled sliding along DNA. The changes in DNA structure coupled to binding result in a high free-energy barrier, thereby retarding 1D diffusion along DNA.
Free-energy costs of rotational sliding and the biological properties of architectural DNA-binding proteins
Architectural DNA-binding proteins, which include prokaryotic “nucleoid-associated” and eukaryotic high-mobility group B chromatin proteins, are present at high levels in vivo and bind prolifically throughout chromosomes [50,79-85]. Indeed, HU and Fis are the most abundant DNA binding proteins in rapidly growing E. coli [85,86], and Nhp6A, together with its paralog Nhp6B, is the most abundant non-histone DNA binding proteins in S. cerevisiae [79]. These proteins participate in many different reactions on DNA and, in some cases, contribute to chromosome packaging [1-3,5,6]. Their diverse biological functions differ from sequence- or reaction-specific DNA binding proteins like transcription or replication factors that are present in much lower abundance and are often targeted to rare sites on chromosomes. Sequence-specific DNA binding proteins face the challenge of rapidly finding their rare binding sites embedded within an overwhelming amount of structurally similar sequence information by processes involving low energy 1D searches along relatively short DNA stretches combined with hopping to distant DNA segments [18,19]. Architectural proteins do not have to search over huge sequence space to find biologically relevant binding sites, whose features are often deformations in DNA structure. For example, HU functions, in part, to stabilize DNA loops involved in regulating transcription and recombination [1,9,51], and Fis molecules are stably bound every few kbs along the bacterial chromosome, often within intergenic regulatory regions [81,82]. HU and Fis also contribute to general compaction of chromosomes by introducing DNA bends at frequent binding sites [53,62-64,87]. The high free-energy cost of rotationally diffusing along DNA is thus compatible with their promiscuous biological functions that require reading DNA conformation over relatively short distances.
Materials and Methods
Preparation of fluorescentlylabeled proteins
Proteins without tags but containing single cysteines were expressed from pET11a-derived plasmids. Nhp6A-A92C, containing a cysteine substituted for the C-terminal alanine residue (Fig. 1a), was purified as described by Yen et al. [47]. Fis-Q21C, containing a cysteine near the tip of the N-terminβ-hairpin arm motif of Fis (Fig. 1c), was purified essentially as described by Stella et al. [17]. HupB (HU β subunit), containing a cysteine added to the C-terminus, and HupA (HU α subunit) were co-expressed from pRJ2601 (constructed by Yana Bernatavichute and R.C.J.). HU was purified from clarified extracts by first removal of nucleic acids by precipitation with polyethyl-enimine followed by ion-exchange (SP-Sepharose) and size-exclusion (Superdex 75, GE Healthcare) chromatography using an FPLC.
Proteins were labeled by Atto488 (ATTO-TEC) using maleimido chemistry. Briefly, proteins were reduced by overnight incubation with 20 mM DTT at 4°C and then exchanged into a solution containing 20 mM Hepes (pH7.2), 1 M NaCl, and 1 mM TCEP by batch chromatography on phosphocellulose (Whatman P11). The reduced proteins were incubated with > 10-fold excess of Atto488-maleimide for 2 h at ~20°C or overnight at 4°C. Unreacted label was removed by phosphocellulose chromatography. Labeling efficiencies were determined from the 500-nm fluorescence absorbance and Bradford protein assays.
Preparation of DNA arrays and single-molecule measurements
The construction of DNA gardens, containing arrays of extended DNA molecules in flow cells for high-throughput analysis, is described in detail by Igarashi et al. [68]. Phage λ DNA (New England Biolabs) annealed with 5′-AGGTCGCCGCCC-bio-tin-3′ (Sigma-Aldrich) for tethering onto the surface of the flow cell was used for this work.
Most of the data measuring 1D movements of proteins along the DNA arrays were collected using an inverted fluorescence microscope (IX-73; Olympus) coupled with a total internal reflection fluorescence (TIRF) unit (IX3-RFAEVAW; Olympus). A488-nm laser (Sapphire LP488-50CW CDRH; Coherent) was illuminated through the TIRF unit and dichroic mirror (FF495-Di03; Semrock) into an objective lens with N.A. = 1.49 (UAPON 100×OTIRF; Olympus) using a highly inclined thin illumination geometry [69]. The angle of the laser in the flow cell was adjusted to selectively illuminate the fluorescent proteins bound on DNA apart from the surface. The laser power was 5mW and the observation area was ~1500 μm2. Fluorescence collected by the objective lens was passed through a band-pass filter (FF01-520/35; Semrock), and was detected by an EM-CCD camera (iXon Ultra 888; Andor). Fluorescent proteins in a solution containing 20 mM Hepes, 1 mM EDTA, 1 mM DTT, 2 mM trolox, 0.1 mg/mL BSA, and various concentrations of KGlu at pH 7.2 were introduced into the flow cell containing the DNA arrays using a syringe pump (Chemyx). The concentrations of Nhp6A, HU, and Fis were 0.1, 0.25, and 0.04 nM, respectively. The protein concentrations used here are kept low to avoid compaction and looping of the DNA by these proteins [62,64,87], and to avoid collisions of protein molecules on DNA or from solution [67,88]. In a control experiment, we added 2 nM unlabeled HU to the labeled HU preparation at 0.25 nM and observed no significant effect on diffusion by the labeled HU molecules. Binned images (2 × 2) with 33-ms integration time were taken at 70-ms intervals at a flow rate of 600 μL/min at 22°C.
The fluorescent spots of single-protein molecules were tracked from a time series of images using ImageJ software with the plugin “Particle track and analysis” developed by Yoshiyuki Arai (Osaka University, Japan). To remove non-specific adsorption of the fluorescent molecules on the surface of the flow cell, we selected trajectories satisfying MSD larger than 1 binned pixel (260 nm×260 nm) at 350-ms time intervals. Also, we selected trajectories that included at least 6 points. The trajectory selection was conducted by an Excel macro. The trajectory selection excluded most of immobile molecules on DNA for Fis. The experimental accuracy of the position of a Fis molecule on DNA was 110 ± 9 nm, determined from the intersect of the MSD plot. The signal-to-noise ratio of a Fis molecule was 15. Most trajectories obtained for the three proteins move at longer distance than the maximum DNA fluctuation (370 ± 40 nm, n = 3) estimated by measuring the free end position of DNA stained by labeled Nhp6A at 0.4 nM along the stretched axis (Supplementary Movie 4). Displacements were calculated from all pairs of positions of a molecule at time intervals of 280 ms in all trajectories. For the fitting of distribution analysis, we used the following equation:
| (2) |
where δt, δx, P(δx), Ai, vi, and Di represent time interval, displacement in the time interval, the occurrence of δx, amplitude of the ith mode, drift velocity of the ith mode, and diffusion coefficient of the ith mode, respectively. For Nhp6A and HU, we used the above equation with A2 = 0, because of their single mode of sliding.
Parameters of DNA conformational changes in protein–DNA complexes
For global DNA bending angles, we used values reported in the references for the high-resolution protein–DNA structures of Nhp6A [49], HU[16], Fis [17], EcoRV [89], LacI [90], MutY [91,92], and hOgg1 [93]. For the other proteins, we obtained global DNA bending angles using the 3D-DART server [94]. For groove-width deviations, we calculated major and minor groove widths for each phosphate pair in the protein–DNA complex structures using w3DNA [95] and obtained the maximum width deviations for the major and minor grooves. We also calculated relative groove-width deviations by summing the maximum width deviations of the major and minor grooves divided by the respective widths in canonical B-DNA (11.6 and 6 Å for major and minor grooves [96]). Buried DNA accessible surface areas in the protein–DNA structures were calculated using the PyMol plugin PDIviz [97]. Structural data from nonspecific DNA complexes were used if available; PDB codes are listed in Supplementary Table S4. For Nhp6A, HU, Fis, and EcoRV, Rg was calculated from PDB files using the following equation:
| (3) |
where mi, ri, and rc represent the mass of ith atom, the coordinate of ith atom, and the coordinate of the center of mass, respectively.
Supplementary Material
Acknowledgment
K.K., E.M., and R.C.J. thank Dr. Sridhar Mandali, Dr. Wenyang Chen, and Dr. Stephen P. Hancock (UCLA) for helpful support; Prof. Margot E. Quinlan (UCLA) for use of her fluorescence microscope; and Prof. Charles M. Schroeder (University of Illinois) for providing data sets of diffusion coefficients and radii for many proteins.
Funding Sources: This work was supported by MEXT/JSPS KAKENHI 16K07313 and 16KK0157 (to K.K.) and by NIGMS grant GM038509 (to R.C.J.).
Abbreviations used:
- TIRF
total internal reflection fluorescence
- KGlu
potassium glutamate
- MSD
mean square displacements
- FRET
fluorescence resonance energy transfer
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2018.01.001.
Supporting Information: Representative single-molecule trajectories of Fis transiting between two sliding modes; tables of experimental parameters determined for architectural binding proteins; table of free-energy barriers and structural parameters of protein–DNA complexes; single-molecule fluorescence imaging movies of architectural DNA-binding proteins binding to DNA; fluorescence imaging movies of DNA molecules.
References
- [1].Johnson RC, Johnson LM, Schmidt JW, Gardner JF, Major nucleoid proteins in the structure and function of the Escherichia coli chromosome, in: Higgins NP (Ed.), The Bacterial Chromosome, ASM Press, Washington, D.C: 2005, pp. 65–132. [Google Scholar]
- [2].Bianchi ME, Agresti A, HMG proteins: dynamic players in gene regulation and differentiation, Curr. Opin. Genet. Dev 15 (2005) 496–506. [DOI] [PubMed] [Google Scholar]
- [3].Dillon SC, Dorman CJ, Bacterial nucleoid-associated proteins, nucleoid structure and gene expression, Nat. Rev. Microbiol 8 (2010) 185–195. [DOI] [PubMed] [Google Scholar]
- [4].Stros M, HMGB proteins: interactions with DNA and chromatin, Biochim. Biophys. Acta 1799 (2010) 101–113. [DOI] [PubMed] [Google Scholar]
- [5].Browning DF, Grainger DC, Busby SJ, Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression, Curr. Opin. Microbiol 13 (2010) 773–780. [DOI] [PubMed] [Google Scholar]
- [6].Reeves R, Nuclear functions of the HMG proteins, Biochim. Biophys. Acta 1799 (2010) 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Murphy FV, Churchill ME, Nonsequence-specificDNA recognition: a structural perspective, Structure 8 (2000) R83–89. [DOI] [PubMed] [Google Scholar]
- [8].Thomas JO, Travers AA, HMG1 and 2, and related ‘architectural’ DNA-binding proteins, Trends Biochem. Sci 26 (2001) 167–174. [DOI] [PubMed] [Google Scholar]
- [9].Swinger KK, Rice PA, IHF and HU: flexible architects of bent DNA, Curr. Opin. Struct. Biol 14 (2004) 28–35. [DOI] [PubMed] [Google Scholar]
- [10].Luijsterburg MS, Noom MC, Wuite GJ, Dame RT, The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective, J. Struct. Biol 156 (2006) 262–272. [DOI] [PubMed] [Google Scholar]
- [11].Dorman CJ, Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation, J. Mol. Microbiol. Biotechnol 24 (2014) 316–331. [DOI] [PubMed] [Google Scholar]
- [12].Song D, Loparo JJ, Building bridges within the bacterial chromosome, Trends Genet. 31 (2015) 164–173. [DOI] [PubMed] [Google Scholar]
- [13].Rice PA, Yang S, Mizuuchi K, Nash HA, Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn, Cell 87 (1996) 1295–1306. [DOI] [PubMed] [Google Scholar]
- [14].Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ, Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins, Nature 399 (1999) 708–712. [DOI] [PubMed] [Google Scholar]
- [15].Allain FH, Yen YM, Masse JE, Schultze P, Dieckmann T, Johnson RC, et al. , Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding, EMBO J. 18 (1999) 2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Swinger KK, Lemberg KM, Zhang Y, Rice PA, Flexible DNA bending in HU-DNA cocrystal structures, EMBO J. 22 (2003) 3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stella S, Cascio D, Johnson RC, The shape of the DNA minor groove directs binding by the DNA-bending protein Fis, Genes Dev. 24 (2010) 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Von Hippel PH, Berg OG, Facilitated target location in biological systems, J. Biol. Chem 264 (1989) 675–678. [PubMed] [Google Scholar]
- [19].Halford SE, Marko JF, How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32 (2004) 3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Florescu AM, Joyeux M, Description of nonspecific DNA–protein interaction and facilitated diffusion with a dynamical model, J. Chem. Phys 130 (2009), 015103. [DOI] [PubMed] [Google Scholar]
- [21].Lomholt MA, van den Broek B, Kalisch S-MJ, Wuite GJL, Metzler R, Facilitated diffusion with DNA coiling, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 8204–8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J, The Lac repressor displays facilitated diffusion in living cells, Science 336 (2012) 1595–1598. [DOI] [PubMed] [Google Scholar]
- [23].Kamagata K, Murata A, Itoh Y, Takahashi S, Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging, J.Photo-chem. Photobiol. C Photochem. Rev 30 (2017) 36–50. [Google Scholar]
- [24].Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, et al. , Visualization of single molecules of RNA-polymerase sliding along DNA, Science 262 (1993) 1561–1563. [DOI] [PubMed] [Google Scholar]
- [25].Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM, Tumor suppressor p53 slides on DNA with low friction and high stability, Biophys. J 95 (2008) L01–03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, et al. , Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6, Mol. Cell 28 (2007) 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, et al. , Nonspecifically bound proteins spin while diffusing along DNA, Nat. Struct. Mol. Biol 16 (2009) 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC, Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice, Nat. Struct. Mol. Biol 17 (2010) 932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, et al. , Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair, Proc. Natl. Acad. Sci. U. S. A 109 (2012) E3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC, Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee JB, Cho WK, Park J, Jeon Y, Kim D, Lee SH, et al. , Single-molecule views of MutS on mismatched DNA, DNA Repair 20 (2014) 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nelson SR, Dunn AR, Kathe SD, Warshaw DM, Wallace SS, Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases, Proc. Natl. Acad. Sci. U. S. A 111 (2014) E2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mangel WF, McGrath WJ, Xiong K, Graziano V, Blainey PC, Molecular sled is an eleven-amino acid vehicle facilitating biochemical interactions via sliding components along DNA, Nat. Commun 7 (2016) 10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jones ND, Lopez MA Jr., Hanne J, Peake MB, Lee JB, Fishel R, et al. , Retroviral intasomes search for a target DNA by 1D diffusion which rarely results in integration, Nat. Commun 7 (2016) 11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cuculis L, Abil Z, Zhao H, Schroeder CM, TALE proteins search DNA using a rotationally decoupled mechanism, Nat. Chem. Biol 12 (2016) 831–837. [DOI] [PubMed] [Google Scholar]
- [36].Itoh Y, Murata A, Sakamoto S, Nanatani K, Wada T, Takahashi S, et al. , Activation of p53 facilitates the target search in DNA by enhancing the target recognition probability, J. Mol. Biol 428 (2016) 2916–2930. [DOI] [PubMed] [Google Scholar]
- [37].Murata A, Itoh Y, Mano E, Kanbayashi S, Igarashi C, Takahashi H, et al. , One-dimensional search dynamics of tumor suppressor p53 regulated by a disordered C-terminal domain, Biophys. J 112 (2017) 2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ranjith P, Yan J, Marko JF, Nucleosome hopping and sliding kinetics determined from dynamics of single chromatin fibers in Xenopus egg extracts, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 13649–13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Visnapuu ML, Greene EC, Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition, Nat. Struct. Mol. Biol 16 (2009) 1056–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tan C, Terakawa T, Takada S, Dynamic coupling among protein binding, sliding, and DNA bending revealed by molecular dynamics, J. Am. Chem. Soc 138 (2016) 1520–5126. [DOI] [PubMed] [Google Scholar]
- [41].Pil PM, Lippard SJ, Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin, Science 256 (1992) 234–237. [DOI] [PubMed] [Google Scholar]
- [42].Pontiggia A, Negri A, Beltrame M, Bianchi ME, Protein HU binds specifically to kinked DNA, Mol. Microbiol 7 (1993) 343–350. [DOI] [PubMed] [Google Scholar]
- [43].Pinson V, Takahashi M, Rouviere-Yaniv J, Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA, J. Mol. Biol 287 (1999) 485–497. [DOI] [PubMed] [Google Scholar]
- [44].Wong B, Masse JE, Yen YM, Giannikopoulos P, Feigon J, Johnson RC, Binding to cisplatin-modified DNA by the Saccharomyces cerevisiaeHMGB protein Nhp6A, Biochemistry 41 (2002)5404–5414. [DOI] [PubMed] [Google Scholar]
- [45].Swinger KK, Rice PA, Structure-based analysis of HU-DNA binding, J. Mol. Biol 365 (2007) 1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Paull TT, Johnson RC, DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation, J. Biol. Chem 270 (1995) 8744–8754. [DOI] [PubMed] [Google Scholar]
- [47].Yen YM, Wong B, Johnson RC, Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine, J. Biol. Chem 273 (1998) 4424–4435. [DOI] [PubMed] [Google Scholar]
- [48].Stillman DJ, Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae, Biochim. Biophys. Acta 1799 (2010) 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Masse JE, Wong B, Yen YM, Allain FH, Johnson RC, Feigon J, The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding, J. Mol. Biol 323 (2002) 263–284. [DOI] [PubMed] [Google Scholar]
- [50].Dowell NL, Sperling AS, Mason MJ, Johnson RC, Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription, Genes Dev. 24 (2010) 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Macvanin M, Adhya S, Architectural organization in E. coli nucleoid, Biochim. Biophys. Acta 1819 (2012) 830–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kamashev D, Balandina A, Rouviere-Yaniv J, The binding motif recognized by HU on both nicked and cruciform DNA, EMBO J. 18 (1999) 5434–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sagi D, Friedman N, Vorgias C, Oppenheim AB, Stavans J, Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes, J. Mol. Biol 341 (2004) 419–428. [DOI] [PubMed] [Google Scholar]
- [54].Yuan HS, Finkel SE, Feng JA, Kaczor-Grzeskowiak M, Johnson RC, Dickerson RE, The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding, Proc. Natl. Acad. Sci. U. S. A 88 (1991) 9558–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kostrewa D, Granzin J, Koch C, Choe HW, Raghunathan S, Wolf W, et al. , Three-dimensional structure of the E. coli DNA-binding protein FIS, Nature 349 (1991) 178–180. [DOI] [PubMed] [Google Scholar]
- [56].Finkel SE, Johnson RC, The Fis protein: it's not just for DNA inversion anymore, Mol. Microbiol 6 (1992) 3257–3265. [DOI] [PubMed] [Google Scholar]
- [57].Hancock SP, Stella S, Cascio D, Johnson RC, DNA sequence determinants controlling affinity, stability and shape of DNA complexes bound by the nucleoid protein Fis, PLoS One 11 (2016), e0150189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hancock SP, Ghane T, Cascio D, Rohs R, Di Felice R, Johnson RC, Control of DNA minor groove width and Fis protein binding by the purine 2-amino group, Nucleic Acids Res. 41 (2013) 6750–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Thompson JF, Landy A, Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes, Nucleic Acids Res. 16 (1988) 9687–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Perkins-Balding D, Dias DP, Glasgow AC, Location, degree, and direction of DNA bending associated with the Hin recombinational enhancer sequence and Fis-enhancer complex, J. Bacteriol 179 (1997) 4747–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pan CQ, Finkel SE, Cramton SE, Feng JA, Sigman DS, Johnson RC, Variable structures of Fis-DNA complexes determined by flanking DNA–protein contacts, J. Mol. Biol 264 (1996) 675–695. [DOI] [PubMed] [Google Scholar]
- [62].Skoko D, Wong B, Johnson RC, Marko JF, Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA–protein complexes, Biochemistry 43 (2004) 13867–13874. [DOI] [PubMed] [Google Scholar]
- [63].van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT, Dual architectural roles of HU: formation of flexible hinges and rigid filaments, Proc. Natl. Acad. Sci. U. S. A 101 (2004) 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Skoko D, Yoo D, Bai H, Schnurr B, Yan J, McLeod SM, et al. , Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis, J. Mol. Biol 364 (2006) 777–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Czapla L, Peters JP, Rueter EM, Olson WK, Maher III LJ, Understanding apparent DNA flexibility enhancement by HU and HMGB architectural proteins, J. Mol. Biol 409 (2011) 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yen YM, Roberts PM, Johnson RC, Nuclear localization of the Saccharomyces cerevisiae HMG protein NHP6A occurs by a Ran-independent nonclassical pathway, Traffic (Copenhagen, Denmark) 2 (2001) 449–464. [DOI] [PubMed] [Google Scholar]
- [67].Graham JS, Johnson RC, Marko JF, Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA, Nucleic Acids Res. 39 (2011) 2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Igarashi C, Murata A, Itoh Y, Subekti DRG, Takahashi S, Kamagata K, DNA garden: a simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins, Bull. Chem. Soc. Jpn 90 (2017) 34–43. [Google Scholar]
- [69].Tokunaga M, Imamoto N, Sakata-Sogawa K, Highly inclined thin illumination enables clear single-molecule imaging in cells, Nat. Methods 5 (2008) 159–161. [DOI] [PubMed] [Google Scholar]
- [70].Bonnet I, Biebricher A, Porte P-L, Loverdo C, Benichou O, Voituriez R, et al. , Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA, Nucleic Acids Res. 36 (2008) 4118–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Komazin-Meredith G, Mirchev R, Golan DE, van Oijen AM, Coen DM, Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 10721–10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Blainey PC, van Oijent AM, Banerjee A, Verdine GL, Xie XS, A base-excisionDNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Murata A, Ito Y, Kashima R, Kanbayashi S, Nanatani K, Igarashi C, et al. , One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca(2+) or Mg(2+) at millimolar concentrations, J. Mol. Biol 427 (2015) 2663–2678. [DOI] [PubMed] [Google Scholar]
- [74].Subekti DRG, Murata A, Itoh Y, Fukuchi S, Takahashi H, Kanbayashi S, et al. , The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements, Biochemistry 56 (2017) 4134–4144. [DOI] [PubMed] [Google Scholar]
- [75].Giuntoli RD, Linzer NB, Banigan EJ, Sing CE, de la Cruz MO, Graham JS, et al. , DNA-segment-facilitated dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response, J. Mol. Biol 427 (2015) 3123–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kochaniak AB, Habuchi S, Loparo JJ, Chang DJ, Cimprich KA, Walter JC, et al. , Proliferating cell nuclear antigen uses two distinct modes to move along DNA, J. Biol. Chem 284 (2009) 17700–17710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Dikic J, Menges C, Clarke S, Kokkinidis M, Pingoud A, Wende W, et al. , The rotation-coupled sliding of EcoRV, Nucleic Acids Res. 40 (2012) 4064–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bagchi B, Blainey PC, Xie XS, Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA, J. Phys. Chem. B 112 (2008) 6282–6284. [DOI] [PubMed] [Google Scholar]
- [79].Paull TT, Carey M, Johnson RC, Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro, Genes Dev. 10 (1996) 2769–2781. [DOI] [PubMed] [Google Scholar]
- [80].Azam T. Ali , Iwata A, Nishimura A, Ueda S, Ishihama A, Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid, J. Bacteriol 181 (1999) 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cho BK, Knight EM, Barrett CL, Palsson BO, Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts, Genome Res. 18 (2008) 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, et al. , Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli, Nucleic Acids Res. 39 (2011) 2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, et al. , Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output, PLoS Biol. 9 (2011), e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM, Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12, Nucleic Acids Res. 40 (2012) 3524–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ishihama A, Kori A, Koshio E, Yamada K, Maeda H, Shimada T, et al. , Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli, J. Bacteriol. 196 (2014) 2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ball CA, Osuna R, Ferguson KC, Johnson RC, Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli, J. Bacteriol 174 (1992) 8043–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Xiao B, Johnson RC, Marko JF, Modulation of HU-DNA interactions by salt concentration and applied force, Nucleic Acids Res. 38 (2010) 6176–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kamar RI, Banigan EJ, Erbas A, Giuntoli RD, Olverade la Cruz M, Johnson RC, et al. , Facilitated dissociation of transcription factors from single DNA binding sites, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E3251–E3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hiller DA, Rodriguez AM, Perona JJ, Non-cognateenzyme-DNA complex: structural and kinetic analysis of EcoRV endonuclease bound to the EcoRI recognition site GAATTC, J. Mol. Biol 354 (2005) 121–136. [DOI] [PubMed] [Google Scholar]
- [90].Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, et al. , Structure and flexibility adaptation in nonspecific and specific protein–DNA complexes, Science 305 (2004) 386–389. [DOI] [PubMed] [Google Scholar]
- [91].Fromme JC, Banerjee A, Huang SJ, Verdine GL, Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase, Nature 427 (2004) 652–656. [DOI] [PubMed] [Google Scholar]
- [92].Wang L, Chakravarthy S, Verdine GL, Structural basis for the lesion-scanning mechanism of the MutY DNA glycosylase, J. Biol. Chem 292 (2017) 5007–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bruner SD, Norman DP, Verdine GL, Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA, Nature 403 (2000) 859–866. [DOI] [PubMed] [Google Scholar]
- [94].van Dijk M, Bonvin AM, 3D-DART: a DNA structure modelling server, Nucleic Acids Res. 37 (2009) W235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zheng G, Lu XJ, Olson WK, Web 3DNA—a web server for the analysis, reconstruction, and visualization of three-dimensionalnucleic-acid structures, Nucleic Acids Res. 37 (2009) W240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chandrasekaran R, Arnott S, The structure of B-DNA in oriented fibers, J. Biomol. Struct. Dyn 13 (1996) 1015–1027. [DOI] [PubMed] [Google Scholar]
- [97].Ribeiro J, Melo F, Schuller A, PDIviz: analysis and visualization of protein–DNA binding interfaces, Bioinformatics 31 (2015) 2751–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.