Abstract
Coronavirus disease 2019 (COVID-19) with the infection of SARS-CoV-2 has become a serious pandemic worldwide. However, only few studies focused on risk factors of prolonged SARS-CoV-2 RNA detection among patients with COVID-19. We included 206 adult patients with laboratory-confirmed COVID-19 from two hospitals between 23 Jan and 1 April 2020. Least absolute shrinkage and selection operator (LASSO) analysis was used to screen out independent risk factors of SARS-CoV-2 RNA detection. By multivariate binomial logistic regression analysis and Cox regression analysis, we further determined the associations between SARS-CoV-2 RNA detection and potential risk factors. All patients had two negative SARS-CoV-2 tests with 33 days of median duration of SARS-CoV-2 RNA detection (interquartile range: 25.2–39 days). LASSO and binomial logistic regression analyses suggested that delayed hospital admission (adjusted OR = 3.70, 95% CI: 1.82–7.50), hypokalemia, and subpleural lesion (adjusted OR = 4.32, 95% CI: 1.10–16.97) were associated with prolonged SARS-CoV-2 RNA detection. By LASSO and multivariate Cox regression analyses, we observed that delayed hospital admission, subpleural lesion, and high-dose corticosteroid use were independent risk factors of prolonged SARS-CoV-2 RNA detection. Early hospital admission shortened 5.73 days of mean duration of SARS-CoV-2 RNA detection than delayed hospital admission after adjusting confounding factors. Our study demonstrated that delayed hospital admission and subpleural lesion were associated with prolonged SARS-CoV-2 RNA detection among patients with COVID-19. The use of high-dose corticosteroids should be interpreted with extreme caution in treating COVID-19.
Keywords: Hospital admission, COVID-19, Corticosteroids, SARS-CoV-2
Introduction
Coronavirus disease 2019 (COVID-19) with the infection of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was initially reported in Wuhan, China [1]. As of 20 April, there were 82,758 laboratory-confirmed COVID-19 patients and 4623 death (5.58%) in China [2]. Compared with previous coronavirus diseases caused by the SARS and the Middle East respiratory syndrome coronavirus (MERS-CoV), COVID-19 for SARS-CoV-2 was associated with lower overall mortality and stronger transmissibility [3]. Few studies focused on risk factors of SARS-CoV-2 RNA detection duration among patients with COVID-19 [4–7]. In a retrospective study from China, COVID-19 survivors have a median duration of about 20 days of SARS-CoV-2 RNA detection after illness onset, but deceased COVID-19 patients have continuously detectable SARS-CoV-2 shedding until their death [4]. The study of Xu and colleagues [5] estimated the risk factors of delayed viral shedding (≥ 15 days after illness onset) and found that male, delayed hospital admission, and invasive mechanical ventilation were positively associated with prolonged SARS-CoV-2 RNA detection duration. Another study for patients with severe COVID-19 found no significant effect of sex and age on SARS-CoV-2 RNA detection duration [7]. Overall, the current studies related to SARS-CoV-2 RNA detection duration were relatively few and reported inconsistent results.
Here, we performed a retrospective study to describe the characteristics of patients with COVID-19 outside of Wuhan in Hubei province. Least absolute shrinkage and selection operator (LASSO) analysis and binomial logistic regression analysis with a generalized additive model were used to determine the independent risk factors of SARS-CoV-2 RNA detection. We also estimated the median duration of SARS-CoV-2 RNA detection and identified its independent risk factors by LASSO analysis, multivariate Cox regression analysis, and restricted mean survival time analysis.
Methods
Study design and population
Our study population came from two designated hospitals for treating patients with COVID-19, which included Yichang Central People’s Hospital and Yichang Third People’s Hospital. A total of 206 adult patients with laboratory-confirmed COVID-19 were included into this retrospective study between 23 Jan and 1 April 2020. All patients had two negative SARS-CoV-2 testing results and were discharged from the hospital. All patients gave written informed consent and the retrospective study was approved by the ethics committee of Yichang Central People’s Hospital.
Data collection and sources
Two physicians extracted all data about epidemiological, demographic, clinical symptoms, laboratory findings, chest imaging, treatment, and outcome from electronic medical records using a standardized data collection. The third physician checked all relevant data and adjudicated any difference in interpretation between the two primary physicians.
The demographic and epidemiological data included the following variables: sex, age, smoking, comorbidity, exposure history, and family clustering occurrence.
We also collected some clinical symptoms as potential risk factors, including fever, cough, sputum, fatigue, diarrhea, nausea or vomiting, muscle soreness, and dyspnea. A large amount of laboratory findings were incorporated into our study, including complete blood counts, liver and renal function, D-dimer, C-reactive protein, procalcitonin, lactate dehydrogenase and creatine kinase, and blood chemistry (serum potassium and sodium). The samples of sputum or nasopharyngeal swab were tested by real-time reverse transcriptase polymerase chain reaction (RT-PCR) with the Chinese Center for Disease Control and Prevention (CDC)–recommended Kit (Shengxiang, Hunan, China). All specimens were performed at the clinical laboratory of Yichang Central People’s Hospital for further testing. Laboratory findings were stratified according to previous studies [5, 6]. All patients received chest computed tomography scan. The following radiological manifestations were considered potential risk factors of SARS-CoV-2 RNA detection: ground-glass opacities, consolidation, interlobular septal thickening, fibrosis, the site of involving pulmonary lobe, the number of involving pulmonary segments, and subpleural lesion. The severity of COVID-19 at admission was evaluated according to the guidelines of the National Health Commission of the People’s Republic of China [8]. To simplify the analysis process, all patients were categorized into mild, severe, and critical groups with reference to the study of Xu and colleagues [5]. Considering the lack of specific drugs for COVID-19 treatment, all patients received different therapeutic options based on the recommendation of the National Health Commission of the People’s Republic of China and brainstorming of senior physicians [8]. We regarded the time from illness onset to admission as hospital admission (≤ 5 days vs > 5 days) and the time from admission with first positive SARS-CoV-2 to two negative SARS-CoV-2 testing as the duration of negative SARS-CoV-2 RNA detection.
Statistical analysis
In our study, all patients were classified into short-term (< 30 days) and long-term (≥ 30 days) positive SARS-CoV-2 groups according to the duration of SARS-CoV-2 RNA detection. All relevant data were merged and compared using Microsoft Excel and SPSS. Categorical variables were presented as counts and percentages (%) and differences between two groups were estimated through a chi-square test. Means and standard deviations were used to describe continuous variables with the Mann-Whitney U test for skewed continuous variables and Student’s t test for normally distributed continuous variables. Chi-square goodness of fit and Kolmogorov-Smirnov tests were used to examine the normality of distribution of the data. Our study included more than 60 variables as the potential risk factors of SARS-CoV-2 RNA detection among patients with COVID-19. For high-dimensional data, the least absolute shrinkage and selection operator (LASSO) method is more available to screen the predictive features from the primary data set compared with conventional regression analysis [9–12]. Tuning parameter (lambda) selection in the LASSO model uses 10-fold cross-validation [9, 11]. By shrinking down to zero coefficient weights, LASSO regression analysis has the ability to eliminate exposures that are non-related to the outcome [9, 11]. In the first step of our study, we identified the independent risk factors of long-term positive SARS-CoV-2 RNA detection by using LASSO logistic regression analysis and SARS-CoV-2 RNA detection duration by using LASSO Cox regression analysis. A generalized additive model with a binomial logistic regression model was used to further determine the associations between long-term SARS-CoV-2 RNA detection and the independent risk factors obtained by LASSO analysis. We further examined the associations between the independent risk factors obtained by LASSO analysis and SARS-CoV-2 RNA detection duration through multivariate Cox regression analysis with a proportional hazards model. Moreover, we applied restricted mean survival time analysis to better assess the effect of the independent risk factors on SARS-CoV-2 RNA detection duration. A valid hazard ratio (HR) in conventional Cox regression analysis requires the proportional hazards assumption [13]. When proportional hazards are not met, HR may lack statistical power to detect a true treatment effect [13]. Restricted mean survival time analysis is an alternative robust and clinically interpretable summary measure of time-to-event outcome, which does not depend on the proportional hazards assumption [14]. Restricted mean survival time analysis also provides crude and adjusted differences between two groups.
All statistical analyses were performed by using Empower(R) (www. empowerstats. com; X&Y solutions, Inc., Boston, MA) and R software, version 3.1.2 (http: //www. r-project. org). The odds ratio (OR) for binomial logistic regression analysis and HR for multivariate Cox regression analysis with 95% confidence intervals (CIs) were used to estimate the differences, and a two-tailed P < 0.05 was considered statistically significant.
Results
The characteristics of patients in this study
Of the 206 adult patients with COVID-19, there was predominantly female (51.9%) and 53.7 ± 15.7 years of mean age that ranged from 18 to 87 years. There were 123 patients with exposure history of SARS-CoV-2 and 61 patients with family clustering occurrence. Fever (92.2%) was the most common symptom of COVID-19, followed by cough (75.7%), fatigue (37.9%), dyspnea (36.4%), sputum (27.7%), muscle soreness (24.7%), diarrhea (8.7%), and nausea or vomiting (8.2%). In total, 61.0% of patients with COVID-19 received early hospital admission (the time from illness onset to admission ≤ 5 days). In terms of laboratory findings, lymphocytopenia and eosinophilia were presented in 75 patients and 62 patients, respectively. The median duration of SARS-CoV-2 RNA detection was 33 days (interquartile range (IQR): 25.2–39 days). About 60.2% of patients were classified into the long-term positive SARS-CoV-2 group. The shortest duration of SARS-CoV-2 RNA detection was 4 days, whereas the longest was 69 days. The most common lobe involved by SARS-CoV-2 infection was the right lower (82.5%), followed by the left lower (76.2%), left upper (63.6%), right upper (62.6%), and right middle (21.4%). There were 124 patients involving > 5 pulmonary segments and 190 patients with subpleural lesion. The majority of patients received combined antiviral treatment, including lopinavir/ritonavir, interferon-α, oseltamivir, and Arbidol. A total of 143 patients (69.4%) received antibiotics and 139 patients (67.4%) received intravenous immunoglobulin. Corticosteroids (methylprednisolone) and thymosin were used in 57.3% and 38.3% of patients with COVID-19, respectively. Fifteen patients with severe and critical COVID-19 (7.3%) required high-flow nasal cannula oxygen therapy. Five patients underwent non-invasive mechanical ventilation treatment and 3 patients with invasive mechanical ventilation (more detailed information are shown in Table 1).
Table 1.
The demographic data, epidemiological data, clinical symptoms, laboratory, chest imaging, treatment, and outcome between the short-term and long-term positive SARS-CoV-2 groups
| Positive SARS-CoV-2 | |||
|---|---|---|---|
| Short-term (≤ 30 days) | Long-term (> 30 days) | P value | |
| N | 82 | 124 | |
| Demographics and epidemiological data | |||
| Age | 51.2 ± 17.3 | 55.3 ± 14.3 | 0.064 |
| < 65 years | 60 (73.2%) | 90 (72.6%) | 0.926 |
| ≥ 65 years | 22 (26.8%) | 34 (27.4%) | |
| Sex | 0.65 | ||
| Female | 41 (50.0%) | 66 (53.2%) | |
| Male | 41 (50.0%) | 58 (46.8%) | |
| Smoking | 0.109 | ||
| Never | 67 (81.7%) | 111 (89.5%) | |
| Ever | 15 (18.3%) | 13 (10.5%) | |
| Hypertension | 17 (20.7%) | 35 (28.2%) | 0.226 |
| Diabetes | 14 (17.1%) | 25 (20.2%) | 0.58 |
| Cardiocerebrovascular diseases | 7 (8.5%) | 10 (8.1%) | 0.904 |
| Chronic kidney diseases | 4 (4.9%) | 1 (0.8%) | 0.063 |
| Chronic lung diseases | 3 (3.7%) | 6 (4.8%) | 0.685 |
| Exposure history | 56 (68.3%) | 67 (54.0%) | 0.041 |
| Family clustering occurrence | 32 (39.0%) | 29 (23.4%) | 0.016 |
| Clinical symptoms | |||
| Fever | 0.344 | ||
| < 37.3 °C | 9 (11.0%) | 7 (5.6%) | |
| 37.3–38 °C | 28 (34.1%) | 38 (30.6%) | |
| 38.1–39.0 °C | 39 (47.6%) | 61 (49.2%) | |
| 39.1–41 °C | 6 (7.3%) | 17 (13.7%) | |
| > 41 °C | 0 (0.0%) | 1 (0.8%) | |
| Cough | 58 (70.7%) | 98 (79.0%) | 0.174 |
| Sputum | 26 (31.7%) | 31 (25.0%) | 0.292 |
| Dyspnea | 20 (24.4%) | 55 (44.4%) | 0.004 |
| Muscle soreness | 20 (24.4%) | 31 (25.0%) | 0.921 |
| Fatigue | 28 (34.1%) | 50 (40.3%) | 0.371 |
| Nausea or vomiting | 9 (11.0%) | 8 (6.5%) | 0.248 |
| Diarrhea | 10 (12.2%) | 8 (6.5%) | 0.153 |
| Laboratory findings | |||
| Red blood cell count | 4.4 ± 0.7 | 4.3 ± 0.6 | 0.289 |
| Hemoglobin | 135.0 ± 18.2 | 132.5 ± 16.0 | 0.288 |
| White blood cell count (* 109/L) | 4.9 ± 1.8 | 4.8 ± 1.9 | 0.636 |
| < 4 * 109/L | 28 (34.1%) | 50 (40.3%) | 0.371 |
| ≥ 4 * 109/L | 54 (65.9%) | 74 (59.7%) | |
| Neutrophil count (* 109/L) | 3.3 ± 1.6 | 3.3 ± 1.7 | 0.866 |
| Lymphocyte count (* 109/L) | 1.1 ± 0.5 | 1.0 ± 0.4 | 0.305 |
| < 1.1 * 109/L | 50 (61.0%) | 81 (65.3%) | 0.526 |
| ≥ 1.1 * 109/L | 32 (39.0%) | 43 (34.7%) | |
| Neutrophil to lymphocyte ratio | 3.8 ± 3.0 | 3.8 ± 2.9 | 0.983 |
| Eosinophil count (* 109/L) | 0.18 | ||
| < 0.02 * 109/L | 53 (64.6%) | 91 (73.4%) | |
| ≥ 0.02 * 109/L | 29 (35.4%) | 33 (26.6%) | |
| Lymphocyte + eosinophil | 0.578 | ||
| Normal | 40 (48.8%) | 68 (54.8%) | |
| Eosinophilia alone | 13 (15.9%) | 23 (18.5%) | |
| Lymphocytopenia + eosinophilia | 19 (23.2%) | 20 (16.1%) | |
| Lymphocytopenia alone | 10 (12.2%) | 13 (10.5%) | |
| Platelet count (* 109/L) | 175.8 ± 65.1 | 175.2 ± 75.4 | 0.954 |
| ALT (U/L) | 30.8 ± 17.7 | 31.5 ± 26.1 | 0.829 |
| ≤ 40 U/L | 74 (90.2%) | 112 (90.3%) | 1 |
| > 40 U/L | 8 (9.8%) | 12 (9.7%) | |
| AST (U/L) | 34.0 ± 15.3 | 34.7 ± 17.3 | 0.771 |
| ≤ 50 U/L | 67 (81.7%) | 99 (79.8%) | 0.985 |
| > 50 U/L | 15 (18.3%) | 25 (20.2%) | |
| Albumin (g/L) | 40.3 ± 6.2 | 38.3 ± 4.7 | 0.009 |
| CRP (mg/L) | 26.6 ± 34.0 | 28.9 ± 32.8 | 0.64 |
| ≤ 10 mg/L | 33 (40.2%) | 43 (34.7%) | 0.601 |
| 10–40 mg/L | 34 (41.5%) | 52 (41.9%) | |
| > 40 mg/L | 15 (18.3%) | 29 (23.4%) | |
| D-dimer (μg/mL) | 0.09 | ||
| ≥ 0.5 μg/mL | 46 (56.1%) | 59 (47.6%) | |
| > 0.5 to ≤ 1 μg/mL | 20 (24.4%) | 48 (38.7%) | |
| > 1 μg/mL | 16 (19.5%) | 17 (13.7%) | |
| PCT (ng/mL) | 0.597 | ||
| < 0.05 ng/mL | 70 (85.4%) | 109 (87.9%) | |
| ≥ 0.05 ng/mL | 12 (14.6%) | 15 (12.1%) | |
| Lactate dehydrogenase (U/L) | 0.698 | ||
| ≤ 245 U/L | 57 (69.5%) | 83 (66.9%) | |
| > 245 U/L | 25 (30.5%) | 41 (33.1%) | |
| Creatine kinase (U/L) | 0.849 | ||
| ≤ 185 U/L | 67 (81.7%) | 100 (80.6%) | |
| > 185 U/L | 15 (18.3%) | 24 (19.4%) | |
| Creatinine (μmol/L) | 80.5 ± 83.7 | 69.4 ± 20.4 | 0.16 |
| Serum sodium (mmol/L) | 0.191 | ||
| < 135 mmol/L | 8 (9.8%) | 20 (16.1%) | |
| ≥ 135 mmol/L | 74 (90.2%) | 104 (83.9%) | |
| Serum potassium (mmol/L) | 0.005 | ||
| < 3.5 mmol/L | 15 (18.3%) | 45 (36.3%) | |
| ≥ 3.5 mmol/L | 67 (81.7%) | 79 (63.7%) | |
| Chest imaging | |||
| Ground-glass opacities | 46 (56.1%) | 81 (65.3%) | 0.183 |
| Consolidation | 40 (48.8%) | 63 (50.8%) | 0.776 |
| Interlobular septal thickening | 5 (6.1%) | 20 (16.1%) | 0.031 |
| Fibrosis | 41 (50.0%) | 66 (53.2%) | 0.65 |
| Involving pulmonary lobe | |||
| Left upper | 43 (52.4%) | 88 (71.0%) | 0.007 |
| Left lower | 57 (69.5%) | 100 (80.6%) | 0.066 |
| Right lower | 40 (48.8%) | 89 (71.8%) | < 0.001 |
| Right middle | 17 (51.5%) | 27 (69.2%) | 0.124 |
| Right lower | 66 (80.5%) | 104 (83.9%) | 0.531 |
| Involving pulmonary segment | 0.064 | ||
| ≤ 5 | 39 (47.6%) | 43 (34.7%) | |
| > 5 | 43 (52.4%) | 81 (65.3%) | |
| Subpleural lesion | 69 (84.1%) | 121 (97.6%) | < 0.001 |
| Treatment | |||
| Lopinavir/ritonavir | 72 (87.8%) | 104 (83.9%) | 0.433 |
| Arbidol | 78 (95.1%) | 121 (97.6%) | 0.34 |
| Oseltamivir | 73 (89.0%) | 118 (95.2%) | 0.097 |
| Interferon-α | 76 (92.7%) | 114 (91.9%) | 0.844 |
| Herbs | 77 (93.9%) | 111 (89.5%) | 0.275 |
| XueBi Jing injection | 49 (59.8%) | 94 (75.8%) | 0.014 |
| Immunoglobulin | 49 (59.8%) | 90 (72.6%) | 0.054 |
| Thymosin | 22 (26.8%) | 57 (46.0%) | 0.006 |
| Corticosteroid | 0.03 | ||
| No | 45 (54.9%) | 43 (34.7%) | |
| Low-dose (40 mg) | 21 (25.6%) | 39 (31.5%) | |
| How-dose (80 mg) | 14 (17.1%) | 36 (29.0%) | |
| Other | 2 (2.4%) | 6 (4.8%) | |
| Antibiotics | 49 (59.8%) | 94 (75.8%) | 0.014 |
| Mask oxygen therapy | 5 (6.1%) | 17 (13.7%) | 0.083 |
| High-flow nasal cannula oxygen therapy | 4 (4.9%) | 11 (8.9%) | 0.28 |
| Non-invasive mechanical ventilation | 0 (0.0%) | 5 (4.0%) | 0.066 |
| Invasive mechanical ventilation | 0 (0.0%) | 3 (2.4%) | 0.156 |
| Outcome | |||
| Hospital admission | < 0.001 | ||
| Early (≤ 5 days) | 64 (78.0%) | 62 (50.0%) | |
| Delayed (> 5 days) | 18 (22.0%) | 62 (50.0%) | |
| The need for ICU | 6 (7.3%) | 7 (5.6%) | 0.629 |
| The severity of COVID-19 | 0.159 | ||
| Mild | 78 (95.1%) | 113 (91.1%) | |
| Severe | 4 (4.9%) | 6 (4.9%) | |
| Critical | 0 (0%) | 5 (4.0%) | |
| Median duration of viral shedding (days) | 24 | 37.5 | |
Risk factors of SARS-CoV-2 RNA detection
LASSO analysis with binomial logistic regression model screened out 7 risk factors of long-term SARS-CoV-2 RNA detection, which included dyspnea, delayed hospital admission, hypokalemia, subpleural lesion, right upper lesion, the use of methylprednisolone, and the use of thymosin. The area under the receiver operating characteristic curve (AUC) was 0.764. Delayed hospital admission, hypokalemia, and subpleural lesion were still the independent risk factors of long-term SARS-CoV-2 RNA detection in multivariate binomial logistic regression analysis with a generalized additive model. Early hospital admission was associated with less probability of long-term SARS-CoV-2 RNA detection compared with delayed hospital admission (49% vs 78%, adjusted OR = 3.70, 95% CI: 1.82–7.50, P < 0.001; see Fig. 1). Patients with hypokalemia (hypokalemia vs normal potassium, adjusted OR = 0.38, 95% CI: 0.17–0.83, P = 0.015) and subpleural lesion (no vs yes, adjusted OR = 4.32, 95% CI: 1.10–16.97, P = 0.015) seemed to prolong SARS-CoV-2 RNA detection.
Fig. 1.
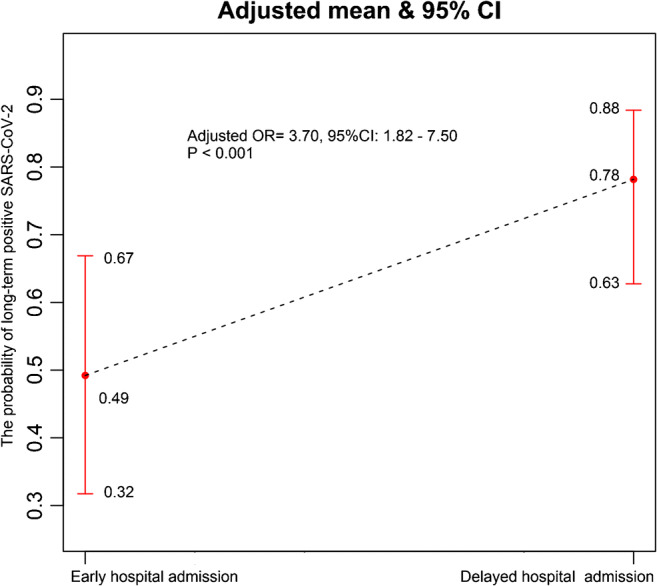
The association between hospital admission and long-term positive SARS-CoV-2 RNA detection in multivariate binomial logistic regression analysis with a generalized additive model
LASSO analysis with Cox regression model found six independent risk factors of prolonged SARS-CoV-2 RNA detection duration, including cough, dyspnea, delayed hospital admission, subpleural lesion, the use of methylprednisolone, and the use of thymosin. The value of AUC was 0.74. Multivariate Cox regression analysis further suggested that delayed hospital admission (adjusted HR = 0.49, 95% CI: 0.36–0.67, P < 0.001; see Fig. 2) and subpleural lesion (adjusted HR = 0.37, 95% CI: 0.21–0.64, P < 0.001) were still the independent risk factors of prolonged SARS-CoV-2 RNA detection duration. In addition, patients with the use of high-dose (80 mg/day vs no, adjusted HR = 0.67, 95% CI: 0.46–0.96, P = 0.031) but not low-dose (40 mg/day vs no, adjusted HR = 0.72, 95% CI: 0.48–1.08, P = 0.11) methylprednisolone seemingly were associated with longer SARS-CoV-2 RNA detection duration than those without methylprednisolone. When we restricted the time point to 42 days, crude mean duration of SARS-CoV-2 RNA detection in the early hospital admission group differed by 6.25 days (95% CI: 4.30–8.20 days) from delayed hospital admission. After adjusting potentially confounding factors, the adjusted mean duration of SARS-CoV-2 RNA detection in the early hospital admission group was 5.73 days (95% CI: 3.91–7.56 days) less than that in the delayed hospital admission group (see Fig. 2).
Fig. 2.
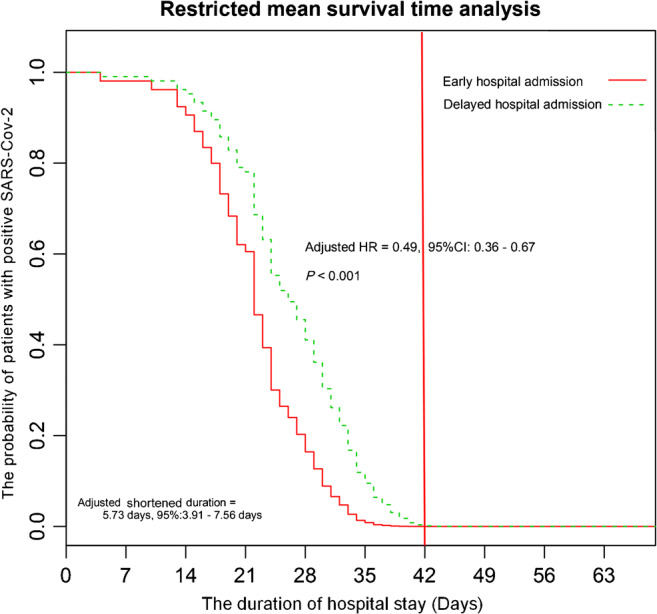
The association between hospital admission and SARS-CoV-2 RNA detection in multivariate Cox regression analysis with a proportional hazards model and restricted mean survival time analysis
Discussion
The majority of current studies about COVID-19 focused on diagnosing this disease, estimating its transmission, assessing its severity, and identifying the risk factors of death [15–17]. Our study provided more detailed information about the epidemiological, demographic, clinical symptoms, laboratory, chest imaging, treatment, and outcome among patients with COVID-19. Compared with patients in Wuhan, our study population had more patients with mild COVID-19. Our study suggested that early hospital admission seemingly had the ability to shorten SARS-CoV-2 RNA detection duration. During 42 days after hospital admission, early hospital admission reduced approximately 5.73 days in mean duration of SARS-CoV-2 RNA detection. In addition, we also observed that high-dose (80 mg/day) but not low-dose (40 mg/day) methylprednisolone use potentially prolonged the mean duration of SARS-CoV-2 RNA detection.
Our study provided another reason for early diagnosis and treatment of COVID-19. Compared with delayed hospital admission, early hospital admission was associated with lower probability of long-term positive SARS-CoV-2 RNA detection and shorter mean duration of SARS-CoV-2 RNA detection. A shorter duration of SARS-CoV-2 RNA detection also means less consumption of medical resources. A possible explanation of this phenomenon is that early hospital admission potentially decreases the probability of the convention of mild to severe COVID-19 and improves the physical conditions to confront SARS-CoV-2 [5]. The study of Xu et al. demonstrated that early hospital admission was associated with a lower probability of severe patients at admission and less frequency of critically severe illness during hospitalization compared with delayed hospital admission [5].
In our study, 92.2% of COVID-19 patients were observed with the involvement of the subpleural area in chest imaging, similar to the result of Zhao et al. [18]. Subpleural lesion was identified to be positively associated with long-term positive SARS-CoV-2 and duration of SARS-CoV-2 RNA detection in our study. Hypokalemia seemingly was associated with higher probability of long-term positive SARS-CoV-2 RNA detection than normal serum potassium. The maintenance of serum potassium balance involves three key elements: gastrointestinal losses, renal excretion, and cellular shifts [19]. SARS-CoV-2 can attack the kidney, gastrointestinal tract, and liver by attaching to angiotensin-converting enzyme 2 receptors of human organs [20]. The occurrence of hypokalemia indicates that multiple human organs may suffer from SARS-CoV-2 viral infection and require long-term recovery. Considering the effect of the cytokine storm syndrome, some patients with COVID-19 received systemic corticosteroids and were recommended 1–2 mg/kg in China [8]. However, the use of systemic corticosteroids in treating coronavirus infection is still controversial. A recent systematic review and meta-analysis showed that the use of systematic corticosteroids is associated with higher mortality and longer length of stay [21]. Several previous studies also suggested that corticosteroid use prolonged the duration of viral RNA shedding in patients with SARS [22] and MERS [23]. Fan et al. [24] reported that the treatment of low-dose corticosteroids does not delay viral shedding in patients with COVID-19, similar to the finding of Xu et al. [5]. Our study demonstrated that the prolonged duration of SARS-CoV-2 RNA detection was shown in high-dose (80 mg) methylprednisolone treatment (adjusted HR = 0.67, 95% CI: 0.46–0.96, P = 0.031), but not in low-dose (40 mg) methylprednisolone treatment (adjusted HR = 0.72, 95% CI: 0.48–1.08, P = 0.11). High-dose but not low-dose corticosteroid treatment was reported to be associated with the increase of mortality in patients with severe COVID-19 [25]. Therefore, the use of high-dose corticosteroids should be interpreted with extreme caution and low-dose corticosteroid use may be considered only for patients with severe COVID-19.
There were the following strengths in our study. This study provided more detailed information about patients with COVID-19. In comparison to the study of Xu et al. [5], we included more than 60 variables as potential risk factors and screened the independent risk factors through comprehensive statistical analyses. Using conventional logistic regression analysis to accurately handle high-dimensional data sets and correlated features is difficult, but LASSO analysis can effectively overcome this obstacle [12]. Restricted mean survival time analysis was initially used to study COVID-19 and confirmed the clinical benefit of early hospital admission in shortening the mean duration of SARS-CoV-2 RNA detection. The major limitation of this work was it being a retrospective study with a relatively small sample size, which can produce selective bias. However, the clinical benefit of early hospital admission in shortening the duration of SARS-CoV-2 RNA detection has been shown in the study from Zhejiang province [5], located 900 km east of Wuhan. Thus, we believe that our conclusions are valid.
In conclusion, our study suggested that early hospital admission seemed to shorten mean duration of SARS-CoV-2 RNA detection among patients with COVID-19, which potentially reduced the severity of COVID-19 and the consumption of medical resources. Moreover, high-dose corticosteroids should be used with extreme caution for treating COVID-19. A recent study showed that the use of dexamethasone (6 mg) potentially decreased 28-day mortality among COVID-19 patients who were receiving either invasive mechanical ventilation or oxygen alone at randomization but not among those without respiratory support [26]. Therefore, low–high corticosteroids can be considered for eligible patients with COVID-19. Large-sample multicenter studies are warranted to further support our conclusions.
Acknowledgments
We thank all our colleagues who helped us during the current study. We are also grateful to the many front-line medical staff for their dedication in the face of this outbreak, despite the potential threat to their own lives and the lives of their families.
Authors’ contributions
All authors contributed to data analysis and drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
The work was supported by the Natural Science Foundation of Hubei Province CN (No. 2018CFB643).
Data availability
The data underlying this study were obtained from four designated hospitals for treating the patients with COVID-19. All relevant data are within the paper and its Supporting Information files.
Compliance with ethical standards
Ethics approval and consent to participate
All patients gave written informed consent and the study was approved by the ethics committee of Yichang Central People’s Hospital.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Zhigang Hu and Xinyu Song are listed as co-corresponding authors.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhigang Hu, Sijia Li, and Ailan Yang contributed equally to this work. They are listed as co-first authors.
Contributor Information
Zhigang Hu, Email: hxq910813@163.com.
Sijia Li, Email: 201910510021022@ctgu.edu.cn.
Ailan Yang, Email: 247982885@qq.com.
Wenxin Li, Email: liwenxin2010@163.com.
Xiaoqi Xiong, Email: glacierxq@126.com.
Jianwu Hu, Email: hu-jianwu@163.com.
Jun Jiang, Email: tqy201307@163.com.
Xinyu Song, Email: songxinyu@ctgu.edu.cn.
References
- 1.Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;26:506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Health Commission of the People’s Republic of China. The outbreaks of COVID-19. http://www.nhc.gov.cn/xcs/yqtb/202004/b504a02486834baf8ed8149701a4175b.shtml. Accessed 21 Apr 2020 [DOI] [PMC free article] [PubMed]
- 3.Peeri NC, Shrestha N, Rahman MS et al (2020) The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol:1–10. 10.1093/ije/dyaa033 [DOI] [PMC free article] [PubMed]
- 4.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu K, Chen Y, Yuan J et al (2020) Factors associated with prolonged viral RNA shedding in patients with COVID-19. Clin Infect Dis. 10.1093/cid/ciaa351 [DOI] [PMC free article] [PubMed]
- 6.Qian GQ, Chen XQ, Lv DF et al (2020) Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect Dis (Lond) 52:511–512 [DOI] [PubMed]
- 7.Zhou B, She J, Wang Y et al (2020) The duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis. 10.1093/cid/ciaa451 [DOI] [PMC free article] [PubMed]
- 8.National Health Commission of the People’s Republic of China. The notice of launching guideline on diagnosis and treatment of the novel coronavirus pneumonia (NCP). Revised version of the 5th edition. http://www.nhc.gov.cn/xcs/zhengcwj/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml (accessed Feb 8, 2020; in Chinese).
- 9.Avalos M, Adroher ND, Lagarde E, et al. Prescription-drug-related risk in driving: comparing conventional and lasso shrinkage logistic regressions. Epidemiology. 2012;23:706–712. doi: 10.1097/EDE.0b013e31825fa528. [DOI] [PubMed] [Google Scholar]
- 10.Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34:2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 11.Mulé S, Galletto Pregliasco A, Tenenhaus A, et al. Multiphase liver MRI for identifying the macrotrabecular-massive subtype of hepatocellular carcinoma. Radiology. 2020;295:562–571. doi: 10.1148/radiol.2020192230. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Carretero R, Vigil-Medina L, Barquero-Perez O, et al. Logistic LASSO and elastic net to characterize vitamin D deficiency in a hypertensive obese population. Metab Syndr Relat Disord. 2020;18:79–85. doi: 10.1089/met.2019.0104. [DOI] [PubMed] [Google Scholar]
- 13.McCaw ZR, Yin G, Wei LJ. Using the restricted mean survival time difference as an alternative to the hazard ratio for analyzing clinical cardiovascular studies. Circulation. 2019;140:1366–1368. doi: 10.1161/CIRCULATIONAHA.119.040680. [DOI] [PubMed] [Google Scholar]
- 14.Huang B, Kuan PF. Comparison of the restricted mean survival time with the hazard ratio in superiority trials with a time-to-event end point. Pharm Stat. 2018;17:202–213. doi: 10.1002/pst.1846. [DOI] [PubMed] [Google Scholar]
- 15.Chen TM, Rui J, Wang QP, Zhao ZY, Cui JA, Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty. 2020;9:24. doi: 10.1186/s40249-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan A, Liu L, Wang C, et al. Association of public health interventions with the epidemiology of the COVID-19 outbreak in Wuhan, China. JAMA. 2020;323:1–9. doi: 10.1001/jama.2020.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wynants L, Van Calster B, Bonten MMJ, et al. Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Liu B, Yu Y, et al. The characteristics and clinical value of chest CT images of novel coronavirus pneumonia. Clin Radiol. 2020;75:335–340. doi: 10.1016/j.crad.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unwin RJ, Luft FC, Shirley DG. Pathophysiology and management of hypokalemia: a clinical perspective. Nat Rev Nephrol. 2011;7:75–84. doi: 10.1038/nrneph.2010.175. [DOI] [PubMed] [Google Scholar]
- 20.Gheblawi M, Wang K, Viveiros A, et al. Angiotensin converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Liu J, Zhou Y, et al. The effect of corticosteroid treatment on patients with coronavirus infection: a systematic review and meta-analysis. J Infect. 2020;81:e13–e20. doi: 10.1016/j.jinf.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee N, Allen Chan KC, Hui DS, et al. Effects of early corticosteroid treatment on plasma SARS-associated coronavirus RNA concentrations in adult patients. J Clin Virol. 2014;31:304–309. doi: 10.1016/j.jcv.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arabi YM, Mandourah Y, Al-Hameed F, et al. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respir Crit Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 24.Fang X, Mei Q, Yang T, et al. Low-dose corticosteroid therapy does not delay viral clearance in patients with COVID-19. J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;30495-4:S0091–S6749. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RECOVERY Collaborative Group, Horby P, Lim WS, et al (2020) Dexamethasone in hospitalized patients with Covid-19 - preliminary report. N Engl J Med. 10.1056/NEJMoa2021436
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this study were obtained from four designated hospitals for treating the patients with COVID-19. All relevant data are within the paper and its Supporting Information files.


