Summary:
The central dogma teaches us that DNA makes RNA which in turn makes proteins, the main building blocks of the cell. But this over simplified linear transmission of information overlooks the vast majority of the genome produces RNAs that don’t encode proteins and the myriad ways that RNA regulates cellular functions. Historically, one of the challenges in illuminating RNA biology has been the lack of tools for visualizing RNA in live cells. But clever approaches for exploiting RNA binding proteins, in vitro RNA evolution, and chemical biology have resulted in significant advances in RNA visualization tools in recent years. This review provides an overview of current tools for tagging RNA with fluorescent probes and tracking their dynamics, localization in function in live mammalian cells.
Graphical Abstract
eTOC blurb
This review article summarizes the current tools that have been developed to light up RNA in live mammalian cells. Such tools permit researchers to dissect the localization, biochemical processing and dynamics of RNA in real time.
ILLUMINATING RNA BIOLOGY
The explosion of genomics has revealed the majority of the human genome is transcribed into RNA, but only a small fraction is translated into protein (Djebali et al., 2012), leading to intense efforts to elucidate the diverse ways non-coding RNAs influence cellular function (Cech and Steitz, 2014). Both mRNAs and ncRNAs associate with other RNAs (Guil and Esteller, 2015) and assemble into macromolecular complexes and granules containing both RNA and proteins (Matera and Wang, 2014; Protter and Parker, 2016; Shay and Wright, 2019), all of which can profoundly influence cellular functions. There is growing appreciation that RNA undergoes complex and dynamic biochemical manipulations, as illustrated by the process of mRNA processing, transport, translation, and degradation (Bertrand et al., 1998; Darzacq et al., 2007; Tutucci et al., 2018a), and transport of mRNAs to sites of activity in neurons (Buxbaum et al., 2014; Ryder and Lerit, 2018). But the full landscape of how RNA localization and dynamics impact function, particularly for ncRNAs, is largely undefined. Here, we review tools that have been developed to tag and track RNA in living cells, with an emphasis on tools that have been applied to mammalian cells. These tools largely fall into two categories: those that rely on a protein to bind an RNA element and those that utilize small molecule probes to bind an RNA element. The RNA element can either be genetically engineered into an RNA of interest or can be an endogenous RNA. Given the diversity of tools and approaches, Figure 1 provides a roadmap of the tools covered in this review. Finally, we also cover a number of applications that illustrate the power and promise of such tools to illuminate different aspects of RNA biology. The majority of these applications focus on mRNA, perhaps because biochemical, cell biology and sequencing efforts have advanced our knowledge about mRNA processes such that sophisticated single mRNA tracking studies provide meaningful information. There are fewer examples of tagging and tracking ncRNA, perhaps because of the diversity of classes, sizes, and structures of ncRNAs and concerns about perturbing ncRNA function when introducing a tag. We suspect that deeper understanding of ncRNA biology is needed as a foundation to design impactful live RNA tracking assays. We highlight several emerging examples of such ncRNA live imaging studies, and provide an outlook of future directions in this field.
Figure 1:
Overview of tools covered in this review
PROTEIN-BASED RECOGNITION OF RNA
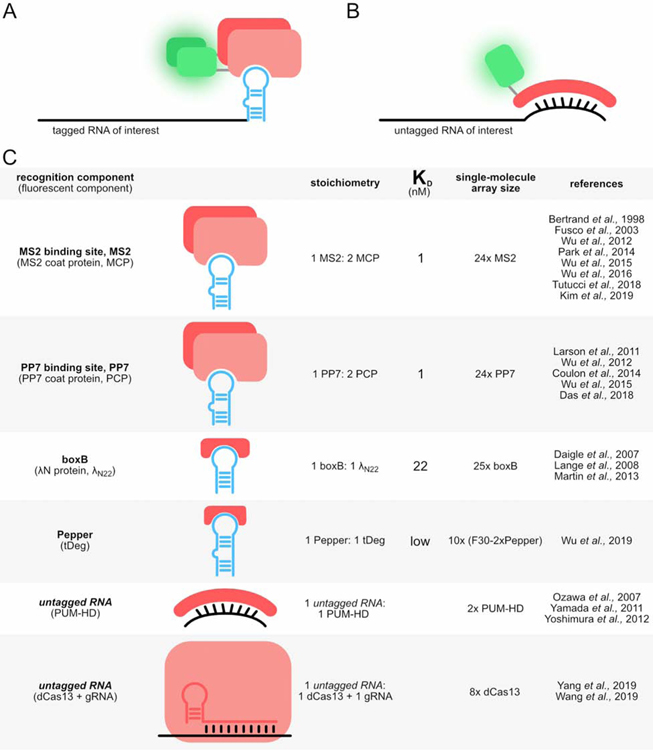
One approach for detection of RNA is to leverage natural protein-RNA interactions, where an RNA binding protein is used to target an RNA of interest (Figure 2). The RNA binding protein is fused to a fluorescent component, which historically has been a fluorescent protein (FP). FPs can be readily exchanged by cloning, providing flexibility in fluorescence properties, including color, photostability, and brightness (Cranfill et al., 2016). More recently FPs have been replaced by Halo or SNAP-tags which can specifically couple to small organic dyes.
Figure 2:
A&B) Genetically encoded tools can visualize tagged or untagged RNAs. C) With FPs, these systems use proteins to image RNA.
MS2
The MS2 system was pioneered by the Singer Lab in the 1990s and remains the most widely used RNA-imaging system today. It was developed from MS2 bacteriophage coat protein dimers that bind sites in the RNA genome (Peabody, 1993). MS2 coat proteins (MCPs) are 129-amino acid proteins that form homodimers (Golmohammadi et al., 1993). MS2 binding sites are 21-nt RNA stem loops (abbreviated MS2) with a conserved loop region and bulge (Johansson et al., 1997; Peabody, 1993; Valegård et al., 1997). The coat protein dimers bind the stem loops specifically and tightly (KD=1 nM (Carey et al., 1983)). Typically, a series of MS2s is appended to the target RNA to serve as the recognition element and each MCP is fused to an FP for visualization (Bertrand et al., 1998). Since its creation, the MS2 system has been iteratively improved and remains the gold standard for live-cell RNA imaging.
Originally six MS2s were appended to the 3’UTR of the target RNA ASH1 and MCP was fused to GFP. This allowed researchers to monitor ASH1 mRNA localization in budding yeast reproduction (Bertrand et al., 1998). Subsequently, twenty-four copies of MS2 were appended to a reporter RNA and a nuclear localization signal was fused to the FP, producing sufficient signal-to-noise for single-molecule imaging. Comparison of the fluorescence intensity of single-molecule puncta with purified GFP was used to estimate that approximately thirty-three fluorescent components were bound to an array of twenty-four stem loops (Fusco et al., 2003). Fluorescence fluctuation spectroscopy revealed that occupancy strongly depends on fluorescent component concentration, saturating at half occupancy. Creation of tandem MCPs eliminated the dependence on concentration but decreased occupancy to thirteen fluorescent components per twenty-four stem loops (Wu et al., 2012). Stable integration by retroviral transduction caused recombination and deletion of MS2 stem loops which lead to significant detection issues. Because recombination relies on sequence homology, divergence of the stem loop sequences significantly increased detection consistency (Wu et al., 2015). Substitution of HaloTag for GFP significantly increased brightness and photostability, allowing researchers to monitor translation dynamics in neurons (Wu et al., 2016). While northern blotting and fluorescence in situ hybridization (FISH) demonstrated that stem loop arrays were stabilized in budding yeast (Garcia and Parker, 2016; Heinrich et al., 2017; Tutucci et al., 2018b), this degradation issue doesn’t occur in mammalian cells. MS2-tagged beta-actin, with or without co-expression of MCP, does not produce stabilized decay intermediates or affect RNA lifetime in primary culture of mouse cells (Kim et al., 2019; Park et al., 2014). This holds true in many tissues (Kim et al., 2019; Lionnet et al., 2011), enabling researchers to monitor single-molecule beta-actin mRNA dynamics in primary mouse culture fibroblasts and hippocampal neurons (Park et al., 2014).
PP7
Analogous to MS2, the PP7 system was developed from PP7 bacteriophage coat protein dimers that bind sites in the RNA genome (Olsthoorn et al., 1995). The PP7 coat protein (PCP) is 127-amino acids and forms homodimers (Olsthoorn et al., 1995; Tars et al., 2000). PP7 binding sites are 25-nt RNA stem loops (abbreviated PP7) with a conserved loop region and bulge (Lim and Peabody, 2002). Coat protein dimers bind PP7 specifically with a KD of 1 nM. Despite their similarities, MS2 and PP7 are orthogonal; coat protein dimers are able to discriminate in favor of their own RNA stem loop by one-thousand-fold (Lim et al., 2001) and form binding clefts that are at right angles to each other (Chao et al., 2008). PP7 was first used in live mammalian cells to track transcription dynamics (Larson et al., 2011). Later PP7 was implemented alongside MS2 to decrease background in a split-GFP system (Wu et al., 2014) and to image multiple regions of a target RNA in splicing (Coulon et al., 2014).
Fluorescence fluctuation spectroscopy again revealed that occupancy of coat proteins on stem loop arrays depends on the fluorescent component concentration, saturating at full occupancy. Tandem coat proteins eliminated the concentration dependence but decreased occupancy to twenty-four fluorescent components per twenty-four stem loops (Wu et al., 2012). By FISH, PP7 shows less perturbation of RNA decay than the original MS2 in budding yeast (Heinrich et al., 2017). FISH, quantitative PCR (qPCR), and western blotting showed that PP7 stem loops do not perturb transcription, translation, or decay in mouse primary culture, and this system enabled detection of the short-lived and tightly controlled mRNA Arc (Das et al., 2018).
λN
The λN imaging system was developed from λ bacteriophage N protein which binds the boxB RNA sequence (Chattopadhyay et al., 1995). To image RNA, the N protein was truncated to its minimal binding motif, the first 22 amino acids denoted λN22. λN22 binds one-to-one with the 15-nt boxB RNA stem loop with a KD of 22 nM (Austin et al., 2002). To attain sufficient signal, a target RNA was appended with four boxB stem loops and four tandem λN22 were fused to three EGFPs with one nuclear localization signal. Although there was high background, fluorescence recovery after photobleaching (FRAP) enabled researchers to monitor target RNA localization (Daigle and Ellenberg, 2007). The λN imaging system is orthogonal to MS2, allowing researchers to monitor the transport of multiple target RNAs to the bud tip during budding yeast reproduction (Lange et al., 2008) and to observe dynamics of single-molecule splicing of beta-globin (Martin et al., 2013). To our knowledge the occupancy of λN22-boxB has not been reported and the system has not been rigorously tested for perturbation of RNA function.
Pepper
The recent Pepper imaging system ((Wu et al., 2019), not to be confused with the dye-binding Peppers described below) was developed from an interaction between the HIV-1 Tat peptide and the trans-activation response element (TAR) (Dingwall et al., 1989). To address the high background fluorescence that many genetically encoded imaging systems face, the 17-amino acid Tat peptide was combined with a short degron to form a 19-amino acid tDeg peptide. When fused to an FP, this tDeg peptide causes degradation of the FP in the absence of the TAR RNA. Upon binding the TAR RNA, the degradation sequence of tDeg is obscured and the fusion protein is not degraded. The TAR RNA variant that caused the highest fluorescence increase was termed “Pepper”. Pepper is a 28-nt stem loop that binds one-to-one with tDeg. Fusing ten (F30–2xPepper) cassettes to a target RNA and fusing tDeg to four mNeonGreen proteins produced sufficient signal-to-noise for single-molecule live-cell imaging. This enabled researchers to monitor recruitment of beta-actin mRNA to stress granules upon treatment with sodium arsenite. Pepper does not perturb target RNA decay or translation efficiency, but does decrease transcription of target RNA. This decreased transcription is comparable to that seen with MS2. Occupancy of the fluorescent component on an array of (F30–2xPepper) cassettes has yet to be measured. The precise binding affinity of the fluorescent component for the cassette is also unknown but is likely close to the low nM KD of a nearly identical TAR variant for the Tat peptide (Smith et al., 1998).
PUM-HD
The PUM-HD imaging system was developed from the human PUMILIO1 protein. The Pumilio homology domain (PUM-HD) binds PUM-HD response elements in RNA (Wang et al., 2001). The PUM-HD contains eight elements each of which binds a specific nucleotide in an 8-nt sequence of RNA; however, nucleotide-contact residues in PUM-HD elements can be engineered to bind any 8-nt RNA sequence (Cheong and Tanaka Hall, 2006). In tandem with another PUM-HD that binds an 8-nt RNA sequence downstream, this imaging system could distinguish between 416 transcripts. To combat background fluorescence, each engineered PUM-HD is fused to one half of split-GFP or split-Venus, only producing fluorescence when both PUM-HDs have bound the target RNA. Because PUM-HD can be engineered to recognize any RNA sequence, this system can detect endogenous RNA. The association of the reconstituted fluorescent protein persists for a long time, so care should be taken to use this system for long-lived RNAs or paired with FRAP, as suggested by the developers (Ozawa et al., 2007). The same system was later used to monitor beta-actin mRNA localization in mammalian cells for short periods of time (Yamada et al., 2011; Yoshimura et al., 2012).
dCas13
The dCas13 system was developed from CRISPR-Cas13 in which the gRNA of Cas13 binds a target RNA by hybridization and the target RNA is cleaved by Cas13 (Abudayyeh et al., 2016). In the imaging system, a variety of catalytically dead Cas13 (dCas13) proteins fused to EGFP were screened with a gRNA for recruitment to a known target RNA location to identify the dCas13-EGFP with the best recruitment and signal-to-background ratio. Similar to PUM-HD, this system enables detection of endogenous untagged RNAs. In addition to general gRNA design guidelines, the developers recommend using sequences that have been validated to work for RNA knockdown. Eight tandem gRNAs for eight tandem dCas13-EGFP fusion proteins is sufficient for single-molecule live-cell imaging. Additionally, dCas13 proteins from two different species can be used orthogonally to monitor two target RNAs or two regions of the same target RNA. By qPCR, the dCas13 imaging system does not decrease transcription of the target RNA unlike MS2 but other perturbations like translational efficiency have not been tested yet (Yang et al., 2019). dCas13 has also been coupled with fluorescently labeled gRNA and electroporated into mammalian cells to monitor transcriptional bursting. This study coupled dCas13 with dCas9 to co-observe the DNA locus and the nascent RNA (Wang et al., 2019).
SMALL MOLECULE-BASED RECOGNITION OF RNA
New small molecule fluorophores have led to significant advances in cellular imaging in the last few years in the same way that fluorescent proteins did in the late 90s. New dye scaffolds (Lavis and Raines, 2014; Lukinavičius et al., 2013) and new synthetic routes to traditional scaffolds (Grimm et al., 2017, 2015) have enabled creative functionalization of dyes, leading to bright, photostable and fluorogenic dyes. While these dyes are readily used with Halo- and SNAP-tags, they have not yet been incorporated into small molecule probes for RNA.
Small molecule-based RNA probes fall into three categories: (1) those that leverage custom-synthesized small molecules that directly bind RNA aptamers and exhibit fluorescence turn-on, (2) aptamers that bind fluorophore-quencher complexes and exhibit turn-on through separation of the quencher from the fluorophore upon binding, and (3) synthetic, nuclease resistant molecular beacon RNAs that hybridize with endogenous RNA of interest.
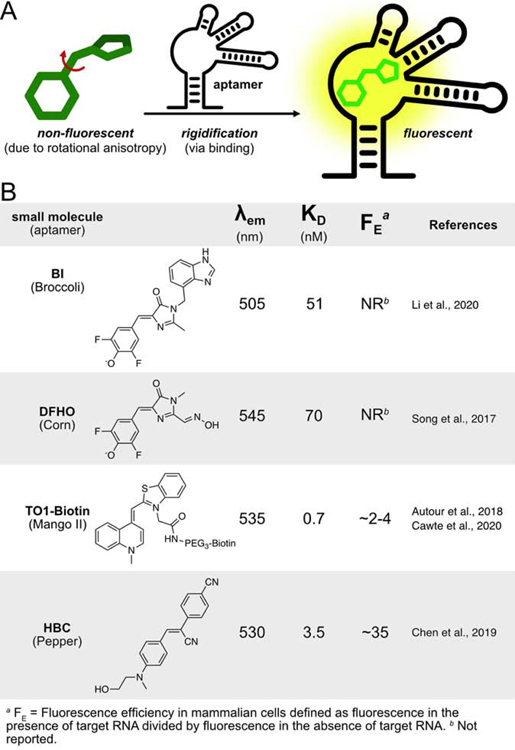
Designed Fluorophore-Aptamer Pairs
The first custom dye-binding aptamer to see wide recognition (Spinach) was developed by Jaffrey and colleagues using a fluorophore that mimicked the structure of green fluorescent protein (GFP) (Paige et al., 2011). The approach exploits the fact that the GFP chromophore is nonfluorescent outside of the protein, but becomes fluorescent upon binding due to rigidification (Figure 3A). Initial aptamers, isolated via 10 rounds of SELEX leading to “Spinach”, exhibited 2,000-fold signal induction. Later, screens that incorporated a fluorescence assay (via bacterial cell sorting) further improved the absolute brightness of the probe, producing a new aptamer, Broccoli (Filonov et al., 2014). A combination of SELEX and fluorescence selection is now standard for the evolution of dye-binding aptamers. More recently, a derivative of the original DFHBI dye, referred to as BI due to a benzimidazole substituent, showed increased photostability and imparted improved thermal stability when bound to Broccoli (Figure 3B, (Li et al., 2020)).
Figure 3:
A) Designed fluorophore-aptamer pairs leverage rigidification upon aptamer binding to product fluorescence turn-on. B) Fluorophores in these structures have a bond capable of cis-trans isomerization, where conformational flexibility renders the molecule nonfluorescent in bulk solution.
Another iteration of the Jaffrey dye-binding vegetables is Corn, which binds DFHO (Song et al., 2017). With the goal of red-shifted emission, the new small molecule draws inspiration from the chromophore of DsRed (Gross et al., 2000). A new aptamer was selected with improved thermal stability relative to Spinach and Broccoli (Filonov et al., 2014; Paige et al., 2011). The improved photostability of Corn enabled imaging of transcription of each promoter subclass by Pol III.
Other groups have built on this concept of fluorophore rigidification by developing their own small molecule with accompanying aptamers (Figure 3B). Unrau and colleagues took advantage of thiazole orange (TO1) that was known to bind double-stranded nucleic acids (Autour et al., 2018; Dolgosheina et al., 2014). The aptamers developed for these fluorophores (termed Mangoes) were tighter binders (KD=0.7 nM) and more red-shifted than Spinach. Mango’s improvement over Spinach is due to its increased thermal stability upon binding the TO1-B substrate (Jeng et al., 2016). Recent research from Unrau and colleagues has shown that arrays of Mango II aptamers can be used to image mRNA granules in live mammalian cells, and single lncRNAs in fixed cells (Cawte et al., 2020). These aptamers outperformed MS2 in signal-to-noise.
One potential limitation of the Broccoli, Spinach, Corn, and Mango is the presence of G-quadruplexes in the aptamer structure. This structure has been found to be crucial for fluorophore rigidification in the aptamer. However, it is increasingly recognized that such secondary structures are actively degraded in mammalian cells (Guo and Bartel, 2016).
The newest family of dye binding aptamers, Peppers, takes a step toward resolving the main issues that plague Spinach/Broccoli/Corn and Mangoes. The Pepper aptamer binds a series of custom small molecules with a stilbene core. Developed by Yang and colleagues (Chen et al., 2019), the structure of the fluorogenic molecular probe was also inspired by GFP. The small molecules were dubbed “HBC” as an abbreviation for the IUPAC name. With the goal of solving outstanding issues of aptamer degradation and fluorophore brightness, authors selected a new aptamer (KD=3.5 nM) that does not contain a G-quadruplex. Through minor modifications to the HBC structure, seven different probes were developed with excitation maxima ranging from 485 to 620 nm. The most well vetted small molecules were HBC (Pepper530) and the red-shifted HBC620 (Pepper620). When compared to other fluorophore-aptamer pairs, Pepper530 demonstrated 9 and 11-fold higher signal than Broccoli and Corn in mammalian cells. mRNA tandem-labeled with 4xPepper and 4xMS2 revealed that Pepper530 outperforms MCP-mCherry, and Pepper620 outperforms MCP-GFP in signal-to-noise. It was also shown that Peppers could be used to image a wide range of different classes of bulk RNAs.
A new class of designed fluorophore-aptamer pairs is emerging that takes cues from recent advances in the field of fluorescent dyes (Lukinavičius et al., 2013; Zheng et al., 2019). Instead of exploiting rotational anisotropy to promote turn-on, lactonization (and ring opening) of xanthene-based probes can control fluorescence. Jäschke and colleagues demonstrated that an aptamer can be selected to promote delactonization (and thus fluorophore turn-on) (Wirth et al., 2019). While this technology has yet to be demonstrated in live mammalian cells, it has the potential to overcome limitations (photostability and brightness) of the other designer fluorophore-aptamer pairs.
Leveraging Modern Fluorophores for RNA Imaging
An alternative approach to imaging RNA with small-molecule fluorophores uses conventional fluorophores to increase the brightness and photostability of the tool. To achieve fluorescence turn-on, these methods quench the fluorescence of the unbound probes through pendant quenching moieties (Figure 4). An advantage of such techniques is the flexibility to incorporate widely-used fluorophores. These molecules benefit from years of vetting in cellular experiments, are well understood by the field, and are compatible with commonly used laser lines and emission filters.
Figure 4:
A) Tethered fluorophore-quencher complexes utilize fluorophore-quencher pairs that primarily undergo contact quenching. This enables an aptamer binding event to reduce quenching and turn on fluorescence. B) Fluorophore-quencher pairs are constructed with modern fluorophores linked (usually via PEG) to a quenching moiety.
Initial reports of RNA aptamers selected to bind small molecule fluorophores came as early as 1998 when Szostak developed an aptamer to bind sulforhodamine B (SRB-2, KD=300 nM) (Holeman et al., 1998). Remarkably, SRB-2 is being incorporated into a few of the newest RNA probes due to its robust binding without a G-quadruplex. Jäschke and colleagues adapted this fluorophore-aptamer pair by engineering a mechanism of fluorophore turn-on (Sunbul and Jäschke, 2013). To ensure that the fluorophore was dark in solution, they appended dinitroaniline (DN), a commonly used quenching motif. In the presence of SRB-2, fluorescence increased 100-fold in vitro (KD=1.3 mM). This probe was estimated to be three-times brighter than Spinach, though cellular imaging was limited to bacteria. Since their initial report, Jäschke and colleagues have further improved the fluorophore-quencher probe by swapping in tetramethyl rhodamine (TMR), which increased binding affinity (KD=35 nM) (Sunbul and Jäschke, 2018). With TMR-DN, they were able to image 5S ribosomal RNA (the most abundant cellular RNA) in mammalian cells. They have also recently reported (via preprint) an optimized SRB-2 aptamer termed RhoBAST, with fast dye association and dissociation kinetics that further improved performance and enabled single molecule localization microscopy (Sunbul et al., 2020).
Another tool that incorporates sulphorhodamine B and the SRB-2 aptamer exploited self-quenching that arises from the interaction of two identical fluorophores (Bouhedda et al., 2020). Developed by Ryckelynck and colleagues, Gemini-561 consists of two sulphorhodamine B moieties linked by lysine. Through a combination of SELEX and fluorogenicity selection in microfluidics, the authors identified a dimer of SRB-2 that bound Gemini-561 with a KD of 73 nM and an in vitro fluorescence turn-on of 13-fold. They termed this aptamer o-Coral. This probe-aptamer pair was effective in imaging mRNA, as well as 5S ribosomal RNA, with only a single copy of the tag, though single molecule detection in cells was not reported. In comparison to Broccoli, Corn, and Mango, Gemini-561 was brighter and more photostable.
An alternative approach for probes that incorporate a pendant quencher is to utilize the quencher as the binding motif. Uesugi and colleagues chose Black Hole Quencher (BHQ1) to quench Cy3 in their fluorophore-quencher pair (Murata et al., 2011; Sato et al., 2015). They developed an aptamer that binds BHQ1 to preclude contact quenching with Cy3. Interestingly, this probe operates in trans, by hybridizing with endogenous target RNAs of interest in live cells. Using their probe, the authors labelled mRNA granules in live cells. They also developed an aptamer for dinitrobenzene and used it in tandem with their BHQ1 probe to label two mRNAs simultaneously (Yatsuzuka et al., 2018). Unfortunately, this system has not been compared with others in the field, so it is difficult to judge its performance.
With localization and turn-on handled by the quencher, the researcher is free to use the fluorophore that best fits their application. We recently reported a multicolor system (Riboglow) that utilized vitamin B12 (cobalamin, Cbl) as the quenching and localization motif (Braselmann et al., 2018). It was previously reported that Cbl can quench various fluorophores (Lee and Grissom, 2009). We recognized that cobalamin-binding riboswitches are prevalent in nature, and hypothesized that we could use a natural aptamer as our probe binder. Naturally evolved aptamers should have increased biostability due to fast and robust folding. Covalent conjugation of Cbl (Chromiński and Gryko, 2013) to a variety of fluorophores resulted in quenching across the visible spectrum, which was relieved upon binding RNA. Our best probe (Cbl-5xPEG-Atto590) bound to its RNA tag with a KD of 34 nM and exhibited a fluorescence induction of 5-fold. A second probe, Cbl-Cy5, also performed well in live-cell assays. Our tool outperformed MS2 and Broccoli in a head-to-head comparison for detection of RNA stress granules in cells. We were also able to observe the non-coding snRNA U1 localized to U-bodies with a single copy of the Riboglow tag – a task not possible with MS2 because of its large size (relative to snRNA). Preliminary data also show that Riboglow is compatible with detection of single RNA transcripts (Braselmann et al., 2019).
Hybridization-based probes to detect endogenous RNA
Molecular beacons are oligonucleotide-based probes with a stem loop where the sequence in the loop is designed to hybridize with endogenous RNA of interest, and the termini are modified with a fluorophore and a quencher (Ma et al., 2017). These probes are dark in the unbound state because the quencher is held in proximity to the fluorophore, but the beacon unzips and fluorescence turn-on occurs upon hybridization with the target RNA. The first molecular beacon was developed by Tyagi and Kramer in 1995 utilizing an EDANS fluorophore quenched by DABCYL (Tyagi and Kramer, 1996). The technology was adapted for use in live cells in 2003 by creating nuclease resistant beacons and Tyagi and colleagues were able to track mRNA in Drosophila oocytes (Bratu et al., 2003). Recent work has shown that such probes can also be used to track single RNAs in live neurons (Turner-Bridger et al., 2018). The authors targeted two different beacons to two unique sites on β-actin mRNA and tracked the distribution of transcripts in axons.
APPLICATION OF TOOLS FOR DETECTION AND TRACKING OF RNA IN LIVE CELLS
Development and improvement of RNA imaging tools requires researchers to assess feasibility of detecting diverse RNA species in live cells using robust metrics for validation. While biophysical features such as brightness, quantum yield, and fluorescence turn-on are straightforward to measure in vitro, it is critical to quantify RNA detection efficacy in live cells as well. Metrics may include signal-to-noise ratio of RNA versus cellular background, sensitivity and efficacy of detecting single RNA molecules, and systematic assessment of optics and microscopy modes. Quantitative comparison with existing platforms or a previous generation of the system of interest can be especially valuable to evaluate a new tagging platform. Several recently developed RNA imaging platforms include such an assessment (Braselmann et al., 2018; Chen et al., 2019). Imaging in live cells will likely reveal that different tools possess different strengths, but no one tool is likely to emerge as a “silver bullet”. Rather, encompassing the breadth of questions in RNA biology will require a toolbox filled with diverse tools, each with unique strengths.
Below we highlight several applications of tagging and tracking RNAs in live cells. mRNAs tagged with the MS2 platform are the most widely studied RNA species (Buxbaum et al., 2014; Tutucci et al., 2018a), perhaps because a critical depth of knowledge about mRNA function and dynamics is available to ensure that tagging does not alter function. While an overview of the life cycle of mRNA biology (reviewed recently (Tutucci et al., 2018a)) is outside the scope of this review, several recently developed multiplexed assays that probe aspects of mRNA biology are discussed.
Visualizing mRNAs in diverse live cell systems has revealed that the subcellular localization of mRNAs is nonuniform, tightly regulated, and dynamic. A classic example of nonuniform localization of different types of mRNA was characterized for embryo development in Drosophila oocytes (Figure 5A, (Abbaszadeh and Gavis, 2016)), where mRNA dynamics were found to be linked to proper development (Weil et al., 2006; Zimyanin et al., 2008). More broadly, localization of mRNAs was found to be regulated by elements within the RNA sequence, leading to asymmetric mRNA distribution in space and time for localized protein translation (Eliscovich et al., 2013). For example, β-actin mRNA is recruited to focal adhesions at the leading edge of fibroblast cells (Figure 5B (Katz et al., 2012)). Localized protein translation is also critical in neurobiology (Holt et al., 2019). Visualization of mRNAs within live dendrites reveals directed movement (Figure 5C (Dynes and Steward, 2008)) and glutamate triggers mRNA recruitment for local protein translation in dendritic spines (Yoon et al., 2016). Finally, a recent study demonstrated that mRNAs can be transferred between cells through nanotubes (Figure 5D (Haimovich et al., 2017)).
Figure 5: Tracking mRNAs in live cells reveals nonuniform distributions in diverse cell types with implications for function.
(A) Two mRNAs were visualized in a live Drosophila oocyte with MS2/MCP and PP7/PCP. bcd mRNA (MS2/MCP-GFP, green) localizes to the anterior of the oocyte, whereas osk mRNA (PCP/PP7-mCherry, red) localizes to the posterior (arrow: autofluorescent yolk granules). Scale bar = 15 µm. Reprinted from (Abbaszadeh and Gavis, 2016) with permission from Elsevier. (B) Endogenous β-actin mRNA (24xMS2, MCP-TagRFPt, red) localizes to the leading edge in mouse embryonic fibroblasts. Free GFP (green) served as a cytoplasmic marker. Scale bar = 10 µm. Reprinted from (Katz et al., 2012), copyright Cold Spring Harbor Laboratory Press. (C) Movement of mRNA particles in dendrites of rat neurons is dynamic and indicative of targeted, bidirectional transport. The reporter 6xMS2-DsRed-3’UTR of Arc mRNA was cotransfected with MCP-GFP into rat cortical neurons to label mRNA (grey scale, upper panel). Intensely fluorescent spots correspond to gold beads used for biolistic transfection. Bottom panels: Kymographs of regions indicated by square and rectangular boxes on the top panel. Blue arrow heads: Moving mRNA particles (5 s intervals). Scale bar = 20 µm (top panel), 2 µm (inserts). Reprinted from (Dynes and Steward, 2008) with permission from Wiley. (D) Endogenous β-actin mRNA (24xMS2/tdMCP-GFP, arrow) is transferred between two mouse embryonic fibroblast cells via passage through nanotubes (right panel = zoom in of yellow box in the left panel). tdMCP-GFP was produced in the donor cell (bottom right), but not the acceptor cell (top left). Both cells are labeled with membrane targeted TagRFP-T (magenta). Scale bar = 5 µm. Reprinted from (Haimovich et al., 2017).
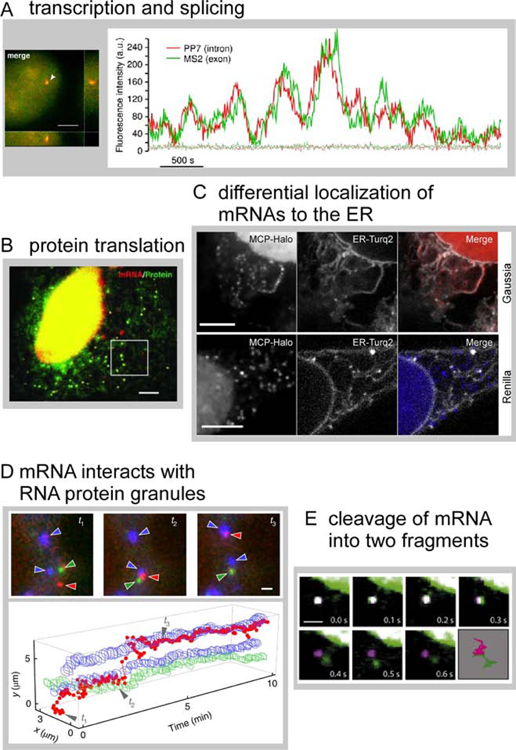
Because the MS2 platform was continuously improved to label mRNAs, robust detection and tracking of single mRNA species is now routinely available, yielding sophisticated assays that cover the entire life cycle of the mRNA. Processes such as transcription bursting and transcription activity during the cell cycle have been quantified (Yunger et al., 2010), as well as pre-mRNA splicing, which is intimately linked to transcription (Coulon et al., 2014; Martin et al., 2013). To image splicing, two orthogonal mRNA tags were used to label an intron and the 3’ untranslated region (UTR) of the same mRNA, yielding two-color spots for the transcript prior to splicing, and loss of the labeled intron upon co-transcriptional splicing (Figure 6A,(Coulon et al., 2014)). A later step in the mRNA life cycle, protein translation, was also achieved on the single mRNA level (Morisaki et al., 2016; Pichon et al., 2016; Wang et al., 2016; Wu et al., 2016; Yan et al., 2016). The mRNA was labeled with MS2, and the protein emerging from the ribosome was labeled with a fluorescent affinity probe that binds an epitope in the nascent chain, yielding dual-colored translation spots (Figure 6B,(Wu et al., 2016)). Together, robust fluorescent labeling and tracking of single mRNAs and multiplexing with additional markers yields a toolkit for insights in mRNA biology and further refinement and expansion of these tools (Boersma et al., 2019; Zhao et al., 2019) promise sophisticated insights into diverse biological questions.
Figure 6: Single mRNA tracking enables insights into mRNA life cycle processes.
(A) β-globin mRNA (24xPP7 in the intron, 24xMS2 in the 3’UTR) shows diffraction limited spots that are both green and red in the nucleus (left panel, arrow, scale bar = 4 µm). Fluorescence signal at the transcription site was recorded over time; fluctuations indicate stochastic transcription events. Reprinted from (Coulon et al., 2014). (B) Epithelial cells stably express the FLAG-SINAPS reporter to monitor protein translation at the single mRNA level. mRNA is labelled with 24xMS2 (red puncta). The encoded protein includes 24xSunTag at the N-terminus which binds scFV-GFP (green puncta represent single translation sites). Scale bar = 5 µm. From (Wu et al., 2016), reprinted with permission from AAAS. (C) NLS-MCP-Halo was produced to label mRNAs and ER-Turq2 is a marker for the ER. Gaussia mRNA (a protein trafficked through the secretory pathway) localizes to the ER, but only a small fraction of Renilla mRNA (a cytosolic protein) localizes to the ER. Scale bars = 5 µm. Reprinted from (Voigt et al., 2017) with permission from Elsevier. (D) Characterization of mRNA interaction dynamics with stress granules (SG) and P-bodies (PB). KDM5B-24xMS2 mRNA was labeled with NLS-MCP-Halo-JF646 (red). SGs and PBs were marked by fluorescent marker proteins (GFP-G3BP1, blue, and mRFP-DCP1a, green, respectively). Shown is a mRNA trajectory between two SGs and a PB over time (bottom panel). Three selected time points are indicated and cropped images are shown (top panel). Scale bar = 1 µm. Reprinted by permission from Springer Nature Customer Service Centre GmbH: (Moon et al., 2019). (E) The ‘TREAT siRNA reporter’ includes the Renilla coding sequence, 24xPP7 in the 3’UTR, followed by a siRNA site, and 24xMS2. Cells expressing the reporter also produce PCP-GFP (green) and MCP-Halo (magenta). Intact mRNA is dual colored (white) and was observed for several frames. Slicing of the RNA at the siRNA sites spatially separates PP7 and MS2 tags. Scale bar = 1 µm. Reprinted from (Horvathova et al., 2017) with permission from Elsevier.
The establishment of single mRNA detection and tracking assays has revealed insights into underlying RNA biology (Chen et al., 2020; Lyon et al., 2019). Localization of mRNA encoding cytosolic versus secretory proteins to the endoplasmic reticulum (ER) revealed that a small fraction of cytosolic proteins localized to the ER and was translated (Figure 6C, (Voigt et al., 2017)). Results were validated by the single mRNA translation assay mentioned above (Tanenbaum et al., 2014). The fate of mRNAs once they exit translation and interact with RNA-protein (RNP) granules (stress granules and P-bodies) was investigated on the single mRNA level (Moon et al., 2019). Multiplexing live cell fluorescence assays to simultaneously detect the mRNA and two different RNPs made it possible to visualize recruitment of mRNA to stress granules and P-bodies (Figure 6D, (Moon et al., 2019)) and revealed rapid and bidirectional movement of mRNA between the two granules. Finally, an assay to quantify mRNA degradation was developed (Figure 6E, (Horvathova et al., 2017)). Single mRNA assays to visualize aspects of the mRNA life cycle provide unprecedented opportunities for future directions, such as expanding to other cell types like neurons, exploring diverse mRNA species, and comparing healthy and disease states.
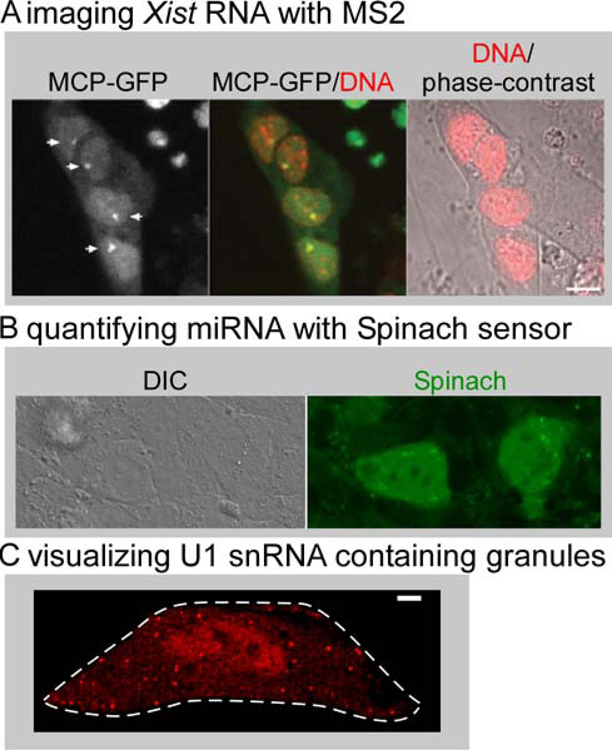
Much less is known about the biology of ncRNAs and there are only a few examples of visualization of ncRNAs with fluorescent tags. We highlight several exciting studies to inspire further development of creative assays for insights in ncRNA biology. The Xist ncRNA is involved in inactivation of one X-chromosome in development and spreads over the chromosome (Borsani et al., 1991; Brown et al., 1991). Visualization of MS2-tagged Xist in embryonic stem cells revealed dynamics of Xist spreading on the chromosome during cell cycle. (Figure 7A, (Ng et al., 2011)). More recently, the Bgl stem-loop system was successfully used to visualize live Xist dynamics (Dossin et al., 2020). microRNAs (miRNA) are a class of ncRNAs that regulate gene expression (O’Brien et al., 2018). Approaches to visualize miRNA dynamics live include microinjecting fluorescent versions of the miRNA of interest for live tracking on the single RNA level (Pitchiaya et al., 2017, 2012). miRNA dynamics were also visualized using genetically encoded fluorescent sensors where the Spinach system was engineered as a turn-on platform that quantifies the presence of miRNA (Figure 7B, (Huang et al., 2017)). We have recently used the Riboglow platform to label and track the non-coding U1 snRNA and visualized recruitment to U-bodies live (Figure 7C, (Braselmann et al., 2018)). While assays to label and track ncRNAs are much less widespread than their mRNA counterparts, much insight into RNA biology can be gained by exploring live tracking systems for these different RNA species. As diverse platforms to visualize RNAs with orthogonal strengths become available, we anticipate that a robust suite of tools for imaging ncRNAs will be established.
Figure 7: Live cell tracking of non-coding RNAs.
(A) The 3’end of Xist was tagged with 24xMS2 stem and stably integrated in mouse embryonic stem cells under control of a Doxycyclin-inducible promoter. Clusters of Xist RNA labeled with MCP-GFP 12 hours after induction are indicated by arrows (left panel). DNA is stained with Hoechst (red). Scale bar = 10 µm. Reproduced from (Ng et al., 2011). (B) The FASTmiR122 sensor consists of a modified Spinach RNA sequence that binds miR122, resulting in turn-on green fluorescence from the Spinach sensor in Huh7 cells (right panel). Reproduced from (Huang et al., 2017) by permission of Oxford University Press / RNA Society. (C) U1 snRNA was tagged at the 5’end with one copy of a minimal Riboglow RNA tag in HeLa cells and Cbl-5xPEG-ATTO 590 was added to live cells. Cytosolic U-bodies were induced by treatment with Thapsigargin. Scale bar = 5 µm. Reproduced from (Braselmann et al., 2018).
OUTLOOK
Looking forward, there are exciting opportunities to expand the breadth of RNA visualization of living systems. The MS2 and PP7 systems have been used to monitor transcription and chromosome-chromosome dynamics in live Drosophila embryos (Garcia et al., 2013; Lim et al., 2018), demonstrating that imaging tools can be used in tissues and organisms. The Schuman lab recently showed that processing of a fluorescent miRNA can report on spatially restricted miRNA maturation in live neurons (Sambandan et al., 2017), indicating that processing of the miRNA itself may be exploited to develop sensors of this important class of ncRNAs. Visualizing RNA dynamics in live mammalian cells provides critical insight into the biology of RNA and relies on the availability of robust and versatile tools to label and track RNA species. The development of diverse tools with complementary strengths is key. Benchmarking these systems to define features including cellular contrast, applicability of single RNA detection, and the potential for perturbing RNA biology will be a critical component of making these new tools broadly accessible within the community. Defining an RNA imaging toolbox rather than searching for a ‘silver bullet’ reflects the true diversity in RNA species and their complex cellular dynamics.
Significance:
While RNA has long been known for its role as an intermediary between DNA and proteins (mRNA), a structural component of ribosomes (rRNA) and a workhorse for protein synthesis (tRNA), it is now emerging as one of the central regulators of cellular processes. This is due in part to the discovery of myriad diverse kinds of non-coding RNAs, as well as increased recognition of the ways in which biochemical processing of mRNA fundamentally regulates gene expression. This rich and evolving landscaping of RNA regulation of cell function has led to intense interest in developing approaches for visualizing RNA in live cells to illuminate RNA biology. Historically, the field has suffered from a lack of tools capable of detecting RNA with the appropriate specificity and sensitivity. But clever approaches for exploiting RNA binding proteins, in vitro RNA evolution, and chemical biology have resulted in significant advances in RNA visualization tools in recent years. Here, we review tools that have been developed to tag and track RNA in living cells, with an emphasis on tools that have been applied to mammalian cells.
Highlights.
Overview of current tools for tagging and tracking RNA in live mammalian cells.
RNA imaging toolkit involves protein-based and small-molecule tools.
Application of tools and development of assays to illuminate RNA biology.
Examples are provided for both mRNA and ncRNA.
Acknowledgements
We would like to thank the following sources of financial support K99-GM127752 to EB, F32-GM131666 to CR, T32-GM065103 training grant to EMR, R01-GM133184 and DP1-GM114863 to AEP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
Amy E Palmer and Esther Braselmann are listed as inventors on a patent (U.S. Application No. 16/526,835, Compositions and Methods for Tagging Ribonucleic Acids) The patent was filed July 31, 2019; it has not been awarded yet. One technology mentioned in the article (Riboglow) is mentioned in this patent.
References
- Abbaszadeh EK,Gavis ER, 2016. Fixed and live visualization of RNAs in Drosophila oocytes and embryos. Methods 98, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO,Gootenberg JS,Konermann S,Joung J,Slaymaker IM,Cox DBT,Shmakov S,Makarova KS,Semenova E,Minakhin L,et al. , 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RJ,Xia T,Ren J,Takahashi TT,Roberts RW, 2002. Designed Arginine-Rich RNA-Binding Peptides with Picomolar Affinity. J. Am. Chem. Soc 124, 10966–10967. [DOI] [PubMed] [Google Scholar]
- Autour A,Jeng SCY,Cawte AD,Abdolahzadeh A,Galli A,Panchapakesan SSS,Rueda D,Ryckelynck M,Unrau PJ, 2018. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun 9, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E,Chartrand P,Schaefer M,Shenoy SM,Singer RH,Long RM, 1998. Localization of ASH1 mRNA Particles in Living Yeast. Mol. Cell 2, 437–445. [DOI] [PubMed] [Google Scholar]
- Boersma S,Khuperkar D,Verhagen BMP,Sonneveld S,Grimm JB,Lavis LD,Tanenbaum ME,Boersma S,Khuperkar D,Verhagen BMP,et al. , 2019. Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding Resource Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding. Cell 178, 458–472.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G,Tonlorenzi R,Simmler MC,Dandolo L,Arnaud D,Capra V,Grompe M,Pizzuti A,Muzny D,Lawrence C,et al. , 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. [DOI] [PubMed] [Google Scholar]
- Bouhedda F,Fam KT,Collot M,Autour A,Marzi S,Klymchenko A,Ryckelynck M, 2020. A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells. Nat. Chem. Biol 16, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselmann E,Stasevich TJ,Lyon K,Batey RT,Palmer AE, 2019. Detection and quantification of single mRNA dynamics with the Riboglow fluorescent RNA tag. Biorxiv 701649 doi: 10.1101/701649. [DOI]
- Braselmann E,Wierzba A,Polaski JT,Chromiński M,Holmes ZE,Hung ST,Batan D,Wheeler JR,Parker R,Jimenez R,et al. , 2018. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol 14, 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu DP,Cha B-J,Mhlanga MM,Kramer FR,Tyagi S, 2003. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. U. S. A 100, 13308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ,Ballabio A,Rupert JL,Lafreniere RG,Grompe M,Tonlorenzi R,Willard HF, 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR,Haimovich G,Singer RH, 2014. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol 16, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J,Cameron V,De Haseth PL,Uhlenbeck OC, 1983. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry 22, 2601–2610. [DOI] [PubMed] [Google Scholar]
- Cawte AD,Unrau PJ,Rueda DS, 2020. Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nat. Commun 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR,Steitz JA, 2014. The Noncoding RNA Revolution — Trashing Old Rules to Forge New Ones. Cell 157, 77–94. [DOI] [PubMed] [Google Scholar]
- Chao J. a,Patskovsky Y,Almo SC,Singer RH, 2008. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol 15, 103–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S,Garcia-Mena J,Devito J,Wolska K,Das A, 1995. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage A. Proc. Natl. Acad. Sci 92, 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Jianbo,Liu Y,Wu B,Nikolaitchik OA,Mohan PR,Chen Jiji,Pathak VK,Hu WS, 2020. Visualizing the translation and packaging of HIV-1 full-length RNA. Proc. Natl. Acad. Sci 117, 6145–6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X,Zhang D,Su N,Bao B,Xie X,Zuo F,Yang L,Wang H,Jiang L,Lin Q,et al. , 2019. Visualizing RNA Dynamics in Live Cells with Bright and Stable Fluorescent RNAs. Nat. Biotechnol 37, 1287–1293. [DOI] [PubMed] [Google Scholar]
- Cheong C,Tanaka Hall T, 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci 103, 13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromiński M,Gryko D, 2013. “Clickable” vitamin B12 derivative. Chem. - A Eur. J 19, 5141–8. [DOI] [PubMed] [Google Scholar]
- Coulon A,Ferguson ML,Turris V. De,Palangat M,Chow CC,Larson DR, 2014. Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife 3, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranfill PJ,Sell BR,Baird MA,Allen JR,Lavagnino Z,de Gruiter HM,Kremers G-J,Davidson MW,Ustione A,Piston DW, 2016. Quantitative assessment of fluorescent proteins. Nat. Methods 13, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N,Ellenberg J, 2007. λN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods 4, 633–636. [DOI] [PubMed] [Google Scholar]
- Darzacq X,Shav-Tal Y,De Turris V,Brody Y,Shenoy SM,Phair RD,Singer RH, 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol 14, 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S,Moon HC,Singer RH,Park HY, 2018. A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Sci. Adv 4, eaar3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C,Ernberg I,Gait MJ,Green SM,Heaphy S,Karn J,Lowe AD,Singh M,Skinner MA,Valerio R, 1989. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci 86, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S,Davis CA,Merkel A,Dobin A,Lassmann T,Mortazavi A,Tanzer A,Lagarde J,Lin W,Schlesinger F,et al. , 2012. Landscape of transcription in human cells. Nature 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgosheina EV,Jeng SCY,Panchapakesan SSS,Cojocaru R,Chen PSK,Wilson PD,Hawkins N,Wiggins PA,Unrau PJ, 2014. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem. Biol 9, 2412–2420. [DOI] [PubMed] [Google Scholar]
- Dossin F,Pinheiro I,Żylicz JJ,Roensch J,Collombet S,Le Saux A,Chelmicki T,Attia M,Kapoor V,Zhan Y,et al. , 2020. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578, 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL,Steward O, 2008. Dynamics of Bidirectional Transport of Arc mRNA in Neuronal Dendrites. J. Comp. Neurol 500, 433–447. [DOI] [PubMed] [Google Scholar]
- Eliscovich C,Buxbaum AR,Katz ZB,Singer RH, 2013. mRNA on the move: The road to its biological destiny. J. Biol. Chem 288, 20361–20368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS,Moon JD,Svensen N,Jaffrey SR, 2014. Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc 136, 16299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D,Accornero N,Lavoie B,Shenoy SM,Blanchard JM,Singer RH,Bertrand E, 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol 13, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG,Tikhonov M,Lin A,Gregor T, 2013. Quantitative Imaging of Transcription in Living Drosophila Embryos Links Polymerase Activity to Patterning. Curr. Biol 23, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF,Parker R, 2016. Ubiquitous accumulation of 3′ mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays. RNA 22, 657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golmohammadi R,Valegard K,Fridborg K,Liljas L, 1993. The Refined Structure of Bacteriophage MS2 at 2.8A Resolution. J. Mol. Biol 234, 620–639. [DOI] [PubMed] [Google Scholar]
- Grimm JB,English BP,Chen J,Slaughter JP,Zhang Z,Revyakin A,Patel R,Macklin JJ,Normanno D,Singer RH,et al. , 2015. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB,Muthusamy AK,Liang Y,Brown TA,Lemon WC,Patel R,Lu R,Macklin JJ,Keller PJ,Ji N,et al. , 2017. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 14, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross LA,Baird GS,Hoffman RC,Baldridge KK,Tsien RY, 2000. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci 97, 11990–11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S,Esteller M, 2015. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem. Sci 40, 248–256. [DOI] [PubMed] [Google Scholar]
- Guo JU,Bartel DP, 2016. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353, aaf5371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G,Ecker CM,Dunagin MC,Eggan E,Raj A,Gerst JE,Singer RH, 2017. Intercellular mRNA trafficking via membrane nanotube-like extensions in mammalian cells. Proc. Natl. Acad. Sci 114, E9873–E9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S,Sidler CL,Azzalin CM,Weis K, 2017. Stem–loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. RNA 23, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeman LA,Robinson SL,Szostak JW,Wilson C, 1998. Isolation and characterization of fluorophore-binding RNA aptamers. Fold. Des 3, 423–431. [DOI] [PubMed] [Google Scholar]
- Holt CE,Martin KC,Schuman EM, 2019. Local translation in neurons: visualization and function. Nat. Struct. Mol. Biol 26, 557–566. [DOI] [PubMed] [Google Scholar]
- Horvathova I,Voigt F,Kotrys AV,Zhan Y,Artus-Revel CG,Eglinger J,Stadler MB,Giorgetti L,Chao JA, 2017. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell 68, 615–625. [DOI] [PubMed] [Google Scholar]
- Huang K,Doyle F,Wurz ZE,Tenenbaum SA,Hammond RK,Caplan JL,Meyers BC, 2017. FASTmiR: an RNA-based sensor for in vitro quantification and live-cell localization of small RNAs. Nucleic Acids Res 45, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng SCY,Chan HHY,Booy EP,McKenna SA,Unrau PJ, 2016. Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers. RNA 22, 1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson HE,Liljas L,Uhlenbeck OC, 1997. RNA Recognition by the MS2 Phage Coat Protein. Semin. Virol 8, 176–185. [Google Scholar]
- Katz ZB,Wells AL,Park HY,Wu B,Shenoy SM,Singer RH, 2012. β-actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev 26, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH,Vieira M,Kim H,Kesawat MS,Park HY, 2019. MS2 Labeling of Endogenous Beta-Actin mRNA Does Not Result in Stabilization of Degradation Intermediates. Mol. Cells 42, 356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S,Katayama Y,Schmid M,Burkacky O,Bruchle C,Lamb DC,Jansen R-P, 2008. Simultaneous Transport of Different Localized mRNA Species Revealed by Live-Cell Imaging. Traffic 9, 1256–1267. [DOI] [PubMed] [Google Scholar]
- Larson DR,Zenklusen D,Wu B,Chao J. a,Singer RH, 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD,Raines RT, 2014. Bright Building Blocks for Chemical Biology. ACS Chem. Biol 9, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M,Grissom CB, 2009. Design, synthesis, and characterization of fluorescent cobalamin analogues with high quantum efficiencies. Org. Lett 11, 2499–502. [DOI] [PubMed] [Google Scholar]
- Li X,Kim H,Litke JL,Wu J,Jaffrey SR, 2020. Fluorophore-Promoted RNA Folding and Photostability Enables Imaging of Single Broccoli-Tagged mRNAs in Live Mammalian Cells. Angew. Chemie Int. Ed 59, 4511–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B,Heist T,Levine M,Fukaya T, 2018. Visualization of Transvection in Living Drosophila Embryos. Mol. Cell 70, 287–296.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F,Downey TP,Peabody DS, 2001. Translational Repression and Specific RNA Binding by the Coat Protein of the Pseudomonas Phage PP7. J. Biol. Chem 276, 22507–22513. [DOI] [PubMed] [Google Scholar]
- Lim F,Peabody DS, 2002. RNA recognition site of PP7 coat protein. Nucleic Acids Res 30, 4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T,Czaplinski K,Darzacq X,Shav-Tal Y,Wells AL,Chao J. a,Park HY,de Turris V,Lopez-Jones M,Singer RH, 2011. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G,Umezawa K,Olivier N,Honigmann A,Yang G,Plass T,Mueller V,Reymond L,Corrêa IR Jr,Luo Z-G,et al. , 2013. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem 5, 132–139. [DOI] [PubMed] [Google Scholar]
- Lyon K,Aguilera LU,Morisaki T,Munsky B,Stasevich TJ, 2019. Live-Cell Single RNA Imaging Reveals Bursts of Translational Frameshifting. Mol. Cell 75, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z,Wu X,Krueger CJ,Chen AK, 2017. Engineering Novel Molecular Beacon Constructs to Study Intracellular RNA Dynamics and Localization. Genomics. Proteomics Bioinformatics, RNA Epigenetics (II) 15, 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RMM,Rino JJ,Carvalho C,Kirchhausen T,Carmo-Fonseca M, 2013. Live-Cell Visualization of Pre-mRNA Splicing with Single-Molecule Sensitivity. Cell Rep. 4, 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG,Wang Z, 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol 15, 108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL,Morisaki T,Khong A,Lyon K,Parker R,Stasevich TJ, 2019. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol 21, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T,Lyon K,DeLuca KF,DeLuca JG,English BP,Zhang Z,Lavis LD,Grimm JB,Viswanathan S,Looger LL,et al. , 2016. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429. [DOI] [PubMed] [Google Scholar]
- Murata A,Sato S,Kawazoe Y,Uesugi M, 2011. Small-molecule fluorescent probes for specific RNA targets. Chem. Commun. (Camb) 47, 4712–4. [DOI] [PubMed] [Google Scholar]
- Ng K,Daigle N,Bancaud A,Ohhata T,Humphreys P,Walker R,Ellenberg J,Wutz A, 2011. A system for imaging the regulatory noncoding Xist RNA in living mouse embryonic stem cells. Mol. Biol. Cell 22, 2634–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J,Hayder H,Zayed Y,Peng C, 2018. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn RCL,Garde G,Dayhuff T,Atkins JF,Van Duin J, 1995. Nucleotide sequence of a single-stranded RNA phage from Pseudomonas aeruginosa:Kinship to coliphages and conservation of regulatory RNA structures. Virology 206, 611–625. [DOI] [PubMed] [Google Scholar]
- Ozawa T,Natori Y,Sato M,Umezawa Y, 2007. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods 4, 413–419. [DOI] [PubMed] [Google Scholar]
- Paige JS,Wu KY,Jaffrey SR, 2011. RNA mimics of green fluorescent protein. Science 333, 642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY,Lim H,Yoon YJ,Follenzi A,Nwokafor C,Lopez-Jones M,Meng X,Singer RH, 2014. Visualization of Dynamics of Single Endogenous mRNA Labeled in Live Mouse. Science 343, 422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS, 1993. The RNA binding site of bacteriophage MS2 coat protein. EMBO J 12, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon X,Bastide A,Safieddine A,Chouaib R,Samacoits A,Basyuk E,Peter M,Mueller F,Bertrand E, 2016. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol 214, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchiaya S,Androsavich JR,Walter NG, 2012. Intracellular single molecule microscopy reveals two kinetically distinct pathways for microRNA assembly. EMBO Rep 13, 709–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchiaya S,Heinicke LA,Park JI,Cameron EL,Walter NG,Pitchiaya S,Heinicke LA,Park JI,Cameron EL,Walter NG, 2017. Resolving Subcellular miRNA Trafficking and Turnover at Single-Molecule Resolution. CellReports 19, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW,Parker R, 2016. Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV,Lerit DA, 2018. RNA localization regulates diverse and dynamic cellular processes. Traffic 19, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan S,Akbalik G,Kochen L,Rinne J,Kahlstatt J,Glock C,Tushev G,Alvarez-Castelao B,Heckel A,Schuman EM, 2017. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science (80-. ) 355, 634–637. [DOI] [PubMed] [Google Scholar]
- Sato SI,Watanabe M,Katsuda Y,Murata A,Wang DO,Uesugi M, 2015. Live-cell imaging of endogenous mRNAs with a small molecule. Angew. Chem. Int. Ed. Engl 54, 1855–1858. [DOI] [PubMed] [Google Scholar]
- Shay JW,Wright WE, 2019. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet 20, 299–309. [DOI] [PubMed] [Google Scholar]
- Smith CA,Crotty S,Harada Y,Frankel AD, 1998. Altering the Context of an RNA Bulge Switches the Binding Specificities of Two Viral Tat Proteins. Biochemistry 37, 10808–14. [DOI] [PubMed] [Google Scholar]
- Song W,Filonov GS,Kim H,Hirsch M,Li X,Moon JD,Jaffrey SR, 2017. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol 13, 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunbul M,Jäschke A, 2018. SRB-2: a promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res 46, e110–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunbul M,Jäschke A, 2013. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew. Chem. Int. Ed. Engl 52, 13401–4. [DOI] [PubMed] [Google Scholar]
- Sunbul M,Lackner J,Martin A,Englert D,Hacene B,Nienhaus K,Nienhaus GU,Jäschke A, 2020. RhoBAST - a rhodamine-binding aptamer for super-resolution RNA imaging. bioRxiv 2020.03.12.988782 doi: 10.1101/2020.03.12.988782. [DOI] [PubMed]
- Tanenbaum ME,Gilbert LA,Qi LS,Weissman JS,Vale RD, 2014. A protein tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tars K,Fridborg K,Bundule M,Liljas L, 2000. The Three-Dimensional Structure of Bacteriophage PP7 from Pseudomonas aeruginosa at 3.7-Å Resolution. Virology 272, 331–337. [DOI] [PubMed] [Google Scholar]
- Turner-Bridger B,Jakobs M,Muresan L,Wong HH-W,Franze K,Harris WA,Holt CE, 2018. Single-molecule analysis of endogenous β-actin mRNA trafficking reveals a mechanism for compartmentalized mRNA localization in axons. Proc. Natl. Acad. Sci 115, E9697–E9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutucci E,Livingston NM,Singer RH,Wu B, 2018a. Imaging mRNA In Vivo, from Birth to Death. Annu. Rev. Biophys May 20, 85–106. [DOI] [PubMed]
- Tutucci E,Vera M,Biswas J,Garcia J,Parker R,Singer RH, 2018b. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods 15, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S,Kramer FR, 1996. Molecular Beacons: Probes that Fluoresce upon Hybridization. Nat. Biotechnol 14, 303–308. [DOI] [PubMed] [Google Scholar]
- Valegård K,Murray JB,Stonehouse NJ,van den Worm S,Stockley PG,Liljas L, 1997. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J. Mol. Biol 270, 724–738. [DOI] [PubMed] [Google Scholar]
- Voigt F,Zhang H,Cui XA,Lee ES,Chao JA,Palazzo AF, 2017. Single-Molecule Quantification of Translation-Dependent Association of mRNAs with the Endoplasmic Reticulum. Cell Rep 21, 3740–3753. [DOI] [PubMed] [Google Scholar]
- Wang C,Han B,Zhou R,Zhuang X, 2016. Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 165, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H,Nakamura M,Abbott TR,Zhao D,Luo K,Yu C,Nguyen CM,Lo A,Daley TP,La Russa M,et al. , 2019. CRISPR-mediated live imaging of genome editing and transcription. Science (80-. ) 365, 1301–1305. [DOI] [PubMed] [Google Scholar]
- Wang X,Zamore PD,Hall TMT, 2001. Crystal Structure of a Pumilio Homology Domain. Mol. Cell 7, 855–865. [DOI] [PubMed] [Google Scholar]
- Weil TT,Forrest KM,Gavis ER, 2006. Localization of bicoid mRNA in Late Oocytes Is Maintained by Continual Active Transport. Dev. Cell 11, 251–262. [DOI] [PubMed] [Google Scholar]
- Wirth R,Gao P,Nienhaus GU,Sunbul M,Jäschke A, 2019. SiRA: A Silicon Rhodamine-Binding Aptamer for Live-Cell Super-Resolution RNA Imaging. J. Am. Chem. Soc 141, 7562–7571. [DOI] [PubMed] [Google Scholar]
- Wu B,Chao JA,Singer RH, 2012. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J 102, 2936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B,Chen J,Singer RH, 2014. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Sci. Rep 4, 3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B,Eliscovich C,Yoon YJ,Singer RH, 2016. Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B,Miskolci V,Sato H,Tutucci E,Kenworthy CA,Donnelly SK,Yoon YJ,Cox D,Singer RH,Hodgson L, 2015. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev 29, 876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J,Zaccara S,Khuperkar D,Kim H,Tanenbaum ME,Jaffrey SR, 2019. Live imaging of mRNA using RNA-stabilized fluorogenic proteins. Nat. Methods 16, 862–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T,Yoshimura H,Inaguma A,Ozawa T, 2011. Visualization of Nonengineered Single mRNAs in Living Cells Using Genetically Encoded Fluorescent Probes. Anal. Chem 83, 5708–5714. [DOI] [PubMed] [Google Scholar]
- Yan X,Hoek TA,Vale RD,Tanenbaum ME, 2016. Dynamics of Translation of Single mRNA Molecules in Vivo. Cell 165, 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L-Z,Wang Y,Li S-Q,Yao R-W,Luan P-F,Wu H,Carmichael GG,Chen L-L, 2019. Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems. Mol. Cell 76, 981–997.e7. [DOI] [PubMed] [Google Scholar]
- Yatsuzuka K,Sato S,Beverly Pe K,Katsuda Y,Takashima I,Watanabe M,Uesugi M, 2018. Live-cell imaging of multiple endogenous mRNAs permits the direct observation of RNA granule dynamics. Chem. Commun 54, 7151–7154. [DOI] [PubMed] [Google Scholar]
- Yoon YJ,Wu B,Buxbaum AR,Das S,Tsai A,English BP,Grimm JB,Lavis LD,Singer RH, 2016. Glutamate-induced RNA localization and translation in neurons. Proc. Natl. Acad. Sci 113, E6877–E6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H,Inaguma A,Yamada T,Ozawa T, 2012. Fluorescent Probes for Imaging Endogenous β-Actin mRNA in Living Cells Using Fluorescent Protein-Tagged Pumilio. ACS Chem. Biol 7, 999–1005. [DOI] [PubMed] [Google Scholar]
- Yunger S,Rosenfeld L,Garini Y,Shav-Tal Y, 2010. Single-allele analysis of transcription kinetics in living mammalian cells. Nat. Methods 7, 631–633. [DOI] [PubMed] [Google Scholar]
- Zhao N,Kamijo K,Fox PD,Oda H,Morisaki T,Kimura H,Stasevich TJ,Sato Y, 2019. A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q,Ayala AX,Chung I,Weigel AV,Ranjan A,Falco N,Grimm JB,Tkachuk AN,Wu C,Lippincott-Schwartz J,et al. , 2019. Rational Design of Fluorogenic and Spontaneously Blinking Labels for Super-Resolution Imaging. ACS Cent. Sci 5, 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL,Belaya K,Pecreaux J,Gilchrist MJ,Clark A,Davis I, 2008. In Vivo Imaging of oskar mRNA Transport Reveals the Mechanism of Posterior Localization. Cell 134, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]