Abstract
The inclusion of health-related traits, or functionally associated genetic markers, in pig breeding programs could contribute to produce more robust and disease resistant animals. The aim of the present work was to study the genetic determinism and genomic regions associated to global immunocompetence and health in a Duroc pig population. For this purpose, a set of 30 health-related traits covering immune (mainly innate), haematological, and stress parameters were measured in 432 healthy Duroc piglets aged 8 weeks. Moderate to high heritabilities were obtained for most traits and significant genetic correlations among them were observed. A genome wide association study pointed out 31 significantly associated SNPs at whole-genome level, located in six chromosomal regions on pig chromosomes SSC4, SSC6, SSC17 and SSCX, for IgG, γδ T-cells, C-reactive protein, lymphocytes phagocytic capacity, total number of lymphocytes, mean corpuscular volume and mean corpuscular haemoglobin. A total of 16 promising functionally-related candidate genes, including CRP, NFATC2, PRDX1, SLA, ST3GAL1, and VPS4A, have been proposed to explain the variation of immune and haematological traits. Our results enhance the knowledge of the genetic control of traits related with immunity and support the possibility of applying effective selection programs to improve immunocompetence in pigs.
Subject terms: Genetics, Animal breeding, Genetic association study, Genetic markers, Immunogenetics, Quantitative trait, Immunology, Immunogenetics, Innate immunity
Introduction
Over the last decades, the genetic selection in commercial pig breeds has greatly improved traits directly related with production performance1, while health-related traits have traditionally played a minor role in breeding programs. Nowadays, the emergence of antibiotic resistance and society demands for healthier livestock products and for more sustainable production systems2 represent new challenges for the pig production industry. Animal health is one of the most important contributors to productivity, profitability, and welfare, with multiple factors being involved in the maintenance of high health herd status such as co-infections of viral or bacterial pathogens, environmental stressors, and management practices. In the midst of strong investment for designing alternatives to antimicrobials in veterinary medicine3, the incorporation of health-related traits in pig breeding programs has become an emerging and challenging trend to produce more resilient, wellbeing and disease resistant pig populations.
Breeding approaches to improve animal robustness and disease resistance have been mainly focused on direct and indirect methods4. Direct methods usually rely on targeting the genetic resistance/susceptibility to specific diseases, requiring exposition to the infectious agents. This approach is expensive, time-consuming and information demanding. An indirect approach focused on the determination of the global immunocompetence of animals with no sign of infection has become a good alternative, but requires detailed knowledge of the different components of the immune system4,5. In this approach, immunity traits (ITs) may be considered as biologically relevant parameters to measure immunocompetence4. These traits may be classified into the two major components of the immune system, innate (or natural) immunity or acquired (adaptive) immunity, although there are also traits which are considered a bridge between both components6.
The innate immune system is the host’s first line of defence against infectious agents. In addition, haematological traits and stress parameters are also important indicators of the physiological and health status of farm animals7–9. During last years, several studies have reported medium to high heritabilities for several immune and haematological traits in pigs, suggesting an important genetic contribution to the phenotypic variability of these traits4,10–14. Since the pioneering work on quantitative trait loci (QTLs) mapping for general immune-capacity performed by15 in a wild boar × Swedish Yorkshire crossbred population, other groups have reported QTLs for traits related to immune-capacity in pigs16–23. More recently, with the development of high-density genotyping SNP chips, analyses applying genome wide association study (GWAS) have been performed to identify genetic markers associated with health-related traits. These studies have been however mainly addressed on haematological traits24–34. To date, genetic information in pigs on stress parameters focused primarily on acute adrenal activity35–38 with little or no genetic study on chronic stress parameters, such as cortisol measured in hair.
The present work aimed to study the genetic architecture of 30 health-related traits covering immune (mainly innate), haematological, and stress parameters associated to immunocompetence in a Duroc commercial line by estimating their genetic parameters and identifying associated genomic regions and candidate genes.
Results
Descriptive statistics and phenotypic correlations
In the present study we measured a set of 30 traits related with immune, haematological and stress parameters on a commercial Duroc pig line comprising 432 individuals. The data on descriptive statistics, as well as the abbreviated name of the analysed measured traits are shown in Table 1. Among the haematological traits, the erythrocyte-related traits presented the lowest phenotypic dispersion, with a coefficient of variation (CV) below 0.1, while the leukocyte and platelet-related traits presented CV ranging from 0.34 (leukocytes count) to 0.63 (eosinophils count). Regarding the ITs, most phagocytosis traits presented limited dispersion (CV ≤ 0.2), whereas the highest CVs were obtained for the acute-phase proteins CRP and HP (CV = 0.73 and 0.67, respectively). Finally, the stress parameters presented a moderate phenotypic variation with a CV = 0.44.
Table 1.
Descriptive statistics of the analysed immunity, haematological and well-being traits.
| Trait | Abrev | N | Mean | SD | CV |
|---|---|---|---|---|---|
| Haematocrit (%) | HCT | 430 | 33.76 | 2.93 | 0.09 |
| Haemoglobin (g/dL) | HB | 430 | 10.64 | 0.94 | 0.09 |
| Erythrocytes count n/µL | ERY | 430 | 6,527,907 | 551,399 | 0.08 |
| Mean corpuscular volume (fL) | MCV | 430 | 51.83 | 3.47 | 0.07 |
| Mean corpuscular haemoglobin (pg/cell) | MCH | 430 | 16.34 | 1.15 | 0.07 |
| Mean corpuscular haemoglobin concentration (g/dL) | MCHC | 430 | 31.54 | 1.02 | 0.03 |
| Platelets count n/µL | PLA | 424 | 277,797 | 133,467 | 0.48 |
| Leukocytes count n/µL | LEU | 430 | 20,541.8 | 6919.3 | 0.34 |
| Eosinophils count n/µL | EOS | 430 | 314.59 | 196.95 | 0.63 |
| Lymphocytes count n/µL | LYM | 430 | 12,203.3 | 4472.4 | 0.37 |
| Monocytes count n/µL | MON | 430 | 532.93 | 279.14 | 0.52 |
| Neutrophils count n/µL | NEU | 430 | 7448.9 | 3309.2 | 0.44 |
| IgA in saliva (mg/dl) | IgAsal | 404 | 5.05 | 3.06 | 0.61 |
| IgA in plasma (mg/ml) | IgA | 432 | 0.65 | 0.31 | 0.48 |
| IgG in plasma (mg/ml) | IgG | 431 | 12.61 | 4.90 | 0.39 |
| IgM in plasma (mg/ml) | IgM | 432 | 2.26 | 0.82 | 0.36 |
| C-reactive protein in serum (µg/ml) | CRP | 428 | 173.01 | 126.01 | 0.73 |
| Haptoglobin in serum (mg/ml) | HP | 432 | 0.99 | 0.67 | 0.67 |
| Nitric oxide in serum (µM) | NO | 427 | 205.98 | 80.29 | 0.39 |
| γδ T-lymphocyte subpopulation (%) | γδ T cells | 396 | 7.97 | 5.03 | 0.63 |
| Phagocytosis (% cells) | PHAGO_% | 432 | 43.32 | 8.50 | 0.20 |
| Granulocytes phagocytosis (%) | GRANU_PHAGO_% | 432 | 91.74 | 3.81 | 0.04 |
| Monocytes phagocytosis (%) | MON_PHAGO_% | 432 | 49.90 | 9.74 | 0.20 |
| Lymphocytes phagocytosis (%) | LYM_PHAGO_% | 432 | 6.35 | 4.11 | 0.65 |
| Phagocytosis FITC | PHAGO_FITC | 432 | 4.69 | 0.33 | 0.07 |
| Granulocytes phagocytosis FITC | GRANU_PHAGO_FITC | 432 | 5.14 | 0.41 | 0.08 |
| Monocytes phagocytosis FITC | MON_PHAGO_FITC | 432 | 3.91 | 0.25 | 0.06 |
| Lymphocytes phagocytosis FITC | LYM_PHAGO_FITC | 432 | 3.16 | 0.13 | 0.04 |
| Cortisol in hair (pg/mg) | CORT | 431 | 166.91 | 72.62 | 0.44 |
| NEU/LYM ratio | NLR | 430 | 0.65 | 0.286 | 0.44 |
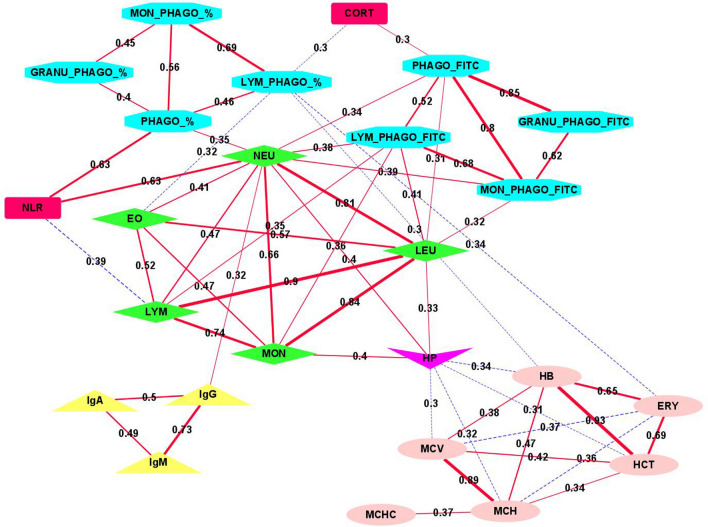
The network based on phenotypic correlation coefficients (Fig. 1) identified five interconnected clusters of correlated traits. The central cluster grouped five hemogram leukocyte-related traits (LEU, NEU, EO, LYM and MON), with rp ranging from 0.41 to 0.90. Another cluster connected with the previous one included traits related to phagocytic capacity (PHAGO_FITC, LYM_PHAGO_FITC, MON_PHAGO_FITC and GRANU_PHAGO_FITC) with rp > 0.5 among them. The phagocytosis traits related to percentage of phagocytic cells (PHAGO_%, GRANU_PHAGO_%, LYM_PHAGO_%, MON_PHAGO_%) clustered also together. Finally, two more clusters were found, the plasma concentration of Ig (IgA, IgM and IgG), with a rp = 0.73 between IgG and IgM, and the haematological erythrocyte-related traits (HB, ERY, MCV, MCH, HCT, MCHC), with negative and positive correlation coefficients. It is worth to highlight that HP generally correlated negatively with this last cluster. More details about the phenotypic correlation coefficients are provided in Supplementary Table S1.
Figure 1.
Network based on phenotypic correlation coefficients (|r| ≥ 0.3) among the immunity-related traits. Red lines indicate positive correlated traits while blue dashed lines indicate negative correlations. The numbers along the lines and the width of the lines indicate correlation coefficients. The shape and color of nodes indicate the different classes of the analysed traits.
Genetic parameters of immunity-related traits: heritability and genetic correlations
The genetic determinism of immunocompetence was first explored by the heritability of these immunity and health-related phenotypes. Heritability (Table 2) ranged between 0.092 and 0.786. Most traits exhibited moderate to high heritability values, 22 out of 30 traits showing h2 values above 0.4. These heritabilities had relatively wide confidence intervals, that in some cases encompasses more than half heritability parameter space. Despite this, the h2 confidence intervals did not overlap zero but for the heritability of MON, MON_PHAGO_FITC and GRANU_PHAGO_%.
Table 2.
Heritability values () for the immunity, haematological and well-being analysed traits, plus standard errors (SE) and confidence intervals at 95% (CI95) of the estimates.
| Trait | SE | CI95 | |
|---|---|---|---|
| Haematocrit (%) | 0.405 | 0.136 | 0.140–0.671 |
| Haemoglobin (g/dL) | 0.419 | 0.135 | 0.154–0.685 |
| Erythrocytes count n/µL | 0.669 | 0.133 | 0.408–0.930 |
| Mean corpuscular volume (fL) | 0.600 | 0.150 | 0.305–0.895 |
| Mean corpuscular haemoglobin (pg) | 0.470 | 0.135 | 0.205–0.736 |
| Mean corpuscular haemoglobin concentration (g/dL) | 0.767 | 0.150 | 0.473–1.061 |
| Platelets count n/µL | 0.651 | 0.153 | 0.351–0.951 |
| Leukocytes count n/µL | 0.281 | 0.128 | 0.030–0.533 |
| Eosinophils count n/µL | 0.594 | 0.143 | 0.314–0.875 |
| Lymphocytes count n/µL | 0.273 | 0.130 | 0.019–0.528 |
| Monocytes count n/µL | 0.092 | 0.080 | − 0.065 to 0.248 |
| Neutrophils count n/µL | 0.640 | 0.149 | 0.348–0.932 |
| IgA in saliva (mg/dl) | 0.467 | 0.161 | 0.151–0.783 |
| IgA in plasma (mg/ml) | 0.671 | 0.132 | 0.412–0.930 |
| IgG in plasma (mg/ml) | 0.652 | 0.136 | 0.517–1.054 |
| IgM in plasma (mg/ml) | 0.786 | 0.137 | 0.386–0.919 |
| C-reactive protein in serum (µg/ml) | 0.245 | 0.114 | 0.119–0.685 |
| Haptoglobin in serum (mg/ml) | 0.402 | 0.144 | 0.021–0.469 |
| Nitric oxide in serum (µM) | 0.256 | 0.120 | 0.330–0.896 |
| γδ T-lymphocytes subpopulation (%) | 0.613 | 0.144 | 0.021–0.490 |
| Phagocytosis (% cells) | 0.425 | 0.146 | 0.139–0.710 |
| Granulocytes phagocytosis (%) | 0.185 | 0.103 | − 0.016 to 0.386 |
| Monocytes phagocytosis (%) | 0.431 | 0.152 | 0.133–0.729 |
| Lymphocytes phagocytosis (%) | 0.495 | 0.149 | 0.202–0.788 |
| Phagocytosis FITC | 0.349 | 0.141 | 0.073–0.626 |
| Granulocytes phagocytosis FITC | 0.474 | 0.151 | 0.179–0.769 |
| Monocytes phagocytosis FITC | 0.118 | 0.106 | − 0.089 to 0.325 |
| Lymphocytes phagocytosis FITC | 0.407 | 0.115 | 0.181–0.633 |
| Cortisol in hair (pg/mg) | 0.456 | 0.142 | 0.177–0.735 |
| NEU/LYM ratio | 0.731 | 0.155 | 0.427–1.035 |
Among analysed ITs, plasma concentrations of Ig showed the highest heritabilities (from 0.652 to 0.786), followed by the percentage of γδ T cells (h2 = 0.613). Focusing traits related to acute phase proteins, HP exhibited a relatively high heritability (h2 > 0.4), whereas more limited genetic contribution was estimated for CRP (h2 = 0.245) and also for NO (h2 = 0.256). For phenotypes related to phagocytosis, low to moderate heritabilities were obtained (from 0.118 to 0.495). Several haematological traits also exhibited high heritabilities, being the MCHC the most heritable among them (h2 = 0.767), followed by the quantity of ERY and PLA in blood and by the MCV (h2 ≥ 0.65 in both cases). Other erythrocyte-related phenotypes, such as total and mean corpuscular HB as well as HCT, also showed significant heritabilities above 0.4. Regarding white blood cells counts, the quantity of NEU and EOS in blood showed heritabilities above 0.55, whereas lowly values (h2 below 0.3) were obtained for the number of LYM and total LEU, and no significant additive genetic contribution to the number of MON could be assessed. Concerning stress parameters, a medium heritability (h2 = 0.456) was obtained for the CORT levels in hair (chronic stress indicator), whereas NLR showed a particularly high heritability (h2 = 0.731).
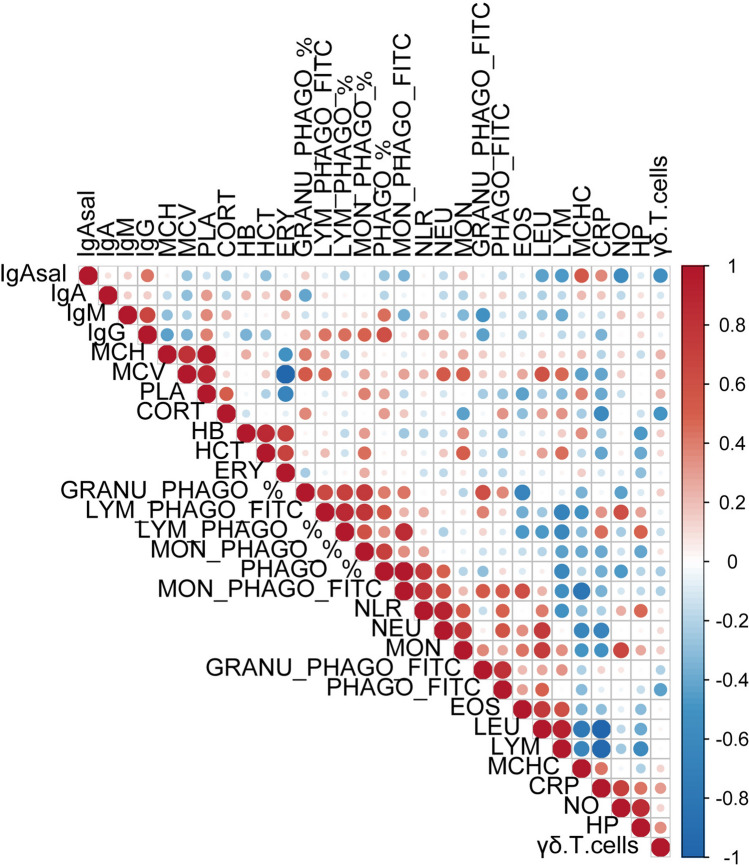
Genetic relationship among the immunity-related phenotypes was analysed through estimating the genetic correlations between each pairwise combination of traits; a heatmap showing the magnitude of the estimated correlations between the 30 traits is presented in Fig. 2. More details about these genetic correlation coefficients and their estimation standard errors are provided in Supplementary Table S2. It should be mentioned that the limited population size resulted in a limited precision of genetic correlations estimates, which generally showed high SE. The reliability of parameter estimation in the bivariate models was particularly compromised when they involved traits with heritabilities close to zero (i.e. MON, MON_PHAGO_FITC and GRANU_PHAGO_%), so these genetic correlations should be taken with caution.
Figure 2.
Heatmap of genetic correlations estimated by pairwise combination among immunity, haematological and stress related traits in pigs.
The three plasma Ig (IgA, IgG and IgM) clustered together, showing positive genetic correlations among them but lower than phenotypic correlations (Fig. 1); only the genetic correlation between plasma IgM and IgG was relevant and significant (rg = 0.662, SE = 0.118). In general, plasma Ig concentrations did not show important genetic correlations with other ITs but positive correlations between IgG and the percentage of phagocytic cells. Scarcely related with plasmatic Ig levels, the acute phase proteins concentrations in serum clustered together with NO and γδ T cells, with moderate to high positive genetic correlations among them but between γδ T cells and NO. CRP exhibited a very strong negative genetic associations with white blood cells counts, particularly LEU and LYM. Weaker but also negative genetic associations with red cells parameters (ERY, HB and HCT) were estimated for both CRP and HP.
A large cluster grouped together white cells counts and most phagocytosis capacity phenotypes quantified as mean fluorescence in FITC among the total phagocytic cells. The different leucocyte types (LYM, EOS, NEU, MON) showed a positive and high genetic correlation with total LEU count (rg from 0.72 to 0.92), whereas genetic correlations between the different LEU types varied largely. Phagocytosis-related traits showed a complex and not always consistent (i.e. with large estimation error) picture of genetic associations. The FITC measurements of phagocytosis correlated positively between them and with leucocytes counts, with some exceptions involving lymphocytes that in turn showed a pattern of genetic associations relatively different from the rest of leukocyte types. Conversely, negative genetic associations between most leukocyte subsets and the proportion of phagocytic cells (PHAGO_%, GRANU_PHAGO_%, LYM_PHAGO_%, MON_PHAGO_%) were obtained. These percentages of phagocytic cells correlated positively between them (rg between 0.35 and 0.75) but also with LYM_PHAGO_FITC and IgG.
As far as erythrocytes-related parameters is concerned, the HTC, HB and ERY constituted a cluster, with rg between them ranging between 0.69 and 0.86. No evidences of relevant genetic associations between these traits and white blood cells were obtained but for HTC, that showed moderate positive genetic correlations with LYM, LEU and MON. Separately and opposed to the previous cluster, PLA, MCH and MCV also grouped together (rg from 0.81 to 0.94); all three traits correlated negatively with ERY count.
Finally and regarding stress-related parameters, the CORT concentration in hair showed negative genetic correlations with CRP and γδ T cells (rg = − 0.59 and − 0.50, respectively) and positive correlation with PLA (rg = 0.50); associations with the rest of haematological and ITs were generally weak and/or no significant. Regarding NLR, expectedly it was strongly correlated to NEU (rg = 0.90), but also in a lesser extent to MON and LEU (rg = 0.55 and 0.41, respectively), and negatively to LYM (rg = − 0.52).
Genomic regions and candidate genes associated with immunity traits, stress indicators and haematological parameters
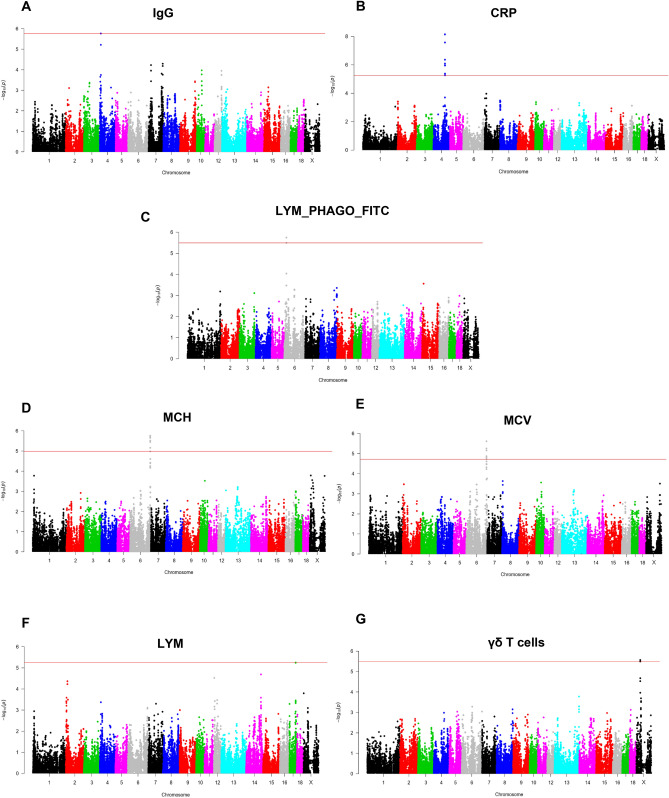
To identify genomic regions associated with health-related traits, a GWAS was performed using the 30 phenotypic traits and the genotypes of 42,641 SNPs of the Porcine GGPSNP70 BeadChip (Illumina) in the 432 Duroc pigs. Significant associations at whole-genome level (FDR ≤ 0.1) were detected for IgG, γδ T cells, LYM_PHAGO_FITC, LYM, CRP, MCV and MCH. A total of 31 significant associated SNPs located in six chromosomal regions on pig chromosomes SSC4, SSC6, SSC17 and SSCX were identified (Table 3). In addition, a genomic region in SSC12 and SSC14 for the total number of LEU and in SSC13 for the total number of NEU passed the FDR threshold of < 0.2. In those regions, we identified nine associated SNPs (Table 3). The full list of associated SNPs, with their predicted consequences, is shown in Supplementary Table S3. In addition, graphical representation of the GWAS results for the traits are displayed in Manhattan plots in Supplementary Figure S1 (FDR ≤ 0.2) and Fig. 3 (FDR ≤ 0.1). It is worth to highlight that almost half of the QTLs identified (4 out of 9) were associated with lymphocytes-related traits.
Table 3.
Description of the nine chromosomal regions associated with health-related traits and the annotated candidate genes.
| Region | Chr | Start (Mbp) | End (Mbp) | N SNPs | Top SNPs | MAF | p-value | FDR | Trait | Candidate genes |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 8.39 | 1 | rs319560097 | 0.44 | 1.71E−06 | 7.29E−02 | IgG | SLA, ST3GAL1 | |
| 2 | 4 | 90.54 | 91.26 | 10 | rs81233340, rs81382318 | 0.15 | 7.17E−09 | 1.53E−04 | CRP | CRP |
| 3 | 6 | 17.11 | 17.18 | 2 | rs338661853 | 0.50 | 1.78E−06 | 6.81E−02 | LYM_PHAGO_FITC | CDH1, COG8, VPS4A |
| 4 | 6 | 164.85 | 165.78 | 10 | rs323656844 | 0.43 | 1.73E−06 | 2.18E−02 | MCV, MCH* | PRDX1, PIK3R3 |
| 5 | 17 | 52.47 | 52.51 | 3 | rs80924885, rs80899023, rs80803525 | 0.33 | 5.68E−06 | 8.07E−02 | LYM | NFATC2 |
| 6 | X | 33.51 | 33.64 | 5 | rs342772739 | 0.13 | 2.71E−06 | 2.71E−02 | γδ T cells | ssc-mir-9786-1 |
| 7 | 12 | 3.25 | 1 | rs323856019 | 0.17 | 3.26E−06 | 1.39E−01 | LEU | SOCS3, BIRC5 | |
| 8 | 13 | 69.03 | 71.96 | 7 | rs81270251 | 0.48 | 9.01E−06 | 1.24E−01 | NEU | GATA2, PPARG, RAF1, SEC61A1 |
| 9 | 14 | 123.89 | 1 | rs343667976 | 0.45 | 9.07E−06 | 1.93E−01 | LEU | – |
* most significant trait
Figure 3.
Manhattan plots representing the association analysis between the health-related traits and SNPs distributed along the pig genome. Red line indicates those SNPs that are below the genome-wide significance threshold (FDR ≤ 0.1).
In SSC4, two regions at 8.3 Mb and 90.5–91.2 Mb associated with IgG plasma levels and CRP (Fig. 3A,B), respectively, were identified. In the proximal region of SSC4, an SNP (rs319560097) was associated with the IgG plasma levels. In this region, the candidate genes Src like adaptor (SLA) and ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (ST3GAL1) related to B cell development, differentiation and function were identified. Ten SNPs in quite linkage disequilibrium (Dʹ = 0.203–0.999) were identified associated with CRP levels in a more distal region of SSC4. It is worth highlighting that two of these associated SNPs (rs81285109 and rs80958253) were located inside the CRP locus (Ensembl gene id: ENSSSCG00000006403), the main candidate gene identified in this region.
In SSC6, two regions at 17.11–17.18 Mb and 164.85–165.78 Mb were identified associated with three traits. In the proximal region of SSC6, two SNPs (rs338661853 and rs81285171) were associated with LYM_PHAGO_FITC (Fig. 3C). In this region, three candidate genes were annotated (CDH1, COG8 and VPS4A), with the vacuolar protein sorting 4 homolog A (VPS4A) gene involved in the endosomal multivesicular bodies (MVB) pathway. In the second region, we identified ten SNPs in strong linkage disequilibrium (Dʹ > 0.9), except the rs81394075 that showed Dʹ > 0.48 with the rest of SNPs, associated with MCH and/or MCV traits (Fig. 3D,E). Remarkably, a strong candidate gene, the peroxiredoxin 1 (PRDX1), associated with haematocrit levels and haemoglobin concentration functions, was mapped in this pleitropic region. Another candidate gene also mapped in this region was the phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3) which was identified as component of multiple canonical pathways of which erythropoietin signalling was among them.
Three SNPs (rs80924885, rs80899023, rs80803525) at 52.46–52.51 Mb of SSC17 were associated with LYM trait (Fig. 3F). In this region, we also identified a promising candidate gene, the nuclear factor of activated T cells 2 (NFATC2), directly related with the quantity of lymphocytes, quantity of T and B lymphocytes and size of thymus cortex functions. For the percentage of γδ T cells, five SNPs located at 33.51–33.64 Mb of SSCX were found to be significantly associated to this trait (Fig. 3G). In this region, we did not identify any candidate gene among the annotated protein coding genes. However, four lncRNA and ssc-mir-9786-1 were annotated in this region. Both types of RNAs have been directly implicated in the innate immune response39,40, although there is still a lack of information about the mechanism of action of lncRNAs. In contrast, miRNAs are better characterized and there are tools and databases that allow to perform an in-silico target prediction. In this way, ssc-mir-9786-1 was predicted to target a total of 528 genes by RNAhybrid. Noteworthy, among the biological functions represented in the list of targeted genes there was T cell differentiation (Supplementary Table S4).
Regarding LEU trait, two SNPs (rs323856019 and rs343667976) at 3.25 Mb and 123.89 Mb on SSC12 and SSC14, respectively, were associated with this trait. Two candidate genes (SOCS3 and BIRC5) located on SSC12 and associated with quantity of leukocytes, lymphocytes, B and T lymphocytes, and peripheral blood leukocytes among other related diseases and functions were identified. On SSC14, no candidate genes were found among the annotated genes according to their functional information. Finally, seven SNPs with a Dʹ > 0.68 and located at 69.03–71.96 Mb on SSC13 were associated with NEU trait. In this region, several candidate genes (GATA2, PPARG, RAF1 and SEC61A1) associated with functions such as quantity of leukocytes or neutropenia were identified.
Comparison with other GWAS studies
A comparison was performed between the QTLs identified in our study and those previously published for health-related traits to identify overlapping chromosomal regions. We only identified two overlapping regions for LYM and MCH traits. Regarding LYM trait, the overlapping region (SSC17: 48.7–66.9 Mb) was identified in a F2 population from a Meishan/Pietrain family after 42 days post-infection with the protozoan pathogen Sarcocystis miescheriana41. The region for MCH at 166.3 Mb on SSC6 was less than 1 Mb away from the region identified in our analysis (164.8–165.8 Mb). This QTL was identified at chromosome-level in a GWAS analysis performed on animals of three breeds, Large-White, Landrace and Songliao Black27.
Discussion
Robustness and resilience, together with well-being, are becoming a priority in livestock breeding. In our study, 30 traits including immunity, haematological and stress parameters have been measured in 432 healthy Duroc pigs to analyse individual’s immunocompetence. Most of these health-related traits presented medium to high heritability values, confirming a relevant genetic determinism in the phenotypic variation of the global immunocompetence in pigs, as had been suggested by several authors10–14. Overall, the heritabilities in our study are in close agreement with results previously reported in other pig populations10–14, very particularly those estimates showing high heritabilities for phagocytosis, γδ T cells, haematological erythrocyte-related traits, and IgA antibody levels, and the lower heritability for CRP. The LEU and LYM heritabilities in our study (low-to-medium) were also similar to those previously reported by10–12,14, but lower to the high heritabilities reported by13. In contrast, other haematological leukocyte-related traits (i.e. EO and NEU counts) together with HP and IgG antibody levels showed in our study higher heritabilities than those reported by10–12, but similar to the values of13. Finally, our heritability for total IgM antibody levels was higher than previously published13. Discrepancies with other studies were however not unexpected; they could be partially attributable to differences regarding age of animals, environmental factors or the lack of protocols standardization between laboratories, but it could also denote differences in the genetic determinism of immune capacity between different pig populations.
Besides former immunity and haematological-related traits, we also analysed the genetic determinism of two stress parameters, CORT and NLR, obtaining for them medium to high heritabilities. To the best of our knowledge, this is the first study reporting a heritability for the cortisol measured in hair, suggesting the existence of genetic determinism in the susceptibility of animals to chronic stress. In line with our results, some studies in humans determined low-to-medium heritabilities for acute plasma and saliva cortisol levels42, and post-ACTH cortisol levels in blood were reported to be highly heritable in pigs43.
Heritabilities previously stated would support the possibility of genetically improving the analysed immune and health-related traits in the studied Duroc pig population. Besides the possibility of applying a multi-trait selection for immune response (ability to respond immunologically) already reported in Yorkshire pigs by44,45, the alternative of applying selection on global immunocompetence of healthy animals is worthy to be explored4,5. The complex map of genetic interactions depicted by the estimated genetic correlations should be considered to design a global strategy to improve global immunocompetence for more robust and resilient pig populations. Estimates of genetic correlations between immunity and haematological traits reported here and in previous studies should be interpreted with caution, given the low estimabitity for such multi-trait models with limited sample sizes. We found however evidences of some relevant (and indisputably different from zero) genetic correlations. They allowed inferring a map of genetic associations among these traits that differed substantially from the phenotypic association map, and was in partial agreement with previous studies in other pig population (e.g.13), that generally found weak and mostly positive genetic correlations among ITs. In our study, we confirmed positive genetic correlations between several ITs, e.g. between IgG and IgM plasmatic concentrations or between leukocyte cells subsets, and of them with their phagocytosis capacity. But we also reported strong negative genetic correlations, for instance between leukocytes counts and CRP levels or between lymphocytes and percentage of phagocytic cells. Interestingly, the chronic stress indicator (CORT levels in hair) showed also genetic antagonism with relevant immunity parameters such as the percentage of γδ T-lymphocytes or CRP basal levels.
As signalled by5, major queries about the possibility and consequences of using genetic variation in immunocompetence in breeding programs should be addressed. According to our results, applying a selection program to increase the immunocompetence of the analysed Duroc population focusing for instance in lymphocyte related traits and/or immunoglobulins is feasible, but could be accompanied by correlated responses in other immunity parameters related to inflammation and stress that are worthy to be further explored. The question about putative correlated responses in (re)production performance should also be raised. Yorkshire pigs selected for high humoral and cellular immune responses had increased weight gains but were also prone to develop more severe arthritis after infection with Mycoplasma hyorhinis44,45. Further studies and functional validations are needed to determine the best combination of ITs and to assess the effects of selecting these ITs on global animal health and well-being, as well as on production performance. In this context and considering the time and cost-demanding phenotyping of ITs, the possibility of identifying genetic variants functionally related with immunity that could be implemented in the breeding schemes assumes paramount relevance.
GWAS analyses followed by gene annotation in the significantly associated genomic regions led to identify 16 promising candidate genes that may be implicated in the phenotypic variation of nine health-related traits. Remarkably, four out of nine of the traits with significant associated signals in the pig genome were related to lymphocytes, performing functions in the innate (percentage of γδ T cells in peripheral blood and lymphocytes phagocytic capacity) or the adaptive (total concentration of IgG in plasma and total number of lymphocytes) immune systems. In the opposite, our study did not allow identifying any SNP associated to CORT stress parameter. Genetic variants associated with plasma cortisol levels have been identified in pigs35–38, but there is a lack of GWAS studies with cortisol level measured in hair samples.
Among genes identified in the lymphocyte-signalled genomic regions, the NFATC2 was mapped in the region associated with the total number of lymphocytes. This gene encodes a transcription factor that is expressed in peripheral blood lymphocytes, among others, and was firstly identified in T cells. NFATC2 plays a critical role in regulating the expression of cytokine genes in T cells during the immune response46,47 and is required for B cell development and function46,48. It is worth mentioning that knockout NFATC2 mouse displayed enhanced immune response49 and hyperproliferation of primary B cells48, which suggest a negative regulatory function in the immune system.
Other two candidate genes, SLA and ST3GAL1, were located in the genomic region associated with the total concentration of IgG, the predominant serum isotype produced by B-lymphocytes. Remarkably, both genes have been implicated in the B cell differentiation process50,51. Specifically, expression of SLA is required to optimally regulate BCR levels and signal strength during B-cell development50, while ST3GAL1 modulates the plant lectin peanut agglutinin (PNA) binding phenotype of activated B-cells, through O-glycan remodelling on CD4551.
As far as lymphocytes phagocytic capacity, three candidate genes were identified: cadherin 1 (CDH1), component of oligomeric golgi complex 8 (COG8) and VPS4A. Several studies have determined the phagocytic capacity of B-cells, mainly B1-cells but also follicular B-cells, playing an important role in innate immunity and the development of a strong humoral response52–54. VPS4A and COG8 have been involved in the generation of multivesicular bodies (MVBs) during phagosome maturation55, and retrograde intracellular membrane trafficking at the Golgi56, respectively. Furthermore, CDH1, a cellular receptor found on epithelial cells that can mediate entry of bacteria, is also expressed in other cells such as macrophages57.
Among the lymphocyte lineage there are cells such as the γδ T cells considered to be a bridge between innate and adaptive immunity58. Unlike in humans and mice, γδ T cells represent a prominent population in pigs’ peripheral blood59. In the genomic region associated with γδ T cells, we have identified a promising miRNA (ssc-mir-9786-1) which was predicted to target genes implicated in the T cell differentiation process. This miRNA was previously identified in porcine milk exosomes60 but there is still a lack of functional validation of the direct ssc-mir-9786-1-target mRNA interaction involving genes related with the immune system.
Also related to white cells-mediated immunity, we identified promising candidate genes annotated in the regions associated with the total number of leukocytes (SSC12 and SSC14) and neutrophils (SSC13). Remarkably, one of the candidate genes selected for the total number of leukocytes, baculoviral IAP repeat containing 5 (BIRC5), also known as Survivin, is essential for T cell maturation and proliferation61. This result is in accordance with the phenotypic and genetic correlations of rp = 0.9 and rg = 0.92 observed between the total number of leukocytes and lymphocytes. In fact, when we look in detail at the regions associated with the number of lymphocytes, the same signals previously observed for leukocytes are identified at chromosome level; therefore, these regions may be more specifically affecting lymphocytes. Among the candidate genes annotated in the region associated with NEU, it is worth to highlight the peroxisome proliferator activated receptor gamma (PPARG) gene. This gene encodes a nuclear hormone receptor with a wide variety of biological functions, including a critical role in modulating inflammatory processes of the innate immune system through regulation of neutrophil trafficking and apoptosis, among other functions62.
A particularly remarkable result arising in this study was the identification of CRP as candidate gene, annotated in the region associated with variation in its traduced protein levels. CRP is highly expressed during the acute-phase response, playing an important role in host defence through activating the complement system and cell-mediated pathways63. CRP is considered a blood biomarker of inflammation, although clinical studies in humans have determined that small elevation in baseline concentration of CRP is a powerful and specific predictor of cardiovascular event risk in healthy adults64. Remarkably, differences in CRP blood level have been associated with polymorphisms in the CRP gene, and some large-scale studies have provided evidence between the relationship of CRP polymorphisms, CRP blood levels and disease risk in humans (reviewed in65). In our study, we identified two associated SNPs in the intron 2 of the isoform ENSSSCT00000083957.1 and the 3′ UTR region (exon 2) of the isoform ENSSSCT00000007016.4. Further studies are warranted to determine the role of CRP polymorphisms in the variation of CRP serum levels in our Duroc population. Moreover, taking into account the higher resemblance of the immune responses of pigs with humans compared to mice66, the present results may contribute to the implementation of pigs as large animal models for cardiovascular diseases.
Finally, two interesting candidate genes (PRDX1 and PIK3R3) were also identified in the region associated with both MCH and MCV. These red cell parameters are highly related, showing positive phenotypic and genetic correlations between them (rp = 0.89, rg = 0.81), which is concordant with the identification of this pleiotropic region. PRDX1 is a member of the peroxiredoxin family of antioxidant enzymes. Severe haemolytic anaemia characterized by marked decrease in haematocrit and haemoglobin in peripheral blood has been observed in mice lacking PRDX167. Remarkably, MCV is among the 15 traits with the highest number of QTLs identified so far, with 546 associations (PigQTLdatabase, release 41, April 26, 2020). Nonetheless, we only identified a previously published QTL region associated with MCH27, which was proximal to the region for MCH/MCV identified in our study. This result agrees with previous studies in which few overlapping QTL regions for health-related traits have been identified so far, reinforcing the specificity of the genomic architecture of immunological parameters depending on the pig population (reviewed in4).
Conclusions
This study focuses on the genetic basis of 30 phenotypes associated to health and well-being in a Duroc pig population. The medium-to-high heritability estimates confirmed the existence of genetic determinism in most traits related to global immunocompetence in pigs. Positive genetic correlations but also strong negative genetic correlations between several immunity traits were reported. We also identified nine chromosomal regions associated with the variation of nine immune traits, highlighting 16 promising candidate genes, including CRP, NFATC2, PRDX1, SLA, ST3GAL1, and VPS4A, functionally related to these traits. Overall, our results provide new insights into the genetic control of traits related with immunity and support the possibility of applying effective selection programs to improve immunocompetence in pigs.
Methods
Ethics statement
All experimental procedures with pigs were performed according to the Spanish Policy for Animal Protection RD1201/05, which meets the European Union Directive 86/609 about the protection of animals used in experimentation. The experimental protocol was approved by the Ethical Committee of the Institut de Recerca i Tecnologia Agroalimentàries (IRTA).
Animal material
A total of 432 animals (217 males and 215 females) from a commercial Duroc pig line were used for this study. The pigs came from six batches (72 ± 1 animals per batch) and belonged to 134 litters (two to four piglets by litter, balancing gender when possible) obtained from 132 sows and 22 boars (all active boars in the commercial population). All animals were raised in the same farm and fed ad libitum with a commercial cereal-based diet. All animals were apparently healthy, without any sign of infection.
Samples of blood, saliva and hair were collected at 60 ± 8 d of age from all animals. Blood was collected via the external jugular vein into vacutainer tubes with or without anti-coagulants (Sangüesa S.A., Spain), according to the requirements for further immunity measurements. Saliva was collected with Salivette tubes (Sarstedt S.A.U., Germany) according to the protocols recommended by the manufacturer. Hair was collected with scissors from the dorsal area of the neck behind the ears and placed in numbered bags. All samples were transported with ice blocks to the laboratory for later processing.
Phenotypic parameters
Haematological parameters
Hemograms were measured in the Laboratory Echevarne (Spain; Barcelona) from blood sampled in 4 ml EDTA tubes. The following haematological traits were included in the genetic analyses: haematocrit (HCT), haemoglobin (HB), mean corpuscular volume (MCV), mean corpuscular haemoglobin (MCH), mean corpuscular haemoglobin concentration (MCHC), total number of leukocytes (LEU), eosinophils (EO), lymphocytes (LYM), monocytes (MON), neutrophils (NEU), erythrocytes (ERY) and platelets (PLA).
Immunity parameters
Immunity parameters were measured from plasma or serum depending on the trait. Blood samples for serum were collected in 6 ml tubes with gel serum separator and centrifuged at 1600g for 10 min at RT. Plasma was collected from blood sampled in 6 ml heparinised tubes and centrifuged at 1300g for 10 min at 4 °C. Plasma and serum samples were collected, aliquoted, and stored a − 80 °C until use.
Immunoglobulins
Total concentrations of immunoglobulins IgA, IgG and IgM in plasma, and IgA in saliva, were measured by ELISA with commercial kits (Bethyl laboratories Inc., Bionova, Spain), following the manufacturer’s instructions. Plasma samples were diluted 1:10,000, 1:50,000 and 1:500,000 to detect IgA, IgG and IgM, respectively, while saliva samples were diluted 1:100 to detect IgA. Samples, in duplicate, were quantified by interpolating their absorbance from the standard curves constructed with known amounts of each pig immunoglobulin class and corrected for sample dilution. Absorbance was read at 450 nm using an ELISA plate reader (Bio-Rad) and analysed using the Microplate manager 5.2.1 software (Bio-Rad).
Acute-phase proteins
C-reactive protein (CRP) levels were measured in serum samples diluted 1:3000 by ELISA kit (Abcam Plc., Spain) following manufacturer’s instructions. Haptoglobin (HP) concentration was measured in undiluted serum samples by colorimetric assay (Tridelta Development Limited, Ireland) following manufacturer’s instructions. All samples were quantified in duplicate using standard curves constructed by plotting absorbance against CRP or HP concentration, respectively. Absorbance was read at 450 nm for CRP and 630 nm for HP using an ELISA plate reader (Bio-Rad) and analysed using the Microplate manager 5.2.1 software.
Gamma-delta T cells (γδ T cells)
Peripheral blood mononuclear cells (PBMCs) were separated from heparinised peripheral blood by density-gradient centrifugation with Histopaque-1077 (Sigma, Spain) at 450 g for 30 min. The cells were resuspended in RPMI 1640 medium supplemented with 5% foetal bovine serum (FBS) (Sigma, Spain), 1% Penicillin–Streptomycin (10,000 U/mL–10 mg/mL) and 1% l-Glutamine (200 mM) (Cultek, Spain). For PBMCs staining, the monoclonal antibody APC Rat Anti-Pig γδ T Lymphocytes (MAC320 clone, BD Pharmigen, Spain) and the APC Rat IgG2a κ isotype control (R35-95 clone, BD Pharmigen, Spain) were used. Briefly, 106 PBMCs were stained with the primary-conjugated antibodies (1:100) in 1×PBS-1% FBS for 20 min at 4 °C. After two washes with 1×PBS-1% FBS at 4 °C, cells were resuspended in 1×PBS-1% FBS and analysed by flow cytometry using the MACSQuant Analyzer 10 Flow cytometer (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) and the MACSQuantify sofware v2.6 (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). For automated flow cytometry analysis, files were imported in R environment (v3.6.1)68 with the read.flowSet function implemented in flowCore package (v1.50.0)69. Fluorescence was transformed using arcsinhTransform function. Doublets were removed using gate_singlet function (flowStats package v3.42.070), margin events using boundaryFilter function (flowCore package v1.50.069). Gatings were then performed using gate_mindensity2 function (openCyto package v1.22.271) on FSC channel to remove FSC low events corresponding to debris, SSC high events corresponding to residual granulocytes and to gate γδ T cells as APC positive. Parameters were adjusted for each day of lab analyses on a representative sample pooling all the data into one using getGlobalFrame function.
Phagocytosis assay
Phagocytosis assay was carried out in heparinized whole blood samples incubated with fluorescein (FITC)-labelled opsonized E. coli bacteria by using the Phagotest kit (BD Pharmigen, Spain) and according to the protocol recommended by the manufacturer. Samples were analysed by flow cytometry using the MACSQuant Analyzer 10 Flow cytometer (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) and the MACSQuantify sofware v2.6 (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). With this assay we analyzed the percentage of cells having performed phagocytosis as well as their mean fluorescence intensity (number of ingested bacteria). Phagocytosis assay analyses were performed in R (v3.6.0)68. Doublets were removed using gate_singlet function (flowStats package v3.42.070), margin events using boundaryFilter function (flowCore package v1.50.069). Gatings were then performed using either gate_mindensity2 or gate_flowClust_2d functions (openCyto package v1.22.071) on propidium iodure (PI) channel to gate blood cells as PIhi and cells having performed phagocytosis as FITC+ . Granulocytes, monocytes, and lymphocytes were gated based on their FSC SSC properties. Parameters were adjusted for each day of lab analyses on a representative sample pooling all the data into one using getGlobalFrame function. The following phagocytosis traits were quantified: percentage of total phagocytic cells (PHAGO_%), percentage of phagocytic cells among granulocytes (GRANU_PHAGO_%), monocytes (MON_PHAGO_%), and lymphocytes (LYM_PHAGO_%), mean fluorescence in FITC among the total phagocytic cells (PHAGO_FITC), and mean fluorescence in FITC among the granulocytes (GRANU_PHAGO_FITC), monocytes (MON_PHAGO_FITC) and lymphocytes (LYM_PHAGO_FITC) that phagocyte.
Nitric oxide
Total concentrations of Nitric Oxide (NO) were measured by colorimetric assay (Thermo Fisher Scientific, Spain) following manufacturer’s instructions. Serum samples were ultrafiltered through a 10,000 molecular weight cut-off (MWCO) and diluted 1:10. Samples were quantified by reference to standard curves constructed with known amounts of Nitrate Standard solution. Absorbance was read at 540 nm using a microplate reader (LUMistar Omega, BMG Labtech) and analysed using the Omega MARS software (BMG Labtech).
Stress indicators
Cortisol
One hundred and fifty mg of hair were weighted from each sample and placed into a 50-ml conical tube. After three washes with 3 ml of isopropanol, all samples were left to dry in an extractor hood during 12 h. Dried hair samples were cut into 2–3 mm pieces using scissors, and 50 mg were transferred into 2 ml eppendorf. One ml of methanol was added to each sample and incubated 18 h at 37 °C under moderate shaking (100 rpm). After incubation, extracted samples were centrifuged at 7000g for 2 min and 700 µl of supernatant was transferred to a new 1.5 ml tube. The supernatant was then placed into a speed vac for 2 h to evaporate the methanol. The dried extracts were stored at − 20 °C until analysis. Total concentrations of cortisol (CORT) were measured by ELISA kit (Cusabio Technology LLC., Bionova, Spain) with dried samples reconstituted with 210 µl of sample diluent. Samples were quantified by reference to standard curves constructed with known concentrations of pig cortisol dilutions of the Standard. Absorbance was read at 450 nm using an ELISA plate reader (Bio-Rad) and analysed using the Microplate manager 5.2.1 software (Bio-Rad).
Neutrophil to lymphocyte ratio
The neutrophil to lymphocyte ratio (NLR) was calculated as a ratio of NEU and LYM.
Exploratory and phenotypic analyses
Descriptive statistics of the formerly described immunity, haematological and well-being traits in our studied Duroc population are shown in Table 1. Exploratory analyses of these phenotypes were carried out for investigating both the raw data distribution and the best fitting model for subsequent analyses. Systematic non-genetic putative effects (sex, batch and day of lab analyses within batch) on each trait were tested by using a linear model (lm) in R. Normal probability plots and Shapiro–Wilk test were performed to investigate the goodness-of-fit of the residuals with the normal distribution. For most phenotypes but the percentage of phagocytic cells, data in raw form and its residuals were quite skewed to the right; in those cases, log-transformation of data corrected these departures from normality. A filtered dataset of log-transformed data (most phenotypes) and raw-data (% of phagocytosis) was employed to perform further analyses. Subsequently, pairwise phenotypic correlations (rp) among all analysed phenotypes were computed after adjusting for significant environmental factors, and a correlation network was built up with Cytoscape72, considering those Pearson’s correlation coefficients with absolute value ≥ 0.3.
Estimation of genetic parameters
The heritability , i.e. the proportion of phenotypic variance attributable to additive genetic effects, was estimated for the 30 immune, haematological and stress traits showed in Table 1. Variance components and the corresponding h2 were estimated from an univariate animal model as follows:
where is the vector of phenotypic observations of all individuals for the health-related trait (either log-transformed or raw data, depending upon the trait); is the vector of systematic (fixed) effects on the trait, including effect of sex (2 levels) plus batch effects (6 levels) for most traits but for phagocytosis-related traits, for which the data of laboratory analysis (12 levels, two by batch) was considered instead; is the corresponding incidence matrix; is the vector of animal’s genetic additive (random) effects on the trait, and the corresponding incidence matrix; and is the vector of random residual terms. The assumed distribution of additive genetic effects was u∼N(0,A ), where A is the numerator relationship matrix computed on the basis of pedigree (1388 individuals, five generations) and is the additive genetic variance; random errors were distributed as e ∼ N(0,I ). Estimation of the model variance components and the corresponding heritability for each trait was performed by REML using the aireml program included in the BGF90 package73; the standard errors (SE) of the heritability estimates were computed by repeated sampling from their asymptotic normal distribution following74, thus obtaining the corresponding confidence intervals at 95% (CI95).
Subsequently, pairwise genetic correlations (for each two traits combination) were estimated in a two-traits animal model described as follows in matrix notation:
where and are the vectors of phenotypic observations for trait 1 and trait 2, respectively; and are the vectors of systematic (fixed) effects on each trait previously described, and and the correspondent incidence matrices; and are the vectors of animal genetic additive effects on trait 1 or trait 2 (random effects), and and the corresponding incidence matrices; finally and are the vectors of residual errors for each trait, and 0 a matrix of zeros, assumed independent. The (co)variance matrix of random genetic effects was defined as:
where and are the additive genetic variance of traits 1 and 2, respectively, is the genetic covariance between the traits, and A is the numerator relationship matrix as defined above. Estimation of the (co)variance components of each pairwise analysis was also performed by REML using the aireml program included in the BGF90 package73. Genetic correlation between traits were obtained as , and the SE of the genetic correlation estimates were also computed following74.
DNA extraction and SNP genotyping
Genomic DNA was extracted from blood samples using the NucleoSpin Blood (Macherey–Nagel). DNA concentration and purity were measured in a Nanodrop ND-1000 spectrophotometer.
The 432 animals were genotyped for 68,516 single nucleotide polymorphisms (SNPs) with the GGP Porcine HD Array (Illumina, San Diego, CA) using the Infinium HD Assay Ultra protocol (Illumina). Plink software75 was used to remove SNPs with a minor allele frequency (MAF) less than 5%, SNPs with more than 10% missing genotype data, and SNPs that did not map to the porcine reference genome (Sscrofa11.1 assembly). After quality control a subset of 42,641 SNPs were retained for subsequent analysis.
Genome-wide association studies (GWAS)
GWAS was carried out between the 42,641 filtered SNPs and the 30 health-related traits described in Table 1. For this purpose, the genome-wide complex trait analysis (GCTA) software76 was employed using the following model for each trait across all SNPs:
where yijk corresponds to the phenotypic trait (either log-transformed or raw data) of the ith individual of sex j in the kth batch; sexj corresponds to the jth sex effect (2 levels); bk corresponds to the kth batch effect (6 levels) for most traits but for phagocytosis related traits, for which the data of laboratory analysis (12 levels, two by batch) was considered instead; ui is the infinitesimal genetic effect of individual i, with u∼N(0,Gσ2u), where G is the genomic relationship matrix (GRM) calculated using the filtered autosomal SNPs based on the methodology of76 and σ2u is the additive genetic variance; sli is the genotype (coded as 0,1,2) for the lth SNP, and al is the allele substitution effect of the SNP on the trait under study; and eijk is the residual term. The false discovery rate (FDR) method of multiple testing described by Benjamini and Hochberg77 was used to measure the statistical significance for association studies at genome-wide level with the p.adjust function of R. The significant association threshold was set at FDR ≤ 0.1. Manhattan plots based on the significance of the associations across the whole genome were generated using the R package qqman78.
Comparative QTL analysis between our results and previous published data was performed by retrieving all pig QTL and association data on SSC11.1 for health traits from the pigQTL database79.
Gene annotation and SNP functional prediction
Biomart software80 was used to retrieve gene annotations from the Ensembl Genes 98 Database using the Scrofa11.1 reference assembly, considering 1 Mb downstream/upstream of around the candidate chromosomal regions. Furthermore, functional predictions of the significantly associated SNPs were performed with Variant Effect Predictor software81.
For functional categorisation of the annotated genes, data were analyzed through the use of IPA82 (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) to obtain gene ontologies (GO), biological functions, gene networks and canonical pathways in which the genes annotated in the associated regions were involved. Orthologous human gene names were retrieved from the Ensembl Genes 98 Database for functional categorisation when a pig gene name was not assigned to the gene stable id. Furthermore, information from Mouse Genome Database83 and Genecards84 was used to identify gene functions affecting the analysed phenotypes.
miRNA target prediction and functional annotation
Porcine mRNA 3′UTR sequences from the whole pig genome (assembly 11.1) were downloaded from Ensembl Gene 100 database85, and Seqkit tool86 was used to search 3′ UTR seed matches with the 7mer seed miRNA sequence. Subsequently, the obtained list of 3′ UTR was used to predict miRNA target using the RNA hybrid software87 with the following criteria: energy threshold of no more than − 25 kcal/mol and perfect match of 2–8 nt in the seed region. Enriched GO terms and pathways of the predicted miRNA target genes was performed with the ClueGO v2.5.7 plug-in of Cytoscape v3.8.0 software88.
Supplementary information
Acknowledgements
The study was funded by grant AGL2016-75432-R awarded by the Spanish Ministry of Economy and Competitivity (MINECO). Maria Ballester is financially supported by a Ramon y Cajal contract (RYC-2013-12573) from the Spanish Ministry of Economy and Competitiveness. YRC was funded by Marie Skłodowska-Curie grant (P-Sphere) agreement No 6655919 530 (EU). The authors belong to Consolidated Research Group AGAUR, ref. 2017SGR-1719. We gratefully acknowledge to technical staff from IRTA and Selección Batallé S.A for their collaboration in the experimental protocols at farm and slaughterhouse, as well as to Albert Bensaid and Virginia Aragón for valuable discussions and comments on the study.
Author contributions
M.B. and R.Q. designed the study. M.B. and J.R. supervised the generation of the material animal used in this work. M.B., Y.R.C., O.G.R., M.P., J.R., M.D., J.T. and R.Q. performed the sampling. M.B., O.G.R. and S.L.S. carried out the laboratory analyses. M.B., Y.R.C., F.B. and R.Q. analyzed the data. M.B. and R.Q. interpreted the results and wrote the manuscript. All the authors read and approved the final version of the manuscript.
Data availability
The results from all data generated or analysed during this study are included in this published article (and its Supplementary Information files). However, the datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Ballester, Email: maria.ballester@irta.cat.
Raquel Quintanilla, Email: raquel.quintanilla@irta.cat.
Supplementary information
is available for this paper at 10.1038/s41598-020-75417-7.
References
- 1.Ernst CW, Steibel JP. Molecular advances in QTL discovery and application in pig breeding. Trends Genet. 2013;29:215–224. doi: 10.1016/j.tig.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Thornton PK. Livestock production: Recent trends, future prospects. Philos. Trans. R. Soc. B Biol. Sci. 2010 doi: 10.1098/rstb.2010.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng G, et al. Antibiotic alternatives: The substitution of antibiotics in animal husbandry? Front. Microbiol. 2014 doi: 10.3389/fmicb.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher AH, Janss LL, Niewold TA, de Greef KH. Disease incidence and immunological traits for the selection of healthy pigs. A review. Vet. Q. 2002;24:29–34. doi: 10.1080/01652176.2002.9695121. [DOI] [PubMed] [Google Scholar]
- 5.Knap PW, Bishop SC. Relationships between genetic change and infectious disease in domestic livestock. Occ. Publ. Br. Soc. Anim. Sci. 2000;27:65–80. [Google Scholar]
- 6.Rich R, et al. Clinical Immunology: Principles and Practice. Amsterdam: Elsevier; 2018. [Google Scholar]
- 7.Kumar B. Stress and its impact on farm animals. Front. Biosci. 2012;E4:1759. doi: 10.2741/e496. [DOI] [PubMed] [Google Scholar]
- 8.Gerner W, Käser T, Saalmüller A. Porcine T lymphocytes and NK cells—An update. Dev. Comp. Immunol. 2009;33:310–320. doi: 10.1016/j.dci.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Schalm’s Veterinary Hematology. (Wiley-Blackwell, Hoboken, 2010). 10.1111/j.1939-165X.2011.00324.x.
- 10.Edfors-Lilja I, Wattrang E, Magnusson U, Fossum C. Genetic variation in parameters reflecting immune competence of swine. Vet. Immunol. Immunopathol. 1994;40:1–16. doi: 10.1016/0165-2427(94)90011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clapperton M, et al. Traits associated with innate and adaptive immunity in pigs: Heritability and associations with performance under different health status conditions. Genet. Sel. Evol. 2009;41:54. doi: 10.1186/1297-9686-41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henryon M, Heegaard PMH, Nielsen J, Berg P, Juul-Madsen HR. Immunological traits have the potential to improve selection of pigs for resistance to clinical and subclinical disease. Anim. Sci. 2006;82:596–606. doi: 10.1079/ASC200671. [DOI] [Google Scholar]
- 13.Flori L, et al. Immunity traits in pigs: Substantial genetic variation and limited covariation. PLoS ONE. 2011;6:e22717. doi: 10.1371/journal.pone.0022717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clapperton M, Glass EJ, Bishop SC. Pig peripheral blood mononuclear leucocyte subsets are heritable and genetically correlated with performance. Animal. 2008;2:1575–1584. doi: 10.1017/S1751731108002929. [DOI] [PubMed] [Google Scholar]
- 15.Edfors-Lilja I, et al. Mapping quantitative trait loci for immune capacity in the pig. J. Immunol. 1998;161:829–835. [PubMed] [Google Scholar]
- 16.Wattrang E, et al. Confirmation of QTL on porcine chromosomes 1 and 8 influencing leukocyte numbers, haematological parameters and leukocyte function. Anim. Genet. 2005;36:337–345. doi: 10.1111/j.1365-2052.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- 17.Zou Z, et al. Quantitative trait loci for porcine baseline erythroid traits at three growth ages in a White Duroc × Erhualian F2 resource population. Mamm. Genome. 2008;19:640–646. doi: 10.1007/s00335-008-9142-9. [DOI] [PubMed] [Google Scholar]
- 18.Yang S, et al. Quantitative trait loci for porcine white blood cells and platelet-related traits in a white duroc × Erhualian F2 resource population. Anim. Genet. 2009;40:273–278. doi: 10.1111/j.1365-2052.2008.01830.x. [DOI] [PubMed] [Google Scholar]
- 19.Wimmers K, Murani E, Schellander K, Ponsuksili S. QTL for traits related to humoral immune response estimated from data of a porcine F2 resource population. Int. J. Immunogenet. 2009;36:141–151. doi: 10.1111/j.1744-313X.2009.00838.x. [DOI] [PubMed] [Google Scholar]
- 20.Uddin MJ, et al. Mapping quantitative trait loci for innate immune response in the pig. Int J Immunogenet. 2011;38:121–131. doi: 10.1111/j.1744-313X.2010.00985.x. [DOI] [PubMed] [Google Scholar]
- 21.Lu X, et al. Mapping quantitative trait loci for cytokines in the pig. Anim. Genet. 2011;42:1–5. doi: 10.1111/j.1365-2052.2010.02071.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, et al. Mapping quantitative trait loci for T lymphocyte subpopulations in peripheral blood in swine. BMC Genet. 2011;12:79. doi: 10.1186/1471-2156-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho IC, et al. QTL analysis of white blood cell, platelet and red blood cell-related traits in an F 2 intercross between Landrace and Korean native pigs. Anim. Genet. 2011;42:621–626. doi: 10.1111/j.1365-2052.2011.02204.x. [DOI] [PubMed] [Google Scholar]
- 24.Luo W, et al. Genome-wide association study of porcine hematological parameters in a large white × Minzhu F2 resource population. Int. J. Biol. Sci. 2012;8:870–881. doi: 10.7150/ijbs.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X, et al. Genome-wide association study for T lymphocyte subpopulations in swine. BMC Genomics. 2012;13:488. doi: 10.1186/1471-2164-13-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bovo S, et al. Genome-wide association studies for 30 haematological and blood clinical-biochemical traits in Large White pigs reveal genomic regions affecting intermediate phenotypes. Sci. Rep. 2019;9:1–7. doi: 10.1038/s41598-019-43297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JY, et al. Genome-wide association studies for hematological traits in swine. Anim. Genet. 2013;44:34–43. doi: 10.1111/j.1365-2052.2012.02366.x. [DOI] [PubMed] [Google Scholar]
- 28.Lu X, et al. Genome-wide association study for cytokines and immunoglobulin G in swine. PLoS ONE. 2013;8:e74846. doi: 10.1371/journal.pone.0074846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, et al. Genome-wide association study reveals constant and specific loci for hematological traits at three time stages in a White Duroc × Erhualian F2 resource population. PLoS ONE. 2013;8:e63665. doi: 10.1371/journal.pone.0063665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung EJ, et al. Genome-wide association study identifies quantitative trait loci affecting hematological traits in an F2 intercross between Landrace and Korean native pigs. Anim. Genet. 2014;45:534–541. doi: 10.1111/age.12175. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F, et al. Genome-wide association studies for hematological traits in Chinese Sutai pigs. BMC Genet. 2014;15:41. doi: 10.1186/1471-2156-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponsuksili S, Reyer H, Trakooljul N, Murani E, Wimmers K. Single- and Bayesian multi-marker genome-wide association for haematological parameters in pigs. PLoS ONE. 2016;11:e0159212. doi: 10.1371/journal.pone.0159212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Genomewide association studies for hematological traits and T lymphocyte subpopulations in a Duroc × Erhualian F2 resource population. J. Anim. Sci. 2016;94:5028–5041. doi: 10.2527/jas.2016-0924. [DOI] [PubMed] [Google Scholar]
- 34.Yan G, et al. Imputation-based whole-genome sequence association study reveals constant and novel loci for hematological traits in a large-scale swine F2 resource population. Front. Genet. 2018;9:401. doi: 10.3389/fgene.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Désautés C, et al. Genetic linkage mapping of quantitative trait loci for behavioral and neuroendocrine stress response traits in pigs. J. Anim. Sci. 2002;80:2276–2285. doi: 10.2527/2002.8092276x. [DOI] [PubMed] [Google Scholar]
- 36.Ousova O, et al. Corticosteroid binding globulin: A new target for cortisol-driven obesity. Mol. Endocrinol. 2004;18:1687–1696. doi: 10.1210/me.2004-0005. [DOI] [PubMed] [Google Scholar]
- 37.Görres A, Ponsuksili S, Wimmers K, Muráni E. Analysis of non-synonymous SNPs of the porcine SERPINA6 gene as potential causal variants for a QTL affecting plasma cortisol levels on SSC7. Anim. Genet. 2015;46:239–246. doi: 10.1111/age.12276. [DOI] [PubMed] [Google Scholar]
- 38.Murani, E., Reyer, H., Ponsuksili, S., Fritschka, S. & Wimmers, K. A substitution in the ligand binding domain of the porcine glucocorticoid receptor affects activity of the adrenal gland. PLoS ONE7, e45518 (2012). [DOI] [PMC free article] [PubMed]
- 39.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016;16:279–294. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 41.Reiner G, et al. Quantitative trait loci for white blood cell numbers in swine. Anim. Genet. 2008;39:163–168. doi: 10.1111/j.1365-2052.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- 42.Neumann A, et al. The low single nucleotide polymorphism heritability of plasma and saliva cortisol levels. Psychoneuroendocrinology. 2017;85:88–95. doi: 10.1016/j.psyneuen.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Larzul C, et al. The cortisol response to ACTH in pigs, heritability and influence of corticosteroid-binding globulin. Animal. 2015;9:1929–1934. doi: 10.1017/S1751731115001767. [DOI] [PubMed] [Google Scholar]
- 44.Mallard, B. A., Wilkie, B. N., Kennedy, B. W., Gibson, J. & Quinton, M. Immune responsiveness in swine: eight generations of selection for high and low immune response in Yorkshire pigs. In Proceedings of the 6th World Congress on Genetics Applied to Livestock Production, Armidale, Australia, January 11–16, 1998. Volume 27: Reproduction; fish breeding; genetics and the environment; genetics in agricultural systems; disease resistance; animal (1998).
- 45.Wilkie B, Mallard B. Selection for high immune response: An alternative approach to animal health maintenance? Vet. Immunol. Immunopathol. 1999;72:231–235. doi: 10.1016/S0165-2427(99)00136-1. [DOI] [PubMed] [Google Scholar]
- 46.Peng SL, Gerth AJ, Ranger AM, Glimcher LH. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity. 2001;14:13–20. doi: 10.1016/S1074-7613(01)00085-1. [DOI] [PubMed] [Google Scholar]
- 47.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 48.Teixeira LK, et al. NFAT1 transcription factor regulates cell cycle progression and cyclin E expression in B lymphocytes. Cell Cycle. 2016;15:2346–2359. doi: 10.1080/15384101.2016.1203485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xanthoudakis S, et al. An enhanced imune respones in mice lackinh the transcription factor NFAT1. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 50.Dragone LL, Myers MD, White C, Sosinowski T, Weiss A. Src-like adaptor protein regulates B cell development and function. J. Immunol. 2006;176:335–345. doi: 10.4049/jimmunol.176.1.335. [DOI] [PubMed] [Google Scholar]
- 51.Giovannone N, et al. Human B cell differentiation is characterized by progressive remodeling of O-linked glycans. Front. Immunol. 2018;9:2857. doi: 10.3389/fimmu.2018.02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martínez-Riaño A, et al. Antigen phagocytosis by B cells is required for a potent humoral response. EMBO Rep. 2018;19:e46016. doi: 10.15252/embr.201846016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, et al. Novel functions of murine B1 cells: Active phagocytic and microbicidal abilities. Eur. J. Immunol. 2012;42:982–992. doi: 10.1002/eji.201141519. [DOI] [PubMed] [Google Scholar]
- 54.Parra D, et al. Pivotal advance: Peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+ T cells. J. Leukoc. Biol. 2012;91:525–536. doi: 10.1189/jlb.0711372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flannagan RS, Cosío G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 56.D’Souza Z, Blackburn JB, Kudlyk T, Pokrovskaya ID, Lupashin VV. Defects in COG-mediated golgi trafficking alter endo-lysosomal system in human cells. Front. Cell Dev. Biol. 2019;7:118. doi: 10.3389/fcell.2019.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119:1623–1633. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 58.Holtmeier, W. & Kabelitz, D. γδ T Cells link innate and adaptive immune responses. In Mechanisms of Epithelial Defense, Vol. 86, 151–183 (KARGER, Basel, 2005). [DOI] [PubMed]
- 59.Mair KH, et al. The porcine innate immune system: An update. Dev. Comp. Immunol. 2014;45:321–343. doi: 10.1016/j.dci.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen T, et al. Exploration of microRNAs in porcine milk exosomes. BMC Genomics. 2014;15:100. doi: 10.1186/1471-2164-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xing Z, Conway EM, Kang C, Winoto A. essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J. Exp. Med. 2004;199:69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Croasdell A, et al. PPARγ and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015;2015:549691. doi: 10.1155/2015/549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szalai, A. J. The biological functions of C-reactive protein. In Vascular Pharmacology (2002). 10.1016/S1537-1891(02)00294-X. [DOI] [PubMed]
- 64.Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N. Engl. J. Med. 1997;336:973–979. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- 65.Hage FG, Szalai AJ. C-reactive protein gene polymorphisms, C-reactive protein blood levels, and cardiovascular disease risk. J. Am. Coll. Cardiol. 2007 doi: 10.1016/j.jacc.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 66.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: A model for human infectious diseases. Trends Microbiol. 2011;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neumann CA, et al. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003 doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 68.R Core Team. R: A Language and Environment for Statistical Computing. (2016).
- 69.Ellis, B. et al. FlowCore: Basic Structures for Flow Cytometry Data. R package version 2.0.1 (2020).
- 70.Hahne, F., Gopalakrishnan, N., Hadj Khodabakhshi, A., Wong, C.-J. & Lee, K. flowStats: Statistical Methods for the Analysis of Flow Cytometry Data. (2019).
- 71.Finak G, et al. OpenCyto: An open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput. Biol. 2014 doi: 10.1371/journal.pcbi.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Misztal, I. et al. 7th World Congress on Genetics Applied to Livestock Production, August 19–23, 2002, Montpellier, France. In 7th World Congress on Genetics Applied to Livestock Production (2002).
- 74.Meyer K, Houle D. Sampling based approximation of confidence intervals for functions of genetic covariance matrices. Proc. Assoc. Advmt. Anim. Breed. Genet. 2013;20:523–526. [Google Scholar]
- 75.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011 doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 78.Turner S. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018 doi: 10.1101/005165. [DOI] [Google Scholar]
- 79.Hu ZL, Park CA, Reecy JM. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smedley D, et al. The BioMart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016 doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eppig, J. T. et al. Mouse genome informatics (MGI): resources for mining mouse genetic, genomic, and biological data in support of primary and translational research. In Methods in Molecular Biology Vol. 1488, 47–73 (2017). [DOI] [PubMed]
- 84.Safran M, et al. GeneCardsTM 2002: Towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18:1542–1543. doi: 10.1093/bioinformatics/18.11.1542. [DOI] [PubMed] [Google Scholar]
- 85.Yates AD, et al. Ensembl 2020. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen W, Le S, Li Y, Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE. 2016 doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bindea H, et al. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results from all data generated or analysed during this study are included in this published article (and its Supplementary Information files). However, the datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.