Abstract
The incidence of oropharyngeal squamous cell carcinoma (OPSCC) has been increasing due to high-risk HPV infection. We explored the significance of genetic alterations in HPV-positive (HPV-P) and HPV-negative (HPV-N) OPSCC patients on long-term outcome. A total of 157 cases of primary resected OPSCC diagnosed from 1978 to 2005 were subjected to a targeted exome sequencing by MSK-IMPACT™ interrogating somatic mutations in 410 cancer-related genes. Mutational profiles were correlated to recurrence and survival outcomes. OPSCC included 47% HPV-positive (HPV-P) and 53% HPV-negative (HPV-N) tumors arising in the base of tongue (BOT, 43%), palatine tonsil (30%) and soft palate (SP, 27%). HPV negative status, SP location and smoking were associated with poorer outcome. Poorer overall survival was found in NOTCH1-mutated HPV-P (p=0.039), and in SOX2-amplified HPV-N cases (p=0.036). Chromosomal arm gains in 8p and 8q, and 16q loss were more common in HPV-P (p=0.005, 0.04 and 0.01, respectively), while 9p, 18q and 21q losses were more frequent in HPV-N OPSCC (p=0.006, 0.002 and 0.01, respectively). Novel, potentially functional JAK3, MYC and EP300 intragenic deletions were found in HPV-P, and FOXP1, CDKN2A, CCND1 and RUNX1 intragenic deletions and one FGFR3 inversion were detected in HPV-N tumors. HPV-N/TP53-wild type OPSCC harbored recurrent mutations in NOTCH1/3/4 (39%), PIK3CA, FAT1, and TERT. In comparison to their oral and laryngeal counterparts, HPV-N OPSCC were genetically distinct. In OPSCC, HPV status, tumor subsite and smoking determine outcome. Risk-stratification can be further refined based on the mutational signature, namely NOTCH1 and SOX2 mutation status.
Keywords: HPV, oropharynx, squamous cell carcinoma, NOTCH1, SOX2
INTRODUCTION
The incidence of oropharyngeal squamous cell carcinoma (OPSCC) has increased over the past several decades, largely due to the increasing number of human papillomavirus (HPV)-induced tumors 1, 2. In comparison to HPV-negative (HPV-N) squamous cell carcinoma cases, which typically occur in older men with a significant history of tobacco exposure and increased alcohol consumption, HPV-related OPSCC affects patients who are younger, have less comorbidities, respond favorably to treatment and have remarkably better overall survival (OS) 3–6. The differences in etiology, clinical presentation and response to treatment suggests that HPV-positive (HPV-P) and HPV-N SCC might be biologically distinct although the main putative mechanisms of early carcinogenesis may be similar in their final result. While the loss of functioning p53 and p16INK4A-Cyclin D1-RB pathways in HPV-negative tumors occur through highly recurrent pathogenic TP53 and CDKN2A mutations, these mutations are relatively uncommon in HPV-P SCC 7 and the inactivation of p53 and RB occurs as a result of binding of these tumor suppressors by HPV E6 and E7, respectively 8, 9. Over the past few years, several genomic studies have emerged highlighting the mutational profiles of HPV-P and HPV-N SCC 7, 10–13. However, the majority of these prior studies include a relatively small number of cases arising from the oropharynx 7, are focused on all HPV-P tumors irrespective of the site of origin 12, and often compare alterations to HPV-N tumors from the oral cavity, larynx, hypopharynx rather than HPV-N oropharynx tumors. In addition, these prior studies lack clinical outcomes data rendering correlations of genomic changes to outcome very difficult if not impossible 13. In the present study, we have overcome these limitations by focusing only on oropharyngeal SCC with an approximately equal number of HPV-P and HPV-N tumors from a large retrospective cohort of surgically resected primary OPSCC. This has allowed a more accurate comparison of the genomic alterations between HPV-P and HPV-N oropharyngeal cancer. The long-term follow-up data on survival and recurrence has also enabled us to identify molecular signatures which impact on survival and recurrence for both HPV-P and HPV-N oropharyngeal cancer.
MATERIALS AND METHODS
I. Study cohort
The study was approved by the institutional review board of Memorial Sloan Kettering Cancer Center (MSKCC, New York, NY). A total of 157 patients who were diagnosed with primary OPSCC and underwent surgical resection with curative intent at out institution between 1978 and 2005 with available archival tumor specimens were included in the present study. Patients with recurrent disease, those who received any prior treatment or with tumors arising from other sites with extension to the oropharynx were excluded from the study. The clinical and pathologic features were reviewed. The high risk human papillomavirus (HR-HPV) status in each case was determined based on a positive p16 cytoplasmic and nuclear immunohistochemical stain in at least 70% tumor cells as previously described 14 and by detection of HPV sequence reads using the bioinformatics algorithm for mapping of off-target reads to the HPV genome 15. HPV RNA ISH was performed in 5 select cases 16.
II. DNA extraction and targeted capture massively parallel sequencing
Genomic DNA from surgically resected tumor and patient matched normal samples were extracted from formalin-fixed paraffin-embedded (FFPE) specimens. Massively parallel sequencing was performed using MSK-IMPACT™ (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets) platform designed for targeted sequencing of exons and select introns of 410 cancer-related genes as previously reported 17, 18, (Supplementary Methods).
III. Functionality of somatic mutations
Potential biological significance of genetic alterations of select genes (NOTHC1/2/3/4) were determined using OncoKB annotation 19 and “Functional impact” as listed on www.cbioprtal.org. Mutations designated as likely/predicted oncogenic and/or probably/possibly damaging were referred to as “pathogenic”.
IV. Copy number analysis (FACETS)
Copy number aberrations were identified by comparing sequence coverage of targeted regions in a tumor sample relative to a standard diploid normal sample. A minimum of 2-fold change was required to consider gene amplification or deletion. The FACETS analysis was performed in 86 (55%) samples (35 HPV-N and 51 HPV-P) with matched normal DNA and in 71 (45%) samples (48 HPV-N and 23 HPV-P) using unmatched/pool normal DNA as previously described 20. In brief, the FACETS algorithm determines the total and allele-specific copy number from the sequence coverage and genotypes of polymorphic SNPs (single nucleotide polymorphisms) across the genome, inferred from both on- and off-target read alignments. Estimates of total copy number from FACETS analysis was used for downstream comparison of copy numbers across samples. Due to the sparsity of targeted regions in MSK-IMPACT™ assay, gains and losses were called at the arm level if at least 50% of the arm is gained or lost respectively. These are then compared across HPV status using Fisher’s exact test and p-values corrected for multiple testing using false discovery rate correction 21.
V. Statistical analysis
Statistical analyses were performed using the SPSS software 24.0 (IBM Corporation, New York, NY, U.S.) and R 22. Clinical characteristics, molecular alteration and prognosis were compared using appropriate statistical tests, i.e. Log rank test for survival analysis, Chi-square test or Fisher’s exact test for nonparametric variables, and two tailed Student’s t test for continuous variables. P values less than 0.05 were considered to be statistically significant. Holm-Bonferroni multiple testing correction was used for comparison of copy number changes in HPV-P and HPV-N OPSCC.
RESULTS
I. Clinicopathologic characteristics and prognosis relative to HPV status
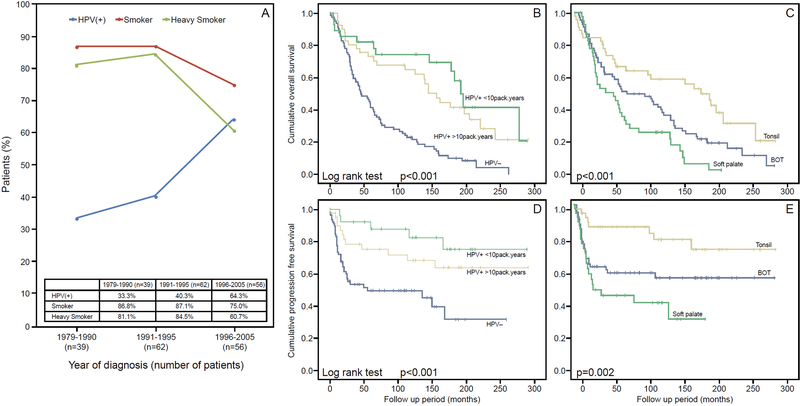
The clinical and pathologic features of the study cohort are summarized in Table 1. In brief, among the 157 patients with OPSCC, 47% (74/157) were positive and 53% (83/157) were negative for HR-HPV. Patients diagnoses rendered over the 26 year-period showed that the proportion of smokers and heavy smokers gradually declined from 87% and 81% to 75% and 61%, respectively, while the proportion on HPV-P tumors showed a gradual temporal increase from 33% to 64% (Figure 1A). Sixty-seven tumors (43%) originated from base of tongue (BOT), 47 (30%) from palatine tonsil, and the remaining 43 (27%) from soft palate (SP). With a median follow up of 67 months overall and 182 months among living patients (range 0.2 – 291 months), 56 patients (36%) exhibited disease progression, including 31 (20%) patients with local recurrence, 19 (12%) patients with regional recurrence and 22 (14%) patients with distant metastases. Compared with HPV-N OPSCC, HPV-P tumors were associated with non-smoker status (p=0.002), a tumor location in the BOT and tonsil rather than SP (p<0.001), a higher frequency of positive neck disease (p=0.005). The remaining clinicopathologic characteristics did not differ significantly based on the HPV status (Table 1). The 5-year and 10-year overall survival (OS) in our cohort was 58% and 44% respectively. Patients harboring HPV-P OPSCC had significantly improved OS and progression-free survival (PFS) compared to their HPV-N counterpart (Figure 1B, 1D, Log rank test, p<0.001 for OS and PFS). The 5-year and 10-year OS were 42% and 23% in HPV-N patients, and 76% and 69% in HPV-P patients. In our cohort, HPV-P OPSCCs patients who smoked showed a nonsignificant trend towards poorer outcome compared with never-smokers (Log rank test, p=0.318 for OS, and p=0.229 for PFS). When comparing tumors from different subsites of the oropharynx, OPSCC from the SP had a poorer OS with OPSCC from the tonsil having the best OS (Figure 1C, 1E, Log rank test, p<0.001 for OS, and p=0.002 for PFS). Such differences in clinical outcomes were significant when comparing among any two different subsites using Log rank pairwise test (BOT vs. SP: p=0.036; BOT vs. tonsil: p=0.023, and SP vs. tonsil: p<0.001).
Table 1.
Clinicopathologic characteristics of the study cohort.
| HPV (−) | HPV (+) | P value | |
|---|---|---|---|
| N | 83 (53%) | 74 (47%) | |
| Sex | 0.126 | ||
| Female | 27 (33%) | 16 (22%) | |
| Male | 56 (67%) | 58 (78%) | |
| Age, median (range) | 61 (27 – 79) | 59 (35–84) | 0.445 |
| Smoking history | 0.002 | ||
| Unknown | 1 (1%) | 0 | |
| No | 7 (8%) | 20 (27%) | |
| Yes | 75 (90%) | 54 (73%) | |
| < 10 pack/years | 2 (2%) | 9 (12%) | |
| > 10 pack/years | 71 (86%) | 42 (57%) | |
| Unknown quantity | 2 (2%) | 3 (4%) | |
| Site of origin | < 0.001 | ||
| Base of tongue | 30 (36%) | 37 (50%) | |
| Tonsil | 18 (22%) | 29 (39%) | |
| Soft palate | 35 (42%) | 8 (11%) | |
| Clinical T staging | 0.183 | ||
| TX | 1 (1%) | 0 | |
| T1/T2 | 49 (59%) | 52 (70%) | |
| T3/T4 | 33 (40%) | 22 (30%) | |
| Clinical N staging | 0.02 | ||
| N0 | 38 (46%) | 20 (27%) | |
| N+ | 45 (54%) | 54 (73%) | |
| Perineural invasion | 0.171 | ||
| Unknown | 23 (28%) | 13 (18%) | |
| Absent | 43 (52%) | 49 (66%) | |
| Present | 17 (20%) | 12 (16%) | |
| Vascular invasion | 0.264 | ||
| Unknown | 21 (25%) | 13 (18%) | |
| Absent | 41 (49%) | 49 (66%) | |
| Present | 17 (25%) | 15 (20%) | |
| Surgical margin status | 0.791 | ||
| Unknown | 1 (1%) | 2 (3%) | |
| Negative | 43 (52%) | 38 (51%) | |
| Positive or close (<5 mm) | 39 (47%) | 34 (46%) | |
| AJCC pT staging | 0.313 | ||
| T1/T2 | 58 (70%) | 57 (77%) | |
| T3/T4 | 25 (30%) | 17 (23%) | |
| AJCC pN staging | 0.005 | ||
| Nx | 1 (1%) | 0 | |
| N0 | 40 (48%) | 19 (26%) | |
| N+ | 42 (51%) | 55 (74%) | |
| Post-operative radiation | 0.008 | ||
| No | 33 (40%) | 15 (20%) | |
| Yes | 50 (60%) | 59 (80%) | |
| Follow up period, months, median (range) | 153 (9 – 213) | 182 (0.2 – 291) | 0.626 |
| Overall survival status | NA | ||
| Deceased | 75 (90%) | 40 (54%) | |
| Alive | 8 10%) | 34 (46%) | |
| Progression free survival status | NA | ||
| Progressed | 38 (46%) | 18 (24%) | |
| Not progressed | 45 (54%) | 56 (76%) | |
| Local recurrence | NA | ||
| Absent | 60 (72%) | 66 (89%) | |
| Present | 23 (28%) | 8 (11%) | |
| Regional recurrence | NA | ||
| Absent | 71 (86%) | 67 (91%) | |
| Present | 12 (14%) | 7 (9%) | |
| Distant metastasis | NA | ||
| Absent | 68 (82%) | 67 (91%) | |
| Present | 15 (18%) | 7 (9%) | |
P values were obtained using Fisher’s Exact test or Chi-square test for nonparametric and two-tailed Student’s t test for continuous variables.
Deceased patients were excluded from the calculation.
Abbreviations: HPV=human papillomavirus; AJCC=American Joint Committee on Cancer, NA=not applicable.
Figure 1. Epidemiology and survival impact of HPV and smoking in OPSCC.
Trend of smoking (red), heavy smoking (>10 pack/years, green) and HPV positivity (blue) in OPSCC over years in the study cohort. The rate of HPV in OPSCC increased, while the proportion of patients who smoked decreased over time (A). Kaplan Meier plots for overall survival and progression free survival according to human papillomavirus (HPV) status, smoking history (B and D) and site of origin (C and E).
II. Genetic characteristics of OPSCC
A. Somatic mutations
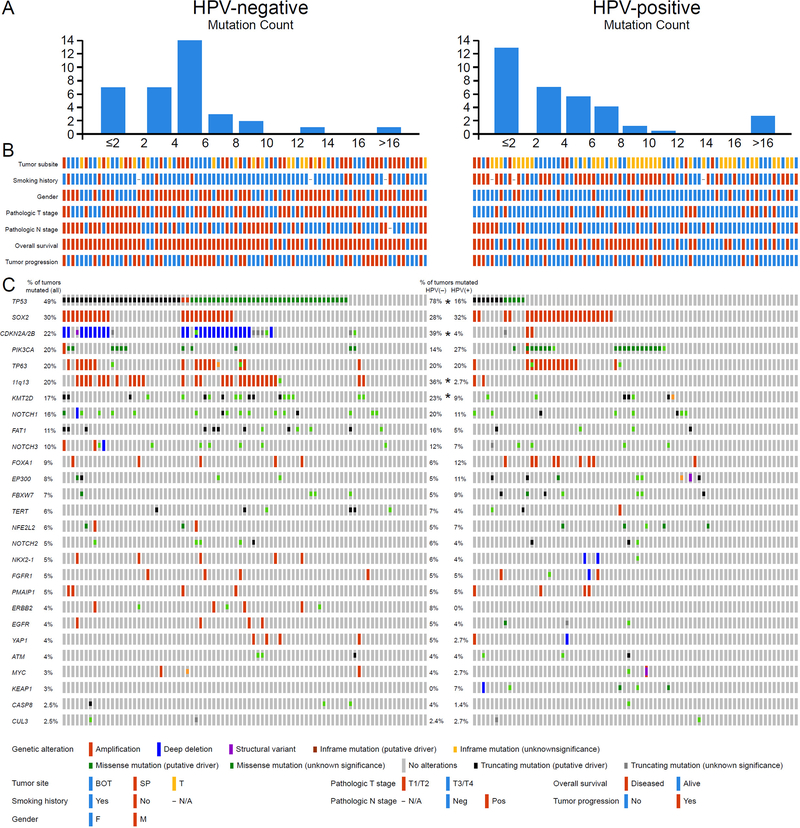
The cases were sequenced at a median depth of coverage of 550-fold (range 36–4765) and 858-fold (range 52–5362) for tumor and normal DNA, respectively. Eighty-four (54%) cases were analyzed with matched normal DNA, and in the remaining cases, a pool of 10 normal DNA samples was used because the matched normal DNA was either unavailable or did not pass QC. In addition to 5 promoter (TERT) mutations, 1236 exonic mutations were detected in 321 of 410 sequenced genes including 998 missense, 196 protein truncating (frameshift indels, nonsense, splice site and stop-loss) variants, and 42 non-frameshift indel mutations. There was no significant difference in tumor mutation burden (TMB), in respect to the HPV status in OPSCC. The median number of mutations was 5 (range 0–18) and 4 (range 0–60) mutations per case, in HPV-N and HPV-P tumors, respectively (Figure 2A).
Figure 2. Common genetic alterations in OPSCC according to HPV status.
(A) Tumor mutation burden (TMB) in OPSCC relative to the HPV status. (B) Clinicopathologic features the study cohort based on HPV status of the tumors, including tumor site (SP: soft palate; BOT: base of tongue; T: tonsil), smoking history, gender, AJCC pT stage, AJCC pN stage, overall survival status and tumor progression status. Color keys are shown in the bottom panel. (C) Oncoprints of HPV-negative (left) and HPV-positive (right) OPSCCs. Middle panel shows percentage of tumors altered for each event. *p<0.05 between HPV-negative and HPV-positive OPSCCs (Fisher’s exact test).
The most frequent and potentially significant genetic alterations in OPSCC were mutations in TP53 (49%), SOX2 (30%), CDKN2A/2B (22%), PIK3CA (20%), TP63 (20%), KMT2D (17%), NOTCH1 (16%), FAT1 (11%), and 11q13 (19%), and FOXA1 amplifications (9%), (Figure 2B).
When comparing HPV-N tumors to HPV-P tumors the most notable genetic differences between the two subsets of OPSCC relative to the HPV/p16 status included higher frequency of mutations in TP53 (78%, 65/83 vs. 16%, 12/74, p<0.001), CDKN2A/2B (39%, 33/83 vs. 4%, 7/74, p<0.001), more 11q13 gene cluster (FGF3/FGF4/FGF19/CCND1) amplifications (35%, 29/83 vs. 3%, 2/74, p<0.001), and more KMT2D mutations (23%, 19/83 vs. 9%, 7/74, Fisher’s exact test, p=0.030) in HPV-N OPSCC (Figure 2). Interestingly, among HPV-P tumors the frequency of TP53 mutations were significantly higher among cases diagnosed from 1979–1995 than among more recent cases diagnosed from 1996–2005 (25%, 10/38 vs. 6%, 2/36, Fisher’s exact test, p=0.020). This correlates with the higher percentage of smokers in earlier years and explains why this study has more TP53 mutants in HPV-P cases compared to more recent studies.
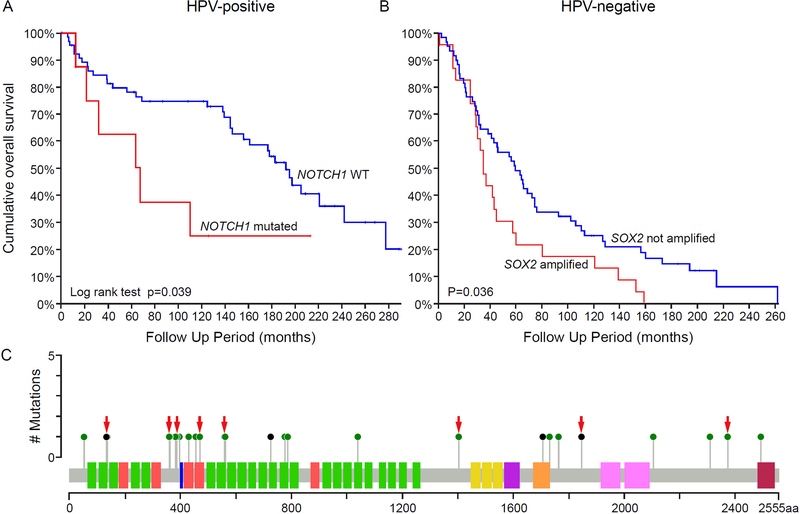
NOTCH pathway genes (NOTCH1, NOTCH2, NOTCH3, NOTCH4, EP300, FBXW7, SPEN, KDM5A) were mutated in 40% (33/83) and 36% (27/74) in HPV-N and HPV-P tumors, respectively. NOTCH1 mutations alone were detected in 20% (17/83) and 11% (8/74) of HPV-N and HPV-P OPSCC, respectively (Fisher’s exact test, p=0.086). NOTCH2, NOTCH3, and NOTCH4 mutations were found in 5% (8/157), 10% (16/157) and 4% (6/157) OPSCC, respectively, with 88% (7/8), 50% (8/16) and 28% (2/7) of these mutations being potentially pathogenic (Supplementary Figure S1). Importantly, HPV-P NOTCH1 mutated cases showed significantly worse OS than those with intact NOTCH1 (Log rank test, p=0.039, Figure 3A). Details of the location of the NOTCH1 mutations are shown in Figure 3C. Two, potentially bi-allelic mutations were detected in 4 cases; 71% (20/29) affected N-terminal EGF-like binding domain, 14% (4/29) were truncating alterations, and 76% (22/29) were likely pathogenic (i.e. likely/predicted oncogenic and/or probably/possibly damaging, Supplementary Table S1).
Figure 3. Prognostic molecular signatures for overall survival in OPSCC.
(A) In HPV-positive OPSCC, the presence of NOTCH1 mutation predicts worse overall survival (OS). (B) In HPV-negative OPSCC, SOX2 amplification is associated with worse OS. (C) NOTCH1 mutations in OPSCC. Red arrows point to the mutations in HPV-P tumors.
SOX2 amplifications were present at comparable frequencies in HPV-N and HPV-P cases (28%, 23/83 and 32%, 24/74). However, among HPV-N cases, SOX2 amplification was found exclusively in TP53 mutated tumors (Fisher’s exact test, p=0.002), and was associated with a worse OS in comparison to SOX2 copy number neutral cases (Log rank test, p=0.036, Figure 3B). In contrast, amplification of SOX2 did not affect OS (Log rank test, p = 0.328) in HPV-P OPSCC.
Histone modifiers (KMT2D, CREBBP, KMT2C, EP300, KMT2A) were overall more frequently altered in HPV-N tumors than in HPV-P SCC (40%, 33/83 vs. 26%, 9/74, Fisher’s exact test, p<0.001, Supplementary Figure S2).
NFE2L2, KEAP1, CUL3 oxidative stress genes mutations were more common in HPV-P tumors (16%,12/74 in HPV-P vs. 6%, 5/83 in HPV-N OPSCC; Fisher’s exact test, p=0.069) but this did not reach statistical significance. Interestingly, the majority of these variants were detected in HPV-P tumors (71%) including all 5 KEAP1 mutations. Four NFE2L2 mutations, including 2 hotspot variants were detected in HPV-P never smokers (Figure 2, Supplementary Figure S2).
Targetable alterations involving EGFR, ERBB2, and FGFR1 amplifications and FGFR3 hotspot mutations (in R249 and S248 codons) were detected in 8% (6/74) and 14% (12/83) HPV-P and HPV-N tumors, respectively (Supplementary Figure S2). Full mutational profiles for all cases are provided in Supplementary Table S2A.
B. Somatic chromosome arm level copy number alterations
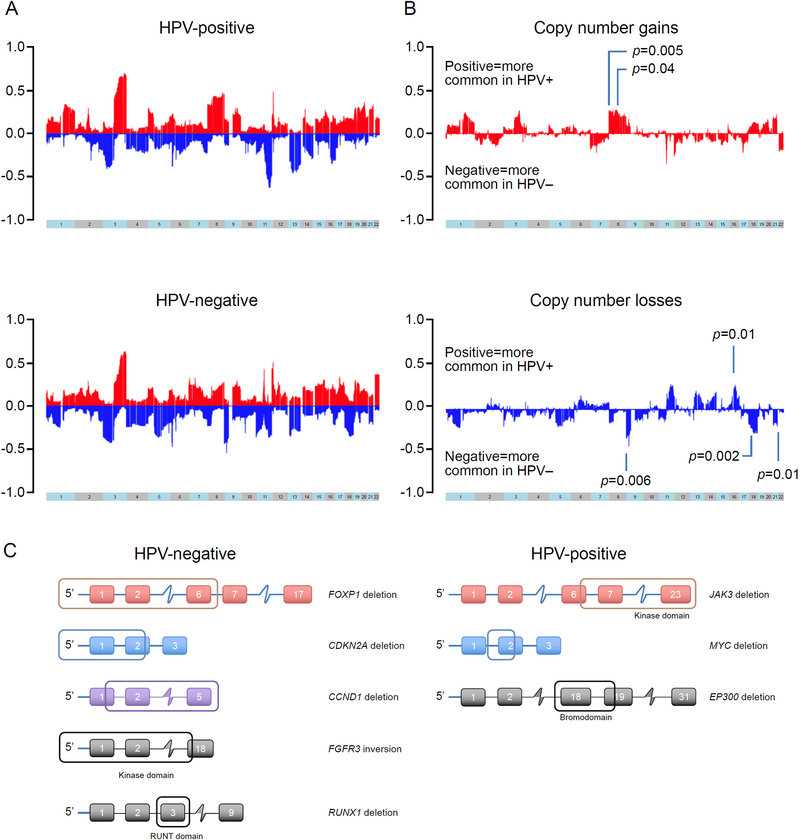
OPSCC were enriched in recurrent gains in chromosomes 1q, 3q, 22q and 20 and losses in 3p, 11q and 13q. HPV-P cases showed more frequent gains of chromosome arms 8p (p=0.005) and 8q (p=0.04) and more frequent losses of 16q (p=0.010). In contrast, HPV-N cases showed more frequent losses of 9p (p=0.006), 18q (p=0.002) and 21q (p=0.010, adjusted p-values, Holm-Bonferroni multiple testing correction), (Figure 4). No significant impact on OS was found relative to CNAs (data not shown). 11q losses and 3q gains tended to be more common in HPV-P OPSCC (43% vs 27%, p=0.03 and 62% vs 40%, p=0.007, respectively, Fisher’s exact test) although no significant difference was found after Holm-Bonferroni’s correction for multiple testing. Details on chromosome arms copy number changes for all cases is provided in Supplementary Table S2B.
Figure 4. Somatic copy number alterations (CNA) and structural variants in OPSCC.
(A) Frequent gains (red) of 1q, 3q, chromosome 20 and 22q, and losses (blue) of 3p, 11q and 13q were detected among all cases, irrespective of the HPV status. (B) Significant differences in copy number gains (red, top) and losses (blue, bottom). A positive value means that more common in HPV-positive patients and negative means more common in HPV-negative patients. HPV positive tumors showed more frequent gains of chromosome arms 8p (p=0.005), and 8q (p=0.040), and losses of 16q (p=0.010), and HPV negative tumors were significant for more losses of 9p (p=0.006), 18q (p=0.002) and 21q (p=0.010) (adjusted p-values, Holm-Bonferroni multiple testing correction). (C) Structural variants in OPSCC. Left side and right-side panels depict structural variants in HPV negative and HPV positive tumors, respectively. Affected regions are framed.
C. Structural variants
We identified 9 structural variants with potential functional significance, with 6 being detected in HPV-N and 3 in HPV-P SCC (Figure 4C). Among HPV-N carcinomas, in two cases there were large deletions including the promoter region which suggested loss of transcriptional activity. In one such case, FOXP1 was affected showing a large deletion involving the 5’ UTR and exons 1–6, and in the other case CDKN2A showed a 44 Kb intragenic deletion involving the 5’ UTR and exon 1 and part of exon 2. In other cases, large deletions suggested a loss of functional domain(s) and/or nearly a complete loss of functional protein. A cell cycle regulator gene CCND1 showed 1.4 Mb intragenic deletion involving part of exon 1 and remaining downstream exons suggesting a near complete loss of the protein. A kinase encoding gene FGFR3 showed an 18 Kb inversion with the breakpoints (1) 5 Kb upstream from exon 1 (intragenic region) and (2) within the exon 18, involving the entire protein kinase domain and suggesting a loss of functional protein. A transcription factor gene RUNX1 showed a 3 Kb inversion involving exon 3 disrupting the RUNT domain. In one case, NTRK2 showed multiple duplications/deletions suggestive of a processed pseudogene 23. Among HPV positive cases, JAK3 was affected in one case showing a 50 Kb deletion involving exon 6 and the remaining downstream exons encoding the entire kinase domain (amino acids 521–777, 822–1095) and FERM domain. In another case, MYC oncogene showed a 2 Kb intragenic deletion with the breakpoints (1) in the intron closely downstream to exon 1 and (2) within the exon 2. In another HPV positive case, an epigenetic modifier gene EP300 showed a 1.2 Kb intragenic deletion of exon 18 and part of exon 19, resulting in the loss of bromodomain. Our findings are in line with previous studies suggesting that most structural variants in head and neck squamous cell carcinoma are more commonly associated with loss of function rather than with protein-coding fusion events 7.
D. Effect of smoking in HPV-P OPSCC
Our clinical outcomes data suggested patients who smoked >10 pack-years had a poorer OS compared to patients who were nonsmokers or smoked <10 pack-years (Figure 1B). Supplementary Figure S3A shows a comparison of the somatic mutations in HPV-P tumors stratified by smoking status. HPV-P patients who smoked >10 pack-years were more likely to have tumors with a high frequency of TP53 mutations and MAP3K13 amplification, as well as a lower frequency of FOXA1 amplification. However, the difference did not reach significant level due to the small sample size (Fisher’s exact test, p = 0.183, 0.140 and 0.146 respectively). Among HPV-P heavy smokers, patients with TP53-mutated tumors tend to have worse OS and shorter PFS than TP53-wild types (p=0.07 and p=0.149, respectively, Supplementary Figures 3SB and S3C).
E. HPV-negative/TP53-wild type oropharyngeal squamous cell carcinomas
Eighteen patients had HPV-negative/TP53 wild type tumors. Six (33%) tumors were found to have other known driver oncogenic alterations involving PIK3CA, HRAS, ATM, MYC, CCND1 and FGFR1 genes. Notably, 7 (39%) cases harbored NOTCH1, NOTCH3 and/or NOTCH4 mutations, and in 2 (11%) cases had TERT promoter mutations (Supplementary Figure S4A). Interestingly, this subset of patients showed a tendency towards poorer outcome than HPV-N/TP53-mutated cases although a statistical significance was not reached (Supplementary Figure S4B).
III. Pathways altered in OPSCC
Supplementary Figure S2 shows the mutations grouped by pathways in HPV-N and HPV-P tumors. The p53 signaling pathway and cell cycle control pathway were more commonly altered in HPV-N tumors whereas genes involved in oxidative stress were more commonly altered in HPV-P tumors. The PIK3/AKT/mTOR pathway was more commonly altered in HPV-P tumors (47% vs. 37%) but this was not statistically significant. NOTCH signaling was altered in 36% HPV-P tumors and 40% HPV-N tumors. As previously mentioned, mutations in NOTCH1 were associated with poorer OS in the HPV-P patients (Figure 3A).
IV. Comparison of the genetic profile of OPSCC to other HNSCC in the literature
Supplementary Table S3 shows a comparison of mutational profiles in HNSCC. Comparisons are made only for 410 genes included in the MSK-IMPACT™ panel (Supplementary Table S4).
A. Comparison to reported genetic profiles of HPV-P OPSCC
No significant genetic differences were observed between the primary and recurrent/metastatic HPV-P tumors. Interestingly, HPV-P OPSCC showed fewer PIK3CA mutations in comparison to the TCGA HPV-P tumors (19% vs. 56%, Fisher’s exact test, p=0.006), (Supplementary Table S3).
B. Comparison to reported genetic profiles of HPV-N SCC: OPSCC vs. oral (OSCC) vs. laryngeal SCC (LSCC)
In comparison to HPV-N OSCC and LSCC, HPV-N OPSCC had less mutations in CDKN2A (p=0.010 and p=0.035), MYC (p=0.035 and p=0.007), EGFR (p=0.013 and 0.053), and KEAP1 (p=0.054 and p=0.004) respectively. HPV-N OPSCC also had less FAT1 (p<0.001), CASP8 (p=0.005), and NFE2L2 (p=0.042) mutations, and more ERBB2 (p=0.049) mutations, and more FOXA1 (p=0.048), SOX2 (p=0.004), and NKX2–1 (p=0.048) amplifications than HPV-N OSCC. In addition, HPV-N OPSCC harbored less TP53 (p=0.013), CUL3 (p=0.024), and TP63 (p=0.031) than HPV-N LSCC (Supplementary Table S3).
Clinical and pathological features of 157 OPSCC are provided in Supplementary Table S5.
DISCUSSION
Here we report the genomic landscape of the largest retrospective OPSCC cohort to date with long-term outcome data which has allowed correlations to be carried out between clinical, pathological and genetic characteristics in both HPV-P and HPV-N OPSCC patients. In contrast to previously published genomic studies on SCC which report on multiple different head and neck sites, we were able to focus specifically on the oropharynx and therefore have been able to carry out a more accurate comparison of the genomic alterations in HPV-P and HPV-N tumors. We found that HPV infection, and tonsil and BOT as primary subsites emerged as favorable prognostic indicators. We found that NOTCH1 mutations were associated with worse OS in HPV-P OPSCC, and SOX2 amplifications were associated with worse OS in HPV-N OPSCC. Smokers with HPV-P OPSCC had a poorer OS compared to nonsmokers, harbored TP53 mutation and MAP3K13 amplification, and had fewer FOXA1 amplifications. We also found that HPV-N tumors which were TP53-wild-type harbored frequent genetic alterations in the NOTCH pathway. In comparison to their oral and laryngeal counterparts, HPV-N OPSCC were genetically distinct and had less CDKN2A, MYC, EGFR, and oxidative stress genes KEAP1 and NFE2L2 mutations, and were relatively enriched in SOX2, FOXA1, ERBB2 and NKX2–1 amplifications.
Given the time period of the patients’ cohort we were able to show the inverse trend of decreasing smoking and increasing HPV infection in OPSCC 1, 3, 4. Although the reported rate of HPV-P OPSCC in the literature is variable reaching 70% 24, a 47% HPV positivity rate in our study may be explained by a high proportion of cases diagnosed prior to 1995. Even though the majority of HPV-P OPSCC arose in smokers, the significant association of HPV infection with the non-smoker status further supports the established independent oncogenic role of high-risk HPV in OPSCC 1, 4. The presence of HPV was associated with significantly better outcome; 5-year and 10-year OS were 76% and 69%, respectively in HPV-P cases compared to 42% and 23%, respectively in HPV-N OPSCCs. A poorer outcome of SCC of the SP may be attributed to the predominance of HPV-N tumors in this subsite. Although anatomically a part of the oropharynx, SP mucosa is notably histologically distinct; it lacks a rich lymphoid stroma and specialized epithelium, which are typically seen in tonsil and considered a suitable host environment for HPV infection 25. The distinct anatomy of SP might contribute to the lower frequency of HPV-P tumors arising in this location in comparison to the BOT and tonsil.
As expected, we found that TP53 mutations were the most common genetic alteration in our cohort 7, 13, 26 affecting 49% of OPSCC patients and that the presence of TP53 mutation was associated with a significantly decreased OS (p<0.001, data not shown). When sub-stratified by the HPV-status, TP53 mutation alone did not significantly impact survival in either HPV-P or HPV-N group suggesting that the survival disadvantage of TP53 mutation by itself is not sufficient to modify the impact of HPV on outcome in OPSCC. However, among HPV-P heavy smokers, those with TP53-mutated tumors tend to fare worse than their TP53-wild type counterparts. This finding emphasizes the importance of the dosage to tobacco carcinogens exposure raising a question if the behavior of HPV-P/TP53-mutated OPSCC in heavy smokers may be more similar to that of HPV-N OPSCC and could argue against de-intensified treatment in this subset of HPV-P patients.
NOTCH1 mutations in OPSCC overall occurred at comparable frequencies to those reported on cohorts comprised of predominantly non-oropharyngeal HNSCC, and most variants were detected in the EGF-like ligand binding domain 10, 7. Importantly, we observed poorer OS in patients with NOTCH1-mutated HPV-P tumors compared to their NOTCH1-wild type counterparts, but no such difference was seen in the HPV-N group. Clinical studies on the prognostic significance of NOTCH1 alterations in OPSCC are limited. Tinhofer et al. studied outcome in a SCC cohort treated with concurrent chemoradiation (CXRT), including 60% OPSCC and overall 21% HPV-P tumors, and found that NOTCH1-mutated SCC were associated with better outcome irrespective of the HPV status 13. They suggested that the absence of high expression and activity of NOTCH1, which is reportedly linked to chemoresistance, might explain the better outcome in CXRT-treated NOTCH1-mutated cases 13. In contrast to the latter study and in line with our findings, in an OSCC cohort in Asian patients (lacking information on HPV status or postoperative treatment modality) mutant NOTCH1 emerged as an independent prognostic indicator of decreased DFS and OS 27. These results may support NOTCH1 as a putative tumor suppressor gene in HNSCC. Furthermore, a recent study on a mouse model showed that inactivation of canonical Notch signaling drives head and neck carcinogenesis irrespective of the presence of HPV oncogenes. However, they also demonstrated that HPV oncogenes synergized with lost Notch signaling to induce more and higher grade cancers than seen in the HPV-N mouse model 28. The latter data may lend support to our observation that NOTCH1 mutated tumors may be a relatively more aggressive subset of HPV-P OPSCC. We conclude that the prognostic significance of NOTCH1 mutations in HNSCC needs further investigation and might depend on the HPV status, treatment modality, and the tumor primary site.
Among HPV-N cases, SOX2 emerged as a gene of prognostic significance. SOX2 amplification was found exclusively among TP53 mutated cases (Fisher’s exact test, p=0.004), and was associated with worse OS than SOX2 copy number-neutral cases. This could suggest that overexpressed SOX2 may act in synergy with loss of p53 function and result in a relatively more aggressive subset of HPV-N OPSCC. SOX2 copy number status and protein expression in HNSCC relative to the HPV status are limited and controversial. While Schrock et al. reported lack of SOX2 amplification in 31 HPV-P HNSCC along with reduced protein expression suggesting SOX2 amplification to be mutually exclusive with HPV infection 29, other studies reported SOX2 protein overexpression in up to 92% high risk HPV-P HNSCC 30, 31. SOX2 protein overexpression was identified as a poor prognostic indicator in HNSCC including sites other than oropharynx 31, 32. Given the low frequency of HPV-P SCC outside the oropharynx these previously published data are in line with our data and imply that SOX2 amplification might be a poor prognostic indicator in HPV-N OPSCC. On the other hand, in the study by Bayo et al. in a multivariate analysis on 287 HNSCC (59% from oropharynx) low SOX2 expression emerged as an independent prognostic marker for HNSCC patients overall irrespective of the tumor site or HPV status 31.
Our study also identified chromosomal and structural variant alterations in both HPV-P and HPV-N tumors. 11q losses have been reported to be frequent in HNSCC irrespective of the tumor site or HPV status 7 and commonly co-occur with 11q13 cluster amplification. Interestingly, in our study 11q loss including ATM gene tended to be more frequent in HPV-P tumors, which were otherwise characterized by very few 11q13 amplifications. Other recurrent chromosome arms losses implied losses of important tumor suppressor genes such as SETD2, VHL and BAP1 on 3p, and RB1 on 13q. In contrast, HPV-N tumors had a higher frequency of 9p losses consistent with frequent deletion of CDKN2A and CDKN2B genes, which is similar to prior reports on HPV-N arising in different head and neck sites 7, 12. Interestingly, in contrast to prior studies, we also noted that HPV-N OPSCC were enriched for losses of 18q including SMAD4, as well as 21q losses. HPV-P had frequent chromosome 8 gains including FGFR1 and MYC, and although gain of 3q including PIK3CA, SOX2 and TP63 was common in both subsets 12, it tended to be more common in HPV-P cases. HPV-P OPSCC also harbored more losses of 16q including CDH1 gene. Loss of functional CDH1 has been reported to be related to tumor infiltration and metastasis in other cancers 33 and raises the question that CDH1 loss may have a similar effect in HPV-P tumors and contribute to the significantly higher number of N positive cases in comparison to HPV-N patients.
Despite the limitations of our molecular assay to detect gene fusions, our data implied that recurrent gene fusions were not common in OPSCC. This is consistent with prior genomic studies on HNSCC. However, we did identify several novel structural variants with possible loss of functional protein involving FOXP1, CDKN2A, CCND1, FGFR3 and RUNX1 in HPV-N, and JAK3, MYC and EP300 in HPV-P tumors. These findings warrant further studies to determine their role in OPSCC carcinogenesis.
In conclusion, our study is unique in that it is the largest genomic study on OPSCC as a single head and neck site to date. By having a cohort of tumors only on the oropharynx subsite with equal numbers of HPV-P and HPV-N tumors we have been able to identify the differences in somatic mutations, chromosomal alterations and structural variants between HPV-P and HPV-N tumors. We were able to observe the differences in respect to the oropharyngeal subsites and identify SP as the location that gives rise to predominantly HPV-N SCC with poor clinical outcome. Our data support the notion that lost Notch signaling in NOTCH1-mutated HPV-P tumors may co-operate with HPV oncogenes in the process of carcinogenesis and characterize a relatively more aggressive subset of HPV-P OPSCC. In addition, patients with HPV-P tumors who smoke have poorer outcome and we identify a higher incidence of TP53 mutations in these patients. Larger studies are therefore needed to assess the risk in heavy smokers with HPV-P/TP53-mutant OPSCC. In HPV-N tumors, we identify the prognostic significance of SOX2 amplification and also show that HPV-N oropharynx tumors have a different genomic profile to HPV-N tumors from other sites of the head and neck such as the oral cavity and larynx. This comparison of tumors shows that HNSCC does not merely comprise two SCC categories that can be either HPV-P or HPV-N but rather, it is a group of subsite-specific SCC that can be further subclassified based on site as well as the HPV status.
Supplementary Material
Supplementary Figure S1. Spectrum and functional significance of NOTCH2/3/4 mutations in OPSCC.
Supplementary Figure S2: Genetic differences in pathway alterations in HPV-P and HPV-N OPSCC.
Supplementary Figure S3. (A) Effects of cigarette smoking on molecular profiles of HPV-P OPSCC. HPV-P OPSCC in heavy smokers (>10 pack-years) are associated with a non-significant trend of higher frequency of TP53 mutation and MAP3K13 amplification, as well as lower frequency of FOXA1 amplifications (Fisher’s exact test, p=0.183, 0.140 and 0.146 respectively). (B) HPV-P with TP53 mutations arising in heavy smokers showed trend towards poorer OS (p=0.07) and (C) poorer progression free survival (p=0.149) than their TP53-wild type counterparts.
Supplementary Figure S4. (A) Mutation profile of HPV-N/TP53-wild-type OPSCC. (B) HPV-N/TP53-wild type cases trended towards worse progression free survival than HPV-N/TP53-mutated ones.
Supplementary Table S1. Spectrum and functional significance of NOTCH1 mutations in OPSCC.
Supplementary Table S2A. Somatic mutations in 157 OPSCC.
Supplementary Table S2B. Chromosome arm changes in 157 OPSCC.
Supplementary Table S3. Comparison of mutational profiles in HNSCC.
Supplementary Table S4. 410 gene list of MSK-IMPACT™ assay.
Supplementary Table S5. Clinical and pathological features of 157 OPSCC.
Novelty & Impact Statement:
NOTCH1 mutations (11%) in HPV-positive and SOX2 amplification (28%) in HPV-negative oropharyngeal squamous cell carcinoma are poor prognostic indicators suggesting that each group can be further risk sub-stratified based on their mutational signatures. Mutational profiles of HPV-negative squamous cell carcinoma arising in different subsites, oropharynx, larynx and oral cavity are distinct suggesting that HPV-negative squamous cell carcinoma is a genetically heterogeneous disease.
ACKNOWLEDGEMENTS
We thank Susan D. Weil for her expert assistance in figures preparation.
Research reported in this publication was supported by the LesLois Hutchison Foundation and in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LIST OF ABBREVIATIONS
- AJCC
American Joint Committee on Cancer
- AWD
alive with disease
- BOT
base of tongue
- damage
damaging
- FFPE
formalin-fixed paraffin-embedded
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- HR
high risk
- HPV-N
HPV-negative
- HPV-P
HPV-positive
- MSK-IMPACT™
Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets
- NA
not applicable
- NED
no evidence of disease
- neg
negative
- onc
oncogenic
- OPSCC
oropharyngeal squamous cell carcinoma
- OS
overall survival
- PFS
progression-free survival
- pos
positive
- SCC
squamous cell carcinoma
- SNP
single nucleotide polymorphism
- SP
soft palate
- T
tonsil
- unk
unknown
Footnotes
Disclosure Statement: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000;92: 709–20. [DOI] [PubMed] [Google Scholar]
- 2.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev 2005;14: 467–75. [DOI] [PubMed] [Google Scholar]
- 3.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008;100: 261–9. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlstrom KR, Calzada G, Hanby JD, Garden AS, Glisson BS, Li G, Roberts DB, Weber RS, Sturgis EM. An evolution in demographics, treatment, and outcomes of oropharyngeal cancer at a major cancer center. Cancer 2013;119: 81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang SH, Xu W, Waldron J, Siu L, Shen X, Tong L, Ringash J, Bayley A, Kim J, Hope A, Cho J, Giuliani M, et al. Refining American Joint Committee on Cancer/Union for International Cancer Control TNM stage and prognostic groups for human papillomavirus-related oropharyngeal carcinomas. J Clin Oncol 2015;33: 836–45. [DOI] [PubMed] [Google Scholar]
- 7.Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517: 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11: 9–22. [DOI] [PubMed] [Google Scholar]
- 9.Smeets SJ, van der Plas M, Schaaij-Visser TB, van Veen EA, van Meerloo J, Braakhuis BJ, Steenbergen RD, Brakenhoff RH. Immortalization of oral keratinocytes by functional inactivation of the p53 and pRb pathways. Int J Cancer 2011;128: 1596–605. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333: 1154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333: 1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, Lawrence MS, Getz G, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res 2015;21: 632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tinhofer I, Stenzinger A, Eder T, Konschak R, Niehr F, Endris V, Distel L, Hautmann MG, Mandic R, Stromberger C, Weichert W, Budach V. Targeted next-generation sequencing identifies molecular subgroups in squamous cell carcinoma of the head and neck with distinct outcome after concurrent chemoradiation. Ann Oncol 2016;27: 2262–8. [DOI] [PubMed] [Google Scholar]
- 14.Iyer NG, Dogan S, Palmer F, Rahmati R, Nixon IJ, Lee N, Patel SG, Shah JP, Ganly I. Detailed analysis of clinicopathologic factors demonstrate distinct difference in outcome and prognostic factors between surgically treated HPV-positive and negative oropharyngeal cancer. Ann Surg Oncol 2015;22: 4411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chad M, Vanderbilt JR, Ahmet Zehir, Arcila Maria E, Snjezana Dogan, Marc Ladanyi, Sumit Middha. Mining Large Panel Hybrid Capture-based Clinical NGS Data for Novel Virus Pathogen-Tumor Associations Based on Mapping of Off-Target Reads to Viral Genomes. Mod Pathol 2018;31: 709. [Google Scholar]
- 16.Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS Jr. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2011;35: 1343–50. [DOI] [PubMed] [Google Scholar]
- 17.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17: 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris LG, Chandramohan R, West L, Zehir A, Chakravarty D, Pfister DG, Wong RJ, Lee NY, Sherman EJ, Baxi SS. The molecular landscape of recurrent and metastatic head and neck cancers: insights from a precision oncology sequencing platform. JAMA oncology 2017;3: 244–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995: 289–300. [Google Scholar]
- 22.Team R, R Core Team (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: In 2012. ISBN 3–900051-07–0. http://www.R-project.org. [Google Scholar]
- 23.Millson A, Lewis T, Pesaran T, Salvador D, Gillespie K, Gau CL, Pont-Kingdon G, Lyon E, Bayrak-Toydemir P. Processed Pseudogene Confounding Deletion/Duplication Assays for SMAD4. J Mol Diagn 2015;17: 576–82. [DOI] [PubMed] [Google Scholar]
- 24.Faraji F, Eisele DW, Fakhry C. Emerging insights into recurrent and metastatic human papillomavirus-related oropharyngeal squamous cell carcinoma. Laryngoscope investigative otolaryngology 2017;2: 10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang SYC, Kannan N, Zhang L, Martinez V, Rosin MP, Eaves CJ. Characterization of epithelial progenitors in normal human palatine tonsils and their HPV16 E6/E7-induced perturbation. Stem cell reports 2015;5: 1210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, Westra W, Sidransky D, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 2007;357: 2552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song X, Xia R, Li J, Long Z, Ren H, Chen W, Mao L. Common and complex Notch1 mutations in Chinese oral squamous cell carcinoma. Clin Cancer Res 2014;20: 701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyman PE, Buehler D, Lambert PF. Loss of Function of Canonical Notch Signaling Drives Head and Neck Carcinogenesis. Clin Cancer Res 2018;24: 6308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrock A, Bode M, Goke FJ, Bareiss PM, Schairer R, Wang H, Weichert W, Franzen A, Kirsten R, van Bremen T, Queisser A, Kristiansen G, et al. Expression and role of the embryonic protein SOX2 in head and neck squamous cell carcinoma. Carcinogenesis 2014;35: 1636–42. [DOI] [PubMed] [Google Scholar]
- 30.Mazibrada J, Longo L, Vatrano S, Cappia S, Giorcelli J, Pentenero M, Gandolfo S, Volante M, dell’Oste V, Lo Cigno I, Biolatti M, Landolfo S, et al. Differential expression of HER2, STAT3, SOX2, IFI16 and cell cycle markers during HPV-related head and neck carcinogenesis. New Microbiol 2014;37: 129–43. [PubMed] [Google Scholar]
- 31.Bayo P, Jou A, Stenzinger A, Shao C, Gross M, Jensen A, Grabe N, Mende CH, Rados PV, Debus J. Loss of SOX2 expression induces cell motility via vimentin up-regulation and is an unfavorable risk factor for survival of head and neck squamous cell carcinoma. Mol Oncol 2015;9: 1704–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Z, Liu G, Huang B, Sun J, Wu D. Prognostic significance of SOX2 in head and neck cancer: a meta-analysis. Int J Clin Exp Med 2014;7: 5010–20. [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SA, Inamura K, Yamauchi M, Nishihara R, Mima K, Sukawa Y, Li T, Yasunari M, Morikawa T, Fitzgerald KC. Loss of CDH1 (E-cadherin) expression is associated with infiltrative tumour growth and lymph node metastasis. Br J Cancer 2016;114: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Spectrum and functional significance of NOTCH2/3/4 mutations in OPSCC.
Supplementary Figure S2: Genetic differences in pathway alterations in HPV-P and HPV-N OPSCC.
Supplementary Figure S3. (A) Effects of cigarette smoking on molecular profiles of HPV-P OPSCC. HPV-P OPSCC in heavy smokers (>10 pack-years) are associated with a non-significant trend of higher frequency of TP53 mutation and MAP3K13 amplification, as well as lower frequency of FOXA1 amplifications (Fisher’s exact test, p=0.183, 0.140 and 0.146 respectively). (B) HPV-P with TP53 mutations arising in heavy smokers showed trend towards poorer OS (p=0.07) and (C) poorer progression free survival (p=0.149) than their TP53-wild type counterparts.
Supplementary Figure S4. (A) Mutation profile of HPV-N/TP53-wild-type OPSCC. (B) HPV-N/TP53-wild type cases trended towards worse progression free survival than HPV-N/TP53-mutated ones.
Supplementary Table S1. Spectrum and functional significance of NOTCH1 mutations in OPSCC.
Supplementary Table S2A. Somatic mutations in 157 OPSCC.
Supplementary Table S2B. Chromosome arm changes in 157 OPSCC.
Supplementary Table S3. Comparison of mutational profiles in HNSCC.
Supplementary Table S4. 410 gene list of MSK-IMPACT™ assay.
Supplementary Table S5. Clinical and pathological features of 157 OPSCC.