Abstract
The two flagella of Chlamydomonas reinhardtii are of the same size and structure but display functional differences, which are critical for flagellar steering movements. However, biochemical differences between the two flagella have not been identified. Here, we show that fluorescence protein-tagged carbonic anhydrase 6 (CAH6-mNG) preferentially localizes to the trans-flagellum, which is organized by the older of the two flagella-bearing basal bodies. The uneven distribution of CAH6-mNG is established early during flagellar assembly and restored after photobleaching, suggesting that it is based on preferred entry or retention of CAH6-mNG in the trans-flagellum. Since CAH6-mNG moves mostly by diffusion, a role of intraflagellar transport (IFT) in establishing its asymmetric distribution is unlikely. Interestingly, CAH6-mNG is present in both flagella of the non-phototactic bardet-biedl syndrome 1 (bbs1) mutant revealing that the BBSome is involved in establishing CAH6-mNG flagellar asymmetry. Using dikaryon rescue experiments, we show that the de novo assembly of CAH6-mNG in flagella is considerably faster than the removal of ectopic CAH6-mNG from bbs flagella. Thus, different rates of flagellar entry of CAH6-mNG rather than its export from flagella is the likely basis for its asymmetric distribution. The data identify a novel role for the C. reinhardtii BBSome in preventing the entry of CAH6-mNG specifically into the cis-flagellum.
Introduction
Cilia function, structure and composition varies considerably between different species and cell types. Multicellular organisms, for example, possess cells with motile cilia, primary cilia and specialized sensory cilia such as the outer segment of photoreceptor neurons [1]. During development, mammalian epithelial cells, such as those of the ependyma, first assemble a single nonmotile cilium (hence the term primary cilium), followed by the development of numerous motile cilia [2]. In many unicellular organisms, distinct types of motile cilia or flagella are present concomitantly on a given cell. In ciliates such a Tetrahymena, the motile cilia in the oral apparatus capture food particles whereas those covering the rest of the cell body serve in cell locomotion. Most heterokonts possess a long flagellum with tripartite mastigonemes to provide propulsion and a short smooth flagellum possibly functioning as a ruder [3, 4]. Some green algae develop flagella of unequal length but apparently otherwise similar ultrastructure [5, 6]. In contrast, Chlamydomonas reinhardtii and other isokont algae have two flagella of equal length and morphology. Nevertheless, the two flagella are distinct in their responses and sensitivity to calcium [7]. These functional differences between the two flagella of a given cell are likely the basis for the stirring movements that underlie directed swimming, for example, during phototaxis, when cells seek light conditions optimal for photosynthesis [8]. Concomitant assembly of distinct cilia on a given cell likely requires selective entry or retention of proteins into the cilia but most details remain to be explored.
The differences between the two flagella of C. reinhardtii and other flagellates are related to the developmental age of the associated basal bodies [5, 9]. In C. reinhardtii, the younger basal body organizes the cis-flagellum and is connected via a four-stranded microtubular root to the sole eyespot apparatus, which is positioned on one side of the cell body (Fig 1A). The trans-flagellum emerges from the older basal body and faces the side opposite of the eyespot (Fig 1B). The differences between the two flagella become even more apparent after mechanical removal of either the cis or the trans-flagellum: the remaining cis-flagellum of such uniflagellate cells beats with the frequency of standard biflagellate cells whereas the beat frequency of a trans-uniflagellate cell is much higher [10, 11]. Thus, the motility of the two flagella, likely synchronized by hydrodynamic coupling when paired, is intrinsically distinct indicating differences in their composition and structure. However, biochemical differences distinguishing the cis and trans-flagellum of C. reinhardtii have not yet been identified.
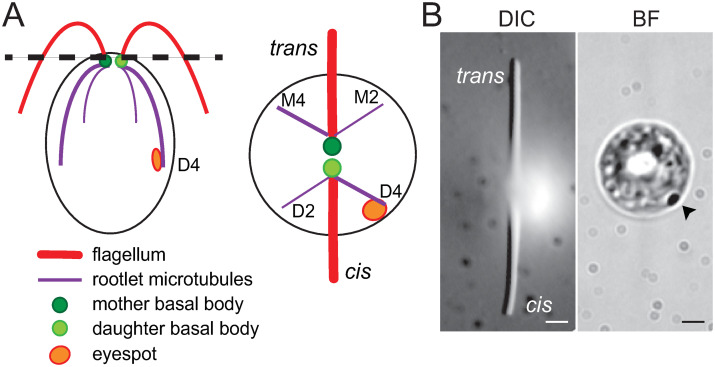
Fig 1. Flagellar asymmetry in C. reinhardtii.
A) Schematic representation of C. reinhardtii viewed from the side (left) and from the top (right). Flagella (red) extend from the two basal bodies (green), each of which is associated with two microtubular rootlets (purple). The eyespot (orange) is associated with the 4-standed microtubular rootlet (D4) linked to the daughter basal body. Thus, the cis-flagellum is closer to the eyespot than the trans-flagellum. The rootlets are named to indicate the number of microtubules (2 or 4) and their association with the mother (M) or daughter (D) basal body [12]. (B) Differential interference contrast (DIC) and brightfield (BF) images of a live cell captured at two different focal planes showing the flagella (left) and eyespot (right) indicated with an arrowhead. Bar = 2μm.
Non-phototactic mutants of C. reinhardtii that lack calcium-dependent differences in flagellar dominance could be instrumental in identifying protein signatures specific for the cis- and trans-flagellum [8, 13, 14]. These include ptx1, which is uncharacterized at the molecular level, and bbs mutants, which are defective in the BBSome, a conserved octameric protein complex that moves via intraflagellar transport (IFT) through flagella [15–17]. In C. reinhardtii, the BBSome is required to remove a subset of membrane-associated proteins from flagella and the abnormal accumulation of the BBSome cargo phospholipase D (PLD) in bbs mutant cilia is likely causative for the loss of phototaxis [18]. However, additional biochemical defects of bbs flagella have been identified and we show here that BBSome loss affects the distribution of carbonic anhydrase 6 (CAH6) in flagella [19].
Carbonic anhydrases, 12 of which are predicted to be encoded in the C. reinhardtii genome [20], reversibly catalyze the conversion of CO2 and H2O into bicarbonate or carbonic acid. Aside from light, carbon dioxide (CO2) is the other rate-limiting substrate of photosynthesis in the low CO2 environments experienced by aquatic algae. CAH activity is critical to capture inorganic carbon and concentrate it near ribulose-1,5-biphosphate carboxylase-oxygenase (Rubisco) in the plastid, for assimilation in a pathway referred to as the carbon concentrating mechanism (CCM) [21]. Interestingly, CAH6 is located in the flagella of C. reinhardtii but its role in flagella and possible contribution to the CCM remain unknown [19, 22].
Here, we show that CAH6 fused to mNeonGreen (mNG) is preferentially localized to the trans-flagellum, making it the first biochemical marker that distinguishes the cis and trans-flagellum of C. reinhardtii. Interestingly, the asymmetric flagellar distribution of CAH6 is lost in bbs mutants. A cah6 insertional mutant displayed normal cell motility and tactic cell behaviors including phototaxis and chemotaxis for bicarbonate phototaxis, suggesting that CAH6 is not linked to flagellar steering movements. In vivo analyses indicate that bbs mutants fail to prevent CAH6 from entering the cis-flagella revealing a novel role for the C. reinhardtii BBSome in controlling protein entry into flagella and establishing cis/trans-flagellar asymmetry.
Materials and methods
Strains and culture conditions
C. reinhardtii was maintained in modified M medium at 24°C with a light/dark cycle of 14:10 h (www.chlamycollection.org/methods/media-recipes/minimal-or-m-medium-and-derivatives-sager-granick/). For gamete generation, cells were resuspended in M without nitrogen (M-N), and aerated for 15–18 h in constant light. Then, cells were incubated in 20% M-N with 10 mM HEPES, pH 7, for 1 h in constant light with agitation. The following strains were used in this study: cah6, cah6 CAH6-mNG, bbs1 cah6 CAH6-mNG, bbs4-1 and g1 as wild-type control strain. All other strains have been previously described and are available at the Chlamydomonas Resource Center (www.chlamycollection.org/).
Transgenic strain generation
To express mNG-tagged CAH6, the coding portions of the gene were amplified via PCR using strain CC-620 genomic DNA as a template; the PCR fragment was inserted into the previously described ipBR vector, placing CAH6 downstream of the HSP70A-rbcS2 fusion promoter and upstream of the mNG gene followed by the 2A sequence, the selectable marker BLE, and the rbcS 3’ terminator sequence (Fig 3A) [18, 23, 24]. For transformation, the plasmid was linearized with KpnI, gel-purified and transformed into C. reinhardtii by electroporation. Transformants were selected on Tris-acetate-phosphate (TAP) plates containing 5 to 10 μg/mL zeocin (Invitrogen), and clones expressing mNG-tagged protein were identified by TIRF microscopy (https://www.chlamycollection.org/methods/media-recipes/tap-and-tris-minimal/). Mating was used to introduce CAH6-mNG into other strains.
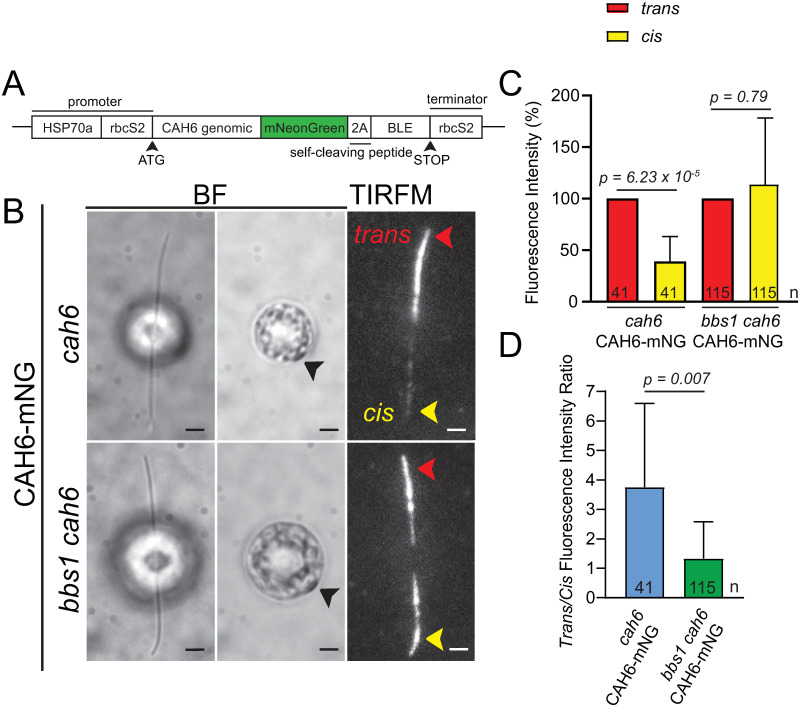
Fig 3. The asymmetric localization of CAH6-mNG in flagella is lost in the bbs1 mutant.
A) Schematic representation of the CAH6-mNG expression vector. The sequence for mNG was inserted at the 3’ end of the genomic CAH6 coding region. B) Brightfield (BF) images of the flagella (left) and the eyespot (right, indicated with black arrowheads) and the corresponding TIRF images of live cah6 CAH6-mNG and bbs1 cah6 CAH6-mNG cells. The trans-flagella are indicated with red arrowheads and the cis-flagella with yellow arrowheads. Bar = 2μm. C) Histograms of the fluorescence intensity of CAH6-mNG in the trans- and cis-flagella of the cah6 CAH6-mNG rescue strain and the bbs1 cah6 CAH6-mNG strain. For each cell, the fluorescence intensity in its trans-flagellum was set to 100% and the fluorescence intensity of the corresponding cis-flagellum is displayed as % of that of the trans-flagellum. One cell, lacking detectable CAH6-mNG in the cis-flagellum and thus having an outlier trans/cis-ration of 158, was ignored for the statistical analyses. Error bars indicate the standard deviation. n = number of cells analyzed. D) Histogram of the ratio of the fluorescence intensity of CAH6-mNG between the trans- and the cis-flagellum in the cah6 CAH6-mNG and the bbs1 cah6 CAH6-mNG strain; shown is the mean of the ratios determined for the individual trans/cis pairs of individual cells. Error bars indicate the standard deviation and n the number of flagellar pairs analyzed. See S3C Fig for the individual data points.
Flagellar isolation and western blot
To isolate flagella for protein analysis, we followed a protocol described by [25]. In brief, cells were concentrated and washed in 10 mM HEPES, pH 7.4, resuspended in HMS at 4°C (10 mM HEPES, 5 mM MgSO4, and 4% sucrose) and deflagellated by adding dibucaine to a final concentration of 4.17 mM (Sigma-Aldrich) and repeated pipetting. Flagella were separated from cell bodies by centrifugation, collected from the supernatant by centrifugation, and resuspended in HMEK (30 mM HEPES, 5 mM MgSO4, 25 mM KCl, and 0.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetra acetic acid [EGTA]) with protease inhibitor cocktail (Sigma-Aldrich). For Western blotting, SDS–PAGE sample buffer was added to flagella samples and samples were incubated at 85°C for 10 min. To raise a polyclonal antibody to CAH6, the C-terminal peptide encoded by the last exon of CAH6 was fused downstream of maltose-binding protein (MBP), the recombinant fusion protein was purified using amylose-sepharose and used to immunize rabbits (Pocono Rabbit Farm). For affinity-purification, we used the fusion protein immobilized on nitrocellulose membrane. The following primary antibodies were used: rabbit anti-CAH6 (1:500), mouse anti-IC2 (1:1,000) [26] rabbit anti-BBS4 (1: 500) [17], mouse anti-α-tubulin (1:10,000; Sigma) and mouse anti-IFT81 (1:1,000) [27]. Western blots were developed using anti-mouse and anti-rabbit IgG conjugated to horseradish peroxidase (Invitrogen) and chemiluminescence substrate (Michigan Diagnostics). Images were captured using a BioRad Gel Doc imaging system.
Flagellar regeneration
For flagellar regeneration, cells in M medium were deflagellated by a pH shock (∼pH 4.3 for ∼45 s), pelleted, and resuspended in fresh M medium. Flagella regrowth was allowed to proceed at room temperature under constant light and agitation. To delay flagellar regeneration, cells were kept on ice until needed.
In vivo microscopy
For in vivo imaging, a Nikon Eclipse Ti-U inverted microscope equipped with a 60×/1.49 numerical aperture (NA) TIRF objective and a through-the-objective TIRF illumination system was used. Excitation light was provided by a 40-mW, 488-nm diode laser (Spectraphysics), and filtered by a Nikon GFP/mCherry TIRF filter cube [28, 29]. The emission was documented at 10 frames/s using an EMCCD camera (Andor iXon X3 DU897) and the Elements software package (Nikon). For photobleaching the entire flagellum, the laser intensity of the 488-nm laser was increased to 10% for 4–10 s. To prepare the observation chamber, 10 μl of cells was placed on a 24 × 60 mm no. 1.5 coverslip previously applied with a ring of petroleum jelly. The cells were allowed to settle for ∼1–10 min, mixed with an equal volume of 10 mM HEPES and 6.25 mM EGTA (pH 7.4) under a 22 × 22 mm no. 1.5 coverslip. The images were analyzed and kymograms and walking averages were generated in FIJI (= ImageJ; National Institutes of Health). Merged images or kymograms were produced using Photoshop and figures were assembled in Illustrator (Adobe).
FRAP and fluorescence intensity analysis
For FRAP analysis, videos were opened in ImageJ, and the region of interest (ROI) was selected using the rectangle tool. The fluorescence intensity inside the selected region was determined using the “Plot Z-axis Profile” tool, and the data were exported into Excel. The fluorescence intensity in the ROI was corrected for the background fluorescence. The highest intensity value before the bleaching event was set to 100%, and the recovery of fluorescence in percentage of the prebleached value was calculated.
Membrane inlet mass spectrometry (MIMS)
18O-labeled dissolved inorganic carbon (DIC) was added to assay buffer [DIC-free water, 20 mM Bicine, pH 8.0] in the MIMS chamber as previously described by [30]. 18O exchange was monitored for ∼10 min before the addition of lysed concentrated cells or isolated flagella to determine the background rate of hydration and dehydration. After addition of cells or flagella, 18O exchange was monitored for an additional 10–20 min to determine the acceleration of 18O removal by CA. The CA-enhanced rate of CO2 hydration (kcf) was determined by fitting the 18O data to a model as described previously (Silverman 1982, [31]) and then normalized to assay protein concentration and the spontaneous CO2 hydration rate (kuf) as a measure of CA activity.
Phototaxis and chemotaxis assays
Population phototaxis assays were performed by placing a cell suspension (∼106 cells per milliliter) into a Petri dish, followed by illumination with light from one side for ∼5 min. Images were taken with a standard digital camera (OM-D; Olympus). For the control, we used g1 (nit1, agg1, mt+), a strain selected for strong negative phototaxis [8]. Cells were washed with fresh M-medium before assaying phototaxis.
Population chemotaxis assays were performed by placing a cell suspension (∼106 cells per milliliter) into a Petri dish, followed by the addition of an agar pellet containing 5% sodium bicarbonate for ∼20 min in complete darkness to prevent potential interference from light [32]. Agar pellets containing 20% tryptone were used as a positive control [32] and agar pellets prepared with medium were used as a negative control. Images were taken with a standard digital camera. Cells were washed with fresh M-medium prior to the chemotaxis assays.
Results
Identification of a cah6 null mutant
From the C. reinhardtii CLiP mutant collection, we obtained strain LMJ.RY0402.174362 that carries an insertion in the CAH6 gene causing a 224bp deletion encompassing the first intron and the first exon including the start codon (Fig 2A). The insertion in the cah6CLIP mutant was verified by PCR, which shows the expected ~2.8-kb size of the amplicon using primers flanking the insertion and a product of ~700bp when a primer specific for the inserted cassette was combined with a CAH6 gene-specific primer (S1A Fig) [33]. To eliminate possible second site mutations in cah6CLIP, we outcrossed the cah6CLIP mutant to the wild-type strain g1 resulting in strain cah6, which was used for all subsequent experiments. Western blot analysis of flagella isolated from control cells (g1) using a polyclonal antibody raised against the peptide encoded by the last exon of C. reinhardtii CAH6 identified a band of ~25kDa, which is close to the predicted size of CAH6 (28 kDa) (Fig 2B). The corresponding band was absent in flagellar samples of cah6 strain indicating that the mutant lacks CAH6 (Fig 2B). For reasons unknown, the antibody failed to detect CAH6 in western blot analyses of the more complex whole cell and cell body samples (S1B Fig). Our data confirm that CAH6 is present in C. reinhardtii flagella [19, 22, 34].
Fig 2. Carbonic anhydrase 6 is active in flagella.
A) Schematic presentation of the CAH6 gene and the insertion in the cah6 mutant. The insertion of the APHVIII cassette caused a deletion (indicated by a red crossed rectangle) of 224 bp encompassing the start codon and first exon of CAH6. The positions of the primers (F1, R1, and R2) used to track the mutation by PCR are indicated (see S1A Fig). B) Western blot analysis of isolated flagella from wild-type (g1) and the cah6 mutant were probed with anti-CAH6. Antibodies to the IFT particle protein IFT81 were used to control for equal loading. C) 18O-exhange measurements (open circles) and models (solid lines) for control (g1) and cah6 flagella. Three 13CO2 isotopologues are tracked and analyzed: 13C16O16O (blue circles), 13C18O16O (green circles) and 13C18O18O (red circles). After determining the background rate of exchange (before time 0), isolated flagella from the strain indicated were added at time 0 and the exchange rate was measured for 400 seconds. CA activity is indicated by the accelerated conversion of 13C18O18O into 13C18O16O and ultimately 13C16O16O. The best fit line (kcf) of the model after addition of isolated flagella is shown. D) Flagellar and whole cell CA activity (kcf/kuf) of g1 and cah6; the values were normalized for the protein concentration (mg/mL) in the respective samples and are based on repeat measurements of the same samples (n = 1). E) Western blot analysis for CAH6 in wild-type (g1) flagella. Cells were treated for 24 hours prior to the flagellar isolation as follows: The cultures were either aerated with high CO2 (0.5%) or maintained without aeration both in the presence (+) or absence (-) of cycloheximide. Anti-IC2 was used as a loading control.
Carbonic anhydrase 6 is active in flagella
Based on the presence of 21 of 23 highly conserved residues, C. reinhardtii CAH6 belongs to the β-family of carbonic anhydrases (CAs), which is present in prokaryotes and the plastids of higher plants and algae (Fig 1C and 1D) [35]. In addition, the N-terminal domain of CAH6 is predicted to encompass a chloroplast transit peptide for import into the plastid (S1C and S1D Fig). Thus, it is possible that some CAH6 is imported into the plastid [36]. However, CAH6 was identified in the C. reinhardtii flagellar proteome, our western blot analyses suggest that only a small pool of CAH6 is present in the cell body and a CAH6-YFP signal was not detected in the plastid [19, 22, 34]. Further, the N-terminal region of CAH6 is predicted to be dual acylated (S1D Fig), a known flagellar localization signal that likely is also responsible for attaching the protein to the flagellar membrane [37]. The data suggest that CAH6 is predominately a flagellar protein.
To analyze the role of flagellar CAH6, we measured carbonic anhydrase (CA) activity in flagella isolated from control and cah6 using membrane inlet mass spectrometry (MIMS) based on 18O removal from 13CO2 isotopologues (S1E Fig) [38]. Compared to the spontaneous conversion observed prior to addition of the sample (T-200 to T0), control flagella significantly accelerated the conversion of 13C18O18O to 13C16O16O with a CA activity of 14.81 kcf/kuf per mg protein (Fig 2C and S1 Table). In contrast, CA activity was more than 5-fold lower in the cah6 mutant flagella (Fig 2D). In whole cell samples of both strains, we measured an approximately equal CA activity (Fig 2D). All other characterized CAs of C. reinhardtii are located in the cell body and in agreement CA activity in whole cell samples was about 40-fold higher than that in wild-type flagella (Fig 2D) [20]. Likely, the residual CA activity in cah6 flagella results from a contamination of the flagellar sample with cell body material. Since CAH6 is the only known CA in C. reinhardtii flagella [20, 22], we conclude that CAH6 is active in flagella.
In C. reinhardtii, the expression of CAs and other proteins associated with the carbon concentrating mechanism (CCM) is upregulated in response to limited supply of inorganic carbon in the environment [39–41]. To test if CAH6 levels in flagella adjust with changes in the environmental CO2 levels, we maintained g1 cells aerated with air containing low (0.04%) or high (0.5%) levels of CO2 and without aeration for 24 hours and analyzed the flagella with anti-CAH6. In comparison to the high CO2 sample, the amount of CAH6 in flagella was increased approximately 2-fold when cells were treated with air (~0.04% CO2) and more than 10-fold in cells maintained without aeration (Fig 2E, S2E Fig). To test whether a low environmental CO2 concentration induces de novo synthesis of CAH6 or its redistribution from the cell body into the flagella, wild-type cells were incubated at high CO2 (0.5%) and without aeration for 24 hours both in the absence or the presence of cycloheximide, an inhibitor of proteins synthesis. An increase of CAH6 in flagella as it occurred in controls without cycloheximide in response to the lack of aeration, was not observed in the cycloheximide treated sample (Fig 2E). Instead, cycloheximide treatment reduced the amount of CAH6 in the flagella probably because the block of protein synthesis prevented the replacement of CAH6 lost by turnover. Thus, the increased presence of CAH6 in flagella at low environmental CO2 likely results from an increased expression and de novo synthesis of CAH6. The data suggest that CAH6 is active in flagella and that CAH6 expression is significantly up-regulated under environmental CO2 limitation as described for other CCM proteins. The cah6 mutant, however, lacks an apparent phenotype as it developed flagella of normal length (S2A Fig), swam with wild-type velocity (S2B Fig), displayed positive chemotaxis for bicarbonate (S2F and S2G Fig), negative phototaxis (S2D Fig) and grew at a similar rate as the g1 control strain (S2C Fig).
The BBSome ensures a preferential localization of CAH6-mNG to the trans-flagellum
When fixed with formaldehyde or -20°C methanol, the flagella of both control and cah6 mutant cells were weakly stained revealing that the antibody is unsuited for immunocytochemistry (not shown); we obtained similar results with previously described antibodies to C. reinhardtii CAH6 [36]. To gain insights into the distribution of CAH6, we fused mNeonGreen to the C-terminus of CAH6 and expressed the fusion protein in the cah6 mutant using a bicistronic vector encompassing the auto-cleavable 2A sequence and the BLE gene conferring resistance to zeocin for selection (Fig 3A) [18, 23]. C-terminal tagging was used because the N-terminal region of CAH6 is predicted to be co-translationally myristoylated and post-translationally palmitoylated, modifications that likely ensure the reported association with the ciliary membrane [42]. Potential caveats of this approach include the loss of the endogenous transcriptional regulation of CAH6 and interference of the C-terminal mNG tag with the subcellular targeting or function of CAH6. In western blots of isolated flagella, anti-CAH6 identified a band of ~60 kD representing the CAH6-mNG fusion protein (shown in S3B Fig for a bbs1 cah6 CAH6-mNG strain).
Surprisingly, in vivo imaging revealed that CAH6-mNG is unequally distributed between the two flagella of a given cell (Fig 3B, S3A Fig). Using the eyespot as a positional marker, we determined that CAH6-mNG preferentially localizes to the trans-flagellum (Fig 3B and 3C and S3A Fig). The average fluorescence intensity of CAH6-mNG in the trans-flagellum of a given cell exceeded that of the corresponding cis-flagellum approximately 4-fold (Fig 3D and, for individual data points, S3C Fig). To our knowledge, CAH6-mNG is the first protein identified to preferentially localize to one of the two flagella of C. reinhardtii.
We previously reported that the amount of CAH6 in bbs4 flagella progressively decreases over several hours after flagellar regeneration suggesting a role of the BBSome in the flagellar maintenance of CAH6 [19]. To investigate the distribution of CAH6-mNG in the absence of intact BBSomes, we generated a bbs1 cah6 CAH6-mNG double mutant by mating. Remarkably, the preferential localization of CAH6-mNG to the trans-flagellum is lost in the bbs1 background and more or less similar amounts of the protein were observed in the cis- and trans-flagellum of a given cell (Fig 3B, 3C and 3D; S1 Movie). The data indicate that intact BBSomes are required to establish the asymmetric distribution of CAH6-mNG in flagella.
CAH6-mNG cis/trans-asymmetry is established early during flagellar regeneration and maintained dynamically
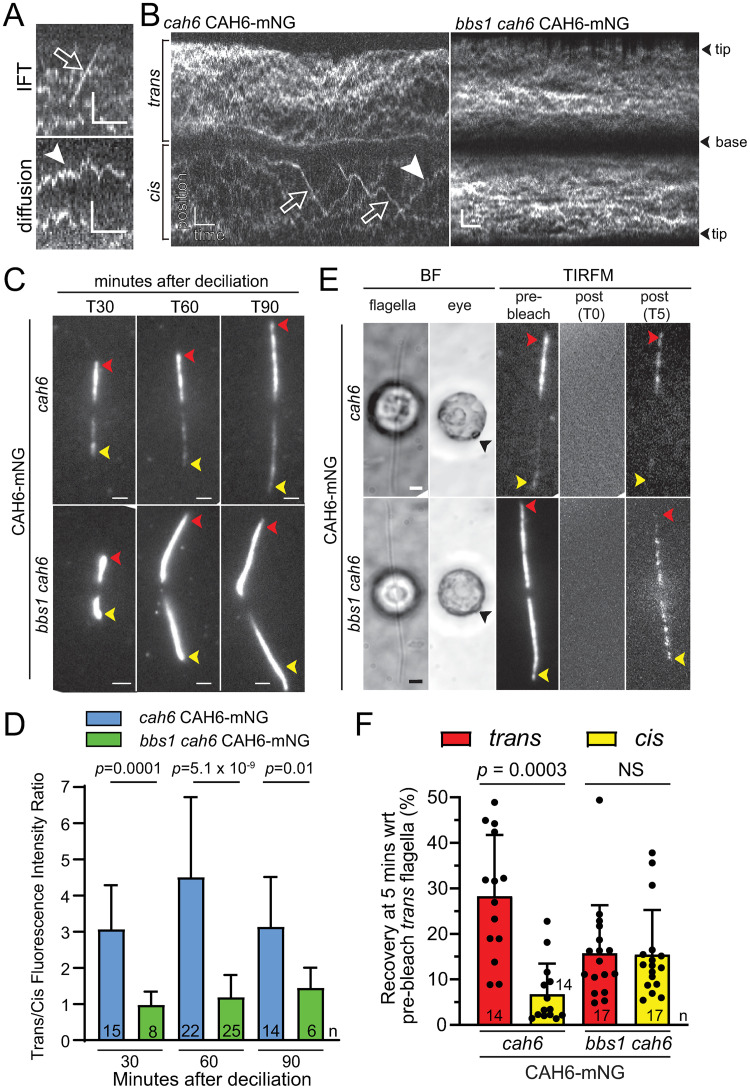
To determine how the asymmetric flagellar localization of CAH6-mNG is established, we analyzed the dynamics of CAH6-mNG in full-length and re-growing flagella. Live imaging showed that CAH6-mNG moved by a slow random walk in the cis- and trans-flagella of both the cah6 CAH6-mNG and bbs1 cah6 CAH6-mNG strains (Fig 4A and 4B). This movement is reminiscent of that of phospholipase D (PLD)-mNG, which, similar to CAH6, is predicted to be associated to the flagellar membrane by dual acylation [18]. In contrast to PLD, transport of CAH6-mNG by IFT was rare in both the control and the bbs1 strain (~0.3 and ~0.4 events/flagellum/minute, respectively) and we did not observe a significant difference in the frequency of CAH6-mNG transport between the cis- and the trans-flagella (Fig 4A and 4B). During flagellar regeneration, the preference of CAH6-mNG for the trans-flagellum was already apparent in short flagella (~4μm) of the cah6 CAH6-mNG strain whereas it was mostly equally distributed in the two short flagella of bbs1 cah6 CAH6-mNG cells (Fig 4C and 4D). IFT of CAH6-mNG remained scarce during flagellar regeneration. The data indicate that CAH6-mNG cis/trans-asymmetry in control cells is established early during flagellar regrowth. CAH6-mNG cis/trans asymmetry could be static with a fixed amount of CAH6-mNG deposited preferentially in trans-flagellum during assembly and remaining trapped. Alternatively, the asymmetry could be maintained dynamically with a continuous exchange of CAH6-mNG between flagella and the cell body and distinct rates of entry and/or retention between cis and trans-flagellum. Fluorescence recovery after photobleaching (FRAP) analyses revealed a partial recovery (25% or less depending on flagellar type and strain) of the CAH6-mNG signals within 5 minutes; the recovery occurred asymmetrically in control cells and symmetrically in bbs1 cells (Fig 4E and 4F). In conclusion, cis/trans-asymmetry of CAH6-mNG is maintained dynamically with the protein continuously entering and exiting the flagella at distinct rates.
Fig 4. The asymmetric distribution of CAH6-mNG is established early during ciliary assembly and restored after photobleaching.
A) Partial kymograms showing IFT (open arrow) and diffusion (white arrowhead) of CAH6-mNG. Scale bars: 2 s and 2 μm. B) Kymograms showing the movements of CAH6-mNG in full-length cah6 CAH6-mNG and bbs1 cah6 CAH6-mNG flagella. CAH6-mNG moved mostly by diffusion (white arrowhead) but occasional transport by IFT (open arrows) was observed as well. Scale bars: 2 s and 2 μm. C, D) Analysis of CAH6-mNG in regenerating flagella. C) Still images showing the distribution of CAH6-mNG during flagella regeneration of cah6 CAH6-mNG and bbs1 cah6 CAH6-mNG cells. TIRFM images of different live cells were obtained at 30 minutes (T30), 60 minutes (T60) and 90 minutes (T90) after deciliation by a pH shock. Bar = 2μm. D) Quantification of the ratios of CAH6-mNG fluorescence intensity between regenerating trans- and cis-flagella at 30, 60 and 90 minutes after deciliation. The error bars, indicating standard deviation, and the number of cells analyzed (n) are indicated. E, F) FRAP analysis of CAH6-mNG in full-length flagella. E) Brightfield (BF) of cah6 CAH6-mNG and bbs1 cah6 CAH6-mNG cells showing optical section through the flagella and the eyespot (indicated with a black arrow). The corresponding TIRF images show the flagella before (pre), immediately after (T0) and 5 minutes after (T5) the photobleaching step. While partial, the recovery pattern indicated that CAH6-mNG cis/trans-asymmetry in wild-type cells is maintained dynamically, i.e., involves continuous entry and exit of CAH6-mNG from flagella. Bar = 2μm. F) Histograms with individual data points comparing recovery of CAH6-mNG fluorescence in trans- and cis-flagella at 5 minutes after photobleaching. Data are presented as percentage of the prebleach intensity of the trans-flagellum and were calculated for each flagella pair. The significance, based on a two-tailed t test, is indicated. n = number of flagella analyzed.
The BBSome restricts entry of CAH6-mNG into the cis-flagellum
Previously, we showed that the C. reinhardtii BBSome functions as a cargo adapter exporting proteins like PLD from flagella by mediating their transport on IFT trains [18]. To determine the kinetics of PLD-mNG removal from bbs flagella, we used dikaryon rescue experiments fusing control and bbs1 PLD-mNG gametes and monitored the removal of PLD-mNG from the bbs1-derived flagella after cell fusion by TIRF microscopy (S4A Fig). Even at the earliest time points (≤ 30 min after mixing of the gametes), an abnormal accumulation of PLD-mNG in bbs1-derived flagella was not observed, indicating that BBSomes provided by the wild-type parent quickly enter the bbs1-derived flagella and rapidly remove PLD-mNG (S4A Fig) [19].
While IFT of CAH6-mNG was rarely observed, this could potentially be explained by the low abundance of CAH6-mNG in the cis-flagellum, i.e., if BBSomes and IFT continuously remove CAH6-mNG from the cis-flagellum, the chance to observe an actual transport during the short periods covered by our recordings is low. To expose BBSomes to higher concentrations of CAH6-mNG, we fused control and bbs1 CAH6-mNG gametes and monitored the removal of CAH6-mNG from the bbs1-derived cis-flagellum after cell fusion by TIRF microscopy (Fig 5A).
Fig 5. Export of CAH6-mNG from bbs1-derived cis-flagella is slow.
A) Schematic representation of a mating between bbs1 cah6 gametes expressing CAH6-mNG (green dots) and cah6 gametes possessing intact BBSomes (indicated by yellow shading). After cell fusion, BBSomes and CAH6-mNG are present in the shared cytoplasm of the zygotes and will enter all four cilia, allowing us to study both the entry of CAH6-mNG into cah6-deficient flagella and the BBSome-dependent repair of CAH6-mNG distribution in the bbs1-derived flagella. B) Differential interference contrast (DIC), brightfield (BF) and TIRFM images of live zygotes from a mating between bbs1 CAH6-mNG and a rsp3 RSP3-mCherry strain; the RSP3-mCherry tag allows us to distinguish bbs1- (arrowheads) and wild type-derived flagella (arrows) after cell fusion. Red arrows and arrowheads indicate the trans-flagella and yellow arrows and arrowheads indicate the cis-flagella. Note that establishing CAH6-mNG cis/trans asymmetry in the rsp3 RSP3-mCherry-derived wild-type cilia precedes correcting the distribution of CAH6-mNG in the bbs1-derived flagella. Bar = 2 μm. C) Brightfield (BF) images of different focal planes showing the flagella and the two eyespots (indicated with arrowheads) and the corresponding TIRF image of live bbs1 cah6 CAH6-mNG × cah6 zygotes; the time passed since mixing of the gametes is indicated. The bbs1-derived flagella are marked by arrowheads (red for trans and yellow for cis) and the cah6-derived flagella are marked correspondingly with arrowheads. Bars = 2 μm. (D) Diagrams of the classification system of zygotes based on the distribution pattern of CAH6-mNG. Early stages possessed one flagellar pair containing ~82% of the total CAH6-mNG signal and a weaker pair accounting for ~18% of the signal. Intermediate stages possessed three flagella with strong signals and one with a weaker signal; accordingly, the signal strength of the stronger cis/trans-pair was reduced to 66% and that of the weaker pair increased to 34% of the total CAH6-mNG fluorescence in a given zygote. Late stages, only a few of which were detected, possessed two flagellar pairs with discernable cis/trans-asymmetry in the CAH6-mNG signal. (E) Cumulative bar chart showing the distribution of the three different patterns of flagellar CAH6-mNG distribution in zygotes (see panel D) at different time points.
First, we mated a CAH6-mNG control strain to the cah6 mutant to test if CAH6-mNG cis/trans-asymmetry is established in zygotes. The resulting zygotes showed CAH6-mNG in the trans-flagellum of each pair indicating that CAH6-mNG cis/trans-asymmetry is preserved and established de novo in zygotic flagella (S4B Fig). This observation agrees with previous data showing that both eyespots are positioned on the same side of the zygote indicating that cell fusion occurs with the flagellar pairs in a parallel configuration with respect to the cis/trans-axis [43]. Next, we mated bbs1 CAH6-mNG cells to rsp3 RSP3-mCherry (mC) cells, which enabled us to identify the bbs1- and the wild type-derived flagella after formation of the zygotes based on the RSP3-mC signal (Fig 5B). Interestingly, the accumulation of CAH6-mNG in the rsp3 RSP3-mC-derived trans-flagellum preceded the removal of CAH6-mNG from the cis-flagellum derived from the bbs1 parent as indicated by zygotes possessing three flagella with a strong CAH6-mNG signal and one flagellum in a cis-position with a weak signal (Fig 5B). The pattern suggests that the repair of bbs1-derived flagella is a rather slow process. To avoid a possible interference from endogenous CAH6, we mated the cah6 mutant to the bbs1 cah6 CAH6-mNG strain (Fig 5C). In the flagella of the resulting zygotes, CAH6-mNG moved mostly by diffusion and only 2 IFT events were observed during the course of this study, resulting in an average of 0.03 IFT events/minute/flagellum (S4C Fig). Early zygotes (≤ 30 min after mixing of the gametes) possessed strong CAH6-mNG signals in the bbs1-derived flagellar pair (containing 85% or more of the total CAH6-mNG signal of a given zygote) and a weakly fluorescent pair derived from the wild type-parent (Fig 5Ca-d, 5D and 5E). A different pattern was prevalent at ~30 minutes or more after mixing of the gametes with many zygotes possessing three flagella with a strong CAH6-mNG signal and one with a weaker signal. The latter was always in a cis position and, considering the above observation using the RSP3-mCherry strain, is likely derived from the cah6 parent (Fig 5Ce-l, 5D and 5E). Only at the later time points (>60 minutes), we observed partial or complete formation of cis/trans-asymmetry in both flagellar pairs in a subset of zygotes (n = 2; Fig 5B bottom, Fig 5Cm-p, 5D and 5E). Thus, establishing cis/trans-asymmetry of CAH6-mNG in the cah6-mutant derived flagella of mosaic zygotes outpaces the removal of CAH6-mNG from the bbs1-derived cis-flagella. We conclude that the rate of CAH6-mNG export during the repair of the bbs1 cis-flagella is substantially slower than the rate of CAH6-mNG entry and accumulation in wild-type or cah6-derived trans-flagella. This observation is best explained by a role of the BBSome in restricting access of CAH6-mNG specifically to the cis-flagellum rather than by BBSome-dependent export of CAH6-mNG from the cis-flagellum.
To test if BBSomes can restore the control of CAH6-mNG entry into bbs-derived cis-flagella of zygotes, we mated the cah6 CAH6-mNG rescue strain to the bbs4-1 mutant strain. Even at early time points (≤30 min), the majority of the analyzed zygotes had 2 trans-flagella with a strong CAH6-mNG signal and 2 cis-flagella with weaker signal (S4D Fig). Thus, the presence of BBSomes ensures that the bbs4-derived flagella of such zygotes directly develop the characteristic cis/trans-pattern of CAH6-mNG. Apparently, the de novo assembly of the CAH6-mNG cis/trans-pattern in bbs-derived flagella is faster than removal of CAH6-mNG from bbs-derived flagella. The data further support the notion that the BBSome restricts the entry of CAH6-mNG into the cis-flagellum to establish the cis/trans-asymmetry of CAH6-mNG in C. reinhardtii flagella.
Discussion
CAH6-mNG is a biochemical marker for the C. reinhardtii trans-flagellum
We showed that CAH6-mNG is enriched in the trans-flagellum of C. reinhardtii. Two polyclonal antibodies raised against CAH6 or parts of the protein equally stained the flagella of both control and cah6 mutant cells. Since western blotting revealed the absence of CAH6 from the mutant flagella, the currently available antibodies to CAH6 are unsuited for immunocytochemistry. Thus, the distribution of the endogenous CAH6 in flagella remains unknown. However, GFP alone and other membrane-associate proteins tagged with mNG distribute equally between the two flagella of a given cell [18]. In an independent study, CAH6-YFP expressed using the PsaD promoter also preferably accumulates in one of the two flagella of C. reinhardtii control cells [22]. Thus, it is likely that the CAH6 portion of the fusion protein mediates its asymmetric flagellar distribution. Remarkably, the asymmetric flagellar distribution of CAH6-mNG is lost in the non-phototactic bbs1 mutant indicating that defects in a known flagellar protein transport pathway affect the localization of the fusion protein. Clearly, CAH6-mNG impinges on a cellular pathway that established biochemical differences between the two flagella. Taken together, we consider it likely that CAH6-mNG mimics the distribution of endogenous CAH6.
CAH6 levels in flagella are regulated by CO2 concentration
CAH6 is apparently active in flagella and its amount in flagella is increased at low levels of CO2 due to upregulated expression (Fig 6A and 6B; [44]). An increase in expression under low CO2 is typical for most CAs participating in the C. reinhardtii carbon concentrating mechanism (CCM) [45]. The other known C. reinhardtii CAs of are located in the periplasm, the cell body cytoplasm and the cell organelles converting CO2 to bicarbonate and vice versa to concentrate CO2 in the chloroplast. This helps aquatic algae to assimilate CO2, which has a low solubility in water. However, endogenous and tagged CAH6 are largely flagellar proteins and it is unclear how CA activity in the flagella could contribute to CO2 capture. C. reinhardtii shows positive chemotaxis for bicarbonate suggesting that its motility is somehow regulated by bicarbonate or CO2 [46]. Fungal CAs function as CO2 sensors and the regulation of sperm motility in a variety of species involves CAs raising the possibility that CAH6 contributes to CO2/bicarbonate chemotaxis of C. reinhardtii [47, 48]. However, the cah6 mutant swam with normal velocity, displayed positive chemotaxis for bicarbonate and grew at similar rates as the wild-type control in high and low CO2 conditions. The asymmetric flagellar distribution of CAH6-mNG was lost in C. reinhardtii bbs1 mutants, which fail to perform the characteristic steering movements required for phototaxis probably because both flagella of bbs-mutants move like wild-type trans-flagella [8, 17]. Biochemically, the defect in bbs flagella has been linked to the abnormal accumulation of signaling proteins including PLD, which appears to be a negative regulator of phototaxis [18]. However, we consider it unlikely that PLD positively promotes the differential motility of the two flagella because PLD does not display an asymmetric flagellar distribution in either control or bbs cells. On the other hand, negative phototaxis was intact in the cah6 mutant indicating that CAH6 or its asymmetric distribution are not required for light-induced flagellar steering movements. While the role of CAH6 remains enigmatic, it is intriguing that both the asymmetric flagellar distribution of CAH6-mNG and the asymmetric flagellar behavior are lost in the bbs mutants. It is possible that other, yet to be identified proteins are also distributed asymmetrically in flagella in a BBSome-dependent manner; such proteins could underly the differential motility of the cis- and the trans-flagellum required for phototaxis.
Fig 6. Schematic summary and model of expression regulation and flagellar localization of CAH6-mNG in C. reinhardtii.
A) CAH6 in flagella is actively converting carbon dioxide to bicarbonate and vice versa. CAH6 is predicted to attach to the flagellar membrane (pink) by a dual fatty acid modification (indicated by two wavy lines). B) In CO2-limiting conditions, the amount of CAH6 in flagella is upregulated 2- to 10-fold. C, D) The BBSome largely prevents the entry of CAH6-mNG into the cis-flagellum. In a hypothetical scenario, the younger basal body could recruit BBSomes to a specific location or activate the BBSome (indicated by the dark red color) to minimize entry of CAH6-mNG into the cis-flagellum; during maturation, the older basal body lost its ability to modulate BBSome function and CAH6-mNG will enter the flagella.
Establishing cis/trans-flagellar asymmetry involves the BBSome
Flagellar asymmetry is linked to the developmental differences between the basal bodies that template them. While of similar size and structure, the trans- and cis-flagellum of C. reinhardtii are organized by the mother and the daughter basal body, respectively, which are of distinct developmental age and inhabit distinct positions within the cell relative to the single eyespot. Differences between flagella organized by basal bodies of distinct developmental age are apparent in many species and often are more pronounced than in C. reinhardtii [5, 9]. In mammalian cells, only the mother centriole assembles a primary cilium whereas the daughter basal body requires an additional cell division to become a mother centriole and gain competency to template a flagellum. Heterokont algae possess a long flagellum with mastigonemes attached to the daughter basal body while the mother basal body forms a short smooth flagellum. In Epipyxis pulchra, the long flagellum is shortened prior to cell division and loses its mastigonemes as its basal body migrates into the position characteristically occupied by mother basal bodies whereas the short smooth flagellum remains unaltered; each of the two new daughter basal bodies template a long flagellum with mastigonemes [49]. Thus, the basal bodies’ developmental age dictates the structure and composition of the attached flagellum. However, the mechanism by which differences between the flagella of a given cell are established remain unknown. Here, we show that in C. reinhardtii, the BBSome is required to establish the asymmetric flagellar distribution of CAH6-mNG providing initial mechanistic insight into the process.
A novel role for the BBSome in limiting protein entry into flagella
In principal, a distinct biochemical composition of the cis and trans-flagella could result from differences in protein entry or retention. Considering the known role of the BBSome in ciliary protein export, an increased removal of CAH6-mNG specifically from the cis-flagellum via the BBS/IFT pathway could establish its asymmetry distribution. However, IFT of CAH6-mNG was rarely observed, the majority of those transports was anterograde, and significant differences in CAH6-mNG IFT frequencies between the cis and trans-flagella or between control and bbs1 flagella were not observed. It could be reasoned that in cells with full-length flagella, CAH6-mNG flagellar asymmetry is already established and thus additional transport events will be exceedingly rare. However, flagellar asymmetry of CAH6-mNG is restored at moderate speed after photobleaching indicating that CAH6-mNG continuously enters and exits flagella. Further, the frequency of CAH6-mNG transport remained low during flagellar regeneration when the protein enters cilia de novo and its asymmetric distribution develops. In zygotes, IFT remained exceedingly rare during the repair of bbs1-derived cis-flagella preloaded with CAH6-mNG. Further, restoring CAH6-mNG cis/trans-asymmetry in bbs1-derived flagella was substantially slower than establishing it de novo in cilia initially lacking CAH6-mNG derived from the wild-type, cah6 or bbs parent. While the former involves lowering the flagellar content of CAH6-mNG, the latter can be explained by a BBSome-dependent reduction of CAH6-mNG entry into cis-flagella. Thus, BBSome-dependent restriction of CAH6-mNG entry into the cis-flagellum could explain both the rapid development of cis/trans-flagellar asymmetry in flagella initially lacking CAH6-mNG and the slow exit of CAH6-mNG form the bbs1-derived cis-flagella, which likely depends on slow CAH6-mNG turnover. Thus, we conclude that the BBSome specifically limits CAH6-mNG entry into cis-flagella of C. reinhardtii. A role of the BBSome in preventing entry of proteins into cilia could also explain defects in other systems such the abnormal presence of numerous distinct non-ciliary proteins in the outer segment of bbs mutant mice [50].
How could the BBSome establish CAH6-mNG cis/trans-asymmetry?
How could the BBSome limit protein entry into flagella, specifically into just one of two flagella in a closely spaced pair? BBS proteins localize to the transition zone, which functions as a ciliary gate, and to the basal body region, which is a platform for the recruitment of ciliary proteins and loading of IFT trains [51–54]. Thus, the BBSome could function as a roadblock preventing CAH6-mNG from approaching the daughter basal body or entering the cis-flagellum (Fig 6C and 6D). In mammalian systems, several biochemical differences between the mother and the daughter centriole have been identified [55, 56]. Recent data indicate that cargo binding by the BBSome and probably the passage of the complex through the transition zone require activation of the BBSome likely by the small GTPase Arl6/BBS3 [57, 58]. Similarly, a daughter basal body-specific protein could activate nearby BBSomes enabling them to bind CAH6-mNG and prevent its flow into the attached cis-flagellum. However, in C. reinhardtii, BBS proteins are present at the base of both flagella and the known structural differences between the two basal bodies are minor, e.g., more cartwheel layers are present in the daughter basal body [17, 59]. In an alternative model, the BBSome could deliver CAH6-mNG selectively to the base of the trans-flagellum, which will ensure its preferred entry into the trans-flagellum. We previously observed that bbs-mutant cilia contain less total endogenous CAH6 than wild-type cilia suggesting a role of the BBSome in the delivery of CAH6 to the flagella [19]. Indeed, the BBSome has been linked to intracellular non-IFT transports such as the dynein-dependent movements of zebrafish melanosomes to the cell center [60, 61]. In C. reinhardtii, IFT dynein appears to promote entry of SAG-1 into flagella of activated gametes by transporting it along the microtubular roots to the flagellar base [62]. It has been shown that the microtubular rootlets are specialized for specific transports related to cis/trans-asymmetry: The MLT1 protein is specifically present along the four-stranded root attached to the daughter basal body of C. reinhardtii [12]. Multiple eyespot 1 (mlt1) mutants develop additional eyespots in aberrant positions, i.e., on the four-stranded rootlet attached to the trans-basal body [63]. MTL1 could specify the four-stranded cis-rootlet for the transport of eyespot components ensuring proper positioning of the eyespot [12]. Similarly, rootlets attached to the trans-basal body could ensure transport of CAH6-mNG to the base of the trans-flagellum in a BBSome-dependent manner whereas BBSome loss diminishes channeling of CAH6-mNG to the trans-flagellum causing its aberrant entry into the cis-flagellum and an overall reduction of its presence in flagella.
Supporting information
The table list the parameters and numerical values of the MIMS experiment for the control (g1) and the cah6 mutant strain.
(DOCX)
A) Agarose gel of PCR products using cah6 genomic DNA as a template; G-beta primers are used as control. See Fig 2A for the positions of the primers. B) Western blot analysis of whole cells, deflagellated cell bodies (Cell Body), and isolated flagella (Flagella) from wild-type g1 and cah6 probed with anti-CAH6. Equivalent amounts of cells and flagella were loaded (i.e., one whole cell, one cell body, and two flagella). Antibodies to the IFT particle protein IFT81 were used to control for equal loading. A part of the same blot is shown in Fig 2B, 2C and 2D) Schematic presentation of CAH6 (C) and amino acid sequence of CAH6 (D). The predicted chloroplast transit peptide (cTP) is indicated in green and the catalytic domain in red. In D, the 23 conserved residues typical for β-type CAs are indicated by red font. Asterisks indicate the two amino acids that are not conserved in CAH6. (E) Schematic representation illustrating the measurement of CA activity.
(TIF)
A) Flagellar length of control (g1) and the cah6 strains. The standard deviations and the number of measurements are indicated. B) Mean swimming velocity of wild-type (g1) and cah6 cells in 0.5% CO2 and 0.04% CO2 (air). C) Liquid growth curves for wild-type g1 (red circles) and cah6 (black squares) in 5% CO2, 0.04% CO2 (air) and <0.04% CO2. D) Phototaxis assay of control (g1) and cah6. The time of light exposure (5 mins) and direction of the light (arrowheads) are indicated. E) Western blot analysis of flagella for CAH6 isolated from wild-type (g1) cells aerated for 24 hours under high (0.5%), low (0.04%) and no aeration with CO2 conditions. Anti-IC2 was used as a loading control. F) Schematic presentation of the chemotaxis assay. Cells in M-medium were placed into a small Petri dish or 6-well cell culture plate and agar plaques (A, B) containing either 5% sodium bicarbonate or 20% tryptone (A) or no attractant (B) were added. G) Chemotaxis assay showing g1, cah6 and bbs4-1 before (T0) and after a 20-minute incubation in the dark (T20). Accumulated cells are indicated by the arrows. The following results were noted in repeat experiments (positive chemotaxis/no chemotaxis): g1 aerated with CO2 (0/9), g1 without aeration (5/5), cah6 (4/3), bbs4-1 (1/2), and bbs1 (2/1).
(TIF)
A) Brightfield (BF) and TIRFM images of live g1 CAH6-mNG cells. The two focal planes show flagella and eyespot (indicated with black arrowheads). Trans-flagella are indicated with red arrowheads and cis-flagella with yellow arrowheads. Bar = 2μm. B) Western blot analysis of isolated flagella (Flagella) from wild-type g1, cah6 and bbs1 cah6 CAH6-mNG were probed with anti-CAH6 and anti-BBS4. Antibodies to acetylated tubulin were used as a loading control. C) Scatter plot showing the ratios between the trans- and cis-flagellum of control and bbs1 cells; see Fig 3D for presentation as a histogram.
(TIF)
A) Brightfield (BF) and TIRFM images of a bbs1 PLD-mNG gametes and live dikaryons from a matings between bbs1 PLD-mNG and control (g1) or bbs4. In bbs1 PLD-mNG x bbs4 zygotes, both parental strains lack intact BBSomes and BBSomes need to be assembled de novo after cell fusion delaying the export of PLD-mNG. Indeed, an abnormal presence of PLD-mNG was observed in two of the four flagella in a subset of the zygotes analyzed at ≤ 30 min whereas PLD-mNG was essentially absent from all four flagella at the later time points (>30 min). Bar = 2 μm. B) Brightfield (BF) and TIRFM images of a live dikaryon from a mating between g1 CAH6-mNG and cah6 gametes. Trans-flagella are indicated with red arrowheads and cis-flagella with yellow arrowheads. Bar = 2 μm. C) Kymograms showing CAH6-mNG dynamics in the flagella of cah6 × bbs1 cah6 CAH6-mNG zygotes. Open arrows indicate transport of CAH6-mNG via anterograde IFT. D) Brightfield (BF) and TIRFM images of live dikaryons from a mating between cah6 CAH6-mNG and bbs4 gametes. The two focal planes show the flagella and the eyespot (indicated with black arrowheads). Trans-flagella are indicated with red arrowheads and cis-flagella with yellow arrowheads. Bar = 2 μm.
(TIF)
TIRF localization of CAH6-mNG in cah6 CAH6-mNG (left) and bbs1 cah6 CAH6-mNG (right) flagella. The beginning of the video was recorded in brightfield mode and shows first the eyespot (eye) and then moves to the focal plane of the flagella before switching to TIRF illumination. The cis- and trans-flagella are indicated. Images were acquired at 10 fps, and playback is set at 20 fps (2× speed). The timer counts seconds.
(AVI)
(DOCX)
Acknowledgments
We thank James V. Moroney (LSU) for the kind gift of CAH6 antibodies and advice.
Abbreviations
- BBS
Bardet-Biedl syndrome
- CA
carbonic anhydrase
- CAH6
carbonic anhydrase 6
- CCM
carbon concentrating mechanism
- FRAP
fluorescence recovery after photobleaching
- IFT
intraflagellar transport
- mC
mCherry
- MIMS
membrane inlet mass spectrometry
- MLT
multiple-eyespot
- mNG
mNeonGreen
- PLD
phospholipase D
- Rubisco
ribulose-1,5-biphosphate carboxylase-oxygenase
- TIRFM
Total Internal Reflection Fluorescence Microscopy
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (https://www.nih.gov/) under Award Number R01GM110413 (to K.L.) and by the National Science Foundation grant (https://www.nsf.gov/) under Award Number OPP 1744760 (to B.M.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Nachury M.V., and Mick D.U. (2019). Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol 20, 389–405. 10.1038/s41580-019-0116-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogino T., Sawada M., Takase H., Nakai C., Herranz-Perez V., Cebrian-Silla A., et al. (2016). Characterization of multiciliated ependymal cells that emerge in the neurogenic niche of the aged zebrafish brain. J Comp Neurol 524, 2982–2992. 10.1002/cne.24001 [DOI] [PubMed] [Google Scholar]
- 3.Beech P., Heimann K., and Melkonian M. (1991). Development of the flagellar apparatus during the cell cycle in unicellular algae In The Cytoskeleton of Flagellate and Ciliate Protists. (Springer; ), pp. 23–37. [Google Scholar]
- 4.Hoek C., Mann D., Jahns H.M., and Jahns M. (1995). Algae: an introduction to phycology, (Cambridge university press; ). [Google Scholar]
- 5.Melkonian M., Reize I.B., and Preisig H.R. (1987). Maturation of a Flagellum/Basal Body Requires More than One Cell Cycle in Algal Flagellates: Studies on Nephroselmis Olivacea (Prasinophyceae). (Berlin, Heidelberg: Springer Berlin Heidelberg; ), pp. 102–113. [Google Scholar]
- 6.Schoppmeier J., and Lechtreck K.F. (2003). Flagellar regeneration in Spermatozopsis similis (Chlorophyta). J Phycol 39, 918–922. [Google Scholar]
- 7.Kamiya R., and Witman G.B. (1984). Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. The Journal of Cell Biology 98, 97–107. 10.1083/jcb.98.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pazour G.J., Sineshchekov O.A., and Witman G.B. (1995). Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J Cell Biol 131, 427–440. 10.1083/jcb.131.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingfield J.L., and Lechtreck K.F. (2018). Chlamydomonas Basal Bodies as Flagella Organizing Centers. Cells 7 10.3390/cells7070079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan K.Y., Leptos K.C., and Goldstein R.E. (2014). Lag, lock, sync, slip: the many ‘phases’ of coupled flagella. J R Soc Interface 11, 20131160. 10.1098/rsif.2013.1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamiya R., and Hasegawa E. (1987). Intrinsic difference in beat frequency between the two flagella of Chlamydomonas reinhardtii. Exp Cell Res 173, 299–304. 10.1016/0014-4827(87)90357-0 [DOI] [PubMed] [Google Scholar]
- 12.Mittelmeier T.M., Thompson M.D., Lamb M.R., Lin H., and Dieckmann C.L. (2015). MLT1 links cytoskeletal asymmetry to organelle placement in chlamydomonas. Cytoskeleton (Hoboken) 72, 113–123. 10.1002/cm.21220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rüffer U., and Nultsch W. (1997). Flagellar photoresponses of ptx1, a nonphototactic mutant of Chlamydomonas. Cell motility and the cytoskeleton 37, 111–119. [DOI] [PubMed] [Google Scholar]
- 14.Leptos K.C., Wan K.Y., Polin M., Tuval I., Pesci A.I., and Goldstein R.E. (2013). Antiphase synchronization in a flagellar-dominance mutant of Chlamydomonas. Phys Rev Lett 111, 158101 10.1103/PhysRevLett.111.158101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horst C.J., and Witman G.B. (1993). ptx1, a nonphototactic mutant of Chlamydomonas, lacks control of flagellar dominance. J Cell Biol 120, 733–741. 10.1083/jcb.120.3.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- 17.Lechtreck K.F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., et al. (2009). The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol 187, 1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu P., and Lechtreck K.F. (2018). The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc Natl Acad Sci U S A. 10.1073/pnas.1713226115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lechtreck K.F., Brown J.M., Sampaio J.L., Craft J.M., Shevchenko A., Evans J.E., et al. (2013). Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol 201, 249–261. 10.1083/jcb.201207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moroney J.V., Ma Y., Frey W.D., Fusilier K.A., Pham T.T., Simms T.A., et al. (2011). The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth Res 109, 133–149. 10.1007/s11120-011-9635-3 [DOI] [PubMed] [Google Scholar]
- 21.Badger M.J.P.R. (2003). The roles of carbonic anhydrases in photosynthetic CO 2 concentrating mechanisms. 77, 83. [DOI] [PubMed] [Google Scholar]
- 22.Mackinder L.C.M., Chen C., Leib R.D., Patena W., Blum S.R., Rodman M., et al. (2017). A Spatial Interactome Reveals the Protein Organization of the Algal CO2-Concentrating Mechanism. Cell 171, 133–147.e114. 10.1016/j.cell.2017.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasala B.A., Barrera D.J., Ng J., Plucinak T.M., Rosenberg J.N., Weeks D.P., et al. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 10.1111/tpj.12165 [DOI] [PubMed] [Google Scholar]
- 24.Schroda M., Blocker D., and Beck C.F. (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J 21, 121–131. 10.1046/j.1365-313x.2000.00652.x [DOI] [PubMed] [Google Scholar]
- 25.Witman G.B. (1986). [28] Isolation of Chlamydomonas flagella and flagellar axonemes In Methods in enzymology, Volume 134 (Elsevier; ), pp. 280–290. [DOI] [PubMed] [Google Scholar]
- 26.King S.M., and Witman G.B. (1990). Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J Biol Chem 265, 19807–19811. [PubMed] [Google Scholar]
- 27.Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., and Rosenbaum J.L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141, 993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lechtreck K.F. (2013). In vivo imaging of IFT in Chlamydomonas flagella In Methods in enzymology, Volume 524 (Elsevier; ), pp. 265–284. [DOI] [PubMed] [Google Scholar]
- 29.Lechtreck K.F. (2016). Methods for studying movement of molecules within cilia In Cilia. (Springer; ), pp. 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkinson B.M., Dupont C.L., Allen A.E., and Morel F.M.M. (2011). Efficiency of the CO2-concentrating mechanism of diatoms. Proceedings of the National Academy of Sciences of the United States of America 108, 3830–3837. 10.1073/pnas.1018062108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopkinson B.M., Tansik A.L., and Fitt W.K. (2015). High internal carbonic anhydrase activity in the tissue of scleractinian corals is sufficient to support proposed roles in photosynthesis and calcification. J. Exp. Biol. 218, 2039–2048. 10.1242/jeb.118182 [DOI] [PubMed] [Google Scholar]
- 32.Arias-Darraz L., Colenso C.K., Veliz L.A., Vivar J.P., Cardenas S., and Brauchi S. (2015). A TRP conductance modulates repolarization after sensory-dependent depolarization in Chlamydomonas reinhardtii. Plant Signal Behav 10, e1052924 10.1080/15592324.2015.1052924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Zhang R., Patena W., Gang S.S., Blum S.R., Ivanova N., et al. (2016). An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387. 10.1105/tpc.15.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pazour G.J., Agrin N., Leszyk J., and Witman G.B. (2005). Proteomic analysis of a eukaryotic cilium. J Cell Biol 170, 103–113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitsuhashi S., Mizushima T., Yamashita E., Yamamoto M., Kumasaka T., Moriyama H., et al. (2000). X-ray structure of β-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO2 hydration. Journal of Biological Chemistry 275, 5521–5526. 10.1074/jbc.275.8.5521 [DOI] [PubMed] [Google Scholar]
- 36.Mitra M., Lato S.M., Ynalvez R.A., Xiao Y., and Moroney J.V. (2004). Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 135, 173–182. 10.1104/pp.103.037283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmer B.T., Maric D., and Engman D.M. (2010). Molecular mechanisms of protein and lipid targeting to ciliary membranes. J Cell Sci 123, 529–536. 10.1242/jcs.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman D.N. (1982). Carbonic anhydrase: oxygen-18 exchange catalyzed by an enzyme with rate-contributing proton-transfer steps. Methods Enzymol 87, 732–752. 10.1016/s0076-6879(82)87037-7 [DOI] [PubMed] [Google Scholar]
- 39.Gee C.W., and Niyogi K.K. (2017). The carbonic anhydrase CAH1 is an essential component of the carbon-concentrating mechanism in Nannochloropsis oceanica. Proc Natl Acad Sci U S A 114, 4537–4542. 10.1073/pnas.1700139114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamano T., and Fukuzawa H. (2009). Carbon-concentrating mechanism in a green alga, Chlamydomonas reinhardtii, revealed by transcriptome analyses. J Basic Microbiol 49, 42–51. 10.1002/jobm.200800352 [DOI] [PubMed] [Google Scholar]
- 41.Yamano T., Miura K., and Fukuzawa H. (2008). Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 147, 340–354. 10.1104/pp.107.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maric D., McGwire B.S., Buchanan K.T., Olson C.L., Emmer B.T., Epting C.L., et al. (2011). Molecular determinants of ciliary membrane localization of Trypanosoma cruzi flagellar calcium-binding protein. J Biol Chem 286, 33109–33117. 10.1074/jbc.M111.240895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes J.A., and Dutcher S.K. (1989). Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci 94 (Pt 2), 273–285. [DOI] [PubMed] [Google Scholar]
- 44.Strenkert D., Schmollinger S., Gallaher S.D., Salome P.A., Purvine S.O., Nicora C.D., et al. (2019). Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc Natl Acad Sci U S A 116, 2374–2383. 10.1073/pnas.1815238116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moroney J.V., and Ynalvez R.A. (2007). Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot Cell 6, 1251–1259. 10.1128/EC.00064-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi H.I., Kim J.Y., Kwak H.S., Sung Y.J., and Sim S.J. (2016). Quantitative analysis of the chemotaxis of a green alga, Chlamydomonas reinhardtii, to bicarbonate using diffusion-based microfluidic device. Biomicrofluidics 10, 014121 10.1063/1.4942756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bahn Y.S., Cox G.M., Perfect J.R., and Heitman J. (2005). Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol 15, 2013–2020. 10.1016/j.cub.2005.09.047 [DOI] [PubMed] [Google Scholar]
- 48.Wandernoth P.M., Raubuch M., Mannowetz N., Becker H.M., Deitmer J.W., Sly W.S., et al. (2010). Role of carbonic anhydrase IV in the bicarbonate-mediated activation of murine and human sperm. PLoS One 5, e15061 10.1371/journal.pone.0015061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wetherbee R., Platt S.J., Beech P.L., and Pickett-Heaps J.D. (1988). Flagellar transformation in the heterokont Epipyxis pulchra (Chrysophyceae): Direct observations using image enhanced light microscopy. Protoplasma 145, 47–54. [Google Scholar]
- 50.Datta P., Allamargot C., Hudson J.S., Andersen E.K., Bhattarai S., Drack A.V., et al. (2015). Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc Natl Acad Sci U S A 112, E4400–4409. 10.1073/pnas.1510111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dean S., Moreira-Leite F., Varga V., and Gull K. (2016). Cilium transition zone proteome reveals compartmentalization and differential dynamics of ciliopathy complexes. Proc Natl Acad Sci U S A 113, E5135–5143. 10.1073/pnas.1604258113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., et al. (2003). Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425, 628–633. 10.1038/nature02030 [DOI] [PubMed] [Google Scholar]
- 53.Wingfield J.L., Mengoni I., Bomberger H., Jiang Y.Y., Walsh J.D., Brown J.M., et al. (2017). IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. Elife 6 10.7554/eLife.26609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deane J.A., Cole D.G., Seeley E.S., Diener D.R., and Rosenbaum J.L. (2001). Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol 11, 1586–1590. 10.1016/s0960-9822(01)00484-5 [DOI] [PubMed] [Google Scholar]
- 55.Lange B.M., and Gull K. (1995). A molecular marker for centriole maturation in the mammalian cell cycle. J Cell Biol 130, 919–927. 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piel M., Meyer P., Khodjakov A., Rieder C.L., and Bornens M. (2000). The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol 149, 317–330. 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye F., Nager A.R., and Nachury M.V. (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol 217, 1847–1868. 10.1083/jcb.201709041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh S.K., Gui M., Koh F., Yip M.C., and Brown A. (2020). Structure and activation mechanism of the BBSome membrane protein trafficking complex. Elife 9 10.7554/eLife.53322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geimer S., and Melkonian M. (2004). The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J Cell Sci 117, 2663–2674. 10.1242/jcs.01120 [DOI] [PubMed] [Google Scholar]
- 60.Yen H.J., Tayeh M.K., Mullins R.F., Stone E.M., Sheffield V.C., and Slusarski D.C. (2006). Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum Mol Genet 15, 667–677. 10.1093/hmg/ddi468 [DOI] [PubMed] [Google Scholar]
- 61.Wingfield J.L., Lechtreck K.F., and Lorentzen E. (2018). Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem 62, 753–763. 10.1042/EBC20180030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao M., Ning J., Hernandez-Lara C.I., Belzile O., Wang Q., Dutcher S.K., et al. (2015). Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife 4 10.7554/eLife.05242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamb M.R., Dutcher S.K., Worley C.K., and Dieckmann C.L. (1999). Eyespot-assembly mutants in Chlamydomonas reinhardtii. Genetics 153, 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table list the parameters and numerical values of the MIMS experiment for the control (g1) and the cah6 mutant strain.
(DOCX)
A) Agarose gel of PCR products using cah6 genomic DNA as a template; G-beta primers are used as control. See Fig 2A for the positions of the primers. B) Western blot analysis of whole cells, deflagellated cell bodies (Cell Body), and isolated flagella (Flagella) from wild-type g1 and cah6 probed with anti-CAH6. Equivalent amounts of cells and flagella were loaded (i.e., one whole cell, one cell body, and two flagella). Antibodies to the IFT particle protein IFT81 were used to control for equal loading. A part of the same blot is shown in Fig 2B, 2C and 2D) Schematic presentation of CAH6 (C) and amino acid sequence of CAH6 (D). The predicted chloroplast transit peptide (cTP) is indicated in green and the catalytic domain in red. In D, the 23 conserved residues typical for β-type CAs are indicated by red font. Asterisks indicate the two amino acids that are not conserved in CAH6. (E) Schematic representation illustrating the measurement of CA activity.
(TIF)
A) Flagellar length of control (g1) and the cah6 strains. The standard deviations and the number of measurements are indicated. B) Mean swimming velocity of wild-type (g1) and cah6 cells in 0.5% CO2 and 0.04% CO2 (air). C) Liquid growth curves for wild-type g1 (red circles) and cah6 (black squares) in 5% CO2, 0.04% CO2 (air) and <0.04% CO2. D) Phototaxis assay of control (g1) and cah6. The time of light exposure (5 mins) and direction of the light (arrowheads) are indicated. E) Western blot analysis of flagella for CAH6 isolated from wild-type (g1) cells aerated for 24 hours under high (0.5%), low (0.04%) and no aeration with CO2 conditions. Anti-IC2 was used as a loading control. F) Schematic presentation of the chemotaxis assay. Cells in M-medium were placed into a small Petri dish or 6-well cell culture plate and agar plaques (A, B) containing either 5% sodium bicarbonate or 20% tryptone (A) or no attractant (B) were added. G) Chemotaxis assay showing g1, cah6 and bbs4-1 before (T0) and after a 20-minute incubation in the dark (T20). Accumulated cells are indicated by the arrows. The following results were noted in repeat experiments (positive chemotaxis/no chemotaxis): g1 aerated with CO2 (0/9), g1 without aeration (5/5), cah6 (4/3), bbs4-1 (1/2), and bbs1 (2/1).
(TIF)
A) Brightfield (BF) and TIRFM images of live g1 CAH6-mNG cells. The two focal planes show flagella and eyespot (indicated with black arrowheads). Trans-flagella are indicated with red arrowheads and cis-flagella with yellow arrowheads. Bar = 2μm. B) Western blot analysis of isolated flagella (Flagella) from wild-type g1, cah6 and bbs1 cah6 CAH6-mNG were probed with anti-CAH6 and anti-BBS4. Antibodies to acetylated tubulin were used as a loading control. C) Scatter plot showing the ratios between the trans- and cis-flagellum of control and bbs1 cells; see Fig 3D for presentation as a histogram.
(TIF)
A) Brightfield (BF) and TIRFM images of a bbs1 PLD-mNG gametes and live dikaryons from a matings between bbs1 PLD-mNG and control (g1) or bbs4. In bbs1 PLD-mNG x bbs4 zygotes, both parental strains lack intact BBSomes and BBSomes need to be assembled de novo after cell fusion delaying the export of PLD-mNG. Indeed, an abnormal presence of PLD-mNG was observed in two of the four flagella in a subset of the zygotes analyzed at ≤ 30 min whereas PLD-mNG was essentially absent from all four flagella at the later time points (>30 min). Bar = 2 μm. B) Brightfield (BF) and TIRFM images of a live dikaryon from a mating between g1 CAH6-mNG and cah6 gametes. Trans-flagella are indicated with red arrowheads and cis-flagella with yellow arrowheads. Bar = 2 μm. C) Kymograms showing CAH6-mNG dynamics in the flagella of cah6 × bbs1 cah6 CAH6-mNG zygotes. Open arrows indicate transport of CAH6-mNG via anterograde IFT. D) Brightfield (BF) and TIRFM images of live dikaryons from a mating between cah6 CAH6-mNG and bbs4 gametes. The two focal planes show the flagella and the eyespot (indicated with black arrowheads). Trans-flagella are indicated with red arrowheads and cis-flagella with yellow arrowheads. Bar = 2 μm.
(TIF)
TIRF localization of CAH6-mNG in cah6 CAH6-mNG (left) and bbs1 cah6 CAH6-mNG (right) flagella. The beginning of the video was recorded in brightfield mode and shows first the eyespot (eye) and then moves to the focal plane of the flagella before switching to TIRF illumination. The cis- and trans-flagella are indicated. Images were acquired at 10 fps, and playback is set at 20 fps (2× speed). The timer counts seconds.
(AVI)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.