Abstract
Expression of virulence factors in non-typhoidal Salmonella enterica depends on a wide variety of general and specific transcriptional factors that act in response to multiple environmental signals. Expression of genes for cellular invasion located in the Salmonella pathogenicity island 1 (SPI-1) is tightly regulated by several transcriptional regulators arrayed in a cascade, while repression of this system is exerted mainly by H-NS. In SPI-1, H-NS represses the expression mainly by binding to the regulatory region of hilA and derepression is exercised mainly by HilD. However, the possible regulatory role of H-NS in genes downstream from HilD and HilA, such as those regulated by InvF, has not been fully explored. Here the role of H-NS on the expression of sopB, an InvF dependent gene encoded in SPI-5, was evaluated. Our data show that InvF is required for the expression of sopB even in the absence of H-NS. Furthermore, in agreement with previous results on other InvF-regulated genes, we found that the expression of sopB requires the InvF/SicA complex. Our results support that SicA is not required for DNA binding nor for increasing affinity of InvF to DNA in vitro. Moreover, by using a bacterial two-hybrid system we were able to identify interactions between SicA and InvF. Lastly, protein-protein interaction assays suggest that InvF functions as a monomer. Derived from these results we postulate that the InvF/SicA complex does not act on sopB as an anti-H-NS factor; instead, it seems to induce the expression of sopB by acting as a classical transcriptional regulator.
Introduction
Salmonella species are widely distributed and are the cause of gastroenteritis, diarrhea and typhoid fever around the globe [1,2]. These bacteria have a plethora of virulence factors mainly grouped in discrete genomic regions called Salmonella pathogenicity islands (SPIs) [3,4]. SPI-1 and SPI-2 each encode for a type three-secretion system (T3SS), T3SS-1 and T3SS-2, respectively. The T3SS-1 is required for invasion as well as for replication of Salmonella in the cytosol of epithelial cells [4–6]. The T3SS-2 is mainly needed for bacterial survival and replication within macrophages, in compartments denoted Salmonella containing vacuole (SCV) [4,7].
Expression of T3SS-1 and T3SS-2 has been widely studied in both in vivo and in vitro conditions, allowing to determine many details of how and when these two virulence apparatuses are expressed. In general, global regulators activate the expression of Salmonella-specific transcriptional factors, in response to environmental signals present in the hosts; then, the specific regulators induce the expression of genes required for the colonization of particular in vivo niches where Salmonella can replicate [8–11]. Among the global regulators, the nucleoid-associated protein H-NS has a pivotal regulatory role in Salmonella by acting as a repressor on most virulence genes, including the SPI-1 and SPI-2 regulons [12–17]. Thus, the expression of most Salmonella virulence genes is induced by the action of a regulator that counteracts H-NS-mediated repression on the respective promoters [14,18,19].
In regard to SPI-1, this island encodes the HilD, HilC, HilA, InvF and SprB regulators; HilD, HilC and InvF belonging to the AraC/XylS family of transcriptional regulators [8,9]. HilD, HilA and InvF form a regulatory cascade that induces expression of the SPI-1 genes and many other genes located outside SPI-1 (Fig 1). This regulatory cascade starts with a positive feed-forward loop between HilD, HilC and RtsA (encoded outside of the SPI-1); HilD being the dominant transcriptional factor in this regulatory loop [8,20], induces the expression of hilA and several other target genes by antagonizing H-NS-mediated repression [8,9,20,21]. Then, HilA activates the expression of genes involved in the biosynthesis of the T3SS-1; additionally, it activates the expression of the InvF transcriptional regulator [22]. Finally, InvF induces the expression of a set of genes encoding effectors that are translocated into host cells through the T3SS-1, including sopB (sigD), which is located in SPI-5 [23,24] (Fig 1). InvF is the only member of the AraC/XylS family of regulators known to form a complex with another protein to activate its cognate genes [25,26]. That is, in order for InvF to be active it needs to interact with the chaperone protein SicA, which also binds to multiple T3SS-1 components and effectors [27–30]. The InvF/SicA complex positively controls the expression of sicA, sopB, sptP, sopE, sopE2, STM1239 and several other genes and, being in the last portion of the regulatory cascade, InvF/SicA controls the timing of effector protein expression [10,28,29]. However, the mechanism by which InvF/SicA induces expression of target genes remains undetermined, i.e. it is unknown if InvF/SicA acts as an anti-H-NS factor, as in the case of HilD, or if it mediates gene expression as a classical transcriptional factor.
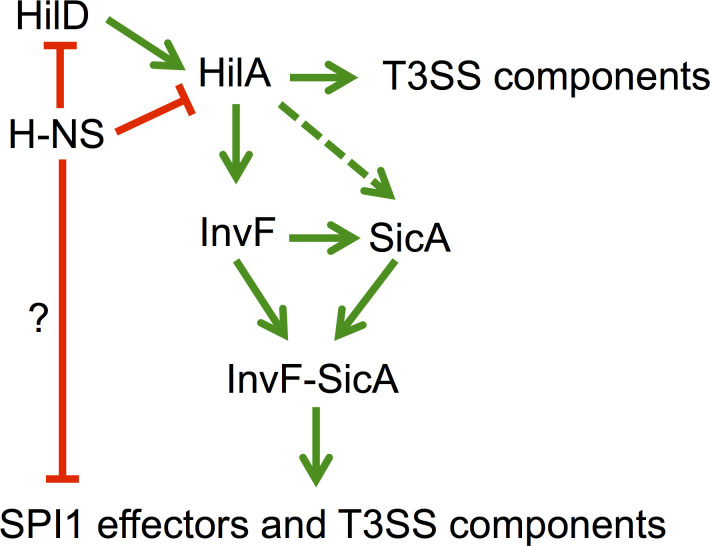
Fig 1. Simplified schematic representation of a section of the SPI-1 regulatory cascade.
H-NS represses the expression of HilD and HilA. As described by Ellermeier et al. [20], HilD, HilC and RtsA activate HilA transcription but HilD has a predominant role and it is the only one illustrated here. HilD removes H-NS and once HilA is translated it activates genes for T3SS-1 biogenesis and also the last transcriptional regulator in this cascade, InvF. This activates some structural genes and the chaperone SicA. Together, the InvF/SicA complex activates the expression of effector proteins. To our knowledge, it is not known whether H-NS represses the genes coding for the effectors and whether InvF acts as a derepressor or as a classical activator. A green line with an arrow indicates activation of gene expression, a green pointed line indicates readthrough activation and a red blunted line indicates repression of expression.
Here we show that, in contrast to HilD and other Salmonella transcriptional regulators involved in virulence, the InvF/SicA complex is required for the expression of target genes even in the absence of the H-NS repressor. Additionally, we show that InvF binds in vitro to the promoter region of sopB independently of SicA. However, our results and those previously reported indicate that transcriptional activation of sopB requires both InvF and SicA. These findings led us to propose that the InvF/SicA complex might be acting as a classical transcriptional regulator to induce the expression of virulence genes.
Materials and methods
Bacterial strains and culture conditions
Salmonella enterica serotype Typhimurium SL1344 strain [31] and the isogenic ΔSPI1, ΔSPI1 ΔrtsA and invF::Tn5 mutants, were previously generated [16,23,32]. Strain SL1344 invF::Flag was obtained by transducing the marked gene from strain 14028 invF::Flag using P22 phage [33]. Escherichia coli MC4100 and its Δhns derivative were used for expression experiments [34,35]. E. coli DH10b and BL21 (DE3) strains were used for genetic constructs and protein expression, respectively. E. coli SU101 and SU202 were used for the dimerization experiments [36]. SPI-1 inducing conditions were used as previously described using LB-Miller broth [37]. Briefly, one or two colonies of the Salmonella strains to be tested were taken from a fresh plate and inoculated into 3 ml LB-Miller broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, pH 7.0) in a 16 x 100 mm, glass round-bottom test tube with loose cap that was incubated at 37 oC in shaking conditions (220 rpm) for 16 to 18 h. Subsequently, a 300 μl of this culture were subcultured into a 125 ml glass flask containing 10 ml of sterile LB-Miller broth with a loose cap or cotton wool stopper, and incubated at 37 oC in shaking conditions (220 rpm) for 3.5 h (aerated conditions). Media for selecting bacterial clones was LB agar supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml), tetracycline (10 μg/ml) or streptomycin (100 μg/ml).
Construction of plasmids
Plasmids used in this work are listed in S1 Table. To construct a plasmid containing invF (pT3-InvF), invF-XH1-Fw and invF-H3-Rv oligos (S1 Table) were used to amplify this gene using S. Typhimurium genomic DNA as a template. The PCR product was purified, digested with HindIII and XhoI and then cloned into the pMPM-T3 vector [38]. A plasmid containing sicA (pTOPO-SicA) was constructed using the oligos sicA-Fw and sicA-His6-Rv (designed to include a His6 tag) to amplify by PCR sicA using S. Typhimurium genomic DNA as template. The obtained amplicon was cloned into the pCRTOPO 2.1 vector (Thermo Scientific) using the TOPO-TA cloning kit.
Plasmids pSR658-InvF and pSR658-SicA [39] were constructed by using the oligos lexA-invF-Fw/lexA-invF-Rv2, and sicA-Lex-Fw/sicA-Lex-Rv2 respectively, then the PCR products were purified and digested with XhoI and PstI and cloned into the plasmid pSR658. To construct the plasmid pSR659-SicA, oligos sicA-Lex-Fw and sicA-Lex-Rv3 were used to amplify sicA, then the PCR product was digested with XhoI and KpnI and cloned into pSR659. All plasmid constructs were sequenced for verification.
CAT assays
Chloramphenicol acetyl transferase (CAT) specific activity from the cat transcriptional fusions was determined as described before [32,35,40,41]. Briefly, samples of 1 ml were taken from bacterial cultures in SPI-1 inducing conditions described above in this section and in a previous study [37]. Samples were lysed by sonication and soluble extracts were obtained by centrifugation. CAT activity and protein quantification to calculate CAT specific activity were determined for the soluble extracts on 96-well microplates as described previously [32,35,40,41].
RT-qPCR assays
Relative expression of sopB in the different Salmonella strains was determined by RT-qPCR as described previously [37,42]. Briefly, RNA was obtained from bacterial cultures grown in SPI-1-inducing conditions. DNA was removed with DNA-Free (Ambion) and then cDNA was obtained with a GoScript kit (Promega). qPCR was performed in a Rotor-gene Q Thermocycler (Qiagen). Relative expression of sopB was calculated with the ΔΔCt method using the expression of the gene coding for the 16S rRNA as a normalizer. Oligos for each gene are listed in S1 Table (sopB-RT-Fw and sopB-RT-Rv for sopB; Eub338F and Eub518R for the 16S gene). Experiments were done in triplicates and the results are the average of three independent experiments.
Expression and purification of MBP-InvF and MBP
Both proteins, chimeric protein MBP-InvF and MBP, were purified from E. coli BL21 harboring plasmids pMal-InvF and pMal-c2xa, respectively, and over-expressed by IPTG induction, lyzed by sonication and by using affinity chromatography with amylose resin as previously described [16,43]. Fractions of the purified proteins were observed in a 12% SDS-PAGE and analyzed by Western blot using anti-MBP antibodies (New England Biolabs). Protein concentrations were determined by following the Bradford protocol and by using an albumin standard curve. In addition to the Western blot analysis, MBP-InvF bands were excised from the Coomassie stained SDS-PAGE and sent for identification by LC-MS/MS to the Centre de Recherche CHU de Québec Proteomics facility. Results were analyzed by using the Scaffold 4 program.
Expression and purification of SicA-His6
SicA-His6 was purified from cultures of E. coli BL21 pTOPO-SicA by affinity chromatography with Ni-NTA (Qiagen) as described previously [44,45]. Fractions of the purified protein were observed in a 12% SDS-PAGE and also analyzed by Western blot using an HRP-conjugated His-probe (Thermo Scientific). As a control, the GlpQ-His10 fusion protein previously reported by our lab was also purified [45]. Protein concentration was determined by Bradford with the use of an albumin standard curve.
Electrophoretic Mobility Shift Assays (EMSAs)
Protein-DNA interactions were observed by a change in the electrophoretic mobility of DNA fragments as described before [43,46]. Briefly, a DNA fragment corresponding to the promoter region of sopB was amplified by PCR with the pair of oligos sopB-200-Fw and sopB-Fus-Rv. A region of fliC was amplified by PCR with primers fliC-Fw and fliC-Rv and used as a negative control in these assays. Protein molar concentrations were calculated by using the following webpage: http://molbiol.edu.ru/eng/scripts/01_04.html. Proteins and DNA fragments were mixed in 1X binding buffer (10X buffer: 400 mM HEPES, 80 mM MgCl2, 500 mM KCl, 10 mM DTT, 0.5% NP40 and 1 mg/ml BSA) [46] to a final volume of 20 to 40 μl and incubated at room temperature for 30 min. DNA fragments were resolved by electrophoresis in a native TBE 6% acrylamide gel and stained with ethidium bromide.
Protein-protein interactions (pulldown)
Pulldown experiments were performed with purified MBP, MBP-InvF and SicA-His6 proteins. Two mixtures were done: a negative control with MBP and SicA-His6 and tests with MBP-InvF and SicA-His6; both were done by using 50 μg of each protein in an 2X interaction buffer (100 mM NaH2PO4, 600 mM NaCl, 40 mM imidazol, 0.5% NP-40 and 20% glycerol, pH 8.0) [47]. Proteins were let to interact for 30 min on ice, then 50 μl of amylose resin or Ni-NTA agarose beads were added to each mixture and let to interact for 2 h in agitation at ~4°C. Beads were centrifuged at 2,000 x g for 2 min, amylose beads were washed three times with cold washing buffer (see above) and Ni-NTA beads were washed with low imidalozole buffer (Qiagen). After the last washing step, supernatant was removed carefully and then 20 μl of Laemmli buffer were added. Samples were resolved in an SDS-PAGE and stained with Coomassie blue. Western blot was performed by transferring the proteins from the SDS-PAGE to a PVDF membrane (Merck) and by following a previously described protocol [45,47]. Western blot was developed by using chemiluminescence kit (Invitrogen) and observed in a Chemidoc imaging system (Biorad).
Pulldown experiments were also performed with a cell free extract containing InvF-Flag and either MBP or MBP-InvF. For this, S. Typhimurium invF::Flag strain was transformed with either pMal-InvF or pMal-c2xa, as a negative control. Then, cell free extracts were obtained from bacterial cultures grown in SPI-1-inducing conditions and the addition of 0.3 mM IPTG. One ml of each cell free extract was mixed with 50 μl of amylose resin and let to interact for 2 h in agitation at ~4°C. Beads were centrifuged at 2,000 x g for 2 min and washed four times with cold washing buffer (see above). After the last washing step supernatant was removed carefully, 30 μl of Laemmli buffer were added to the beads and samples were boiled for 10 min. Samples were resolved in a SDS-PAGE, Western blot was performed by transferring the proteins to a PVDF membrane (Merck) and using anti-MBP antibodies (New England Biolabs) and anti-FLAG antibodies (Sigma) as suggested by the manufacturers. Membrane developing was done as described above in this section.
Dimerization assays
A LexA-based two hybrid system was used to evaluate protein-protein interactions between InvF and SicA [36,39]. In order to verify the integrity of the LexA-derived proteins a Western blot was performed by using the primary antibody anti-LexA (Millipore) (1;2,000) and the secondary antibody anti-rabbit IgG-HRP (Sigma-Aldrich) (1;10,000) (S4 Fig). To test InvF and SicA homodimerization competent cells of E. coli SU101 were transformed with pSR658-InvF or pSR658-SicA and selected in LB plates with the corresponding antibiotic (S1 Table). Selected transformants were grown in LB supplemented with 1 mM IPTG and let them grow to an OD of 0.6. Aliquots were taken to assess β-galactosidase activity as described before [48]. To test InvF and SicA heterodimerization, E. coli SU202 was co-transformed with the plasmids pSR658-InvF and pSR659-SicA and tested as described above. The plasmids pSR658-HNS, pSR658-HilD and pSR659-HilE were used as controls in the experiments [49].
Statistical analysis
Statistical analysis was performed in Excel (Microsoft) or GraphPad Prism version 5 (GraphPad Software) by using a Student’s t-test. A significant difference was considered when P < 0.05.
Results
The InvF/SicA complex does not act as an anti-HNS factor on sopB
The global transcriptional regulator H-NS has been shown to play an important role as a repressor of the expression of many virulence genes in Salmonella [8,9,20,21] (Fig 1). As for genes depending on InvF, several evidences have shown that this regulator, together with SicA, is necessary for expression [28,29] (Fig 1). Here we questioned whether InvF induces gene expression by antagonizing H-NS-mediated repression on target genes, for which sopB was used as a probe gene. Our reasoning was that if H-NS represses sopB and InvF just acts as an anti-H-NS factor, then the expression of this gene should be induced in the absence of H-NS independently of InvF similarly as it has been observed with other virulence genes in Salmonella. To further investigate the action mechanism of InvF, we first analyzed the activity of a sopB-cat transcriptional fusion in the WT S. Typhimurium strain and its isogenic ΔinvF mutant carrying the pT3-InvF plasmid, which expresses InvF from a constitutive promoter, or carrying the pMPM-T3 vector. As expected, the sopB-cat fusion was not expressed in the invF mutant; furthermore, its activity was restored in the invF::Tn5 mutant by the presence of the pT3-InvF plasmid, to levels even higher than those reached by this fusion in the WT strain (Fig 2A).
Fig 2. InvF and SicA are required for expression of sopB.
sopB expression in Salmonella Typhimurium (A) and E. coli (B) strains was determined by means of a transcriptional fusion to the cat reporter gene (psopB-cat1) in SPI-1 inducing conditions, as mentioned in the Methods section. The pT3-InvF and pTOPO-SicA plasmids constitutively express InvF and SicA, respectively, and the corresponding empty vectors are indicated (pT3 is pMPM-T3 and pTOPO is pCR2.1TOPO-TA, see S1 Table). Bars represent the average of at least three different experiments and error bas are standard deviations.
Additionally, since previous studies indicate that InvF needs to interact with the SicA chaperone to be active [28,29], we analyzed the activity of the sopB-cat fusion in a Salmonella strain lacking SPI-1 and RtsA (ΔSPI1 ΔrtsA). As expected, the pT3-InvF plasmid was not able to induce expression of sopB-cat in this mutant strain (Fig 2A). Similar results were observed when an Escherichia coli K-12 surrogate system was used. Expression of sopB-cat was only induced when both the pT3-InvF plasmid expressing InvF and the pTOPO-SicA plasmid expressing SicA constitutively, were present (Fig 2B). These results corroborate previous reports showing that expression of sopB requires the InvF/SicA complex.
Then, the effect of the inactivation of H-NS on the activity of the sopB-cat fusion was analyzed when the InvF/SicA complex is not present. Ideally, a Salmonella hns mutant should be used for these experiments, but this mutation has been shown to be unstable because it causes the overexpression of SPI-1 and other related genes, and suppressor mutations are generated [50]. Thus, we decided to use a surrogate system, an E. coli Δhns mutant, which has been useful to study the effect of H-NS on other Salmonella virulence genes [51]. As a positive control, the hilA-cat transcriptional fusion was assessed in WT E. coli and its derivative Δhns mutant; H-NS represses expression of hilA [52,53]. As seen in Fig 3, while the expression of the hilA-cat transcriptional fusion was induced in the E. coli Δhns mutant (Fig 3B), the expression of sopB-cat was not (Fig 3A). As observed in the WT E. coli strain, the sopB-cat fusion was expressed in the E. coli Δhns mutant only in the presence of both InvF and SicA (Fig 3A). These results confirm previous reports made in Salmonella indicating that both InvF and the chaperon SicA are required in vivo for expression of InvF-dependent genes and that both proteins are necessary for expression of sopB even in the absence of the global repressor H-NS when using an E. coli surrogate system.
Fig 3. sopB expression is dependent of InvF/SicA in an E. coli hns mutant.
sopB (A) and hilA (B) expression was determined in wt and Δhns E. coli strains using transcriptional fusions to cat (psopB-cat1 and philA-cat-410+66, respectively) in SPI-1 inducing conditions as mentioned in the Methods section. The pT3-InvF and pTOPO-SicA plasmids constitutively express InvF and SicA, respectively and the corresponding empty vectors are indicated (pT3 is pMPM-T3 and pTOPO is pCR2.1TOPO, see S1 Table). Bars represent the average of at least three different experiments and error bas are standard deviations.
SicA is not required for InvF binding to the sopB promoter
To find possible interactions between InvF and the sopB regulatory region the MBP-InvF protein was used for electrophoretic motility shift assays (EMSAs). First, in order to determine whether this construct encoding the protein fusion was able to complement a Salmonella invF::Tn5 mutant sopB mRNA levels were measured and the result showed that indeed it was able to partially complement the mutation (S1 Fig). In order to corroborate the interaction between the tagged versions of InvF and SicA both proteins were purified by using their corresponding tags: MBP and His6, respectively (S2 and S3 Figs). Both proteins were purified but MBP-InvF was very unstable once purified protein fractions were collected; multiple lower bands were observed in the collected fractions that contained the MBP tag (S1B Fig). A band corresponding to the expected size for the MBP-InvF fusion (~ 77 kDa) and those underneath this one were excised and analyzed through LC-MS/MS. Peptides corresponding mainly to MBP (MALE_ECOLI, P0AEX9) and a few corresponding to InvF (P69343) were detected. These data suggest that the MBP-InvF fusion protein is being degraded after purification. Despite these issues, the two purified proteins were used to detect interactions by pull down experiments (Fig 4). Both proteins were able to interact with each other by either using amylose, as a negative control purified MBP was not able to interact with SicA-His6. This result further corroborates previous results showing that InvF interacts with SicA [29,54] and shows that proteins used in this study are functional. Later, this interaction was further corroborated with a bacterial two-hybrid system (see below). Next, MBP-InvF was tested for its ability to bind to the sopB regulatory region comprised between -200 to +79 with respect to the transcriptional start site (Fig 5A). As a negative control, a DNA fragment of a similar size from the coding region of fliC was also assessed. MBP-InvF was able to bind to the sopB DNA fragment and it did not shift the fliC fragment. In these experiments we were unable to detect DNA-protein complexes in the gel, but the disappearance of free DNA was observed only with the sopB regulatory region and not with a non-related DNA fragment (Fig 5A). Some faint bands were observed but they were not clear and were also observed in the lanes without protein, so these were discarded. These results showed that MBP-InvF is able to bind specifically to the sopB regulatory region and suggests that SicA is not needed for this role in vitro.
Fig 4. InvF interacts with SicA in vitro.
Purified versions of MBP, MBP-InvF and SicA-His6 in the combinations: MBP + SicA-His6 and MBP-InvF + SicA-His6, were let to interact in solution and then pulled down with amylose resin. Interactions were detected by Western blot with anti-MBP (A) or anti-His6 (B) (labeled “Affinity purified”). Proteins alone are shown in the first three lanes of each gel as controls and the interactions are indicated.
Fig 5. MBP-InvF interaction with sopB regulatory region is not dependent on SicA.
EMSA experiments were performed with purified MBP-InvF or SicA-His6 or both with the regulatory regions of sopB. (A and B) Increasing amounts of MBP-InvF protein were used as shown. Increasing amounts of SicA-His6 protein and MBP-InvF proteins were used as shown in the image. (C) In order to determine the effect of both proteins in DNA binding, increasing amounts of MBP-InvF (dark triangle) and constant amount of SicA-His6 (grey rectangle) were used. Protein-DNA complexes were resolved by electrophoresis in native acrilamyde gels and stained with ethidium bromide. Images were reverted to improve visualization.
Next we asked whether SicA is able to independently bind to DNA. For this, increasing amounts of SicA-His6 were used with a DNA fragment containing the sopB regulatory region. As seen in Fig 5B, a DNA shift was not observed even when a large amount of protein was used, while a control DNA shift with MBP-InvF required a low amount of protein. To explore the possibility that SicA increases MBP-InvF affinity for its binding site, both proteins were tested in vitro (Fig 5C). Results showed that an increase in MBP-InvF affinity for the sopB regulatory region was not observed. Altogether, these results support that InvF binds in vitro to the regulatory region of sopB independently of SicA.
InvF does not form dimers
Some members of the AraC/XylS family of transcriptional regulators, such as AraC, HilD, etc., are able to form dimers both in solution and when binding to their target DNA [25,49]; dimerization is, in some cases, important for the role as a transcriptional regulator. Mutants defective in this dimer formation are not able to either repress or activate. In order to determine whether InvF is able to form dimers, a bacterial LexA based two-hybrid system was used [39,49]. In this system the LexA DNA-binding domain (LexADBDwt) is fused to the protein of interest and then tested with a sulA-lacZ chromosomal transcriptional fusion. When there is no interaction there is β-galactosidase activity; in contrast, when there is protein-protein interaction then there is no enzymatic activity from the reporter fusion [36]. As shown in Fig 6A this system demonstrated to be useful to detect SicA dimerization; in contrast, InvF did not form dimers. Considering the possibility that SicA affects InvF stability or that the chaperone was needed for InvF to form dimers, SicA was supplied in trans. Results showed that even when SicA was present dimer formation for InvF was not detected. One possibility was that LexADBDwt coupled to InvF impedes the InvF-SicA interaction. In order to test this latter, the interaction between InvF and SicA was tested when both proteins are fused to LexADBDwt and LexADBDmut in the heterodimer system. As observed in Fig 6B, control tests using HilD and HilE and also SicA-SicA showed interactions as expected. When InvF and SicA were tested the expression of the reporter fusion was also diminished, demonstrating an interaction among these proteins. Unexpectedly, LexADBDwt-SicA combined with the empty vector encoding LexADBDmut also showed a lower activity than that compared to the HilD-HilE control but it was not as low as that observed with the SicA-SicA control and with InvF-SicA. Thus, our results suggest that InvF does not form dimers.
Fig 6. LexA-based two-hybrid analysis of InvF and SicA interactions.
LexA-derivatives were introduced either in strain SU101 (A) or SU202 (B) and grown and processed as described in the Methods section. Constructs or combinations of them are indicated below each bar. Bars represent the average of at least three independent experiments and the error bars represent the standard deviation. *, indicates statically significance difference (P < 0.01) as follows: For (A), LexADBDwt-H-NS vs. LexADBDwt; LexADBDwt-SicA vs.LexADBDwt; LexADBDwt-InvF vs. LexADBDwt-H-NS; and LexADBDwt-InvF/pTOPO-SicA vs. LexADBDwt-H-NS; For (B) LexADBDwt/LexADBDmut vs. LexADBDwt-SicA/LexADBDmut-SicA; and LexADBDwt/LexADBDmut vs. LexADBDwt-InvF/LexADBDmut-SicA.
In order to corroborate that InvF is not able to interact with itself the pMal-InvF plasmid was transformed in a Salmonella invF::Flag strain. Once induction was reached by adding IPTG in SPI-1 inducing conditions, cells were obtained and sonicated to obtain a cellular extract which was used for a pulldown experiment with amylose resin. Results showed that MBP-InvF did not interact with the genomic InvF-Flag version (Fig 7B). Together, these results demonstrate that InvF does not form dimers and suggest that it is likely acting as a monomer.
Fig 7. InvF does not form dimers.
Plasmids expressing either MBP or MBP-InvF were transformed into a Salmonella invF::Flag strain and samples were collected from IPTG and SPI-1 induced cultures. Cell free extracts were prepared, pulled down with amylose resin to detect interactions of MBP and MBP-InvF and tested by Western blot. In (A) pull downs were tested to detect production of both MBP versions with an anti-MBP antibody (Input). In (B) an anti-Flag antibody was used to detect possible interactions of InvF-Flag with MBP and MBP-InvF (Affinity purified). As a control for the expression of InvF-Flag in the tested conditions the last lane shows a whole cell extract (Input).
Discussion
Expression of the SPI-1 virulence genes requires the concerted action of several regulators, including InvF and HilD, both encoded in SPI-1 [8,9,20]. In this regulatory cascade HilD induces the expression of these genes mainly by antagonizing H-NS mediated repression, which means that in the absence of H-NS the SPI-1 genes are expressed independently of HilD (Fig 1) [12,13,14,16,51]. In this work, by analyzing the regulation of the sopB gene by InvF, we further define the mechanism by which InvF induces expression of target genes. First, our results further confirm that InvF needs to act together with the SicA chaperone to be active and that both proteins interact in vitro. Additionally, we found that the InvF/SicA complex is required for the expression of sopB even in the absence of H-NS, which supports that InvF/SicA acts as a classical transcriptional activator. Classical activators recruit the RNA polymerase to the promoters they activate, while derepressors remove the negative effect of a repressor [19,55,56]. Alternatively, InvF/SicA could act on sopB by antagonizing repression mediated by an unknown factor, other than H-NS.
In agreement with our results indicating that InvF does not act as an anti-H-NS factor on sopB, we previously demonstrated that purified H-NS does not bind to the sopB regulatory region [16,51]. Moreover, ChIP-on-chip experiments by Lucchini et al. [12] and Navarre et al. [13] did not show a significant signal for binding of H-NS to the sopB promoter nor they showed overexpression of sopB in the absence of H-NS. These findings strongly suggest that H-NS is not directly involved in the regulation of sopB. In any case, what it is clear from our results is that the InvF/SicA complex is requiered to initiate the transcription of sopB in the absence of H-NS. This contrasts with other SPI-1 and SPI-2 regulated genes in which H-NS plays a repressor role that is removed under inducing conditions by dedicated transcriptional regulators [12,13,18,51].
Our data further corroborates the previously shown dimerization of SicA [29,57] and the interaction between InvF and SicA described by Darwin and Miller [29] and Kim et al. [54]. Here, in contrast to results reported by Darwin and Miller [29], we were able to observe the InvF-SicA interaction using a bacterial two-hybrid system based on LexA. Though this interaction was not as clear as that observed for the SicA-SicA interaction, InvF-SicA interaction was corroborated as mentioned above. Moreover, results using pulldown experiments and the LexA-based system have shown that InvF is not able to form homodimers, indicating that it acts as a monomer. In general, the transcriptional regulators in the AraC/XylS family form dimers, but it is not uncommon that some of them act as monomers. This is the case for PerA and Rns [43,58], which are two classical regulators involved in the activation of virulence genes in E. coli. Previously, Darwin and Miller [29] suggested a putative binding site for InvF which overlaps the suggested -35 box in the regulatory regions of sicA, sopB and sopE. Moreover, they showed experimentally that the proposed sequence in sicA is indeed a binding site for InvF. The length of these putative binding sites suggests that only one monomer is able to bind. Thus, it is possible that only one InvF molecule binds to it as we suggest. Ideally, in order to precisely define the InvF binding sites a footprinting should be done; in this case Darwin and Miller did a point-mutation analysis for the sicA promoter region and corrobotrated that the InvF binding site was indeed the one they suggested [29]. This latter supports the idea that InvF binds as a monomer to its biding site and, given the fact that this site overlaps with the -35 site, it is likely that it makes contact with the RNA polymerase. This possibility is currently being tested in our laboratory.
Here, in order to observe the ability of InvF to bind to DNA a chimeric MBP-InvF protein was used. AraC/XylS regulators are very unstable as documented elsewhere, but previous reports from our labs and others have shown that MBP stabilizes these proteins [43,59,60]. One of the initial steps was to demonstrate that the addition of MBP to InvF was not a hindrance for the regulator protein. When using sopB as a probe, we observed that its expression was almost recovered when complementing an invF mutant with a plasmid encoding the chimera. Then, by using the purified MBP-InvF protein our results indicate that it can bind to the regulatory region of sopB in the absence of SicA, which is consistent with a previous report showing InvF binding to sicA regulatory region independently of SicA [29].
Our data also confirms that SicA does not bind to DNA. Moreover, when MBP-InvF and SicA-His6 were tested in combination for binding on sopB, an increase in the affinity of InvF was not detected. Given that SicA is needed in vivo for InvF function we cannot discard the likelihood that an increase in DNA affinity happens when it is expressed in Salmonella. Darwin and Miller [29] also reported that the role of SicA does not increase InvF stability; however, we observed that purified MBP-InvF is unstable as it seems to be degraded even in the presence of commercial proteinase inhibitors.
Conclusions
In summary, our study supports the idea that the InvF/SicA complex could act as a classical transcriptional activator and not as an anti-H-NS factor. Additionally, our results suggest that InvF acts as a monomer in a similar fashion as other AraC/XylS regulators that are also classical regulators.
Supporting information
Expression of sopB was detected by qRT-PCR using the gene coding for rRNA 16S as a normalizer. Indicated strains were grown in SPI-1 inducing conditions and samples were taken for RNA extraction. Bars represent the average of at least three independent experiments and the error bars represent +/- SD. **, indicates statically significance difference (P < 0.05).
(TIFF)
MBP-InvF was purified as described in the Methods section and fractions of this process were taken and observed in a 12% SDS-PAGE (A). Lanes: 1, non-induced cells; 2, IPTG-induced cells; 3–8, eluted fractions obtained from the amylose column. (B) Immune detection (WB) of MBP (lane 1) and MBP-InvF (lanes 2–4) with an anti-MBP antibody. Lines in both panels indicate variants of MBP-InvF.
(TIFF)
SicA-His6 was purified and samples of this process were taken and observed in a 12% SDS-PAGE (A). Lanes: 1, non-induced cells; 2, IPTG-induced cells; 3–8, eluted fractions obtained from the Ni-NTA column. (B) Immune detection (WB) using a HRP-His probe: Control protein GlpQ-His10 (lane 1), non-induced cells (lane 2), and eluted fractions (3–6).
(TIFF)
Samples of the indicated proteins were obtained after inducing with IPTG and subjected to electrophoresis. Proteins were transfered to PVDF and developed using an anti-LexA antibody as described in the Methods section. Controls included the non-fused wild type LexA DNA binding domain encoded in pRS658 (LexADBDwt), LexADBDwt fused to repressor H-NS, and non-fused mutated LexA DNA binding domain encoded in pRS659 (LexADBDmut) [39]. *, indicates the expected LexADBDwt or LexADBDmut fusion product.
(TIFF)
(DOCX)
(ZIP)
(PDF)
Acknowledgments
JAI is in deep gratitude with Dr. Virginia L. Miller (UNC-Chapel Hill) for the gift of the plasmid encoding the MBP-InvF fusion and for her advice in the initial steps in this project. We would like to thank all the members in the research group (Ms. Michelle Vázquez-Pelaez is especially appreciated for her technical support). Moreover, we thank Cristina Lara-Ochoa (BUAP), Dr. Elba Reyes-Maldonado (ENCB-IPN), Dr. Isabel Salazar-Sánchez (ENCB-IPN) and Dr. Blanca E. García-Pérez (ENCB-IPN) for their helpful support in many aspects. Francisco J. Santana, Jorge A. Yañez, Paul Gaytán and Eugenio López-Bustos from IBT-UNAM are appreciated for their technical assistance. Special thanks to Dr. Rodolfo Garcia-Contreras (UNAM) for his tenacity on gathering us for a fight to obtain support from CONACyT. Lastly, JAI would like to thank J. Antonio Ibarra-Olivares for proofreading this manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by grants from Consejo Nacional de Ciencia y Tecnología (Proyecto Apoyado por el Fondo Sectorial de Investigación para la Educación CONACYT A1-S-25438), Secretaría de Investigación y Posgrado (SIP 2020-0728) and COFAA to JAI, from Consejo Nacional de Ciencia y Tecnología (CONACYT 254531) and Dirección General de Asuntos del Personal Académico de la UNAM (IN202418) to VHB. LER-G, DC-A and EV-G obtained a grant for graduate studies from CONACYT (619402, 935269 and 618908, respectively). LER-G, DC-A and DAP-H enjoyed a BEIFI-IPN fellowship.
References
- 1.WHO, 2016. Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. Available from: https://www.who.int/foodsafety/publications/foodborne_disease/fergreport/en/.
- 2.Haselbeck AH, Panzner U, Im J, Baker S, Meyer CG, Marks F. 2017. Current perspectives on invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis. 30(5): 498–503. 10.1097/QCO.0000000000000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ilyas B, Tsai CN and Coombes BK. 2017. Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. 7: 428 10.3389/fcimb.2017.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dos Santos AMP, Ferrari RG, & Conte-Junior CA. 2019. Virulence Factors in Salmonella Typhimurium: The sagacity of a bacterium. Curr Microbiol. 76(6): 762–773. 10.1007/s00284-018-1510-4 [DOI] [PubMed] [Google Scholar]
- 5.Knodler LA, Nair V, Steele-Mortimer O. 2014. Quantitative Assessment of Cytosolic Salmonella in Epithelial Cells. PLoS One. 9(1): e84681 10.1371/journal.pone.0084681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knodler LA. 2015. Salmonella enterica: living a double life in epithelial cells. Curr Opin Microbiol. 23: 23–31. 10.1016/j.mib.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 7.Ibarra JA, Steele-Mortimer O. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 11(11): 1579–1586. 10.1111/j.1462-5822.2009.01368.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubeva YA, Sadik AY, Ellermeier JR, and Slauch JM. 2012. Integrating Global Regulatory Input Into the Salmonella Pathogenicity Island 1 Type III Secretion System. Genetics. 190(1): 79–90. 10.1534/genetics.111.132779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fàbrega A, and Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev. 26(2): 308–341. 10.1128/CMR.00066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith C, Stringer AM, Mao C, Palumbo MJ, Wade JT. 2016. Mapping the regulatory network for Salmonella enterica serovar Typhimurium invasion. MBio. 7(5). pii: e01024–16. 10.1128/mBio.01024-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgan AM, Kröger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, et al. 2016. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12(8): e1006258 10.1371/journal.pgen.1006258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2(8): e81 10.1371/journal.ppat.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, et al. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 313(5784): 236–238. 10.1126/science.1128794 [DOI] [PubMed] [Google Scholar]
- 14.Baños RC, Vivero A, Aznar S, García J, Pons M, Madrid C, et al. 2009. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet. 5(6): e1000513 10.1371/journal.pgen.1000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorman CJ. 2013. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol. 11(5): 349–355. 10.1038/nrmicro3007 [DOI] [PubMed] [Google Scholar]
- 16.Martínez LC, Banda MM, Fernández-Mora M, Santana FJ, Bustamante VH. 2014. HilD induces expression of Salmonella pathogenicity island 2 genes by displacing the global negative regulator H-NS from ssrAB. J Bacteriol. 196(21): 3746–3755. 10.1128/JB.01799-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh K, Milstein JN, Navarre WW. 2016. Xenogeneic silencing and its impact on bacterial genomes. Annu Rev Microbiol. 70: 199–213. 10.1146/annurev-micro-102215-095301 [DOI] [PubMed] [Google Scholar]
- 18.Stoebel DM, Free A, Dorman CJ. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 154(Pt 9): 2533–2545. 10.1099/mic.0.2008/020693-0 [DOI] [PubMed] [Google Scholar]
- 19.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nature Microbiol. Revs. 8: 185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- 20.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 57(3): 691–705. 10.1111/j.1365-2958.2005.04737.x [DOI] [PubMed] [Google Scholar]
- 21.Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI-1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 10(1): 24–29. 10.1016/j.mib.2006.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Schechter LM, Jain S, Akbar S, Lee CA. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun. 71(9): 5432–5435. 10.1128/iai.71.9.5432-5435.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaniga K, Bossio JC, and Galán JE. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 13(4): 355–368. 10.1111/j.1365-2958.1994.tb00450.x [DOI] [PubMed] [Google Scholar]
- 24.Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181(16): 4949–4954. 10.1128/JB.181.16.4949-4954.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallegos M.-T, Schleif R, Bairoch A, Hofman K, Ramos JL.1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol Biol Rev. 61: 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan SM (2002) Growing repertoire of AraC/XylS activators. J Bacteriol 184: 5529–5532. 10.1128/jb.184.20.5529-5532.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tucker SC and Galán JE. 2000. Complex function for SicA, a Salmonella enterica serovar typhimurium type III secretion-associated chaperone. J Bacteriol. 182(8): 2262–2268. 10.1128/jb.182.8.2262-2268.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darwin KH, Miller VL. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35(4): 949–960. 10.1046/j.1365-2958.2000.01772.x [DOI] [PubMed] [Google Scholar]
- 29.Darwin KH, Miller VL. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella Typhimurium. EMBO J. 20(8): 1850–1862. 10.1093/emboj/20.8.1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahiri C, Pawar S, Sabarinathan R, Ashraf MI, Chand Y, Chakravortty D. 2014. Interactome analyses of Salmonella pathogenicity islands reveal SicA indispensable for virulence. J Theor Biol. 363: 188–197. 10.1016/j.jtbi.2014.08.013 [DOI] [PubMed] [Google Scholar]
- 31.Hoiseth SK and Stocker BA. 1981. Aromatic dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 291(5812): 238–239. 10.1038/291238a0 [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Morales D, Banda MM, Chau NYE, Salgado H, Martínez-Flores I, Ibarra JA, et al. 2017. The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLoS Pathog. 13(7): e1006497 10.1371/journal.ppat.1006497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De la Cruz MA, Pérez-Morelas D, Palacios IJ, Fernández- Mora M, Calva E, Bustamante VH. 2015. The two-component system CpxR/A represses the expression of Salmonella virulence genes by affecting the stability of the transcriptional regulator HilD. Front. Microbiol. 6: 807 10.3389/fmicb.2015.00807 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Casadaban M.J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104: 541–555. 10.1016/0022-2836(76)90119-4 [DOI] [PubMed] [Google Scholar]
- 35.Barba J, Bustamante VH, Flores-Valdez MA, Deng W, Finlay BB, Puente JL. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol. 187(23): 7918–7930. 10.1128/JB.187.23.7918-7930.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dmitrova M, Younès-Cauet G, Oertel-Buchheit P, Porte D, Schnarr M, Granger-Schnarr M. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol Gen Genet 257: 205–212. 10.1007/s004380050640 [DOI] [PubMed] [Google Scholar]
- 37.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, et al. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interaction in vitro. Microbiology. 156(pt4): 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer MP. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163(1): 41–46. 10.1016/0378-1119(95)00389-n [DOI] [PubMed] [Google Scholar]
- 39.Daines DA y RP Silver. 2000. Evidence for multimerization of neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J Bacteriol 182: 5267–5270. 10.1128/jb.182.18.5267-5270.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustamante VH, Martínez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci USA. 105(38): 14591–14596. 10.1073/pnas.0801205105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibarra JA, García-Zacarias CM, Lara-Ochoa C, Carabarin-Lima A, Tecpanecatl-Xihuitl JS, Pérez-Rueda E, et al. 2013. Further characterization of functional domains of PerA, role of amino and carboxy terminal domains in DNA binding. PLoS One. 8(2): e56977 10.1371/journal.pone.0056977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortez-Sánchez JL, Cortés-Acosta E, Cueto-Hernández VM, Reyes-Maldonado E, Hernández-Rodríguez C, Villa-Tanaca L, et al. 2018. Activity and expression of Candida glabrata vacuolar proteases in autophagy-like conditions. FEMS Yeast Res. 18(2): foy006. 10.1093/femsyr/foy006 [DOI] [PubMed] [Google Scholar]
- 43.Ibarra JA, Villalba MI, Puente JL. 2003. Identification of DNA Binding Sites of PerA, the Transcriptional Activator of the bfp and per operons in the enteropathogenic Escherichia coli. J Bacteriol. 185(9): 2835–2847. 10.1128/jb.185.9.2835-2847.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, Jarillo-Quijada MD, von Bargen K, Torres J, et al. 2016. H-NS Nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front Cell Infect Microbiol. 9(6): 13 10.3389/fcimb.2016.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vázquez-Guerrero E, Adan-Bante NP, Mercado-Uribe MC, Hernández-Rodríguez C, Villa-Tanaca L, Lopez JE, et al. 2019. Case report: A retrospective serological analysis indicating human exposure to tick-borne relapsing fever spirochetes in Sonora, Mexico. PLoS Negl Trop Dis. 13(4): e0007215 10.1371/journal.pntd.0007215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De la Cruz MA, Fernández-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vázquez A, et al. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol. 66(3): 727–743. 10.1111/j.1365-2958.2007.05958.x [DOI] [PubMed] [Google Scholar]
- 47.Knodler LA, Ibarra JA, Perez-Rueda E, Yip CK, and Steele-Mortimer O. 2011. Coiled-coil domains enhance the membrane association of Salmonella type III effectors. Cell. Microbiol. 13(10): 1497–1517. 10.1111/j.1462-5822.2011.01635.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JH. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press. Cold Spring Harbor; New York. [Google Scholar]
- 49.Paredes-Amaya CC, Valdés-García G, Juárez-González VR, Rudiño-Piñera y BH Bustamante E. 2018. The Hcp-like protein HilE inhibits homodimerization and DNA binding of the virulence-associated transcriptional regulator HilD in Salmonella. J Biol Chem 293: 6578–6592. 10.1074/jbc.RA117.001421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ali SS, Soo J, Rao C, Leung AS, Ngai DH-M, Ensminger AW, et al. 2014. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 10(11): e1004500 10.1371/journal.ppat.1004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banda MM, Manzo R, Bustamante VH. 2019. HilD induces expression of a novel Salmonella Typhimurium invasion factor, YobH, through a regulatory cascade involving SprB. Sci Rep 9: 12725 10.1038/s41598-019-49192-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olekhnovich IN, Kadner RJ. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J Mol Biol. 357(2): 373–386. 10.1016/j.jmb.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 53.Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 189: 6882–6890. 10.1128/JB.00905-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JS, Kim BH, Jang JI, Eom JS, Kim HG, et al. 2014. Functional insight from the tetratricopeptide repeat-like motifs of the type III secretion chaperone SicA in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 350(2): 146–153. 10.1111/1574-6968.12315 [DOI] [PubMed] [Google Scholar]
- 55.Lee DJ, Minchin SD, and Busby SJW. 2012. Activating Transcription in Bacteria. Annu Rev Microbiol. 66: 125–152. 10.1146/annurev-micro-092611-150012 [DOI] [PubMed] [Google Scholar]
- 56.Browning DFand Busby SJW. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol. 14(10): 638–650. 10.1038/nrmicro.2016.103 [DOI] [PubMed] [Google Scholar]
- 57.Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. 2009. IpaB–IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci USA 106 (24): 9661–9666. 10.1073/pnas.0812900106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basturea GN, Bodero MD, Moreno ME, Munson GP. 2008. Residues near the amino terminus of Rns are essential for positive autoregulation and DNA binding. J Bacteriol. 190: 2279–2285. 10.1128/JB.01705-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hovey AK and Frank DW. 1995. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177:4427–4436. 10.1128/jb.177.15.4427-4436.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munson GP, Holcomb LG, and Scott JR. 2001. Novel group of virulence activators within the AraC family that are not restricted to up- stream binding sites. Infect. Immun. 69:186–193. 10.1128/IAI.69.1.186-193.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of sopB was detected by qRT-PCR using the gene coding for rRNA 16S as a normalizer. Indicated strains were grown in SPI-1 inducing conditions and samples were taken for RNA extraction. Bars represent the average of at least three independent experiments and the error bars represent +/- SD. **, indicates statically significance difference (P < 0.05).
(TIFF)
MBP-InvF was purified as described in the Methods section and fractions of this process were taken and observed in a 12% SDS-PAGE (A). Lanes: 1, non-induced cells; 2, IPTG-induced cells; 3–8, eluted fractions obtained from the amylose column. (B) Immune detection (WB) of MBP (lane 1) and MBP-InvF (lanes 2–4) with an anti-MBP antibody. Lines in both panels indicate variants of MBP-InvF.
(TIFF)
SicA-His6 was purified and samples of this process were taken and observed in a 12% SDS-PAGE (A). Lanes: 1, non-induced cells; 2, IPTG-induced cells; 3–8, eluted fractions obtained from the Ni-NTA column. (B) Immune detection (WB) using a HRP-His probe: Control protein GlpQ-His10 (lane 1), non-induced cells (lane 2), and eluted fractions (3–6).
(TIFF)
Samples of the indicated proteins were obtained after inducing with IPTG and subjected to electrophoresis. Proteins were transfered to PVDF and developed using an anti-LexA antibody as described in the Methods section. Controls included the non-fused wild type LexA DNA binding domain encoded in pRS658 (LexADBDwt), LexADBDwt fused to repressor H-NS, and non-fused mutated LexA DNA binding domain encoded in pRS659 (LexADBDmut) [39]. *, indicates the expected LexADBDwt or LexADBDmut fusion product.
(TIFF)
(DOCX)
(ZIP)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.