ABSTRACT
Group I catalytic introns are widespread in bacterial, archaeal, viral, organellar, and some eukaryotic genomes, where they are reported to provide regulatory functions. The group I introns are currently divided into five types (A-E), which are themselves distributed into several subtypes, with the exception of group I type D intron (GI-D). GI-D introns belong to the rarest group with only 17 described to date, including only one with a putative role reported in fungi, where it would interfere with an adaptive response in the cytochrome b (COB) gene to quinone outside inhibitor (QoI) fungicide resistance. Using homology search methods taking into account both conserved sequences and RNA secondary structures, we analysed the mitochondrial genomes or COB genes of 169 fungal species, including some frequently under QoI selection pressure. These analyses have led to the identification of 216 novel GI-D introns, and the definition of three distinct subtypes, one of which being linked with a functional activity. We have further uncovered a homing site for this GI-D intron type, which helps refine the accepted model of quinone outside inhibitor resistance, whereby mobility of the intron across fungal mitochondrial genomes, would influence a fungus ability to develop resistance to QoIs.
KEYWORDS: QoI resistance, genome evolution, fungi, group I intron
Introduction
Non-coding RNA molecules are well known to have functions of genome regulation [1,2]. Among them, Group I introns are self-splicing catalytic RNA elements [3,4] that are abundantly reported in organellar genomes from a wide variety of organisms [5,6]. Mitochondrial group I introns are often inserted within conserved encoding genes [7,8]. Except for a few group I introns, their functional impacts on the host genes are still enigmatic and they are often thought to be benign selfish elements [9,10].
Scalley-Kim et al. [11] suggested that group I introns were mainly homed into nucleotide domains that encode functionally critical amino-acid or RNA sequences of their host gene, by securing it from deleterious mutation. This idea parallels the widely accepted hypothesis that, in eukaryotes, the splicing of mitochondrial group I introns may affect their host gene evolution by blocking mutation in their homing sites [12,13]. This DNA homing site (HS) has usually a length size ≥14 base pairs and is often found in functionally important codons in up- and down-stream exons [11,14,15].
The potential implication of group I intron in gene evolution was initially reported in fungi in 2006 [16]. In their hypothesis, Grasso et al. [16] suggested that the presence of one group I mitochondrial intron blocked the main mutation involved in the resistance against the Quinone outside Inhibitors (QoIs) fungicides. This mutation (G143A), caused by a single amino-acid change of Glycine into Alanine at the position 143 of the cytochrome b protein [17], is mainly responsible for QoI resistance. In some fungal species, one intron interrupts the cytochrome b gene (COB) directly after its 143th codon (i.e. the 429th base pair into the nucleic coding sequence, CDS) in a highly important domain involved in respiratory metabolism [18]. The main hypothesis explaining why the cob_429 intron would block resistance is that the G143A mutation may prevent proper splicing of the intron and therefore lead to a non-functional protein [19,20]. Under the current model ‘presence/absence of the intron’, all species having this intron are unable to acquire the mutation and, therefore, to develop resistance against QoIs.
In the context of this hypothesis, many studies have prospected for the presence of this intron across a large number of pathogenic fungal species in the predictive context of QoI resistance. However, some fungi present unexplained exceptions to the Grasso’s model [21–25]. For instance, Botrytis cinerea [21,22], some Moniliniaspp. [26,27] were found to have two different COB genotypes, including one with, and one without the group I intron after the 429th COB CDS position. Conversely, Phyllosticta capitalensis, Phyllosticta citriasiana [23] and Podosphaera fusca [25] are species for which no phenotypes of QoI resistance have been observed, in spite of lacking the cob_429 intron.
Many group I introns have the ability to move within and between genomes from a donor gene into an intronless acceptor gene (see reviews [5] and [28], for example). The mobility of group I introns has been speculated to be a compensation mechanism to restore the potential for mutation, a phenomenon which would explain why B. cinerea and, as shown experimentally, Saccharomyces cerevisiae can display resistance against QoIs [20,21]. Under this ‘intron mobility model’, species with intron can develop resistance if the HS in the intron acceptor genes exists elsewhere in the mitochondrial genome. By contrast, if COB is a potentially important acceptor gene for the intron of other donor genes, the cob_429 HS will be conserved and resistance to QoIs will not appear.
Because group I introns have little primary sequence conservation and are interrupted by long homing endonuclease genes (HEG), making them difficult to detect with standard sequence similarity searches [29], one method to identify them is to search for their conserved RNA structural features [30,31]. Computational approaches using probabilistic covariance models (CMs) have recently allowed to efficiently detect intron homologs by combining multiple sequence alignments and consensus secondary structures [29,32].
Using the well-studied QoI resistance model in fungi, this study aimed to address the hypothesis that group I introns can impact the adaptive process against a selective pressure. If intron mobility plays a role in QoI resistance, the HS associated with this COB gene intron should exist elsewhere in mitochondrial genomes. As a result, we expect to find these conserved HS in other mitochondrial genes associated with secondary structures of cob_429. The main objectives to validate the hypothesis were: (i) to characterize the structural type of cob_429 within Group I introns using a CM approach; (ii) to identify the HS associated with cob_429, (iii) to perform a whole mitochondrial genome search for cob_429 homologs that possess cob_429-associated HS, and (iv) to compare and associate the structure of cob_429 inherent to QoI resistance with fungi known to develop or not resistance to QoIs.
Materials and methods
QoI resistant species and sequence dataset composition
Among fungal pathogens, those belonging to the Pezizomycotina taxa within the Ascomycota phylum are the most numerous and economically important, and the most frequently targeted by QoIs for the control of plant diseases [33]. Several cases of resistance in field populations of many important Pezizomycotina plant pathogens have been observed as early as within the first year of application [24,34]. In 2018, the Fungicide Resistance Action Committee (FRAC) reported more than 50 fungal plant pathogen species resistant to QoIs. Among them, 44 belonged to the Pezizomycotina. The list of fungal species with established status for their sensitivity to strobilurin fungicides was obtained from a compilation of the FRAC 2018 report on resistant plant pathogens [35] combined with a search of scientific studies on QoI resistant risks. The bibliographical review was conducted from 1996 to 2018 with a focus on Pezizomycotina species.
The full mitochondrial genomes of Pezizomycotina fungi entered in the NCBI GenBank were searched in December 2018 with the ‘Entrez’ query command ‘txid147538’[Organism] AND (biomol_genomic[PROP] AND mitochondrion[filter]) AND (‘complete’[Title] NOT ‘Plasmid’[Title]) AND (‘20000’[SLEN]: ‘999999’[SLEN]))” (Supp. Dataset SD 1).
The list of Pezizomycotina species known to have the cob_429 intron was retrieved via the FRAC report [35] and the bibliographical review. This information was used for extracting COB genes from mitochondrial genomes of Pezizomycotina dataset (Supp. Dataset SD 1) or for downloading the COB gene sequence records from the National Center for Biotechnology Information (NCBI) GenBank when the corresponding species had not a full mitochondrial genome available. In addition, mRNA sequences from NCBI GenBank for 11 COB genes (Table S1) were used to perform a manual alignment of all the sequences previously identified. The sequence files were downloaded in GenBank format (Table S2) and aligned using CLC Genomic Workbench v.20.0.2 (Qiagen, Aarthus A/S). Subsequently, the cob_429 introns and their flanking 50 nt exon flanking sequences were extracted following the common splicing mechanism for group I introns: after a U at the 5ʹ splicing site, and after a G at the 3ʹ splicing site [4]. This rule was employed for all intron extraction in this study.
Characterization and whole mitochondrial genome detection of group I introns
Group I introns are classified into five types (GI-A, GI-B, GI-C, GI-D and GI-E) and subdivided into 14 subtypes (from GI-A1 to GI-E3) based on variations in conserved consensus secondary structure [36,37]. Following the approach of Lang et al. [30], the subtype-specific models were used to properly detect and characterize the diversity of introns across COB genes and mitochondrial genomes. The 14 curated type-specific group I intron alignments from the Group I Intron Sequence and Structure Database (GISSD; 39) and the family-specific group I intron CM (RF00028) from Rfam database [38] were obtained in December 2018 from supplementary data of Nawrocki et al. [29] (Supp. Dataset SD 6).
Infernal v.1.1.2 [39] was used to characterize the type and subtypes of cob_429 introns, as well as to perform whole-genome detection of group I introns. The programs cmbuilt and cmcalibrate with default parameters were used to build and calibrate CMs from the 14 group I alignments. The cmpress program was used to compress and index a concatenation of individual CM files into a CM database flatfile (GroupIfam.cm.il{m,i,f,p}). The cmscan and cmsearch programs were used to scan each cob_429 sequences with GroupIfam.cm database and to search the Group I intron CMs against the Pezizomycotina mitochondrial genomes dataset, respectively. The two programs were used with the – anytrunc option, which improves performance on interrupted group I introns by HEGs [29]. Hits with an E-value ≤0.01 were kept, and overlapping hits were removed, keeping the hit with the lowest E-value.
Cluster analysis of Pezizomycotina mitochondrial group I introns
Intron catalytic core (ICC) and homing site (HS). The common secondary structure for group I introns includes nine base-paired regions designated by P1, P3–P10 [36,40]. P1 and P10 regions contain the 5ʹ and 3ʹ splice sites in up- and downstream exons, while the other conserved regions constitute the ICC composed by the two typical helical domains comprised between base-pairing P4–P6 and P3–P9 [36,41]. By following the method proposed by Zhou et al. [32], the ICC was extracted for each intron detected by Infernal and a secondary structure-favoured alignment of the ICC sequences were then manually performed [32,37].
The HS is an intron-specific target site usually centred on the intron-insertion site [5,42]. To detect putative HS associated with the cob_429, both up- and downstream exonic sequences were used to build the 5ʹ and 3ʹ parts, respectively, of the potential HS motif for each intron detected by Infernal search.
Two k-mers-based clustering reconstructions with an alignment-free approach [43] were used to construct cladograms among ICC and HS (Supplementary Information Text STx 1). Distance between pairwise sequences was estimated using the fractional common k-mer count [44] with k = 7 and a distance-based tree was built with the UPGMA clustering method [45]. Distance calculation and tree construction were performed with R v. 3.6.1 using packages kmer [46] and dendextend [47], respectively.
Intron homing endonuclease gene characterization
Homing endonuclease gene (HEG) homologs were detected by searching all translated open reading frames (ORFs) present in detected introns, against Pfam v.32.0 [48] using the hmmscan program (HMMER v.3.1b2 [49]) with an E-value threshold of ≤10−3. The translation of ORFs ≥100 amino-acids was obtained using the genetic code translation table 4 (Mould, Protozoan, Coelenterate Mitochondrial code).
Results
Constitution of the datasets
Following NCBI queries, a total of 229 non-redundant mitochondrial genome sequences from 157 fungal species within the Pezizomycotina were obtained (Supp. Data SD1), and the COB genes were extracted. From the FRAC report and the literature review, the sequences only for the COB gene were retrieved in 12 additional fungal species. Among these 169 COB genes, the cob _429 intron was found in 39 sequences of 19 species (Table S2, and Supp. Dataset SD2).
Structure identification of cob_429 intron
The Infernal scan on the 39 cob_429 sequences detected significant hits at a threshold of E-value <0.01 for three GISSD CMs (GI-A3, GI-C1, and GI-D subgroups) and the complete G-I intron family Rfam-CM (RF00028). After suppressing overlapping hits and keeping the ones with the lowest E-value, all 39 cob_429 had the best alignment with the GISSD-GI-D CM with a strongly significant E-value <10−10 (Table S2). As a result, only the group I-D intron GISSD-CM was considered in subsequent searches.
To control the good correspondence to GI-D introns, the 39 cob_429 intron sequences were compared with the well-known intron 1 of the COB gene found in Saccharomyces douglasii (SdCob.1, NCBI-accession X59280) used as a structure reference [50]. Although the consensus secondary structure of GI-D intron was found in each intron, many distinctions from the structure of SdCob.1 were observed, mainly in the sequence delimited by the P9 base-pairing.
Pezizomycotina group I D introns
Whole-genome detection and host genes
The whole mitochondrial genome search in 157 species for GI-D CM detected initially a total of 648 significant hits with an E-value ≤0.01, after removal of overlapping, lower Infernal scoring hits. The examination of remaining hits resulted in the suppression of additional 296 that corresponded to redundant identical sequences in the mitochondrial genome dataset (Supp. Dataset SD1), 34 sequence records with incomplete intron and 17 sequences where we were not confident that the sequence folded into a complete group I consensus intron structure. Because Infernal may identify a single intron with large non-conserved insertions (e.g. HEGs) in more than one local alignment piece, the remaining hits were assigned to 216 single non-redundant full length putative GI-D introns. These ones were kept for further analyses (Supp. Dataset SD3).
In addition, the GI-D introns were found in three positions in the COB gene; three, one and two positions in genes for cytochrome c oxydase subunits 1, 2 and 3 (COX1, COX2 and COX3), respectively; four and one positions in genes for NADH deshydrogenase subunits 5 and 6 (ND5 and ND6), respectively; and three positions in the ribosomal RNA small subunit gene (RNS) (Table 1). The number of introns at insertion positions demonstrated a strong insertion bias to positions 393 of COB, 715 of COX1, and 414 of ND5. Of the 157 fungal species analysed, 82 possessed GI-D introns. Interestingly, many economically important plant pathogenic taxa were found not to carry GI-D introns (e.g. Aspergillus spp. and Penicilium spp.). Of the 82 species with GI-D, 65 did not possess the typical cob_429 intron involved in QoI resistance.
Table 1.
Host genes and positions for the GI-D introns detected in fungal mitochondrial genomes
| Gene namesa |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G-ID subtypes | COB | COX1 | COX2 | COX3 | ND5 | ND6 | RNS | ||||||||
| GI-D1 | [65] | 393 | [60] | 414 | [5] | ||||||||||
| GI-D2 | [67] | 429 | [1]b | 284 | [4] | 372 | [11] | 428 | [5] | 248 | [10] | 239 | [7] | 1a | [2] |
| 824 | [3] | 414 | [9] | 1b | [3] | ||||||||||
| 698 | [12] | ||||||||||||||
| GI-D3 | [84] | 374 | [5] | 715 | [38] | 447 | [2] | 260 | [1] | 2 | [2] | ||||
| 393 | [13] | ||||||||||||||
| 429 | [23] | ||||||||||||||
aCOB: cytochrome b gene; COX1, COX2 and COX3: genes for cytochrome c oxydase subunits 1, 2 and 3, respectively; ND5 and ND6: genes for NADH dehydrogenase subunits 5 and 6, respectively; RNS: Ribosomal RNA small subunit gene.
bAfter structure verification, cob_429 for KC788404 may have been displaced in subtype GI-D3.
This table gives intron GI-D subtypes defined in this study (see results, Fig. 2), the intron insertion position in the corresponding gene, and the number of occurrences for each position into square brackets. Position corresponds to the nucleotide position in coding sequence (CDS) directly before the intron insertion, excepted for RNS gene where position is according with the order of intron apparition.
GI-D subtypes and subtype distribution in Pezizomycotina species
When the nucleotide sequences in the ICC (Supp. Dataset SD4) were used to compute the clustering of the GI-D introns, it was found that almost all introns fell into one of the three well-separated clusters (Fig. 1A), representing three major subtypes, named here GI-D1, GI-D2 and GI-D3. The three main different secondary structures identified in GI-D subtypes are illustrated as an example by folding of cob_393 intron for Usnea mutabilis (NCBI-accession NC_039633), cob_429 for Botrytis cinerea (NCBI-accession KC832409) and nd5_698 for Hirsutella minnesotensis (NCBI-accession NC_027660) corresponding to GI-D1, GI-D2 and GI-D3, respectively (Fig. 2).
Figure 1.
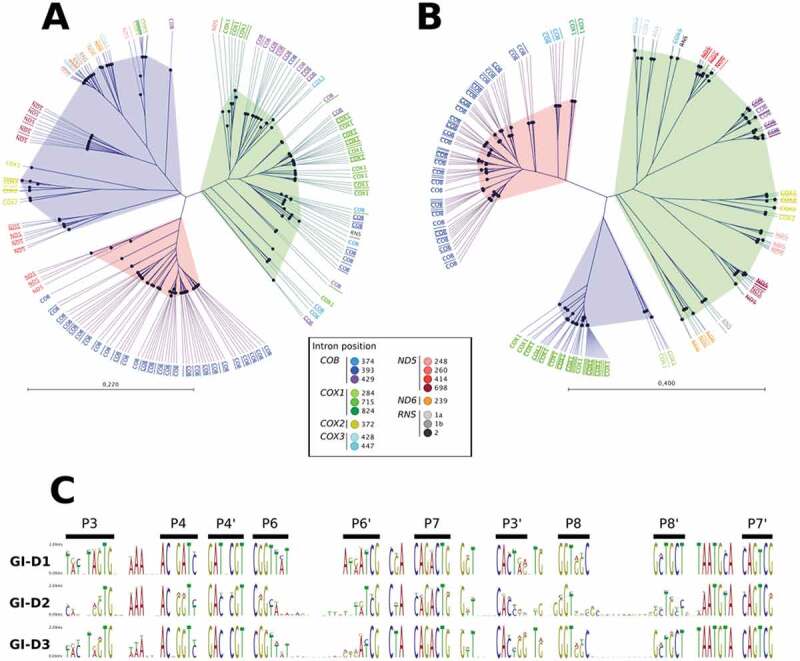
The intron catalytic core (ICC) and the homing site (HS) variations result in the Pezizomycotina GI-D intron classification into three subtypes. Clustering of ICC (A) and HS (B) sequences was estimated using k-mers (k = 7) cluster reconstructions for the 216 introns detected by Infernal search, including the 39 cob_429 identified. The clusters proposed in this study are represented by different colours: GI-D1 and HS1 in red; GI-D2 and HS2 in blue, and GI-D3 and HS3 in green. (C) Sequence logo representations for the ICC regions of all GI-D introns and of each subtype
Figure 2.
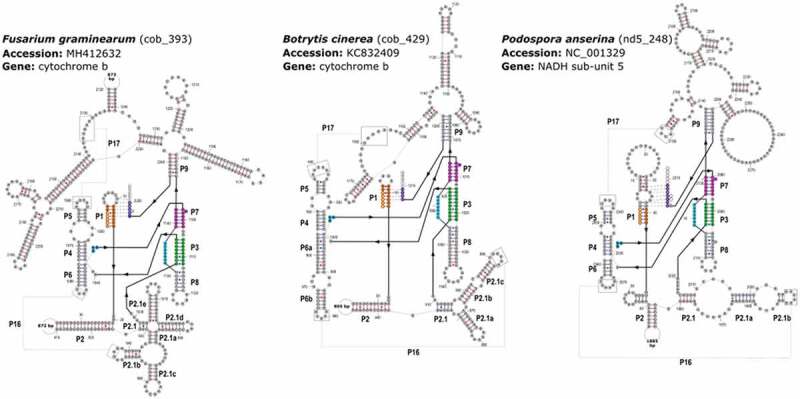
Examples of predicted secondary structures for each the GI-D intron subtypes proposed in this study. Base pairs shown in blue lines (or dots, for G-U pairs) were predicted by Infernal, those shown by red lines (or dots) were manually predicted. Exonic residues are shown in lower case. Colours correspond to the main base-pairings found in GI-D introns according to Zhou et al. [31] and Lehnert et al. [32]. Intron positions are numbered starting at 2; by convention of Cech et al. (S1). The secondary structures were folded by the software RNAstructure v.6.1 (S2, see Supp. References SR 1)
Multiple sequence patterns in the conserved core of GI-D introns were observed and they were associated with the three subtypes (Fig. 1C). Each subtype presents a homogeneous composition of intron gene positions (Table 1). Note that the introns found at the 429 position of the COB gene represent the GI-D3 subtype with the introns mainly present at the 715 position of the COX1 gene and a part of the introns found at position 393 of the COB gene. Interestingly, however, for all the eight species possessing both the cob_429 and the cob_393 introns, the cob_393 introns are systematically in the GI-D1 subtypes (Fig. 1A and Table 1).
Putative homing sites of GI-D
The alignment of the 39 sequences of cob_429 introns (Supp. Dataset SD2) showed 24 and eight common nucleotides among the 19 species in 5ʹ and 3ʹ exonic flanking regions, respectively. These two regions were considered as the 5ʹ and 3ʹ parts of the putative HS of the cob_429 introns and compared with length equivalent HS for each group I-D introns found in the 229 Pezizomycotina mitochondrial genome sequences (Supp. Dataset SD4). Among the 216 GI-D introns, the cluster construction showed a repartition of the HS in three distinct clusters (Fig. 1B).
Homing endonuclease genes in GI-D intron
A total of 460 ORFs (≥100 aa) were detected in the 216 GI-D intron sequences. Homology search for translated ORFs found against Pfam protein domains resulted in significant similarities for LAGLI-DADG homing endonucleases (Pfam LAGLIDADG_1 and LAGLIDADG_2; PF00961 and PF03161, respectively), GIY-YIG endonuclease (Pfam GIY-YIG; PF01541) and acetyltransferase (Pfam Acetyltransf_3; PF13302) in 176 of the 216 introns (Table S3).
Among the 39 cob_429 sequences analysed, two types of HEG allowed to differentiate these introns into two groups. One group had the LAGLI-DADG endonuclease gene and the other possessed the acetyltransferase gene. Both genes were found in the intron sequence delimited by the P2-P2ʹ (Fig. 2). Interestingly, the cob_429 intron of B. cinerea expected to be mobile [20] housed the type 2 of the LAGLIDADG-like gene.
QoI resistant species and GI-D intron distribution
In 2018, from FRAC report and literature review, a total of 70 species were found with a reported status on QoI resistance (Table S3), 17 of which having a full mitochondrial genome or the full COB genes available in NCBI database. Of these 17 species, 11 were found to contain GI-D introns, and only two were found with the cob_429 intron. Interestingly, the two species with the cob_429 intron had a reported resistance for QoIs. On the other hand, two species lacked the cob_429 intron but were not reported as having developed resistance to QoIs (Tables 2 and S3).
Table 2.
Pezizomycotina species with a QoI-resistant status reported in 2018 and with an available mitochondrial genome in NCBI database
| Species | NCBI Accession | Referencea | QoIResistance | GI-D Detection | cob_429 Presence |
|---|---|---|---|---|---|
| Alternaria alternata | MF669499 | (S1) | Yes | No | No |
| Beauveria bassiana | NC_010652 | (S2) | No | Yes | No |
| Bipolaris maydis | AC277286 | (S3) | Yes | Yes | Yes |
| Botrytis cinerea | KC832409 | (S4) | Yes | Yes | Yes |
| Colletotrichum acutatum | NC_027280 | (S5) | Yes | No | No |
| Colletotrichum gloeosporioides | KX885104 | (S6) | Yes | Yes | No |
| Colletotrichum graminicola | NW_007361658 | (S6) | Yes | No | No |
| Colletotrichum siamense | KX885098 | (S7) | Yes | Yes | No |
| Fusarium culmorum | NC_026993 | (S8) | Yes | Yes | No |
| Fusarium graminearum | NC_009493 | (S9) | Yes | Yes | No |
| Fusarium temperatum | KP742837 | (S10) | Yes | Yes | No |
| Parastagonospora nodorum | NC_009746 | (S6) | Yes | No | No |
| Penicillium digitatum | NC_015080 | (S11) | Yes | No | No |
| Rhynchosporium commune | NC_023126 | (S12) | Yes | Yes | No |
| Rhynchosporium secalis | NC_023128 | (S6) | Yes | Yes | No |
| Sclerotinia sclerotiorum | NC_035155 | (S13) | No | Yes | No |
| Zymoseptoria tritici | NC_010222 | (S14) | Yes | No | No |
aSee Supp. References SR 2
For species with more than one mitochondrial genome recorded in NCBI, only the accession for the reference sequence is reported (See Supp. References SR 2). For each species, the table gives if GI-D introns and the cob_429 were detected in this study. For more information see Table S2.
Discussion
Group I intron cob_429 is reported to have an important impact on QoI resistance of plant pathogenic fungi, namely among species belonging to the Pezizomycotina [18]. This phenomenon was hypothesized to be based on a dysfunction of the splicing process caused by the mutation responsible for the resistance [16]. In simpler terms, presence of the intron cob_429 in the COB gene of fungi was believed to prevent mutation by interfering with the splicing process. However, as more fungi were reported to develop resistance to QoIs, some species were found to develop resistance against QoIs in spite of harbouring the cob_429 intron [21,51]. Because the cob_429 intron was reported as GI-D in several Pezizomycotina species, this study highlights a unique diversity among GI-D introns that explains the intricate adaptive process of fungi in response to selection pressure.
At the time of its discovery in Pezizomycotina species, the cob_429 intron was reported to belong to the D type of group I introns [9,10]. This intron type was previously described in bacteria, mitochondria of plants and fungi, as well as nuclear genomes of a few green algae [5]. It is nevertheless considered the rarest type of group I introns with no subtypes reported to date. Only 17 sequences for GI-D introns are recorded in GISSD [32], comparative RNA web (CRW) site [52] and RNArchitecture database [53], including seven found in Pezizomycotina species. Surprisingly, by performing whole mitochondrial genome searches on 157 species, our study uncovered 216 additional GI-D introns. This result suggests that introns of type D may not be as rare as currently assumed in spite of the scarcity of reference sequences available. One possible explanation is the difficulty to model the secondary and tertiary structures of GI-D introns, due to long sequence insertions, usually used to perform detection of non-coding RNA molecules [15]. As such, the analytical tools described here can certainly be exploited to expand our understanding of the presence and role of GI-D introns in other organisms. Alternatively, our study may reveal that fungi are particularly prone to harbour GI-D introns for evolutive purposes yet to be defined.
Based on the recent literature, group I introns are mostly homed in structural RNA genes (rRNA or tRNA), and much less common into protein-coding genes [5]. However, in fungi, the D type was mainly reported in mitochondrial coding genes [7,8]. In Pezizomycotina species, of the 216 introns reported here, more than 90% were found in nucleic sequences coding for important catalytic domains of the mitochondrial constitutive proteins. For instance, cob_429 will splice the COB gene in the codon sequence translated in the outside binding site that catalyses the quinone oxidative reaction [18]. These observations may alter the current concepts defining the role of group I introns in general and group I-D introns in particular. Indeed, the propensity of the latter to be localized in functional protein domains suggests they may play a role in mechanisms altering the activity of proteins. Previous studies suggested similar regulatory functions for intron in fungi [15], plants [54], animals [55], and humans [56]. In this context, one would expect that the first target of natural selection for introns would be regulatory system involved in the respiratory metabolism rather than the acquisition of resistance. This suggests that the role of cob_429 in QoI resistance is fortuitous.
In fungi, group I introns vary greatly in their primary sequences [7,8], while the nucleotide sequences in the core region (ICC) are relatively conserved [36,37]. On the other hand, the ICC sequences vary among the subgroups of group I introns, which is the main criterion used to categorize group I introns into different types [5,36]. For instance, ICC sequence variation has been used previously as a criterion to distinguish the GI-E intron subtypes [37]. Basically, the ICC is composed of small base-paired regions relatively conserved (≈10 bp) constituting a part of the intron secondary structure. As reported here, when the ICCs were used to discriminate GI-D subtypes, all the 216 GI-D introns were clustered into three different subtypes. This clustering grouped the cob_429 with seven other introns, localized in five genes (Table 1), suggesting different origins for GI-D introns present in COB gene. Under this expectation, the GI-D2 subtype (including cob_429) may be viewed as an intron family directly involved in QoI resistance. Although few species have cob_429 intron, more species have GI-D2 subtype introns. Under the selective pressure exercised by QoIs on fungal species, this result suggests progressive loss of cob_429, allowing the development of resistance.
Commonly, the HEG has coevolved with its group I intron [5,11]. Each intron-encoded nuclease recognizes and cleaves one specific target site (homing site) in intronless acceptor genomic sequences [5,57]. The HEG found in the GI-D introns reported in this study were as GIY-YIG, LAGLI-DADG and NAT endonucleases. Some of them have already been reported to have the capacity to displace genomic elements [20,21], although never previously in Pezizomycotina species [21,22]. The cob_429 introns found in this study have mainly the LAGLI-DADG and NAT endonuclease genes, which further supports the implication of the mobility of this intron in the acquisition of resistance.
The homing site (HS) is an intron-specific target site and is centred on the intron-insertion positions [5,42]. In this study, we have found different Pezizomycotina species, reported to be resistant against QoIs, with the cob_429 intron, an observation that goes against the concept that it prevents resistance. The best example for this observation is B. cinerea, an important plant pathogen able to develop resistance quickly despite being observed with the presence of the cob_429 intron [21]. This means that when compatible homing sites exist in other parts of the mitochondrial genome, a transient displacement of the cob_429 intron may result in resistance. This would explain why some isolates of B. cinerea were also found lacking the intron at the COB_429 position. On the other hand, some fungal species have no reported occurrence of resistance to QoIs and, yet, do not appear to possess a cob_429 intron. Interestingly, these species do have an HS corresponding to cob_429 which is conserved, and GI-D introns in other parts of their genome, which suggests that mobility of the intron at the target site for resistance, may hamper the development of the phenotype (Fig. 3).
Figure 3.
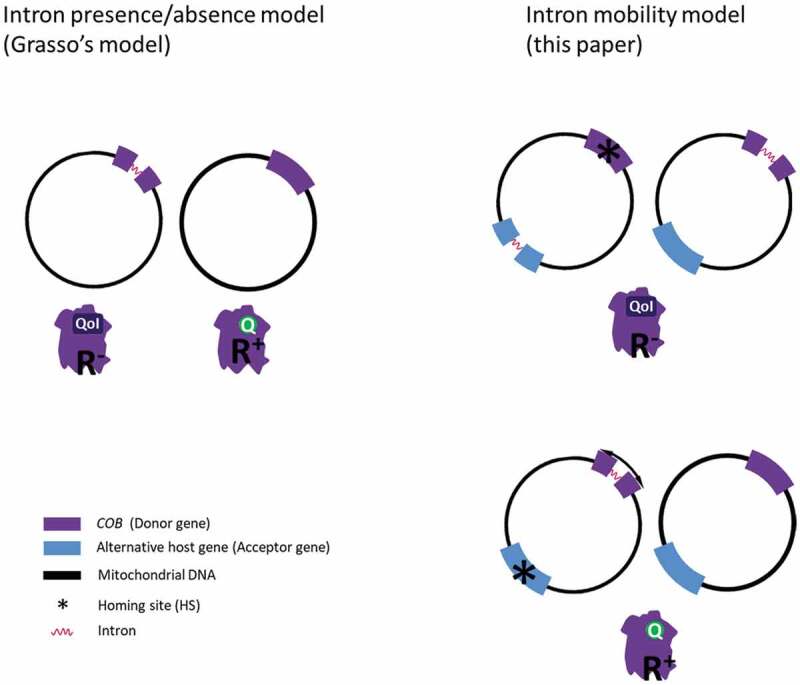
Conceptual representation of the models of the expected intron role in mitochondrial genome regulation involved in fungicide resistance. Left panels depict the intron presence/absence-based model [18] in which the presence of the intron only blocks the mutation capacity of its host gene. Right panels portray the intron mobility-based model where the mutation capacity is thought to be related to intron mobility
Grasso et al. [16] inferred that the group I intron (cob_429), situated directly after codon 143 in the mitochondrial COB gene, prevented QoI resistance by blocking the mutation involved in the resistance in many plant pathogenic fungi. While this hypothesis remains essentially valid, the involvement of the GI-D intron in the mechanism of QoI resistance seems more complex than a simple case of absence/presence. As shown here, despite the absence of the cob_429 intron, some species do not seem to be able to develop resistance, while others do in spite of the presence of the intron. Consequently, it appears that the mobility of the Group I cob_429 intron provides a mechanism enabling the mutation capacity in species where a corresponding HS is present. This phenomenon was reported in B. cinerea and Saccharomyces cerevisiae [20,21]. By contrast, the species with GI-D subtype 2 introns (homologous to cob_429) presented a conserved HS in the COB gene just after the CDS 429th base pair. Therefore, for these species, which have not been reported as resistant yet, the GI-D subtype 2 introns may play one role more evolutionally important, like genome regulation. As cases of resistance to QoIs expand in more fungal species, this will provide the opportunity to validate the extent of the role of GI-D introns and in particular cob_429 in fungi.
Supplementary Material
Acknowledgments
We thank Jérôme Laroche (IBIS, Université Laval) and Caroline Labbé (Université Laval) for their technical assistance.
Funding Statement
This work was supported by the Natural Sciences and Engineering Research Council of Canada [CRDPJ 508525-17] to Richard R. Bélanger.
Author contributions
B.C. and R.R.B designed research and wrote the paper; B.C. performed research and analysed data.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Rinn JL, Chang HY.. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Atkins JF, Gesteland RF, Cech TR. The RNA world. 3rd ed Atkins JF, Gesteland RF, Cech TR, editors. Long Island (NY): Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- [3].Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31(1):147–157. . [DOI] [PubMed] [Google Scholar]
- [4].Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. [DOI] [PubMed] [Google Scholar]
- [5].Hausner G, Hafez M, Edgell DR. Bacterial group I introns: mobile RNA catalysts. Mob DNA. 2014;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nielsen H, Johansen SD. Group I introns: moving in new directions. RNA Biol. 2009;6(4):375–383. [DOI] [PubMed] [Google Scholar]
- [7].Guha TK, Wai A, Mullineux S-T, et al. The intron landscape of the mtDNA cytb gene among the Ascomycota: introns and intron-encoded open reading frames. Mitochondrial DNA Part A DNA Mapping Seq Anal. 2018;29(7):1015–1024. [DOI] [PubMed] [Google Scholar]
- [8].Yin L-F, Hu M-J, Wang F, et al. Frequent gain and loss of introns in fungal cytochrome b genes. PLoS One. 2012;7(11):1–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koonin EV, Senkevich TG, Dolja VV. The ancient virus world and evolution of cells. Biol Direct. 2006;1:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gogarten JP, Hilario E. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol Biol. 2006;6:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J Mol Biol. 2007;372:1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rogers JH. The role of introns in evolution. FEBS Lett. 1990;268:339–343. [DOI] [PubMed] [Google Scholar]
- [13].Edgell DR, Chalamcharla VR, Belfort M. Learning to live together: mutualism between self-splicing introns and their hosts. BMC Biol. 2011;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Edgell DR, Stanger MJ, Belfort M. Importance of a single base pair for discrimination between intron-containing and intronless alleles by endonuclease I-BmoI. Curr Biol. 2003;13:973–978. [DOI] [PubMed] [Google Scholar]
- [15].Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J Mol Biol. 2004;343:1231–1241. [DOI] [PubMed] [Google Scholar]
- [16].Grasso V, Sierotzki H, Garibaldi A, et al. Characterization of the cytochrome b gene fragment of Puccinia species responsible for the binding site of QoI fungicides. Pestic Biochem Physiol. 2006;84: 72–82. [Google Scholar]
- [17].Birla K, Rivera-Varas V, Secor GA, et al. Characterization of cytochrome b from European field isolates of Cercospora beticola with quinone outside inhibitor resistance. Eur J Plant Pathol. 2012;134: 475–488. [Google Scholar]
- [18].Bartlett DW, Clough JM, Godwin JR, et al. The strobilurin fungicides. Pest Manag. Sci. 2002;58(7):649–662. . [DOI] [PubMed] [Google Scholar]
- [19].Grasso V, Palermo S, Sierotzki H, et al. Allelopathy – a natural alternative for weed control. Pest Manag Sci. 2006;62:465–472.16688790 [Google Scholar]
- [20].Vallières C, Trouillard M, Dujardin G, et al. Deleterious effect of the Qo inhibitor compound resistance-conferring mutation G143A in the intron-containing cytochrome b gene and mechanisms for bypassing it. Appl Environ Microbiol. 2011;77:2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Banno S, Yamashita K, Fukumori F, et al. Characterization of QoI resistance in Botrytis cinerea and identification of two types of mitochondrial cytochrome b gene. Plant Pathol. 2009;58:120–129. [Google Scholar]
- [22].Leroux P, Gredt M, Leroch M, et al. Exploring mechanisms of resistance to respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of gray mold. Appl Environ Microbiol. 2010;76:6615–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stammler G, Schutte GC, Speakman J, et al. Phyllosticta species on citrus: risk estimation of resistance to QoI fungicides and identification of species with cytochrome b gene sequences. Crop Prot. 2013;48:6–12. [Google Scholar]
- [24].Hily JM, Singer SD, Villani SM, et al. Characterization of the cytochrome b (cyt b) gene from Monilinia species causing brown rot of stone and pome fruit and its significance in the development of QoI resistance. Pest Manag Sci. 2011;67:385–396. [DOI] [PubMed] [Google Scholar]
- [25].Fernández-Ortuño D, Torés JA, De Vicente A, et al. Field resistance to QoI fungicides in Podosphaera fusca is not supported by typical mutations in the mitochondrial cytochrome b gene. Pest Manag Sci. 2008;64:694–702. [DOI] [PubMed] [Google Scholar]
- [26].Luo C-X, Hu M-J, Jin X, et al. An intron in the cytochrome b gene of Monilinia fructicola mitigates the risk of resistance development to QoI fungicides. Pest Manag Sci. 2010;66:1308–1315. [DOI] [PubMed] [Google Scholar]
- [27].Miessner S, Stammler G. Monilinia laxa, M. fructigena and M. fructicola: risk estimation of resistance to QoI fungicides and identification of species with cytochrome b gene sequences. J Plant Dis Prot. 2010;117: 162–167. [Google Scholar]
- [28].Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations - a review. Gene. 1989;82:91–114. [DOI] [PubMed] [Google Scholar]
- [29].Nawrocki EP, Jones TA, Eddy SR. Group I introns are widespread in archaea. Nucleic Acids Res. 2018;46:7970–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. [DOI] [PubMed] [Google Scholar]
- [31].Lisacek F, Diaz Y, Michel F. Automatic identification of group I intron cores in genomic DNA sequences. J Mol Biol. 1994;235:1206–1217. [DOI] [PubMed] [Google Scholar]
- [32].Zhou Y, Lu C, Wu Q-J, et al. GISSD: group I intron sequence and structure database. Nucleic Acids Res. 2008;36(suppl_1):31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brent KJ, Hollomon DW. Fungicide resistance: the assessment of risk. 2nd ed. . Brent KJ, Hollomon DW, editors. Brussels: Fungicide Resistance Action Committee; 2007. [Google Scholar]
- [34].Zeng F, Arnao E, Zhang G, et al. Characterization of quinone outside inhibitor fungicide resistance in Cercospora sojina and development of diagnostic tools for its identification. Plant Dis. 2015;99(4):544–550. . [DOI] [PubMed] [Google Scholar]
- [35].Fungicide Resistance Action Commitee , “List of plant pathogenic organisms resistant to disease control agents” (2018).
- [36].Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. [DOI] [PubMed] [Google Scholar]
- [37].Li Z, Zhang Y. Predicting the secondary structures and tertiary interactions of 211 group I introns in IE subgroup. Nucleic Acids Res. 2005;33:2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kalvari I, Argasinska J, Quinones-Olvera N, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46(D1):D335–D342. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Burke JM, Belfort M, Cech TR, et al. Structural conventions for group I introns. Nucleic Acids Res. 1987;15(18):7217–7221. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cech TR. Conserved sequences and structures of group I introns: building an active site for RNA catalysis - a review. Gene. 1988;73:259–271. [DOI] [PubMed] [Google Scholar]
- [42].Belfort M. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25(17):3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Höhl M, Rigoutsos I, Ragan MA. Pattern-based phylogenetic distance estimation and tree reconstruction. Evol Bioinforma. 2017;2: 117693430600200. [PMC free article] [PubMed] [Google Scholar]
- [44].Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. Univ. Kansas, Sci. Bull. 1958;38, 1409–1438. [Google Scholar]
- [46].Wilkinson SP, kmer: an R package for fast alignment-free clustering of biological sequences. 2018. doi: 10.5281/zenodo.1227690. [DOI]
- [47].Galili T. Dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015;31:3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7. DOI: 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lehnert V, Jaeger L, Michele F, et al. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol. 1996;3:993–1009. [DOI] [PubMed] [Google Scholar]
- [51].Fillinger S, Walker A-S. Chemical control and resistance management of botrytis diseases. In: Fillinger S, Elad Y, editors. Botrytis – the fungus, the pathogen and its management in agricultural systems. Cham (Switzerland): Springer International Publishing; 2016. p. 189–216. [Google Scholar]
- [52].Cannone JJ, Subramanian S, Schnare MN, et al. The comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3(1):2. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Boccaletto P, Magnus M, Almeida C, et al. RNArchitecture: a database and a classification system of RNA families, with a focus on structural information. Nucleic Acids Res. 2017;46:D202–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].McNellis TW, von Arnim AG, Araki T, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6(4):487–500. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lin K, Dorman JB, Rodan A, et al. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. [DOI] [PubMed] [Google Scholar]
- [56].Fox JW, Lamperti ED, Ekşioğlu YZ, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21(6):1315–1325. . [DOI] [PubMed] [Google Scholar]
- [57].Belfort M, Derbyshire V, Parker MM, et al. Mobile introns: pathways and proteins. In: Craig N, Craigie R, Gellert M, et al., editors. Mobile DNA II. Washington (DC): American Society of Microbiology; 2002. p. 761–783. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


