Abstract
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of the mannose phosphate isomerase (mpi) gene was applied to 134 skin samples collected from patients with cutaneous leishmaniasis (CL) in Peru for identification of the infecting parasite at the species level, and the results were compared with those of cytochrome b (cyt b) gene sequencing obtained in previous studies. Although most results (121/134) including 4 hybrids of Leishmania (Viannia) braziliensis and L. (V.) peruviana corresponded to those obtained in the previous study, PCR-RFLP analyses revealed the distribution of putative hybrid strains between L. (V.) peruviana and L. (V.) lainsoni in two samples, which has never been reported. Moreover, parasite strains showing discordance between kinetoplast and nuclear genes (kDNA and nDNA), so-called mito-nuclear discordance, were identified in 11 samples. Of these, six strains had the kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) guyanensis, and three strains had the kDNAs of L. (V.) shawi and nDNAs of L. (V.) braziliensis. The rest were identified as mito-nuclear discordance strains having kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) lainsoni, and kDNAs of L. (V.) lainsoni and nDNAs of L. (V.) braziliensis. The results demonstrate that Leishmania strains in Peru are genetically more complex than previously considered.
Author summary
Protozoan parasites of the genus Leishmania are able to undergo genetic exchange during their growth. The previous description of hybrids in Peru and the recent discovery of unexpected genetically complex strains having characteristics of both hybrid and mito-nuclear discordance in its neighbouring country (Ecuador) with a similar eco-epidemiological situation led us to consider that the genetic structure of Leishmania strains in Peru is more complicated than previously thought. In an effort to revise the data on Leishmania strain dispersion in Peru and to search for evidence of genetic recombination, the present study was conducted. A polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis targeting the mannose phosphate isomerase (mpi) gene sequence was performed to identify the infecting parasite at the species level in 134 skin samples collected from patients with cutaneous leishmaniasis (CL) in Peru, and the results were compared with those of cytochrome b (cyt b) gene sequencing obtained in previous studies. Most results (121/134) including 4 hybrids between L. (V.) braziliensis and L. (V.) peruviana showed agreement between PCR-RFLP of the mpi gene and cyt b gene sequence analysis; however, 13 of 134 samples revealed the distribution of strains with hybrids and mito-nuclear discordance. The results demonstrate that genetically complex Leishmania strains are present in Peru. These findings indicate that Leishmania strain dispersion in Peru is genetically more complex than previously considered. Further prospective studies including larger samples and the isolation of parasite strains are required to update the available data.
Introduction
Parasitic protozoa belonging to the genus Leishmania show marked epidemiologic and clinical diversity, causing a wide-range of human and animal diseases extending from localized, self-limiting cutaneous lesions, and severe diffuse and destructive mucocutaneous lesions, to disseminating visceral infection that is fatal in the absence of treatment. The clinical heterogeneity, together with reservoir host spectrum and vector species compatibilities, lead to different parasite species associations, with over 20 species related to human infections. The cause of this heterogeneity, whether by continuous accumulation of mutations via mitotic cell division and/or via sexual recombination promoting admixtures of divergent genomes, is still a matter of debate [1]. Even if we are far from explaining how Leishmania parasites evolve and emerge in natural populations, correct species identification and searching for evidence of genetic recombination can provide key clues to the ecology and transmission patterns of these organisms. Correct diagnosis of leishmaniasis will also help to determine the clinical prognosis and an appropriate species-specific therapeutic regimen [2].
Multi-locus enzyme electrophoresis (MLEE) has been the gold standard for species characterization [3, 4]. However, this classification has been challenged based on genetic analysis of molecular targets, as they were much simpler, easier, and more practical [5–14]. Kinetoplast DNA (kDNA) is frequently used as a target for detection and typing of Leishmania due to its multicopy nature and high sensitivity [15, 16]. However, combining nuclear DNA (nDNA) and kDNA markers has improved the power of molecular data to detect unexpected genetically complex strains with characteristics of hybrid and mito-nuclear discordance widely distributed in Ecuador [17]. These recent findings have shown the necessity of updating the available data in its neighbouring country of Peru, with a similar eco-epidemiological situation, and searching for genetic recombination in Peruvian strains that were identified at the species level by kinetoplast cytochrome b (cyt b) gene sequence analysis in a previous study. The latter led to the identification of 7 species and 1 hybrid: Leishmania (Viannia) braziliensis, L. (V.) peruviana, L. (V.) guyanensis, L. (V.) lainsoni, L. (V.) shawi, Leishmania (Leishmania) mexicana, L. (L.) amazonensis, and a hybrid of L. (V.) braziliensis/L. (V.) peruviana [18]. Previous results on the current epidemiological situation in Peru showed that the predominant species were L. (V.) peruviana, L. (V.) braziliensis, and L. (V.) guyanensis in the Andean highlands, tropical lowland rainforests, and northern to central rainforest areas, respectively [19–22]. These studies also demonstrated the presence of L. (V.) lainsoni and L. (L.) amazonensis in lower-altitude rainforest areas [20–22], and the current prevalence in the same areas was confirmed [18]. The identification of hybrid forms was performed using a polymerase chain reaction-restriction fragment length polymorphism (PCR- RFLP) analysis of a short mannose phosphate isomerase (mpi) gene fragment. Hybrid genotypes have been rarely revealed among natural isolates of Leishmania parasites, which conventionally indicated that members of the genus possess the machinery for genetic exchange [17, 19]. A hybrid of L. (V.) braziliensis and L. (V.) peruviana was first reported in the Department of Huanuco [19, 23], and was recently identified in the Department of Cajamarca [18]. Additionally, just four cases of infection by L. (V.) shawi were detected in rainforest areas by the Departments of Junin, Madre de Dios, Cusco, and Puno, as previously reported [18, 22], and the question remains unanswered about the low rate of infection caused by this species. Verification of the full details of gene property variants will help to understand the eco-epidemiological situation in this endemic area.
The present study utilized PCR-RFLP analysis targeting the mpi gene sequence to identify the infecting parasite at the species level in 134 skin samples collected from patients with cutaneous leishmaniasis (CL) in Peru, and the results were compared with those of cyt b gene sequencing obtained in previous studies. The results demonstrate that genetically complex Leishmania strains are present in Peru, highlighting the need to combine both nDNA and kDNA targets to improve the validity of species identifications for full details of gene property variants.
Materials and methods
Sample collection
The clinical samples employed in this study were collected from patients suspected of having CL from 58 sites in 38 provinces administered by 17 departments in Peru using FTA cards (Whatman, Newton Center, MA, USA) and Giemsa-stained smears in previous studies [18, 22] (S1 Fig). Only identified specimens in previous studies were used in this study to avoid selection bias. The preparation of FTA Card samples was done as previously described [18]. Briefly, two-mm-diameter disks were punched out from each filter paper. The disks were then transferred to separate tubes, washed two times with FTA Purification Reagent (Whatman) and one time with Tris-EDTA buffer, and then air dried. The filter paper disks were directly subjected to PCR amplification. DNA extraction from Giemsa-stained smears obtained from skin lesions (ulcers and/or nodules) on CL patients was performed as previously described [18]. Briefly, 50 μL of DNA extraction buffer [150 mM NaCl, 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.1% sodium dodecyl sulfate (SDS)] containing 100 μg/mL of proteinase K was spotted onto each smear sample and mixed well. The samples were transferred to 1.5-mL tubes and incubated at 37°C for 12 hours. After being heat-inactivated at 95°C for 5 min, 0.5-μL portions were directly used as the templates for PCR amplification.
PCR and sequence analysis
Multiple targets including mpi, 6-phosphogluconate dehydrogenase (6pgd), heat shock protein 70 (hsp70), and cytochrome oxidase subunit II-NADH dehydrogenase subunit I (COII-ND1) gene fragments were used to confirm species identifications for full details of gene property variants. The primers and PCR conditions were described previously [17] except for the mpi gene fragment, where new primers were designed to amplify a shorter fragment (S2 and S3 Figs). For the mpi gene fragment, PCR amplification with a pair of specific primers, L.MPI-AS (5’- TCGATTCGCACGGCTCTGTC-3’) and L.MPI-OR (5’-CTCAAGTCGTTGGTCGACGC-3’), was performed with 30 cycles of denaturation (95°C, 1 min), annealing (55°C, 1 min), and polymerization (72°C, 1 min) using Ampdirect Plus reagent (Shimadzu Biotech, Tsukuba, Japan) and high-fidelity DNA polymerase, KOD plus (Toyobo, Osaka, Japan), or PrimeSTAR HS (Takara Bio, Shiga, Japan). Each 0.5-μL portion of the PCR product was reamplified with L.MPI-IR2 (5’-GCCGTACGGYACCGCAAAGC-3’) and L.MPI-BS (5’-AACCACAAGCCWGAGCTCAT-3’).
PCR-RFLP analysis
PCR products of the mpi gene fragment were digested with the restriction enzymes HaeIII, VpaK11BI, and BstXI. The restriction enzymes were selected with the support program BioEdit. The digested samples were separated by electrophoresis in 2% agarose gel (STAR Agarose RSV-AGRP from Rikaken, Aichi, Japan) for VpaK11BI and BstXI to separate longer fragments and 3% gel for HaeIII to separate shorter fragments in order to produce DNA fragment patterns using the GeneRuler 100 bp Plus DNA ladder (Thermo Fisher Scientific, Waltham, MA, USA) as molecular size markers. The gel was stained with GelRed Nucleic Acid Gel Stain (Biotium, Hayward, CA, USA), and DNA fragments were visualized with a UV transilluminator.
Ethics statement
Informed consent was obtained from all participating adults; for children under the age of 18 years, consent was also obtained from their parents or guardians. Verbal explanations and written information sheets were provided, following the guidelines of the Ethics Committee of the Ministry of Health, Peru. Clinical samples were collected by well-trained local doctors, biologists, and laboratory technicians from health posts and centers of the ministry. The subjects studied were volunteers in routine diagnosis/screening and treatment programs promoted by the ministry. All routine laboratory examinations were carried out free of charge, and treatment with a specific drug, sodium stibogluconate (Pentostan), was also offered free of charge at each health center. The study was approved by the ethics committee of the Graduate School of Veterinary Medicine, Hokkaido University (approval number: vet26-4) and Jichi Medical University (approval number: 17–080) [17, 18]. A case report form (CRF) was collected for each sample and unique codes were assigned to ensure confidentiality. All subjects and guardians consented to lesions being photographed anonymously. Permission was obtained from the Ministry of Health and from community leaders in Peru.
Results
Identification of Leishmania species using PCR-RFLP
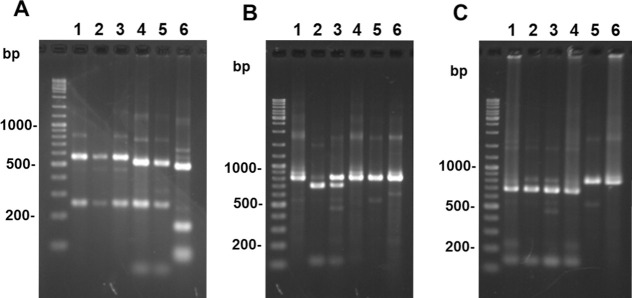
In a recent study, PCR-RFLP analyses of a long mpi gene fragment (1,130 bp) using the restriction enzymes HaeIII and HpaI differentiated Leishmania species in Ecuador, except for two very closely-related species, L. (V.) guyanensis and L. (V.) panamensis [17]. In addition, using a short mpi gene fragment (460 bp), 4 Leishmania species, L. (V.) braziliensis, L. (V.) peruviana, L. (V.) guyanensis, and L. (V.) lainsoni, and a hybrid of L. (V.) braziliensis and L. (V.) peruviana, could be differentiated in the Department of Huanuco, Peru [23]. In the present study, new primers were designed to amplify a 807 bp-mpi fragment (S2 Fig), and the PCR-RFLP technique was optimized using three diagnostic restriction enzymes (HaeIII, VpaK11BI, and BstXI) for species identifications in the studied area. The RFLP patterns were predicted according to the sequences obtained from the GenBank database, and were tested on the Leishmania species listed in S1 Table (Fig 1). The application of PCR-RFLP to 134 skin samples collected from patients with CL in Peru (Fig 2) identified 5 Leishmania species [L. (V.) braziliensis, L. (V.) peruviana, L. (V.) guyanensis, L. (V.) lainsoni, and L (L.) amazonensis] and two putative hybrids [L. (V.) braziliensis/L. (V.) peruviana and L. (V.) peruviana/L. (V.) lainsoni] (Table 1). However, an unexpected RFLP pattern appeared in one sample, in which a single polymorphism nucleotide prevented the enzymes from cutting the target position, corresponding to L. (V.) braziliensis based on sequence analysis.
Fig 1. PCR-RFLP analyses of mpi gene fragments from 6 Leishmania species in Peru.
PCR amplification was performed with leishmanial mannose phosphate isomerase (mpi) gene-specific primers, and PCR products were digested with HaeIII (A), Vpak11BI (B), or BstXI (C), and resulting restriction fragment patterns were analyzed by agarose gel electrophoresis. 1. L. (V.) braziliensis, 2. L. (V.) peruviana, 3. a hybrid of L. (V.) braziliensis/L. (V.) peruviana, 4. L. (V.) guyanensis, 5. L. (V.) lainsoni, 6. L. (L.) amazonensis.
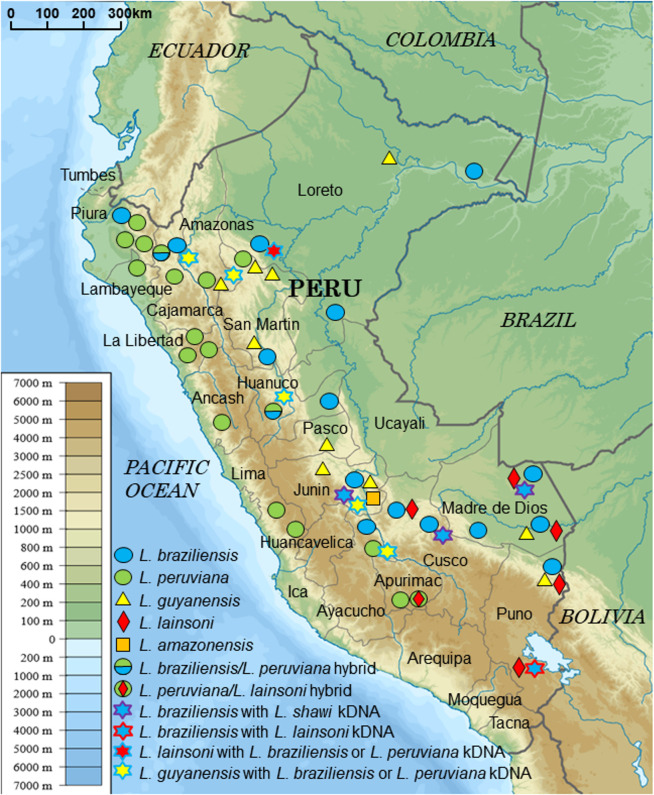
Fig 2. Geographic distribution of Leishmania species in Peru identified by PCR-RFLP of mpi and sequence analyses of kinetoplast and nuclear DNAs.
(Adapted from a map available at https://commons.wikimedia.org/wiki/File%3APeru_physical_map.svg).
Table 1. Distribution of Leishmania species by department in Peru.
| Department | Leishmania species* | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| Lb | Lp | Lg | Ll | La | Lb/Lp | Lp/Ll | ||
| Amazonas | 0 | 1 | 3 (2#) | 0 | 0 | 0 | 0 | 4 |
| Ancash | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Apurimac | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 3 |
| Ayacucho | 2 | 1 | 2# | 0 | 0 | 0 | 0 | 5 |
| Cajamarca | 1 | 1 | 0 | 0 | 0 | 3 | 0 | 5 |
| Cusco | 4 (1#) | 0 | 0 | 1 | 0 | 0 | 0 | 5 |
| Huanuco | 3 | 0 | 1# | 0 | 0 | 1 | 0 | 5 |
| Junin | 2 (1#) | 0 | 3 (1#) | 0 | 1 | 0 | 0 | 6 |
| La Libertad | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 8 |
| Lambayeque | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 18 |
| Lima | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Loreto | 3 | 0 | 1 | 1# | 0 | 0 | 0 | 5 |
| Madre de Dios | 24 (1#) | 0 | 1 | 2 | 0 | 0 | 0 | 27 |
| Pasco | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| Piura | 1 | 10 | 0 | 0 | 0 | 0 | 0 | 11 |
| Puno | 6 (1#) | 0 | 1 | 12 | 0 | 0 | 0 | 19 |
| San Martin | 1 | 1 | 6 | 0 | 0 | 0 | 0 | 8 |
| Total | 47 | 44 | 20 | 16 | 1 | 4 | 2 | 134 |
*Parasite species were identified by the PCR-RFLP analysis targeting 807bp mpi gene fragments (S2 Fig) with restriction enzymes HaeIII, VpaK11BI, and BstXI. Lb, L. (V.) braziliensis; Lp, L. (V.) peruviana; Lg, L. (V.) guyanensis; Ll, L. (V.) lainsoni; La, L (L.) amazonensis; Lb/Lp, a putative hybrid of L. (V.) braziliensis and L. (V.) peruviana; Lb/Ll, a putative hybrid of L. (V.) peruviana and L. (V.) lainsoni
#Mito-nuclear discordance was determined by the comparative analysis of nuclear (mpi, hsp70, and 6pgd) and kinetoplast (cyt b and COII-ND1) DNA fragments.
Comparative analysis of nDNA and kDNA
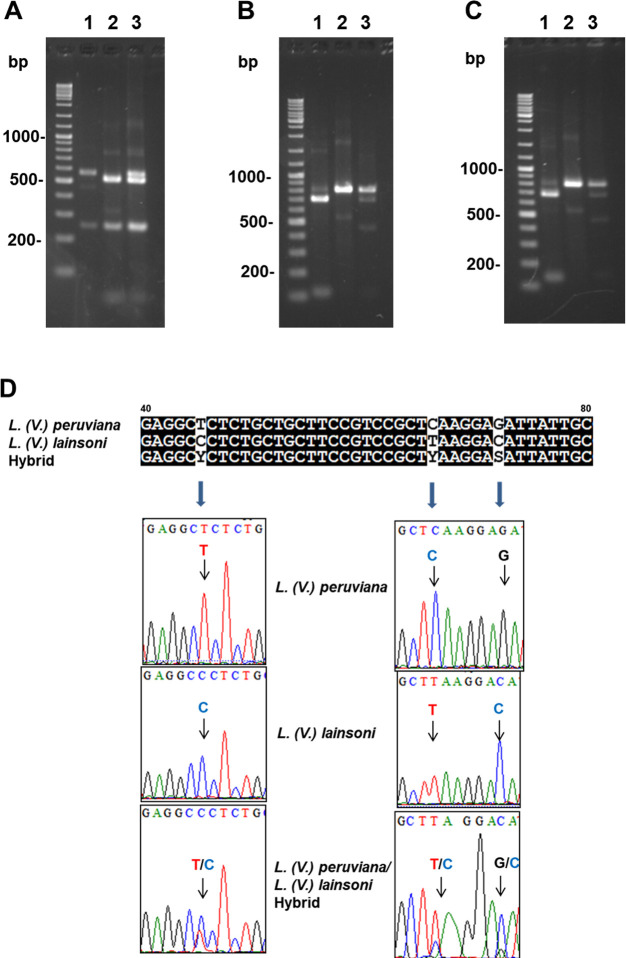
The results obtained by PCR-RFLP analyses were compared with those obtained by cyt b gene sequence analysis. In the previous studies, L. (V.) braziliensis was differentiated from L. (V.) peruviana by PCR-RFLP of the 460bp mpi gene fragment (S2 Fig) since the two species cannot be differentiated by kDNA sequence analysis [18,22]. Most results including 4 hybrids between L. (V.) braziliensis and L. (V.) peruviana agreed with previous ones obtained by cyt b gene sequence analysis; however, 13 of 134 samples showed discordance between the present and previous analyses (Tables 1 and 2) [18, 22]. In detail, of the 100 samples identified as L. (V.) braziliensis or L. (V.) peruviana by cyt b gene sequence analysis, 43, 44, and 4 samples were identified as L. (V.) braziliensis, L. (V.) peruviana, and a hybrid between L. (V.) braziliensis and L. (V.) peruviana by PCR-RFLP analyses of the mpi gene, corresponding to previous studies [18, 22]. On the other hand, six samples from wide areas of Peru and one sample from the north area, all of which were identified as L. (V.) braziliensis or L. (V.) peruviana by the cyt b gene sequence analysis, were L. (V.) guyanensis and L. (V.) lainsoni, respectively, by PCR-RFLP analyses of the mpi gene (Table 1). Sequence analyses of the mpi gene fragment supported the results of PCR-RFLP. In addition, two samples identified as L. (V.) peruviana using sequence analysis of the cyt b gene and PCR-RFLP analysis of the short mpi gene fragment in the previous studies showed hybrid patterns with L. (V.) lainsoni using PCR-RFLP analyses targeting 807bp-mpi gene fragments in the present study (Fig 3A–3C). Direct sequencing of the mpi fragments identified double peaks at the corresponding positions (Figs 3D and S3). According to our knowledge, this is the first reported evidence of putative hybridization between L. (V.) peruviana and L. (V.) lainsoni. On the other hand, of the 16 samples identified as L. (V.) lainsoni by cyt b gene analysis, 15 samples were identified as L. (V.) lainsoni whereas one sample was L. (V.) braziliensis by PCR-RFLP analyses of the mpi gene. In the same way, all three samples identified as L. (V.) shawi by sequence analysis of the cyt b gene were identified as L. (V.) braziliensis by PCR-RFLP analysis of the mpi gene. The mito-nuclear discordances having kDNA of L. (V.) braziliensis or L. (V.) peruviana and nDNA of L. (V.) lainsoni, and kDNA of L. (V.) braziliensis or L. (V.) peruviana and nDNA of L. (V.) shawi have not been reported in Peru or elsewhere. This is also the first reported evidence of mito-nuclear discordance having kDNA of L. (V.) braziliensis or L. (V.) peruviana and nDNA of L. (V.) guyanensis in Peru. Eleven samples showing discordance between mpi gene and cyt b gene analyses were systematically analyzed using multiple targets including mpi, hsp70, 6pgd, and COII-ND1 genes to confirm mito-nuclear discordance (Tables 1 and 2). Sequence analysis confirmed the discordance between nDNAs and kDNAs in these samples. The nucleotide sequence data reported were deposited in GenBank databases under the accession numbers LC517842-LC517879.
Table 2. Comparison of Leishmania species identification in Peru targeting kinetoplast and nuclear DNAs.
| Target DNA1) | Identification2): numbers | ||||||
|---|---|---|---|---|---|---|---|
| kDNA | Lb or Lp3): 100 | Lg: 14 | Ll: 16 | Ls: 3 | La: 1 | ||
| nDNA | Lb: 43 | Lp: 44 | Lb/Lp: 4 | Lg: 14 | Ll: 15 | Lb: 3 | La: 1 |
| Lg: 6 | Ll: 1 | Lp/Ll: 2 | Lb: 1 | ||||
| kDNA + nDNA | Lb: 43 | Lp: 44 | Lb/Lp: 4 | Lg: 14 | Ll: 15 | Ls-Lb: 3 | La: 1 |
| Lb,p-Lg: 6 | Lp/Ll: 2 | Ll-Lb: 1 | |||||
| Lb,p-Ll: 1 | |||||||
1)kDNA: kinetoplast DNA, nDNA: nuclear DNA. Parasite species were identified by the sequencing analysis of cyt b gene (kDNA) and by the PCR-RFLP analysis of mpi gene (nDNA) fragments with restriction enzymes HaeIII, VpaK11BI, and BstXI. Mito-nuclear discordance was further determined by the comparative analysis of nDNA (hsp70, and 6pgd) and kDNA (COII-ND1) fragments.
2)Lb: L. (V.) braziliensis, Lp: L. (V.) peruviana, Lg: L. (V.) guyanensis, Ll: L. (V.) lainsoni, Ls: L. (V.) shawi, La: L. (L.) amazonensis, Lb/Lp: a putative hybrid of L. (V.) braziliensis and L. (V.) peruviana, Lp/Ll: a putative hybrid of L. (V.) peruviana and L. (V.) lainsoni, Lb,p-Lg: L. (V.) guyanensis with kDNAs of L. (V.) braziliensis or L. (V.) peruviana, Lb,p-Ll: L. (V.) lainsoni with kDNAs of L. (V.) braziliensis or L. (V.) peruviana, Ll-Lb: L. (V.) braziliensis with L. (V.) lainsoni kDNA, Ls-Lb: L. (V.) braziliensis with L. (V.) shawi kDNA
3)L. (V.) braziliensis and L. (V.) peruviana cannot be differentiated by kDNA sequence analysis.
Fig 3. Differentiation between L. (V.) peruviana and L. (V.) lainsoni by PCR-RFLP and direct sequence analyses of mpi gene fragment.
A, B, C. PCR amplification was performed with mpi gene-specific primers and the PCR products were digested with HaeIII (A), Vpak11BI (B), or BstXI (C). 1. L. (V.) peruviana, 2. L. (V.) lainsoni, 3. a hybrid of L. (V.) peruviana and L. (V.) lainsoni. D. Direct sequence analysis showing three among several species-specific polymorphic sites of the Leishmania mpi gene fragment.
Discussion
Comparative analysis of nuclear and kinetoplast DNA fragments revealed the occurrence of 5 Leishmania species in the study areas: L. (V.) braziliensis, L. (V.) peruviana, L. (V.) guyanensis, L. (V.) lainsoni, and L. (L.) amazonensis, and two putative hybrids: L. (V.) braziliensis/L. (V.) peruviana and L. (V.) peruviana/L. (V.) lainsoni, of which the latter was firstly identified in Peru. We also identified mito-nuclear discordances having kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) lainsoni, and kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) shawi, which have never been reported in Peru or elsewhere. In addition, we reported here the first evidence of mito-nuclear discordance having kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) guyanensis in Peru. Based on the geographic distribution, kDNAs of these mito-nuclear discordance strains probably originate from L. (V.) braziliensis since L. (V.) peruviana is prevalent mostly in Andean highland areas. Unexpectedly, approximately 10% of the samples were identified as strains with mito-nuclear discordance or hybrids caused by genetic exchange, indicating that the genetic structure of Leishmania strains in Peru is more complicated than previously recorded [18].
Of the 134 studied samples, 2 and 4 samples were identified as putative hybrids of L. (V.) peruviana/L. (V.) lainsoni and L. (V.) braziliensis/L. (V.) peruviana, respectively. Regarding its capacity to identify hybrids of these species, the PCR-RFLP method should be considered as a fundamental tool to be applied. Here, we report the first putative hybrid between L. (V.) peruviana and L. (V.) lainsoni. Natural hybrids were reported to occur episodically [17, 24–33]. In this context, it is important to note that most hybridization reported was either intra-specific or involved very closely related species: L. (V.) braziliensis and L. (V.) peruviana, although hybrids of more distant species have also been reported, such as L. (L). major/L. (L.) infantum [33] and L. (L.) major/L. (L.) arabica [31]. The compatibility of different species to construct hybrids is unknown, and our finding of a putative L. (V.) peruviana/L. (V.) lainsoni hybrid expands the known natural compatibility between species. Both species are quite different with respect to these characteristics, and the impact of hybridization between such divergent species is currently unknown. Although the possibility of co-infection cannot be excluded completely, both alleles were proportionately amplified in PCR-RFLP analysis, strongly suggesting infection by hybrid strains in these patients. It is important to note here that isolation of putative hybrid strains as a culture is required for further detailed characterization of these parasites, since the possibility of co-infection cannot be excluded completely. Lutzomyia ayacuchensis is a proven vector of L. (V.) peruviana in the Andean area of southern Peru [34], whereas no transmission of L. (V.) lainsoni by sand fly species has been reported in these areas. Further investigation in relation to isolation of natural hybrids strains may be necessary for basic parasitological research on how and where genetic exchange occurs among Leishmania species. The genetic exchange may affect vector susceptibility, and hybrid strains may be transmitted by both vectors of parental species/strains [35]. In addition, using an animal model, hybrids between L. (V.) braziliensis and L. (V.) peruviana have been suggested to increase the disease severity when compared with the parental strains [36]. The pathological impact of recombinant genotypes in human infections associated with virulence, transmissibility, and drug susceptibility warrants further investigation in relation to improving methods of identification and isolation of natural hybrid strains.
Our results are the first reported evidence of mito-nuclear discordance among Leishmania species from Peru, highlighting the importance of combining both nDNA and kDNA to improve the validity of species identification for full details of gene property variants. Here, we report the first evidence of mito-nuclear discordance having kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) guyanensis in Peru, which was reported in Ecuador [17]. In this context, it is important to note that the most mito-nuclear discordant strains were found in the species identified as L. (V.) braziliensis or L. (V.) peruviana using cyt b gene analysis. The present study showed the occurrence of mito-nuclear discordance having kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) shawi, never detected in Peru or elsewhere. In this context, it is important to note that just four cases infected by L. (V.) shawi were detected in rainforest areas by the Departments of Junin, Madre de Dios, Cusco, and Puno, as reported previously [18, 22] and the question remains unanswered about the rarity of infection caused by this species. L. (V.) shawi was originally identified as a parasite of wild animals, and sand fly species that transmit it may prefer to feed on animals rather than humans, resulting in a steady rate of infections in wild animals via sand fly bites and a lower risk of infection for humans. However, further investigations using both and nDNA and kDNA genes may provide more clarifications about its epidemiological roles and geographical distributions including mito-nuclear discordance, as reported in the present work. Another unexpected finding was the identification of mito-nuclear discordant strains having kDNAs of L. (V.) braziliensis or L. (V.) peruviana and nDNAs of L. (V.) lainsoni, and kDNAs of L. (V.) lainsoni and nDNAs of L. (V.) braziliensis, never reported in Peru or elsewhere. Only one mito-nuclear discordant strain was identified in the species identified as L. (V.) lainsoni by cyt b gene analysis, which may confuse the previous results on identifications [18]. Further investigations are required to elucidate the mechanism of mito-nuclear discordance in protozoa. Several mechanisms have been shown to cause mito-nuclear discordance: incomplete lineage sorting, sex-biased asymmetries, introgression, natural selection, and genetic sweeps mediated by Wolbachia infection [37]. Mito-nuclear discordance was firstly reported in systems where early genetic tools were more developed compared with other taxa such us mitochondrial introgression between two species of fruit fly (Drosophila pseudoobscura and D. persimilis) and two species of mouse (Mus domesticus and M. musculus) [38, 39]. The number of cases continued to increase slowly until 2001. Following these early discoveries, methods for assaying numerous individuals for their nuclear genotype became more widely available to researchers and, subsequently, the number of cases increased markedly in animals such as mammals, birds, reptiles, amphibians, fish, and insects [37]. More recent studies revealed that mito-nuclear discordance occurs in helminth parasites [40–44].
In the present study, PCR- RFLP targeting leishmanial mpi gene fragments was performed, and the results were compared with those obtained by kinetoplast cyt b gene sequence analysis. The PCR-RFLP method was shown to be practical for the identification of Leishmania species in Peru, revealing unexpected genetically complex Leishmania strains with characteristics of hybrid and mito-nuclear discordance. Since hybrid strains were proposed to aggravate the severity of disease and may be transmitted by a larger range of sand fly species, many questions have been raised about the occurrence and frequency of such cross-species genetic exchange under natural conditions, modalities of hybrid transmission, their long-term maintenance, and the consequences of these transfers on phenotypes such as drug resistance or pathogenicity. Further country-wide epidemiological studies will be needed to reveal the characteristics of hybrid and mito-nuclear discordance and provide further insight into the mechanism of genetic exchanges of these parasites.
Supporting information
1, Utcubamba, Amazonas; 2, Rodriguez de Mendoza, Amazonas; 3, Luya, Amazonas; 4, Huarmey, Ancash; 5, Aymaraes, Apurimac; 6, La Mar, Ayacucho; 7, Huanta, Ayacucho; 8, Jaen, Cajamarca; 9, Cutervo, Cajamarca; 10, Calca, Cusco; 11, La Convención, Cusco; 12, Puerto Inca, Huanuco; 13, Huanuco, Huanuco; 14, Leoncio Prado, Huanuco; 15, Chanchamayo, Junin; 16, Satipo, Junin; 17, Otuzco, La Libertad; 18, Santiago de Chuco, La Libertad; 19, Gran Chimú, La Libertad; 20, Lambayeque, Lambayeque; 21, Yauyos, Lima; 22, Huarochirí, Lima; 23, Alto Amazonas, Loreto; 24, Maynas, Loreto; 25, Mariscal Ramón Castilla, Loreto; 26, Ucayali, Loreto; 27, Tahuamanu, Madre de Dios; 28, Tambopata, Madre de Dios; 29, Manu, Madre de Dios; 30, Oxapampa, Pasco; 31, Ayabaca, Piura; 32, Huancabamba, Piura; 33, Morropon, Piura; 34, Sandia, Puno; 35, Puno, Puno; 36, Moyobamba, San Martin; 37, Lamas, San Martin; 38, Tocache, San Martin. (Adapted from a map available at http://english.freemap.jp/).
(TIF)
Triangles denote primers used for amplification of each mpi gene fragment, and nucleotide positions of each primer are in parentheses. An asterisk at nucleotide position 1,082 shows a polymorphic nucleotide between L. (V.) peruviana and L. (V.) braziliensis.
(TIF)
(TIF)
(DOCX)
Acknowledgments
We would like to thank all the Regional Health Directors, Directors of the Sub-Regions of Health, and Directors of Epidemiology of the various Health Regions in Peru. Thanks also go to the doctors and auxiliary personnel, including biologists, nurses, medical technologists, laboratory technicians, and health promoters from the various Health Posts and Health Centers of Peru who helped achieve the research objectives.
Data Availability
All relevant data are within the manuscript and its Supporting Information files. The nucleotide sequence data reported are deposited in DNA Data Bank of Japan (DDBJ http://getentry.ddbj.nig.ac.jp/top-e.html) under the accession numbers LC517842-LC517879.
Funding Statement
This study was financially supported by the Ministry of Education, Culture and Sports, Science and Technology (MEXT) of Japan (Grant Nos. 25257501 and 17H01685). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rougeron V, De Meeus T, Banuls AL. A primer for Leishmania population genetic studies. Trends in parasitology. 2015;31(2):52–9. 10.1016/j.pt.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 2.Berman JD. Human leishmaniasis: clinical, diagnostic, and chemotherapeutic developments in the last 10 years. Clin Infect Dis. 1997;24:684–703. 10.1093/clind/24.4.684 [DOI] [PubMed] [Google Scholar]
- 3.Cupolillo E, Grimaldi G Jr, Momen H. A general classification of New World Leishmania using numerical zymotaxonomy. Am J Trop Med Hyg. 1994;50:296–311. 10.4269/ajtmh.1994.50.296 [DOI] [PubMed] [Google Scholar]
- 4.Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp. 1990;65:111–125. 10.1051/parasite/1990653111 [DOI] [PubMed] [Google Scholar]
- 5.Dedet JP. Current status of epidemiology of leishmaniases Farrell JP, ed. Leishmania, World-Class Parasites: Volume 4 Boston, MA: Kluwer Academic Publishers, 2002;1–10. [Google Scholar]
- 6.Laurent T, Van der Auwera G, Hide M, Mertens P, Quispe-Tintaya W, Deborggraeve S, De Doncker S, Leclipteux T, Bañuls AL, Büscher P, Dujardin JC. Identification of Old World Leishmania spp. by specific polymerase chain reaction amplification of cysteine proteinase B genes and rapid dipstick detection. Diagn Microbiol Infect Dis. 2009;63:173–181. 10.1016/j.diagmicrobio.2008.10.015 [DOI] [PubMed] [Google Scholar]
- 7.da Silva LA, de Sousa C, dos S, da Grac GC, Porrozzi R, Cupolillo E. Sequence analysis and PCR RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect Genet Evol. 2010;10:77–83. 10.1016/j.meegid.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 8.Kato H, Caceres AG, Mimori T, Ishimaru Y, Sayed AS, Fujita M, Iwata H, Uezato H, Velez LN, Gomez EA, Hashiguchi Y. Use of FTA cards for direct sampling of patient’s lesions in the ecological study of cutaneous leishmaniasis. J Clin Microbiol. 2010;48:3661–3665. 10.1128/JCM.00498-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, Gomez EA, Martini-Robles L, Muzzio J, Velez L, Calvopiña M, Romero-Alvarez D, Mimori T, Uezato H, Hashiguchi Y. Geographic distribution of Leishmania species in Ecuador based on the cytochrome b gene sequence analysis. PLoS Negl Trop Dis. 2016. a;10:e0004844 10.1371/journal.pntd.0004844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talmi-Frank D, Nasereddin A, Schnur LF, Schönian G, Töz SO, Jaffe CL, Baneth G. Detection and identification of Old World Leishmania by high resolution melt analysis. PLoS Negl Trop Dis. 2010;4:e581 10.1371/journal.pntd.0000581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Almeida ME, Steurer FJ, Koru O, Herwaldt BL, Pieniazek NJ, da Silva AJ. Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J Clin Microbiol. 2011;49:3143–3149. 10.1128/JCM.01177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraga J, Veland N, Montalvo AM, Praet N, Boggild AK, Valencia BM, Arévalo J, Llanos-Cuentas A, Dujardin JC, Van der Auwera G. Accurate and rapid species typing from cutaneous and mucocutaneous leishmaniasis lesions of the New World. Diagn Microbiol Infect Dis. 2012;74:142–150. 10.1016/j.diagmicrobio.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 13.Montalvo AM, Fraga J, Maes I, Dujardin JC, Van der Auwera G. Three new sensitive and specific heatshock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis. 2012;31:1453–1461. 10.1007/s10096-011-1463-z [DOI] [PubMed] [Google Scholar]
- 14.Chaouch M, Fathallah-Mili A, Driss M, Lahmadi R, Ayari C, Guizani I, Ben Said M, Benabderrazak S. Identification of Tunisian Leishmania spp. by PCR amplification of cysteine proteinase B (cpb) genes and phylogenetic analysis. Acta Trop. 2013;125:357–365. 10.1016/j.actatropica.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 15.Jara M, Adaui V, Valencia BM, Martinez D, Alba M, Castrillon C, Cruz M, Cruz I, Van der Auwera G, Llanos-Cuentas A, Dujardin JC, Arevalo J. 2013. Real-time PCR assay for detection and quantification of Leishmania (viannia) organisms in skin and mucosal lesions: exploratory study of parasite load and clinical parameters. J. Clin. Microbiol. 2013;51: 1826e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato H, Uezato H, Katakura K, Calvopiña M, Marco JD, Bar- roso PA, Gomez EA, Mimori T, Korenaga M, Iwata H, Nonaka S, Hashiguchi Y. Detection and identification of Leishmania species within naturally infected sand flies in the Andean areas of Ecuador by a polymerase chain reaction. Am J Trop Med Hyg. 2005;72: 87–93. [PubMed] [Google Scholar]
- 17.Kato H, Gomez EA, Seki C, Furumoto H, Martini-Robles L, Muzzio J, Calvopiña M, Velez L, Kubo M, Tabbabi A, Yamamoto DS, Hashiguchi Y. PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. PLoS Negl Trop Dis. 2019;13(5):e0007403 10.1371/journal.pntd.0007403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H, Cáceres AG, Seki C, Silupu García CR, Holguín Mauricci C, Castro Martínez SC, Moreno Paico D, Castro Muniz JL, Troyes Rivera LD, Villegas Briones ZI, Guerrero Quincho S, Sulca Jayo GL, Tineo Villafuerte E, Manrique de Lara Estrada C, Arias FR, Passara FS, Ruelas Llerena N, Kubo M, Tabbabi A, Yamamoto DS, Hashiguchi Y. Further insight into the geographic distribution of Leishmania species in Peru by cytochrome b and mannose phosphate isomerase gene analyses. PLoS Negl Trop Dis. 2019;13(6):e0007496 10.1371/journal.pntd.0007496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dujardin JC, Banuls AL, Llanos-Cuentas A, Alvarez E, DeDoncker S, Jacquet D, et al. Putative Leishmania hybrids in the Eastern Andean valley of Huanuco, Peru. Acta Trop. 1995;59(4):293–307. 10.1016/0001-706x(95)00094-u [DOI] [PubMed] [Google Scholar]
- 20.Lucas CM, Franke ED, Cachay MI, Tejada A, Cruz ME, Kreutzer RD, Barker DC, McCann SH, Watts DM. Geographic distribution and clinical description of leishmaniasis cases in Peru. Am J Trop Med Hyg. 1998;59:312–317. 10.4269/ajtmh.1998.59.312 [DOI] [PubMed] [Google Scholar]
- 21.Arevalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, Miranda-Vera stegui C, Lazo M, Loayza-Muro R, De Doncker S, Maurer A, Chappuis F, Dujardin JC, Llanos-Cuentas A. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis. 2007;195:1846–1851. 10.1086/518041 [DOI] [PubMed] [Google Scholar]
- 22.Kato H, Caceres AG, Mimori T, Ishimaru Y, Sayed AS, Fujita M, Iwata H, Uezato H, Velez LN, Gomez EA, Hashiguchi Y. Use of FTA cards for direct sampling of patients’ lesions in the ecological study of cutaneous leishmaniasis. J Clin Microbiol. 2010;48:3661–3665. 10.1128/JCM.00498-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koarashi Y, Cáceres AG, Saca FMZ, Flores EEP, Trujillo AC, Alvares JLA, Yoshimatsu K, Arikawa J, Katakura K, Hashiguchi Y, Kato H. Identification of causative Leishmania species in Giemsa-stained smears prepared from patients with cutaneous leishmaniasis in Peru using PCR-RFLP. Acta Trop. 2016;158:83–87. 10.1016/j.actatropica.2016.02.024 [DOI] [PubMed] [Google Scholar]
- 24.Banuls AL, Guerrini F, Le Pont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol. 1997;44:408–411. 10.1111/j.1550-7408.1997.tb05716.x [DOI] [PubMed] [Google Scholar]
- 25.Belli AA, Miles MA, Kelly JM. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology. 1994;109:435–442. 10.1017/s0031182000080689 [DOI] [PubMed] [Google Scholar]
- 26.Cruz AK, Titus R, Beverley SM. 1993. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc Natl Acad Sci USA. 1993;90:1599–1603. 10.1073/pnas.90.4.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado O, Cupolillo E, Bonfante-Garrido R, Silva S, Belfort E, Grimaldi JG, Momen H. Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem Inst Oswaldo Cruz. 1997;92:581–582. 10.1590/s0074-02761997000500002 [DOI] [PubMed] [Google Scholar]
- 28.Dujardin JC, Arevalo J, Llanos-Cuentas A, Caceres A, Le Ray D. From pulsed field to field: contribution of molecular karyotyping to epidemiological studies on New World leishmaniasis. Arch Inst Pasteur Tunis. 1993. a;70:505–515. [PubMed] [Google Scholar]
- 29.Dujardin JC, Gajendran N, Arevalo J, Llanos-Cuentas A, Guerra H, Gomez J, Arroyo J, De Doncker S, Jacquet D, Hamers R. Karyotype polymorphism and conserved characters in the Leishmania (Viannia) braziliensis complex explored with chromosome-derived probes. Ann Soc Belg Med Trop. 1993. b;73:101–118. [PubMed] [Google Scholar]
- 30.Dujardin JC, Llanos-Cuentas A, Caceres A, Arana M, Dujardin JP, Guerrini F, Gomez J, Arroyo J, De Doncker S, Jacquet D. 1993c. Molecular karyotype variation in Leishmania (Viannia) peruviana: indication of geographical populations in Peru distributed along a north-south cline. Ann Trop Med Parasitol. 1993. b;87:335–347. 10.1080/00034983.1993.11812777 [DOI] [PubMed] [Google Scholar]
- 31.Kelly JM, Law JM, Chapman CJ, van Eys GJ, Evans DA. 1991. Evidence of genetic recombination in Leishmania. Mol Biochem Parasitol. 1991. 46:253–263. 10.1016/0166-6851(91)90049-c [DOI] [PubMed] [Google Scholar]
- 32.Nolder D, Roncal N, Davies CR, Llanos-Cuentas A, Miles MA. Multiple hybrid genotypes of Leishmania (Viannia) in a focus of mucocutaneous leishmaniasis. Am J Trop Med Hyg. 2007;76:573–578. [PubMed] [Google Scholar]
- 33.Ravel C, Cortes S, Pratlong F, Morio F, Dedet JP, Campino L. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int J Parasitol. 2006;36:1383–1388. 10.1016/j.ijpara.2006.06.019 [DOI] [PubMed] [Google Scholar]
- 34.Caceres AG, Villaseca P, Dujardin JC, Banuls AL, Inga R, Lopez M, Arana M, Le Ray D, Arevalo J. 2004. Epidemiology of Andean cutaneous leishmaniasis: incrimination of Lutzomyia ayacuchensis (Diptera: psychodidae) as a vector of Leishmania in geographically isolated, upland valleys of Peru. Am J Trop Med Hyg. 2004;70:607–612. [PubMed] [Google Scholar]
- 35.Volf P, Benkova I, Myskova J, Sadlova J, Campino L, Christophe Ravel C. Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int J Parasitol. 2007;37:589–593. 10.1016/j.ijpara.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortes S, Esteves C, Mauricio I, Maia C, Cristovao JM, Miles M, Campino L. 2012. In vitro and in vivo behaviour of sympatric Leishmania (V.) braziliensis L. (V.) peruviana and their hybrids. Parasitology. 2012;139:191–199. 10.1017/S0031182011001909 [DOI] [PubMed] [Google Scholar]
- 37.Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012;21:3907–3930 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- 38.Powell JR. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc Natl Acad Sci USA. 1983;80:492–495. 10.1073/pnas.80.2.492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris SD, Sage RD, Huang CM, Nielsen JT, Ritte UR, Wilson AC (1983) Flow of mitochondrial DNA across a species boundary. Proc Natl Acad Sci USA.1983;80:2290–2294. 10.1073/pnas.80.8.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawton SP, Bowen LI, Emery AM, Majoros G. Signatures of mito-nuclear discordance in Schistosoma turkestanicum indicate a complex evolutionary history of emergence in Europe. Parasitology. 2017;144:1752–1762. 10.1017/S0031182017000920 [DOI] [PubMed] [Google Scholar]
- 41.Yanagida T, Carod JF, Sako Y, Nakao M, Hoberg EP, Ito A. Genetics of the pig tapeworm in Madagascar reveal a history of human dispersal and colonization. PLoS One. 2014;9:e109002 10.1371/journal.pone.0109002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamane K, Suzuki Y, Tachi E, Li T, Chen X, Nakao M, Nkouawa A, Yanagida T, Sako Y, Ito A, Sato H, Okamoto M. Recent hybridization between Taenia asiatica and Taenia saginata. Parasitol Int. 2012;61:351–355. 10.1016/j.parint.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 43.Yamane K, Yanagida T, Li T, Chen X, Dekumyoy P, Waikagul J, Nkouawa A, Nakao M, Sako Y, Ito A, Sato H, Okamoto M. Genotypic relationships between Taenia saginata, Taenia asiatica and their hybrids. Parasitology. 2013;140:1595–1601. 10.1017/S0031182013001273 [DOI] [PubMed] [Google Scholar]
- 44.Sato MO, Sato M, Yanagida T, Waikagul J, Pongvongsa T, Sako Y, Sanguankiat S, Yoonuan T, Kounnavang S, Kawai S, Ito A, Okamoto M, Moji K. Taenia solium, Taenia saginata, Taenia asiatica, their hybrids and other helminthic infections occurring in a neglected tropical diseases’ highly endemic area in Lao PDR. PLoS Negl Trop Dis. 2018;12:e0006260 10.1371/journal.pntd.0006260 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1, Utcubamba, Amazonas; 2, Rodriguez de Mendoza, Amazonas; 3, Luya, Amazonas; 4, Huarmey, Ancash; 5, Aymaraes, Apurimac; 6, La Mar, Ayacucho; 7, Huanta, Ayacucho; 8, Jaen, Cajamarca; 9, Cutervo, Cajamarca; 10, Calca, Cusco; 11, La Convención, Cusco; 12, Puerto Inca, Huanuco; 13, Huanuco, Huanuco; 14, Leoncio Prado, Huanuco; 15, Chanchamayo, Junin; 16, Satipo, Junin; 17, Otuzco, La Libertad; 18, Santiago de Chuco, La Libertad; 19, Gran Chimú, La Libertad; 20, Lambayeque, Lambayeque; 21, Yauyos, Lima; 22, Huarochirí, Lima; 23, Alto Amazonas, Loreto; 24, Maynas, Loreto; 25, Mariscal Ramón Castilla, Loreto; 26, Ucayali, Loreto; 27, Tahuamanu, Madre de Dios; 28, Tambopata, Madre de Dios; 29, Manu, Madre de Dios; 30, Oxapampa, Pasco; 31, Ayabaca, Piura; 32, Huancabamba, Piura; 33, Morropon, Piura; 34, Sandia, Puno; 35, Puno, Puno; 36, Moyobamba, San Martin; 37, Lamas, San Martin; 38, Tocache, San Martin. (Adapted from a map available at http://english.freemap.jp/).
(TIF)
Triangles denote primers used for amplification of each mpi gene fragment, and nucleotide positions of each primer are in parentheses. An asterisk at nucleotide position 1,082 shows a polymorphic nucleotide between L. (V.) peruviana and L. (V.) braziliensis.
(TIF)
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. The nucleotide sequence data reported are deposited in DNA Data Bank of Japan (DDBJ http://getentry.ddbj.nig.ac.jp/top-e.html) under the accession numbers LC517842-LC517879.