Abstract
Acinetobacter baumannii is an emerging pathogen that poses a global health threat due to a lack of therapeutic options for treating drug-resistant strains. In addition to acquiring resistance to last-resort antibiotics, the success of A. baumannii is partially due to its ability to effectively compete with the host for essential metals. Iron is fundamental in shaping host-pathogen interactions, where the host restricts availability of this nutrient in an effort to curtail bacterial proliferation. To circumvent restriction, pathogens possess numerous mechanisms to obtain iron, including through the use of iron-scavenging siderophores. A. baumannii elaborates up to ten distinct siderophores, encoded from three different loci: acinetobactin and pre-acinetobactin (collectively, acinetobactin), baumannoferrins A and B, and fimsbactins A-F. The expression of multiple siderophores is common amongst bacterial pathogens and often linked to virulence, yet the collective contribution of these siderophores to A. baumannii survival and pathogenesis has not been investigated. Here we begin dissecting functional redundancy in the siderophore-based iron acquisition pathways of A. baumannii. Excess iron inhibits overall siderophore production by the bacterium, and the siderophore-associated loci are uniformly upregulated during iron restriction in vitro and in vivo. Further, disrupting all of the siderophore biosynthetic pathways is necessary to drastically reduce total siderophore production by A. baumannii, together suggesting a high degree of functional redundancy between the metabolites. By contrast, inactivation of acinetobactin biosynthesis alone impairs growth on human serum, transferrin, and lactoferrin, and severely attenuates survival of A. baumannii in a murine bacteremia model. These results suggest that whilst A. baumannii synthesizes multiple iron chelators, acinetobactin is critical to supporting growth of the pathogen on host iron sources. Given the acinetobactin locus is highly conserved and required for virulence of A. baumannii, designing therapeutics targeting the biosynthesis and/or transport of this siderophore may represent an effective means of combating this pathogen.
Author summary
Acinetobacter baumannii is an emerging pathogen that is gaining notoriety as a major global health threat due to its extensive drug resistance. Accordingly, the World Health Organization has placed A. baumannii at the top of its list of bacteria urgently requiring research and development into novel therapeutic approaches, designating it a priority “critical” pathogen. Unfortunately, little is known about how A. baumannii causes infection. Iron is an essential nutrient to almost all forms of life and plays a key role in shaping host-pathogen interactions. A. baumannii secretes multiple low molecular weight iron chelators (siderophores) to help fulfill its need for the metal. Here we demonstrate that one of these siderophores, acinetobactin, is required for growth using host iron sources, whilst two others (baumannoferrin and fimsbactins) are dispensable to this function. Although all three siderophores are expressed under iron-restriction and in the infected host, disrupting acinetobactin production alone severely attenuates the survival of A. baumannii in a mouse model of bacteremia. Given the high degree of conservation of acinetobactin in Acinetobacter spp., and its essentiality to virulence, we propose that targeting production or uptake of this siderophore represents an attractive option for the development of novel therapeutics.
Introduction
Acinetobacter baumannii is an opportunistic pathogen that is capable of causing a wide variety of diseases ranging from burn, wound, and urinary tract infections to more serious conditions such as ventilator-associated pneumonia and sepsis [1]. Initially regarded as a relatively innocuous nosocomial pathogen, A. baumannii has gained increased notoriety through the appearance of severe community-acquired infections [2–4], and concerningly, through the rapid emergence of multidrug-resistance [5–8]. As a result, the World Health Organization has placed A. baumannii at the top of its list of bacteria urgently requiring research and development into novel therapeutic approaches, designating it as a priority “critical” pathogen [9]. More recently, the Centers for Disease Control increased the threat level categorization of carbapenem-resistant A. baumannii from serious to urgent, due in part to a lack of drugs in the development pipeline [10]. Unfortunately, little is known about the factors that contribute to the survival and proliferation of A. baumannii within the host, and this knowledge is necessary for the informed design of new treatment options.
The success of A. baumannii as an emerging pathogen is not thought to be due to the evolution of traditional virulence factors, like toxins, but rather through a strategy referred to as “persist and resist” [11]. In addition to avoiding antibiotic-mediated killing, A. baumannii endures a wide variety of environmental insults including desiccation [12–14], oxidative stress [15–18], and micronutrient limitation [19–21]. Iron is one trace element that is essential to the vertebrate host and nearly all bacterial pathogens [22]. However, the concentration of free iron at the host-pathogen interface falls orders of magnitude below what is required to support microbial growth, due both to the formation of insoluble ferric oxyhydroxide precipitates under physiological conditions, and to the host maintaining stringent control over the availability of iron [23]. The withholding of metals, such as iron, to curtail bacterial growth in the host is a facet of the innate immune response referred to as “nutritional immunity” [24–26]. Host mechanisms of iron-withholding include maintaining it in complex with heme in hemoproteins, reducing ferroportin expression to limit its release from macrophages, and sequestering it within glycoproteins such as transferrin and lactoferrin, found predominantly in the blood and bodily secretions, respectively [27]. As such, accessing iron is perhaps one of the most formidable challenges faced by invading pathogens and is required for replication within the host.
To overcome the barrier imposed by nutritional immunity, bacteria express a multitude of strategies to obtain metals. Broadly, the mechanisms of bacterial iron acquisition include capturing and extracting heme-iron from hemoproteins, expressing transporters for the uptake of free ferrous iron, and stealing iron from transferrin and lactoferrin by way of surface-bound receptor proteins and/or through secretion of small iron-binding molecules known as siderophores [22]. Many pathogens produce more than one type of siderophore, which are characterized based on the moieties used to coordinate ferric iron: catecholate, hydroxamate, and α-hydroxycarboxylate [28,29]. A fourth class includes siderophores possessing more than one of the aforementioned iron-coordinating groups and thus is referred to as “mixed-type” [28]. At least eight gene clusters have been identified to function in iron acquisition in A. baumannii, with the number of clusters present often differing between strains [30–32]. These clusters include a conserved ferrous iron acquisition system (feoABC) [33,34], two heme uptake loci (heme uptake cluster 1, and hemO cluster) [35], and five clusters involved in endogenous siderophore biosynthesis and utilization [30,31,36,37]. Of the siderophore biosynthetic clusters, one consists of two genes, entA and entB, which are used in the synthesis of 2,3-dihydroxybenzoic acid, a siderophore precursor molecule and weak iron chelator [31]. Another cluster, found only in A. baumannii strain 8399 is involved in the synthesis of an uncharacterized catecholate siderophore [36,37]. The remaining clusters have been more extensively investigated and encode for as many as ten structurally distinct siderophores, expressed from three loci and include acinetobactin and pre-acinetobactin [38–41], baumannoferrins A and B [42], and fimsbactins A through F [43] (Fig 1).
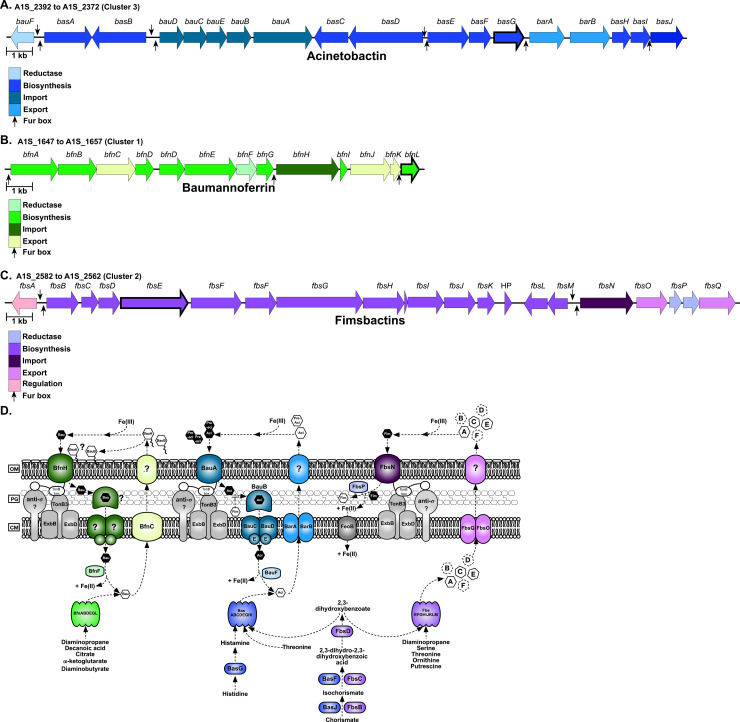
Fig 1. Genetic loci associated with siderophore biosynthesis and transport in A. baumannii ATCC 17978.
Gene clusters for acinetobactin (A), baumannoferrin (B), and fimsbactins (C) are shown. Genes for the reductive release of iron from the siderophore, biosynthesis, import, export, and regulation are shown in the colors indicated in the corresponding legend. Putative Fur boxes are represented by black arrows, and the scale bar for each panel represents 1 kb. Key biosynthetic genes that were disrupted to elucidate siderophore function in this study are highlighted with thick black borders. A schematic highlighting the known and predicted siderophore biosynthetic and transport proteins in A. baumannii ATCC 17978, using the same color scheme as in A-C (D).
Acinetobactin is often referred to as being the major siderophore produced by A. baumannii, largely due to the high degree of conservation of the encoding locus amongst Acinetobacter clinical isolates [30,31,44]. The siderophore exists as two pH-dependent isomers, pre-acinetobactin which is released by the bacteria and is prevalent under acidic conditions, and acinetobactin which is favored under neutral and basic conditions [38,39,45]. All of the genes required for the biosynthesis (basA-J), efflux (barAB) and uptake (bauA-E) of pre-acinetobactin and acinetobactin (herein collectively referred to as acinetobactin) are encoded from the same locus, with the exception of an entA homologue, found elsewhere in the chromosome (Fig 1A and 1D; S1 and S3 Tables [41]). Expression of acinetobactin is required for virulence of A. baumannii strain ATCC 19606 in vertebrate and invertebrate models of infection [41,46]. It is important to note, however, that acinetobactin-mediated iron acquisition is the only high affinity iron uptake system that is functional in ATCC 19606 [41,47], and thus the importance of this siderophore in the context of multiple iron utilization systems is unknown. Indeed, the prevalence of at least two siderophore-encoding loci amongst clinical A. baumannii isolates highlights the importance of elucidating their individual contributions to infection [30].
In addition to acinetobactin, in silico analyses predict that most A. baumannii strains also express two hydroxamate-type siderophores, baumannoferrins A and B [30,31,42]. Twelve genes are thought to be involved in the biosynthesis (bfnA, B, D, E, G, I and L), transport (bfnC, H, J, and K), and utilization (bfnF) of the baumannoferrins and they are confined to a single locus (Fig 1B and 1D; S2 Table [42]). The bfn gene cluster bears homology to other loci associated with the nonribosomal peptide synthase (NRPS)-independent assembly of hydroxamate or carboxylate-type siderophores [42,48]. Purification and characterization of the baumannoferrins was performed using A. baumannii strain AYE [42], a commonly used clinical isolate which does not produce acinetobactin or other siderophores [41,49]. Structural elucidation revealed that baumannoferrins A and B differ only by one double-bond, but the significance of this is unknown [42]. Both baumannoferrins are able to chelate iron and facilitate growth of A. baumannii when supplied as an exogenous iron source [42,50]. Notably, A. baumannii strain AYE proliferates robustly under iron limitation [42], and is virulent in Galleria mellonella and mouse models of infection [51,52], suggesting that the expression of the baumannoferrins may play an important but underappreciated role in the survival and pathogenicity of A. baumannii.
Unlike with acinetobactin and baumannoferrin, whose loci are broadly represented in A. baumannii strains, the locus for fimsbactins biosynthesis and transport only appears to be present in a small fraction of sequenced A. baumannii isolates (~2%) [30,31,43,53]. Although the siderophores were initially isolated from the non-pathogenic species Acinetobacter baylyi [43], the fimsbactins cluster has been identified in pathogenic Acinetobacter isolates, including A. baumannii strain ATCC 17978 which is a commonly used laboratory strain that was initially isolated from a case of fatal meningitis [54], and AbPK1 which was identified as the causative agent of a deadly outbreak of pneumonia in sheep [53]. In the aforementioned species, the fimsbactins locus ((fbsA-Q) Fig 1C; S3 Table) is flanked by transposases, suggesting that it may have been acquired by horizontal gene transfer [31]. Like acinetobactin, the fimsbactins A-F are mixed catechol-hydroxamate type siderophores, where fimsbactin A represents the most abundant product of synthesis pathway, and fimsbactins B through F are likely biosynthetic intermediates or shunt products [43]. Given their structural similarity, it is perhaps not surprising that genes within the fimsbactins pathway (fbsB, fbsC, and fbsH) appear to be functionally redundant with those in the acinetobactin pathway ((basJ, basF, and basE, respectively) Fig 1D; S1 and S3 Tables [41]). Further, the sole entA homologue in A. baumannii strain ATCC 17978, required for both acinetobactin and fimsbactins production, appears to be located in the fimsbactins biosynthetic cluster (fbsD), and the siderophores are thought to compete for the same periplasmic binding protein [41,55]. The high degree of functional redundancy not only between acinetobactin and fimsbactins biosynthesis, but between the siderophore biosynthetic pathways overall in A. baumannii has confounded the ability to elucidate the individual function of the siderophores. Indeed, whilst purified fimsbactins A and B are capable of supporting the iron-dependent growth of A. baumannii [55,56], it is not known how endogenous expression of this siderophore influences survival of the pathogen in vitro or in vivo.
The expression of multiple pathways for the synthesis and utilization of siderophores is a common feature of bacterial pathogens, although the evolutionary reasons for having these seemingly redundant iron acquisition systems is not always known. With A. baumannii, the functional characterization of siderophores has historically been assessed using strains that inherently express only one siderophore, where determinations of overall importance to A. baumannii survival and pathogenesis can be difficult to make. In this study, we sought to address the complexity of A. baumannii siderophore-based iron acquisition systems by using a strain that encodes for acinetobactin, baumannoferrin, and fimsbactins [30,31], but does not utilize heme as an iron source [35]. A panel of A. baumannii ATCC 17978 mutants was generated where one, two, or all three siderophore biosynthetic pathways were genetically inactivated, such that the contributions of the siderophores could be assessed, both independently and in combination. The siderophores were found to be largely functionally redundant in their iron chelation ability in vitro, where disrupting all three biosynthetic pathways was required to substantially reduce total siderophore activity and to abolish growth under iron restriction. These findings were consistent with the observation that all three pathways are upregulated during iron-restriction in vitro and in the metal-restricted host. By contrast, disruption to acinetobactin biosynthesis alone was sufficient to attenuate growth on human serum, transferrin, or lactoferrin as a sole iron source. Strikingly, the acinetobactin biosynthetic mutant was severely attenuated for survival in the murine host, where reduced bacterial burdens were recovered from every major organ during infection. However, in the same model of murine bacteremia, the baumannoferrin and fimsbactins biosynthetic mutants did not exhibit a defect in survival, and a siderophore-deficient strain was no more attenuated for virulence than the acinetobactin mutant alone. Together these results suggest that whilst all three siderophores contribute to iron acquisition by A. baumannii, acinetobactin is the major siderophore required by this pathogen in vivo due to its ability to mobilize iron from host sources and its requirement for survival in a murine model of bacteremia.
Results
Acinetobacter baumannii siderophore biosynthesis and transport are upregulated in response to iron limitation
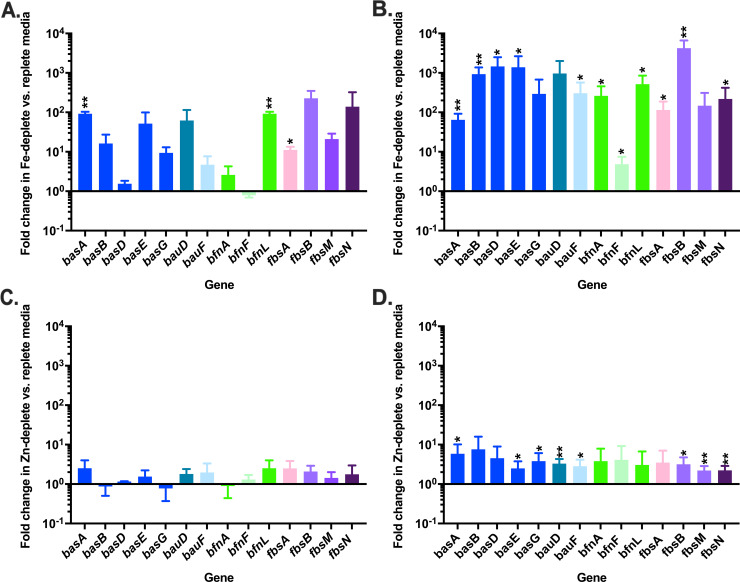
A. baumannii strain ATCC 17978 encodes for ten structurally distinct siderophores encoded from three different biosynthetic loci (Fig 1; S1, S2 and S3 Tables). We previously observed that expression of the loci encoding for the biosynthesis and transport of acinetobactin, baumannoferrin, and fimsbactins is upregulated in the presence of the multi-metal sequestering innate immune protein, calprotectin [57]. Given the apparent functional redundancies in the siderophore pathways of A. baumannii, we hypothesized that these systems are differentially regulated, thus allowing for expression in response to distinct environmental cues, such as differences in the availability of essential transition metals over time. To address this, wild-type (WT) A. baumannii was grown in metal-chelated Tris minimal succinate media (chelex-treated; cTMS) with and without the addition of exogenous iron or zinc. Following 4 or 12 hours (h) growth, RNA was extracted and the expression of various siderophore-associated transcripts from each of the three loci was assessed by quantitative reverse transcription PCR (qRT-PCR). Consistent with identification of putative Fur boxes upstream of many genes or operons within these loci (Fig 1A and 1B, and 1C and reference [31]), key biosynthetic, transport, and regulatory genes in all three siderophore-associated loci are strongly upregulated in iron-deplete versus replete conditions (Fig 2A and 2B). By contrast, and with the exception of bfnF, which encodes a putative oxidoreductase thought to release iron from the siderophore, the change in gene expression in zinc-deplete versus -replete conditions is 10 to 1000-fold lower than that observed with iron (Fig 2C and 2D), suggesting that the expression of genes encoding for siderophore biosynthesis and transport in A. baumannii is predominantly iron-regulated. Notably, gene expression under zinc limitation increased between 4 and 12 h, suggesting that zinc may play a role in the regulation of the siderophore-associated loci during prolonged metal restriction. To determine if these genes are directly regulated by the zinc uptake regulator, Zur, we performed qRT-PCR on a representative gene from each locus in WT A. baumannii and a Δzur mutant [58]. Additionally, we assessed whether Zur can directly regulate fur by similarly looking at expression of this transcriptional regulator in the zur mutant. We found that although the expression of fur was unaltered in Δzur, basA and bfnA were slightly but significantly upregulated in this background (S1 Fig). Together these results suggest that whilst Zur does not appear to directly regulate fur in A. baumannii, it may play a small but unappreciated role in the expression of known fur-regulated genes.
Fig 2. Siderophore biosynthetic and transport genes are upregulated in metal deplete conditions.
WT A. baumannii was grown in metal-restricted media with or without the addition of exogenous iron or zinc. RNA was extracted and transcriptional changes in the expression of siderophore-associated genes were assessed by qRT-PCR in iron-deplete vs. replete conditions at 4 (A) and 12 h (B), and zinc-deplete vs. replete at 4 (C) and 12 (D) conditions, where expression was normalized to the expression of rpoB. * p < 0.05 and ** p < 0.01, as determined by Student’s t test relative to a hypothetical value of 1. Data are the means combined from two independent experiments, each with three biological replicates.
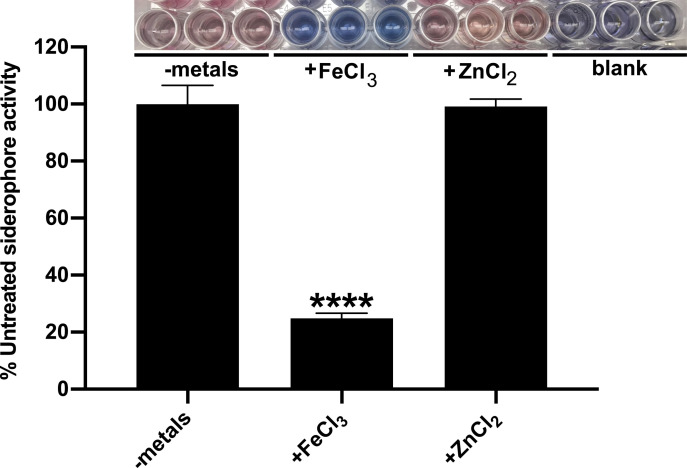
To determine if the transcriptional changes observed above translate to overall differences in siderophore production by A. baumannii, WT bacteria were grown in cTMS with and without the addition of exogenous metals. Following 12 h growth, assessment of siderophore activity in the A. baumannii supernatants was assessed by Chrome Azurol S (CAS) assay. CAS is an iron-binding dye where mobilization of iron from CAS to a chelator within the media can be detected through a colorimetric change of the dye from blue to orange [59,60]. Consistent with the impact of iron on gene transcription, the addition of iron to A. baumannii cultures strongly represses overall siderophore production by the bacteria (Fig 3), whereas the addition of zinc does not appear to impact this activity. These observations indicate that derepression of siderophore-associated genes during zinc limitation may not lead to increased siderophore production overall, and that acinetobactin, baumannoferrin, and fimsbactins biosynthesis and utilization in A. baumannii is primarily influenced by iron availability. It is possible, however, that the semi-quantitative nature of assessing total siderophore activity by CAS assay is not sensitive enough to detect subtle changes in siderophore production upon zinc limitation over time, as the accumulation of siderophores and/or siderophore biosynthetic enzymes may mask these effects.
Fig 3. Iron, but not zinc, represses the overall siderophore activity of A. baumannii.
Chrome Azurol S (CAS) assays to assess for total siderophore activity were performed on the spent culture supernatants of WT A. baumannii grown in cTMS media for 12 h with or without the addition of exogenous metals, as indicated. The colorimetric changes observed in the CAS assay are shown, where blue indicates an absence of siderophore activity and orange indicates that an iron chelator capable of mobilizing iron from CAS is present. Total siderophore activity is expressed as the percent activity of WT A. baumannii grown in the absence of exogenously added metals. **** p < 0.0001, as determined by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test. Data are the means of three biological replicates and are representative of three independent assays.
A. baumannii siderophores exhibit functional redundancy in vitro
To begin identifying the contribution of acinetobactin, baumannoferrin, and fimsbactins production to A. baumannii iron acquisition, key genes in each of the siderophore biosynthetic pathways were disrupted using recombineering [61]. The details of all strains employed in this study can be found in Materials and Methods, and the genes disrupted are highlighted in Fig 1. Genes targeted for inactivation were selected based on predicted function, as well as a lack of any potentially redundant homologues present elsewhere in the A. baumannii ATCC 17978 genome. The disrupted genes include basG, encoding a putative histidine decarboxylase predicted to synthesize histamine as an essential precursor molecule to acinetobactin biosynthesis [44,62], bfnL, encoding a putative acetyltransferase predicted to facilitate the conversion of N3-hydroxy-1,3-diaminopropane to N3-decanoyl-hydroxy-1,3-diaminopropane as an early step in baumannoferrin biosynthesis [42], and fbsE, encoding a putative nonribosomal peptide synthase (NRPS) and the first part of the multi-modular unit required to assemble fimsbactins [43].
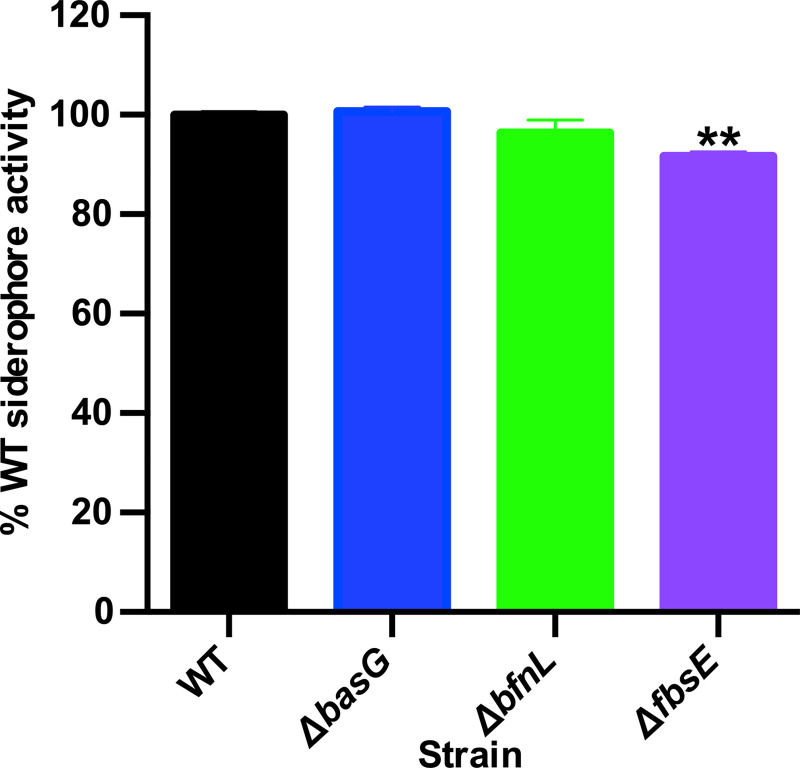
To determine if disruption to any one siderophore biosynthetic pathway impacts the ability of A. baumannii to chelate iron, WT A. baumannii and the strains inactivated for acinetobactin (ΔbasG), baumannoferrin (ΔbfnL), or fimsbactins (ΔfbsE) biosynthesis were grown in cTMS for 12 h and CAS assays were performed on the culture supernatants. Aside from a small but reproducible reduction in siderophore activity in the ΔfbsE mutant, inactivating one siderophore pathway in A. baumannii does not drastically impact total siderophore activity (Fig 4). These results suggest that functional redundancy exists between the siderophore-based iron chelation activities of A. baumannii in vitro.
Fig 4. Disrupting a single siderophore biosynthetic pathway does not drastically impact overall siderophore activity in A. baumannii.
Wild-type (WT) A. baumannii and its isogenic acinetobactin (ΔbasG), baumannoferrin (ΔbfnL) and fimsbactins (ΔfbsE) biosynthetic mutants were grown for 12 h in cTMS, and CAS assays to assess overall siderophore activity were performed on the spent culture supernatants. Total siderophore activity is expressed as the percent activity of WT A. baumannii, where data are the means of three biological replicates and are representative of three independent assays. ** p < 0.01, as determined by one-way analysis ANOVA with Dunnett’s multiple comparisons test.
Acinetobactin is required for maximal growth on host iron sources
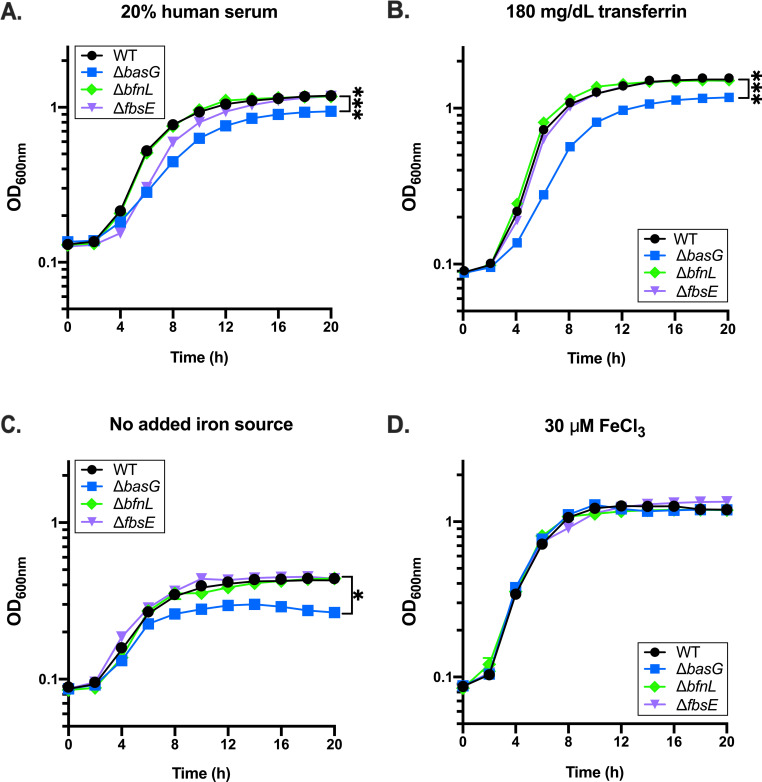
In addition to sequestering small amounts of free ferric iron from the extracellular milieu, siderophores are capable of pirating iron from host glycoproteins such as transferrin and lactoferrin. To determine if acinetobactin, baumannoferrin, or fimsbactins are essential for iron acquisition from host iron sources, WT A. baumannii and the three siderophore biosynthetic mutants, ΔbasG, ΔbfnL, and ΔfbsE, were grown in cTMS media with 20% human serum as the sole iron source. Bacterial growth was monitored by optical density (OD6oonm) over 20 hours. In contrast to the results of the CAS assay indicating that none of the mutations strongly impact overall siderophore production, the ΔbasG mutant was found to be impaired for growth in serum relative to WT A. baumannii and the ΔbfnL and ΔfbsE mutants (Fig 5A). The defect in growth of ΔbasG in serum is likely due to reduced acquisition of iron from transferrin, as this acinetobactin-deficient mutant is similarly impaired for growth on human transferrin as a sole iron source (Fig 5B). In addition to growing to a lower maximal OD6oonm than WT when grown under iron restriction (Fig 5 and S2 Fig), further analysis of the growth kinetics of the basG-deficient strain revealed that that it also exhibits a statistically significant decrease in growth rate, and an increase in lag time under these conditions (S2 Fig). Although the ΔfbsE mutant exhibits a slight lag and decrease in growth rate in serum, it ultimately reaches an OD6oonm comparable to WT and the ΔbfnL mutant (Fig 5A and S2 Fig). These effects were confirmed to be iron-dependent, as none of the strains are capable of robust growth in cTMS without an exogenous iron source (Fig 5C) but grow equivalently in the same media in the presence of excess free ferric iron (Fig 5D). The aforementioned phenotypes could be complemented by expression of basG in trans (S3 Fig), and are not specific to A. baumannii ATCC 17978, as similar results were observed for a basG-deficient strain of AB5075 (S4 Fig), a multidrug-resistant isolate that possesses the loci to express both acinetobactin and baumannoferrins [63]. Additionally, the basG-deficient strain also exhibited a similar growth defect when lactoferrin was provided as the sole iron source (S5 Fig). Together these findings show that iron is essential to the proliferation of A. baumannii, and that acinetobactin is required for maximal acquisition of iron from host sources such as serum transferrin, and lactoferrin.
Fig 5. Acinetobactin biosynthetic mutants are impaired for growth under iron restriction.
Wild-type (WT) A. baumannii and its isogenic acinetobactin (ΔbasG), baumannoferrin (ΔbfnL) and fimsbactins (ΔfbsE) biosynthetic mutants were grown in cTMS media with 20% human serum (A), 180 mg/dL human transferrin (B), no added iron source (C), or 30 μM FeCl3 (D). Bacterial growth was assessed by determining the optical density at 600 nm (OD600), at the time points indicated. Data are representative of three independent experiments, and error bars represent the standard error of the mean. Where error bars are not visible, they are shorter than the height of the symbol. Statistical analysis is given for the endpoint growth, as performed by repeated measures two-way ANOVA, where *p < 0.05 and *** p < 0.001. Additional analyses of growth kinetics for these data can be found in S2 Fig.
Expression of a single siderophore biosynthetic pathway is sufficient to promote WT siderophore activity
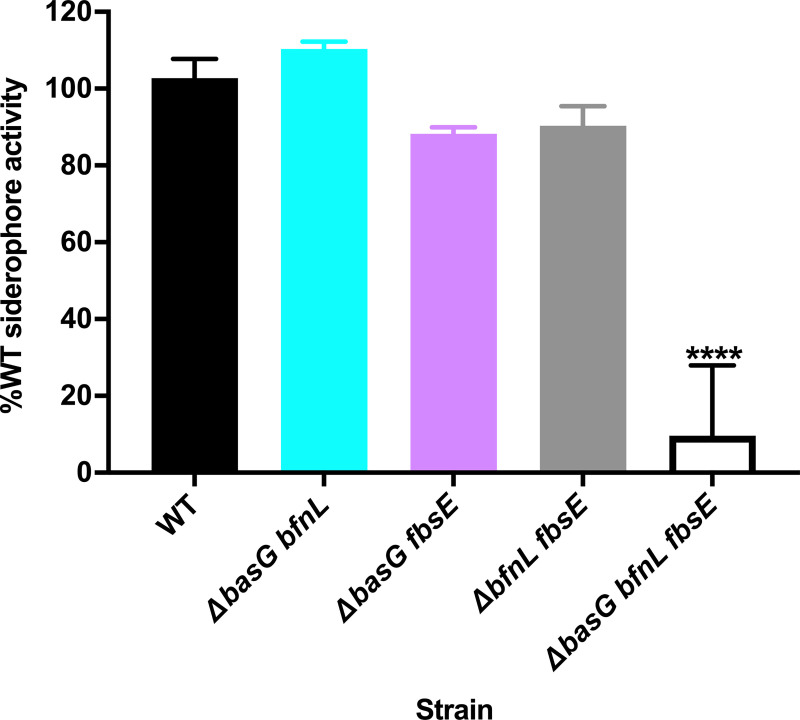
To ascertain the role of each individual siderophore in A. baumannii iron acquisition, it was necessary to generate strains with only one siderophore biosynthetic pathway intact. To this end, double mutants were generated, such that a panel of strains capable of synthesizing only acinetobactin (ΔbfnL fbsE), baumannoferrin (ΔbasG fbsE), or fimsbactins (ΔbasG bfnL) was available. To assess the overall role of siderophore production on A. baumannii growth and pathogenesis, a strain with disruptions in each of the three biosynthetic pathways (ΔbasG bfnL fbsE) was produced. With these strains in hand, the CAS assays were revisited, assessing overall siderophore activity in the culture supernatants of WT A. baumannii and its isogenic mutants grown in cTMS for 12 h. Interestingly, whilst the ΔbasG fbsE and ΔbfnL fbsE mutants exhibited a small but reproducible reduction in overall siderophore activity relative to the WT, none of the double mutants were drastically impaired overall (Fig 6). In fact, the ΔbasG bfnL mutant consistently exhibited increased CAS activity relative to the WT, suggesting that fimsbactins may be overexpressed in the absence of the other two siderophores, although the difference at 12 h was not statistically significant. We did find that genes within the fimsbactins-encoding locus are upregulated approximately 6.6 to 10-fold under the same conditions (S6 Fig), indicating that the loss of acinetobactin and baumannoferrin may induce further iron starvation in the bacteria leading to further transcriptional derepression of fimsbactins. In contrast to the robust siderophore production by the double mutants, when all three siderophore pathways were disrupted in the ΔbasG bfnL fbsE mutant, siderophore activity was severely attenuated (Fig 6). Overall, these findings demonstrate that acinetobactin, baumannoferrin, and fimsbactins appear to be the predominant iron chelators produced by A. baumannii, and that expression of just one of these siderophores is sufficient to confer robust iron chelating activity in vitro.
Fig 6. Disruption to all three siderophore biosynthetic pathways in A. baumannii is required to severely attenuate overall siderophore activity.
Wild-type (WT) A. baumannii and its isogenic combinatorial mutants were grown for 12 h in cTMS, and CAS assays to assess overall siderophore production were performed on the spent culture supernatants. Total siderophore activity is expressed as the percent activity of WT A. baumannii, where data are the means of three biological replicates and are representative of three independent assays. **** p < 0.0001, as determined by one-way ANOVA with Dunnett’s multiple comparisons test.
Siderophores are essential for A. baumannii growth when using serum transferrin as a sole iron source
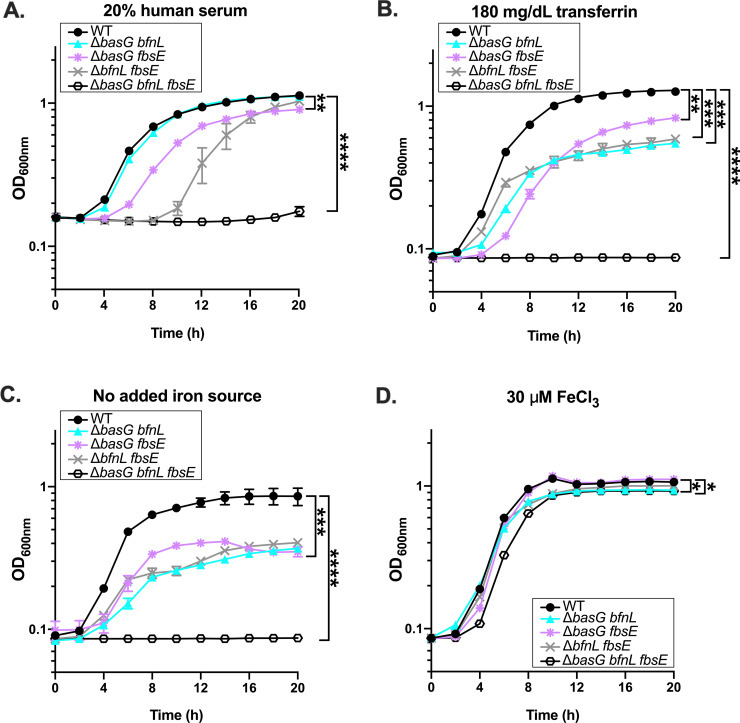
Given that the ΔbasG mutant alone was impaired for growth on serum and transferrin, we next sought to determine if more than just acinetobactin contributes to the acquisition of iron from these sources. To reveal the overall contribution of individual siderophores to A. baumannii growth under iron restriction, WT and the combinatorial mutant strains were grown in cTMS media with 20% human serum as the sole iron source. As previously described, growth was assessed by determining the OD600nm over 20 h. Under these conditions, it was observed that all three mutants were capable of growing on serum as a sole iron source (Fig 7A), with varying degrees of impairment relative to the WT. The strains expressing baumannoferrin (ΔbasG fbsE) or acinetobactin alone (ΔbfnL fbsE) exhibited a substantial growth lag relative to WT, while the strain expressing fimsbactins alone (ΔbasG bfnL) grew comparably (Fig 7A and S7 Fig). Although it is unclear how a strain expressing fimsbactins alone is capable of growing on serum as a sole iron source, possible explanations include increased expression of fimsbactins biosynthesis (S6 Fig) or the utilization of other weak chelators such as siderophore biosynthetic intermediates or shunt products (Fig 6). Notably, the complete siderophore mutant was abolished for growth on 20% serum as a sole iron source (Fig 7A). Similar results were observed when 180 mg/dL human transferrin was provided as the iron source (Fig 7B), except that the ΔbasG bfnL mutant grew poorly under these conditions, indicating that the fimsbactins or another chelator may scavenge residual iron from serum, but are less efficient at acquiring iron from unsaturated human transferrin. Again, all of the strains grew poorly in the absence of an iron source (Fig 7C), but growth was restored in the double mutants with the addition of exogenous iron (Fig 7D). Even with excess iron added, the siderophore-deficient strain exhibited a reproducible growth lag and reduced final biomass relative to WT (Fig 7 and S7 Fig), likely due to the absence of a high affinity means of facilitating iron uptake. Together these results highlight a high degree of functional redundancy between acinetobactin, baumannoferrins, and fimsbactins, and show that siderophore expression overall is essential to growth of A. baumannii on host iron-binding glycoproteins.
Fig 7. Siderophores are required for A. baumannii to utilize human serum and transferrin as iron sources to support growth.
Wild-type (WT) A. baumannii and its isogenic combinatorial mutants were grown in cTMS media with 20% human serum (A), 180 mg/dL human transferrin (B), no added iron source (C), or 30 μM FeCl3 (D). Bacterial growth was assessed by determining OD600nm, at the time points indicated. Data are representative of three independent experiments, and error bars represent the standard error of the mean. Statistical analysis is given for the endpoint growth, as performed by repeated measures two-way ANOVA, where *p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. Additional analyses of growth kinetics for these data can be found in S7 Fig.
Acinetobactin, baumannoferrin, and fimsbactins biosynthesis and transport genes are upregulated during infection
To begin dissecting the potential functional redundancy of acinetobactin, baumannoferrin, and fimsbactins in vivo, a murine model of systemic infection was employed to assess the expression of the siderophore biosynthesis and utilization pathways during infection using NanoString technology. NanoString is a digital, amplification-free means of quantifying targeted transcripts using color-coded molecular probes [64,65], and has been shown to reliably detect pathogen gene expression in complex samples [66,67]. In preparation for NanoString, female C57BL6/J mice were infected systemically with 2 x 108 to 5 x 108 colony forming units (CFUs) of WT A. baumannii by retroorbital injection. After 24 h, mice were humanely euthanized, and the organs were harvested. Following homogenization, RNA was extracted from the infected tissues, hybridized to the custom oligonucleotide probe set, and analyzed as per manufacturer instructions. As the in vitro comparator, RNA was similarly extracted and analyzed from bacteria grown in the same manner as used to prepare the inoculum for infection.
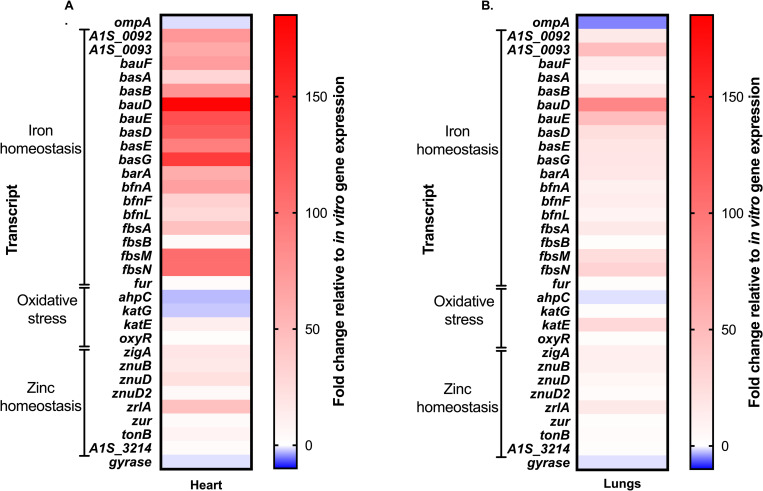
Consistent with the in vitro qRT-PCR results (Fig 2A), all three siderophore loci were found to be broadly upregulated in the heart (Fig 8A) and lungs (Fig 8B) of the A. baumannii infected host, relative to bacteria grown in vitro. Similar results were observed in the kidneys, liver, and spleen (S8 Fig). Although the overall magnitude of gene expression varied between organs, likely due to the bioavailability of iron to the pathogen, no other major differences were observed in the expression of the three siderophore loci. These results suggest that the siderophore pathways are upregulated overall in the iron-restricted host and that the presence of functionally redundant siderophores cannot be readily explained by differential expression to facilitate growth in specific host niches.
Fig 8. Siderophore biosynthetic and transport genes are upregulated in vivo.
Mice were systemically infected with WT A. baumannii and sacrificed at 24 h post-infection. Organs were harvested, RNA extracted, and gene expression changes relative to growth in vitro were determined in the heart (A) and lungs (B) using NanoString technology. Genes are clustered by known or predicted function, as indicated.
Acinetobactin biosynthesis is essential for the survival and proliferation of A. baumannii within the host
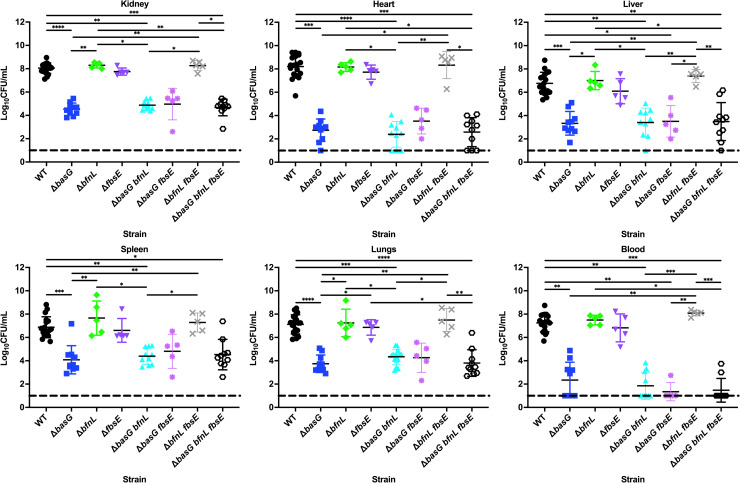
Since all three siderophore pathways were found to be upregulated during infection, it remained unclear as to what system, if any, is important to A. baumannii pathogenesis and whether the siderophores exhibit differential utility in specific host tissues. To elucidate the function of the individual siderophores in vivo, the same murine model of infection described above was employed, but instead mice were infected with WT A. baumannii or one of the single or combinatorial biosynthetic mutants. At 24 h post-infection the mice were humanely euthanized, the kidneys, hearts, livers, spleens, lungs, and blood were harvested, and the bacterial burdens of each were determined.
In support of the results demonstrating that acinetobactin is required for acquisition of iron from host sources such as serum transferrin and lactoferrin, the ΔbasG mutant was found to be severely attenuated in vivo. Relative to WT, the acinetobactin-deficient ΔbasG mutant had reduced bacterial burdens recovered from every organ and the blood, in the range of 2.8 to 5.0 log10 (Fig 9). Conversely, the ΔbfnL and ΔfbsE mutants did not exhibit a statistically significant reduction in burdens in any organ or in the blood relative to WT. Although the strain producing fimsbactins alone was capable of supporting growth of A. baumannii in serum (Fig 7A), neither this siderophore nor baumannoferrin were required for survival during A. baumannii bacteremia, as both ΔbasG bfnL and ΔbasG fbsE were recovered in similar numbers to the ΔbasG mutant alone. Conversely, the strain producing acinetobactin alone (ΔbfnL fbsE) was not statistically different from WT. Lastly, disrupting all three siderophore biosynthetic pathways in the ΔbasG bfnL fbsE mutant did not appreciably decrease the survival of the bacteria in vivo relative to ΔbasG. Together, these results suggest that acinetobactin alone is required for the survival and pathogenesis of A. baumannii during bacteremia, whilst the function of the baumannoferrins and fimsbactins in vivo remains unclear.
Fig 9. Acinetobactin biosynthetic mutant is severely attenuated for survival and proliferation within the host.
Mice were systemically infected with WT A. baumannii or its isogenic siderophore biosynthetic mutants, as indicated. After 24 h, mice were sacrificed and the bacterial burdens of the kidneys, heart, liver, spleen, lungs, and blood were determined by plating for viable cell counts on lysogeny agar. Each symbol represents the A. baumannii count in the corresponding organ of one animal. Data are compiled from three independent experiments. Statistical significance was determined by Kruskal-Wallis with Dunn’s multiple comparisons test, where *p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
Discussion
The expression of multiple, structurally distinct but functionally similar siderophores is a common feature of many bacteria [29]. For bacterial pathogens, the utilization of more than one siderophore class may fulfill a number of purposes, such as avoiding siderophore piracy by other bacteria and promoting competitiveness, facilitating iron acquisition under different physiological conditions and/or iron source composition, promoting uptake of metals other than iron, and most importantly, helping to evade nutritional immunity [29,68]. Although most A. baumannii clinical isolates possess the loci for acinetobactin and baumannoferrin biosynthesis and utilization, the individual contributions of these siderophores in vitro and in vivo have not been fully elucidated. Furthermore, although the synthesis pathway and structures of the fimsbactins have been characterized [43,55,56], their function in A. baumannii growth and survival has never been interrogated using strains that are specifically inactivated for fimsbactins biosynthesis. Here we investigate the function of the individual A. baumannii siderophores using a series of single and combinatorial biosynthetic mutants. We demonstrate that all three siderophore biosynthetic pathways contribute to iron chelation in vitro and functional redundancy exists in their ability to facilitate iron mobilization from host sources. However, acinetobactin is essential to the efficient use of iron from host iron-binding glycoproteins, and to the survival and dissemination of A. baumannii in a murine model of bacteremia. These findings support an often stated, but not fully substantiated assertion that acinetobactin is the major, or most important, siderophore produced by A. baumannii [44].
In the context of A. baumannii bacteremia, it remains unclear why acinetobactin biosynthesis is indispensable to colonization and survival, especially with the availability of multiple alternative mechanisms for iron acquisition. Acinetobactin has been proposed to function as a “two-for-one” siderophore allowing for iron chelation over an expanded pH range, with pre-acinetobactin predominating under acidic conditions, and acinetobactin under neutral and basic conditions [38]. Thus, the critical importance of acinetobactin may be explained by its ability to bind iron under different physiological conditions. As acinetobactin biosynthesis is required for efficient utilization of transferrin and lactoferrin as sole iron sources (Fig 5 and S5 Fig, respectively), it is also possible that this siderophore has a higher affinity for iron than the baumannoferrins and fimsbactins, and is therefore better able to appropriate iron from the host. However, whilst the affinity of the baumannoferrins for iron has not been elucidated, pre-acinetobactin binds iron with a comparable affinity to fimsbactins A (log KFe = 27.1 ± 0.2 versus 27.4 ± 0.2) and acinetobactin binds iron with a lower affinity (log KFe = 26.2 ± 0.2) [38,55], suggesting that iron affinity alone does not readily explain the importance of acinetobactin. Alternatively, ferric-acinetobactin may be captured and utilized more efficiently than the other two siderophores, supported partly by the observation that the receptor protein for acinetobactin (BauA) binds a heterotrimeric acinetobactin:preacinetobactin:Fe(III) complex or a preacinetobactin2:Fe(III) complex with nanomolar affinity prior to uptake of the siderophore [69]. The specificity and affinity of the predicted baumannoferrin (BfnH) and fimsbactins (FbsN) receptors have not been defined, and thus further research is required to determine if efficacy in siderophore transport contributes to the utility of the siderophores.
In addition to intrinsic factors controlling the uptake of siderophores, it is also known that during infection neutrophils release siderocalin (also known as neutrophil gelatinase associated lipocalin (NGAL), 24p3, and lipocalin-2), an acute phase protein that is capable of binding and sequestering extracellular ferric catecholate and carboxymycobactin-type siderophores, thus preventing their capture by invading pathogens [70–72]. To circumvent the effects of siderocalin, some bacteria secrete structurally modified siderophores that are incompatible with the binding site of the protein, and thus are referred to as “stealth siderophores” [73]. The interactions between A. baumannii siderophores and siderocalin has not been interrogated, but it is possible their redundancy may be explained by differing susceptibilities to sequestration by the vertebrate host. Indeed, hydroxamate siderophores such as baumannoferrins A and B are unlikely to be bound by siderocalin and thus would be free to facilitate iron acquisition in vivo, although our findings suggest that baumannoferrin expression is dispensable to pathogenesis in a murine bacteremia model (Fig 9). Notably, the baumannoferrins possess a lipophilic decanoic side chain, which may anchor them to the cell envelope [55,74] and potentially minimize their role in the capture of iron from the extracellular milieu. Unlike the baumannoferrins, the fimsbactins are mixed ligand, bis-catecholate monohydroxamate siderophores that are also reminiscent of tris-catecholate siderophores such as enterobactin and vibriobactin, known ligands of siderocalin [55,70,75,76]. Although not experimentally validated, the fimsbactins are predicted to bind siderocalin [55], which would effectively inactivate them in vivo and would be consistent with our findings that disruption to fimsbactins biosynthesis does not significantly impact the outcome of infection (Fig 9). Research by the Wencewicz group has revealed that apo-fimsbactins and holo-acinetobactin compete for the same periplasmic binding protein (BauB; Fig 1D), and hence are antagonistic in vitro [77]. Reductive release of iron from fimsbactins is thought to be facilitated by a putative periplasmic ferric iron reductase, FbsP, allowing for accumulation of the apo-siderophore in this compartment whilst free ferrous iron is transported into the cytoplasm by FeoABC (Fig 1D) [55]. It is possible that if the fimsbactins are sequestered by siderocalin in vivo that this competition is alleviated, and there may be a trade-off between an in vivo benefit and an in vitro detriment to the bacteria. However, further research is required to determine what, if any, interaction exists between the fimsbactins and siderocalin. Lastly, the essentiality of acinetobactin to A. baumannii pathogenesis suggests that this siderophore is capable of evading or overwhelming siderocalin to facilitate iron acquisition by the bacteria within the host.
In assessing the role of metals on the expression of siderophore biosynthetic clusters, our results are consistent with previous findings that the acinetobactin, baumannoferrins, and fimsbactins-encoding loci are all upregulated during iron restriction [31]. Zinc starvation could also induce expression of siderophore-associated genes (Fig 2), but these transcriptional changes were 10 to 1000-fold lower than that seen with iron. It is unclear if the influence of zinc on the expression of siderophore-associated transcripts is biologically relevant, and could be explained by similarities between the Fur and Zur box consensus sequences leading to accidental crosstalk between the two regulators [20,31]. Alternatively, if Fur binds structural zinc in A. baumannii as has been reported in other bacteria [78], prolonged zinc starvation could result in dissociation of the regulator from its cognate DNA, resulting in derepression of the locus. Whilst our results strongly support that iron limitation induces the expression of acinetobactin, baumannoferrin, and fimsbactins, the role for zinc remains unknown.
Consistent with a previous study demonstrating that all three siderophore biosynthetic clusters are upregulated in the blood [79], we found each locus to be upregulated in the heart, lungs, kidneys, liver and spleen of mice during A. baumannii bacteremia (Fig 8 and S8 Fig). Notably, we did not detect niche-dependent variation in transcription of the different siderophore loci in distinct organs, although by assessing expression in whole organ homogenates, these assays may not have had the resolution required to observe these differences. Additionally, it is possible that whilst the genes for baumannoferrin and fimsbactins biosynthesis were expressed, the siderophores were not produced. Recently, high-performance matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) was used to reveal that Staphylococcus aureus exhibits differential production of its siderophores, staphyloferrin A and staphyloferrin B, between abscesses within the same infected tissue [80]. Utilization of higher resolution techniques such as MALDI-IMS that incorporate spatial detection may help confirm not only the production and relative abundances of acinetobactin, baumannoferrins, and fimsbactins but may also be used to reveal niche-specific expression of the siderophores in vivo. Notably, in assessing total mass recovery following the in vitro purification of acinetobactin and fimsbactins, Bohac et al observed greater mass production of the former, whilst the baumannoferrins were not detected using the methods employed [55]. The aforementioned results suggest that there may indeed be differences in overall siderophore abundances that may help to explain why acinetobactin is critically important to infection. Whilst the use of mass spectrometry to identify and quantify the production of each of the siderophores was outside the scope of this study, use of these technologies represent powerful tools going forward.
Together, the conservation of the acinetobactin locus amongst clinical isolates of A. baumannii and essentiality to survival of the pathogen A. baumannii in vivo [31,46,51], even when other siderophores are expressed, leads us to propose that developing novel therapeutics that specifically target acinetobactin biosynthesis or transport may represent a viable strategy to combat this extensively drug-resistant pathogen. Several approaches exist for targeting iron homeostasis pathways in bacterial pathogens, but two main methods include developing vaccines against iron-regulated surface-exposed antigens and coopting iron acquisition systems to deliver a toxic payload to the bacterium [22]. Antibodies raised against iron-regulated TonB-dependent transporters (TBDTs) often function both to promote osponophagocytosis and to inhibit iron uptake [81], and indeed the receptor proteins for both acinetobactin and baumannoferrins, BauA and BfnH, respectively, have shown promise as antigens for vaccine development against A. baumannii [82–84]. Although the expression of bfnH was not specifically assessed by NanoString in this study, both bauA and fbsN are upregulated during infection, with the latter possessing some of the highest transcript abundances in most organs (Fig 8 and S8 Fig), again providing support for the use of TBDTs as vaccine antigens. A potential limitation to vaccination against A. baumannii, is that patients afflicted by the disease are often critically ill and immunocompromised and thus unable to mount a sufficient antibody response. Passive immunization, using monoclonal antibodies, therefore represents a potential option for protecting patients in high risk environments, such as intensive care units.
An alternative strategy for exploiting iron acquisition systems to the detriment of bacterial pathogens is to covalently link either a natural or synthetic siderophore to an antibiotic and utilize the siderophore uptake systems of the bacteria to help internalize this compound, known as a sideromycin. This “Trojan horse” approach to treating bacterial infections recently gained traction with the development of cefiderocol (Fetroja), a catechol-cephalosporin conjugate that recently became the first sideromycin-type antibiotic approved by the United States Food and Drug Administration (FDA) [85]. Indicated for the treatment of complicated urinary tract infections with few or no other treatment options, cefiderocol is active against Gram-negative, multidrug-resistant pathogens such as A. baumannii [86,87]. As with other sideromycins, cefiderocol is actively transported into the cell by coopting a siderophore-TBDT [88], and its minimum inhibitory concentration decreases under iron restriction [86]. Additionally, fimsbactins A and fimsbactins-like analogues conjugated with daptomycin or cephalosporins are highly effective and selective in killing A. baumannii in vitro, showing that antibiotics typically reserved for Gram-positives may be repurposed by using a siderophore delivery mechanism to gain access to the periplasm of Gram-negative bacteria [89,90]. These findings highlight that sideromycins can be effective as antibiotics against A. baumannii. Importantly, the essential features required for iron coordination by, and cellular uptake of acinetobactin have been identified, along with a promising sites for the conjugation of an antibiotic [38,44,91]. Interestingly, in assessing the development of resistance to another experimental sideromycin (BAL30072), A. baumannii developed mutations in genes encoding the TonB machinery required to energize siderophore uptake systems, tonB3 and exbD3 [33,88]. Although these mutants had decreased sensitivity to the sideromycin, the tonB3 and exbD3 mutants exhibited decreased growth under iron restriction, suggesting that should these mutations evolve in the iron-restricted host, the bacteria may still be compromised for survival [88]. Indeed, Runci et al. have found that tonB3 is required for maximal virulence of A. baumannii ATCC 19606 in insect and murine models of infection [33].
The results of this study have revealed that acinetobactin is essential for iron acquisition in vivo, whereas other siderophores, the baumannoferrins and fimsbactins, are dispensable. Moreover, we have demonstrated that the genes for acinetobactin biosynthesis and transport are broadly upregulated in a systemic murine infection model. Collectively these data, coupled with the observation that the acinetobactin locus is highly conserved amongst clinical isolates, presents the targeting of acinetobactin biosynthesis or transport as an attractive candidate for the development of novel therapeutics to combat multidrug-resistant A. baumannii.
Materials and methods
Bacterial strains and growth conditions
All bacterial strains and plasmids employed in this study can be found in Table 1. Unless otherwise indicated, experiments were performed with A. baumannii strain ATCC 17978 and its isogenic mutants. For routine cultivation and genetic manipulation, A. baumannii was cultured in lysogeny broth (LB) or on LB with 1.5% w/v agar (LBA). Antibiotics, when required for selection of recombinants or maintenance of plasmids in A. baumannii, were supplied at the following concentrations: ampicillin, 500 μg/mL; carbenicillin, 75 μg/mL; kanamycin, 15 μg/mL; sulfamethoxazole, 100 μg/mL; as indicated. For selection and maintenance of plasmids in E. coli, 100 μg/mL of ampicillin was used. Plasmid gene expression was induced using 1–2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), as detailed in the text.
Table 1. Strains and plasmids employed in this study.
| Strain | Description | Reference |
|---|---|---|
| A. baumannii ATCC 17978 | WT A. baumannii fatal meningitis isolate from 1951 | American Type Culture Collection (ATCC) strain 5377 [54] |
| WT/pWH1266 | WT A. baumannii ATCC 17978 with empty pWH1266 plasmid | This study |
| A. baumannii AB5075-UW | Multidrug-resistant WT A. baumannii AB5075 isolate from a wound infection | [92,93] |
| ΔbasG | Acinetobactin biosynthetic mutant where basG (A1S_2379) was replaced by a kanamycin resistance determinant, and then the cassette was excised, to leave a markerless mutant | This study |
| basG102::Tn26 | Transposon mutant of basG in A. baumannii AB5075-UW from three-allele mutant library; identifier tnab1_kr121128p06q102 | [92] |
| basG142::Tn26 | Transposon mutant of basG in A. baumannii AB5075-UW from three-allele mutant library; identifier tnab1_kr130909p02q142 |
[92] |
| ΔbasG/pWH1266::basG | basG mutant complemented with pWH1266 expressing basG from its native promoter | This study |
| ΔbfnL | Baumannoferrin biosynthetic mutant where bfnL (A1S_1657) was replaced by a kanamycin resistance determinant, and then the cassette was excised, to leave a markerless mutant | This study |
| ΔfbsE | Fimsbactins biosynthetic mutant where fbsE (A1S_2578) was replaced by a kanamycin resistance determinant; KanR | This study |
| ΔbasG bfnL | Acinetobactin and baumannoferrin biosynthetic mutant. Proficient in fimsbactins biosynthesis; KanR | This study |
| ΔbasG fbsE | Acinetobactin and fimsbactins biosynthetic mutant. Proficient in baumannoferrin biosynthesis; KanR | This study |
| ΔbfnL fbsE | Baumannoferrin and fimsbactins biosynthetic mutant. Proficient in acinetobactin biosynthesis; KanR | This study |
| ΔbasG bfnL fbsE | Complete siderophore biosynthetic mutant with disruptions in acinetobactin, baumannoferrin, and fimsbactins production; KanR | This study |
| Δzur | Zur mutant; KanR | [58] |
| Plasmid | ||
| pKD4 | Template for amplification of the FRT-flanked kanamycin resistance determinant; KanR, AmpR | [94] |
| pAT02 | A. baumannii recombinase expressing plasmid; CarbR/AmpR | [61] |
| pAT03 | Expresses an IPTG-inducible flipase to remove the kanamycin resistance determinant; CarbR/AmpR | [61] |
| pWH1266 | A. baumannii/E.coli shuttle and complementation vector; CarbR/AmpR | [95] |
| pWH1266::basG | pWH1266 expressing basG from its native promoter; CarbR/AmpR | This study |
For bacterial growth under metal-restriction, media were prepared in sterile polypropylene vessels or in glassware that was washed overnight with 0.1 M HCl and then subsequently rinsed extensively with ultrapure water to remove contaminating iron. Media and all components were prepared using water purified with a Nanopure Diamond filtration system (Thermo Scientific). When required, base media and additives were Chelex-treated using 5% w/v Chelex-100 resin (Sigma-Aldrich) stirring overnight at 4°C. Tris Minimal Succinate (TMS) media were employed in zinc and iron-restriction studies, prepared as previously described [96]. For growth curves, TMS was treated with Chelex-100 (cTMS) to remove residual iron.
RNA extraction and quantitative reverse transcription PCR
To assess in vitro gene expression, WT A. baumannii or its isogenic mutants were grown overnight in TMS media prior to subculturing 1:50 in 10 mL cTMS media with or without the addition of 30 μM ZnCl2 or FeCl3. Bacteria were grown until exponential (4 h) or stationary phase (12 h), pelleted by centrifugation, and siderophore activity was confirmed in metal-deplete culture supernatants (see “Determination of siderophore production by Chrome Azurol S assay”). Bacterial pellets were stored in a 1:1 solution of acetone and ethanol at -80°C until RNA extraction. Prior to extraction, the acetate:ethanol was removed by aspiration following centrifugation for 15 min at maximum speed. The bacterial pellets were resuspended in 750 μL of LETS buffer (0.1 M LiCl, 10 mM EDTA, 10 mM Tris HCl (pH 7.4), 1% sodium dodecyl sulphate) and lysed in Lysing Matrix B tubes using a FastPrep-24 bead beater (MP Biomedicals) with two 45 s pulses at 6 M/s separated by a 5-min rest. Following incubation at 55°C for 5 min, the samples were centrifuged at 21,130 x g for 10 min and then transferred to new 2 mL RNAase-free tubes. RNA was solubilized by mixing lysates with 1 mL TRI reagent (Sigma-Aldrich) and incubated at room temperature for 5 min. To each sample, 0.2 mL of chloroform was added and mixed by rapid inversion for 15 s. Following incubation at room temperature for 2–3 min, the samples were centrifuged at 21,130 x g for 10 min, and the upper aqueous phase was transferred to a new tube and mixed with 1 mL of isopropanol. RNA was precipitated at room temperature for 10 min, pelleted by centrifugation, washed with 200 μL of 70% ethanol, and dried at room temperature for 1 min. The RNA pellet was resuspended in 100 μL of nuclease-free water and treated with RNA Qualified (RQ1) RNase-Free DNase for 2 h at 37°C (Promega). RNA was then purified further using an RNeasy miniprep kit, as per instructions from the manufacturer (Qiagen). RNA quality and quantity were assessed using a NanoDrop 8000 Spectrophotometer (Fisher Scientific).
cDNA was synthesized from 1 μg RNA using random primers and M-MLV reverse transcriptase, using instructions from the supplier (Promega). The cDNA was diluted 1:50 and used in quantitative reverse transcription PCR (qRT-PCR) using the primer pairs denoted in Table 2 and iQ SYBR Green Supermix, as directed (Bio-Rad). qRT-PCR was performed using a two-step melt curve program on a CFX96 Touch Real-Time PCR detection system (Bio-Rad). Target gene expression was normalized to the expression of rpoB and presented as the fold change (2-ΔΔCT) relative to expression under metal-replete conditions, or to WT when gene expression was assessed in the Δzur or ΔbasG bfnL mutants.
Table 2. Primers employed in this study.
| Designation | a,bSequence | Function |
|---|---|---|
| basE-F RT | CGCGCATTTTAACGGTAGGG | basE qRT-PCR |
| basE-R RT | CCCTTGCCCAAGCCTTTTTC | basE qRT-PCR |
| basD-F RT | GGCAACGCAACTTTAGGTGG | basD qRT-PCR |
| basD-R RT | AGCCATGATGTTTGCGATGC | basD qRT-PCR |
| bauD-F RT | GTATCGGGTTGGCGGTATGT | bauD qRT-PCR |
| bauD-R RT | ATCATCTTGCTCAGCGTGCT | bauD qRT-PCR |
| basB-F RT | CAAAATGCCAAAGGTCGCCA | basB qRT-PCR |
| basB-R RT | GCTTTCCAGTTTGGGGCTTG | basB qRT-PCR |
| basA-F RT | TATGGATTCTCCGCCATCGC | basA qRT-PCR |
| basA-R RT | AGCCGGACGTCTGTTGATTT | basA qRT-PCR |
| bauF-F RT | ATTGTCTCGATCCAGACGCC | bauF qRT-PCR |
| bauF-R RT | TTGCCAAATTGGGAGCTTGC | bauF qRT-PCR |
| bfnA-F RT | TCGGTTTACGTGGATGGCAA | bfnA qRT-PCR |
| bfnA-R RT | TCGCCACGATGACACTCTTT | bfnA qRT-PCR |
| bfnF-F RT | AGTCAAAAGCTGCCGAAAGT | bfnF qRT-PCR |
| bfnF-R RT | ACGGCATAAGGTCTGCTCAG | bfnF qRT-PCR |
| bfnL-F RT | TACACCGCTGGATGCATGAG | bfnL qRT-PCR |
| bfnL-R RT | AATTTCGGCATACCCCACGT | bfnL qRT-PCR |
| fbsA-F RT | TCACCTCTCCCCAACCATCA | fbsA qRT-PCR |
| fbsA-R RT | GGCGACTCAGCACTCATCTT | fbsA qRT-PCR |
| fbsB-F RT | AGCCTTGGCATTGTCTCTCC | fbsB qRT-PCR |
| fbsB-R RT | ATCCATCCACCCGACCAAAC | fbsB qRT-PCR |
| fbsM-F RT | GTGTTCCCTCTGCAGGTAGC | fbsM qRT-PCR |
| fbsM-R RT | CTTTCGTCCATGACCAGGGT | fbsM qRT-PCR |
| fbsN-F RT | ACCGTTATTTGGGTTCGGCT | fbsN qRT-PCR |
| fbsN-R RT | AATCGCACCACGTGTTTTGG | fbsN qRT-PCR |
| basG-F RT | AGCGCAAATCGGAATCATGC | basG qRT-PCR |
| basG-R RT | TGGCCAGACACACAAATCGA | basG qRT-PCR |
| fur-F RT | TGCGCAAAGCTGGACTTAAA | fur qRT-PCR |
| fur-R RT | TCGCAAGTCCGACATCTTCC | fur qRT-PCR |
| zur-F RT | GTCACCCACGTGAAGGTCAT | zur qRT-PCR |
| zur-R RT | CTGTGTTGTGCTGCGAAATC | zur qRT-PCR |
| zigA-F RT | GATCAGGCTCAGCAAGACCAG | zigA qRT-PCR |
| zigA-R RT | GTGCTTGGACAGCTTCATCA | zigA qRT-PCR |
| rpoB-F RT | ATGCCGCCTGAAAAAGTAAC | rpoB qRT-PCR (housekeeping) |
| rpoB-R RT | TCCGCACGTAAAGTAGGAAC | rpoB qRT-PCR (housekeeping) |
| gyrA-F RT | GACGACGGTACCGGTTTACA | gyrA qRT-PCR (housekeeping) |
| gyrA-R RT | ACCGCGGCCATTATCTGAAA | gyrA qRT-PCR (housekeeping) |
| basG-F | CAGGGTAGAGGGTTGCCATCATAAGGCATACACAAATCCTGGTCGCTAAATTTATACATAAAATTTATATCTTTGATAGCGACTCCTTAGACGACTTGTAATCGTCATATTAACGGAGCAAAAAGTGTAGGCTGGAGCTGCTTC | Recombineering to replace basG with KmR |
| basG-R | AGTATTTTTTTAAAAACTTTAATTGCATCAAAAACTTCCTAACCACCCTATCAAAATATTTTTAGATCATTTTCTAGACTGTAAAAATTAATATGGATTTGTCTAAATCATCTTGGTTGATATGACATATGAATATC CTCCTTAG | Recombineering to replace basG with KmR |
| Full basG-F | GTATCTTCACGTTGCGGTCAGGTC | Checking recombineering of basG |
| Full basG-R | AATGCTGGCTTGAGTGCAGGT | Checking recombineering of basG |
| bfnL-F | GGGGAAAGCTGAATTTTTTGTCCATGATTTATGCAATAAAAAAAATAGTAAAAATAAGCTTGCATCTTAAATAAAAATGATTATCATTATCAATTATAGGTTATGTCATAGCAAGGACCGTTATAGTGTAGGCTGGAGCTGCTTC | Recombineering to replace bfnL with KmR |
| bfnL-R | AACTCCTGCCCAATGAGTTCTTGAGATGCCTGTTGCAATGTTTGAAACCGCAACCGTTCAGTATCAAACATTGTCCCATCCATATCGAAGATAGCTCCATGAACAGGTTTTCCATGAAAAATAAGCATATGAATATCCTCCTTAG | Recombineering to replace bfnL with KmR |
| Full bfnL-F | TGTCGATCTGGCGCTCATAC | Checking recombineering of bfnL |
| Full bfnL-R | GGTTCCTGTATCTCTGGCGT | Checking recombineering of bfnL |
| fbsE-F | ATGCAATTACAACAAGAAATTACCGCAGACATTCACCATGTTCTGCAAATTGATAGCATTCAAATTCAACCTGAAGATAATTTGATTGAACATGGGCTGCATTCATTGGCGATCATGCAGTTAGTGTGTAGGCTGGAGCTGCTTC | Recombineering to replace fbsE with KmR |
| fbsE-R | ATCACTTGCATCAATTGCATTTCATCGATGGTGGTACGTAATGCAGGATATAATGCAATCAATTGAGTCAAACGCTGAGTCAGTGTTTCAACATCCATTCGCCCATGAAATTCTTGAAAGTCATGCATATGAATATCCTCCTTAG | Recombineering to replace fbsE with KmR |
| Full fbsE-F | AACTGTGGGGTTGGAACTCG | Checking recombineering of fbsE |
| Full fbsE-R | TGAATGGTCGCTTCCATGCT | Checking recombineering of fbsE |
| basG Gibson-F | gcgaccacacccgtcctgtgTTGGACTCATTACGAATTATG | Complementation of basG |
| basG Gibson-R | aaggctctcaagggcatcggCTAAAAGCCAACTGTACG | Complementation of basG |
aUnderlined areas denote regions of homology with the kanamycin resistance determinant
bLower case letters denote regions of homology with pWH1266
Generation of A. baumannii siderophore biosynthetic mutants
Oligonucleotides used in generating, confirming, or complementing the siderophore biosynthetic mutants detailed below can be found in Table 2. All mutants were generated using recombineering, as previously described [61]. In brief, the FRT-flanked kanamycin resistance determinant of pKD4 [94] was amplified using primers bearing 120 bp of homology to the region flanking the gene of interest (basG, bfnL, or fbsE, using primers basG-F/R, bfnL-F/R, and fbsE-F/R, respectively). The PCR product was purified and subsequently concentrated to ~1–2 μg/μL. Approximately 5 μg of linear recombineering product was introduced to 100 μL (~108 colony forming units (CFU)) of competent A. baumannii containing the recombinase-expressing plasmid pAT02 [61] by electroporation at 1800 V. Cells were recovered immediately in pre-warmed LB, and recombinase expression was induced through the addition of 2 mM of IPTG to the media. After 4 h of growth at 37°C, the transformants were plated to LBA with kanamycin. The putative ΔbasG::km, ΔbfnL::km or ΔfbsE::km colonies that arose on the transformation plates were screened by PCR using primer pairs Full basG-F/R, Full bfnL-F/R, or Full fbsE-F/R, respectively, each of which flank the site of recombination. Colonies were screened for the loss of pAT02 through patching to LBA supplemented with carbenicillin, and maintenance of the native plasmid pAB3 was confirmed through resistance to sulfamethoxazole [97]. The basG mutants obtained from the A. baumannii AB5075-UW three-allele mutant library [92] were confirmed through PCR using primer pairs Full basG-F/R.
In order to generate combinatorial mutants, it was necessary to excise the kanamycin cassette from the single siderophore biosynthetic mutants to yield unmarked mutations. The strains ΔbasG::km and ΔbfnL::km were transformed with the Flp recombinase plasmid, pAT03 [61]. Expression of the flippase was induced by growth of ΔbasG::km/pAT03 and ΔbfnL::km/pAT03 in LB with 1 mM IPTG for 4 h. The cells were harvested by centrifugation, plated to LBA containing 1 mM IPTG, and incubated overnight at 37°C. Biomass was scraped from the induction plates and streaked for isolated colonies on LBA without antibiotics. Colonies were screened for excision of the kanamycin cassette and simultaneous loss of the pAT03 plasmid through identifying isolates that were both kanamycin and carbenicillin sensitive. Excision of the kanamycin cassette was confirmed through colony PCR using primer pairs Full basG-F/R or Full bfnL-F/R.
Upon generation of the markerless strains, combinatorial mutants were generated in the ΔbasG and ΔbfnL backgrounds. The procedure was performed as described above, except that the linear DNA fragment bearing homology to fbsE was introduced to both ΔbasG/pAT02 and ΔbfnL/pAT02, and the linear DNA fragment bearing homology to bfnL was also introduced to ΔbasG/pAT02. Screening and confirmation of the ΔbasG bfnL::km, ΔbasG fbsE::km, and ΔbfnL fbsE::km mutants was performed as detailed above. A complete siderophore biosynthetic mutant was generated by excising the kanamycin cassette from ΔbasG bfnL::km, and subsequently disrupting fbsE. All mutations were confirmed through PCR and maintenance of pAB3 was assured through resistance to sulfamethoxazole.
Complementation of basG
Complementation of basG was performed in trans using the vector pWH1266 [95]. The basG gene and its native promoter were amplified using the primers basG Gibson-F/R (Table 2). pWH1266 was digested with BamHI and SalI and the PCR product and plasmid were joined using NEB Builder HiFi Assembly (New England BioLabs) at 50°C for 1 h. The resulting assembly was transformed into chemically competent DH5α by heat shock, and successful transformants were selected for on LBA with ampicillin following overnight growth at 37°C. The pWH1266::basG construct was confirmed by PCR and sequencing prior to introduction of both this vector, as well as the native empty vector, to ΔbasG by electroporation. Successful transformants were selected on LBA with carbenicillin.
Determination of siderophore production by Chrome Azurol S assay
To assess for overall siderophore production, Chrome Azurol S (CAS) assays were performed. In preparation for the assays, bacteria were grown in biological triplicate overnight in TMS media, pelleted by centrifugation at 1073 x g and washed three times with 1X phosphate buffered saline (PBS). The OD600 of each resuspension was adjusted to 1.0 and used to inoculate 10 mL of cTMS media to an OD600 of 0.005 in 50 mL conical tubes. The cultures were grown for 12 h at 37°C with shaking at 180 rpm. Following growth, the OD600nm of each culture was determined and bacteria were pelleted by centrifugation. The supernatants were removed and sterilized using a 0.2 μm filter prior to use in the CAS assays.
CAS reagent was prepared, and the assays performed, essentially as described previously [59,60]. In brief, CAS is an iron-binding dye complex that detects mobilization of iron from the dye to a chelator through a colorimetric change from blue (negative reaction) to pink/orange (positive reaction) and/or through a decrease in absorbance at 630 nm (A630). Prepared CAS reagent was mixed 1:1 with the filtered culture supernatants and incubated in the dark at room temperature for 30 min. The A630 was determined, each value was normalized to the final OD600nm of the corresponding culture, and siderophore activity was expressed as a percentage of WT A. baumannii.
Assessing the ability of A. baumannii to utilize host iron sources under iron-restricted growth
To assess growth of A. baumannii WT and the siderophore biosynthetic mutants under iron-restriction, single-isolated colonies of bacteria grown on LBA were used to inoculate 3 mL of TMS media and incubated overnight at 37°C with shaking at 180 rpm. The overnight cultures were pelleted by centrifugation at 604 x g for 15 min and washed once with sterile 1x PBS. The pellets were resuspended in PBS and subcultured 1:100 in cTMS media. Cultures were grown to an OD600nm of ~1, pelleted by centrifugation, washed three times with sterile 1 x PBS and resuspended to an OD600nm of 0.1. Growth curves were set-up in cTMS in 96-well plates using 20% v/v heat inactivated and filter sterilized human serum (Sigma-Aldrich; H4522), 180 mg/dL partially saturated human transferrin (Sigma-Aldrich; T3309), or 15 mg/mL lactoferrin from human milk (Sigma-Aldrich; L0520). Concentrations were selected to be physiologically relevant and to support growth of WT A. baumannii in the absence of another iron source. For zinc and iron-replete conditions, 30 μM of ZnCl2 or FeCl3 were added, respectively. Carbenicillin was added when required to maintain pWH1266 or its derivatives. Plates were inoculated using 5 μL of the prepared cells in 200 μL of media and grown in an Epoch 2 microplate reader (BioTek) at 37°C with medium amplitude linear shaking. Cell density was assessed by OD600nm every 30 min, but for graphical clarity only data collected at 4 h intervals is shown. Estimations of the asymptote of the maximal OD600nm (A), maximum specific growth rate (μm) and lag time (λ) were modeled using single Gompertz growth curve fitting and open source code available at https://scott-h-saunders.shinyapps.io/gompertz_fitting_0v2/ [98–100].
Murine model of A. baumannii bacteremia
All infection experiments were approved by the Vanderbilt University Institutional Animal Care and Use Committee and are in compliance with guidelines set by the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association. Six-to-eight- week old female C57BL/6J mice (Jackson Laboratories) were injected retro-orbitally with 100 μL of bacterial cell suspensions containing ~2 x 108 to 5 x 108 colony forming units (CFU) of WT A. baumannii or its isogenic mutants in PBS. In preparation for infection, overnight cultures of bacteria were subcultured 1:100 in fresh LB and grown to mid-to-late log phase (OD600 = 2.0–2.5) at 37°C with shaking. The bacteria were pelleted by centrifugation, washed twice with PBS, and normalized to an OD6oonm of 0.35 at a 1:20 dilution. Following injection, the mice were humanely euthanized at 24 h, and the kidneys, heart, liver, spleen, lungs, and blood of each mouse were harvested. The organs were homogenized in PBS using a Bullet Blender (Next Advance), and all samples were serially diluted in PBS prior to plating on LBA to determine the bacterial burdens.
RNA extraction and in vivo gene expression analysis by NanoString
For assessing in vivo gene expression, mice were infected as detailed above. At 24 h, mice were humanely euthanized, and the harvested organs were placed in RNAase-free navy bullet blender tubes (Next Advance) containing 600 μL of RLT buffer (Qiagen) with 1% (v/v) β-mercaptoethanol. Following homogenization in the bullet blender for 2 x 5 min (speed 8 for livers, speed 10 for all other organs; Next Advance), the homogenate was transferred to a fresh bullet blender tube containing 600 μL of phenol:chloroform:isoamyl alcohol (25:24:1, Fisher Scientific). After an additional 5 min homogenization cycle, the samples were centrifuged at 21,139 x g for 10 min. The upper aqueous phase was transferred to a 2-mL nuclease-free tube containing 600 μL of 70% ethanol and inverted until a visible mass of RNA formed. The extracted RNA was subsequently purified using an RNeasy kit (Qiagen), as per manufacturer’s instructions. As an in vitro comparator, WT A. baumannii was grown in LB using the same methodology used to prepare for infection (see section “Murine model of A. baumannii bacteremia”). RNA was extracted from in vitro grown bacteria using the method described in “RNA extraction and quantitative reverse transcription PCR”. RNA quality and concentration were determined using a NanoDrop 8000 Spectrophotometer (Fisher Scientific).
In preparation for NanoString, 100 ng of RNA in 5 μL of nuclease-free water was aliquoted to 12-well tube strips and mixed with 8 μL of the Reporter Probe Set in hybridization buffer (NanoString). To each well, 2 μL of the Capture Probe Set (NanoString) were added and the samples were hybridized at 65°C in a preheated thermocycler for 18 h. Following hybridization, samples were processed by Vanderbilt Technologies for Advanced Genomics (VANTAGE) on an nCounter FLEX analysis system (NanoString). Data were analyzed using nSolver Analysis software (NanoString), where A. baumannii gene expression was normalized to the expression of housekeeping genes rpsA. Using negative control probes included by the manufacturer in the Reporter Probe Set and designed not to interact with biological samples (NanoString), the minimum threshold for gene expression was determined and all target genes included in the analysis exhibited expression above background. Expression changes are given as the fold change of in vivo A. baumannii gene expression versus gene expression in in vitro grown bacteria.
Supporting information
(DOCX)
(DOCX)
(DOCX)
WT A. baumannii and its isogenic Δzur mutant were grown in metal-restricted media for 12 h. RNA was extracted and transcriptional changes in the expression of siderophore-associated genes and fur were assessed by qRT-PCR and normalized to the expression of rpoB. Expression of a known zur-regulated gene, zigA, was assessed as a positive control, whereas zur was run as a negative control. * p < 0.05, *** p < 0.001, and **** p < 0.0001 as determined by Student’s t test relative to a hypothetical value of 1. Data are representative of two experiments performed in biological quadruplicate.
(TIFF)
Growth kinetics of WT A. baumannii ATCC 17978 and its isogenic acinetobactin (ΔbasG), baumannoferrin (ΔbfnL) and fimsbactins (ΔfbsE) biosynthetic mutants were analyzed from the data presented in Fig 5. Estimates of the maximal OD600 (asymptote (A) A-D), maximum specific growth rate (μm, E-H) and lag time (λ, I-L) are given where *p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
(TIFF)
WT A. baumannii ATCC 17978 with empty vector (WT/pWH1266), the acinetobactin-deficient mutant with empty vector (ΔbasG/pWH1266), and a mutant complemented with basG expressed from pWH1266 (ΔbasG/pWH1266::basG) were grown in cTMS media with human serum added at the concentrations indicated. Bacterial growth was assessed by determining the optical density at 600 nm (OD600nm) at the timepoints indicated. Data are the average of technical triplicates and represent the results of two independent experiments.
(TIFF)
WT A. baumannii AB5075-UW and two unique basG transposon mutants were grown in cTMS media with human serum added at the concentration indicated. Bacterial growth was assessed by determining the OD600nm at the timepoints indicated. Data are the average of technical triplicates and represent the results of two independent experiments. Where error bars are not visible, they are shorter than the height of the symbol.
(TIFF)
Wild-type (WT) A. baumannii and its isogenic acinetobactin (ΔbasG), baumannoferrin (ΔbfnL) and fimsbactins (ΔfbsE) biosynthetic mutants were grown in cTMS media with lactoferrin, no added iron source, or 30 μM FeCl3, as indicated. Bacterial growth was assessed by determining the OD600nm, at the time points indicated. Data are representative of two independent experiments, and error bars represent the standard error of the mean. Where error bars are not visible, they are shorter than the height of the symbol.
(TIFF)
WT A. baumannii and its isogenic ΔbasG bfnL mutant were grown in metal-restricted media for 12 h. RNA was extracted and transcriptional changes in genes of the fimsbactins locus were assessed by qRT-PCR and normalized to the expression of rpoB. Expression of bfnL was used as a negative control. * p < 0.05, ** p < 0.01 and **** p < 0.0001, as determined by Student’s t test relative to a hypothetical value of 1. Data are representative of two experiments performed in biological quadruplicate.
(TIFF)
Growth kinetics of WT A. baumannii ATCC 17978 and its isogenic siderophore biosynthetic mutants, as indicated, were analyzed from the data presented in Fig 7. Estimates of the maximal OD600 (asymptote (A) A-D), maximum specific growth rate (μm, E-H) and lag time (λ, I-L) are given for the conditions listed where are given where *p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. When growth was insufficient to calculate the parameter, no data is given (ND).
(TIFF)
Organs were harvested, RNA extracted, and gene expression changes relative to growth in vitro were determined in the kidney (A), liver (B), and spleen (C) using NanoString technology. Genes are clustered by known or predicted function, as indicated.
(TIFF)
Acknowledgments
The authors thank members of the Skaar Lab for critical review of the manuscript. We also thank Zachery R. Lonergan for construction of the ΔbasG::km mutant and Dr. Luis Actis for providing reagents and expertise required to molecularly manipulate A. baumannii.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship to JRS (MFE-171179) and National Institutes of Health awards R01 AI069233, R01 Al073843, R01 AI101171, R01 AI138581 to EPS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin Microbiol Rev. 2017;30: 409–447. 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dexter C, Murray GL, Paulsen IT, Peleg AY. Community-acquired Acinetobacter baumannii: Clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther. 2015;13: 567–573. 10.1586/14787210.2015.1025055 [DOI] [PubMed] [Google Scholar]
- 3.Serota DP, Sexton ME, Kraft CS, Palacio F. Severe community-acquired pneumonia due to Acinetobacter baumannii in North America: Case report and review of the literature. Open Forum Infect Dis. 2018;5: ofy044 10.1093/ofid/ofy044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas ME, Karveli EA, Kelesidis I, Kelesidis T. Community-acquired Acinetobacter infections. Eur J Clin Microbiol Infect Dis. 2007;26: 857–868. 10.1007/s10096-007-0365-6 [DOI] [PubMed] [Google Scholar]
- 5.Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genomics. 2019;5: e000306 10.1099/mgen.0.000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5: 939–51. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- 7.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51: 3471–3484. 10.1128/AAC.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark NM, Zhanel GG, Lynch JP. Emergence of antimicrobial resistance among Acinetobacter species: A global threat. Curr Opin Crit Care. 2016;22: 491–499. 10.1097/MCC.0000000000000337 [DOI] [PubMed] [Google Scholar]
- 9.Press WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: World Health Organization; 2013. 10.1590/S0100-15742013000100018 [DOI] [Google Scholar]
- 10.CDC. Antibiotic resistance threats in the United States. Atlanta, Georgia; 2019. Available: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
- 11.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16: 91–102. 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wendt C, Dietze B, Dietz E, Rüden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol. 1997;35: 1394–1397. 10.1128/JCM.35.6.1394-1397.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrow JM, Wells G, Pesci EC. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS One. 2018;13 10.1371/journal.pone.0205638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: Comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998;36: 1938–1941. 10.1128/JCM.36.7.1938-1941.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green ER, Juttukonda LJ, Skaar EP. The manganese-responsive transcriptional regulator MumR protects Acinetobacter baumannii from oxidative stress. Infect Immun. 2019;88: e00762–19. 10.1128/IAI.00762-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CL, Singh SS, Alamneh Y, Casella LG, Ernst RK, Lesho EP, et al. In vivo fitness adaptations of colistin-resistant Acinetobacter baumannii isolates to oxidative stress. Antimicrob Agents Chemother. 2017;61: e00598–16. 10.1128/AAC.00598-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soares NC, Cabral MP, Gayoso C, Mallo S, Rodriguez-Velo P, Fernández-Moreira E, et al. Associating growth-phase-related changes in the proteome of Acinetobacter baumannii with increased resistance to oxidative stress. J Proteome Res. 2010;9: 1951–1964. 10.1021/pr901116r [DOI] [PubMed] [Google Scholar]
- 18.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun. 2019;87: e00413–18. 10.1128/IAI.00413-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortensen BL, Skaar EP. The contribution of nutrient metal acquisition and metabolism to Acinetobacter baumannii survival within the host. Front Cell Infect Microbiol. 2013;3 10.3389/fcimb.2013.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, et al. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 2012;8: e1003068 10.1371/journal.ppat.1003068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nairn BL, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, et al. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe. 2016;19: 826–836. 10.1016/j.chom.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheldon JR, Laakso HA, Heinrichs DE. Iron acquisition strategies of bacterial pathogens. Microbiol Spectr. 2016;4: VMBF-0010-2015. 10.1128/microbiolspec.VMBF-0010-2015 [DOI] [PubMed] [Google Scholar]
- 23.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54: 881–941. 10.1146/annurev.micro.54.1.881 [DOI] [PubMed] [Google Scholar]
- 24.Weinberg ED. Nutritional immunity: Host’s attempt to withhold iron from microbial invaders. JAMA J Am Med Assoc. 1975;231: 39–41. 10.1001/jama.1975.03240130021018 [DOI] [PubMed] [Google Scholar]
- 25.Weinberg ED. Iron and susceptibility to infectious disease. Science. 1974;184: 952–956. 10.1126/science.184.4140.952 [DOI] [PubMed] [Google Scholar]
- 26.Kochan I. The role of iron in bacterial infections, with special consideration of host-tubercle bacillus interaction. Curr Top Microbiol Immunol. 1973;60: 1–30. 10.1007/978-3-642-65502-9_1 [DOI] [PubMed] [Google Scholar]
- 27.Ganz T. Iron and infection. Int J Hematol. 2018;107: 7–15. 10.1007/s12185-017-2366-2 [DOI] [PubMed] [Google Scholar]
- 28.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71: 413–451. 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McRose DL, Seyedsayamdost MR, Morel FMM. Multiple siderophores: bug or feature? J Biol Inorg Chem. 2018;23: 983–993. 10.1007/s00775-018-1617-x [DOI] [PubMed] [Google Scholar]
- 30.Antunes LCS, Imperi F, Towner KJ, Visca P. Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res Microbiol. 2011;162: 279–84. 10.1016/j.resmic.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 31.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics. 2011;12: 126 10.1186/1471-2164-12-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez MS, Penwell WF, Traglia GM, Zimbler DL, Gaddy JA, Nikolaidis N, et al. Identification of potential virulence factors in the model strain Acinetobacter baumannii A118. Front Microbiol. 2019;10 10.3389/fmicb.2019.01599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Runci F, Gentile V, Frangipani E, Rampioni G, Leoni L, Lucidi M, et al. Contribution of active iron uptake to Acinetobacter baumannii pathogenicity. Infect Immun. 2019;87: e00755–18. 10.1128/IAI.00755-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Álvarez-Fraga L, Vázquez-Ucha JC, Martínez-Guitián M, Vallejo JA, Bou G, Beceiro A, et al. Pneumonia infection in mice reveals the involvement of the feoA gene in the pathogenesis of Acinetobacter baumannii. Virulence. 2018;9: 496–509. 10.1080/21505594.2017.1420451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giardina BJ, Shahzad S, Huang W, Wilks A. Heme uptake and utilization by hypervirulent Acinetobacter baumannii LAC-4 is dependent on a canonical heme oxygenase (abHemO). Arch Biochem Biophys. 2019;672 10.1016/j.abb.2019.108066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Echenique JR, Arienti H, Tolmasky ME, Read RR, Staneloni RJ, Crosa JH, et al. Characterization of a high-affinity iron transport system in Acinetobacter baumannii. J Bacteriol. 1992;174: 7670–7679. 10.1128/jb.174.23.7670-7679.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorsey CW, Tolmasky ME, Crosa JH, Actis LA. Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology. 2003. pp. 1227–1238. 10.1099/mic.0.26204-0 [DOI] [PubMed] [Google Scholar]
- 38.Shapiro JA, Wencewicz TA. Acinetobactin isomerization enables adaptive iron acquisition in Acinetobacter baumannii through pH-Triggered siderophore swapping. ACS Infect Dis. 2016;2: 157–168. 10.1021/acsinfecdis.5b00145 [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto S, Okujo N, Sakakibara Y. Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch Microbiol. 1994;162: 249–54. 10.1007/BF00301846 [DOI] [PubMed] [Google Scholar]
- 40.Hasan T, Choi CH, Oh MH. Genes involved in the biosynthesis and transport of acinetobactin in Acinetobacter baumannii. Genomics Inform. 2015;13: 2–6. 10.5808/GI.2015.13.1.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penwell WF, Arivett BA, Actis LA. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS One. 2012;7: e36493 10.1371/journal.pone.0036493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penwell WF, Degrace N, Tentarelli S, Gauthier L, Gilbert CM, Arivett BA, et al. Discovery and characterization of new hydroxamate siderophores, baumannoferrin A and B, produced by Acinetobacter baumannii. ChemBioChem. 2015;16: 1896–1904. 10.1002/cbic.201500147 [DOI] [PubMed] [Google Scholar]
- 43.Proschak A, Lubuta P, Grün P, Löhr F, Wilharm G, De Berardinis V, et al. Structure and biosynthesis of fimsbactins A-F, siderophores from Acinetobacter baumannii and Acinetobacter baylyi. ChemBioChem. 2013;14: 633–638. 10.1002/cbic.201200764 [DOI] [PubMed] [Google Scholar]
- 44.Song WY, Kim HJ. Current biochemical understanding regarding the metabolism of acinetobactin, the major siderophore of the human pathogen Acinetobacter baumannii, and outlook for discovery of novel anti-infectious agents based thereon. Nat Prod Rep. 2019. 10.1039/C9NP00046A [DOI] [PubMed] [Google Scholar]
- 45.Wuest WM, Sattely ES, Walsh CT. Three siderophores from one bacterial enzymatic assembly line. J Am Chem Soc. 2009;131: 5056–5057. 10.1021/ja900815w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaddy JA, Actis LA, Arivett BA, Mcconnell MJ, Rafael LR, Pachón J. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80: 1015–1024. 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorsey CW, Tomaras AP, Connerly PL, Tolmasky ME, Crosa JH, Actis LA. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology. 2004;150: 3657–3667. 10.1099/mic.0.27371-0 [DOI] [PubMed] [Google Scholar]
- 48.Berti AD, Thomas MG. Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J Bacteriol. 2009;191: 4594–4604. 10.1128/JB.00457-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, et al. The success of Acinetobacter species; Genetic, metabolic and virulence attributes. PLoS One. 2012;7: e46984 10.1371/journal.pone.0046984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penwell WF, Actis LA. Isolation and characterization of the acinetobactin and baumannoferrin siderophores produced by Acinetobacter baumannii. Methods Mol Biol. 2019;1946: 259–270. 10.1007/978-1-4939-9118-1_24 [DOI] [PubMed] [Google Scholar]
- 51.Antunes LCS, Imperi F, Carattoli A, Visca P. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One. 2011;6: e22674 10.1371/journal.pone.0022674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris G, Lee RK, Lam CK, Kanzaki G, Patel GB, Xu HH, et al. A Mouse model of Acinetobacter baumannii-associated pneumonia using a clinically isolated hypervirulent strain. Antimicrob Agents Chemother. 2013;57: 3601–3613. 10.1128/AAC.00944-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linz B, Mukhtar N, Shabbir MZ, Rivera I, Ivanov Y V., Tahir Z, et al. Virulent epidemic pneumonia in sheep caused by the human pathogen Acinetobacter baumannii. Front Microbiol. 2018;9 10.3389/fmicb.2018.02616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piechaud M, Second L. [Studies of 26 strains of Moraxella iwoffi]. Ann Inst Pasteur (Paris). 1951;80: 97–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/14829885 [PubMed]
- 55.Bohac TJ, Fang L, Giblin DE, Wencewicz TA. Fimsbactin and acinetobactin compete for the periplasmic siderophore binding protein BauB in pathogenic Acinetobacter baumannii. ACS Chem Biol. 2019;14: 674–687. 10.1021/acschembio.8b01051 [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Lee H, Song WY, Kim HJ. Total syntheses of fimsbactin A and B and their stereoisomers to probe the stereoselectivity of the fimsbactin uptake machinery in Acinetobacter baumannii. Org Lett. 2020;22: 2806–2810. 10.1021/acs.orglett.0c00790 [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Lonergan ZR, Gonzalez-Gutierrez G, Nairn BL, Maxwell CN, Zhang Y, et al. Multi-metal restriction by calprotectin impacts de novo flavin biosynthesis in Acinetobacter baumannii. Cell Chem Biol. 2019;26: 745–755.e7. 10.1016/j.chembiol.2019.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mortensen BL, Rathi S, Chazin WJ, Skaar EP. Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator zur. J Bacteriol. 2014;196: 2616–2626. 10.1128/JB.01650-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwyn B, Neilands JBB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160: 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 60.Alexander DB, Zuberer DA. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol Fertil Soils. 1991;12: 39–45. 10.1007/BF00369386 [DOI] [Google Scholar]
- 61.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, et al. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. MBio. 2014;5: 1–9. 10.1128/mBio.01313-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mihara K, Tanabe T, Yamakawa Y, Funahashi T, Nakao H, Narimatsu S, et al. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology. 2004;150: 2587–2597. 10.1099/mic.0.27141-0 [DOI] [PubMed] [Google Scholar]
- 63.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol. 2008;190: 8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26: 317–325. 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- 65.Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, et al. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes. 2009;2: 80 10.1186/1756-0500-2-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu W, Solis N V., Filler SG, Mitchell AP. Pathogen gene expression profiling during infection using a nanostring nCounter platform. Methods in Molecular Biology. 2016. pp. 57–65. 10.1007/978-1-4939-3079-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gifford AH, Willger SD, Dolben EL, Moulton LA, Dorman DB, Bean H, et al. Use of a multiplex transcript method for analysis of Pseudomonas aeruginosa gene expression profiles in the cystic fibrosis lung. Infect Immun. 2016;84: 2995–3006. 10.1128/IAI.00437-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnstone TC, Nolan EM. Beyond iron: Non-classical biological functions of bacterial siderophores. Dalt Trans. 2015;44: 6320–6339. 10.1039/c4dt03559c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moynié L, Serra I, Scorciapino MA, Oueis E, Page MGP, Ceccarelli M, et al. Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogen Acinetobacter baumannii. Elife. 2018;7 10.7554/eLife.42270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10: 1033–43. 10.1016/s1097-2765(02)00708-6 [DOI] [PubMed] [Google Scholar]
- 71.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432: 917–921. 10.1038/nature03104 [DOI] [PubMed] [Google Scholar]
- 72.Clifton MC, Rupert PB, Hoette TM, Raymond KN, Abergel RJ, Strong RK. Parsing the functional specificity of siderocalin/lipocalin 2/NGAL for siderophores and related small-molecule ligands. J Struct Biol. 2019;2 10.1016/j.yjsbx.2019.100008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abergel RJ, Wilson MK, Arceneaux JEL, Hoette TM, Strong RK, Byers BR, et al. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc Natl Acad Sci U S A. 2006;103: 18499–18503. 10.1073/pnas.0607055103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo M, Fadeev EA, Groves JT. Membrane dynamics of the amphiphilic siderophore, acinetoferrin. J Am Chem Soc. 2005;127: 1726–1736. 10.1021/ja044230f [DOI] [PubMed] [Google Scholar]
- 75.Griffiths GL, Sigel SP, Payne SM, Neilands JB. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259: 383–385. Available: https://www.jbc.org/content/259/1/383.long [PubMed] [Google Scholar]
- 76.Allred BE, Correnti C, Clifton MC, Strong RK, Raymond KN. Siderocalin outwits the coordination chemistry of vibriobactin, a siderophore of Vibrio cholerae. ACS Chem Biol. 2013;8: 1882–1887. 10.1021/cb4002552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bohac TJ, Shapiro JA, Wencewicz TA. Rigid oxazole acinetobactin analog blocks siderophore cycling in Acinetobacter baumannii. ACS Infect Dis. 2017;3: 802–806. 10.1021/acsinfecdis.7b00146 [DOI] [PubMed] [Google Scholar]
- 78.Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: Crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47: 903–915. 10.1046/j.1365-2958.2003.03337.x [DOI] [PubMed] [Google Scholar]
- 79.Murray GL, Tsyganov K, Kostoulias XP, Bulach DM, Powell D, Creek DJ, et al. Global gene expression profile of Acinetobacter baumannii during bacteremia. J Infect Dis. 2017;215: S52–S57. 10.1093/infdis/jiw529 [DOI] [PubMed] [Google Scholar]
- 80.Perry WJ, Spraggins JM, Sheldon JR, Grunenwald CM, Heinrichs DE, Cassat JE, et al. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc Natl Acad Sci U S A. 2019;116: 21980–21982. 10.1073/pnas.1913991116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goel VK, Kapil A. Monoclonal antibodies against the iron regulated outer membrane proteins of Acinetobacter baumannii are bactericidal. BMC Microbiol. 2001;1: 1–8. 10.1186/1471-2180-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Esmaeilkhani H, Rasooli I, Hashemi M, Nazarian S, Sefid F. Immunogenicity of cork and loop domains of recombinant baumannii acinetobactin utilization protein in murine model. Avicenna J Med Biotechnol. 2015;11: 180–186. Available: http://www.ncbi.nlm.nih.gov/pubmed/31057721 [PMC free article] [PubMed] [Google Scholar]
- 83.Esmaeilkhani H, Rasooli I, Nazarian S, Sefid F. In vivo validation of the immunogenicity of recombinant baumannii acinetobactin utilization A protein (rBauA). Microb Pathog. 2016;98: 77–81. 10.1016/j.micpath.2016.06.032 [DOI] [PubMed] [Google Scholar]
- 84.Aghajani Z, Rasooli I, Mousavi Gargari SL. Exploitation of two siderophore receptors, BauA and BfnH, for protection against Acinetobacter baumannii infection. APMIS. 2019;127: 753–763. 10.1111/apm.12992 [DOI] [PubMed] [Google Scholar]
- 85.U.S. Food and Drug Administration. FDA approves new antibacterial drug to treat complicated urinary tract infections as part of ongoing efforts to address antimicrobial resistance | FDA. Available: https://www.fda.gov/news-events/press-announcements/fda-approves-new-antibacterial-drug-treat-complicated-urinary-tract-infections-part-ongoing-efforts
- 86.Ito A, Nishikawa T, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, et al. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60: 7396–7401. 10.1128/AAC.01405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.U.S. Food and Drug Administration. Drugs@FDA: FDA-Approved Drugs. US Food Drug Adm. 2019. Available: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event = overview.process&varApplNo = 209445
- 88.Moynié L, Luscher A, Rolo D, Pletzer D, Tortajada A, Weingart H, et al. Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother. 2017;61: e02531–16. 10.1128/AAC.02531-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghosh M, Miller PA, Möllmann U, Claypool WD, Schroeder VA, Wolter WR, et al. Targeted antibiotic delivery: selective siderophore conjugation with daptomycin confers potent activity against multidrug resistant Acinetobacter baumannii both in vitro and in vivo. J Med Chem. 2017;60: 4577–4583. 10.1021/acs.jmedchem.7b00102 [DOI] [PubMed] [Google Scholar]
- 90.Wencewicz TA, Miller MJ. Biscatecholate-monohydroxamate mixed ligand siderophore-carbacephalosporin conjugates are selective sideromycin antibiotics that target Acinetobacter baumannii. J Med Chem. 2013;56: 4044–4052. 10.1021/jm400265k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song WY, Jeong D, Kim J, Lee MW, Oh MH, Kim HJ. Key structural elements for cellular uptake of acinetobactin, a major siderophore of Acinetobacter baumannii. Org Lett. 2017;19: 500–503. 10.1021/acs.orglett.6b03671 [DOI] [PubMed] [Google Scholar]
- 92.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, et al. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol. 2015;197: 2027–2035. 10.1128/JB.00131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio. 2014;5: e01076–14. 10.1128/mBio.01076-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87: 45–51. 10.1016/0378-1119(90)90494-c [DOI] [PubMed] [Google Scholar]
- 96.Sebulsky MT, Speziali CD, Shilton BH, Edgell DR, Heinrichs DE. FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: A case of gene duplication and lateral transfer. J Biol Chem. 2004;279: 53152–53159. 10.1074/jbc.M409793200 [DOI] [PubMed] [Google Scholar]
- 97.Moon KH, Weber BS, Feldman MF. Subinhibitory concentrations of trimethoprim and sulfamethoxazole prevent biofilm formation by Acinetobacter baumannii through inhibition of Csu pilus expression. Antimicrob Agents Chemother. 2017;61: e00778–17. 10.1128/AAC.00778-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zwietering MH, Jongenburger I, Rombouts FM, Van’t Riet K. Modeling of the bacterial growth curve. Appl Environ Microbiol. 1990;56: 1875–1881. 10.1128/AEM.56.6.1875-1881.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tjørve KMC, Tjørve E. The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS One. 2017;12 10.1371/journal.pone.0178691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saunders S. Growth curve fitting. GitHub; 2020. Available: https://scott-h-saunders.shinyapps.io/gompertz_fitting_0v2/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
WT A. baumannii and its isogenic Δzur mutant were grown in metal-restricted media for 12 h. RNA was extracted and transcriptional changes in the expression of siderophore-associated genes and fur were assessed by qRT-PCR and normalized to the expression of rpoB. Expression of a known zur-regulated gene, zigA, was assessed as a positive control, whereas zur was run as a negative control. * p < 0.05, *** p < 0.001, and **** p < 0.0001 as determined by Student’s t test relative to a hypothetical value of 1. Data are representative of two experiments performed in biological quadruplicate.
(TIFF)
Growth kinetics of WT A. baumannii ATCC 17978 and its isogenic acinetobactin (ΔbasG), baumannoferrin (ΔbfnL) and fimsbactins (ΔfbsE) biosynthetic mutants were analyzed from the data presented in Fig 5. Estimates of the maximal OD600 (asymptote (A) A-D), maximum specific growth rate (μm, E-H) and lag time (λ, I-L) are given where *p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
(TIFF)
WT A. baumannii ATCC 17978 with empty vector (WT/pWH1266), the acinetobactin-deficient mutant with empty vector (ΔbasG/pWH1266), and a mutant complemented with basG expressed from pWH1266 (ΔbasG/pWH1266::basG) were grown in cTMS media with human serum added at the concentrations indicated. Bacterial growth was assessed by determining the optical density at 600 nm (OD600nm) at the timepoints indicated. Data are the average of technical triplicates and represent the results of two independent experiments.
(TIFF)
WT A. baumannii AB5075-UW and two unique basG transposon mutants were grown in cTMS media with human serum added at the concentration indicated. Bacterial growth was assessed by determining the OD600nm at the timepoints indicated. Data are the average of technical triplicates and represent the results of two independent experiments. Where error bars are not visible, they are shorter than the height of the symbol.
(TIFF)
Wild-type (WT) A. baumannii and its isogenic acinetobactin (ΔbasG), baumannoferrin (ΔbfnL) and fimsbactins (ΔfbsE) biosynthetic mutants were grown in cTMS media with lactoferrin, no added iron source, or 30 μM FeCl3, as indicated. Bacterial growth was assessed by determining the OD600nm, at the time points indicated. Data are representative of two independent experiments, and error bars represent the standard error of the mean. Where error bars are not visible, they are shorter than the height of the symbol.
(TIFF)
WT A. baumannii and its isogenic ΔbasG bfnL mutant were grown in metal-restricted media for 12 h. RNA was extracted and transcriptional changes in genes of the fimsbactins locus were assessed by qRT-PCR and normalized to the expression of rpoB. Expression of bfnL was used as a negative control. * p < 0.05, ** p < 0.01 and **** p < 0.0001, as determined by Student’s t test relative to a hypothetical value of 1. Data are representative of two experiments performed in biological quadruplicate.
(TIFF)
Growth kinetics of WT A. baumannii ATCC 17978 and its isogenic siderophore biosynthetic mutants, as indicated, were analyzed from the data presented in Fig 7. Estimates of the maximal OD600 (asymptote (A) A-D), maximum specific growth rate (μm, E-H) and lag time (λ, I-L) are given for the conditions listed where are given where *p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. When growth was insufficient to calculate the parameter, no data is given (ND).
(TIFF)
Organs were harvested, RNA extracted, and gene expression changes relative to growth in vitro were determined in the kidney (A), liver (B), and spleen (C) using NanoString technology. Genes are clustered by known or predicted function, as indicated.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.