Abstract
Circadian rhythm plays an important role in diverse physiological processes. Abnormal expression of circadian rhythm genes is associated with increased risk of disease, including different types of cancer. The cancer stem cell (CSC) hypothesis suggests that there is a small subset of stem-like cells within tumors that are responsible for tumor initiation. However, the biological effect of circadian rhythm on CSCs remains largely unknown. Studies have highlighted that the circadian rhythm protein CLOCK controls key aspects of various diseases. In the present study, lung cancer stem-like cells were successfully enriched using a sphere formation assay. Next, it was observed that CLOCK mRNA and protein expression levels in the A549 and H1299 sphere cells were notably increased compared with those in the corresponding parental cells. In addition, flow cytometry was performed to isolate CD133+ cells and, consistently, CLOCK expression was also found to be markedly upregulated in CD133+ lung cancer cells. Subsequently, to determine the effect of CLOCK on lung cancer stem cells in detail, CLOCK was knocked down using targeted short inhibiting RNA and the results demonstrated that the sphere-forming ability of the A549 and H1299 cell lines was reduced. In addition, CSC-like properties, including the expression of CD133, CD44, sex determining region Y-box 2, Nanog and octamer-binding transcription factor 4, were markedly decreased in the A549 and H1299 sphere cells following knockdown of CLOCK. Epigallocatechin-3-gallate (EGCG), a green tea polyphenol, has been reported to be a potential anticancer phytochemical. EGCG was found to repress CLOCK expression in A549 and H1299 sphere cells. In addition, EGCG also decreased the ratio of CD133+ cells. The Wnt/β-catenin pathway was notably inactivated by the knockdown of CLOCK in A549 and H1299 sphere cells. Subsequently, using a xenograft model, it was demonstrated that EGCG suppressed the CSC-like characteristics of lung cancer cells by targeting CLOCK. In conclusion, the present study demonstrated that EGCG inhibited the self-renewal ability of lung cancer stem-like cells by targeting CLOCK.
Keywords: cancer stem cells, CLOCK, epigallocatechin-3-gallate, Wnt/β-catenin, lung cancer
Introduction
Lung cancer remains the leading cause of cancer-associated mortality worldwide and non-small cell lung cancer (NSCLC) accounts for ~85% of lung cancer cases (1). Significant improvements have been made in the treatment of lung cancer; however, its 5-year survival rate remains low, primarily due to treatment resistance, which may be present before or develop during the course of treatment (2).
Recently, circadian rhythm disruption by shiftwork has been reported in tumorigenesis. Previous epidemiological studies have revealed that individuals working night shifts are at higher risk of developing cancer or exhibit poorer cancer prognosis (3-5). Systemic disruption of the circadian machinery may result in changes in cellular functions that are highly associated with cancer (6-8). An important role of the core circadian genes has been reported in carcinogenesis (9,10). Under normal conditions, the key circadian genes, such as Bmal1, CLOCK, period, cryptochrome and casein kinase Iε, function in tightly regulated feedback loops (11,12). For example, the circadian gene CLOCK may contribute to glioma progression, which is directly modulated by microRNA (miR)-124 (13). Chronic shift-lag may alter CLOCK expression in natural killer cells, which notably induces lung cancer growth in vivo (14). However, the detailed role of CLOCK in lung cancer remains to be further investigated.
The cancer stem cell (CSC) hypothesis suggests that a small population of cancer cells with self-renewing ability are responsible for tumor relapse (14,15). The existence of CSCs has been verified in various types of tumors (16,17). A recent report revealed that the circadian dynamics of CSCs are modulated by the tumor microenvironment and provide a principle for the treatment of breast cancer (18). Disruption of CLOCK may also affect glioblastoma stem cells (19).
The aim of the present study was to determine whether CLOCK can regulate lung CSCs. CLOCK was induced and knocked down in lung CSCs to determine its effects on CSC-like properties and whether these were mediated by the Wnt signaling pathway. Furthermore, it was investigated whether epigallocatechin-3-gallate (EGCG) can inhibit the stemness of lung cancer cells by regulating CLOCK expression, in order to determine whether CLOCK is a potential target for suppressing CSC-like characteristics in lung cancer cells.
Materials and methods
Cell culture and reagents
The A549 and H1299 cell lines were purchased from the Chinese Academy of Sciences Committee on Type Culture Collection Cell Bank (Shanghai, China) and cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were cultured at 37°C in a humidified incubator with 5% CO2. EGCG (purity ≥95%) powder was purchased from Sigma-Aldrich; Merck KGaA.
Western blot analysis
Total protein was extracted from the cell samples following lysis using RIPA buffer (Beyotime Institute of Biotechnology), supplemented with protease and phosphatase inhibitors, and was quantified using a BCA protein assay. The proteins were then separated on a 10% SDS-PAGE (Invitrogen; Thermo Fisher Scientific, Inc.), transferred onto nitrocellulose filter membranes (Cytiva Bioscience) and incubated with primary antibodies overnight at 4°C. After 24 h, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. The following primary antibodies were used: Anti-CLOCK (1:1,000; cat no. ab3517, Abcam), anti-CD133 (1:1,000; cat no. ab216323, Abcam), anti-CD44 (1:1,000; cat no. ab189524, Abcam), anti-sex determining region Y-box (Sox)2 (1:1,000; cat no. ab92494, Abcam), anti-Nanog (1:1,000; cat no. ab109250, Abcam), anti-octamer-binding transcription factor (Oct)4 (1:1,000; cat no. ab181557, Abcam), anti-glycogen synthase kinase (GSK)3β (1:1,000; cat no. ab32391, Abcam), anti-phosphorylated (p)-GSK3β (1:1,000; ab131097, Abcam), anti-β-catenin (1:1,000; cat no. ab32572, Abcam), anti-p-β-catenin (1:1,000; cat no. ab27798, Abcam), anti-β-actin (1:1,000; cat no. ab179467, Abcam) and anti-GAPDH (1:1,000; cat no. A00227-1, Boster Biological Technology, Ltd.). The following secondary antibodies were used: HRP-conjugated AffiniPure goat anti-rabbit IgG (1:2,000; cat no. TA130015, OriGene Technologies, Inc.) and HRP-conjugated AffiniPure goat anti-mouse IgG (1:2,000; cat no. TA130001, OriGene Technologies, Inc.). The immunoreactive proteins were then detected using an enhanced chemiluminescence kit (Cell Signaling Technology, Inc.).
Reverse transcription-quantitative PCR analysis
Total RNA was extracted using a RNAiso Plus kit (Takara Biotechnology Co., Ltd.) and the Prime Script™ RT Master mix (Takara Biotechnology Co., Ltd.) was utilized to reverse-transcribe RNA into cDNA according to the manufacturer's instructions, while qPCR was performed using the SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. Reactions were carried out using the following thermocycling conditions: Pre-denaturation at 95°C for 1 min; 40 cycles at 95°C for 5 sec, 60°C for 15 sec, and a final step at 72°C for 15 sec. The RT-qPCR primers are provided in Table I. GAPDH was used as an internal mRNA control. qPCR was performed using the Applied Biosystems 7,300 Real Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The fold change was calculated using the 2−∆∆Cq method (20).
Table I.
Primers used for quantitative PCR analysis.
| Genes | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| GAPDH | CAAGGTCATCCATGACAACTTTG | GTCCACCACCCTGTTGCTGTAG |
| CLOCK | ATGGATTGGTGGAAGAAG | ACCATCAAGAGCCTCTAAC |
Sphere formation assay
Cells were treated with 10 ng/ml of human recombinant basic fibroblast growth factor (FGF; R&D Systems, Inc.) and 20 ng/ml of epidermal growth factor (EGF; R&D Systems, Inc.) in serum-free DMEM-F12 (Gibco; Thermo Fisher Scientific, Inc.). The medium was changed every 48 h and the cells were cultured for 7 days. Tumor spheres were observed using a MOTIC inverted microscope (Olympus Corporation) at a magnification of ×100.
Flow cytometry analysis
For CD133+ cell analyses, the cells were first washed, resuspended in RPMI-1640 medium with 10% FBS, and then incubated at 4°C in the freezer for 30 min with fluorescence-conjugated monoclonal antibodies against human CD133 PE (1:100; cat no. 566593, BD Biosciences) and its isotype IgG.
Short inhibiting (si)RNA and plasmid transfection
siRNA targeting CLOCK at a concentration of 100 nmol or a corresponding negative control (Guangzhou RiboBio, Co., Ltd.) were transfected into NSCLC/CSCs using Lipofectamine™ 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions at room temperature. After transfection for 48 h, cells were collected for the subsequent experiments.
Xenograft studies
A total of 12 female BALB/c nude mice, aged 5-6 weeks and weighing 18-20 g, were purchased from the Shanghai Animal Laboratory Center and maintained at the Experimental Animal Center at Nanjing Medical University, with a temperature and humidity of 22±1°C and 55±5%, respectively. The mice were daily observed for abnormal behavior, including inability to eat or drink, or lack of response upon stimulation or touch. All aspects of animal welfare were considered, and measures were taken to minimize the suffering and distress of the animals. Each mouse was subcutaneously injected with exponentially growing A549 sphere cells (5×106) on the back and the animals were randomly divided into EGCG and control groups. After 2 weeks, 20 mg/kg EGCG was administered to the mice by intraperitoneal injection weekly. The length and width of the tumors were measured using a caliper, and the volumes were calculated using the following formula: Volume (mm3)=(length × width2)/2. The maximum diameter of the tumor was 12 mm and the minimum 5 mm. The maximum weight loss observed in mice from start to endpoint was 1.5 g and the maximum percentage of weight loss was <10%. After receiving treatment with EGCG for 4 weeks, the mice were euthanized using cervical dislocation. Death was confirmed by observing the eyes turn pale and by monitoring lack of heartbeat and breathing and lack of response to external stimuli after cervical dislocation. The study protocol was based on the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Immunohistochemistry
Immunohistochemical staining for Ki67 was performed on xenograft tumor tissues using antibodies against Ki67 (1:100; cat. no. ab15580, Abcam) at the Department of Pathology of The First Affiliated Hospital of Nanjing Medical University. Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.) was used to analyze the staining results. The ratios of positively stained tumor cells were classified into four groups with scores from 0 to 3 (<10, 0; >10, 1; >25, 2; and >50%, 3). The staining intensities were scored as follows: No staining, 0; low, 1; medium, 2; and high, 3). A final IHC score was calculated by adding the two scores; a score >3 was considered to indicate positive expression, and a score ≤3 negative expression.
Histopathology
Histopathological examination of xenograft tissues was performed using hematoxylin-eosin (HE) staining. The samples were placed in 10% formaldehyde solution overnight, dehydrated through a graded ethanol series every 5 min, embedded in paraffin and then cut into 4-µm sections. Subsequently, the sections were stained using HE (Beijing Solarbio Science & Technology Co., Ltd.) at room temperature according to the manufacturer's instructions and observed under a light microscope (Leica Microsystems GmbH) at a magnification of ×100.
Statistical analysis
All data were recorded as the mean ± standard deviation of at least three independent experiments. Comparisons between quantitative variables were performed using a Student's t-test and one-way ANOVA followed by Dunnett's post hoc test. For comparisons among all groups, one-way ANOVA followed by Tukey's post hoc test was used. P<0.05 was considered to indicate a statistically significant difference. SPSS v17.0 (SPSS Inc.) and GraphPad Prism v5.0 (GraphPad Software, Inc.) were used for statistical analysis.
Results
Successfully enrichment of CSC-like cells from parental lung cancer cells
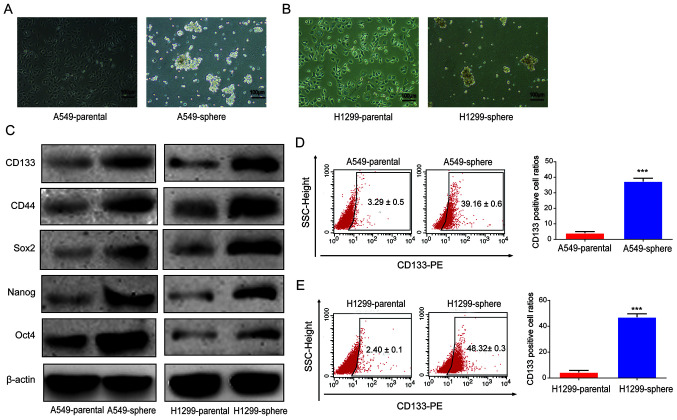
Serum-free medium was used to form spheroid populations to enrich CSCs (21). In the present study, A549 and H1299 cells were cultured using a serum-free suspension medium to induce CSCs for 7 days, and the formation of tumor spheres was observed (Fig. 1A and B). Subsequently, to verify the stemness of sphere-forming cells, the protein expression levels of CD133, CD44, Sox2, Nanog and Oct4 were determined using western blot analysis. A shown in Fig. 1C, the protein expression levels of these CSC markers were notably upregulated in the A549 and H1299 sphere cells. Next, as shown in Fig. 1D and E, the percentage of CD133+ cells was found to be markedly increased among the A549 and H1299 sphere cells compared with that in the corresponding parental A549 and H1299 cells, as indicated by flow cytometry analysis. These data suggested that CSC-like cells were successfully obtained from parental lung cancer cells.
Figure 1.
Self-renewal ability was enriched in lung cancer stem cells. Microscopic observation of sphere and parental (A) A549 and (B) H1299 cells formed tumor spheres following culture for 7 days in serum-free medium. Bar, 100 µm. (C) Protein expression levels of CD133, CD44, Sox2, Nanog and Oct4. NSCLC parental and NSCLC sphere cells were lysed for the detection of protein levels using western blot analysis. β-actin served as a loading control. Flow cytometry analyses of CD133+ cell percentage in (D) A549 and (E) H1299 parental and sphere cells. Bar, 100 µm. Data are presented as the mean ± SD from at least triplicate experiments. ***P<0.001. NSCLC, non-small cell lung cancer; Sox2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4.
Circadian rhythm-related CLOCK gene is upregulated in lung CSCs
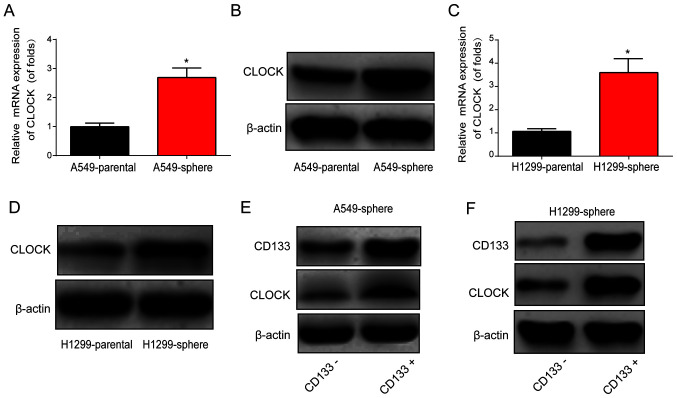
Subsequently, qPCR and western blot assays were performed, and CLOCK mRNA and protein expression levels were found to be markedly upregulated in A549 and H1299 sphere cells, as shown in Fig. 2A-D. Furthermore, cell sorting was performed using flow cytometry to isolate CD133− and CD133+ cells from the A549 and H1299 sphere cells. As shown in Fig. 2E and F, CD133+ cells exhibited an upregulation of CLOCK protein expression in the A549 and H1299 sphere cells, suggesting a role for CLOCK in lung CSCs.
Figure 2.
Expression of circadian rhythm-related CLOCK gene. CLOCK was induced in lung cancer stem cells. CLOCK (A) mRNA and (B) protein expression level in A549 parental and sphere cells. CLOCK (C) mRNA and (D) protein expression level in H1299 parental and sphere cells. CD133 and CLOCK protein expression level in CD133+ and CD133− cells isolated from (E) A549 and (F) H1299 sphere cells. Data are presented as the mean ± SD from at least triplicate experiments. *P<0.05.
Knockdown of CLOCK represses the stemness of lung CSCs
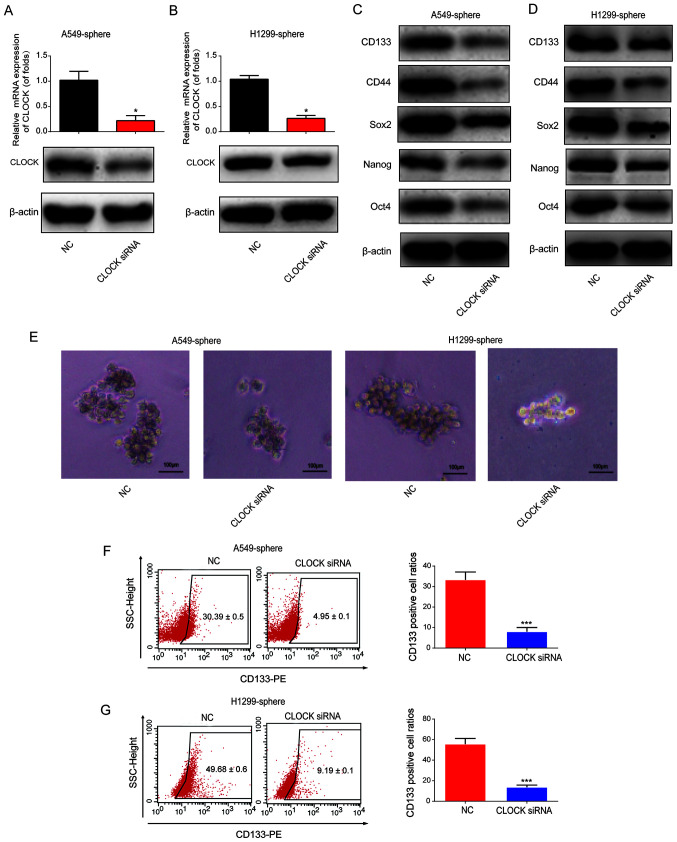
To determine the effect of CLOCK on the stemness of lung CSCs, CLOCK siRNA or siRNA control was transfected into the A549 and H1299 sphere cells for 24 h, after which time the mRNA and protein expression level of CLOCK was found to be significantly downregulated in the A549 and H1299 sphere cells (Fig. 3A and B). It was then observed that the protein expression levels of CD133, CD44, Sox2, Nanog and Oct4 were notably reduced by CLOCK siRNA (Fig. 3C and D). As shown in Fig. 3E, the volume of the tumor spheres was reduced following CLOCK knockdown, which indicated that CLOCK siRNA reduced the number of lung CSCs. Subsequently, flow cytometry was used to determine the ratio of CD133+ cells. As shown in Fig. 3F and G, the percentage of CD133+ cells was markedly decreased following CLOCK knockdown in the A549 and H1299 sphere cells. These data indicated that knockdown of CLOCK was able to inhibit the stemness of lung CSCs.
Figure 3.
Effect of CLOCK siRNA on CSC-like properties of lung cancer cells. Expression of CLOCK in (A) A549 and (B) H1299 sphere cells. Cells were transfected with CLOCK siRNA. Protein levels of CD133, CD44, Sox2, Nanog and Oct4 in (C) A549 and (D) H1299 sphere cells. (E) Tumor volume of A549 and H1299 sphere cells transfected with CLOCK siRNA. Flow cytometry analyses of CD133+ cell ratios in (F) A549 and (G) H1299 sphere cells. Cells were transfected with CLOCK siRNA or siRNA negative control. Data are presented as the mean ± SD from at least triplicate experiments. *P<0.05, ***P<0.001. CSC, cancer stem cell; Sox2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4.
EGCG represses the CSC-like properties of lung CSCs by targeting CLOCK
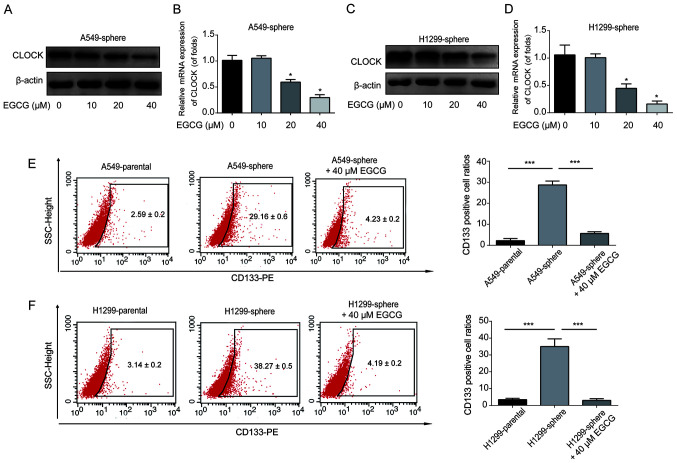
To investigate whether EGCG could regulate CLOCK expression in lung CSCs, the A549 and H1299 sphere cells were treated with the indicated doses of EGCG for 48 h. CLOCK protein and mRNA expression in the A549 sphere cells was notably downregulated by EGCG in a dose-dependent manner (Fig. 4A and B). Consistently, it was observed that 40 µM EGCG could significantly reduce CLOCK expression in H1299 sphere cells (Fig. 4C and D). In addition, the ratio of CD133+ cells among A549 and H1299 sphere cells was reduced by 40 µM EGCG (Fig. 4E and F). These results demonstrated that EGCG was able to reduce NSCLC/CSC stemness by reducing the expression of CLOCK.
Figure 4.
EGCG repressed CSC-like properties of lung cancer cells via inhibiting CLOCK. CLOCK (A) protein and (B) mRNA expression level in A549 sphere cells. Cells were treated with 0, 10, 20 or 40 µM of EGCG for 48 h. CLOCK (C) protein and (D) mRNA expression level in H1299 sphere cells. Cells were treated with 0, 10, 20 or 40 µM of EGCG for 48 h. Flow cytometry analyses of CD133+ cell ratios in (E) A549 and (F) H1299 sphere cells treated with or without 40 µM EGCG. Data are presented as the mean ± SD from at least triplicate experiments. *P<0.05, ***P<0.001. CSC, cancer stem cell; EGCG, epigallocatechin-3-gallate.
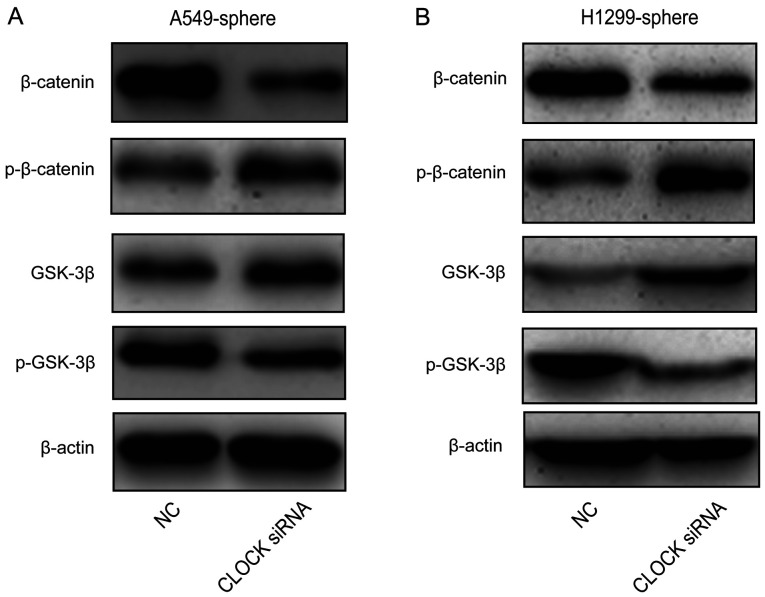
Wnt/β-catenin signaling is blocked by the knockdown of CLOCK in lung CSCs
Next, the effect of CLOCK on Wnt/β-catenin activity was investigated. As shown in Fig. 5A and B, β-catenin and p-GSK-3β protein expression levels were decreased, while p-β-catenin and GSK-3β levels were increased by CLOCK siRNA in the A549 and H1299 sphere cells. These data suggested that the Wnt signaling pathway was inhibited by the knockdown of CLOCK in lung CSCs.
Figure 5.
Knockdown of CLOCK inactivated the Wnt/β-catenin signaling pathway in lung cancer stem cells. β-catenin, p-β-catenin, GSK-3β and p-GSK-3β protein expression in (A) A549 and (B) H1299 sphere cells. Three independent experiments were performed. Data are presented as the mean ± SD from at least triplicate experiments. p, phosphorylated; GSK, glycogen synthase kinase.
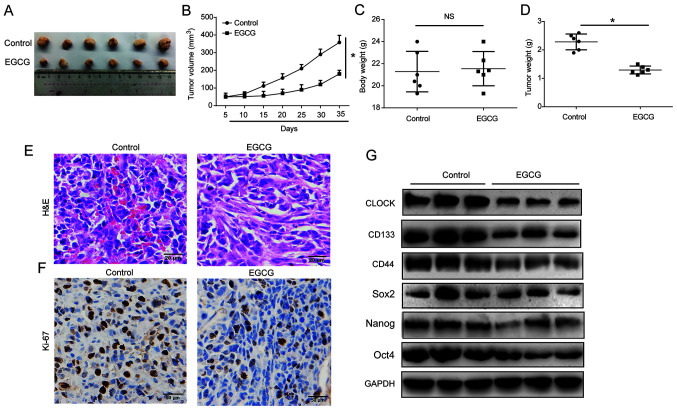
EGCG represses CSC-like characteristics of lung cancer cells by targeting CLOCK in vivo
An A549/CSCs nude mouse xenograft model was established to investigate whether EGCG reduced CSC-like phenotype by targeting CLOCK in vivo. The mice were divided into control and EGCG groups. The subcutaneous tumors were excised from the nude mice and are presented in Fig. 6A. EGCG effectively reduced tumor volume in a time-dependent manner, as shown in Fig. 6B. Body weight was not altered after EGCG treatment, while tumor weight was obviously reduced by EGCG (Fig. 6C and D). The HE and immunohistochemistry staining results are shown in Fig. 6E and F. Subsequently, western blot analysis revealed that EGCG decreased CD133, CD44, Sox2, Nanog, and Oct4 protein expression levels by targeting CLOCK (Fig. 6G). These results suggested that EGCG may target CSC-like properties of lung cancer cells by modulating CLOCK in vivo.
Figure 6.
EGCG inhibited CSC-like characteristics of lung cancer via targeting CLOCK in vivo. A total of 12 5-week-old female BALB/c nude mice were injected with 5×106 A549 sphere cells. Two treatment groups were established as control and EGCG, with 6 mice in each. (A) Solid tumors were excised from the subcutaneous tissue. (B) Tumor volume. (C) Body weight. (D) Tumor weight. (E) Hematoxylin and eosin staining (bar, 20 µm) and (F) immunohis-tochemistry staining (bar, 50 µm) of Ki-67 in the tumor tissues. (G) CLOCK, CD133, CD44, Sox2, Nanog and Oct4 protein expression levels in the tumor tissues. GAPDH was used as the loading control. Three independent experiments were performed. Data are presented as the mean ± SD from at least triplicate experiments. *P<0.05. CSC, cancer stem cell; EGCG, epigallocatechin-3-gallate; Sox2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; NS, not significant.
Discussion
The role of CLOCK in lung CSCs was extensively investigated in the present study. CLOCK was found to be markedly increased in the A549 and H1299 sphere cells, while knockdown of CLOCK markedly reduced the CSC-like properties of lung CSCs. In addition, EGCG reduced stemness by targeting CLOCK in a dose-dependent manner. Furthermore, it was observed that the Wnt signaling pathway was inactivated by the knockdown of CLOCK, and the in vitro data were confirmed using in vivo assays. These findings revealed a novel mechanism involving EGCG-mediated repression of CSC-like properties by CLOCK regulation.
CSCs represent a small cell population that can differentiate into cancerous cells (22). CSCs have been widely investigated, due to their crucial role in cancer progression. Strategies targeting CSCs are becoming increasingly significant for new approaches to cancer therapy (23). Assessing the role of CSCs in tumor recurrence relies heavily on the use of specific CSC markers, including CD133, CD44, Sox2, Nanog and Oct4 (24). In the present study, lung CSCs were enriched from the A549 and H1299 sphere cells. In addition, the sphere cells exhibited higher levels of CD133, CD44, Sox2, Nanog and Oct4, as well as an increased ratio of CD133+ cells, following culture for 7 days in a serum-free suspension medium. It was previously confirmed that the proportion of CD133+ cells reached 80% in a 6-week culture (25). Freshly dissociated lung cancer cells were cultured at low density in serum-free medium with EGF and FGF to determine whether lung cancer CD133+ cells can expand long-term cultures in vitro. It was herein demonstrated the proportion of CD133+ cells gradually increased (3.29 vs. 39.16% in A549 cells, 2.4 vs. 48.32% in H1299 cells) over 7 days and a higher percentage of CD133+ cells may be enriched during if cultured for >7 days. Fresh lung cancer tissues will be used to obtain lung CSCs and longer culture time will be considered in future studies.
Circadian CLOCK is a conserved timekeeper, which can adapt the body's physiology to diurnal cycles. Perturbation of circadian CLOCK participates in the development of various diseases, including cancer (26). PER3 is a circadian CLOCK gene and its overexpression inhibited colorectal cancer stem-like cells by inactivating the Notch and β-catenin signaling pathways (27,28). Circadian CLOCK in colon tumor tissues may promote tumor progression by regulating intracellular iron levels (28), while the circadian gene CLOCK may affect ovarian cancer drug-resistant genes and cell proliferation through autophagy (29). The present study demonstrated that CLOCK was increased in lung CSCs and knockdown of CLOCK notably reduced the stemness of lung cancer cells. The detailed mechanism of action of CLOCK in the proliferation of lung CSCs, involving cell apoptosis or cell cycle arrest, will be investigated in our future study.
EGCG has been widely investigated as a chemopreventive agent with potential anticancer effects (30,31). Our previously study reported that EGCG inhibited lung CSC-like properties by targeting miR-485 and RXRα (32). In addition, EGCG was found to suppress CSC-like characteristics by regulating the miR-485 and CD44 axis in A549-cisplatin resistant cells (33). The aim of the present study was to confirm the inhibitory effect of EGCG on lung CSCs and further investigate the underlying mechanism. CLOCK was found to be notably inhibited by EGCG in a dose-dependent manner. In addition, EGCG was able to reduce CSC-like properties in A549 and H1299 sphere cells via repressing CLOCK expression.
The canonical Wnt/β-catenin pathway is crucial for maintaining CSCs. Dysregulation of this pathway has been identified in various types of human cancer (34,35). The Wnt/β-catenin signaling pathway in lung CSCs is a crucial target for developing novel anticancer drugs (36) and may mediate the inhibitory effects of EGCG on lung CSCs (37). Blocking the Wnt/β-catenin pathway markedly inhibited the proliferation of lung CSCs (38). The present study uncovered that CLOCK is the key regulatory molecule mediating EGCG inhibition of lung cancer stem-like cells, and knockdown of CLOCK was shown to markedly reduce the activity of the Wnt/β-catenin pathway. The focus of future studies will be to investigate the intervention effects of EGCG and the Wnt/β-catenin signaling pathway in a CLOCK overexpression model in vitro and in vivo in order to verify the data of the present study.
To summarize, we herein reported the role of CLOCK in promoting CSC-like characteristics in lung cancer cells. CLOCK is a significant regulator mediating the suppressive effects of EGCG on the stemness characteristics of lung cancer stem-like cells, and EGCG may suppress stemness by targeting CLOCK in A549 and H1299 sphere cells.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions and The National Key Research and Development Program of China (grant no. 2018YFC1313600).
Availability of materials and data
All data generated or analyzed during the present study are included in this article.
Authors' contributions
QF and PJ conceived and designed the experiments. PJ, CX, PZ, JR, FM, XW, LC and FZ conducted the experiments. QF, SL and PJ analyzed the data and revised the manuscript. All the authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
All animal experiments were performed according to the requirements outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and all the protocols were approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 3.Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74–77. doi: 10.1097/00001648-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 4.Kloog I, Haim A, Stevens RG, Portnov BA. Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men. Chronobiol Int. 2009;26:108–125. doi: 10.1080/07420520802694020. [DOI] [PubMed] [Google Scholar]
- 5.Viswanathan AN, Hankinson SE, Schernhammer ES. Night shift work and the risk of endometrial cancer. Cancer Res. 2007;67:10618–10622. doi: 10.1158/0008-5472.CAN-07-2485. [DOI] [PubMed] [Google Scholar]
- 6.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Masri S, Papagiannakopoulos T, Kinouchi K, Liu Y, Cervantes M, Baldi P, Jacks T, Sassone-Corsi P. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell. 2016;165:896–909. doi: 10.1016/j.cell.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, Tothova Z, Tejero H, Heckl D, Järås M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165:303–316. doi: 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Dycke KC, Rodenburg W, van Oostrom CT, van Kerkhof LW, Pennings JL, Roenneberg T, van Steeg H, van der Horst GT. Chronically alternating light cycles increase breast cancer risk in mice. Curr Biol. 2015;25:1932–1937. doi: 10.1016/j.cub.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 12.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/S0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 13.Li A, Lin X, Tan X, Yin B, Han W, Zhao J, Yuan J, Qiang B, Peng X. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett. 2013;587:2455–2460. doi: 10.1016/j.febslet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Ghotra VP, Puigvert JC, Danen EH. The cancer stem cell microenvironment and anti-cancer therapy. Int J Radiat Biol. 2009;85:955–962. doi: 10.3109/09553000903242164. [DOI] [PubMed] [Google Scholar]
- 15.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Florian S, Sonneck K, Hauswirth AW, Krauth MT, Schernthaner GH, Sperr WR, Valent P. Detection of molecular targets on the surface of CD34+/CD38-stem cells in various myeloid malignancies. Leuk Lymphoma. 2006;47:207–222. doi: 10.1080/10428190500272507. [DOI] [PubMed] [Google Scholar]
- 17.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunaga N, Ogino T, Hara Y, Tanaka T, Koyanagi S, Ohdo S. Optimized dosing schedule based on circadian dynamics of mouse breast cancer stem cells improves the antitumor effects of aldehyde dehydrogenase inhibitor. Cancer Res. 2018;78:3698–3708. doi: 10.1158/0008-5472.CAN-17-4034. [DOI] [PubMed] [Google Scholar]
- 19.Dong Z, Zhang G, Qu M, Gimple RC, Wu Q, Qiu Z, Prager BC, Wang X, Kim LJY, Morton AR, et al. Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. 2019;9:1556–1573. doi: 10.1158/2159-8290.CD-19-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Zhang DG, Jiang AG, Lu HY, Zhang LX, Gao XY. Isolation, cultivation and identification of human lung adenocarcinoma stem cells. Oncol Lett. 2015;9:47–54. doi: 10.3892/ol.2014.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: An evolving concept. Nat Rev Cancer. 2012;12:133–13. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 23.Leon G, MacDonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther. 2016;158:71–90. doi: 10.1016/j.pharmthera.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Suresh R, Ali S, Ahmad A, Philip PA, Sarkar FH. The role of cancer stem cells in recurrent and drug-resistant lung cancer. Adv Exp Med Biol. 2016;890:57–74. doi: 10.1007/978-3-319-24932-2_4. [DOI] [PubMed] [Google Scholar]
- 25.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–114. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 26.Dierickx P, Van Laake LW, Geijsen N. Circadian clocks: From stem cells to tissue homeostasis and regeneration. EMBO Rep. 2018;19:18–28. doi: 10.15252/embr.201745130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Sun H, Zhang S, Yang X, Zhang G, Su T. Overexpression of PER3 inhibits self-renewal capability and chemoresistance of colorectal cancer stem-like cells via inhibition of notch and β-catenin signaling. Oncol Res. 2017;25:709–719. doi: 10.3727/096504016X14772331883976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okazaki F, Matsunaga N, Okazaki H, Azuma H, Hamamura K, Tsuruta A, Tsurudome Y, Ogino T, Hara Y, Suzuki T, et al. Circadian clock in a mouse colon tumor regulates intracellular iron levels to promote tumor progression. J Biol Chem. 2016;291:7017–1028. doi: 10.1074/jbc.M115.713412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Jin L, Sui YX, Han LL, Liu JH. Circadian gene CLOCK affects drug-resistant gene expression and cell proliferation in ovarian cancer SKOV3/DDP cell lines through autophagy. Cancer Biother Radiopharm. 2017;32:139–16. doi: 10.1089/cbr.2016.2153. [DOI] [PubMed] [Google Scholar]
- 30.Zaveri NT. Green tea and its polyphenolic catechins: Medicinal uses in cancer and noncancer applications. Life sci. 2006;78:2073–2080. doi: 10.1016/j.lfs.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Kweon MH, Adhami VM, Lee JS, Mukhtar H. Constitutive overexpression of Nrf2-dependent heme oxygenase-1 in A549 cells contributes to resistance to apoptosis induced by epigallocatechin 3-gallate. J Biol Chem. 2006;281:33761–33772. doi: 10.1074/jbc.M604748200. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P, Xu C, Chen L, Chen A, Wu X, Zhou M, Haq IU, Mariyam Z, Feng Q. Epigallocatechin-3-gallate inhibited cancer stem cell-like properties by targeting hsa-mir-485-5p/RXRα in lung cancer. J Cell Biochem. 2018;119:8623–8635. doi: 10.1002/jcb.27117. [DOI] [PubMed] [Google Scholar]
- 33.Jiang P, Xu C, Chen L, Chen A, Wu X, Zhou M, Haq IU, Mariyam Z, Feng Q. EGCG inhibits CSC-like properties through targeting miR-485/CD44 axis in A549-cisplatin resistant cells. Mol Carcinog. 2018;57:1835–1844. doi: 10.1002/mc.22901. [DOI] [PubMed] [Google Scholar]
- 34.Jang GB, Kim JY, Cho SD, Park KS, Jung JY, Lee HY, Hong IS, Nam JS. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep. 2015;5:12465. doi: 10.1038/srep12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shapiro M, Akiri G, Chin C, Wisnivesky JP, Beasley MB, Weiser TS, Swanson SJ, Aaronson SA. Wnt pathway activation predicts increased risk of tumor recurrence in patients with stage I nonsmall cell lung cancer. Ann Surg. 2013;257:548–554. doi: 10.1097/SLA.0b013e31826d81fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang HL, Jiang LM, Han WD. Wnt/β-catenin signaling pathway in lung cancer stem cells is a potential target for the development of novel anticancer drugs. J BUON. 2015;20:1094–1100. [PubMed] [Google Scholar]
- 37.Zhu J, Jiang Y, Yang X, Wang S, Xie C, Li X, Li Y, Chen Y, Wang X, Meng Y, et al. Wnt/β-catenin pathway mediates (-)-Epigallocatechin-3-gallate (EGCG) inhibition of lung cancer stem cells. Biochem Biophys Res Commun. 2017;482:15–21. doi: 10.1016/j.bbrc.2016.11.038. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Lou Y, Zheng X, Wang H, Sun J, Dong Q, Han B. Wnt blockers inhibit the proliferation of lung cancer stem cells. Drug Des Devel Ther. 2015;9:2399–2407. doi: 10.2147/DDDT.S76602. [DOI] [PMC free article] [PubMed] [Google Scholar]