Abstract
Single-cell next-generation sequencing assays are powerful tools to understand the nature of immune cells that drive disease pathogenesis. In this brief review, we explain the value of performing assays at single-cell resolution to better understand the pathogenesis of allergy, asthma and other lung diseases. We explain the challenges in performing single-cell studies of airways and lung samples from patients with lung diseases. A major limitation comes from the amount of diseased tissue that can be utilized for research purposes. Finally, we discuss which sequencing strategies can be utilized for successfully investigate airway and lung diseases at single-cell resolution.
Keywords: Single-cell RNA-seq, airway disease, smart-seq2
Genetic variants associated with risk of many lung diseases affect gene expression in a context-dependent manner in a specific subset of immune cell types in the human body1-7. The immune system contains diverse cell types required to defend from infectious agents. At the same time, immune cells can over-respond, resulting in allergy, asthma, autoimmune and inflammatory diseases. There is probably no other organ system that has a broader influence on human health. However, a complete understanding of the diversity of immune cell types and their properties that help drive disease pathogenesis is still lacking.
Next-generation sequencing (NGS) is one of the newest and most powerful tools to understand biological processes in health and disease. The study of genomes, epigenomes and transcriptomes provide unbiased information on how genetic variations and environmental signals perturb gene expression and chromatin structure in specific cell types to drive disease pathogenesis5, 8-14. Genomic assays like RNA-seq and microarrays are routinely performed on a population of cells under the assumption that cells of a particular type are highly similar. However, recent evidences from studies of single cells reveals that this assumption is incorrect; individual cells within the same population may differ dramatically, and these differences may have important consequences for the function of the entire cell population15-22. The application of NGS to the analysis of single-cell transcriptomes and epigenomes has enabled biologists to collect an unprecedented amount of information from each and every cell present in healthy and diseased tissues. The study of individual immune cells in blood, airways and lung samples from patients with lung diseases (e.g. allergy, asthma, COPD, pulmonary fibrosis, tuberculosis, bronchiectasis Figure 1) will lead to a better definition of the immune cell types driving disease pathogenesis, and also help understand the extent of cellular heterogeneity and molecular plasticity in a “hypothesis free” manner 12, 17, 18, 20, 21, 23-27. These approaches can address significant challenges that currently exist with regard to systematically describing the “molecular status” of pathological or protective cell populations.
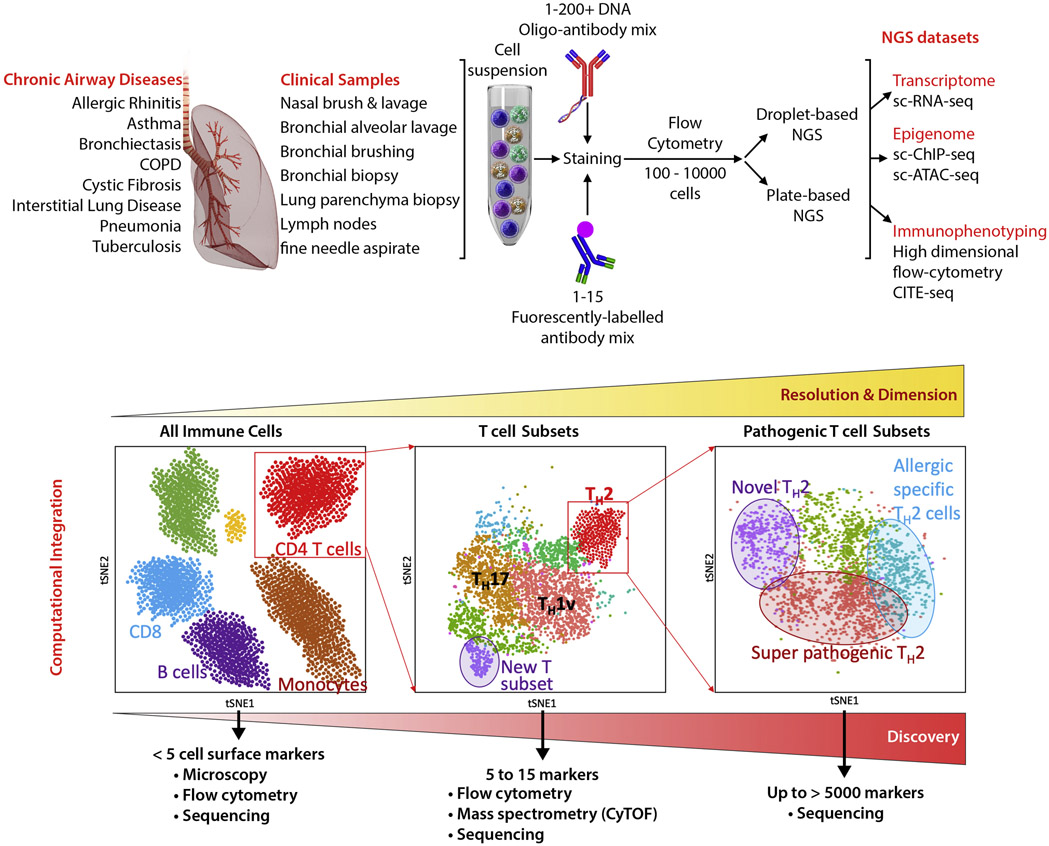
Figure 1. Single-cell next-generation sequencing in airway and lung diseases.
Top panel provides an overview of the processes the clinical samples can go through. Patient samples will be processed by isolating cells and staining with both a mix of antibodies recognizing cell surface markers conjugated with fluorochrome (for flow cytometry analysis) and/or with DNA-barcoded-oligos (for immunophenotyping analysis by sequencing). Depending on the number of cells and output dataset requested, stained cells will be sorted and processed through two single-cell sequencing platforms: the PCR plate-based sorted cells with Smart-seq2 library preparations or the droplet-based 10X platform. The lower panel shows the importance to select the single-cell sequencing methodology that provides the highest resolution. If working with a limited number of cells, higher is the number of genes detected per cell, higher will be clustering resolution and the possibility to separate cells with new molecular features.
Is dysregulate immune response in disease associated with the development of new cell subset with pathogenic properties or a preferential expansion/contraction of pre-existing subsets of immune cells, already physiologically present in blood/lung tissue (Figure 2)? Several studies of chronic diseases in humans and in model organisms have reported the existence of immune cells that acquire a mixed phenotypes. For instance, in patients with latent tuberculosis, T cells responding specifically to mycobacterium tuberculosis antigens display both TH1 and TH17 features8, 28. Similarly, TH2 cells with pathogenic properties have been identified from patients with allergy and asthma 29-33. However, only studies performed at single-cell resolution will enable scientists to determine if those “disease-specific” functional features are the result of an expansion of a pre-existing population of cells or the result of differentiation in response to environmental signals34, 35.
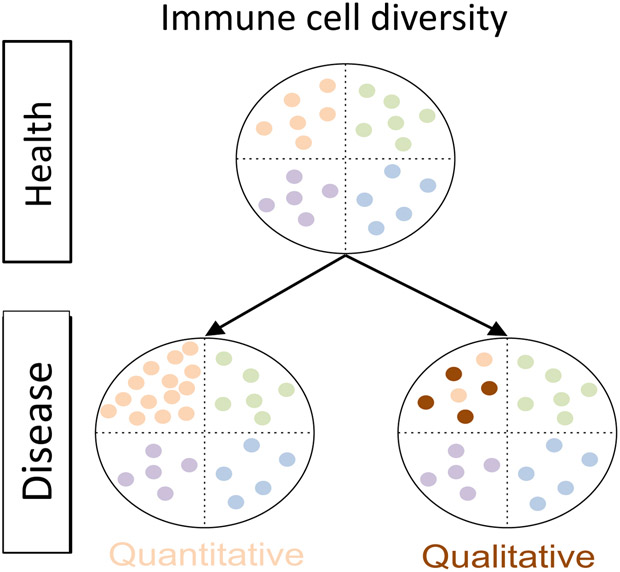
Figure 2. Schematic representation of the information provided by single-cell analysis.
Top panel shows the distribution of four immune cell subsets in a healthy individual. The bottom panel illustrates the possibilities that can happen in diseased states: either the relative proportion of one cell subset is increased without changes in their molecular program (quantitative change) and/or cells in one subset change their molecular program (qualitative change); such differences can be captured by single-cell analyses.
The power of single-cell omics analysis as a discovery tool has been well illustrated in the setting of cancer immunology. Just in the past 2 years, many studies have reported on single-cell omics analysis of the tumor microenvironment in several human cancer types. These studies have led to the identification of new immune cell subsets, new immune evasion and effector mechanisms, and potentially important molecular targets for cancer immunotherapy24, 36-44. For example, our recent single-cell study in lung cancer has led to the identification of a highly functional subset of tissue-resident memory T cells, which is likely to be the key cellular target of anti-PD1 therapy. Furthermore, it has defined a number of novel molecules that are likely to be important for modulating the function of this subset to achieve better anti-tumor immune responses45, 46. Single-cell analysis of tumor samples before and after anti-PD1 treatment have yielded insights into potential mechanisms and biomarkers that can distinguish responders from non-responders to immunotherapy45, 47. The studies in the cancer field highlight that single-cell analyses of diseased tissue samples can provide information relating to key pathogenic mechanisms driving disease as well as help understand mechanisms of action of novel interventions.
The relatively easy access to excess tumor tissue samples from standard diagnostic or therapeutic procedures has enabled researches to rapidly conduct these single-cell studies in large numbers of cells and patient samples. However, for lung diseases such as asthma and COPD it is not routine clinical practice to obtain airway tissue samples. This issue coupled with the limited amount of tissue that can be obtained for research purposes may explain the relative paucity of single-cell studies in lung diseases. One way to address this limitation is to study lung diseases in model organisms. For instance, single-cell analysis of the development of tracheal epithelial cells in vivo has led to the identification of new cell types and a better understanding of cell lineages in the context of pulmonary fibrosis48. More recently, in a mouse model of asthma driven by house dust mite allergen exposure, Tibitt et al. showed that the repertoire of T helper cell in the inflamed lung tissue is much more heterogeneous than usually accepted49. However, airway and lung disease are long-lasting diseases and mouse models are, by definition, only capturing part of the complex human biology. Thus, human studies are likely to provide deeper insights into disease pathology.
An important technical limitation for studying human tissue samples with low cell numbers is that the most commonly used high-throughput platforms for single-cell analysis requires several thousand cells as input material. Sample with just tens to hundreds of cells would require a different approach that relies on single-cell sorting by flow-cytometry into 96-well plate following by single-cell library preparation (Figure 1, upper panel). By far the most successful method is the optimized single-cell RNA-seq library preparation adapted from the Smart-seq2 protocol 50-53 (also commercially developed by Takara, ICell8). When combined with index-sorting, high-dimensional protein expression data also can be obtained in parallel54. This method, although laborious when prepared manually, allows researchers to review the complete surface phenotype of every single cell sorted into a plate and associate that event with its transcriptome. The other method is the new generation of droplet-based single-cell RNA-seq assay55, 56, commercialized by 10X Genomics. As shown by a comparative study57, Drop-seq assays generate lower resolution dataset, making computational analysis difficult, especially when analyzing highly similar cell subsets such as T cell subsets, as opposed to more diverse cell types such as peripheral blood mononuclear cells (Figure 1, lower panel). Our analysis showed that smart-Seq2 assay detected twice the number of genes compared to commercial Drop-Seq assay (10X Genomics)58. At the level of individual genes, the sensitivity of both assays was similar for highly-expressed genes such as B2M. However, for several genes of interest, only 30-40% of the cells showed expression using 10X Genomics whereas 85-95% of the cells showed expression when using Smart-seq2 assay58. This contrast is more important when working with resting immune cells, such as T cells, that contain a much lesser amount of RNA. In early 2019, 10X Genomics has released a newer version with a higher rate of mRNA capture (resolution). This assay when coupled with sample multiplexing using DNA oligo-barcoded-antibodies59 or DNA oligo-barcoded-lipid anchoring in cell membrane60 will enable the study of airway and lung samples at much higher resolution in a cost-effective manner.
Recently, NGS assays that profile the epigenetic state of cells are also available for studying clinical samples with limited cell numbers. Such single-cell epigenetic methods include, histone modification profiling by immunoprecipitation followed by sequencing (sc-ChIP-seq)61, 62 or the more accessible and cost-effective profiling of Transposase Accessible Chromatin with high-throughput sequencing (sc-ATAC-seq)63. These assays will complete the arsenal of tools required for understanding cellular heterogeneity and the mechanisms of cellular differentiation or activation in response to pathogenic perturbations64.
In summary, single-cell high-dimensional immune-phenotyping, RNA-sequencing, and chromatin accessibility profiling analyses of immune cells isolated from blood, airway and lung tissue samples from patients with lung diseases will provide insights into the molecular and cellular mechanisms that are driving health and diseased states. These approaches are likely to constitute the cardinal measures for the future development of diagnostic tools, novel biomarkers and therapeutic interventions in the emerging field of precision genomic medicine.
Acknowledgments
Funding: This work was supported by the National Institutes of Health [grant numbers R01HL114093, U19AI135731, U19AI142742-01, U19AI118626].
List of abbreviations:
- NGS
next-generation sequencing
- COPD
chronic obstructive pulmonary disease
- RNA-seq
RNA sequencing
- ATAC-seq
assay for transposase-accessible chromatin using sequencing
- ChIP-seq
chromatin immunoprecipitation followed by sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare that they have no relevant conflicts of interest. P.V receives research funding unrelated to this work from Pfizer.
REFERENCES
- 1.Schmiedel Benjamin J. DS, Madrigal Ariel, Valdovino-Gonzalez Alan G., White Brandie M., Zapardiel-Gonzalo Jose, Ha Brendan, Altay Gokmen, Greenbaum Jason A., McVicker Graham, Seumois Grégory, Rao Anjana, Kronenberg Mitchell, Peters Bjoern, Vijayanand Pandurangan. Impact of genetic polymorphisms on human immune cell gene expression. CELL 2018; In Press 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve NF, Guyatt AL, Jackson VE, et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet 2019; 51:494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Y, Biancotto A, Cheung F, Remmers E, Shah N, McCoy JP, et al. Systematic Analysis of Cell-to-Cell Expression Variation of T Lymphocytes in a Human Cohort Identifies Aging and Genetic Associations. Immunity 2016; 45:1162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson A, Rask-Andersen M, Karlsson T, Ek WE. Genome-wide association analysis of 350 000 Caucasians from the UK Biobank identifies novel loci for asthma, hay fever and eczema. Hum Mol Genet 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet 2009; 10:184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448:470–3. [DOI] [PubMed] [Google Scholar]
- 7.Jonkers IH, Wijmenga C. Context-specific effects of genetic variants associated with autoimmune disease. Hum Mol Genet 2017; 26:R185–R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, et al. Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J Immunol 2014; 193:2931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nature immunology 2014; 15:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seumois G, Zapardiel-Gonzalo J, White B, Singh D, Schulten V, Dillon M, et al. Transcriptional Profiling of Th2 Cells Identifies Pathogenic Features Associated with Asthma. Journal of Immunology 2016; 197:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayanand P, Seumois G, Simpson LJ, Abdul-Wajid S, Baumjohann D, Panduro M, et al. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity 2012; 36:175–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature biotechnology 2014; 32:1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jirtle RL. Epigenome: the program for human health and disease. Epigenomics 2009; 1:13–6. [DOI] [PubMed] [Google Scholar]
- 14.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007; 8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, Regev A. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol 2014; 107:4 22 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spaethling JM, Piel D, Dueck H, Buckley PT, Morris JF, Fisher SA, et al. Serotonergic neuron regulation informed by in vivo single-cell transcriptomics. FASEB J 2014; 28:771–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 2014; 510:363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods 2014; 11:22–4. [DOI] [PubMed] [Google Scholar]
- 19.Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014; 344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014; 343:193–6. [DOI] [PubMed] [Google Scholar]
- 22.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018; 18:35–45. [DOI] [PubMed] [Google Scholar]
- 23.Satija R, Shalek AK. Heterogeneity in immune responses: from populations to single cells. Trends Immunol 2014; 35:219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017; 169:1342–56 e16. [DOI] [PubMed] [Google Scholar]
- 25.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017; 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 2019; 101:207–23 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miragaia RJ, Gomes T, Chomka A, Jardine L, Riedel A, Hegazy AN, et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 2019; 50:493–504 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burel JG, Lindestam Arlehamn CS, Khan N, Seumois G, Greenbaum JA, Taplitz R, et al. Transcriptomic Analysis of CD4(+) T Cells Reveals Novel Immune Signatures of Latent Tuberculosis. J Immunol 2018; 200:3283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirahara K, Nakayama T. CD4+ T-cell subsets in inflammatory diseases: beyond the Th1/Th2 paradigm. Int Immunol 2016; 28:163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol 2011; 187:3111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo Y, Hirahara K, Yagi R, Tumes DJ, Nakayama T. Pathogenic memory type Th2 cells in allergic inflammation. Trends Immunol 2014; 35:69–78. [DOI] [PubMed] [Google Scholar]
- 32.Wambre E, Bajzik V, DeLong JH, O'Brien K, Nguyen QA, Speake C, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seumois G, Zapardiel-Gonzalo J, White B, Singh D, Schulten V, Dillon M, et al. Transcriptional Profiling of Th2 Cells Identifies Pathogenic Features Associated with Asthma. J Immunol 2016; 197:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, et al. Plasticity of human CD4 T cell subsets. Front Immunol 2014; 5:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat Rev Immunol 2016; 16:149–63. [DOI] [PubMed] [Google Scholar]
- 36.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018; 24:978–85. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018; 564:268–72. [DOI] [PubMed] [Google Scholar]
- 38.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2019; 176:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 2018; 24:1277–89. [DOI] [PubMed] [Google Scholar]
- 40.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017; 171:1611–24 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018; 175:984–97 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018; 24:986–93. [DOI] [PubMed] [Google Scholar]
- 43.Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 2019; 176:775–89 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019; 25:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 2018; 24:144–53. [DOI] [PubMed] [Google Scholar]
- 46.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer 2016; 16:525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prat A, Navarro A, Pare L, Reguart N, Galvan P, Pascual T, et al. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non-Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res 2017; 77:3540–50. [DOI] [PubMed] [Google Scholar]
- 48.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018; 560:319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tibbitt CA, Stark JM, Martens L, Ma J, Mold JE, Deswarte K, et al. Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity 2019; 51:169–84 e5. [DOI] [PubMed] [Google Scholar]
- 50.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013; 10:1096–8. [DOI] [PubMed] [Google Scholar]
- 51.Rosales SL, Liang S, Engel I, Schmiedel BJ, Kronenberg M, Vijayanand P, et al. A Sensitive and Integrated Approach to Profile Messenger RNA from Samples with Low Cell Numbers. Methods Mol Biol 2018; 1799:275–301. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen A, Khoo WH, Moran I, Croucher PI, Phan TG. Single Cell RNA Sequencing of Rare Immune Cell Populations. Front Immunol 2018; 9:1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mould KJ, Jackson ND, Henson PM, Seibold M, Janssen WJ. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell 2015; 16:712–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein AM, Macosko E. InDrops and Drop-seq technologies for single-cell sequencing. Lab Chip 2017; 17:2540–1. [DOI] [PubMed] [Google Scholar]
- 56.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015; 161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 2017; 65:631–43 e4. [DOI] [PubMed] [Google Scholar]
- 58.Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O'Rourke P, de Silva AD, et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol 2018; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017; 14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods 2019; 16:619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 2019; 51:1060–6. [DOI] [PubMed] [Google Scholar]
- 62.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 2015; 33:1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun 2018; 9:3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satpathy AT, Saligrama N, Buenrostro JD, Wei Y, Wu B, Rubin AJ, et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat Med 2018; 24:580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]