Abstract
Objectives:
To investigate the prevalence and molecular characteristics of defective DNA mismatch repair (dMMR) in small-bowel carcinoma (SBC) in a Japanese-hospital population.
Methods:
Immunohistochemistry was performed to evaluate the expression of MMR proteins (MLH1, MSH2, MSH6, and PMS2) in formalin-fixed paraffin-embedded sections prepared from surgically resected primary SBCs from 30 patients during March 2002 to March 2017. Genetic testing for Lynch syndrome was performed in patients who demonstrated MMR protein loss.
Results:
Two of 30 patients (6.7%) demonstrated concomitant loss of MSH2/MSH6 protein expression. Further genetic testing identified a pathogenic MSH2 variant in one of these patients.
Conclusions:
The prevalence of dMMR SBCs in a Japanese hospital-based population seems lower than that reported in previous studies. To determine whether dMMR SBCs might be strongly linked to Lynch syndrome, there is a need for further investigation with a larger sample size.
Keywords: small-bowel carcinoma, DNA mismatch repair, Lynch syndrome, microsatellite instability, universal tumor screening
Introduction
Defective mismatch repair (dMMR) in tumors leads to abnormal functions of MMR proteins because of either genetic[1] or epigenetic[2] events, resulting in the accumulation of errors during DNA replication, especially in the repetitive sequences known as microsatellites. The hallmarks of dMMR tumors are loss of MMR protein expression and/or high-level (high-frequency) microsatellite instability (MSI-H)[3]. Pembrolizumab, an anti-PD-1 antibody, was shown to improve overall survival in a subset of cancer patients with loss of MMR protein expression and/or MSI-H; therefore, it was approved by the US Food and Drug Administration in 2017, for the treatment of such tumors[4]. Thus, evaluation of MMR protein expression and/or MSI in cancer patients is expected to be helpful to stratify them into groups that will benefit from treatment with the anti-PD-1 antibody.
Small-bowel carcinomas (SBCs) are relatively uncommon neoplasms and account for < 2% of all gastrointestinal cancers[5]. As a chemotherapy regimen for patients with advanced SBC has not yet been established because of the rarity of the disease, screening for MMR protein loss by immunohistochemistry (IHC) and/or MSI-H is indicated in patients with SBC, for the identification of patients who could benefit from treatment with anti-PD-1 antibody agents. As documented in the revised Amsterdam criteria[6] and revised Bethesda guidelines[7], SBC is known to be associated with the Lynch syndrome (LS), an inherited autosomal dominant disease, caused by deleterious germline variants for one of the MMR genes, namely, MLH1, MSH2, MSH6, and PMS2, and the more recently identified, 3' deletion of EPCAM located upstream of MSH2[8].
Hypermethylation of the MLH1 promoter region is one of the three molecular mechanisms involved in tumors with dMMR and is a well-known epigenetic mechanism underlying dMMR function in various cancers, including SBCs[9]. In LS, somatic mutation or inactivation of the wild-type allele of the causative MMR gene along with a pathogenic germline variant results in a dysfunctional MMR system, thereby promoting cancer development[10]. Recently, somatic variants in MMR genes in the absence of germline variants have been revealed as a novel mechanism responsible for the development of tumors with dMMR in a subset of patients initially considered as having LS[11-13]. This condition is termed “Lynch-like syndrome (LLS)” or “LS mimic.” Although the occurrence of this syndrome has been extensively examined in colorectal cancers and endometrial cancers, it has been investigated scarcely in other LS-associated tumors; to the best of our knowledge, LLS has not been investigated in patients with SBC. Therefore, this study aimed to investigate the prevalence and molecular characteristics of dMMR in SBCs among a Japanese hospital-based population through a universal tumor screening approach with IHC for MMR protein expression.
Methods
Patient selection
Between March 2002 and March 2017, a total of 38 consecutive patients with primary SBC who underwent surgical resection of the primary tumor at the Saitama Medical Center, Saitama Medical University, Japan were enrolled in this study. The demographic/clinicopathologic data and personal/family histories of the patients were obtained from their medical records. This research was approved by the Institutional Review Board at the Saitama Medical Center, Saitama Medical University (Nos. 924, 925, and 926), and by the Ethics Committee at the Saitama Medical University and was conducted according to the guidelines put forth in the Declaration of Helsinki. Informed consent was obtained from each patient for the genetic testing of MMR genes. For deceased cases, consent was obtained from the next-of-kin (family members).
IHC for MMR proteins
The experimental protocol for performing IHC for MMR proteins and the primary antibodies used for the detection of MMR proteins (MLH1, MSH2, MSH6, and PMS2) have been described previously[14]. Briefly, 4-μm-thick sections prepared from formalin-fixed paraffin-embedded (FFPE) specimens of SBC samples were stained for the detection of four MMR proteins, as per the manufacturer's protocol. The normal staining pattern for MMR proteins is nuclear. Complete loss of nuclear staining in tumor cells with the presence of nuclear staining in non-neoplastic cells, such as normal small-bowel epithelial cells, lymphocytes, or stromal cells was considered to represent an abnormal pattern.
MSI testing
The MSI was tested using the MSI Analysis System, Version 1.2 (Promega Corporation, Madison, WI, US), as described previously[15]. The analytical system evaluated the MSI status of five mononucleotide microsatellite markers recommended by the revised Bethesda guidelines[7] and included BAT25, BAT26, NR21, NR22, and NR24. When two or more markers derived from the tumor DNA showed altered numbers of repeats compared to the markers derived from normal tissue or blood DNA, the tumor was considered to be showing MSI-H, and the MSI test was selectively performed in SBCs by evaluating the loss of MMR protein expression in IHC.
Detection of germline variants and copy number variations
Direct sequencing was used to perform the full-sequence analysis of the germline MMR genes in DNA extracted from the blood or FFPE specimens of normal mucosa, as described previously[14,16]. Pathogenic variants were detected by multi-gene panel analysis including 26 genes[16], and the identified variants were assessed using the InSiGHT classification criteria (http://insight-group.org/variants/classifications/). When no germline MMR variant was detected in the DNA samples, the samples were subjected to RNA sequencing to identify structural alterations of the MMR genes as essentially described previously[17].
Results
Patient background data
Thirty-eight patients were screened during the study period; of these, patients with SBCs associated with apparently cancer-predisposing diseases such as familial adenomatous polyposis (n = 7) or Crohn's disease (n = 1) were excluded. Therefore, 30 patients were evaluated in this study. The demographic and clinicopathologic characteristics of these 30 patients with SBC are shown in Table 1. The median age of the patients (16 males and 14 females) was 64 years (range: 17-87 years). The tumor location varied between duodenal (n = 11), jejunal (n = 14), and ileal (n = 5). AJCC-TNM staging of tumors[18] revealed that five patients had stage I, nine patients had stage II, six patients had stage III, and 10 patients had stage IV tumors. Histological examination revealed well-differentiated adenocarcinoma in 10 patients, moderately differentiated adenocarcinoma in 17 patients, poorly differentiated adenocarcinoma in one patient, mucinous adenocarcinoma in one patient, and adenosquamous cell carcinoma in one patient.
Table 1.
Demographic and Clinicopathologic Characteristics of Patients with Small Bowel Carcinoma.
| Number of patients (%) | n = 30 | ||
|---|---|---|---|
| Mean Age (range) (years) | 64 (17-87) | ||
| Sex | Male | 16 (53.3%) | |
| Female | 14 (46.7%) | ||
| Location of tumor | Duodenum | 11 (36.7%) | |
| Jejunum | 14 (46.7%) | ||
| Ileum | 5 (16.7%) | ||
| Stage | I | 5 (16.7%) | |
| II | 9 (30.0%) | ||
| IIA | 7 (23.3%) | ||
| IIB | 2 (6.7%) | ||
| III | 6 (20.0%) | ||
| IIIA | 5 (16.7%) | ||
| IIIB | 1 (3.3%) | ||
| IV | 10 (33.3%) | ||
| Histology | Well-differentiated adenocarcinoma | 10 (33.3%) | |
| Moderately differentiated adenocarcinoma | 17 (56.7%) | ||
| Poorly differentiated adenocarcinoma | 1 (3.3%) | ||
| Mucinous adenocarcinoma | 1 (3.3%) | ||
| Squamous cell carcinoma | 1 (3.3%) |
IHC for MMR proteins and MSI testing
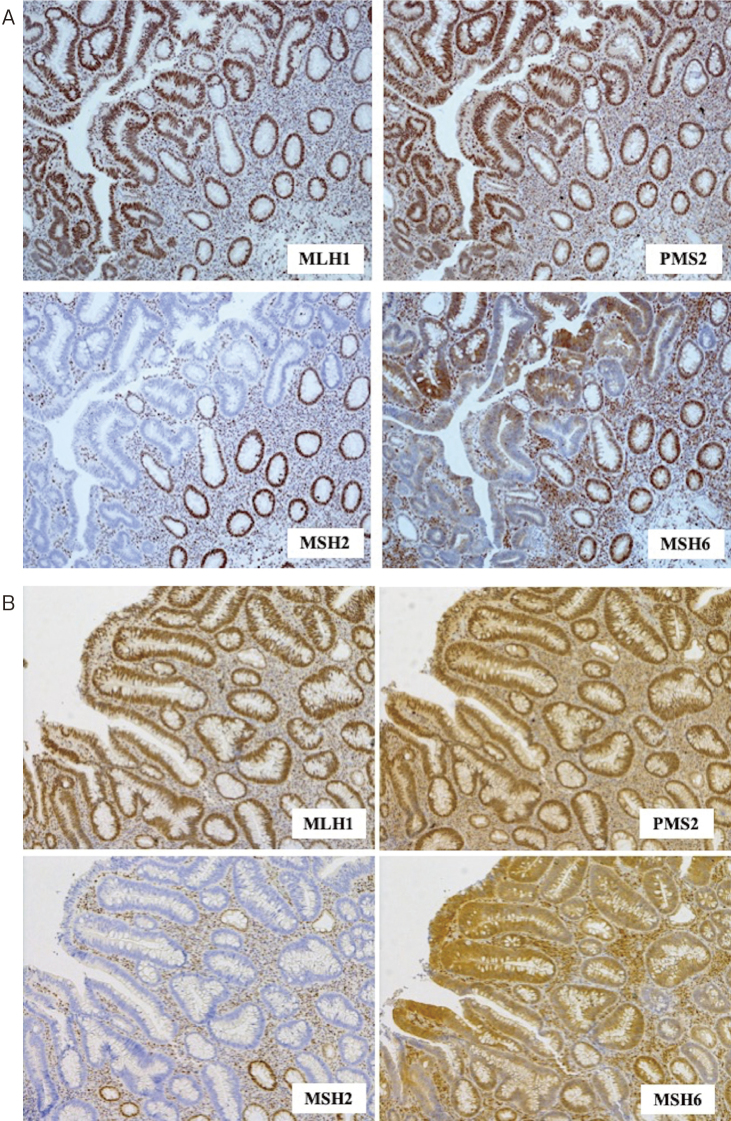
Of the 30 patients with SBC, two (6.7%) patients showed concomitant loss of MSH2 and MSH6 expression in their tumor samples (Figure 1A, 2A). Further, MSI-H was observed in both cases. No other patient showed MMR protein loss.
Figure 1.

(A) Immunohistochemistry for mismatch repair proteins in the jejunal carcinoma specimen of case 1, demonstrating concomitant loss of MSH2 and MSH6 expression. (B) Immunohistochemistry for mismatch repair proteins in the transverse colon carcinoma of case 1, demonstrating concomitant loss of MSH2 and MSH6 expression. (C) Immunohistochemistry for mismatch repair proteins in the sigmoid colon carcinoma specimen of case 1, demonstrating concomitant loss of MSH2 and MSH6 expression.
Figure 2.
(A) Immunohistochemistry for mismatch repair proteins in the ileal carcinoma specimens of case 2 demonstrating concomitant loss of MSH2 and MSH6 expression. (B) Immunohistochemistry for mismatch repair proteins in the ascending colon carcinoma of case 2 demonstrating concomitant loss of MSH2 and MSH6 expression.
Clinicopathologic/molecular characteristics of patients with dMMR SBC
The clinicopathologic/molecular characteristics of the two patients with dMMR SBC are summarized in Table 2. As both of these patients showed concomitant loss of MSH2 and MSH6 expression, we performed genetic testing for MSH2 in them (Figure 1A, 1B, 1C, 2A, and 2B). As a result of genetic testing, we identified a pathogenic germline variant, c.2038C>T/p.Arg 680 Ter in MSH2 (NM000251.2) in the 51-year-old man (case 1) with stage II A jejunal carcinoma. He had synchronously associated stage II transverse colon cancer and stage II sigmoid colon cancer, both of which were resected with curative intent. In case 2 (a 57-year-old man with stage IIIB ileal carcinoma synchronously associated with stage 0 ascending colon carcinoma, which had been endoscopically removed preoperatively), no germline MSH2 variant including copy number variants could be identified using the panel sequencing analysis of 26 genes related to hereditary gastrointestinal cancer / polyposis. Due to severe fragmentation of DNA extracted from the FFPE specimen of the SBC tissue, the somatic events leading to the inactivation of MSH2 in case 2 could not be analyzed. Further, IHC for MMR proteins was performed in colon cancer specimens from these two patients. All three tumors showed concomitant expression loss of MSH2 and MSH6. These two patients did not fulfill the revised Amsterdam criteria but met at least one item of the revised Bethesda guidelines. Notably, the elder brother of the case 2 had died of upper urothelial carcinoma, one of the LS-associated rare neoplasms, at the age of 45 years; however, MMR protein expression could not be evaluated by IHC in his case.
Table 2.
Clinicopathologic Characteristics and Evaluation of Germline Alterations in the MMR Genes of Two Patients with MMR Deficiency.
| Case No. | Age (years) | Sex | Number of rBG criteria fulfilled | Fulfillment of the rAMC | Tumor location | Histology | Stage | Pattern of mismatch repair protein loss | MSI status | Germline alterations of MMR genes | Class* | History of LS- associated tumor | Cancer family history |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 51 | M | 1 | No | Jejunum | Well-differentiated | IIA | MSH2/MSH6 | High (5/5) | MSH2 c.2038C>T | 5 | Synchronous T/C and S/C Ca. | Unknown |
| 2 | 57 | M | 2 | No | Ileum | Moderately differentiated | IIIB | MSH2/MSH6 | High (5/5) | ND | ND | Synchronous A/C Ca. | Elder brother: Upper urothelial carcinoma (45 y.o.) |
*Pathogenicity of the identified variants was confirmed based on the InSiGHT and ClinVar databases
MMR: Mismatch repair; LS: Lynch syndrome; Ca: carcinoma; A/C: Ascending colon; T/C: Transverse colon; S/C: Sigmoid colon; rBG: revised Bethesda guidelines; rAMC: revised Amsterdam criteria; y.o.: years old
Discussion
Immunohistochemistry-based analysis of MMR protein expression is increasingly replacing MSI testing as a preferred screening method for identifying MMR status in colorectal cancer[19], as well as in other LS-associated tumors. The concordance rate of the IHC and MSI testing results is more than 90% in cases of colorectal[20] and endometrial cancers[21], whereas such information is not yet available for SBCs. Michel et al.[22] reported a perfect concordance in the results from MSI-H and loss of MMR protein expression identified by IHC in 56 cases of SBC. Compared to MSI testing, IHC offers the distinct advantages of being cost-effective and widely available. In addition, it can also identify the genetic/epigenetic alterations in MMR genes[23]. Thus, in the present study, we selected IHC for analyzing the expression of MMR proteins in dMMR SBCs, where the frequency of MMR expression loss in SBCs is reported to vary widely (8.5%-33.3%)[9,24-26]. This could be partially explained by ethnic differences, use of different antibody clones, or different cutoffs for defining the loss of MMR expression. For example, Overmann et al.[26] considered immunoreactivity in <10% of tumor cells as loss of MMR expression and reported its frequency at 33.3% in SBCs. In contrast, the majority of recent studies, including the present one, define complete loss of nuclear staining as an indicator of the loss of MMR expression[14,23,27,28]. Furthermore, most of the earlier studies included patient criteria being based on cancer-related and/or a family history of LS, thereby potentially biasing their conclusions. We believe that the present study shows the prevalence of dMMR tumors among unselected SBCs more accurately than previous studies[9,24-26].
There is a scarcity of scientific data[9,24] examining dMMR status in SBCs. Xia et al.[24] analyzed a consecutive series of 71 surgically resected SBCs in six hospitals within the University of Pittsburgh Medical Center system over a period of eight years. The overall prevalence of SBCs was increased from 8.5% to 10.7% (6/56) after patients with known cancer-predisposing conditions such as Crohn's disease (n = 11), familial adenomatous polyposis (n = 3), and celiac disease (n = 1) were excluded from the study. Loss of MMR expression was concomitant with the loss of MSH2 and MSH6 (n = 3), followed by loss of MLH1 and PMS2, and isolated loss of MSH6 (n = 1). However, they did not analyze the germline MMR and therefore, the molecular mechanisms involved in dMMR in SBCs were not explained. In another study, Jun et al.[9] from the Korean Small Intestinal Cancer Center group analyzed LS-related SBCs in 195 consecutive cases of surgically resected SBCs collected from 22 Korean institutions. Although they reported the loss of MMR expression in 26% (51/195) of the SBC cases, they did not show the pattern of MMR loss in their samples. Their study focused on 40 patients with SBCs associated with synchronous or metachronous LS-related tumors (suspected LS group). Loss of MMR expression, both in SBCs and the matched synchronous or metachronous LS-related tumors, was identified in 15 cases. They excluded seven cases with MLH1 promoter methylation and analyzed germline MMR in the remaining eight cases. Loss of MMR expression was concomitant with the loss of MSH2 and MSH6 (n = 5), followed by the loss of PMS2, MSH2, and MSH6 (n = 2), and loss of MLH1 and PMS2 (n = 1). As they did not perform the germline MMR analysis in 35 dMMR SBC patients without suspected LS, the exact proportion of dMMR SBCs remains unclear. Our results based on the universal tumor screening of SBCs show a lower prevalence of dMMR SBCs compared to the earlier two reports[9,24]. However, due to severe degradation of tumor tissue DNA from case 2, we were unable to analyze the somatic inactivation of MSH2, and we thus regarded it as a case of LLS.
Relatively new and with the understanding of its pathogenesis still evolving, the term LLS in association with CRC was first coined in 2013[29] and, where it has not been described in patients with SBC, the term has come to be used for patients with LS-associated tumors such as CRC[14], epithelial ovarian cancer[30], sebaceous tumors[31], and upper urothelial cancer[32]. LLS may account for as many as 70% of all cases with suspected LS[29]. Analysis of colorectal cancers (CRCs) suggested that the LLS was a heterogeneous group including sporadic (non-hereditary) colorectal cancers and undetected LS, and was the intermediate risk for CRC[33]. Some investigators have reported that 50%-60% of LLS-associated CRCs exhibit biallelic somatic inactivation of MMR genes[11,12,33-35] through somatic mutations (nonsense, missense, frameshift, or splicing site deletions or loss of heterozygosity) and double somatic MMR pathogenic variants in their tumors. Although bi-allelic MMR gene inactivation has not been convincingly proven in these tumors, tumor phenotypes such as MSI-H and/or MMR expression loss detected by IHC support this notion. We could not exactly explain the mechanisms involved in the same loss (MSH2/MSH6) of MMR protein expression in both SBC and colon cancer in case 2, of which DNA extracted from FFPE tumor tissue showed a severe degradation. Because we conducted both DNA and RNA next-generation sequencing analysis of the MMR genes using peripheral blood cells of case 2, we could exclude the possibility of LS, even for rare cases with mosaic mutations, pathogenic variant within the deep intron region, or structural alterations such as inversion and translocation. However, considering the development of synchronous cancers in these organs with deficiency of MSH2 and MSH6 proteins, we speculate a possible involvement of some genetic background other than the MMR genes in the development of these cancers in case 2. In fact, several studies have already reported germline variants of candidate genes including BUB1, SETD2, FAN1, BARD1, WRN, MCPH1, and REV3L in the confirmed LLS cases[36,37].
Universal screening for LS using IHC for MMR proteins and/or the MSI test is a cost-effective method for CRC and endometrial cancer assessment[38,39]. It is difficult to estimate the efficacy of universal screening for LS in patients with SBCs, primarily because of their rarity and lack of sufficient scientific literature. Furthermore, in contrast with 82%-87.2% of dMMR colorectal and 77.%-93.5% of endometrial cancers that are caused by MLH1 hypermethylation[14,40-43], results of earlier studies and those of the present study have shown that the molecular mechanisms involved in the majority of dMMR SBCs are not epigenetic but rather suggest germline or somatic MMR gene inactivation[23,25]. Thus, routine IHC for MMR proteins may effectively screen LS (and occasionally LLS) and stratify the potential candidates for effective anti-PD-I-based therapy.
Although this study presents some interesting and convincing results, it has some limitations. For instance, these results are from a single institution, derived from the retrospective data of a relatively small sample size. Nevertheless, this study adds to the scant scientific literature describing the prevalence and molecular mechanisms involved in dMMR in SBCs. Given its clinical implications, the efficacy of universal screening for LS in patients with SBCs and the molecular mechanisms involved in dMMR in SBCs should be validated in further investigations with a larger sample.
Conflicts of Interest
There are no conflicts of interest.
Source of Funding
This work was supported in part by a grant-in-aid for the Support Project of the Strategic Research Center in Private Universities from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan awarded to the Saitama Medical University Research Center for Genomic Medicine [S1311002] and AMED [grant number JP18kk0205004].
Author Contributions
Tetsuya Ito, Hideyuki Ishida, and Okihide Suzuki drafted the manuscript. Tetsuya Ito, Hideyuki Ishida, Okihide Suzuki, Noriyasu Chika, Kunihiko Amano, and Keiichiro Ishibashi performed clinical management of these patients. Nao Kamae performed genetic counseling. Nao Kamae, Yuhki Tada, Kiwamu Akagi, Hidetaka Eguchi, and Yasushi Okazaki performed genetic testing in patients who demonstrated MMR protein loss. Hideyuki Ishida conceived and designed the sturdy and edited the manuscript. All authors read and approved the final manuscript. Tetsuya Ito and Okihide Suzuki contributed equally to this study.
Approval by Institutional Review Board (IRB)
This research was approved by the Institutional Review Board at Saitama Medical Center, Saitama Medical University (Nos. 924, 925, and 926), and the Ethics Committee at the Saitama Medical University and was conducted according to the guidelines put forth in the Declaration of Helsinki.
Informed Consent
Informed consent was obtained from each patient for the genetic testing of MMR genes. For deceased cases, consent was obtained from the next-of-kin (family members).
Acknowledgements
We thank Ms. Aya Saitoh, Fumiyo Fukui, and Akemi Takahashi for their skillful technical assistance.
References
- 1.Peltomäki P. Update on Lynch syndrome genomics. Fam Cancer. 2016 Jul; 15(3): 385-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pal T, Permuth-Wey J, Sellers TA. A review of the clinical relevance of mismatch-repair deficiency in ovarian cancer. Cancer. 2008 Aug; 113(4): 733-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canard G, Lefevre JH, Colas C, et al. Screening for Lynch syndrome in colorectal cancer: are we doing enough? Ann Surg Oncol. 2012 Mar; 19(3): 809-16. [DOI] [PubMed] [Google Scholar]
- 4.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015 Jun; 372(26): 2509-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashley SW, Wells SA. Tumors of the small intestine. Semin Oncol. 1988 Apr; 15(2): 116-28. [PubMed] [Google Scholar]
- 6.Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology. 1999 Jun; 116(6): 1453-6. [DOI] [PubMed] [Google Scholar]
- 7.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004 Feb; 96(4): 261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sehgal R, Sheahan K, O'Connell PR, et al. Lynch syndrome: an updated review. Genes (Basel). 2014 Sep; 5(3): 497-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jun SY, Lee EJ, Kim MJ, et al. Lynch syndrome-related small intestinal adenocarcinomas. Oncotarget. 2017 Mar; 8(13): 21483-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colas C, Coulet F, Svrcek M, et al. Lynch or not Lynch? Is that always a question? Adv Cancer Res. 2012 Jan; 113: 121-66. [DOI] [PubMed] [Google Scholar]
- 11.Geurts-Giele WR, Leenen CH, Dubbink HJ, et al. Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol. 2014 Dec; 234(4): 548-59. [DOI] [PubMed] [Google Scholar]
- 12.Haraldsdottir S, Hampel H, Tomsic J, et al. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014 Dec; 147(6): 1308-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014 Mar; 146(3): 643-6. [DOI] [PubMed] [Google Scholar]
- 14.Chika N, Eguchi H, Kumamoto K, et al. Prevalence of Lynch syndrome and Lynch-like syndrome among patients with colorectal cancer in a Japanese hospital-based population. Jpn J Clin Oncol. 2017 Feb; 47(2): 108-17. [DOI] [PubMed] [Google Scholar]
- 15.Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 2002 Dec; 123(6): 1804-11. [DOI] [PubMed] [Google Scholar]
- 16.Kohda M, Kumamoto K, Eguchi H, et al. Rapid detection of germline mutations for hereditary gastrointestinal polyposis/cancers using HaloPlex target enrichment and high-throughput sequencing technologies. Fam Cancer. 2016 Oct; 15(4): 553-62. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Yamaguchi T, Wakatsuki T, et al. The single-base-pair deletion, MSH2 c.2635-3delC affecting intron 15 splicing can be a cause of Lynch syndrome. Jpn J Clin Oncol. 2019 May; 49(5): 477-80. [DOI] [PubMed] [Google Scholar]
- 18.Edge S, Byrd DR, Compton C, et al. AJCC cancer staging manual. 7th ed. New York: Springer, 2010. [Google Scholar]
- 19.De la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010 Jul; 28(20): 3380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002 Feb; 20(4): 1043-8. [DOI] [PubMed] [Google Scholar]
- 21.Mills AM, Longacre TA. Lynch syndrome: female genital tract cancer diagnosis and screening. Surg Pathol Clin. 2016 Jun; 9(2): 201-14. [DOI] [PubMed] [Google Scholar]
- 22.Michel S, Kloor M, Singh S, et al. Coding microsatellite instability analysis in microsatellite unstable small intestinal adenocarcinomas identifies MARCKS as a common target of inactivation. Mol Carcinog. 2010 Feb; 49(2): 175-82. [DOI] [PubMed] [Google Scholar]
- 23.Shia J, Tang LH, Vakiani E, et al. Immunohistochemistry as first-line screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: a 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol. 2009 Nov; 33(11): 1639-45. [DOI] [PubMed] [Google Scholar]
- 24.Xia M, Singhi AD, Dudley B, et al. Small bowel adenocarcinoma frequently exhibits Lynch syndrome-associated mismatch repair protein deficiency but does not harbor sporadic MLH1 deficiency. Appl Immunohistochem Mol Morphol, 2017 Jul; 25(6): 399-406. [DOI] [PubMed] [Google Scholar]
- 25.Aparicio T, Svrcek M, Zaanan A, et al. Small bowel adenocarcinoma phenotyping, a clinicobiological prognostic study. Br J Cancer. 2013 Dec; 109(12): 3057-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overman MJ, Pozadzides J, Kopetz S, et al. Immunophenotype and molecular characterisation of adenocarcinoma of the small intestine. Br J Cancer. 2010 Jan; 102(1): 144-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garg K, Leitao MM, Jr., Kauff ND, et al. Selection of endometrial carcinomas for DNA mismatch repair protein immunohistochemistry using patient age and tumor morphology enhances detection of mismatch repair abnormalities. Am J Surg Pathol. 2009 Jun; 33(6): 925-33. [DOI] [PubMed] [Google Scholar]
- 28.Gu MJ, Bae YK, Kim A, et al. Expression of hMLH1, hMSH2 and hMSH6 in small intestinal carcinomas. Hepatogastroenterology. 2012 Oct; 59(119): 2228-32. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Soler M, Pérez-Carbonell L, Guarinos C, et al. Risk of cancer in cases of suspected lynch syndrome without germline mutation. Gastroenterology. 2013 May; 144(5): 926-32; quiz e13-4. [DOI] [PubMed] [Google Scholar]
- 30.Tajima Y, Eguchi H, Chika N, et al. Prevalence and molecular characteristics of defective mismatch repair epithelial ovarian cancer in a Japanese hospital-based population. Jpn J Clin Oncol. 2018 Aug; 48(8): 728-35. [DOI] [PubMed] [Google Scholar]
- 31.Kuwabara K, Suzuki O, Chika N, et al. Prevalence and molecular characteristics of DNA mismatch repair protein-deficient sebaceous neoplasms and keratoacanthomas in a Japanese hospital-based population. Jpn J Clin Oncol. 2018 Jun; 48(6): 514-21. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, Kono K, Eguchi H, et al. Prevalence of Lynch syndrome among patients with upper urinary tract carcinoma in a Japanese hospital-based population. Jpn J Clin Oncol. 2020 Jan; 50(1): 80-8. [DOI] [PubMed] [Google Scholar]
- 33.Boland CR. The mystery of mismatch repair deficiency: lynch or lynch-like? Gastroenterology. 2013 May; 144(5): 868-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sourrouille I, Coulet F, Lefevre JH, et al. Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam Cancer. 2013 Mar; 12(1): 27-33. [DOI] [PubMed] [Google Scholar]
- 35.Kang SY, Park CK, Chang DK, et al. Lynch-like syndrome: characterization and comparison with EPCAM deletion carriers. Int J Cancer. 2015 Apr; 136(7): 1568-78. [DOI] [PubMed] [Google Scholar]
- 36.Vargas-Parra GM, González-Acosta M, Thompson BA, et al. Elucidating the molecular basis of MSH2-deficient tumors by combined germline and somatic analysis. Int J Cancer. 2017 Oct; 141(7): 1365-80. [DOI] [PubMed] [Google Scholar]
- 37.Xicola RM, Clark JR, Carroll T, et al. Implication of DNA repair genes in Lynch-like syndrome. Fam Cancer. 2019 Jul; 18(3): 331-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruegl AS, Djordjevic B, Batte B, et al. Evaluation of clinical criteria for the identification of Lynch syndrome among unselected patients with endometrial cancer. Cancer Prev Res (Phila). 2014 Jul; 7(7): 686-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gudgeon JM, Williams JL, Burt RW, et al. Lynch syndrome screening implementation: business analysis by a healthcare system. Am J Manag Care. 2011 Aug; 17(8): e288-300. [PubMed] [Google Scholar]
- 40.Adar T, Rodgers LH, Shannon KM, et al. Universal screening of both endometrial and colon cancers increases the detection of Lynch syndrome. Cancer. 2018 Aug; 124(15): 3145-53. [DOI] [PubMed] [Google Scholar]
- 41.Bruegl AS, Ring KL, Daniels M, et al. Clinical challenges associated with universal screening for Lynch syndrome associated endometrial cancer. Cancer Prev Res (Phila). 2017 Feb; 10(2): 108-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins JC, Yang EJ, Muto MG, et al. Universal screening for mismatch-repair deficiency in endometrial cancers to identify patients with Lynch syndrome and Lynch-like syndrome. Int J Gynecol Pathol. 2017 Mar; 36(2): 115-27. [DOI] [PubMed] [Google Scholar]
- 43.Dillon JL, Gonzalez JL, DeMars L, et al. Universal screening for Lynch syndrome in endometrial cancers: frequency of germline mutations and identification of patients with Lynch-like syndrome. Hum Pathol. 2017 Dec; 70: 121-8. [DOI] [PubMed] [Google Scholar]