ABSTRACT
Since its discovery 150 years ago, the neural crest has intrigued investigators owing to its remarkable developmental potential and extensive migratory ability. Cell lineage analysis has been an essential tool for exploring neural crest cell fate and migration routes. By marking progenitor cells, one can observe their subsequent locations and the cell types into which they differentiate. Here, we review major discoveries in neural crest lineage tracing from a historical perspective. We discuss how advancing technologies have refined lineage-tracing studies, and how clonal analysis can be applied to questions regarding multipotency. We also highlight how effective progenitor cell tracing, when combined with recently developed molecular and imaging tools, such as single-cell transcriptomics, single-molecule fluorescence in situ hybridization and high-resolution imaging, can extend the scope of neural crest lineage studies beyond development to regeneration and cancer initiation.
KEY WORDS: Neural crest, Lineage tracing, Clonal analysis, Multipotency, Tumor development, Regeneration
Summary: This Review outlines the major discoveries in neural crest lineage tracing and how these have been refined to extend beyond development to regeneration and cancer initiation.
Introduction
The neural crest is an embryonic stem cell population unique to vertebrates. In 1868, Wilhelm His first described the ‘Zwischenstrang’ as a band of cells residing between the neural tube and epidermis that later migrates laterally to form the spinal ganglion (His, 1868). Today, this cell population is known as the neural crest. Induced at the neural plate border, neural crest cells undergo an epithelial-to-mesenchymal transition (EMT) to leave the neural tube. They then migrate laterally and ventrally, eventually giving rise to cell types as diverse as cartilage of the head, connective tissue, pigment cells, cardiac outflow septum and the peripheral nervous system. As cells arising from different levels of the rostrocaudal body axis give rise to different cell types, the neural crest can be subdivided into cranial, vagal, trunk and sacral subpopulations.
Although experimental embryologists over the past century have mapped neural crest cells at all of these axial levels, recent studies using refined tools have revealed previously unrecognized neural crest derivatives and behaviors. Thus, there are many interesting unanswered questions about neural crest development. For example, how do migrating neural crest cells interact with each other, both molecularly and mechanically? How are lineage decisions coupled with migration, and how do neural crest cells interact with the rapidly developing surrounding tissue? As neural crest cells have stem cell properties, what is their degree of fate restriction versus multipotency to form diverse cell types? How does this vary along the rostrocaudal body axis? In the adult, do neural crest-derived cells participate in tissue repair? How and why do things go wrong, resulting in neural crest-derived tumor formation? These questions require the means to indelibly label the progeny of single cells or specific groups of neural crest cells in vivo, while enabling assessment of their transcriptomic landscape, spatial characteristics and cellular dynamics.
In this Review, we describe how the neural crest has been studied from a chronological perspective, beginning with methods for lineage tracing at the population level, followed by recent advances in clonal analysis to explore the developmental potential of individual cells. We comment on the advantages and limitations of different approaches, and the adaptation of new tools to questions in the field as technology proceeded. Last, we extend the discussion to the broader application of lineage tracing neural crest-derived cells during regeneration and in cancer.
Fate-mapping neural crest cells at a population level: where do they go, what do they become?
In the first half of the 20th century, the neural crest was studied intensely by experimental embryologists who primarily used amphibian embryos and took advantage of vital dye labeling, interspecies/heterotopic grafting and cell-ablation techniques. In 1950, Sven Hörstadius presented a thorough overview of the state of knowledge regarding neural crest induction, migration and differentiation (Hörstadius, 1950). Although the neural crest then was ‘only sparsely treated by the authors of text books’ (Hörstadius, 1950), many of the results and perspectives in Hörstadius' book remain incredibly insightful. The broadness and accuracy of knowledge obtained by removing, grafting or rotating pieces of tissue is remarkable from today's perspective.
The dawn of experimental embryology
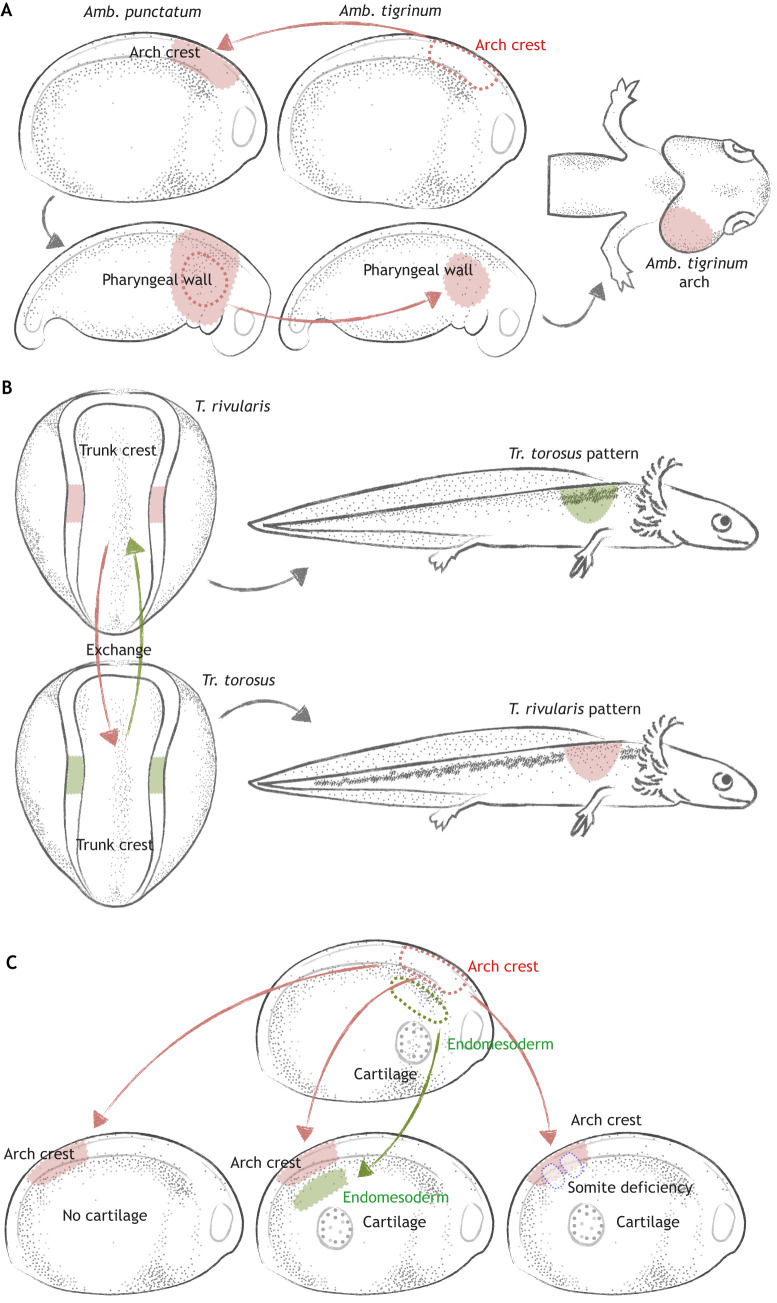
One lineage-tracing experiment Hörstadius addressed as ‘exceedingly beautiful’ was performed by Ross Harrison in 1935. To understand whether neural crest cells contribute to the gill arch skeleton, Harrison exchanged hindbrain regions between two amphibian species, Amblystoma punctatum and Amb. tigrinum or mexicanum, the gill arches of which vary in size. After neural crest cells had migrated to the arch, the branchial arch itself – including both donor and host cells – was transplanted to another host of the same species. In this way, only branchial neural crest cells were derived from the donor. Gill size in the resulting chimera matched that of the donor on the transplanted side, suggesting that intrinsic information in the neural crest contributed to the arch skeleton (Harrison, 1935) (Fig. 1A). The migratory properties of neural crest cells were examined in a similar way by Victor Twitty, who interchanged trunk neural crest cells between two species of salamander, Triturus rivularis and Trogulus torosus, with distinct pigment patterns. He discovered that grafted cells undergo migratory patterns characteristic of the donor, indicating that intrinsic properties of the pigment cells, rather than the environment, affected cell migration (Twitty, 1945) (Fig. 1B). A caveat of such interspecific grafts, however, is that they may alter normal tissue interactions, thus modulating signaling and timing events between donor and host tissues in unpredictable ways (Hörstadius, 1950).
Fig. 1.
Understanding neural crest development through transplantation experiments. (A) A neural crest contribution to the gill arch skeleton was mapped by interchanging hindbrain and ectoderm (arch crest, red) between Amb. punctatum and Amb. tigrinum. After migration, the pharyngeal wall (outlined in red) containing neural crest cells from the donor (Amb. tigrinum) was transplanted to another host. In the chimeric animal, the gill arch skeleton on the transplanted side was larger than that on the host side owing to different growth rates between the two species, demonstrating that the gill arch skeleton originates from hindbrain neural crest. (B) Neural crest cells give rise to melanophores. To understand whether patterning information is intrinsic to neural crest cells or imparted by the environment, trunk neural crest was reciprocally exchanged between two species: T. rivularis (red) and Tr. torosus (green). When grafted into other species, pigment cells continued to form patterns characteristic of the donor, indicating that pigment distribution is dependent on the intrinsic ability of neural crest cells to migrate and aggregate. (C) Heterotopic grafts were used to explore the role of tissue interactions during neural crest chondrogenesis. Usually, cranial neural crest (arch crest, red) cells cannot give rise to cartilage when grafted to the trunk. However, when endomesodermal cells (green) were grafted from the gill arch to the trunk, or when the somites in the trunk were removed (blue dashed lines), the grafted cranial crest cells differentiated into cartilage, suggesting that interaction with branchial endomesodermal cells, or signals released by somite removal, are required for chondrogenic differentiation. Colored arrows represent grafting; gray arrows represent development.
Thanks to the ability to combine different tissue types, grafting experiments have also provided insights into cell-cell interactions. To distinguish whether cranial neural crest cells can differentiate into cartilage autonomously, or whether tissue interactions are required, Hörstadius and others transferred cranial neural crest cells into the trunk region (Hörstadius and Sellman, 1946). Unless other manipulations, such as removing the somite or grafting endomesodermal cells from the gill arch were conducted concomitantly, cranial crest cells could not differentiate into cartilage in the trunk. This suggested that, at the stage grafted, neural crest cells were not yet fated toward chondrogenic lineages. Moreover, differentiation into cartilage required interaction with adjacent tissue at the cranial region or signals from the environment released at the wound where the somite was removed (Hörstadius and Sellman, 1946) (Fig. 1C). Interestingly, these results differ from similar experiments carried out in birds, which suggest that some avian cranial neural crest cells can form cartilage nodules when transplanted to the trunk (Le Douarin and Kalcheim, 1999). This may be due to species-specific differences or to evolutionary changes that render the amniote cranial neural crest gene regulatory circuitry more hard-wired to form craniofacial cartilage (Martik et al., 2019).
While amphibian transplantations elegantly pioneered the field of neural crest experimental embryology, several controversies arose because only partial answers could be obtained, as acknowledged by the authors. In the following decades, neural crest researchers expanded lineage analysis methods to adapt to various model organisms. As predicted, these efforts greatly revolutionized the field by ‘attack[ing] the problems on sufficiently broad lines’ (Hörstadius, 1950).
Quail-chick and other chimerae
Two decades after the publication of Hörstadius' treatise, Nicole Le Douarin extended grafting techniques to avian models using chick-quail chimerae. She took advantage of the fact that quail and chick embryos resemble one another morphologically but can be distinguished at the cellular level: quail cells have condensed heterochromatin in their nucleoli, whereas chick cells are euchromatic, such that cells of the two species can be distinguished by staining for DNA. In these chimerae, the dorsal neural tube or neural folds from quail donors are grafted to replace chick host tissue of equivalent stages for lineage tracing, or of different stages for exploring the developmental potential of the grafted cells. Using this technique, Le Douarin and colleagues identified several populations of neural crest cells along the body axis. The most rostral ‘cranial’ neural crest cells formed the dermis of the face and neck, large arteries, craniofacial bone and cartilage, as well as cranial ganglia (Le Lièvre and Le Douarin, 1975). Vagal neural crest from the caudal hindbrain formed the enteric neurons of the gut, including the large intestines and rectum (Le Douarin and Teillet, 1973). Trunk neural crest cells formed dorsal root and sympathetic ganglia, as well as contributing to the adrenal gland (Le Douarin and Kalcheim, 1999). Like the vagal neural crest, the sacral neural crest also contributed to the enteric nervous system but was restricted to the post-umbilical region (Le Douarin and Teillet, 1973). Neural crest cells from all axial levels were found to contribute to melanocytes (Le Douarin and Kalcheim, 1999).
Quail-chick chimerae were particularly useful for fine resolution fate mapping of the skull, revealing that cranial neural crest derived from each rhombomere contributed to distinct segments of the skull (Couly et al., 1993; Kontges and Lumsden, 1996). Similarly, the caudal hindbrain or ‘cardiac’ neural crest was shown to contribute to the aorticopulmonary septum of the heart (Phillips et al., 1987; Miyagawa-Tomita et al., 1991). Moreover, surgical removal of the cardiac neural folds resulted in defects in the formation of the outflow tract septum, phenocopying the human congenital defect persistent truncus arteriosus (Kirby et al., 1983), which results in mixing of blood flowing to and from the lungs, and transposition of the vessels.
Quail-chick grafting approaches also made it possible to challenge cells by exposing them to diverse environments at distinct times. To analyze the developmental potential of the cranial neural crest, Le Lievre and Le Douarin performed ‘back-transplantation’ experiments by placing pieces of the nodose ganglion, the glia of which are neural crest derived, onto the neural crest migration pathway of a younger chick embryo. The results showed that some quail cells resumed migration and contributed to autonomic structures, suggesting that environmental factors can turn back the differentiation clock (Ayer-Le Lievre and Le Douarin, 1982). Typically, the first neural crest cells to emigrate from the neural tube move the furthest ventrally, e.g. forming craniofacial cartilage, whereas later migrating cells remain closer to the neural tube. However, when grafted to replace early migrating cells, late migrating cells still gave rise to jaw skeleton, thus demonstrating that the early and late migrating populations have similar developmental potential (Baker et al., 1997). Whereas these studies demonstrated developmental plasticity, others suggested that axial level-specific patterning information may be inherent to some neural crest populations prior to emigration. For example, in 1983, Drew Noden replaced cells that would normally migrate into the second or third arches with cells of the first arch, resulting in ectopic beak formation, suggesting that some neural crest cells retain axial level-specific patterning information (Noden, 1983).
Neural crest formation is controlled by a feed-forward gene regulatory network (GRN), which comprises transcription factors and signaling pathways, and is partitioned into developmental modules that control events from induction at the neural plate border to differentiation into numerous cell types (Sauka-Spengler and Bronner-Fraser, 2008). Although all neural crest cells share common GRN components, there are also regional differences along the body axis. This raises the important question of which transcription factors underlie the different neural crest axial identities as distinct as cranial, vagal or trunk? With recent progress in transcriptomics, it has been possible to revisit classical grafting techniques to identify axial level-specific GRNs that determine neural crest identity. For example, trunk neural crest, when grafted in place of cranial or cardiac neural crest, maintains its original axial identity and cannot give rise to cranial cartilage or contribute to the cardiac outflow septum. However, when genes of a cranial-specific subcircuit of the GRN, namely Sox8, Tfap2b and Ets1, are expressed in the trunk neural tube, this is sufficient to activate a cranial-specific promoter in trunk neural crest cells. Moreover, when this ‘reprogrammed’ trunk neural tube is grafted into the head, these cells can differentiate into cartilage, something that trunk neural crest cells normally cannot do (Simoes-Costa and Bronner, 2016). Similarly, ectopic expression in the trunk neural tube of genes comprising a cardiac crest-specific subcircuit, Sox8, Tgif1 and Ets1, can redirect trunk neural crest cells grafted in place of ablated cardiac neural crest to migrate into the cardiac outflow tract, thus rescuing persistent truncus arteriosus (Gandhi et al., 2020). These two examples demonstrate that subtle differences in gene regulatory architectures can alter neural crest cell fate and account for axial level-specific differences between neural crest subpopulations; a more detailed discussion of neural crest regionalization along the AP axis is provided in the accompanying review by Rocha et al. (2020).
While quail and chick were chosen for chimeric analyses due to their similarities, other chimeric combinations have been chosen for their distinct characteristics. For example, because ducks have different head and jaw structures compared with quails, the role of neural crest cells in morphogenesis of the cranial skeleton can be analyzed through quail-duck (quck) chimerae (Lwigale and Schneider, 2008). In these qucks, mesenchymal neural crest forms a beak (Schneider and Helms, 2003) and branchial skeleton (Tucker and Lumsden, 2004) that is characteristic of the donor. These results suggest that, despite environmental and tissue interactions, neural crest-derived cells follow the patterning information of the donor species. More recent studies using the quck chimeric system have explored molecular aspects of species-dependent morphogenesis, including cell cycle regulators (Hall et al., 2014), rates of bone resorption (Ealba et al., 2015) and FGF/TGFβ signaling (Woronowicz et al., 2018).
The mouse-chick chimeric system offers a similar advantage to studying morphological features, such as the development of teeth. Whereas teeth have many neural crest-derived components, including condensed dental mesenchyme, dental papilla, odontoblasts and dentine matrix, chickens lack teeth. Interestingly, combining molar mesenchyme from the mouse with chick epithelium can trigger tooth development in the chick (Kollar and Fisher, 1980; Mitsiadis et al., 2003), suggesting that loss of odontogenesis during avian evolution is associated with the loss of a signal from the chick mesenchyme. Another advantage of rodent-avian chimerae is that this approach enables the use of mammalian genetics in a surgically accessible environment. For example, neural crest cells from splotch mutant mice (which exhibit neural crest defects) can migrate normally after being grafted into chick embryos, suggesting that an abnormal tissue interaction is the underlying cause of defective neural crest migration in the mutant rather than a defect intrinsic to the neural crest (Serbedzija and McMahon, 1997). Recently, rat-chick chimerae were used to study the developmental potential of enteric precursor cells (White and Anderson, 1999). In addition, by transplanting mouse hindgut into a chick-quail host before neural crest colonization, another study was able to show that components of the extracellular matrix, such as collagen 18 and agrin, secreted by enteric neural crest precursors direct their migration through a cell-specific microenvironment (Nagy and Goldstein, 2006; Nagy et al., 2018). These methods have also revealed that a limited number of progenitor cells can give rise to a vast number of enteric neurons; such enteric precursor cells were termed ‘superstars’ due to their extensive proliferative and colonization ability (Cheeseman et al., 2014).
Despite their utility, interspecies grafts have some disadvantages. For example, grafts require healing time, such that cell migration might be delayed, potentially causing some early migrating neural crest derivatives to have been missed. In addition, differences in developmental timing and cellular environment between distinct, albeit related, species may change cell behavior. Thus, many interspecific chimerae have been or are being reproduced with intraspecific grafts using GFP chick donors to circumvent the challenges of using two different species. A useful complement to these experiments is the direct labeling of neural crest cells in situ through non-invasive approaches such as dye or viral labeling, as discussed below.
Dye labeling
In the 1980s and 1990s, vital dyes such as fluorescent dextrans like lysinated rhodamine dextran (LRD) or the lipophilic dye DiI were widely used for labeling single or populations of neural crest cells. These approaches have the advantage of enabling direct observation of the labeled cell or population using a fluorescence microscope immediately after injection; however, these labels are transient, and the signal remains visible for only a few days due to progressive dilution as neural crest cells divide. Because vital dye is less invasive, flexible and requires almost no healing time compared with grafting experiments, it is an ideal tool for analyzing early neural crest migration events. For example, using DiI injection, Serbedzija and co-workers traced dorsolateral and ventral pathways of trunk neural crest migration. By injecting at distinct time points, the authors concluded that cells that migrate first give rise to the most ventral structures (Serbedzija et al., 1989). Similarly, another study took advantage of focal injection into a small group of cells to map the migration of cranial neural crest from particular rhombomeres into the branchial arches (Lumsden et al., 1991). Combined with ex utero culturing technique, vital dye also has been applied to study trunk (Serbedzija et al., 1990), cranial (Serbedzija et al., 1992; Trainor et al., 2002) and sacral (Serbedzija et al., 1991) neural crest cells in mammalian systems.
Dye injection is an ideal complementary approach to other lineage-tracing methods, such as grafting (Krotoski et al., 1988), retroviral lineage tracing (Pomeranz et al., 1991) or neural crest-specific genetic lines (Yoshida et al., 2008). For example, one study combined the use of Wnt1-Cre mice with DiI injection at specific axial levels to study lineage contributions to craniofacial mesenchyme. The results showed that the frontal and parietal bones of the skull originate primarily from proliferation of cranial neural crest-derived cells, with only a minor contribution from the adjacent tissue (Yoshida et al., 2008).
Retroviruses for lineage tracing
Genetic cell labeling via retroviral infection was developed in birds in the 1980s and 1990s, and was applied to studies of the central and peripheral nervous system (Sanes, 1989). To prevent horizontal infection of adjacent cells, envelope protein was removed from the viral genome, creating a recombinant replication incompetent retrovirus. By removing its ability to transmit infection to other cells, an infected progenitor cell can faithfully pass the reporter signal only to its descendants (Chen et al., 1999). Using LacZ as a reporter, retroviral labeling studies have been applied to map the contributions of cardiac neural crest cells to smooth muscle and neurons of the cardiovascular system (Poelmann et al., 1998), and to study vagal neural crest migration into the enteric nervous system (Epstein et al., 1994). This type of lineage tracing also confirmed trunk neural crest contributions to the sympathetic chain ganglia and melanocytes (Stocker et al., 1993). The results were consistent with those obtained through vital dye labeling and quail-chick chimera. Because retroviruses can permanently integrate reporters such as LacZ into the host genome, a unique advantage of this approach is that it supports long-term analysis without signal dilution through time. On the other hand, retrovirus-based labeling is not ideal for short-term analysis, because viral reporters require at least 12 h after initial contact with the cells to be expressed at sufficient levels for detection.
Neural crest-specific driver lines
Unlike amphibians and birds, mammalian embryos are less accessible to surgical manipulations. On the other hand, injection- and transplantation-based approaches only label a subpopulation of cells, raising the possibility that some contributions may have been missed. Using the power of genetics, the Cre-loxP system in combination with a neural crest-specific driver line was established to label the neural crest population in mammalian embryos for long-term lineage analysis. Commonly used Cre-driver lines, recently reviewed in detail by (Debbache et al. 2018) include P3Pro-Cre (Li et al., 2000) and Wnt1-Cre (Danielian et al., 1998) for premigratory neural crest, and Ht-PA-Cre (Pietri et al., 2003), P0-Cre (Yamauchi et al., 1999), Sox10-Cre (Matsuoka et al., 2005) and Mef2c-F10N-Cre (Aoto et al., 2015) for migratory neural crest. Wnt1-Cre has been successfully used in fate mapping of cranial (Chai et al., 2000) and cardiac (Jiang et al., 2000) neural crest cells. The comprehensive nature of Wnt1-Cre driver lines can uncover patterns of morphogenesis, which sparse labeling cannot achieve. For example, during tooth and mandibular development, Wnt1-Cre labeled neural crest cells are dynamically located at the chondrogenic front, while mesodermal tissue contributes to the interior of the cartilage (Chai et al., 2000). To circumvent the potential caveat caused by Wnt1-Cre driver lines due to ectopic activation of Wnt signaling, a new Wnt1-Cre2 transgenic mouse line was introduced to label the neural crest in a similar manner but without ectopic Wnt expression (Lewis et al., 2013).
A number of reporter lines have also been developed to label neural crest cells in zebrafish. One such line that is widely used is Tg(-4.9sox10:egfp) (Carney et al., 2006), in which a Sox10 regulatory element drives GFP in neural crest and otic placode cells (Betancur et al., 2010, 2011). Foxd3 is also an early neural crest specifier that is essential for maintaining some neural crest fates (Teng et al., 2008; Lukoseviciute et al., 2018). Expressed in both cranial and vagal/trunk regions, its enhancers can be used to drive reporter expression in the entire neural crest lineage (Simões-Costa et al., 2012). Additionally, snai1b (Jimenez et al., 2016), crestin (Kaufman et al., 2016) and fli1a (Das and Crump, 2012) reporter lines have been harnessed to understand neural crest-related events such as EMT, melanoma initiation or ectomesenchymal fate decision, respectively.
Taken together, neural crest driver lines in mammalian and zebrafish models provide a stable and reproducible approach for lineage tracing. In addition, neural crest lines with inducible Cre allow for time-specific recombination and clonal analysis, as discussed in the following section. One issue, however, is that these lines can be ‘leaky’ and thus prone to ectopic expression in some cases.
Stem cell properties of the neural crest: are they multipotent?
Because neural crest cells are highly migratory, it is challenging to determine whether individual progenitors can give rise to multiple cell types. Indeed, a matter of interest has been whether individual neural crest cells are multipotent (i.e. able to differentiate into several cell types) or fate restricted (i.e. only able to form single or limited cell types) prior to or during their migration. To distinguish between these possibilities, one needs to trace the progeny of individual progenitors at clonal resolution. In the following sections, we discuss both classical and recently developed methods that enable clonal analyses of neural crest stem cells.
In vitro clonal analysis
Neural crest clonal analysis was first performed by isolating quail neural crest cells that migrated away from neural tube explants, dissociating them into a single cell suspension, seeding them at low density into multi-well plates, and screening for wells with only one cell. Differentiation of clones was subsequently analyzed by immunostaining with cell fate markers. Using this approach, Sieber-Blum and Cohen showed that single neural crest progenitors give rise to three types of clones: pigmented clones differentiating into melanocytes, unpigmented clones giving rise to adrenergic cells, and mixed clones. Differentiation occurred without direct interaction with non-neural crest cells, suggesting intrinsic multipotency of these progenitors (Sieber-Blum and Cohen, 1980). After 7-10 days in culture, clonal progeny exhibited migratory behavior and differentiation potential when injected back to chick embryos, suggesting that some progenitors remained multipotent (Bronner-Fraser et al., 1980).
In the early 1990s, Stemple and Anderson showed that individual mammalian neural crest cell clones in vitro were able to self-renew and give rise to both neuronal and glia fates (Stemple and Anderson, 1992). However, single derivative clones were also isolated (Ito et al., 1993), with TrkC marking neuronal-only clones while cKit marked melanocytic-only clones (Luo et al., 2003). Clonal analysis of quail cranial neural crest cells revealed broad developmental potential for them to differentiate into both neuronal (neurons, glia) and mesenchymal (cartilage, connective tissue) cells of the head (Calloni et al., 2007; Dupin et al., 2010). This multipotent characteristic was amplified by addition of sonic hedgehog (Calloni et al., 2007; Dupin et al., 2010). Taken together, these studies suggested that migratory neural crest cells in amniotes appear to consist of a mixture of multipotent progenitors and progenitors that contribute to a single derivative. Whether the latter are restricted to a particular fate (i.e. unipotent) is unclear as these experiments can assay only cell fate and not developmental potential. Moreover, multipotent premigratory neural crest cells could not be maintained long term until the discovery of neuroepithelial ‘crestospheres’ obtained from neural tubes prior to neural crest emigration (Kerosuo et al., 2015). These culture conditions enable neural crest self-renewal and maintenance of multipotency for weeks. When placed in differentiation media or injected back into the embryo, crestosphere cells gave rise to all neural crest derivatives, including glia, neurons, smooth muscle, cartilage and melanocytes (Kerosuo et al., 2015).
Given the limited tools available for clonal analysis within an embryo, these in vitro systems provided invaluable information about intrinsic neural crest stem cell properties. However, in vitro cultures are typically maintained in a rich medium containing many growth factors as well as on extracellular matrix-rich substrates. Addition of specific grown factors can drive clones toward different pathways of differentiation, with NGF promoting neuronal fate and GGF/neuregulin promoting glial fate (Sieber-Blum and Cohen, 1980). In serum-deprived medium, neural crest cells tend to differentiate towards neuronal fates, whereas the presence of serum facilitates proliferation (Ziller et al., 1983). The influence of such factors in the culture medium suggests that environmental cues may be important for cell fate determination, thus necessitating studying the stem cell properties and developmental potential of individual neural crest cells within an in vivo context.
Vital dye labeling of a single neural crest progenitor
Using microinjection of vital dye into a single premigratory neural crest cell, researchers further extended clonal analysis from cell culture into the embryo. By injecting the fluorescent dextran LRD into individual dorsal neural tube cells, as well as into some early migrating cells of chick embryos, Bronner-Fraser and Fraser found that descendants of individual precursors localized to distinct sites and formed cell types ranging from sensory and sympathetic neurons to non-neuronal cells and melanocyte precursors. They concluded that some premigratory neural crest cells are multipotent and that multipotency is maintained at migratory stages (Bronner-Fraser and Fraser, 1989; Fraser and Bronner-Fraser, 1991). Supporting results were also obtained from studies in Xenopus laevis (Collazo et al., 1993) and mouse (Serbedzija et al., 1994). By adjusting the stage of vital dye injection, Serbedzija et al. showed that contributions to neural crest derivatives vary with time; earlier migrating cells give rise to both dorsal and ventral structures, such as dorsal root ganglia, sympathetic ganglia and Schwann cells along the ventral root, while clones of late migrating cells are restricted to dorsal structures (Serbedzija et al., 1994).
Vital dye injection into single progenitor cells was the first approach to provide clonal resolution in vivo. Although conceptually simple, this approach is technically challenging and has its limitations. For example, because a limited amount of dye is injected, the fluorescence signal becomes diluted by cell division and is undetectable after a few days. Additionally, for clonal analysis, only one cell is injected per embryo. Thus, it can be challenging to retrieve information as the injected cell may die in some cases or its progeny might be missed during tissue processing. As a result, there was a need for new tools with increased ease of manipulation and the ability to provide permanent labeling at a multiplex level. These shortcomings necessitated the genesis of genetic clonal analysis, as made possible by the advent of Confetti transgenic lines, multicolor retroviral analysis and genome editing coupled with in situ hybridization.
Clonal analysis using multicolor reporters
In genetic systems, clonal analysis can be achieved by site-specific recombination using neural crest lineage-tracing lines that express Cre recombinase under the control of a neural crest-specific promoter, such as Wnt1, Plp or Sox10. When crossed with a reporter line, the genetic marker is permanently expressed in cells in which the promoter is active. Particularly useful for clonal analysis are inducible CreER lines that can be activated by tamoxifen treatment.
The multicolor Brainbow reporter (Livet et al., 2007), initially created for synaptic mapping in the brain, uses random recombination of three fluorescent proteins to generate more than 90 colors, each color uniquely assigned as a clone. This multicolor reporter, also called Confetti (Baggiolini et al., 2015), has been broadly applied for clonal analysis in developmental and stem cell biology. By crossing inducible Wnt1-CreERT and Sox10-CreERT2 with an R26R-Confetti reporter line, it was shown that premigratory and migratory trunk neural crest cells are multipotent, contributing to cell types as diverse as sensory neurons, sympathetic neurons, glia and melanocytes within one clone (Baggiolini et al., 2015). In conjunction with this, the Confetti system has uncovered the clonal organization of cranial neural crest during craniofacial morphogenesis (Kaucka et al., 2016) and has revealed the developmental potential of enteric precursor cells in the intestine to form neuronal-only, glial-only and neuronal-glial bipotent clones (Lasrado et al., 2017).
Subsequently, a zebrafish version of Brainbow, called Zebrabow became available (Albert Pan et al., 2013). Owing to their transparency, quick development and accessibility, zebrafish embryos and larvae are ideal models in which to combine multicolor clonal analysis with dynamic imaging. Clonal analysis using Zebrabow has contributed to our understanding of various aspects of neural crest cell behavior, such as intercalation and oriented cell proliferation in Meckel's cartilage morphogenesis (Rochard et al., 2015) and the role of Schwann cell precursors in cranial and trunk skeletal development (Xie et al., 2019). Multicolor Zebrabow has also been adapted to explore coordinated cell death during brain development (Brockway et al., 2019).
Brainbow has recently contributed greatly to our understanding of stem cells and regeneration in both fish and mice. For example, it has been used to elucidate the mechanism of aging through clonal selection of epidermal stem cells (Liu et al., 2019), ploidy reduction during liver regeneration (Matsumoto et al., 2020) and dynamics in clonal complexity after rounds of muscle injury (Tierney et al., 2018). In addition, this transgenic system has been invaluable in cancer studies, revealing clonal outgrowth of colorectal cancer (Lamprecht et al., 2017), clonal restriction in mammary tumors (Rios et al., 2019), and Lgr5+ intestinal cancer stem cell proliferation and transition to polyclonal clusters with tumor progression (Yanai et al., 2017). Despite its wide application, multicolor clonal analysis requires careful consideration. First, tamoxifen dose has to be optimized to achieve the desired recombination frequency. Without titration, labeling density might be too low to visualize multicolor clones, or too high such that it renders clonal relationships hard to interpret. In addition, the presence of multicolor fluorescent proteins that occupy imaging channels limits the number of antibodies that can be used for downstream analysis, which is also an issue for the retroviral multicolor systems described in the next section.
Retroviral clonal analysis
Retroviruses can integrate several kilobases of sequence (e.g. encoding barcodes, histological markers and fluorescent proteins) into the host genome and thus are very useful for lineage tracing. Clonal resolution is achieved by sparse labeling of locally restricted clones, such as in the retina (Khalili et al., 2018; Turner and Cepko, 1987; Fields-Berry et al., 1992), or multiple rounds of infection followed by barcode sequencing, which is more efficient for retrieving clonal information than sequencing for integration sites (Adair et al., 2020). Combined with laser capture microscopy for spatially distinct cell populations, barcode sequencing can provide highly complex lineage information through multiple infection with up to 105 tags. Accordingly, one study has shown that neural stem cells in mice become regionalized and diverge from other lineages by E11.5 (Fuentealba et al., 2015). Although the method provides clonal resolution while preserving spatial information, laser capture microscopy can be time-consuming, thus limiting the number of samples that can be assayed.
In the neural crest field, histological markers and fluorescent proteins have been preferable because spatial information is essential for examining lineage relationships in this highly migratory cell type. Clonal analysis using replication incompetent avian (RIA) retroviruses was made possible by multiple simultaneous infection events, which occur very rarely. When a viral mixture infects the cells, the probability of a cell being infected by two, three or four viral particles concomitantly can be calculated, assuming each viral particle has equal opportunity to enter the cell (Figliozzi et al., 2016). As the probability of multiple infections is extremely low, two or more viruses marking the same cell tends to occur once per embryo. Therefore, progeny carrying the same set of histological/fluorescent markers can be considered a clone. On this basis, Frank and Sanes used two viruses encoding LacZ in the cytosol or nucleus to map clonally related cells in the dorsal root ganglia, providing evidence for neural crest multipotency (Frank and Sanes, 1991).
This method was recently extended by incorporating five fluorescent proteins, targeted to the nucleus, membrane, mitochondria or actin filaments, into RIA retroviruses, allowing multiplex clonal analysis of trunk neural crest cells (Tang et al., 2019a). Infection of a single cell by two or more viruses encoding unique colors and/or subcellular localizations yields easily uniquely identifiable clonal descendants. Applying this to trunk neural crest cells confirmed their multipotency, similar to results from single cell microinjections of LRD in chick and Confetti in the mouse (Bronner-Fraser and Fraser, 1989; Fraser and Bronner-Fraser, 1991; Baggiolini et al., 2015; Tang et al., 2019a). Given that three different methods in two different species generated identical results, this is a nice example of ‘reproducibility’ in science.
Retroviral cell labeling also can be used to achieve stable molecular perturbation. For example, retroviral overexpression of Id2 was shown to convert ectodermal cells into neural crest cells, indicating that regulation of Id2 is required for maintaining ectodermal fate (Martinsen and Bronner-Fraser, 1998). Moreover, progress in viral engineering has enabled molecular perturbations to be combined with clonal analysis to understand cell fate decisions within clonally related cells. Compared with the Confetti system, multicolor retroviral lineage analysis provides more flexible spatiotemporal control. However, one should note that most retroviruses can only infect actively dividing cells (Temin and Rubin, 1958; Yamashita and Emerman, 2006). Additionally, as with tamoxifen titration in the Confetti system, precise titration is required to achieve the proper level of co-infection to guarantee clonality.
Single cell spatial transcriptomics and genome editing-based lineage reconstruction
All clonal analysis approaches mentioned so far obtain clonal information based on cell counts, locations and cell fates observed at the last time point, thus providing a single snapshot of cell fate. However, a complete lineage tree delineating each branching point or round of cell division is far more complex than that observed at the endpoint. Much of the information occurring during neural crest induction, delamination, migration, condensation, differentiation and morphogenesis is therefore lost. This contrasts with elegant lineage analysis in C. elegans by Sulston and colleagues, which allowed direct visualization of the entire cell lineage of the organism (Sulston, 1983).
Single cell spatial transcriptomics, by dissecting cell trajectories at spatiotemporal level, aims to identify transient states and branch points in early cell fate decisions (Soldatov et al., 2019). Using this approach, Soldatov et al. identified transcription factors that bias neural crest cells towards sensory versus other cell fates, and autonomic versus mesenchymal fates, across multiple axial levels and migration states, therefore providing a comprehensive spatiotemporal map of murine neural crest cell fate decisions (Soldatov et al., 2019).
Another approach termed ‘memory by engineered mutagenesis with optical in situ readout’ (MEMOIR) holds the promise of achieving complete lineage reconstruction in complex systems (Frieda et al., 2017). In this approach, CRISPR-Cas9 randomly deletes a series of barcoded sequences during cell division. The deletion is inherited by all daughter cells, with each daughter cell undergoing another round of random deletion, and so forth. In this way, a cell-specific deletion signature will record lineage information within its genome, which subsequently can be detected by single molecule fluorescence in situ hybridization (smFISH). As proof of principle, this method was applied in embryonic stem cells and was shown to be compatible with simultaneous detection of the pluripotency regulator Esrrb (Frieda et al., 2017). Prior to the development of MEMOIR, a technique called ‘genome editing of synthetic target arrays for lineage tracing’ (GESTALT) used DNA/RNA sequencing to retrieve edited barcodes and reconstructed a lineage tree with about 200,000 cells in zebrafish embryos (McKenna et al., 2016). Based on a similar principle, another approach used CRISPR-Cas9 to generate random insertions and deletions in tandem GFP or RFP sequences, which can be retrieved with single-cell RNA sequencing to obtain developmental history (Alemany et al., 2018; Spanjaard et al., 2018). However, these approaches require generation of a transgenic line prior to lineage analysis. Recently, a new method has been developed to identify and edit endogenous 5′G sequence in non-functional genomic region for CRISPR-Cas9-based phylogenetic fate mapping. This approach is useful in species in which transgenic animals cannot be easily obtained (Cotterell et al., 2020).
It is worth noting that, GESTALT and other methods based on deep sequencing obtain clonal information of a large number of cells at the expense of spatial information. Unfortunately, such spatial information is essential for neural crest lineage tracing. Thus, MEMOIR, although tracking fewer clones and genes, is complementary to sequencing-based GESTALT. Built upon MEMOIR, a technique termed ZOMBIE combines the advantage of high-density barcoding through in vitro transcription with preservation of spatial information (Askary et al., 2020). Future efforts toward a complete neural crest lineage tree hold the promise of applying these genome-editing methods to uncover previously inaccessible information, including but not limited to: How do neural crest precursors make lineage choices? Which derivatives are clonally diverse, and which are derived from a limited number of progenitor cells? Is there a clonal selection process after neural crest cells aggregate to form derivatives like ganglia or cartilage?
Tissue-specific roles: tracing neural crest cells in adults
Although the neural crest is a transient structure during development, neural crest-derived cells persist throughout life (Achilleos and Trainor, 2012). In addition, neural crest-derived cells that harbor stem cell properties reside within many adult tissues (Achilleos and Trainor, 2012). This raises the possibility that their unique developmental history can be harnessed in the adult during injury or disease. Neural crest-derived cells in the adult could be either undifferentiated cells or differentiated cells that exhibit plasticity and can dedifferentiate under some circumstances. Here, we use two examples – the adult heart and cancer – to demonstrate how lineage- tracing tools can help uncover the role of neural crest cells beyond embryogenesis. Other examples implicating neural crest-derived cells in regeneration include a role for Schwann cell precursors (SCPs) in limb regeneration (Kumar et al., 2007) and chromaffin cell formation (Furlan et al., 2017). Although studies of heart regeneration and NF1-associated tumor formation both suggest a role for already differentiated cells, the function of undifferentiated neural crest cells remains to be explored and may be important in other situations.
Tracing neural crest cells during heart development and regeneration
The adult zebrafish heart has the remarkable capacity to regenerate when up to 20% of the ventricle is surgically removed (Poss et al., 2002). This has generated great interest in understanding the gene regulation events and cell lineage decisions contributing to new cardiomyocytes during repair. A key question is whether new cardiomyocytes are derived from a resident stem cell population, or from dedifferentiation of existing cardiomyocytes. Using the inducible transgenic line tg-cmlc2a-cre-Ert2:tg-cmlc2a-LnL-GFP to trace cardiomyocytes, Jopling et al. discovered that new cardiomyocytes are derived by dedifferentiation and proliferation of pre-existing cardiomyocytes (Jopling et al., 2010). However, whether dedifferentiation-proliferation was restricted to a specific subpopulation of cardiomyocytes remained unclear. Multiple lineage analyses suggest that the adult zebrafish ventricle is indeed heterogeneous, comprising multiple subpopulations with distinct developmental origins. For example, in addition to the first and second heart fields, which are derived from mesoderm, neural crest cells migrate into the bulbus arteriosus, atrium and ventricle, as shown by uncaging fluorescein (Li et al., 2003; Sato and Yost, 2003), cell transplantation of fluorescently injected donors and laser induction of hsp70-eGFP zebrafish (Sato and Yost, 2003). This result was recently confirmed using advanced genetic lines, specifically NC:NfsB-mCherry for short-term and Tg(NC:mCherry) for long-term tracing (Cavanaugh et al., 2015). This cardiac neural crest contribution is essential for normal heart function, as surgical ablation of the neural crest in zebrafish leads to decreased stroke volume, ejection fraction and cardiac output (Li et al., 2003). In addition, genetically induced removal of neural crest-derived cardiomyocytes causes hypertrophic cardiomyopathy (Abdul-Wajid et al., 2018). Together, these results demonstrate that neural crest-derived cardiomyocytes are functionally important in the zebrafish heart.
There is also evidence suggesting a neural crest contribution to the mammalian heart, which exhibits limited regeneration during a narrow window up to postnatal day 7 (Porrello et al., 2011). Cardiospheres isolated from mammalian hearts, when injected into chick embryos, migrate like neural crest cells into the cardiac outflow tract (Tomita et al., 2005). Another study, using KitCreERT2/+ and Wnt1::Flpe mouse lineage-tracing lines, showed that neural crest cells give rise to a cKit+ stem cell population in the heart (Hatzistergos et al., 2015). Analysis of the P0Cre-GFP transgenic mouse line also showed the presence of neural crest-derived cells as ventricular cardiomyocytes (Tomita et al., 2005). Moreover, these cells migrated toward the injury site through MCP-1 mediated chemoattraction (Tamura et al., 2011), indicating a role for cardiac neural crest-derived cells in heart repair.
Recently, the contribution of neural crest cells to heart repair in amniotes was confirmed using Wnt1-Cre mice and retroviral lineage analysis in chick embryos (Tang et al., 2019b) (Fig. 2A). Consistent with this, the genetic ablation of cardiac neural crest cells in zebrafish was shown to impair heart regeneration (Sande-Melón et al., 2019). These studies also revealed that Sox10 and other neural crest GRN components are upregulated upon injury (Tang et al., 2019b; Sande-Melón et al., 2019) (Fig. 2B,C), suggesting that neural crest-derived cardiomyocytes contribute to heart regeneration. However, further studies are needed to elucidate the precise mechanisms underlying the migration and proliferation of neural crest-derived cells during heart regeneration. Molecularly, CXCL12a expressed in the epicardium can chemoattract cardiac neural crest-derived cardiomyocytes through the receptor CXCR4 (Itou et al., 2012), making this a candidate ligand-receptor pair worthy of further analysis.
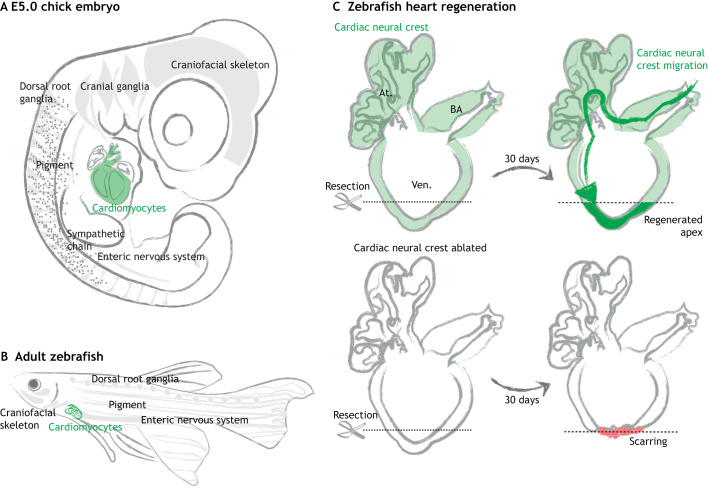
Fig. 2.
Neural crest-derived cardiomyocytes contribute to zebrafish heart regeneration. (A,B) Neural crest-derived cells persist as differentiated cells or stem cells residing in multiple organs, such as the craniofacial skeleton, peripheral nervous system, pigment of the skin (gray) and cardiovascular systems (green) in both amniotes (A) and zebrafish (B). (C) In the normal zebrafish heart, neural crest cells give rise to heart muscle (green) in the bulbus arteriosus (BA), atria (At.) and ventricles (Ven.). When the apex is surgically removed, these neural crest-derived cells participate in heart regeneration by migrating towards the injury site and redeploying genes that regulate neural crest development, such as Sox10. Genetic ablation of these cardiac neural crest cells disrupts heart regeneration, leading to scarring (red) (Tang et al., 2019b; Sande-Melón et al., 2019). Gray arrows represent regeneration period.
Tracing neural crest cells during tumor development
Neural crest-derived cells, including melanocytes and cells of the peripheral nervous system (PNS), can develop into benign or malignant tumors upon genetic mutation (Fig. 3A). Neurofibromatosis type 1 (NF1), which is characterized by café-au-lait spots, plexiform or cutaneous neurofibromas, gliomas and cardiovascular abnormalities, is caused by loss of Nf1, which encodes a tumor suppressor in the Ras signaling pathway. A longstanding issue was whether tumor-forming cells arise from undifferentiated neural crest stem cells or from differentiated PNS cells. Conditional deletion of Nf1 in the PNS (using P0a-Cre: Nf1fl/−) suggested that tumors formed from differentiated neural crest-derived Schwann cells (Joseph et al., 2008). Similarly, it was shown that Nf1-deficient neural crest stem cells developed into normal PNS derivatives, but tumors occurred later in the abnormally differentiated Remak bundle (Zheng et al., 2008). Taken together, these results suggested that Nf1 deficiency predisposes neural crest progenitor cells to tumor development after differentiation (Fig. 3B), highlighting a role for differentiated cells in tumor formation. Finally, using a Schwann cell precursor (SCP) driver line (Dhh-Cre) to delete Nf1 in SCPs resulted in plexiform neurofibroma, thus narrowing down the embryonic origin of neurofibroma-forming cells to SCPs (Wu et al., 2008); these neurofibroma-forming SCPs are localized in the dorsal root ganglia (Chen et al., 2014). Moreover, analyses using PLP-CreERT revealed that the most susceptible time window of Nf1 loss occurs before SCP maturation (Le et al., 2011).
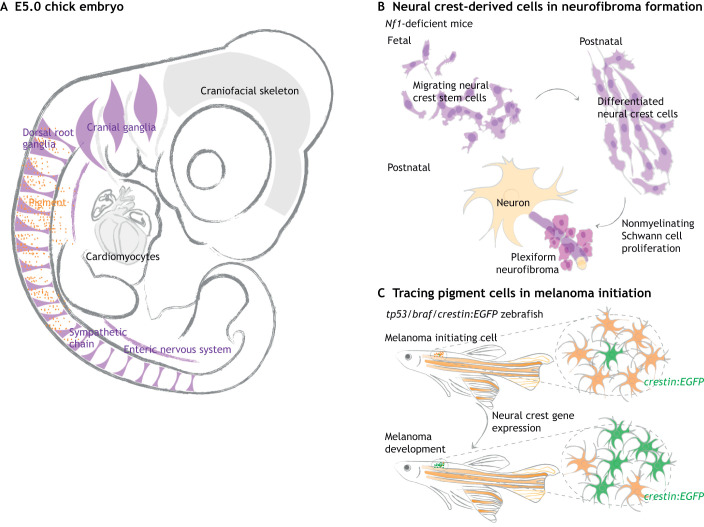
Fig. 3.
Tracing neural crest cells in tumor development. (A) Neural crest-derived cells contribute to the peripheral nervous system (PNS, purple) and pigment of the skin (orange), as illustrated in a chick embryo at E5. (B) Plexiform neurofibroma is a tumor of the PNS caused by mutations in the tumor suppressor gene Nf1. By tracing neural crest cells throughout the tumorigenesis process in Nf1 knockout mice, studies found that Nf1-deficient neural crest cells differentiate normally; tumors subsequently form by proliferation of differentiated Schwann cells. Thus, neural crest stem cells are not directly responsible for plexiform neurofibroma. (C) Neural crest lineage tracing can help identify early signs of melanoma. In a zebrafish model with oncogenic mutations in tp53 and braf, melanoma initiation is characterized by a normal pigment cell (orange) re-expressing neural crest-related genes, as visualized by crestin:EGFP (green). This particular cell transforms into a neural crest progenitor-like cell and later expands into a tumor. Gray arrows represent development.
Embryonic neural crest cells share many properties with cancer cells, including multipotency, migratory ability, general invasiveness and transcriptional/epigenetic signatures (Angeles Rabadán et al., 2013; Dravis et al., 2018). A recent study used the crestin:EGFP reporter line to create a zebrafish model of melanoma initiation in which neural crest cells could be visualized and compared during embryogenic development and melanoma initiation. In braf mutant and tp53-deficient cancerized fish, Sox10 expression correlates with melanoma formation, activating the same regulatory elements as in neural crest cells (Kaufman et al., 2016) (Fig. 3C). In another zebrafish model, mcr:NRAS, the melanoma gene expression pattern was highly reminiscent of that observed in embryonic neural crest cells, as shown by upregulation of crestin, sox10 and dlx2a (McConnell et al., 2019). Yy1, which encodes a transcription factor, has also been shown to regulate both neural crest development and melanoma initiation by controlling MITF and MYC expression (Varum et al., 2019). In addition, a mutant model of melanoma developed in Xiphophorus hybrids, which form malignant melanoma under the control of oncogenic Xmrk, has provided insight into the signaling pathways and adhesion molecules involved in melanoma, especially in comparison with human skin cancer (Schartl et al., 1995; Schartl, 2014; Lu et al., 2018). Together, these studies highlight how exploring the similarities and differences between normal developmental processes and tumor formation can inform our understanding of the ontogeny of neural crest-derived tumors. They also emphasize that neural crest lineage tracing is an indispensable approach for identifying tumor-initiating cells that are present only transiently and are challenging to detect.
Conclusions
Lineage tracing at the population and clonal level, applied to a multitude of vertebrate embryos, has made essential contributions to discoveries in the neural crest field. Although several reports have shown that many newly formed neural crest cells at cranial and trunk levels are multipotent, it is likely that that they become progressively fate restricted as they migrate and reach their final destinations (Soldatov et al., 2019). Thus, the embryonic neural crest is a transient stem cell type. Interestingly, neural crest-derived cells persist into adulthood and may have important roles in tissue regeneration. As such, abnormalities in these cells can lead to birth defects and several adult cancers. Thus, improved lineage-tracing methods will no doubt extend our understanding of the neural crest lineage in both the early embryo and in the context of disease and regeneration.
It is worth noting that only vertebrates have bona fide neural crest cells. However, neural crest-like cells have been identified in non-vertebrate chordates such as ascidians, which have pigment cells (Jeffery et al., 2004) and peripheral neuroendocrine cells (Powell et al., 1996). Even the basal chordate amphioxus has migratory ectodermal cells (Holland et al., 1996); however, these are not derived from the neural tube or neural plate borders. A major difference is that the precursors of these chordate cells tend to give rise to single cell types rather than being multipotent. In contrast, basal jawless vertebrates like the sea lamprey do possess migratory and multipotent neural crest cells, although these are less regionalized along the body axis than in jawed vertebrates. Accordingly, the lamprey neural crest GRN is most similar to the trunk neural crest GRN of amniotes. Indeed, region-specific neural crest GRN components appear to have been progressively added to the neural crest GRN during the course of vertebrate evolution (Martik et al., 2019).
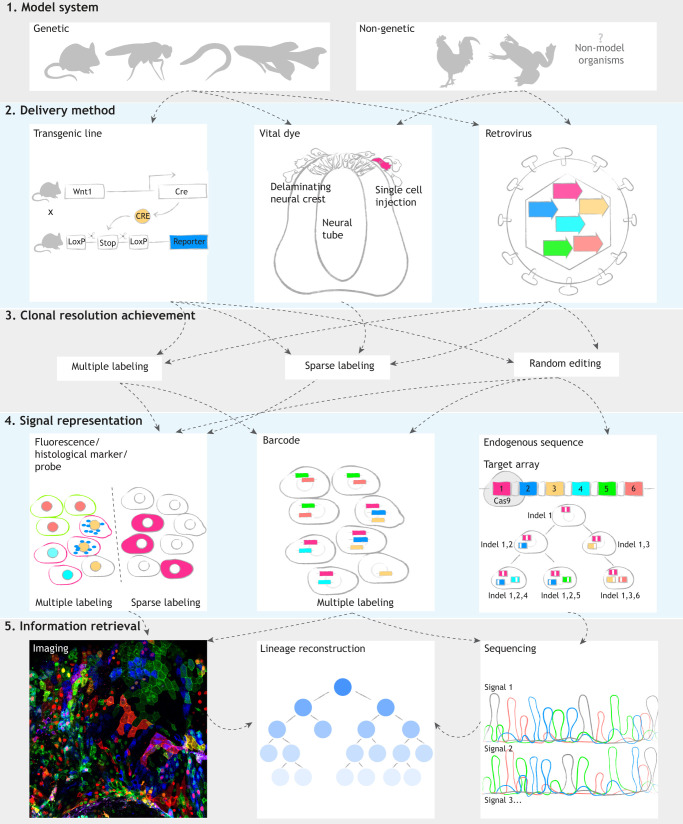
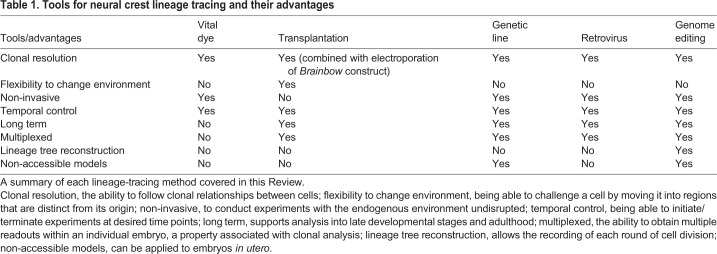
All of the lineage-tracing methods described herein can be mixed and matched (Fig. 4), making the advantages adaptable to numerous model systems (Table 1). Future studies are likely to refine current tools towards ever higher resolution and hold the promise of providing transcriptional and spatial lineage information coupled with identifying clonal relationships. Through genome editing or barcoding, clonal analysis approaches can be combined with RNA-seq to understand changes in transcriptional landscapes during crucial cell fate decisions. Clonal analysis coupled with molecular perturbations will provide a deeper understanding of the ways that differential gene expression can drive clonal diversification. At the same time, high-resolution imaging can be used to dynamically assess cell behavior during clonal expansion, providing new opportunities to manipulate cell fates in stem cell, regenerative and cancer studies. These combined approaches are likely to fulfill Hörstadius' expectations some 70 years ago that ‘further investigations with a variety of methods and giving due consideration to possible sources of error will certainly give us a deeper insight into…the developing neural crest’ (Hörstadius, 1950).
Fig. 4.
Conceptual summary of clonal analysis workflow. Clonal analysis approaches can involve the following five elements: model systems (1), delivery methods (2), ways to achieve clonal resolution (3), signal representation (4) and information retrieval approaches (5). (1 and 2) Stable transgenic lineage-tracing lines can only be obtained with genetic systems. However, non-genetic and non-model organisms are also accessible through vital dye injection and retroviral transduction. (3) Clonal resolution is achieved by rare events such as multiple labeling, sparse labeling and random editing with the CRISPR/Cas9 system. (4) Depending on how clonal resolution is achieved, one can use materials that can be imaged (such as fluorescent proteins, histological markers and probes) or sequences (such as barcodes and endogenous CRISPR/Cas9 target arrays) for downstream analysis. (5) All imaging and sequencing-based methods can obtain endpoint lineage information. Phylogenetic lineage reconstruction can be provided by multiplexed genome editing. Sets of fluorescent markers, barcodes and endogenous target arrays are color-coded to indicate heterogeneity (magenta, blue, yellow, cyan, green and red). Dashed arrows suggest available choices in the following step.
Table 1.
Tools for neural crest lineage tracing and their advantages
Acknowledgements
We thank Dr Vicky Prince for helpful suggestions on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The authors’ research was funded by the National Institutes of Health (DE027568, R35NS111564 and NIHRO1HL14058 to M.E.B.). Deposited in PMC for release after 12 months.
References
- Abdul-Wajid S., Demarest B. L. and Yost H. J. (2018). Loss of embryonic neural crest derived cardiomyocytes causes adult onset hypertrophic cardiomyopathy in zebrafish. Nat. Commun. 9, 4603 10.1038/s41467-018-07054-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilleos A. and Trainor P. A. (2012). Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 22, 288-304. 10.1038/cr.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair J. E., Enstrom M. R., Haworth K. G., Schefter L. E., Shahbazi R., Humphrys D. R., Porter S., Tam K., Porteus M. H. and Kiem H.-P. (2020). DNA barcoding in nonhuman primates reveals important limitations in retrovirus integration site analysis. Mol. Ther. Methods Clin. Dev. 17, 796-809. 10.1016/j.omtm.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert Pan Y., Freundlich T., Weissman T. A., Schoppik D., Wang X. C., Zimmerman S., Ciruna B., Sanes J. R., Lichtman J. W. and Schier A. F. (2013). Zebrabow: Multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Dev. 140, 2835-2846. 10.1242/dev.094631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany A., Florescu M., Baron C. S., Peterson-Maduro J. and Van Oudenaarden A. (2018). Whole-organism clone tracing using single-cell sequencing. Nature 556, 108-112. 10.1038/nature25969 [DOI] [PubMed] [Google Scholar]
- Angeles Rabadán M., Usieto S., Lavarino C. and Martí E. (2013). Identification of a putative transcriptome signature common to neuroblastoma and neural crest cells. Dev. Neurobiol. 73, 815-827. 10.1002/dneu.22099 [DOI] [PubMed] [Google Scholar]
- Aoto K., Sandell L. L., Butler Tjaden N. E., Yuen K. C., Watt K. E. N., Black B. L., Durnin M. and Trainor P. A. (2015). Mef2c-F10N enhancer driven β-galactosidase (LacZ) and Cre recombinase mice facilitate analyses of gene function and lineage fate in neural crest cells. Dev. Biol. 402, 3-16. 10.1016/j.ydbio.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A., Sanchez-Guardado L., Linton J. M., Chadly D. M., Budde M. W., Cai L., Lois C. and Elowitz M. B. (2020). In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat. Biotechnol. 38, 66-75. 10.1038/s41587-019-0299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer-Le Lievre C. S. and Le Douarin N. M. (1982). The early development of cranial sensory ganglia and the potentialities of their component cells studied in quail-chick chimeras. Dev. Biol. 94, 291-310. 10.1016/0012-1606(82)90349-9 [DOI] [PubMed] [Google Scholar]
- Baggiolini A., Varum S., Mateos J. M., Bettosini D., John N., Bonalli M., Ziegler U., Dimou L., Clevers H., Furrer R. et al. (2015). Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell 16, 314-322. 10.1016/j.stem.2015.02.017 [DOI] [PubMed] [Google Scholar]
- Baker C. V., Bronner-Fraser M., Le Douarin N. M. and Teillet M. A. (1997). Early- and late-migrating cranial neural crest cell populations have equivalent developmental potential in vivo. Development 124, 3077-3087. [DOI] [PubMed] [Google Scholar]
- Betancur P., Bronner-Fraser M. and Sauka-Spengler T. (2010). Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. USA 107, 3570-3575. 10.1073/pnas.0906596107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P., Sauka-Spengler T. and Bronner M. (2011). A sox10 enhancer element common to the otic placode and neural crest is activated by tissue-specific paralogs. Development 138, 3689-3698. 10.1242/dev.057836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway N. L., Cook Z. T., O'gallagher M. J., Tobias Z. J. C., Gedi M., Carey K. M., Unni V. K., Pan Y. A., Metz M. R. and Weissman T. A. (2019). Multicolor lineage tracing using in vivo time-lapse imaging reveals coordinated death of clonally related cells in the developing vertebrate brain. Dev. Biol. 453, 130-140. 10.1016/j.ydbio.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M. and Fraser S. (1989). Developmental potential of avian trunk neural crest cells in situ. Neuron 3, 755-766. 10.1016/0896-6273(89)90244-4 [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M., Sieber-Blum M. and Cohen A. M. (1980). Clonal analysis of the avian neural crest: Migration and maturation of mixed neural crest clones injected into host chicken embryos. J. Comp. Neurol. 193, 423-434. 10.1002/cne.901930209 [DOI] [PubMed] [Google Scholar]
- Calloni G. W., Glavieux-Pardanaud C., Le Douarin N. M. and Dupin E. (2007). Sonic Hedgehog promotes the development of multipotent neural crest progenitors endowed with both mesenchymal and neural potentials. Proc. Natl. Acad. Sci. USA 104, 19879-19884. 10.1073/pnas.0708806104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney T. J., Dutton K. A., Greenhill E., Delfino-Machin M., Dufourcq P., Blader P. and Kelsh R. N. (2006). A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 133, 4619-4630. 10.1242/dev.02668 [DOI] [PubMed] [Google Scholar]
- Cavanaugh A. M., Huang J. and Chen J.-N. (2015). Two developmentally distinct populations of neural crest cells contribute to the zebrafish heart. Dev. Biol. 404, 103-112. 10.1016/j.ydbio.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Jiang X., Ito Y., Bringas P. Jr., Han J., Rowitch D. H., Soriano P., McMahon A. P. et al. Sucov H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671-1679. [DOI] [PubMed] [Google Scholar]
- Cheeseman B. L., Zhang D., Binder B. J., Newgreen D. F. and Landman K. A. (2014). Cell lineage tracing in the developing enteric nervous system: superstars revealed by experiment and simulation. J. R. Soc. Interface 11, 20130815 10.1098/rsif.2013.0815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-M. A., Smith D. M., Peters M. A., Samson M. E. S., Zitz J., Tabin C. J. and Cepko C. L. (1999). Production and design of more effective avian replication-incompetent retroviral vectors. Dev. Biol. 214, 370-384. 10.1006/dbio.1999.9432 [DOI] [PubMed] [Google Scholar]
- Chen Z., Liu C., Patel A. J., Liao C.-P., Wang Y. and Le L. Q. (2014). Cells of Origin in the Embryonic Nerve Roots for NF1-Associated Plexiform Neurofibroma. Cancer Cell 26, 695-706. 10.1016/j.ccell.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo A., Bronner-Fraser M. and Fraser S. E. (1993). Vital dye labelling of Xenopus laevis trunk neural crest reveals multipotency and novel pathways of migration. Development 118, 363-376. [DOI] [PubMed] [Google Scholar]
- Cotterell J., Vila-Cejudo M., Batlle-Morera L. and Sharpe J. (2020). Endogenous CRISPR/Cas9 arrays for scalable whole-organism lineage tracing. Development 147, dev184481 10.1242/dev.184481 [DOI] [PubMed] [Google Scholar]
- Couly G. F., Coltey P. M. and Le Douarin N. M. (1993). The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development 117, 409-429. [DOI] [PubMed] [Google Scholar]
- Danielian P. S., Muccino D., Rowitch D. H., Michael S. K. and Mcmahon A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323-1326. 10.1016/S0960-9822(07)00562-3 [DOI] [PubMed] [Google Scholar]
- Das A. and Crump J. G. (2012). Bmps and Id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8, e1002710 10.1371/journal.pgen.1002710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbache J., Parfejevs V. and Sommer L. (2018). Cre-driver lines used for genetic fate mapping of neural crest cells in the mouse: An overview. Genesis 56, e23105 10.1002/dvg.23105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis C., Chung C.-Y., Lytle N. K., Herrera-Valdez J., Luna G., Trejo C. L., Reya T. and Wahl G. M. (2018). Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466-482.e6. 10.1016/j.ccell.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin E., Calloni G. W. and Le Douarin N. M. (2010). The cephalic neural crest of amniote vertebrates is composed of a large majority of precursors endowed with neural, melanocytic, chondrogenic and osteogenic potentialities. Cell Cycle 9, 238-249. 10.4161/cc.9.2.10491 [DOI] [PubMed] [Google Scholar]
- Ealba E. L., Jheon A. H., Hall J., Curantz C., Butcher K. D. and Schneider R. A. (2015). Neural crest-mediated bone resorption is a determinant of species-specific jaw length. Dev. Biol. 408, 151-163. 10.1016/j.ydbio.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M. L., Mikawa T., Brown A. M. C. and Mcfarlin D. R. (1994). Mapping the origin of the avian enteric nervous system with a retroviral marker. Dev. Dyn. 201, 236-244. 10.1002/aja.1002010307 [DOI] [PubMed] [Google Scholar]
- Fields-Berry S. C., Halliday A. L. and Cepko C. L. (1992). A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. USA 89, 693-697. 10.1073/pnas.89.2.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figliozzi R. W., Chen F., Chi A. and Hsia S.-C. V. (2016). Using the inverse Poisson distribution to calculate multiplicity of infection and viral replication by a high-throughput fluorescent imaging system. Virol. Sin. 31, 180-183. 10.1007/s12250-015-3662-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E. and Sanes J. R. (1991). Lineage of neurons and glia in chick dorsal root ganglia: analysis in vivo with a recombinant retrovirus. Development 111, 895-908. [DOI] [PubMed] [Google Scholar]
- Fraser S. E. and Bronner-Fraser M. (1991). Migrating neural crest cells in the trunk of the avian embryo are multipotent. Development 112, 913-920. [DOI] [PubMed] [Google Scholar]
- Frieda K. L., Linton J. M., Hormoz S., Choi J., Chow K.-H. K., Singer Z. S., Budde M. W., Elowitz M. B. and Cai L. (2017). Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107-111. 10.1038/nature20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba L. C., Rompani S. B., Parraguez J. I., Obernier K., Romero R., Cepko C. L. and Alvarez-Buylla A. (2015). Embryonic origin of postnatal neural stem cells. Cell 161, 1644-1655. 10.1016/j.cell.2015.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan A., Dyachuk V., Kastriti M. E., Calvo-Enrique L., Abdo H., Hadjab S., Chontorotzea T., Akkuratova N., Usoskin D., Kamenev D. et al. (2017). Multipotent peripheral glial cells generate neuroendocrine cells of the adrenal medulla. Science 357, eaal3753 10.1126/science.aal3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Ezin M. and Bronner M. E. (2020). Reprogramming axial level identity to rescue neural-crest-related congenital heart defects. Dev. Cell 53, 300-315.e4. 10.1016/j.devcel.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Jheon A. H., Ealba E. L., Eames B. F., Butcher K. D., Mak S.-S., Ladher R., Alliston T. and Schneider R. A. (2014). Evolution of a developmental mechanism: Species-specific regulation of the cell cycle and the timing of events during craniofacial osteogenesis. Dev. Biol. 385, 380-395. 10.1016/j.ydbio.2013.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. (1935). Heteroplastic grafting in embryology. Harvey Lect. Balt. 29, 116-157. [Google Scholar]
- Hatzistergos K. E., Takeuchi L. M., Saur D., Seidler B., Dymecki S. M., Mai J. J., White I. A., Balkan W., Kanashiro-Takeuchi R. M., Schally A. V. et al. (2015). CKit+ cardiac progenitors of neural crest origin. Proc. Natl. Acad. Sci. USA 112, 13051-13056. 10.1073/pnas.1517201112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- His W. (1868). Untersuchungen über die erste Anlage des Wirbelthierleibes: die erste Entwickelung des Hühnchens im Ei. Vogel. [Google Scholar]
- Holland N. D., Panganiban G., Henyey E. L. and Holland L. Z. (1996). Sequence and developmental expression of AmphiDll, an amphioxus Distal-less gene transcribed in the ectoderm, epidermis and nervous system: insights into evolution of craniate forebrain and neural crest. Development 122, 2911-2920. [DOI] [PubMed] [Google Scholar]
- Hörstadius S. (1950). The neural crest, its properties and derivatives in the light of experimental research. Oxford University Press.
- Hörstadius S. and Sellman S. (1946). Experimentelle Untersuchungen über die Determination des knorpeligen Kopfskelletes bei Urodelen. Stockholm: Almquist & Wiksell.
- Ito K., Morita T. and Sieber-Blum M. (1993). In Vitro clonal analysis of mouse neural crest development. Dev. Biol. 157, 517-525. 10.1006/dbio.1993.1154 [DOI] [PubMed] [Google Scholar]
- Itou J., Oishi I., Kawakami H., Glass T. J., Richter J., Johnson A., Lund T. C. and Kawakami Y. (2012). Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Dev. 139, 4133-4142. 10.1242/dev.079756 [DOI] [PubMed] [Google Scholar]
- Jeffery W. R., Strickler A. G. and Yamamoto Y. (2004). Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature 431, 696-699. 10.1038/nature02975 [DOI] [PubMed] [Google Scholar]
- Jiang X., Rowitch D. H., Soriano P., Mcmahon A. P. and Sucov H. M. (2000). Fate of the mammalian cardiac neural crest. Development 127, 1607-1616. [DOI] [PubMed] [Google Scholar]
- Jimenez L., Wang J., Morrison M. A., Whatcott C., Soh K. K., Warner S., Bearss D., Jette C. A. and Stewart R. A. (2016). Phenotypic chemical screening using a zebrafish neural crest EMT reporter identifies retinoic acid as an inhibitor of epithelial morphogenesis. Dis. Model. Mech. 9, 389-400. 10.1242/dmm.021790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A. and Belmonte J. C. I. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609. 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N. M., Mosher J. T., Buchstaller J., Snider P., Mckeever P. E., Lim M., Conway S. J., Parada L. F., Zhu Y. and Morrison S. J. (2008). The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell 13, 129-140. 10.1016/j.ccr.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaucka M., Ivashkin E., Gyllborg D., Zikmund T., Tesarova M., Kaiser J., Xie M., Petersen J., Pachnis V., Nicolis S. K. et al. (2016). Analysis of neural crest–derived clones reveals novel aspects of facial development. Sci. Adv. 2, e1600060 10.1126/sciadv.1600060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman C. K., Mosimann C., Fan Z. P., Yang S., Thomas A. J., Ablain J., Tan J. L., Fogley R. D., Van Rooijen E., Hagedorn E. J. et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerosuo L., Nie S., Bajpai R. and Bronner M. E. (2015). Crestospheres: long-term maintenance of multipotent, premigratory neural crest stem cells. Stem Cell Rep. 5, 499-507. 10.1016/j.stemcr.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili S., Ballios B. G., Belair-Hickey J., Donaldson L., Liu J., Coles B. L. K., Grisé K. N., Baakdhah T., Bader G. D., Wallace V. A. et al. (2018). Induction of rod versus cone photoreceptor-specific progenitors from retinal precursor cells. Stem Cell Res. 33, 215-227. 10.1016/j.scr.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Kirby M. L., Gale T. F. and Stewart D. E. (1983). Neural crest cells contribute to normal aorticopulmonary septation. Science (80-.). 220, 1059-1061. 10.1126/science.6844926 [DOI] [PubMed] [Google Scholar]
- Kollar E. J. and Fisher C. (1980). Tooth induction in chick epithelium: Expression of quiescent genes for enamel synthesis. Science (80-.). 207, 993-995. 10.1126/science.7352302 [DOI] [PubMed] [Google Scholar]
- Kontges G. and Lumsden A. (1996). Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development 122,3229-3242. [DOI] [PubMed] [Google Scholar]
- Krotoski D. M., Fraser S. E. and Bronner-Fraser M. (1988). Mapping of neural crest pathways in Xenopus laevis using inter- and intra-specific cell markers. Dev. Biol. 127, 119-132. 10.1016/0012-1606(88)90194-7 [DOI] [PubMed] [Google Scholar]
- Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A. and Brockes J. P. (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772-777. 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht S., Schmidt E. M., Blaj C., Hermeking H., Jung A., Kirchner T. and Horst D. (2017). Multicolor lineage tracing reveals clonal architecture and dynamics in colon cancer. Nat. Commun. 8, 1406 10.1038/s41467-017-00976-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasrado R., Boesmans W., Kleinjung J., Pin C., Bell D., Bhaw L., Mccallum S., Zong H., Luo L., Clevers H. et al. (2017). Lineage-dependent spatial and functional organization of the mammalian enteric nervous system. Science 356, 722-726. 10.1126/science.aam7511 [DOI] [PubMed] [Google Scholar]
- Le L. Q., Liu C., Shipman T., Chen Z., Suter U. and Parada L. F. (2011). Susceptible stages in Schwann cells for NF1-associated plexiform neurofibroma development. Cancer Res. 71, 4686-4695. 10.1158/0008-5472.CAN-10-4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. and Kalcheim C. (1999). The Neural Crest: Cambridge University Press; 10.1017/cbo9780511897948 [DOI] [Google Scholar]
- Le Douarin N. M. and Teillet M. A. (1973). The migration of neural crest cells to the wall of the digestive tract in avian embryo. J. Embryol. Exp. Morphol. 30, 31-48. [PubMed] [Google Scholar]
- Le Lièvre C. S. and Le Douarin N. M. (1975). Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol . 34, 125-154. [PubMed] [Google Scholar]
- Lewis A. E., Vasudevan H. N., O'neill A. K., Soriano P. and Bush J. O. (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 379, 229-234. 10.1016/j.ydbio.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen F. and Epstein J. A. (2000). Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis 26, 162-164. [DOI] [PubMed] [Google Scholar]
- Li Y.-X., Zdanowicz M., Young L., Kumiski D., Leatherbury L. and Kirby M. L. (2003). Cardiac neural crest in zebrafish embryos contributes to myocardial cell lineage and early heart function. Dev. Dyn. 226, 540-550. 10.1002/dvdy.10264 [DOI] [PubMed] [Google Scholar]
- Liu N., Matsumura H., Kato T., Ichinose S., Takada A., Namiki T., Asakawa K., Morinaga H., Mohri Y., De Arcangelis A. et al. (2019). Stem cell competition orchestrates skin homeostasis and ageing. Nature 568, 344-350. 10.1038/s41586-019-1085-7 [DOI] [PubMed] [Google Scholar]
- Livet J., Weissman T. A., Kang H., Draft R. W., Lu J., Bennis R. A., Sanes J. R. and Lichtman J. W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Lu Y., Boswell M., Boswell W., Kneitz S., Hausmann M., Klotz B., Regneri J., Savage M., Amores A., Postlethwait J. et al. (2018). Comparison of Xiphophorus and human melanoma transcriptomes reveals conserved pathway interactions. Pigment Cell Melanoma Res. 31, 496-508. 10.1111/pcmr.12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoseviciute M., Gavriouchkina D., Williams R. M., Hochgreb-Hagele T., Senanayake U., Chong-Morrison V., Thongjuea S., Repapi E., Mead A. and Sauka-Spengler T. (2018). From pioneer to repressor: bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Dev. Cell 47, 608-628.e6. 10.1016/j.devcel.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A., Sprawson N. and Graham A. (1991). Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development 113, 1281-1291. [DOI] [PubMed] [Google Scholar]
- Luo R., Gao J., Wehrle-Haller B. and Henion P. D. (2003). Molecular identification of distinct neurogenic and melanogenic neural crest sublineages. Development 130, 321-330. 10.1242/dev.00213 [DOI] [PubMed] [Google Scholar]
- Lwigale P. Y. and Schneider R. A. (2008). Chapter 3 other chimeras: quail-duck and mouse-chick. Methods Cell Biol. 87, 59-74. 10.1016/S0091-679X(08)00203-3 [DOI] [PubMed] [Google Scholar]
- Martik M. L., Gandhi S., Uy B. R., Gillis J. A., Green S. A., Simoes-Costa M. and Bronner M. E. (2019). Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 574, 675-678. 10.1038/s41586-019-1691-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen B. J. and Bronner-Fraser M. (1998). Neural crest specification regulated by the helix-loop-helix repressor Id2. Science 281, 988-991. 10.1126/science.281.5379.988 [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Wakefield L., Tarlow B. D. and Grompe M. (2020). In vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell 26, 34-47.e3. 10.1016/j.stem.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T., Ahlberg P. E., Kessaris N., Iannarelli P., Dennehy U., Richardson W. D., Mcmahon A. P. and Koentges G. (2005). Neural crest origins of the neck and shoulder. Nature 436, 347-355. 10.1038/nature03837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell A. M., Mito J. K., Ablain J., Dang M., Formichella L., Fisher D. E. and Zon L. I. (2019). Neural crest state activation in NRAS driven melanoma, but not in NRAS-driven melanocyte expansion. Dev. Biol. 449, 107-114. 10.1016/j.ydbio.2018.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Findlay G. M., Gagnon J. A., Horwitz M. S., Schier A. F. and Shendure J. (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis T. A., Chéraud Y., Sharpe P. and Fontaine-Pérus J. (2003). Development of teeth in chick embryos after mouse neural crest transplantations. Proc. Natl. Acad. Sci. USA 100, 6541-6545. 10.1073/pnas.1137104100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa-Tomita S., Waldo K., Tomita H. and Kirby M. L. (1991). Temporospatial study of the migration and distribution of cardiac neural crest in quail-chick chimeras. Am. J. Anat. 192, 79-88. 10.1002/aja.1001920109 [DOI] [PubMed] [Google Scholar]
- Nagy N. and Goldstein A. M. (2006). Intestinal coelomic transplants: A novel method for studying enteric nervous system development. Cell Tissue Res. 326, 43-55. 10.1007/s00441-006-0207-3 [DOI] [PubMed] [Google Scholar]
- Nagy N., Barad C., Hotta R., Bhave S., Arciero E., Dora D. and Goldstein A. M. (2018). Collagen 18 and agrin are secreted by neural crest cells to remodel their microenvironment and regulate their migration during enteric nervous system development. Development 145, dev160317 10.1242/dev.160317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden D. M. (1983). The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev. Biol. 96, 144-165. 10.1016/0012-1606(83)90318-4 [DOI] [PubMed] [Google Scholar]
- Phillips M. T., Kirby M. L. and Forbes G. (1987). Analysis of cranial neural crest distribution in the developing heart using quail-chick chimeras. Circ. Res. 60, 27-30. 10.1161/01.RES.60.1.27 [DOI] [PubMed] [Google Scholar]
- Pietri T., Eder O., Blanche M., Thiery J. P. and Dufour S. (2003). The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev. Biol. 259, 176-187. 10.1016/S0012-1606(03)00175-1 [DOI] [PubMed] [Google Scholar]
- Poelmann R. E., Mikawa T. and Groot A. C. G. (1998). Neural crest cells in outflow tract septation of the embryonic chicken heart: Differentiation and apoptosis. Dev. Dyn. 212, 373-384. [DOI] [PubMed] [Google Scholar]
- Pomeranz H. D., Rothman T. P. and Gershon M. D. (1991). Colonization of the post-umbilical bowel by cells derived from the sacral neural crest: direct tracing of cell migration using an intercalating probe and a replication-deficient retrovirus. Development 111, 647-655. [DOI] [PubMed] [Google Scholar]
- Porrello E. R., Mahmoud A. I., Simpson E., Hill J. A., Richardson J. A., Olson E. N. and Sadek H. A. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078-1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G. and Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190. 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Powell J. F., Reska-Skinner S. M., Prakash M. O., Fischer W. H., Park M., Rivier J. E., Craig A. G., Mackie G. O. and Sherwood N. M. (1996). Two new forms of gonadotropin-releasing hormone in a protochordate and the evolutionary implications. Proc. Natl. Acad. Sci. USA 93, 10461-10464. 10.1073/pnas.93.19.10461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios A. C., Capaldo B. D., Vaillant F., Pal B., Van Ineveld R., Dawson C. A., Chen Y., Nolan E., Fu N. Y., Jackling F. C. et al. (2019). Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35, 618-632.e6. 10.1016/j.ccell.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Rocha M., Beiriger A., Kushkowski E. E., Miyashita T., Singh N., Venkataraman, V. and Prince, V. E. (2020). From head to tail: regionalization of the neural crest. Development 147, dev193888 10.1242/dev.193888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochard L. J., Ling I. T. C., Kong Y. and Liao E. C. (2015). Visualization of chondrocyte intercalation and directional proliferation via zebrabow clonal cell analysis in the embryonic Meckel's cartilage. J. Vis. Exp. 105, e52935 10.3791/52935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande-Melón M., Marques I. J., Galardi-Castilla M., Langa X., Pérez-Löpez M., Botos M.-A., Sánchez-Iranzo H., Guzmán-Martínez G., Ferreira Francisco D. M., Pavlinic D. et al. (2019). Adult sox10+ cardiomyocytes contribute to myocardial regeneration in the zebrafish. Cell Rep. 29, 1041-1054.e5. 10.1016/j.celrep.2019.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J. R. (1989). Analysing cell lineage with a recombinant retrovirus. Trends Neurosci. 12, 21-28. 10.1016/0166-2236(89)90152-5 [DOI] [PubMed] [Google Scholar]
- Sato M. and Yost H. J. (2003). Cardiac neural crest contributes to cardiomyogenesis in zebrafish. Dev. Biol. 257, 127-139. 10.1016/S0012-1606(03)00037-X [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T. and Bronner-Fraser M. (2008). A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557-568. 10.1038/nrm2428 [DOI] [PubMed] [Google Scholar]
- Schartl M. (2014). Beyond the zebrafish: diverse fish species for modeling human disease. Dis. Model. Mech. 7, 181-192. 10.1242/dmm.012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl A., Malitschek B., Kazianis S., Borowsky R. and Schartl M. (1995). Spontaneous melanoma formation in nonhybrid Xiphophorus. Cancer Res. 55, 159-165. [PubMed] [Google Scholar]
- Schneider R. A. and Helms J. A. (2003). The cellular and molecular origins of beak morphology. Science 299, 565-568. 10.1126/science.1077827 [DOI] [PubMed] [Google Scholar]
- Serbedzija G. N. and Mcmahon A. P. (1997). Analysis of neural crest cell migration in splotch mice using a neural Crest-Specific LacZ reporter. Dev. Biol. 185, 139-147. 10.1006/dbio.1997.8551 [DOI] [PubMed] [Google Scholar]
- Serbedzija G. N., Bronner-Fraser M. and Fraser S. E. (1989). A vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Development 106, 809-816. [DOI] [PubMed] [Google Scholar]
- Serbedzija G. N., Fraser S. E. and Bronner-Fraser M. (1990). Pathways of trunk neural crest cell migration in the mouse embryo as revealed by vital dye labelling. Development 108, 605-612. [DOI] [PubMed] [Google Scholar]
- Serbedzija G. N., Burgan S., Fraser S. E. and Bronner-Fraser M. (1991). Vital dye labelling demonstrates a sacral neural crest contribution to the enteric nervous system of chick and mouse embryos. Development 111, 857-866. [DOI] [PubMed] [Google Scholar]
- Serbedzija G. N., Bronner-Fraser M. and Fraser S. E. (1992). Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development 116, 297-307. [DOI] [PubMed] [Google Scholar]
- Serbedzija G. N., Bronner-Fraser M. and Fraser S. E. (1994). Developmental potential of trunk neural crest cells in the mouse. Development 120, 1709-1718. [DOI] [PubMed] [Google Scholar]
- Sieber-Blum M. and Cohen A. M. (1980). Clonal analysis of quail neural crest cells: they are pluripotent and differentiate in vitro in the absence of noncrest cells. Dev. Biol. 80, 96-106. 10.1016/0012-1606(80)90501-1 [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M. and Bronner M. E. (2016). Reprogramming of avian neural crest axial identity and cell fate. Science 352, 1570-1573. 10.1126/science.aaf2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M. S., McKeown S. J., Tan-Cabugao J., Sauka-Spengler T. and Bronner M. E. (2012). Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is encrypted in the genome. PLoS Genet. 8, e1003142 10.1371/journal.pgen.1003142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov R., Kaucka M., Kastriti M. E., Petersen J., Chontorotzea T., Englmaier L., Akkuratova N., Yang Y., Häring M., Dyachuk V. et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Spanjaard B., Hu B., Mitic N., Olivares-Chauvet P., Janjuha S., Ninov N. and Junker J. P. (2018). Simultaneous lineage tracing and cell-type identification using CrIsPr-Cas9-induced genetic scars. Nat. Biotechnol. 36, 469-473. 10.1038/nbt.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemple D. L. and Anderson D. J. (1992). Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell 71, 973-985. 10.1016/0092-8674(92)90393-Q [DOI] [PubMed] [Google Scholar]
- Stocker K. M., Brown A. M. C. and Ciment G. (1993). Gene transfer oflacZ into avian neural tube and neural crest cells by retroviral infection of grafted embryonic tissues. J. Neurosci. Res. 34, 135-145. 10.1002/jnr.490340114 [DOI] [PubMed] [Google Scholar]
- Sulston J. E. (1983). Neuronal cell lineages in the nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 48, 443-452. 10.1101/SQB.1983.048.01.049 [DOI] [PubMed] [Google Scholar]
- Tamura Y., Matsumura K., Sano M., Tabata H., Kimura K., Ieda M., Arai T., Ohno Y., Kanazawa H., Yuasa S. et al. (2011). Neural crest–derived stem cells migrate and differentiate into cardiomyocytes after myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 31, 582-589. 10.1161/ATVBAHA.110.214726 [DOI] [PubMed] [Google Scholar]
- Tang W., Li Y., Gandhi S. and Bronner M. E. (2019a). Multiplex clonal analysis in the chick embryo using retrovirally-mediated combinatorial labeling. Dev. Biol. 450, 1-8. 10.1016/j.ydbio.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Martik M. L., Li Y. and Bronner M. E. (2019b). Cardiac neural crest contributes to cardiomyocytes in amniotes and heart regeneration in zebrafish. eLife 8, e47929 10.7554/eLife.47929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. and Rubin H. (1958). Characteristics of an assay for Rous sarcoma virus and Rous sarcoma cells in tissue culture. Virology 6, 669-688. 10.1016/0042-6822(58)90114-4 [DOI] [PubMed] [Google Scholar]
- Teng L., Mundel N. A., Frist A. Y., Wang Q. and Labosky P. A. (2008). Requirement for Foxd3 in the maintenance of neural crest progenitors. Development 135, 1615-1624. 10.1242/dev.012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney M. T., Stec M. J., Rulands S., Simons B. D. and Sacco A. (2018). Muscle stem cells exhibit distinct clonal dynamics in response to tissue repair and homeostatic aging. Cell Stem Cell 22, 119-127.e3. 10.1016/j.stem.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y., Matsumura K., Wakamatsu Y., Matsuzaki Y., Shibuya I., Kawaguchi H., Ieda M., Kanakubo S., Shimazaki T., Ogawa S. et al. (2005). Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J. Cell Biol. 170, 1135-1146. 10.1083/jcb.200504061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor P. A., Sobieszczuk D., Wilkinson D. and Krumlauf R. (2002). Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development 129, 433-442. [DOI] [PubMed] [Google Scholar]
- Tucker A. S. and Lumsden A. (2004). Neural crest cells provide species-specific patterning information in the developing branchial skeleton. Evol. Dev. 6, 32-40. 10.1111/j.1525-142X.2004.04004.x [DOI] [PubMed] [Google Scholar]
- Turner D. L. and Cepko C. L. (1987). A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131-136. 10.1038/328131a0 [DOI] [PubMed] [Google Scholar]
- Twitty V. C. (1945). The developmental analysis of specific pigment patterns. J. Exp. Zool. 100, 141-178. 10.1002/jez.1401000108 [DOI] [Google Scholar]
- Varum S., Baggiolini A., Zurkirchen L., Atak Z. K., Cantù C., Marzorati E., Bossart R., Wouters J., Häusel J., Tuncer E. et al. (2019). Yin Yang 1 orchestrates a metabolic program required for both neural crest development and melanoma formation. Cell Stem Cell 24, 637-653.e9. 10.1016/j.stem.2019.03.011 [DOI] [PubMed] [Google Scholar]
- White P. M. and Anderson D. J. (1999). In vivo transplantation of mammalian neural crest cells into chick hosts reveals a new autonomic sublineage restriction. Development 126, 4351-4363. [DOI] [PubMed] [Google Scholar]
- Woronowicz K. C., Gline S. E., Herfat S. T., Fields A. J. and Schneider R. A. (2018). FGF and TGFβ signaling link form and function during jaw development and evolution. Dev. Biol. 444, S219-S236. 10.1016/j.ydbio.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Williams J. P., Rizvi T. A., Kordich J. J., Witte D., Meijer D., Stemmer-Rachamimov A. O., Cancelas J. A. and Ratner N. (2008). Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert Hedgehog-expressing cells. Cancer Cell 13, 105-116. 10.1016/j.ccr.2007.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Kamenev D., Kaucka M., Kastriti M. E., Zhou B., Artemov A. V., Storer M., Fried K., Adameyko I., Dyachuk V. et al. (2019). Schwann cell precursors contribute to skeletal formation during embryonic development in mice and zebrafish. Proc. Natl. Acad. Sci. USA 116, 15068-15073. 10.1073/pnas.1900038116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M. and Emerman M. (2006). Retroviral infection of non-dividing cells: Old and new perspectives. Virology 344, 88-93. 10.1016/j.virol.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Abe K., Mantani A., Hitoshi Y., Suzuki M., Osuzu F., Kuratani S. and Yamamura K. (1999). A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev. Biol. 212, 191-203. 10.1006/dbio.1999.9323 [DOI] [PubMed] [Google Scholar]
- Yanai H., Atsumi N., Tanaka T., Nakamura N., Komai Y., Omachi T., Tanaka K., Ishigaki K., Saiga K., Ohsugi H. et al. (2017). Intestinal cancer stem cells marked by Bmi1 or Lgr5 expression contribute to tumor propagation via clonal expansion. Sci. Rep. 7, 41838 10.1038/srep41838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Vivatbutsiri P., Morriss-Kay G., Saga Y. and Iseki S. (2008). Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 125, 797-808. 10.1016/j.mod.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Zheng H., Chang L., Patel N., Yang J., Lowe L., Burns D. K. and Zhu Y. (2008). Induction of abnormal proliferation by nonmyelinating Schwann cells triggers neurofibroma formation. Cancer Cell 13, 117-128. 10.1016/j.ccr.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Ziller C., Dupin E., Brazeau P., Paulin D. and Le Douarin N. M. (1983). Early segregation of a neuronal precursor cell line in the neural crest as revealed by culture in a chemically defined medium. Cell 32, 627-638. 10.1016/0092-8674(83)90482-8 [DOI] [PubMed] [Google Scholar]