Abstract
The subset of the proteome that contains enzymes in their catalytically active form can be interrogated by using probes targeted towards individual specific enzymes. A subset of such enzymes are proteases that are frequently studied with activity-based probes, small inhibitors equipped with a detectable tag, commonly a fluorophore. Due to the spectral overlap of these commonly used fluorophores, multiplex analysis becomes limited. To overcome this, we developed a series of protease-selective lanthanide-labeled probes compatible with mass cytometry giving us the ability to monitor the activity of multiple proteases in parallel. Using these probes we were able to identify the distribution of four proteases with different active site geometries in three cell lines and peripheral blood mononuclear cells. This provides a framework for the use of mass cytometry for multiplexed enzyme activity detection.
INTRODUCTION
Proteases play critical roles in multiple processes in health and disease.1 Since proteases are characterized by the ability to stimulate peptide bond breakdown, their analysis through genomics or transcriptomics is limited as those do not consider whether the enzyme is active or not.2–3 Even proteomic tools such as antibodies that rely on mass spectrometry are often insufficient to indicate which particular protease is active within a proteome.4 The investigation of protease activity becomes more complicated because these enzymes are regulated on several posttranslational levels.5 Efforts to explore the universe of proteolytic events must include the identification and activity status of protease(s) in question. Here, we define the activome as the functional readout of the proteome, which therefore holds information about the complete activity status of a sample (Figure 1A). The activome can be interrogated by utilizing reagents such as specific inhibitors and activity-based probes.2, 6 The activome does not account for enzymes that were not expressed or are not active due to inhibition, degradation, or lack of proenzyme activation; thus, it extracts information of specific biological relevance. The most convenient approach to study the activome is to use activity-based probes (ABPs) which are small-molecule inhibitors equipped with a detectable tag (biotin, fluorophore, or radioisotope) (Figure 1B).7–8 Fluorescently tagged ABPs provided a breakthrough in revealing activomes in cells and whole organisms.9–10 However, the overlap of fluorescence emission spectra limits the number of enzymes that can be detected and visualized in parallel11, thus limiting their application for multi-parametric analysis. To increase the number of enzymes that can be detected simultaneously we developed metal (lanthanide) isotope tagged ABPs that can be investigated by both, mass and imaging mass cytometry (IMC). These technologies combine flow cytometry with high precision mass markers, thus overcoming the problem of signal overlap and provide a spatial resolution of 1μm.2, 12–14 As elemental mass spectrometry allows for metal isotope discrimination with high precision, it can be applied for simultaneous assessment and quantification of cellular processes at the single cell and protein level.15 CyTOF (Cytometry by Time-Of-Flight) applications incorporate lanthanide metals to label antibodies, proteins, or nucleic acids to visualize and characterize cell types12, 14. Mass cytometry has been also adapted for small molecule chemical probes to monitor whole-cell biological processes such as hypoxia, senescence or protein synthesis 16–21. To further develop this concept, we combined the power of CyTOF with activity-based protein profiling (ABPP) to develop mass cytometry compatible probes for the visualization of individual enzymes in cell populations. This might provide a methodological advancement over currently used enzyme visualization techniques. As proof of concept we focused on proteases.
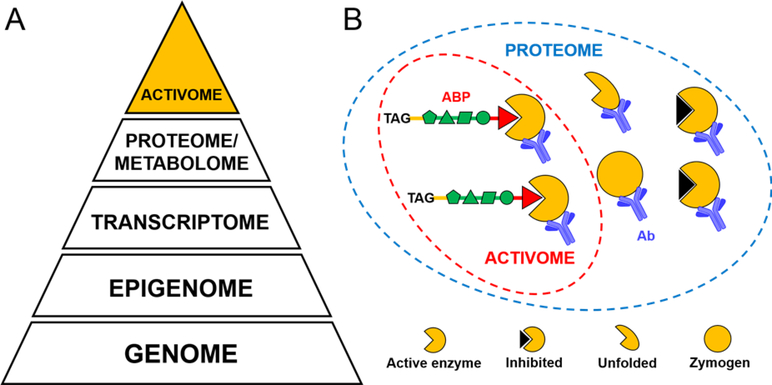
Figure 1. The activome concept.
Panel A The pyramid represents the hierarchy of experimentally verifiable properties of biological systems, where the apex is defined by the activome: the result of supporting events including translation, expression, and posttranslational modifications. Panel B While the proteome is the whole set of proteins regardless of their activity status, the activome is the functional part of the proteome, and can be dissected by selective activity-based probes (ABPs).
The cysteine proteases cathepsin B, L and legumain have unique activities and are each involved in protein processing and antigen presentation. Therefore, they constitute an important component of human peripheral blood mononuclear cells (PBMCs). Human neutrophil elastase (HNE), on the other hand, is a major serine protease in neutrophils, and is responsible for the host defense against pathogens. Our previous studies have defined enzyme-selective sequences that can be utilized to derive metal-mass tagged ABPs.22–25 Accordingly, we used these four proteases, displaying two distinct catalytic mechanisms, as targets for the development of enzyme-specific TOF-probes (metal-tagged, time of flight activity-based probes). The newly developed probes enabled us to investigate the cellular activity and localization of three lysosomal proteases, cathepsin B, cathepsin L and legumain and one neutrophil serine protease, namely neutrophil elastase, using standard mass cytometry and imaging mass cytometry. This work aims to show solid chemistry work that describes the development of the first-in class CyTOF selective activity-based probes for individual enzyme visualization.
RESULTS AND DISCUSSION
In our previous work we had deployed HyCoSuL to develop highly selective peptide recognition sequences for cathepsin L22, cathepsin B24, legumain23 and elastase25. Figure S1 validates the fluorescent probe structures and concentration (1 μM) sufficient for selective and effective labeling. Next we aimed to integrate these sequences into a mass cytometry platform by incorporating an N-terminal tetracarboxylic acid (DOTA)-chelated stable isotope of lanthanides, and a C-terminal acyloxymethylketone (AOMK) and diphenyl phosphonate warhead (Figure 2A, B). TOF-probes were synthesized by coupling a warhead moiety to a DOTA-peptide fragment and chelating corresponding metal particle (Figure 2C). To validate our chemical approach, we selected more than one stable metal isotope for each TOF-probe targeting particular enzyme and created a panel of chemical compounds (Figure 2D). We decided to use two stable and isotopically pure metals-terbium and lutetium (159Tb and 175Lu), and naturally occurring gadolinium, which is a mixture of six stable isotopes (154Gd, 155Gd, 156Gd, 157Gd, 158Gd, 160Gd). The mixture of gadolinium isotopes was used in order to evaluate whether the nature of the isotope can influence enzyme binding specificity. We performed kinetic and cellular analysis of cathepsins B, L and legumain as these enzymes share the same catalytic mechanisms and cellular localization.
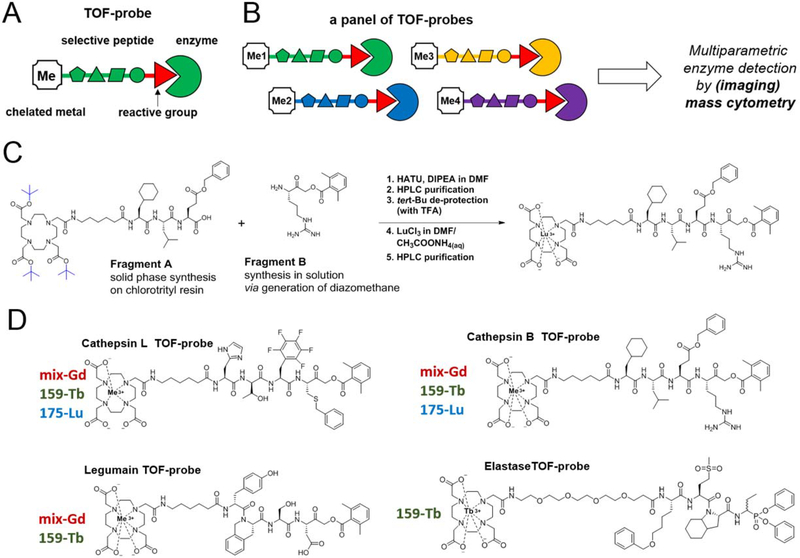
Figure 2. The concept of TOF-probes for the selective detection of enzymes with mass cytometry.
Panel A The general architecture of a TOF-probe. A protease-selective peptide is tagged with a stable lanthanide isotope making it suitable for mass cytometry. Panel B A set of protease-selective probes labeled with different metal tags for the parallel visualization of several enzymes. Panel C An outline of the chemical procedure for the high yield synthesis of TOF-probes. Panel D Examples of protease-selective TOF-probes labeled with different lanthanides. In this work we synthesized a panel of eight TOF-probes for four proteases displaying two distinct catalytic mechanisms (legumain, cathepsin L, cathepsin B and human neutrophil elastase) based on previously developed selective peptide sequences.
Fluorescent ABPs are valuable tools for the detection of enzyme activity, however, one of their main limitations is the fluorescent spectral overlap, which largely reduces the number of enzymes that can be visualized simultaneously (Figure S2).11 In a quest to develop a new method for multiplexed enzyme imaging, we designed ABPs that are compatible with mass cytometry, a method that uses stable isotopes of lanthanide metals as chemical tags.12 DOTA-metal tagged TOF-probes provided good activity and specificity for the individual enzymes (Figure 3). Moreover, regardless of the metal tag used (Lu, Gd, Tb), TOF-probes retained their binding potency (Figure 3A). This strongly suggests that all available metals can be interchangeably used for TOF-probe labeling, making them compatible with commercially available metal-tagged antibodies. We determined the utility of TOF-probes in cells by competition against fluorescent probes previously determined to be selective for the individual proteases22–24. Gd-tagged TOF-probes for legumain, cathepsin L or cathepsin B were incubated at various concentrations in either MDA-MB-231 or HCT-116 cell lines, followed by measuring the residual activity of selected enzymes (Figure 3B, C; Figure S3). This experiment revealed that 2 μM legumain TOF-probe was sufficient to selectively block legumain activity in both cell lines. Cathepsin L and B TOF-probes were also highly potent and although we observed cross-reactivity between them at high concentrations, at 2 μM probes were selective for their target enzymes.
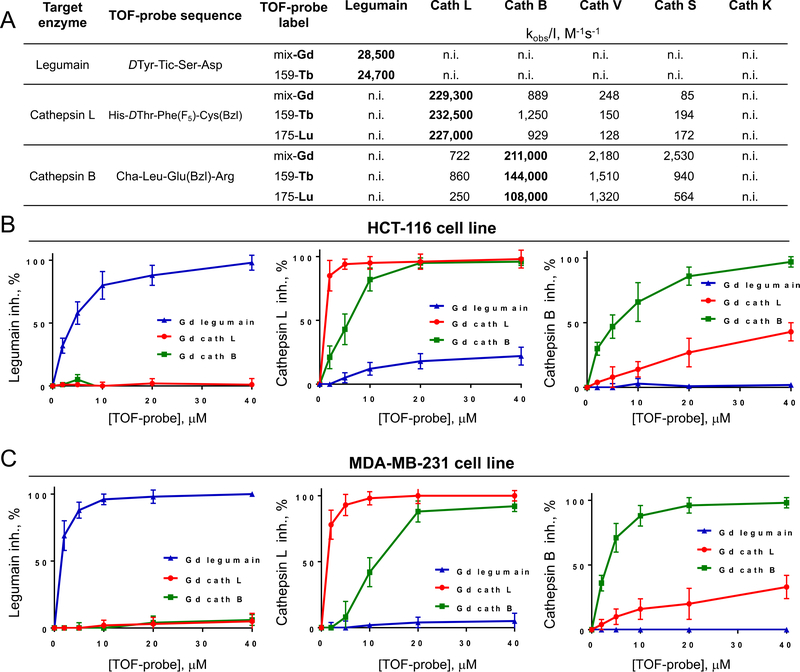
Figure 3. Specificity of TOF-probes toward recombinant proteases and cancer cell lines.
Panel A Kinetic parameters (kobs/[I]) of eight TOF-probes measured against three human recombinant proteases (legumain, cathepsin L and cathepsin B). Panel B and C Selectivity of TOF-probes toward target proteases in cancer cell lines HCT-116 (B) and MDA-MB-231 (C). To assess the selectivity of TOF-probes, they were incubated at various concentration ranges with two cancer cell lines, and residual protease activity was detected with selective, Cy5-labeled probe (Cy5-MP-L01 for legumain, Cy5-MP-cL3 for cathepsin L and Cy5-cB2 for cathepsin B). Based on signal intensity, TOF-probe inhibition profiles were determined. The data demonstrated that regardless of metal tag, all TOF-probes are selective toward targeted proteases.
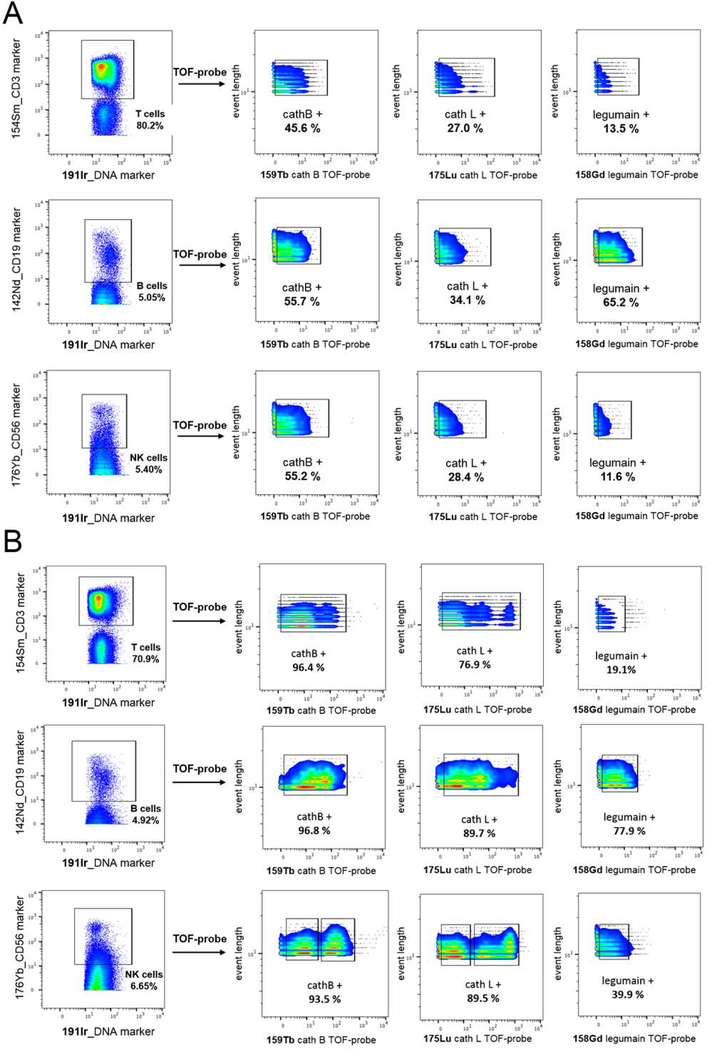
Having confirmed the potency, selectivity and cellular uptake of TOF-probes, we employed these probes to explore the activome by using mass cytometry. After incubation with probes, cells were washed and fixed to remove excess probe. We observed the individual signals from simultaneously labelled samples (Figure S4). Specificity of the TOF-probes in this experiment was confirmed by competition with inhibitors, demonstrating>10-fold signal-to-noise ratio (Figure S4, C). All cells were stained with Ir191/Ir193 DNA intercalator to detect cell events. By using the cathepsin B-directed, Gd-labeled-TOF-probe, which can be detected in several channels, we demonstrated that the individual probe is evenly distributed across the cells (Figure S5). We found that the efficiency of cell labeling is time-dependent, as after 1 hour of incubation around 20% of cells were probe positive, increasing to over 80% after an additional 3 hours (Figure S5). In the next experiment, we stained HCT-116 cells with three TOF-probes (Gd, Tb, and Lu) in various metal combinations to verify whether these metal tags can be used interchangeably in TOF-probes. The results revealed that the pattern of protease labeling was similar across all TOF-probe variants (Figure S6). These data validate the principle of using TOF-probes to investigate subsets of the active proteases in parallel (in this case cathepsin B, L and legumain). Given the fact that the metal tags had no impact on the cellular uptake of TOF-probes and the corresponding protease detection, our approach is highly compatible with other mass cytometry labeling reagents. Detailed gating strategy is shown in Figure S7 and Figure S8.
THP-1, a non-adherent monocyte-like cell line, a model of mononuclear cells, is predicted to transcribe low levels of legumain (Figure S9) and therefore serves as a good platform for determining the sensitivity of TOF-probes. Using the same conditions as in previous experiments (Figure S4, S5, S6) on THP-1 cell line, we showed that cathepsin B and L are abundant, mirroring the transcription data shown in Figure S9. On the other hand, the staining of legumain was poor (only 15.6%), which reflects the low expression of this enzyme. Pre-incubation of THP-1 cells with inhibitors substantially reduced the signals, demonstrating that TOF-probes displayed very little off-target labeling or non-selective accumulation in the lysosomal compartments (Figure S10, S11). Having demonstrated that TOF-probes reveal that different cells in a population can have different amounts of active enzyme we wondered whether this technology would be able to reveal subcellular structures of the cathepsin and legumain activome. Accordingly, we imaged TOF-probe treated THP-1 cells by high-resolution laser ablation coupled to mass cytometry (IMC) (Figure 4A).26 Since the cells are dried during sample preparation, membrane and cytoplasmic components coincide with the nucleus, which defines the segmented area. The images reveal a punctate staining pattern for each of the probes consistent with a lysosomal location for the target proteases, as observed previously using fluorescent ABPs22–23. Moreover, this interpretation is verified by IMC experiments previously performed with antibodies coupled to lysosomal membrane proteins (LAMP-2).27 Thus, we propose that the application of TOF-probes into imaging mass cytometry is a valuable technology for revealing the spatial distribution of the cellular activome (Figure 4B).
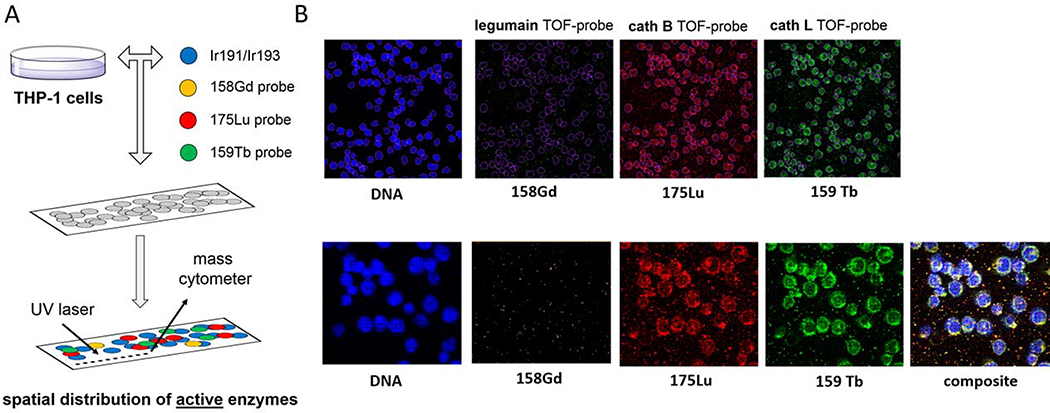
Figure 4. Spatial distribution of active proteases in THP-1 cells.
Panel A THP-1 cells were attached to slides and incubated with a panel of TOF-probes. After TOF-probe treatment, cells were fixed, permeabilized and incubated with 191Ir/193Ir to label DNA. Next, slides were subjected for laser ablation with imaging mass cytometry (IMC) to reveal the spatial distribution of active proteases. Panel B Detection of active cathepsin B and cathepsin L in THP-1 cells with TOF-probes. No active legumain was detected.
To develop the utility and demonstrate the application of our approach we explored the cathepsin B, L and legumain activomes in peripheral blood mononuclear cells (PBMC). PBMCs were incubated with TOF-probes, and metal-tagged cell surface receptor antibodies to detect individual cell populations (T cells, B cells, natural killer (NK) cells) (Figure 5). The data showed that T, B, and NK cells contain equivalent amounts of active cathepsins B and L. What is interesting, we found substantial quantities of active form of legumain in B cells compared to other cell types. The data is consistent across different PBMC preparations from one donor and is not dependent on the metal used in the TOF-probe structure (Figure 5A, Figure S12, S13). To test whether TOF-probes were being taken up non-specifically we increased the concentration of each to 10 μM (Figure 5B). Although the portion of cells labelled with cathepsin B and L probes increased substantially (by almost 2-fold), the portion of cells labelled with a legumain probe increased by only 5.4% in T cells. We interpret this to signify that most PBMCs contain relatively large amounts of cathepsin B and L, but small amounts of legumain, and that our sample washing protocol is sufficient to remove excess probe. Cathepsin L and cathepsin B TOF-probes enabled us to divide the NK cells into two distinct populations: positive, and high positive, demonstrating that the protease activome may vary across the same type of cells as previously seen with neutrophils.11 The labeling of PMBCs with 10 μM legumain TOF-probe confirmed that, in case of this donor, this protease is active in B cells (77.9% positive), moderately active in NK cells (39.9%), but poorly active in T cells (19.1%). Whether this differential is of biological significance is unclear, but merits further investigation.
Figure 5.
Protease detection in Peripheral Blood Mononuclear Cells (PBMCs). Panel A Active proteases were labeled with TOF-probes (2 μM) in PBMCs. The data show that cathepsin B is the most active enzyme in all type of cells, cathepsin L is also present in all cell populations, but less active than cathepsin B, and legumain activity is B cells specific. Panel B Active proteases were labeled with TOF-probes (10 μM) in PBMCs. The results show that cathepsin B and cathepsin L were highly active in all types of cells, whereas legumain was mainly found in B cells, and to a lesser extent in NK cells.
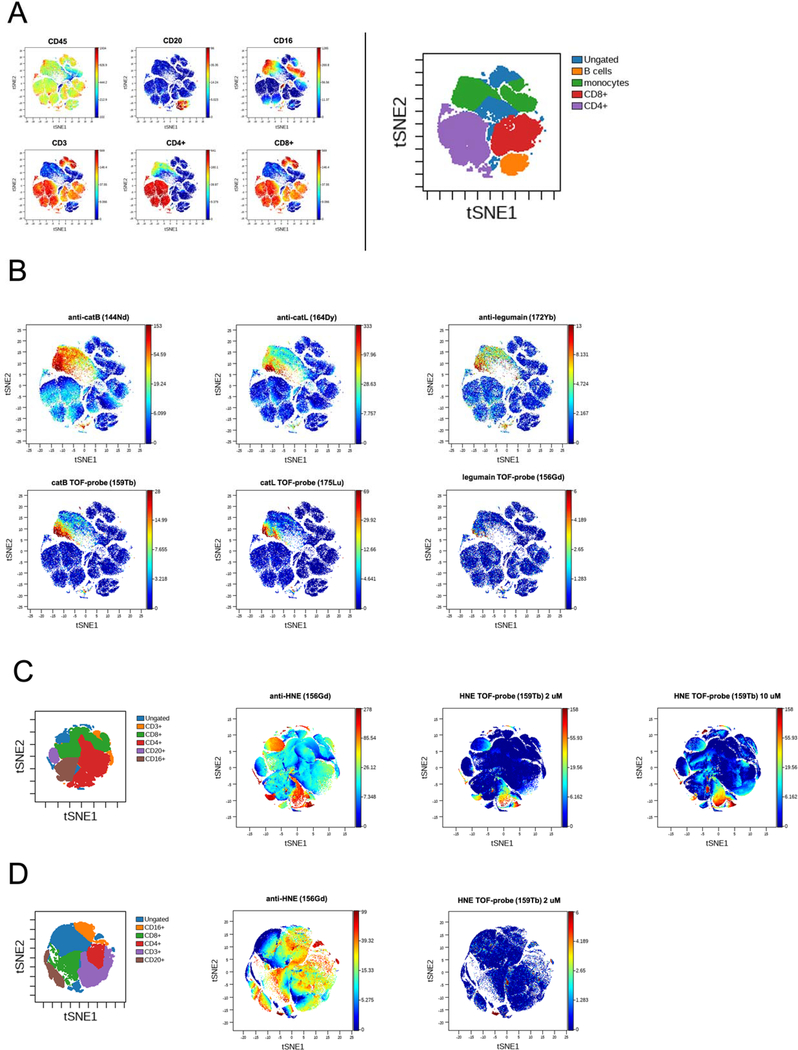
Next, we aimed to perform multiplexed mass cytometry analysis, that include the application of TOF-probes (for enzyme activity) along with metal-labeled antibodies (for enzyme expression). Therefore, we designed a 13-parameter panel (7 cellular markers, 3 antibody markers and 3 TOF-probes) to investigate the target proteases within human PBMCs. As presented in Figure 6, we used viSNE algorithm that enables mapping high-dimensional data onto a 2D plot (see Experimental Section) while preserving high-dimensional structure. We mapped PBMCs cell populations onto key figure (Figure 6A) and subsequently analyzed protease expression and activity status in different subsets. Our analysis revealed that cathepsin B and L can be found mainly in monocytes/macrophages (CD16+ cells) and also, in small portion of CD4+ cells. The active forms of these proteases colocalize with antibodies, however, there also exist subsets of CD16+ cells that do contain cathepsin B and L but are not labeled by TOF-probes. (Figure 6B). This in turn supports the activome concept, where the presence of the enzyme does not indicate its biological activity. The level of legumain expression is low in comparison to cathepsins and also, we did not observe its activity with the TOF-probe. This data is in accordance with our previous findings (Figure 5) and allows us to hypothesize that elevated activity of legumain within B cells may constitute a valuable biological marker.
Figure 6.
viSNE analysis of protease expression and activity status within human PBMCs. Panel A Cells were categorized and defined for analysis according to CD markers. Panel B To decipher the landscape of key lysosomal proteases in PBMCs, protease antibodies along with TOF-probes were employed Cathepsin L, cathepsin B and legumain protein are mainly found in CD16+CD4+ cells but only a small portion of these enzymes is present in the active form. Panel C The analysis of human neutrophil elastase. HNE protein is found in all cell subsets, but only CD4+ and CD16+ cells contain the active form of enzyme. A small CD16+ subset, that seems to be distinct from other CD16+ cells, contains the majority of HNE active enzyme. Panel D PBMCs were stimulated with PMA and analyzed by CyTOF. the highest amount of protein is in the ungated fraction, but this fraction contains no detectable active enzyme.
Next, we targeted one of the most abundant serine proteases- human neutrophil elastase. Its primary residence its within neutrophils and therefore, its expression and activity status within PBMCs it often neglected. We showed, that the use of metal-conjugated antibody revealed abundant distribution of HNE protein within all subsets of PBMCs, but according to our elastase-selective TOF-probe staining only small portion of CD4+ and CD16+ cells carried active form of this protease (Figure 6C). The 5-fold increase in probe concentration (from 2uM to 10uM) did not change the experiment outcome. Moreover, we next treated freshly isolated PBMCs with PMA (50 ng/mL) and observed the lack of elastase activity within CD16+ cells, which is in line with the observation that upon PMA stimulation elastase-rich cells (such as neutrophils) secrete this enzyme into the extracellular space (Figure 6D).25
Our experiments demonstrate that multiplexed analysis that combines cellular markers such as CD antigens with protease metal-tagged antibodies and TOF-probes provided a detailed insight in the proteolytic landscape of human PBMCs. What is more, this approach might be successfully broaden with new proteomic and metal-conjugated chemical tools, to investigate other enzymes in order to build a comprehensive activome profile of a sample.
CONCLUSIONS
The development of selective probes to dissect the activity of individual proteases within a complex system from the test-tube all the way to in vivo imaging in whole animals has witnessed substantial progress over the last 20 years.9, 28–29 The development of increasingly selective probes has enabled the tracking of multiple proteases that together form an activome, a functional component of the proteome. However, one of the major remaining limitations for the parallel monitoring of individual proteases is the lack of appropriate tagging strategies. Currently, most ABPs contain fluorophores that allow for the visualization of only up to 4–5 proteases in parallel to avoid substantial spectral overlap.11 We reasoned that employing lanthanide-based MS-tags for ABP labeling would enable us to study multiple proteases at the same time and expand our ability to interrogate cellular activomes. In this study, we developed metal-tagged ABPs for four proteases displaying various cellular localization and catalytic mechanism, for which selective probe-targeting peptide sequences had been already reported22–23, 25. Our analysis revealed that these new probes, which we called TOF-probes, display high potency and selectivity towards their enzyme targets. To validate their utility, we used them for simultaneous detection of individual proteases in three cell lines using mass cytometry. Multiple technical replicates under various experimental setups provided solid validation for the application of TOF-probes in living cells. Moreover, we determined that the metal tags can be used interchangeably, as the nature of the tag does not affect the TOF-probes potency and selectivity. This makes the approach compatible with metal-tagged antibodies or DNA probes that are currently used for tagging cells in mass cytometry. One difference between TOF-probes and metal-tagged antibodies is that former are able to chelate only one metal particle per probe, whereas antibodies labelled with lanthanides contain up to dozen metal particles attached to the polymer. Therefore, metal-labelled antibodies generate significantly higher signals that may provide for more evident interpretation. Nevertheless, by introducing appropriate controls we are able to dissect positive signals in the samples treated with TOF-probes. Going one step further, we also detected protease activity in cells using imaging mass cytometry (IMC), a technique that enables the simultaneous, multi-parametric analysis of a sample. We showed that active cathepsin B and cathepsin L strongly overlap in THP-1 cells, whereas active legumain is very low to undetectable in these cells. In this experiment, we also demonstrated that TOF-probes can be integrated with IMC settings to create a chemical platform to resolve the spatial distribution of multiple active enzymes within cells or tissue microenvironments.
In our proof of concept study, we deciphered the cathepsins, legumain and human neutrophil elastase activome fingerprint in individual populations of PBMCs. Metal-tagged cell antibody markers were used in combination with metal-tagged protease antibodies and our selective TOF-probes. Antibody analysis showed that cathepsins B and L are abundantly expressed in CD16+ and CD14+ cells which correlated with their activity detected by our TOF-probes. As expected, human neutrophil elastase was found in CD16+ cells (which contain neutrophils) Interestingly, after PMA stimulation no labeling by the TOF-probe was detected, indicating inactivation of the enzyme as previously reported25. The distribution of active cathepsin L, B and elastase within PBMCs subsets revealed by TOF-probes, and correlating with antibody staining, was consistent across multiple blood samples. On the other hand, we found that the legumain activity was low (<20%) across all cell subtypes except B cells, where it varied significantly depending on the donor (10.1–65.2%). We hypothesize that this might be important marker as the elevated legumain activity in B cells may be linked to ongoing infection or cancer30.
Knowledge of the activity of individual proteases, or enzymes in general, in specific cell populations has the potential to translate into therapeutic, theragnostic, or biomarker settings. For example, the analysis of legumain activity in overall PBMC populations is misleading, as T-cells are the most abundant cells in the PBMC population, and they display only negligible legumain activity. However, by combining TOF-probes with cell markers it becomes possible to integrate proteolytic activity across heterogenous cell populations to generate a more holistic understanding of the activome and more informative protease readouts. With the increased number of deployable metal isotopes it will be possible to gain insight into the complexity of enzyme activities within healthy tissues and disease lesions at a single cell level to provide system-wide views of their activation and their role in complex biological networks.
EXPERIMENTAL SECTION
Chemical reagents.
All chemicals used for the synthesis of TOF-probes were purchased from commercial suppliers and used without purification unless otherwise noted. The 2-cholotorityl chloride resin (1.59 mmol/g, 100–200 mesh) was used for the synthesis of peptides that were further converted into TOF-probes (Iris Biotech GmbH, Germany). Fmoc-protected amino acids were purchased from various suppliers: Iris Biotech GmbH, P3 BioSystems (Louisville, USA), QM Bio (Shanghai, China), and Bachem (Bubendorf, Switzerland). Diisopropylcarbodiimide ( peptide grade), N,N-diisopropylethylamine (DIPEA, peptide grade), piperidine (PIP, peptide grade), and trifluoroacetic acid (purity 99%) were all from Iris Biotech. 2,4,6-trimethylpyridine (2,4,6-collidine, peptide grade), triisopropylsilane (TIPS, purity 99%), 2,2,2-trifluoroethanol (TFE), anhydrous tetrahydrofuran (THF), hydrogen bromide (30% wt. in AcOH), 4-methylmorpholine (NMM), isobutylchloroformate (IBCF), and 2,6-dimethylbenzoic acid (2,6-DMBA) were all purchased from Sigma Aldrich. N-hydroxybenzotriazole (HOBt, monohydrate) was from Creosalus (Louisville, USA). HATU and HBTU (both peptide grade) were from ChemPep Inc.. N,N`-dimethylformamide (DMF, peptide grade) and acetonitrile (ACN, HPLC pure) were from WITKO (Lodz, Poland). Methanol (MeOH, pure for analysis), dichloromethane (DCM, pure for analysis), diethyl ether (Et2O, pure for analysis), acetic acid (AcOH, 98% pure) and phosphorus pentoxide (P2O5, 98% pure) were from POCh (Gliwice, Poland). Fluorescent tags (Cyanine-5 NHS and Cyanine-7 NHS) were purchased from Lumiprobe (Hannover, Germany). Diazomethane was generated according to the Aldrich Technical Bulletin (AL-180) protocol.
Synthesis of fluorescent activity-based probes.
The detailed protocol for the synthesis of Cy5-labeled ABPs for legumain, cathepsin L and cathepsin B is published elsewhere.22 All synthesized ABPs were purified on HPLC, and their MS was confirmed via HR-MS.
Synthesis of TOF-probes.
All the amino acids in this procedure are L-enantiomers, unless otherwise stated. Metals were incorporated into probe structures via chelation with DOTA (1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetraacetic acid). The probes utilized an AOMK (acyloxymethyl ketone) as irreversible warhead for cathepsins B, L and legumain and diphenyl phosphonate warhead for neutrophil elastase. The detailed procedure of TOF-probes synthesis is exemplified by DOTA(175Lu)-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK for cathepsin B, Figure 2C, and TOF-probes for legumain and cathepsin L utilized in this work were obtained in a similar manner. The synthesis of 175Lu-tagged cathepsin B probe included five sequential steps: (1) Boc-Arg(Boc)2-was coupled to the AOMK warhead, (2) the synthesis of selective peptide sequence with a 6-aminohexanoic acid spacer to yield Boc-Ahx-Cha-Leu-Glu(Bzl)-OH; (3) coupling of warhead with the peptide sequence and de-protection of amino acids side chains to yield H2N-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK; (4) conjugation of a DOTA chelating group to obtain DOTA-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK; (5) incorporation of metal isotope to yield DOTA(175Lu)-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK. The irreversible reactive group Boc-Arg(Boc)2-AOMK was obtained according to the procedure described previously 23, 31. The Cathepsin B selective peptide sequence was synthesized on the solid support (2-chlorotrityl chloride resin).32 In brief, Fmoc-Glu(Bzl)-OH (3 eq) was coupled to the resin (1 eq, 250 mg) in anhydrous DCM using DIPEA (3 eq) within 3 h. Then, Fmoc-protecting group was removed with 20% piperidine in DMF (5, 5, 25 min cycles). In the next steps Fmoc-Leu-OH, Fmoc-Cha-OH and Boc-6-Ahx-OH (2.5 eq) were attached using HATU (2.5 eq) and 2,4,6-collidine (2.5 eq) in DMF as coupling reagents. Each amino acid coupling was carried out for 2.5 h. In the last step, the peptide was removed from the resin with the mixture of DCM/TFE/AcOH (v/v/v, 8:1:1) within 45 min. Then, the solution was filtered, and solvents were removed under reduced pressure. The crude peptide was dissolved in acetonitrile:H2O (v/v, 3:1) and lyophilized to obtain Boc-6-Ahx-Cha-Leu-Glu(Bzl)-OH as a white powder (purity ≥ 95%). In the next stage H2N-Arg-AOMK was coupled to Boc-6-Ahx-Cha-Leu-Glu(Bzl)-OH. To a small flask containing 1 eq of Boc-Arg(Boc)2-AOMK the mixture of DCM:TFA (v/v, 2:1) was added to remove protecting groups. After 30 min TFA and DCM were evaporated under reduced pressure. To freshly obtained H2N-Arg-AOMK (yellow oil) Boc-6-Ahx-Cha-Leu-Glu(Bzl)-OH (1.2 eq), HATU (1.2 eq) in DMF and 2,4,6-collidine (3 eq) were added. The coupling reaction was monitored by HPLC. After the reaction was completed (3 h) the mixture was diluted with ethyl acetate, transferred to a separatory funnel and extracted with 5% citric acid (once), 5% NaHCO3 (once) and brine (once). The organic fraction was dried over MgSO4 and evaporated under reduced pressure. Obtained Boc-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK was treated with the mixture of DCM:TFA (1:1, v/v) for 30 min and the solvents were removed under reduced pressure. Crude product was purified on HPLC and lyophilized (purity ≥ 95%). In the next stage, DOTA(tBu)3 (1.2 eq) and HATU (1.2 eq) were dissolved in a small volume of DMF. Then, collidine (3 eq) was added to this solution. The mixture was activated for 1 min and added to a small flask containing 1eq of H2N-6-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK in DMF. The reaction mixture was agitated at room temperature until HPLC indicated that the reaction was complete (one hour). Then, obtained DOTA(tBu)3-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK was purified on HPLC and lyophilized (purity ≥ 95%). In the last stage, the conjugation of DOTA-peptide with metal isotope was carried out according to the method described by Sasabowski and Mather 33. To remove tBu-protecting groups DOTA(tBu)3-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK (1 eq) was added to a solution of 50% TFA in DCM and stirred for 30 min. After this time, solvents were removed under reduce pressure. Lutetium (III) chloride (5 eq) was dissolved in minimal volume of 0.1 M ammonium acetate buffer (pH= 5.0) and added to a small flask containing DOTA-Ahx-Cha-Leu-Glu(Bzl)-Arg-AOMK dissolved in minimal volume of DMF. The pH of metal conjugation was in the range of 4 to 6 to avoid reducing the rate of metal complexation (pH<4) or formation of insoluble metal hydroxides (pH>6) [4]. The reaction flask was placed in water bath (55°C) and stirred for 1 h (radiolabeling was monitored by HPLC). Subsequently, the crude product was purified on HPLC and lyophilized (purity ≥ 95%). The TOF-probe for elastase was synthesized in a similar manner by combining a metal-containing peptide fragment with the elastase-selective sequence H2N-AbuP(OPh)2 (Abu is homoalanine)25.
Cysteine cathepsins and legumain.
Human recombinant cathepsins B, L, V, S, and K were expressed and purified as published previously34–35 and active site titrated using E64 inhibitor (Peptide Institute, Japan). Human recombinant pro-legumain was purchased from R&D Systems (2199-CY), activated per the manufacturer’s instructions and active site titrated using MP-L01 inhibitor as described previously23.
Enzymatic kinetic studies.
Inhibitors kinetic experiments were performed using a CLARIOStar (BMG LABTECH) plate reader operating in a fluorescence kinetic mode using 96-well plates (Corning®, Costar®). AMC-labeled fluorescent substrates were screened at 360 nm (excitation) and 460 nm (emission) wavelengths (gain 650). The cathepsin substrate (Z-FR-AMC) was from R&D Systems and legumain substrate (Z-AAN-AMC) was from Bachem. Assay conditions for cathepsin L22, cathepsin B24, and legumain23 were as previously described the second order inhibition rate constant (kobs/I expressed in M−1s−1) towards human cathepsins B, K, L, S, and V and legumain was measured under pseudo-first order kinetic conditions.36 enzyme velocities at each inhibitor concentration were determined by curve fitting using non-linear regression analysis in (GraphPadPrism). and plotted against inhibitor concentration (I) to yield the apparent second order rate constant for inhibition, kapp/I. The absolute value of the second order rate constant kobs/I was calculated as kobs/I =kapp/I *(1+[S]/KM). Experiments were repeated at least three times, and results are presented as an average (S.D. values were always below 20%). The kinetic parameters of the elastase-selective TOF-probe toward human neutrophil proteases were determined using the same methodology and demonstrated a selectivity factor of over 200 compared to the closest paralog NSP3.
Cell culture.
In this study we used low passage cell lines HCT-116, MDA-MB-231, and THP-1 purchased from ATCC. The genomes of all cell lines were authenticated in the Genomics Core at Sanford Burnham Prebys Medical Discovery Institute. HCT-116 cells were cultured in McCoy`s 5A medium, MDA-MB-231 cells were cultured in Dulbeccòs Modification of Eaglès Medium (DMEM) medium, and THP-1 cells were cultured in RPMI-1640 medium. Each medium was supplemented with 10% of Fetal Bovine Serum (FBS), 2 mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin. When appropriate, adherent cells (HCT-116 and MDA-MB-231) were detached from culture plate using trypsin-EDTA (0.25%) – phenol red solution.
Protease labeling in cells using fluorescent ABPs.
50,000 of HCT-116 or MDA-MB-231 cells were seeded into 12-well plates and allow to attach overnight. The next day, 1 μM Cy5-labeled probes for either legumain (Cy5-MP-L01), cathepsin L (MP-cL3) or cathepsin B (MP-cB2) was added to the cells and incubated for 4 hours. Next, cells were harvested, centrifuged at 500 × g for 5 min, cell pellets were washed with 1xDPBS, cells were centrifuged again at 500 × g for 5 min, and the supernatant was discarded. Finally, cell pellets were solubilized into 100 μL of 1x SDS/DTT and boiled for 5 min, cooled to room temperature and sonicated. Subsequently, each sample (30 μL) was subjected for SDS-PAGE analysis (4–12% Bis-Tris Plus 10-well gels, 200 V, 30 min; 3 μL of pre-stained protein ladder was used in the first lane). Gels were scanned at 700 nm (red channel for Cy5 detection) using the Odyssey fluorescence imaging system (LI-COR). Images were analyzed in Image Studio software. This verified that each Cy5 probe specifically labeled its target protease. In a similar manner, we labeled proteases in THP-1 cells. In brief, 100,000 of non-adherent THP-1 cells were placed into 12-well plates, followed by incubation with 1 μM Cy5 probes (4 hours). After this time cells were subjected for SDS-PAGE analysis, and gels were scanned as described above.
Assessment of TOF-probes selectivity and potency in cancer cells.
100,000 of HCT-116 or MDA-MB-231 cells were seeded into 12-well plates and allowed to attach overnight. The next day, selected TOF-probes were incubated with cells at five different concentrations (0, 2, 5, 10, 20, and 40 μM) for 4 hours. After this time, 1 μM Cy5-labeled probe was added and incubated with cells for additional four hours to label residual protease activity (Cy5-MP-L01 for legumain, MP-cL3 for cathepsin L and MP-cB2 for cathepsin B). Next, cells were harvested and subjected to SDS-PAGE as described above. Gels were scanned at 700 nm (red channel for Cy5 detection) as described previously and data were processed in Image Studio software. Each fluorescence band was quantified, and enzyme inhibition profiles were created in GraphPad Prism 7 software. Fluorescent band with no TOF-probe (0 μM) served as control (0% inhibition), and dark black/background band showed no protease activity (100% inhibition). In a similar manner, TOF-probes selectivity was assessed in THP-1 cells. In brief, 100,000 of non-adherent THP-1 cells were placed into 12-well plates, and incubated with various concentrations of TOF-probes, followed by additional incubation with Cy5-labeled probes. For THP-1 only cathepsin L and cathepsin B Cy5 ABPs were used, as this cell line display almost no legumain activity. All experiments were performed in duplicate and data on graphs was presented as averages.
Flow cytometry analysis.
50,000 of HCT-116 and MDA-MB-231 cancer cells were seeded in 12-well plates and allowed to attach overnight. In parallel, 50,000 of non-adherent THP-1 cells were placed in 12-well plates. The following day, 1 μM Cy5-labeleld ABPs for cathepsin L (MP-cL3), cathepsin B (MP-cB2) and legumain (Cy5-MP-L01) were added and incubated with cells for four hours. Control samples were pre-incubated with E64D (25 μM, 2 hours). Two experiments were performed: (1) all three probes were incubated with cells and (2) in each well one probe was added and after four hours of incubation, samples were combined prior to flow cytometry analysis. After incubation, cells were washed twice with DPBS and harvested. In case of separate ABPs incubation, cells from three wells (cathepsin B, cathepsin L and legumain) were combined. Cells were then pelleted by centrifuging at 500 x g, washed with DPBS and fixed with 4% PFA in DPBS for 20 minutes at 4°C. Next, samples were washed once with DPBS, re-suspended in CyFACS buffer and kept on ice until flow cytometry acquisition, but no longer than one hour. Multicolor flow cytometry was performed with single cell suspension (106 cells) on LSRFortessa (14 color) at Sanford Burnham Prebys Flow Cytometry Core Facility and 10,000 events were collected for each sample. Data were analyzed using FlowJo software.
Mass cytometry analysis of cancer cell lines with the use of cysteine protease TOF-probes.
In order to apply our TOF-probes in Helios mass cytometry system, 200,000 cells/well of each cell line (HCT-116 and MDA-MB-231) were seeded onto 12-well plate and allowed to attach overnight. In parallel, 50,000 of non-adherent THP-1 cells were placed in 12-well plates. The next day, medium was exchanged and 2 μM TOF-probes were incubated with cells for four hours. After this time, cells were harvested for mass cytometry analysis. As sample preparation prior to CyTOF acquisition requires multiple washes, cell loss is greater than in case of flow cytometry. Therefore, the cells incubated with three probes were prepared in duplicate and combined after incubation in order to obtain more cells. In case of separate incubation three wells were combined, each containing cells stained with different TOF-probes. Cells were placed in 1.5 mL Eppendorf tubes, pelleted via 5 min centrifugation at 500 x g and washed once with DPBS. Next, cells were fixed with the use of 4% PFA/DPBS for 20 minutes at 4°C and washed again with DPBS. In order to stain DNA, samples were pelleted, supernatant was aspirated, and cells were first washed once with 1xPermeabilization Buffer and then re-suspended in 100 μL of 250 nM Ir191/193 intercalator/1xPermBuffer. After 30 minutes of incubation at room temperature samples were spun down, supernatant was aspirated, cells were re-suspended in 500 μL of CyFACS and kept in the fridge until CyTOF acquisition, but no longer than 12 hours. Prior to acquisition, cells were washed twice with CyFACS buffer and twice with diH2O.
Antibody labeling.
Antibodies toward targeted proteases (cathepsin B, cathepsin L, legumain and human neutrophil elastase) were purchased from R&D Systems. Antibody labeling with stable metal isotopes was performed according to protocol described elsewhere (https://web.stanford.edu/group/nolan/protocols.html). All necessary reagents were purchased from Fluidigm®.
Peripheral blood mononuclear cell (PBMC) analysis.
Fresh blood was obtained from Scripps Research Institute/Scripps Hospital (IRB-16–6789). 60 mL of fresh blood was transferred to storage vessel. An equal volume of 2% FBS/DPBS was added to blood and mixed by swirling. Next, four 50 mL conical tubes were filled with 15 mL of Lymphoprep™ and 30 mL of blood was carefully layered on top of Lymphoprep™. Tubes were centrifuged at 800 x g for 30 minutes. After centrifugation the PBMCs was transferred to a sterile 50 mL tube, an equal volume of 2% FBS/PBS was added and mixed carefully. Tubes were centrifuged at 400 x g for 5 minutes and the supernatants were discarded. Cells were re-suspended in 45 mL of RBC Lysis Buffer (diluted at 4.5 mL of lysis buffer in 40.5 mL H2O) and incubated at room temperature for 7 minutes followed by centrifugation (400 x g for 5 minutes). Supernatant was aspirated, and cells were washed with 2% FBS/PBS, centrifuged using the same settings, and re-suspended in 45–50 mL RPMI + 10% FBS + antibiotics (100 units/mL penicillin and 100 μg/streptomycin). Cells were counted and 50,000 cells per sample were pelleted, re-suspended in media containing 2 μM TOF-probes (various sets of probes, incubated together or separately) and plated onto 12-well dishes. Fresh PBMC’s were incubated with TOF-probes for six hours, harvested and washed twice with DPBS. Next, each sample was fixed with 4% PFA/DPBS for 20 minutes at 4°C and washed once with DPBS and once with CyFACS buffer. Primary antibodies were purchased from Fluidigm® and used as described below. Antibody cocktail was prepared by adding 1 μL of each pre-titrated antibody (anti-hCD3, CD19, CD56, CD45, CD11c, and CD14) into 50 μL of CyFACS buffer. Next, cells were re-suspended in 50 μL of CyFACS buffer and 50 μL of antibody cocktail was added to each sample. Cells were mixed carefully by pipetting and incubated in the cocktail for one hour on ice. After this time, cells were pelleted (800 x g), supernatant was aspirated, cells were washed once, centrifuged and re-suspended in 500 μL of CyFACS buffer. Samples were kept in the fridge (4°C) overnight. The following day, cells were pelleted and re-suspended in 100 μL of 1x Permeabilization Buffer (ThermoScientific) containing 250 nM Ir191/193 DNA intercalator. After 30 minutes of incubation, samples were washed twice with CyFACS buffer, twice with ddH2O and kept on ice until CyTOF acquisition (no longer than 2 hours). Prior to CyTOF run, pellet was adjusted to 2.5–5 × 105/mL. Data was acquired on a CyTOF (Fluidigm®). Obtained data were converted to an FCS file and analyzed by FlowJo software (Treestar Inc.) and Cytobank. The correlation of cell markers, antibodies and TOF-probes was analyzed with the viSNE algorithm. The viSNE optimization algorithm searches for a projection of events revealed in high-dimensional space resulting from multi-parametric analyses into 2 or 3 dimensions for visualization between the events are best conserved between the high- and low-dimensional space37.
Detection of active proteases using Hyperion Imaging System.
To visualize active cathepsin B, cathepsin L and legumain in THP-1 cells we employed a Hyperion imaging mass cytometry system (Fluidigm®). Customized three-well adhesion slides were purchased from Marienfield, Germany. THP-1 cells were suspended in DBPS, attached on slides (20,000 cells/well) and left for an hour in the incubator at 37°C until adhered. Next, cells were washed once with DPBS and fixed with 100 μL of 2%PFA/DPBS for 20 minutes at room temperature. Slides were washed twice with DPBS in coplin jar and cells were permeabilized with 100 μL of 0.1% Triton-X for 10 minutes. Again, slides were washed twice with DPBS followed by blocking with 100 μL of 1% BSA in PBS and incubated for 40 minutes at room temperature in order to avoid unspecific binding of the probes. Afterward, THP-1 cells were washed twice and 100 μL of 2 μM probe preparation was incubated for four hours in 37°C, 5% CO2. Cells were then washed with DBPS and 250 nM Ir191/193 solution was added in order to label DNA within the cells. After 30 minutes of incubation, cells were again washed, rinsed with water for 5 seconds and air dried at room temperature. The prepared slides were subjected to IMC analysis, respectively. At least three Region of Interests (ROIs) of area 400*400 mm were ablated with a laser frequency 200 Hz and ion count was measured by CyTOF Helios mass cytometer as described earlier.26 DNA intercalator 191Ir/193Ir was used for cell segmentation.
Supplementary Material
ACKNOWLEDGMENTS
This project has received funding from the European Union`s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 661187 (to M.P.), National Institutes of Health grant GM99040 and NCI Cancer Center Support Grant P30CA030199 (to G.S.S.). M.D. lab is supported by National Science Centre in Poland (grant Harmonia 2018/30/M/ST5/00440). The “Fix NeutRopenia (FIXNET): focusing on neutrophil proteases defects which serve as novel diagnostic and therapeutic options” project is carried out within the TEAM-NET program of the Foundation for Polish Science financed by the European Union under the European Regional Development Fund (to M.D. and M.P.). The B.T. lab is supported by Slovene Research Agency (gran P1–0140). We thank Yoav Altman, for his support with flow cytometry analysis. WR is a beneficiary of a START scholarship from the Foundation for Polish Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
Footnotes
The authors declare that they have no competing interests.
Material request should be sent to M.P, M.D. or G.S.S.
ASSOCIATED CONTENT
Supporting Information
Selective Cy5-labeled ABPs for the detection of lysosomal proteases in different cell types;
Detection of legumain and cathepsins in HCT116 cells with fluorescent probes and flow cytometry;
Whole gels demonstrating the potency and selectivity of TOF-probes for the labeling/inhibition of legumain, cathepsin L and cathepsin B in human cancer cell lines, HCT-116 and MDA-MB-231;
Detection of proteases by TOF-probes in HCT-116 cells;
Selective detection of legumain, cathepsin L, and cathepsin B in HCT-116 cell line;
Selective labeling of proteases in HCT-116 cells with TOF-probes;
Detailed gating strategy for non-TOF-probes treated and TOF-probes treated HCT-116 cells; Whole gels showing determination of TOF-probes selectivity toward cathepsin L and cathepsin B in THP-1 cells;
Expression level of legumain, cathepsin L and cathepsin B in three human cell lines: HCT-116, MDA-MB-231 and THP-1;
Detection of protease activity in THP-1 cells using TOF-probes;
Proteases detection in Peripheral Blood Mononuclear Cells;
Experimental methods and characterization of all compounds (all in one PDF)
REFERENCES
- 1.Rawlings ND; Barrett AJ; Finn R, Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 2016, 44 (D1), D343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fonovic M; Bogyo M, Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev Proteomics 2008, 5 (5), 721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam GC; Sorensen EJ; Cravatt BF, Chemical strategies for functional proteomics. Mol Cell Proteomics 2002, 1 (10), 781–90. [DOI] [PubMed] [Google Scholar]
- 4.Vizovisek M; Vidmar R; Drag M; Fonovic M; Salvesen GS; Turk B, Protease Specificity: Towards In Vivo Imaging Applications and Biomarker Discovery. Trends Biochem Sci 2018, 43 (10), 829–844. [DOI] [PubMed] [Google Scholar]
- 5.Ryslava H; Doubnerova V; Kavan D; Vanek O, Effect of posttranslational modifications on enzyme function and assembly. J Proteomics 2013, 92, 80–109. [DOI] [PubMed] [Google Scholar]
- 6.Barglow KT; Cravatt BF, Activity-based protein profiling for the functional annotation of enzymes. Nat Methods 2007, 4 (10), 822–7. [DOI] [PubMed] [Google Scholar]
- 7.Willems LI; Overkleeft HS; van Kasteren SI, Current developments in activity-based protein profiling. Bioconjug Chem 2014, 25 (7), 1181–91. [DOI] [PubMed] [Google Scholar]
- 8.Cravatt BF; Wright AT; Kozarich JW, Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem 2008, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- 9.Sanman LE; Bogyo M, Activity-based profiling of proteases. Annu Rev Biochem 2014, 83, 249–73. [DOI] [PubMed] [Google Scholar]
- 10.Blum G; von Degenfeld G; Merchant MJ; Blau HM; Bogyo M, Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol 2007, 3 (10), 668–77. [DOI] [PubMed] [Google Scholar]
- 11.Kasperkiewicz P; Altman Y; D’Angelo M; Salvesen GS; Drag M, Toolbox of Fluorescent Probes for Parallel Imaging Reveals Uneven Location of Serine Proteases in Neutrophils. J Am Chem Soc 2017, 139 (29), 10115–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spitzer MH; Nolan GP, Mass Cytometry: Single Cells, Many Features. Cell 2016, 165 (4), 780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenmiller B; Zunder ER; Finck R; Chen TJ; Savig ES; Bruggner RV; Simonds EF; Bendall SC; Sachs K; Krutzik PO; Nolan GP, Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 2012, 30 (9), 858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendall SC; Simonds EF; Qiu P; Amir el AD; Krutzik PO; Finck R; Bruggner RV; Melamed R; Trejo A; Ornatsky OI; Balderas RS; Plevritis SK; Sachs K; Pe’er D; Tanner SD; Nolan GP, Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332 (6030), 687–96.21551058 [Google Scholar]
- 15.Schulz D; Zanotelli VRT; Fischer JR; Schapiro D; Engler S; Lun XK; Jackson HW; Bodenmiller B, Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell Syst 2018, 6 (4), 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lumba MA; Willis LM; Santra S; Rana R; Schito L; Rey S; Wouters BG; Nitz M, A beta-galactosidase probe for the detection of cellular senescence by mass cytometry. Org Biomol Chem 2017, 15 (30), 6388–6392. [DOI] [PubMed] [Google Scholar]
- 17.Lathia US; Ornatsky O; Baranov V; Nitz M, Multiplexed protease assays using element-tagged substrates. Anal Biochem 2011, 408 (1), 157–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lathia US; Ornatsky O; Baranov V; Nitz M, Development of inductively coupled plasma-mass spectrometry-based protease assays. Anal Biochem 2010, 398 (1), 93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razumienko E; Ornatsky O; Kinach R; Milyavsky M; Lechman E; Baranov V; Winnik MA; Tanner SD, Element-tagged immunoassay with ICP-MS detection: evaluation and comparison to conventional immunoassays. J Immunol Methods 2008, 336 (1), 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar LJ; Vellanki RN; Halupa A; Hedley D; Wouters BG; Nitz M, Identification of hypoxic cells using an organotellurium tag compatible with mass cytometry. Angew Chem Int Ed Engl 2014, 53 (43), 11473–7. [DOI] [PubMed] [Google Scholar]
- 21.Edgar LJ; Vellanki RN; McKee TD; Hedley D; Wouters BG; Nitz M, Isotopologous Organotellurium Probes Reveal Dynamic Hypoxia In Vivo with Cellular Resolution. Angew Chem Int Ed Engl 2016, 55 (42), 13159–13163. [DOI] [PubMed] [Google Scholar]
- 22.Poreba M; Rut W; Vizovisek M; Groborz K; Kasperkiewicz P; Finlay D; Vuori K; Turk D; Turk B; Salvesen GS; Drag M, Selective imaging of cathepsin L in breast cancer by fluorescent activity-based probes. Chem Sci 2018, 9 (8), 2113–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poreba M; Solberg R; Rut W; Lunde NN; Kasperkiewicz P; Snipas SJ; Mihelic M; Turk D; Turk B; Salvesen GS; Drag M, Counter Selection Substrate Library Strategy for Developing Specific Protease Substrates and Probes. Cell Chem Biol 2016, 23 (8), 1023–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poreba M; Groborz K; Vizovisek M; Maruggi M; Turk D; Turk B; Powis G; Drag M; Salvesen GS, Fluorescent probes towards selective cathepsin B detection and visualization in cancer cells and patient samples. Chem Sci 2019, 10 (36), 8461–8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasperkiewicz P; Poreba M; Snipas SJ; Parker H; Winterbourn CC; Salvesen GS; Drag M, Design of ultrasensitive probes for human neutrophil elastase through hybrid combinatorial substrate library profiling. Proc Natl Acad Sci U S A 2014, 111 (7), 2518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giesen C; Wang HA; Schapiro D; Zivanovic N; Jacobs A; Hattendorf B; Schuffler PJ; Grolimund D; Buhmann JM; Brandt S; Varga Z; Wild PJ; Gunther D; Bodenmiller B, Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014, 11 (4), 417–22. [DOI] [PubMed] [Google Scholar]
- 27.Bouzekri A; Esch A; Ornatsky O, Multidimensional profiling of drug-treated cells by Imaging Mass Cytometry. FEBS Open Bio 2019, 9 (9), 1652–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasperkiewicz P; Poreba M; Groborz K; Drag M, Emerging challenges in the design of selective substrates, inhibitors and activity-based probes for indistinguishable proteases. FEBS J 2017, 284 (10), 1518–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarty S; Kahler JP; van de Plassche MAT; Vanhoutte R; Verhelst SHL, Recent Advances in Activity-Based Protein Profiling of Proteases. Curr Top Microbiol Immunol 2018. [DOI] [PubMed] [Google Scholar]
- 30.Shen L; Li H; Shi Y; Wang D; Gong J; Xun J; Zhou S; Xiang R; Tan X, M2 tumour-associated macrophages contribute to tumour progression via legumain remodelling the extracellular matrix in diffuse large B cell lymphoma. Sci Rep 2016, 6, 30347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato D; Boatright KM; Berger AB; Nazif T; Blum G; Ryan C; Chehade KA; Salvesen GS; Bogyo M, Activity-based probes that target diverse cysteine protease families. Nat Chem Biol 2005, 1 (1), 33–8. [DOI] [PubMed] [Google Scholar]
- 32.Maly DJ; Leonetti F; Backes BJ; Dauber DS; Harris JL; Craik CS; Ellman JA, Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem 2002, 67 (3), 910–5. [DOI] [PubMed] [Google Scholar]
- 33.Sosabowski JK; Mather SJ, Conjugation of DOTA-like chelating agents to peptides and radiolabeling with trivalent metallic isotopes. Nat Protoc 2006, 1 (2), 972–6. [DOI] [PubMed] [Google Scholar]
- 34.Mihelic M; Dobersek A; Guncar G; Turk D, Inhibitory fragment from the p41 form of invariant chain can regulate activity of cysteine cathepsins in antigen presentation. J Biol Chem 2008, 283 (21), 14453–60. [DOI] [PubMed] [Google Scholar]
- 35.Bromme D; Nallaseth FS; Turk B, Production and activation of recombinant papain-like cysteine proteases. Methods 2004, 32 (2), 199–206. [DOI] [PubMed] [Google Scholar]
- 36.Poreba M; Groborz K; Navarro M; Snipas SJ; Drag M; Salvesen GS, Caspase selective reagents for diagnosing apoptotic mechanisms. Cell Death Differ 2019, 26 (2), 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amir el AD; Davis KL; Tadmor MD; Simonds EF; Levine JH; Bendall SC; Shenfeld DK; Krishnaswamy S; Nolan GP; Pe’er D, viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013, 31 (6), 545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.