Abstract
Objective
To explore the characteristics of coagulase‐negative Staphylococci other than Staphylococci epidermidis (Nse‐CoNS) meningitis and to apply cerebrospinal fluid (CSF) times to positivity culture (TTPC) for the precise differentiation of meningitis from contamination.
Methods
We conducted a case‐control study to accomplish the following: First, we retrospectively reviewed records of post‐neurosurgical patients’ CSF that yielded Nse‐CoNS from January to October 2019 at the Beijing Tiantan Hospital; 17 clinical and 12 laboratory characteristics were reviewed. Second, we investigated the TTPC of the Nse‐CoNS, the cutoffs, and corresponding parameters to differentiate Nse‐CoNS meningitis from contamination.
Results
In this study, a total of 146 patients with Nse‐CoNS CSF culture positive were enrolled. The average TTPC in the Nse‐CoNS meningitis group was significantly shorter than in the contamination group (20.2 ± 5.0 hours and 30.2 ± 12.6 hours, respectively, P < .05). The area under curve (AUC) of the model was 0.802. A TTPC of 20.0 hours had 94.3% sensitivity and a negative value of 90.2% for predicting Nse‐CoNS meningitis.
Conclusions
Nse‐CoNS meningitis often causes confusion in clinical diagnosis. In this study, we evaluated the clinical predictive factors of Nse‐CoNS meningitis and confirmed that the median TTPC in the Nse‐CoNS meningitis group was significantly shorter than in the contamination group. A TTPC shorter than 20.0 hours was associated with Nse‐CoNS meningitis, while a TTPC longer than 20.0 hours was associated with Nse‐CoNS contamination. This information will be helpful for the rapid diagnosis of Nse‐CoNS meningitis.
Keywords: contamination, meningitis, Nse‐CoNS, time to culture positivity
Cerebrospinal fluids times to positivity culture(TTPC) can differentiate coagulase‐negative Staphylococci other thanStaphylococci epidermidis (Nse‐CoNS) meningitis from contamination. A TTPC shorter than 20.0 h was associated with Nse‐CoNS meningitis, while a TTPC longer than 20.0 h was associated with Nse‐CoNS contamination.
1. INTRODUCTION
Coagulase‐negative Staphylococci (CoNS) are considered to exhibit little virulence and are common contaminants isolated from a fluid specimen such as blood, cerebrospinal fluid (CSF), hydrothorax, and ascites. One study reported that 81.9% of CoNS isolates from fluid specimens were classified as contamination. 1 In recent years, they have become increasingly recognized as microorganisms of clinically significant nosocomial infections, especially in patients with catheter‐related meningitis. 2 CoNS also accounts for significant morbidity and mortality in patients with neurosurgical‐related meningitis. These infections are inherently difficult to treat given the frequent presence of foreign materials and the blood‐brain barrier.
The most common isolate in the CSF of post‐neurosurgical patients is Staphylococcus epidermidis, 3 but several other species (coagulase‐negative Staphylococci other than S epidermidis, Nse‐CoNS), such as Staphylococcus hominis, Staphylococcus haemolyticus, and Staphylococcus capitis are found in the human environment albeit less frequently. S epidermidis accounts for more than 50% of all CoNS isolates in our hospital. There is a report that Nse‐CoNS was responsible for 33% (n = 161/494) of CoNS bloodstream infections in a German population. 4 Considering S epidermidis, the most frequent pathogen related to catheter‐associated infection (CRI), 5 Nse‐CoNS is more likely to lead to contamination than S epidermidis, which may confuse the clinical therapy given. Additionally, little information is available on the clinical relevance of Nse‐CoNS, although evidence suggests that virulence varies among different CoNS species.
Nse‐CoNS isolates from the CSF of post‐neurosurgical patients are often complex and induce some patients to abuse antibiotics while others are misdiagnosed. Therefore, it is a recurrent challenge to differentiate between clinically relevant infections and possible contaminations in Nse‐CoNS isolates. If they are accurately identified, appropriate antibiotic treatment can effectively shorten the hospitalization time and improve the effectiveness of therapy. Moreover, it can reduce the excessive use of antibiotics and thereby help to curb bacterial resistance to antimicrobial agents.
Currently, several studies aimed at differentiating between infections and contamination of Nse‐CoNS, for example, used multiple cultures 6 and carrying out additional biomarker‐assisted diagnosis. 7 However, this approach to the diagnosis of post‐neurosurgical meningitis is insufficient. Multiple cultures require an additional lumbar puncture, which is associated with an increased risk of trauma and infection opportunities. Therefore, employing biomarker detection is an important approach, but it requires the collection of additional specimens, which increases the cost of testing. TTPC can be employed as an effective marker for the diagnosis of meningitis and/or contamination. Because the bacterial concentration in infected patients with meningitis is higher than for contaminating organisms, the higher the initial bacterial concentration of patients, the shorter the TTPC. It has been reported that the time to positivity culture (TTPC) had good diagnostic efficacy in diagnosing bloodstream infections. A TTPC of <16 hours was associated with true CoNS bacteremia requiring an active therapeutic approach, while that >20 hours was associated with CoNS contamination. 8 In the present investigation, to enable the accurate diagnosis of Nse‐CoNS‐related meningitis, we conducted a case‐control study to explore the characteristics of Nse‐CoNS meningitis and to apply CSF TTPC for precise differentiation between meningitis and contamination. To the best of our knowledge, this is the first study to identify Nse‐CoNS infection and contamination using TTPC.
2. MATERIALS AND METHODS
2.1. Patients
This study was conducted at the Beijing Tiantan Hospital & Capital Medical University between January 2019 and October 2019. Beijing Tiantan Hospital & Capital Medical University is a 1850‐bed tertiary teaching hospital with the largest neurosurgical center in northern China. All major neurosurgical departments participated in this study. We retrospectively evaluated the results of CSF cultures that yielded Nse‐CoNS. Information obtained from post‐neurosurgical patients included age (years), male (%), length of hospitalization stay (LOS), post‐neurosurgical infection/contamination time, hypertension, diabetes mellitus, site of surgery, surgical wound classification, extra ventricular drainage (EVD), lumbar drainage (LD), ventilation rate, CSF leakage, craniotomy, ICU admission, malignancy, duration of surgery, and the time required to cure an infection. The study also investigated certain infection‐related clinical laboratory testing parameters, namely CSF leukocyte count (C‐Leu), CSF neutrophil proportions (C‐Neu), CSF cell count (C‐Cell), CSF protein concentration (C‐Pro), CSF chloride ion concentration (C‐Cl‐), CSF glucose concentration (C‐Glu), blood glucose concentration (B‐Glu), blood leukocyte count (B‐Leu), blood neutrophil proportions (B‐Neu), C‐reactive protein (CRP), CSF lactate (C‐Lac), and CSF/blood glucose ratio (C/B‐Glu). Accurate information regarding clinical and laboratory parameters was stored in the electronic files of each patient, which were later retrieved for analysis.
2.2. Inclusion and exclusion criteria
All of the patients with CSF culture Nse‐CoNS during the research time‐interval were included. Criteria for patient exclusion were incomplete medical records, <18 years old, hospitalized for <7 days, discharged from hospital directly without any treatment or died within 7 days.
2.3. Definition of Nse‐CoNS meningitis
All the records of the enrolled hospitalized patients were reviewed in detail. The clinical characteristics of the positive CSF culture patients were retrieved from available copies of discharge letters. Because Nse‐CoNS is one of a special group of microorganisms, the diagnosis of Nse‐CoNS meningitis was defined using much more rigorous criteria. Only patients who met the following criteria simultaneously were classified into the meningitis group: (a) at least two or more CSF bacterial cultures were positive for Nse‐CoNS; treatment with antibiotics inhibited Nse‐CoNS or was associated with removal of the material if it was considered necessary; (b) Nse‐CoNS was cultivated from patient CSF isolates and clinical symptoms such as headache, fever, and mental symptoms changed; and (c) at least one of the following markers of significant inflammation was present: (1) a leukocyte count >0.25 × 109/L containing predominantly polymorphonuclear cells; (2) a lactate concentration >3.5 mmol/L; (3) a concentration of CSF/serum glucose of < 0.4 mmol/L; or a glucose concentration < 2.5 mmol/L if no simultaneous blood glucose concentration was measured. 9 , 10
A positive CSF culture was to be contaminated when: (1) Nse‐CoNS positive cultures were not mentioned in the discharge diagnosis certificate; (2) Nse‐CoNS was rated as not clinically significant by relevant doctors; (3) if the above‐mentioned criteria for post‐neurosurgical infection were not met; (4) if an isolate of Nse‐CoNS was identified in one out of multiple sets of CSF cultures drawn simultaneously; (5) patients who had received antibiotic therapy active against Staphylococcus, ahead of neurosurgery. A patient was considered to be cured when they had provided two consecutive negative CSF microbial cultures together with the alleviation of meningitis symptoms (fever, neck stiffness, etc). TTPC of the CSF in the present study were recorded for the first culture results of all patients.
2.4. CSF bacterial cultures
CSF was collected from all patients when meningitis‐like symptoms presented (temperature >38°C, neck stiffness, etc). At the bedside, CSF was obtained by well‐trained neurosurgical doctors using 70% isopropyl alcohol followed by 10% povidone‐iodine as antiseptics. They withdrew 1‐3 mL of CSF at spinal L3‐L4, L4‐L5 or L5‐S1 levels. All CSF specimens were immediately transferred to the clinical laboratory and were incubated until flagged as positive, or for 5 days in BacT/ALERT 3D (bioMérieux, Marcy l’Etoile, France). The broth was analyzed automatically every 10 minute bacteria from positive bottles, was Gram‐stained, and sub‐cultured onto solid medium using standard protocols. 11 Various antimicrobial drug categories were tested for activity against the isolated bacteria using disk diffusion and broth micro‐dilution methods, according to the Clinical and Laboratory Standards Institute 2018. Susceptibility testing of the isolates was performed according to the guidelines established by the National Committee on Clinical Laboratory Standards. Clinical and Laboratory Standards Institute breakpoint values (CLSI 2018, M100‐S23) were employed.
2.5. Statistical analysis
All the patients enrolled in the study were divided into 2 groups on the basis of the definition described above (vide supra), namely an Nse‐CoNS meningitis group and a contamination group. We determined the clinical significance of differences between the 2 study groups using the Mann‐Whitney U test for continuous variables and Fisher's exact test for categorical variables. A P‐value < .05 was considered to be statistically significant. We constructed a receiver operator characteristic (ROC) curve by plotting the true‐positive rate (sensitivity) against the false‐positive rate (1‐specificity) over a range of cutoff values of TTPC. We estimated the area under curve (AUC) with a parametric procedure and the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio(PLR), negative likelihood ratio(NLR), Youden score, and cutoff values were evaluated. Subsequently, some patients were re‐enrolled during the next 4 months and included in meningitis or contamination groups according to the above inclusion and exclusion criteria for verification. When the TTPC of a patient's CSF was shorter than cutoff, they were classified into the meningitis group, and if it was longer than cutoff, they were classified into the contamination group. Statistical analyses were performed carried using SPSS version 20.0 (IBM, New York, US). All figures were constructed using Prism 7.0 (Graphpad, San Diego, US).
3. RESULTS
3.1. Microbiology
From January 2019 to October 2019, a total of 974 patient samples were collected from CSF, which yielded an 8.1% (974/12 025) positive culture ratio. Of these, 431 patients with CoNS isolates were enrolled in the study, which accounted for 37.8% of all the microbial isolates. In total, 146 patient cases with CSF culture positive for Nse‐CoNS were to be potentially enrolled in the study according to our inclusion and exclusions criteria. Of these, 58 patients with Nse‐CoNS growth in their CSF were classified into the meningitis group and 88 patients were classified into the contamination group. The flow chart for differentiating Nse‐CoNS is shown in Figure 1. The distribution of the respective microorganisms is presented in Table 1 and Figure 2‐A S. hominis (61/146, 41.8%), S haemolyticus (39/146, 26.7%), and S capitis (26/146, 17.8%), together accounted for more than 85% of all relevant Nse‐CoNS CSF culture isolates. Multiple organism infections were excluded, and the results of the antimicrobial susceptibility test for the meningitis group are shown in Table 2. The antimicrobial susceptibility test for Nse‐CoNS in the meningitis group is shown in Figure 2‐B. All Nse‐CoNS were sensitive to vancomycin, tigecycline and linezolid, and 64.47% of Nse‐CoNS were resistant to oxacillin.
FIGURE 1.
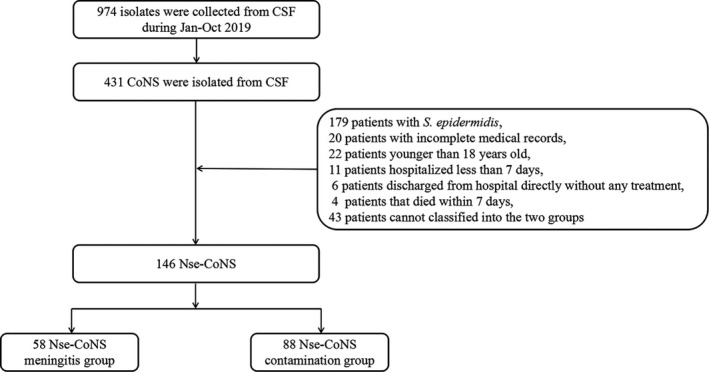
Flow chart of the enrolled patients in the two groups
TABLE 1.
Distribution of the respective microorganisms from the Beijing Tiantan Hospital
| Isolates | Meningitis | Contamination | Total |
|---|---|---|---|
| S hominis | 19 | 42 | 61 |
| S haemolyticus | 25 | 14 | 39 |
| S capitis | 6 | 20 | 26 |
| S warneri | 4 | 5 | 9 |
| S cohnii | 3 | 4 | 7 |
| S simulans | 1 | 0 | 1 |
| S lugdunensis | 0 | 1 | 1 |
| S pettenkoferi | 0 | 1 | 1 |
| S xylosus | 0 | 1 | 1 |
| Total | 58 | 88 | 146 |
FIGURE 2.
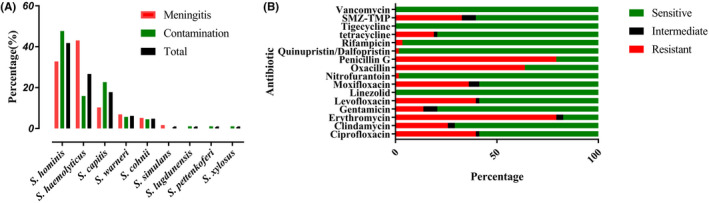
Distribution of the Nse‐CoNS(A) and antimicrobial susceptibility test results for Nse‐CoNS meningitis(B)
TABLE 2.
Clinical characteristics of the patients with Nse‐CoNS meningitis and contamination
| Variables | Meningitis (58) | Contamination (88) | P |
|---|---|---|---|
| Age (years) | 46 (34, 52) | 45 (27, 56) | .962 |
| Male(%) | 38 (65.5%) | 55 (62.5%) | .753 |
| Length of hospitalization Stay (LOS) | 20 (16, 25) | 18 (14, 27) | .563 |
| Post‐neurosurgical infection/contamination | 5 (2.5, 10.5) | 7 (5, 11) | .978 |
| time | |||
| Hypertension | 6 (10.3%) | 14 (15.9%) | .462 |
| Diabetes mellitus | 3 (5.2%) | 4 (4.5%) | .862 |
| Site of surgery | .686 | ||
| Head | 55 (94.8%) | 82 (93.2%) | |
| Spinal | 3 (5.2%) | 6 (6.8%) | |
| Surgical wound classification | .197 | ||
| Clean | 38 (65.5%) | 46 (52.3%) | |
| Clean‐contaminated | 12(20.7%) | 30 (34.1%) | |
| Contaminated | 8(13.8%) | 12 (13.6%) | |
| Extra ventricular drainage(EVD) | 25 (43.1%) | 20 (22.7%) | .009 |
| Lumbar drainage (LD) | 21 (36.2%) | 16 (18.2%) | .014 |
| Ventilation rate | 11 (19.0%) | 14 (15.9%) | .631 |
| CSF leakage | 7 (12.1%) | 11 (12.5%) | .938 |
| Craniotomy | 45 (77.6%) | 42 (47.7%) | .001 |
| ICU admission | 17 (29.3%) | 21 (23.9%) | .463 |
| Malignancy | 33 (56.9%) | 25 (28.4%) | .001 |
| Time duration of surgery | 291 (210, 360) | 244 (168, 300) | .017 |
| Time to cure infection | 9 (7, 13) | ‐ | ‐ |
3.2. Patient demographics and clinical characteristics
Patient demographics and clinical characteristics are shown in Table 2. In univariate analysis of factors in the meningitis and contamination groups, 4 were clinically significantly different (P < .05), including: EVD (P = .009), LD (P = .014), craniotomy (P < .001), and malignancy (P = .001). Twelve meningitis‐related laboratory tests in the meningitis and contamination groups of patients are shown in Table 3, of which C‐Leu [826 (184, 2263) × 106/L, 23 (6, 146) × 106/L], C‐Neu [79.1% (56.6%, 87.7%), 50.0% (21.4, 77.4)], C‐Cl [121.0 (117.2, 123.5) mmol/L, 124.00 (119.4, 127.0) mmol/L], and Lac [4.2 (2.4, 6.1) μM, 2.5 (1.8, 4.0) μM] exhibited significant differences between the Nse‐CoNS meningitis and contamination groups.
TABLE 3.
Clinical and laboratory variables of Nse‐CoNS meningitis and contamination at Beijing Tiantan Hospital in Jan‐Oct 2019
| Variables | Meningitis (58) | Contamination (88) | P |
|---|---|---|---|
| C‐Leu (106/ L) | 826 (184, 2263) | 23 (6, 146) | <.001 |
| C‐Neu(%) | 79.1 (56.6, 87.7) | 50 (21.4, 77.4) | <.001 |
| C‐cell (106/ L) | 2807 (714, 12 874) | 207.5 (46.8, 1541) | .102 |
| C‐Pro (g/L) | 1.20 (0.74, 2.12) | 0.53 (0.32, 1.00) | .836 |
| C‐Cl (mmol/L) | 121 (117.2, 123.5) | 124.0 (119.4, 127.0) | .011 |
| C‐Glu (mmol/L) | 3.17 (2.36, 4.18) | 3.47 (3.00, 4.00) | .280 |
| Glu (mmol/L) | 6.52 (5.48, 7.70) | 5.65 (5.01, 6.56) | .634 |
| Leu (109/L) | 9.84 (8.46, 12.33) | 9.34 (7.01, 11.85) | .501 |
| Neu (%) | 78.85 (71.40, 83.88) | 74.6 (67.3, 82.3) | .272 |
| CRP (mg/L) | 33.06 (12.06, 76.26) | 12.52 (8.47, 26.72) | .487 |
| Lac (μM) | 4.2 (2.4, 6.1) | 2.4 (1.8, 4.0) | <.001 |
| C/B‐Glu | 0.551 (0.418, 0.679) | 0.588 (0.472, 0.704) | 0.219 |
3.3. TTPC
A scatter plot and the proportion of patients with different TTPC intervals are shown in Figure 3. Figure 4 and Table 4 show TTPC and ROC curve parameters in the meningitis and contamination groups. The median TTPC in the meningitis group was significantly shorter than in the contamination group (20.2 ± 5.0 hours versus 30.2 ± 12.6 hours, respectively, P < .05). AUC of the ROC curve was 0.802. A cutoff value of 20.0 hours was associated with the highest diagnostic accuracy with 94.3% sensitivity and 52.5% specificity to differentiate Nse‐CoNS meningitis from contamination. In addition, the model had good NPV (90.2%) and NLR (0.108).
FIGURE 3.
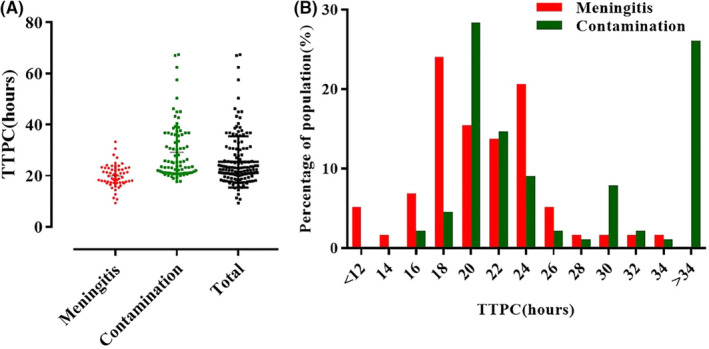
Scatter plot(A) and percentage of population(B) of the CSF TTPC of the two groups
FIGURE 4.
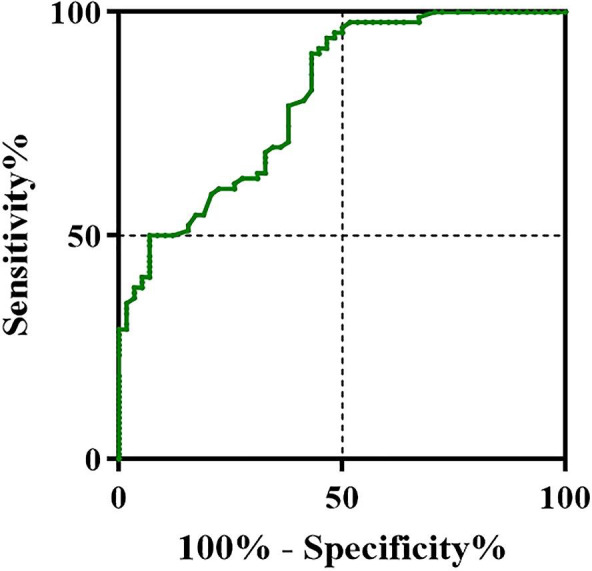
ROC curves of TTPC to differentiate post‐neurosurgical Nse‐CoNS meningitis from contamination
TABLE 4.
CSF TTPC of the patients with Nse‐CoNS meningitis and contamination groups
| Meningitis(hours) | Contamination(hours) | AUC | Cutoff (hours) | SEN | SPE | PPV | NPV | PLR | NLR |
YONDEN Index |
|---|---|---|---|---|---|---|---|---|---|---|
| 20.2 ± 5.0 | 30.2 ± 12.6 | 0.802 | 20.0 | 94.3% | 52.5% | 66.5% | 90.2% | 1.985 | 0.108 | 0.469 |
Abbreviations: SEN, sensitivity; SPE, specificity; PPV, positive predictive value; NPV, negative predictive value; PLR, positive likelihood ratio; NLR, negative likelihood ratio.
3.4. Verification
We selected all Nse‐CoNS from November 2019 to February 2020 and categorized them into meningitis and contamination groups according to the cutoff value of TTPC. There were 33 cases of Nse‐CoNS meningitis and 41 cases of Nse‐CoNS contamination enrolled in the verification cohort. Positive and negative verification was performed on the two groups of patients. Among them, the meningitis group had 26 cases, which were <20.0 hours and the consistency rate of positive verification was 78.8%. In the contamination group, 38 cases were >20 hours, the consistency rate of negative verification being 92.7% and the overall consistency rate 86.5%.
4. DISCUSSION
In the present study, we analyzed Nse‐CoNS isolated from the CSF of patients after neurosurgery, and evaluated its TTPC. It was found that TTPC could distinguish between infection and contamination with Nse‐CoNS (AUC = 0.802), and through verification, the total agreement rate was as high as 86.4%, which is excellent help in accurately diagnosing meningitis. As far as we are aware, this is the first study to evaluate the clinical relevance of Nse‐CoNS in post‐neurosurgical meningitis.
CoNS play an important role in nosocomial infections and are frequently associated with bacteremia bloodstream infections, 12 surgical site infections, catheter‐related infections 13 and it is widely present in hospitals and clinical environments. It has been reported that this type of bacteria is the most common cause of nosocomial bloodstream infections. 14 Nevertheless, most of the Nse‐CoNS detected were usually the consequence of skin contamination. Therefore, whenever Nse‐CoNS is cultured from a patient's sterile fluid sample, such as blood, CSF, hydrothorax and so on, the clinician normally classifies it as a contaminating bacterium. 15 However, this simple classification easily leads to escape diagnosis or misdiagnosis. Similarly, isolation of Nse‐CoNS from the patient's CSF also troubles neurosurgical physicians; they are often notified of a positive Nse‐CoNS CSF culture result and they must make a clinical decision on whether the positive finding is likely to be a pathogen or a contaminant. In the diagnosis of bloodstream infections caused by Nse‐CoNS, patients will provide multiple bottles of blood culture in a short period of time. If only one bottle of CoNS is isolated, it is classified into the contaminant group; if multiple bottles of CoNS are isolated at the same time, it is classified into the bacteremia group. 16 However, we cannot differentiate neurosurgical meningitis from contamination, because CSF bacterial culture always employs a single bottle for their low volume (1‐3 mL), and in addition, multiple cultures may increase the risk of infection.
Indeed, since the first study which demonstrated the value of TTPC of sterile body fluid cultures for the diagnosis, 17 and this method is now considered a useful weapon and recommended in some guidelines. 18 TTPC is an important indicator for differentiating between infection and contamination. Nevertheless, most of the published reports deal with bloodstream infections and do not exclude S epidermidis. Because S epidermidis is the most common pathogen related to CRI, with high infectious properties, differentiating S epidermidis from other CoNS has important clinical significance. In our study, post‐neurosurgical Nse‐CoNS meningitis was chosen as the target; the diagnosis of post‐neurosurgical meningitis from contamination is of clinical great importance. Our results show that 20.0 hours is the cutoff and a patient can be classified into the meningitis or contamination group with sensitivity of 94.3% and 90.2% NPV, and the PLR and NLR are 1.985 and 0.108, respectively, which reflects excellent diagnostic importance. There is one report that the TTPC of cerebral infantile meningitis caused by CoNS is 28.6 ± 16.8 hours, 19 which is slightly higher than the results of our study, and the probably reason may be that compared with infants, adult meningitis patients have a higher CSF inoculation volume, which contains a higher initial bacterial concentration, and leads to a shorter TTPC.
TTPC of post‐neurosurgical Nse‐CoNS meningitis is much longer than that of bloodstream infections, 14 which may be due to various factors: (1) Inoculation volume. CSF collected for patients is generally 1‐3 mL while for bacteremia patients it is generally 8‐10 mL per sample, and the higher the amount of specimen inoculated, the higher the positive culture rate, and the shorter the culture time; (2) Since fewer immune molecules exist in patients’ CSF, when post‐neurosurgical meningitis occurs, even a low bacterial concentration can cause severe meningitis leading to a prolonged incubation time; (3) Patients are often given antibiotic prophylaxis before surgery, which will also lead to prolonged TTPC.
Distinct studies always possess different results on the diagnostic value and efficacy of TTPC. 20 Observed variability can be explained by the different distribution among the researches of variables that may influence TTPC, such as the volume of inoculated, antimicrobial therapy ahead of the specimen collection, 21 delay until the onset of incubation, different incubation system and some clinical conditions such as bacteremia. 22 So, it may be difficult to apply directly what is a useful cutoff TTPC value in a particular study to other settings due to these confounders. We suggest that neurosurgeons should strictly follow the CSF specimen collection procedure, ensure the volume of specimens inoculated, and therefore of more general application.
In the present study, S hominis and S haemolyticus accounted for the majority of meningitis and contaminated group infections. Among them, S hominis had the highest isolation rates of the two groups, which may be due to S hominis being part of the normal human microbiome. 23 Therefore, when patients underwent neurosurgery, lumbar puncture or CSF drainage, bacterial growth may well have been inadvertently introduced due to inadequate disinfection. Consequently, when S hominis was isolated from CSF culture, it is more likely to be classified into contaminated bacteria (32.8% versus 47.7%). S haemolyticus is a skin deposit, which has gained increased attention as an opportunistic pathogen, particularly in surgical patients with a reduced immune defense. It was the second most frequently isolated CoNS from body fluid cultures after S epidermidis. 24 In addition, clinical isolates of S haemolyticus are usually multidrug resistant, and their main virulence factor is formation of biofilms, both factors leading to infections that are difficult to treat. 25 In comparison with S hominis, a higher percentage of S haemolyticus existed in the meningitis group.
Similar to other reports, 26 , 27 resistance against the usual anti‐staphylococcal β‐lactam antibiotics was high in our study, with >60% of the isolates in the meningitis group being resistant to oxacillin, with erythromycin having a lesser effect on Nse‐CoNS (17.11%). Agents active against methicillin resistance Staphylococcus, such as vancomycin, tigecycline, and linezolid should therefore be considered as the best treatment empirical choice in suspected meningitis caused by Nse‐CoNS.
Although our results are exciting, there are a number of shortcomings. First, it was a retrospective study in a single neurosurgical center, and the inoculation volume of the CSF (1‐3 mL) was a range, which cannot make every patient's vaccination volume exactly the same, and may have slightly affected the results of TTPC. All the patients enrolled in the study had experienced a neurosurgical operation. To evaluate the generalizability and the breadth of applications, future multi‐center studies with a larger cohort of patients will be required. Second, the samples in our study were analyzed with only one specific instrument, the BacT/ALERT 3D (bioMérieux, Marcy l’Etoile, France), and further studies are needed with other blood culture equipment. Third, through verification experiments, TTPC can better diagnose the contamination group, but is slightly insufficient for the meningitis group (84.6% versus 75.0%).
5. CONCLUSIONS
We evaluated the clinical predictive factors of Nse‐CoNS meningitis and confirmed that the median TTPC in the Nse‐CoNS meningitis group was significantly shorter than in the contamination group. TTPC of patients CSF culture was <20.0 hours and was associated with Nse‐CoNS meningitis, while a value >20.0 hours was associated with Nse‐CoNS contamination. The verification results revealed that the consistent rate of diagnosis of Nse‐CoNS meningitis by TTPC was 86.5%. This information will be very helpful for the rapid diagnosis of Nse‐CoNS‐induced meningitis.
CONFLICT OF INTEREST
The author declares no conflict of interest.
ETHICAL APPROVAL
No ethical approval was needed for this study.
INFORMED CONSENT
Informed consent was obtained from all individual patients included in the study.
ACKNOWLEDGMENTS
I would like to take this opportunity to thank Anqi Jia, Wenxuan Fu, Siye Liang, and Yu Wang who gave me many academic and precious suggestions and helped to check the manuscript.
Zheng G, Li S, Zhao M, et al. Time to positive culture can differentiate post‐neurosurgical coagulase‐negative Staphylococci other than S epidermidis meningitis from contamination: A case‐control observational study. J Clin Lab Anal. 2020;34:e23447 10.1002/jcla.23447
Guanghui Zheng and Siwen Li contributed equally to this study.
Funding information
This study was supported by research grant: QML20180502, Beijing Municipal Administration of Hospitals.
REFERENCES
- 1. Weinstein MP, Towns ML, Quartey SM, et al. The clinical significance of positive blood cultures in the 1990s: A prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis. 1990s;24(4):584‐602. [DOI] [PubMed] [Google Scholar]
- 2. Raza MS, Das BK, Goyal V, et al. Emerging multidrug resistance isolates of hospital‐acquired bacterial meningitis in a tertiary care centre in North India. J Med Microbiol. 2019;68(11):1585‐1590. [DOI] [PubMed] [Google Scholar]
- 3. Peixoto PB, Massinhani FH, Netto dos Santos KR, et al. Methicillin‐resistant Staphylococcus epidermidis isolates with reduced vancomycin susceptibility from bloodstream infections in a neonatal intensive care unit. J Med Microbiol. 2019;69(1):41‐45. [DOI] [PubMed] [Google Scholar]
- 4. Gatermann SG, Koschinski T, Friedrich S. Distribution and expression of macrolide resistance genes in coagulase‐negative staphylococci. Clin Microbiol Infect. 2007;13(8):777‐781. [DOI] [PubMed] [Google Scholar]
- 5. Santarpia L, Buonomo A, Pagano MC, et al. Central venous catheter related bloodstream infections in adult patients on home parenteral nutrition: Prevalence, predictive factors, therapeutic outcome. Clin Nutr. 2016;35(6):1394‐1398. [DOI] [PubMed] [Google Scholar]
- 6. Yodoshi T, Ueda S, Goldman RD. Skin preparation for prevention of peripheral blood culture contamination in children. Pediatr Int. 2019;61(7):647‐651. [DOI] [PubMed] [Google Scholar]
- 7. San‐Juan R, Martínez‐Redondo I, Fernández‐Ruiz M, et al. A short course of antibiotic treatment is safe after catheter withdrawal in catheter‐related bloodstream infections due to coagulase‐negative staphylococci. Eur J Clin Microbiol Infect Dis. 2019;38(5):977‐983. [DOI] [PubMed] [Google Scholar]
- 8. Kassis C, Rangaraj G, Jiang Y, Hachem RY, Raad I. Differentiating culture samples representing coagulase‐negative staphylococcal bacteremia from those representing contamination by use of time‐to‐positivity and quantitative blood culture methods. J Clin Microbiol. 2009;47(10):3255‐3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang W, Lu C, Huang C, et al. Epidemiology of adult staphylococcal meningitis in southern Taiwan: A clinical comparison of Staphylococcus aureus infection and coagulase‐negative staphylococcal infection. Jpn J Infect Dis. 2007;60(5):262. [PubMed] [Google Scholar]
- 10. Huang C‐R, Lu C‐H, Wu J‐J, et al. Coagulase‐negative staphylococcal meningitis in adults: clinical characteristics and therapeutic outcomes. Infection. 2005;33(2):56. [DOI] [PubMed] [Google Scholar]
- 11. Anevlavis S, Petroglou N, Tzavaras A, et al. A prospective study of the diagnostic utility of sputum Gram stain in pneumonia. J Infect. 2009;59(2):83‐89. [DOI] [PubMed] [Google Scholar]
- 12. Boland L, Streel C, De Wolf H, Rodriguez H, Verroken A. Rapid antimicrobial susceptibility testing on positive blood cultures through an innovative light scattering technology: Performances and turnaround time evaluation. BMC Infect Dis. 2019;19(1):989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng C, Cai J, Liu H, et al. Clinical characteristics and risk factors in mixed‐enterococcal bloodstream infections. Infect Drug Resist. 2019;12:3397‐3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Orihuela‐Martín J, Rodríguez‐Núñez O, Morata L, et al. Performance of differential time to positivity as a routine diagnostic test for catheter‐related bloodstream infections: a single‐centre experience. Clin Microbiol Infect. 2019;3(26):383–390. [DOI] [PubMed] [Google Scholar]
- 15. Rogers KL, Fey PD, Rupp ME. Coagulase‐negative staphylococcal infections. Infect Dis Clin North Am. 2009;23(1):73‐98. [DOI] [PubMed] [Google Scholar]
- 16. Hitzenbichler F, Simon M, Salzberger B, Hanses F. Clinical significance of coagulase‐negative staphylococci other than S. epidermidis blood stream isolates at a tertiary care hospital. Infection. 2017;45(2):179‐186. [DOI] [PubMed] [Google Scholar]
- 17. Catton JA, Dobbins BM, Kite P, et al. In situ diagnosis of intravascular catheter‐related bloodstream infection: a comparison of quantitative culture, differential time to positivity, and endoluminal brushing. Crit Care Med. 2005;33(4):787‐791. [DOI] [PubMed] [Google Scholar]
- 18. Mermel LA, Farr BM, Sherertz RJ, et al. Guidelines for the management of intravascular catheter‐related infections. Clin Infect Dis. 2001;32(9):1249‐1272. [DOI] [PubMed] [Google Scholar]
- 19. Leazer R, Erickson N, Paulson J, et al. Epidemiology of cerebrospinal fluid cultures and time to detection in term infants. Pediatrics. . 2017;139(5):e20163268. [DOI] [PubMed] [Google Scholar]
- 20. Cobos‐Trigueros N, Morata L, Torres J, et al. Usefulness of time‐to‐positivity in aerobic and anaerobic vials to predict the presence of Candida glabrata in patients with candidaemia. J Antimicrob Chemother. 2013;68(12):2839‐2841. [DOI] [PubMed] [Google Scholar]
- 21. Martinez JA, Pozo L, Almela M, et al. Microbial and clinical determinants of time‐to‐positivity in patients with bacteraemia. Clin Microbiol Infect. 2007;13(7):709‐716. [DOI] [PubMed] [Google Scholar]
- 22. Horvath LL, George BJ, Murray CK, Harrison LS, Hospenthal DR. Direct comparison of the BACTEC 9240 and BacT/ALERT 3D automated blood culture systems for candida growth detection. J Clin Microbiol. 2004;42(1):115‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pereira EM, de Mattos CS, dos Santos OC, et al. Staphylococcus hominis subspecies can be identified by SDS‐PAGE or MALDI‐TOF MS profiles. Sci Rep. 2019;9(1):11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bouchami O, Achour W, Mekni MA, Rolo J, Ben HA. Antibiotic resistance and molecular characterization of clinical isolates of methicillin‐resistant coagulase‐negative staphylococci isolated from bacteremic patients in oncohematology. Folia Microbiol (Praha). 2011;56(2):122‐130. [DOI] [PubMed] [Google Scholar]
- 25. Cavanagh JP, Pain M, Askarian F, et al. Comparative exoproteome profiling of an invasive and a commensal Staphylococcus haemolyticus isolate. J Proteomics. 2019;197:106‐114. [DOI] [PubMed] [Google Scholar]
- 26. El‐Zamkan MA, Mubarak AG, Ali AO. Prevalence and phylogenetic relationship among methicillin‐ and vancomycin‐resistant Staphylococci isolated from hospital's dairy food, food handlers, and patients. J Adv Vet Anim Res. 2019;6(4):463‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mamtora D, Saseedharan S, Bhalekar P, Katakdhond S. Microbiological profile and antibiotic susceptibility pattern of Gram‐positive isolates at a tertiary care hospital. J Lab Physicians. 2019;11(2):144‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]


