Abstract
Background
Candida species are considered as the cause of one of the most important opportunistic fungal diseases. Accurate identification of Candida species is important because of antifungal susceptibility patterns are different among these species, so proper identification helps in the selection of antifungal drugs for the prevention and treatment. Phenotypic methods for identification of Candida species, which are widely used in clinical microbiology laboratories, have some limitations. Real‐time PCR followed by the high‐resolution melting analysis (HRMA) is a novel approach for the rapid recognition of pathogenic fungi. Molecular phylogeny is essential for obtaining a better understanding of the evolution of the genus Candida and the identification of the relative degree of the Candida species. The purpose of this study was molecular identification of Candida isolates by Real‐time PCR‐high‐resolution melting analysis and investigation of the genetic diversity of Candida species.
Methods
Two hundred and thirty‐two Candida isolates including 111 Candida isolates obtained from 96 HIV/AIDS patients and 121 Candida isolates obtained from 98 non‐HIV persons were identified by real‐time PCR and high‐resolution melting curve analysis. To evaluate genetic diversity and relationships among Candida species, PCR products of nine clinical Candida isolates, as a representative of each kind of species, were randomly selected for DNA sequence analysis.
Results
In HIV/AIDS patients, six species of Candida spp. were identified as follows: C albicans (n = 64; 57.7%), C glabrata (n = 31; 27.92%), C parapsilosis (n = 9; 8.1%), C tropicalis (n = 4; 3.6%), C krusei (n = 2; 1.8%), and C kefyr (n = 1; 0.90%). In non‐HIV persons, we identified eight species of Candida including C albicans (n = 46; 38.33%) followed by C glabrata and C krusei (each one, n = 18; 15%), C tropicalis (n = 13; 10.83%), C lusitaniae (n = 12; 5.17%), C parapsilosis (n = 10; 4.31%), and C kefyr and C guillermondii (each one, n = 2; 1.66%). Also, the phylogenetic analysis showed the presence of two main clades and six separate subclades. Accordingly, about 88.9% of the isolates were located in clade I and 11.10% of the studied isolates were in clade II.
Conclusions
Real‐time PCR followed by high‐resolution melting analysis (HRMA) is known as a reliable, fast, and simple approach for detection and accurate identification of Candida species, especially in clinical samples.
Keywords: Candida spp, high resolution melting, HIV/AIDS patients, non‐HIV persons, phylogeny
Two hundred and thirty‐two Candida isolates including 111 Candida isolates from 96 HIV/AIDS patients and 121 Candida isolates from 98 non‐HIV persons were used in this study. Clinical Candida isolates and eight Candida reference strains were grown on Sabouraud dextrose agar media for 18‐24 hours at 37°C. The DNA of Candida isolates was extracted. Real‐time PCR followed by HRMA was performed on clinical Candida isolates. The mean Tm ± 4 SD and the dMelt curve shape of clinical Candida isolates were compared to the Candida reference strains panel. To evaluate genetic diversity and relationships among Candida species, PCR products of nine clinical Candida isolates, as a representative of any species, were randomly selected for DNA sequence analysis.
![]()
Abbreviations
- HIV/AIDS
Human immunodeficiency virus infection and acquired immune deficiency syndrome
- HRM
High‐resolution melting
- HRMA
High‐resolution melting analysis
- OC
Oral candidiasis
- PCR
Polymerase chain reaction
- SDA
Sabouraud dextrose agar
- SD
Standard deviation
- Tm
Melting temperature
1. INTRODUCTION
The prevalence of fungal infections has been increased, especially in immunocompromised hosts such as consumers of corticosteroids and antibiotics, the patients with diabetes, malnutrition, severe malnutrition, alcoholics, HIV/ AIDS, cancer, and certain genetic disorders. 1 Candidiasis is considered as the most important opportunistic fungal diseases worldwide and remains a clinical problem, predominantly among the immunocompromised patients. 2 , 3
Oral candidiasis (OC) is the commonest human fungal infection presented in the oral cavity among the HIV/ AIDS patients. 4 The different manifestations of OC in the immunocompromised patients especially among the HIV/ADIS patients involved oral thrush (pseudomembranous candidiasis), denture stomatitis, median rhomboid glossitis, hyperplastic candidiasis, erythematous candidiasis, linear gingival erythema, perleche or angular cheilitis, salivary gland swellings, sore formation in the oral cavity, and oral hairy leukoplakia. 5 Many people with OC can stay without any specific clinical symptoms for a long time. However, several symptoms in these people may include burning sensation and pain in the mouth, changes in taste sensation, and difficulty in swallowing liquids or solids, and or white creamy or creamy plaques in different parts of the oral cavity. 6 , 7
C albicans is the major species in the development of oral candidiasis. Other Candida species including C glabrata, C kefyr, C tropicalis, C parapsilosis, C krusei, and C guilliermondii are found in different parts of the oral cavity. 6 The frequency of Candida species in the oral cavity was 40%‐60% in healthy subjects, 8 while it was between 62% and 93% in the HIV/ AIDS patients. 9
Accurate identification of Candida species is important, because antifungal susceptibility patterns are vary among these species, and also a proper identification helps in the selection of antifungal drugs for the prevention and treatment. 10 The methods used to identify Candida species are phenotypic and genotypic assays. Phenotypic assays for identification of Candida species involve carbohydrate assimilation and fermentation test, investigation of the morphological characteristics of yeasts on malt extract agar (MEA) and corn meal agar (CMA), production of germ tube and chlamydoconidia, used as a commercial chromogenic media such as CHROMAgar™ Candida, growth at different temperatures, cultured in Tween 80 agar, and serological methods. 11 These methods, which are widely used in clinical microbiology laboratories, have some limitations such as insensitive, expensive, time‐consuming, and inability to identify important species, such as C albicans and C dubliniensis. Therefore, finding the reliable and fast approaches for their identification is needed, and proper identification helps in the selection of effective antifungal drugs. 12 , 13
Molecular techniques are known as suitable approaches for identification of Candida species. These methods are expensive; however, their accuracy and speed are undeniable. Molecular techniques such as polymerase chain reaction (PCR), amplified fragment length polymorphism (AFLP), PCR‐restriction length fragment polymorphisms (PCR‐RFLP), random amplification of polymorphic DNA (RAPD), multiplex PCR, nested polymerase chain reaction (Nested PCR), DNA sequencing, matrix‐associated laser desorption/ionization time of flight mass spectrometry (MALDI‐TOF MS), DNA fingerprinting, single‐strand conformational polymorphism (SSCP) analysis, and real‐time polymerase chain reaction (real‐time PCR) are suitable for diagnosing and identifying Candida species. 10 , 14 , 15
High‐resolution melt analysis (HRMA) is a powerful molecular method to detect mutations, polymorphisms, epigenetic information, and also to identify species of different organisms by comparing relative positions and shapes of melting curves. 16 Real‐time PCR followed by HRMA is a basic, fast, accurate, and closed‐tube technique with the susceptibility for the single nucleotide. 17 Phylogenetic analysis is essential for obtaining a better understanding of the evolution of the genus Candida and detection of the relative degree of Candida species as well as infer evolutionary pathways of some of gene families. 18 Therefore, the aim of this study was molecular identification of Candida isolates by Real‐time PCR high‐resolution melting analysis and investigation of the genetic diversity of Candida species.
2. MATERIAL AND METHODS
2.1. Study design, population, and clinical specimens
The present cross‐sectional study was performed in the Department of Medical Parasitology and Mycology, Kerman University of Medical Sciences, Kerman (southeast of Iran), from November 2018 to September 2019. Two hundred and thirty‐two Candida isolates including 111 Candida isolates from 96 HIV/AIDS patients and 121 Candida isolates from 98 non‐HIV persons were used in this study. The oral specimens were taken from the HIV/AIDS patients who were referred to the Kerman Counseling Resource Center for behavioral disorders for taking periodic checkups and/or drugs or solving their health difficulties. As well, 121 Candida isolates from 98 non‐HIV persons were collected from the different parts of the oral cavity of non‐HIV subjects, who were referred to the Medical Mycology Laboratory of Afzalipoor Faculty of Medicine in Kerman. All the participants of this study completed the questionnaire and filled an informed consent form. Samples from different parts of the oral cavity such as the oral mucosa and tongue were collected with a sterile swab. The reason for the differences in the number of the individuals in these two groups was the lack of cooperation of some of the individuals in sampling. The obtained oral swabs were cultured on to chromo Agar Candida (HiMedia, Mumbai, India) and then incubated for 48 hours at 35°C to create specific colony colors.
2.2. Candida reference strains
Eight species of Candida including C albicans, C glabrata, C krusei, C tropicalis, C lusitaniae, C parapsilosis, C kefyr, and C guillermondii were used in this study for optimizing the ITS2‐melting curve analysis. These Candida species were characterized previously in the research mycological laboratory of our department using the standard methods such as the specific color the colony created on CHROMagar Candida media and PCR‐RFLP with Msp I enzyme. 5 , 10 Bln I restriction enzyme was used for discrimination of between C albicans and C dubliniensis.
2.3. DNA isolation
Eight Candida reference strains and clinical Candida isolates were grown on Sabouraud dextrose agar media for 18‐24 hours at 37°C. The DNA of Candida isolates was extracted by the use of Favor Prep™ Blood/Cultured Cell Genomic DNA Extraction Mini Kit (Favorgen Biotech Co, Taiwan) in terms of the manufacturer's protocol. 19 DNA concentrations and A 260/A 280 ratios of the extracted DNA were calculated by a ND‐1000 spectrophotometer (NanoDrop, Fisher Thermo). An A 260/A 280 ratio of 1.8 to 2.1 was selected for subsequent experiments, and all the extracted DNAs were stored at −20°C until use.
2.4. Real‐time PCR assay and HRMA analysis
Real‐time PCR followed by HRMA was performed on eight Candida reference strains. The primer's sequences used for this study were ITS86 as forward primer [5′‐ GTGCATCATCGAATCTTTGAAC‐3′] and ITS4 as a reversed primer [5′‐TCCTCCGCTTATTAGGAC‐3′], which were previously described by Gutzmer et al. 20 For gene amplification by HRM, volume of 20 µL comprising 4 µL 5× HOT FIREPol® EvaGreen® HRM Mix (Solis BioDyne Co), 4 µL of the extracted DNA from each Candida isolates, 1 µL of forward primer (10 pmol/L) (Macrogen Co), 1 µL of reverse primer (10 pmol/L) (Macrogen Co), and 10 µL of distilled water was prepared. HRM was done using Rotor‐Gene 6000 (Corbett Research) using the following program: an initial denaturation step at 95°C for 12 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 2 seconds, and 72°C for 20 seconds. The amplification products for HRMA were cooled 1 minutes at 50°C and then heated from 80°C to 92°C monitoring fluorescence at the rate of 0.1°C/2 s. Melting data were normalized, and the temperature was shifted using Rotor‐Gene 6000. Normalization regions for HRMA were set at 80.5°C to 90.9‐91.3°C. The obtained results were analyzed using HRM Software Rotor‐Gene series software ver. 2.3.1 (Corbett Research). All the experiments were done in duplicate for eight Candida reference strains and 232 clinical Candida isolates. All the reference Candida strains were used in each run. The mean Tm ± 4 SD and the dMelt curve shape of clinical Candida isolates were compared to the Candida reference strains panel.
2.5. PCR Assay
To perform the accurate identification of the species using the results of HRM with the confidence level lower than 50% and for some species with closed melting Tm, for example, C albicans (87.385°C), C lusitaniae (87.84°C), and C krusei (87.04°C), PCR assay with ITS 86 and ITS4 primers, were used.
2.6. Phylogenetic analysis
To evaluate genetic diversity and relationships among Candida species, PCR products of nine clinical Candida isolates, as a representative of any species, were randomly selected for DNA sequence analysis. The PCR products were submitted to Fazapajo Company. The obtained nucleotide sequences were aligned with the sequences registered in NCBI GenBank. All nucleotide sequences were trimmed by Sequence Scanner version 1.0 (Applied Biosystems). Afterward, the similarity percentage was determined by the online BLAST tool and Sequence Alignment software; BioEdit ver. 7.2.5. The phylogenetic analysis was performed following the investigation of the best nucleotide substitution's model using MEGA 7 software. The final phylogenetic tree was drawn in the same software with 1000 replicates using the Bootstrap topologies reliability test.
2.7. Statistical analysis
After data collection, Graph Pad Prism version 8 (Graph Pad Software In, San Diego, USA) was used for descriptive statistical analysis.
3. RESULTS
3.1. Frequency of clinical Candida isolates according to CHROMagar Candida medium
The 232 Candida isolates were directly incubated on CHROMagar Candida medium. The number of Candida isolates observed based on colony color on CHROMagar Candida after 48 hours of incubation including 110 green colonies (47.4%), 46 pink colonies (19.32%), 38 purple colonies (16.37%), 17 dark blue and/or blue gray colonies (7.32%), 11 pinkish‐purple colonies (4.74%), and 10 white colonies (4.31%), respectively.
3.2. HRMA in Candida reference strains
Melting temperature (Tm) ± standard deviation (SD) of Candida reference strains are presented in Table 1. The results show reproducible melting peaks for each species. Most of the Candida reference strains presented single peak, whereas three Candida species including C parapsilosis, C tropicalis, and C guilliermondii melted in two domains (see Table 1). Figure 1 shows the melting curve analysis of Candida reference strains that was used in this study.
TABLE 1.
Melting temperature (Tm) ± standard deviation (SD) of Candida reference strains
| Candida reference strains | Melting peak 1 | Melting peak 2 |
|---|---|---|
| Mean Tm (4 SD)°C | Mean Tm (4 SD)°C | |
| C glabrata | 85.915 (0.348712) | |
| C parapsilosis | 83.985 (0.279544) | 85.625 (0.315008) |
| C albicans | 87.385 (0.440023) | |
| C guilliermondii | 83.34 (0.085323) | 84.725 (0.129692) |
| C lusitaniae | 87.84 (0.106536) | |
| C kefyr | 86.975 (0.188627) | |
| C krusei | 87.04 (0.129538) | |
| C tropicalis | 85.79 (0.141315) | 87.059 (0.156141) |
FIGURE 1.
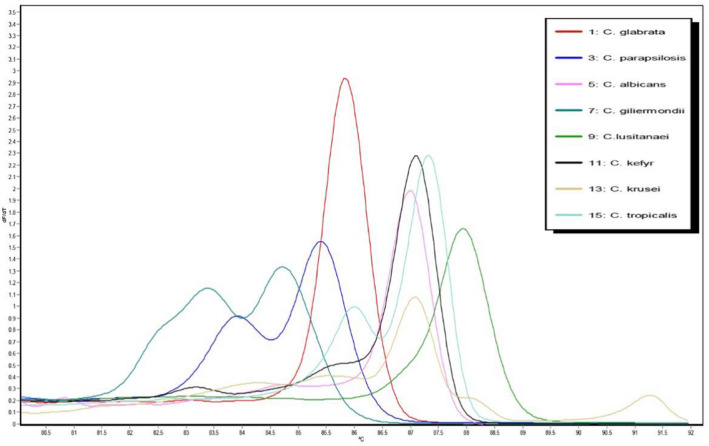
Melting temperature curve of amplicon of Candida reference strains used in this study
3.3. Identification of clinical Candida isolates by high‐resolution melting analysis
In patients with HIV/AIDS, six species of Candida spp. were identified as follows: C albicans (n = 64; 57.7%), C glabrata (n = 31; 27.92%), C parapsilosis (n = 9; 8.1%), C tropicalis (n = 4; 3.6%), C krusei (n = 2; 1.8%), and C kefyr (n = 1; 0.90%). In non‐HIV persons, we identified eight species of Candida including C albicans (n = 46; 38.33%) followed by C glabrata and C krusei (each one, n = 18; 15%), C tropicalis (n = 13; 10.83%), C lusitaniae (n = 12; 5.17%), C parapsilosis (n = 10; 4.31%), and C kefyr and C guillermondii (each one, n = 2; 1.66%). The distribution of Candida clinical isolates in HIV/AIDS patients and non‐HIV persons by HRMA in this study is presented in Table 2. The difference in the number of clinical Candida isolates in these two studied groups was related to the separation of several isolates from one individual in some cases.
TABLE 2.
Distribution of Candida clinical isolates in HIV/AIDS patients and non‐HIV persons by HRMA in this study
| Candida species | HIV/AIDS patients, No. (%) | Non‐HIV persons No. (%) | Total No. (%) |
|---|---|---|---|
| C parapsilosis | 9 (3.87) | 10 (4.31) | 19 (8.18) |
| C glabrata | 31 (13.36) | 18 (7.75) | 49 (21.12) |
| C kefyr | 1 (0.43) | 2 (0.86) | 3 (1.29) |
| C albicans | 64 (27.58) | 46 (19.82) | 110 (47.41) |
| C tropicalis | 4 (1.72) | 13 (5.60) | 17 (7.32) |
| C lusitaniae | 0 (0) | 12 (5.17) | 12 (5.17) |
| C guilliermondii | 0 (0) | 2 (0.86) | 2 (0.86) |
| C krusei | 2 (0.86) | 18 (7.75) | 20 (7.75) |
| Total | 111 (47.84) | 121 (52.15) | 232 (100) |
3.4. Phylogenetic analysis
Figure 2 presents the phylogenetic tree based on DNA sequenced isolate analysis. Phylogenetic analysis showed the presence of two main clades and six separate subclades. Accordingly, about 88.9% of the isolates were located in clade I and 11. 10% of the studied isolates were in clade II. Subclades were numbered from I to VI, with subclade I consisting of six subclades and isolates of H40, H60, H6, H65, H40, H599, and S7. Also, isolates H41, H60, and H63 isolated from the HIV/ AIDS patients were C kefyr, C glabrata, and C tropicalis, respectively. Isolates H6 and H65 obtained from the HIV/AIDS patients were C albicans. H59 and H40 isolates collected from the HIV/AIDS patients were C parapsilosis. Isolate S7 that was obtained from non‐HIV individuals was C guilliermondii. The second clade consisting isolate S9 was isolated from non‐HIV persons and was C lusitaniae. Origins, sources, and accession numbers of DNA sequenced Candida isolates registered in NCBI are presented in Table 3.
FIGURE 2.
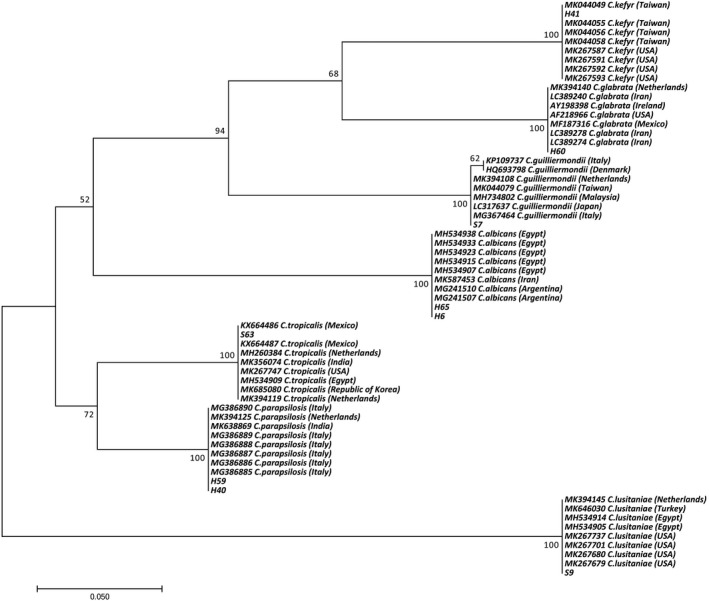
Phylogenetic tree based on sequencing results. The evolutionary history was conducted in MEGA7 by using the maximum likelihood method based on the Tamura 3‐parameter model. All positions containing gaps and missing data were eliminated
TABLE 3.
Origins, sources, and accession numbers of DNA sequenced Candida isolates registered in NCBI
| Species name | Origin | Source | GenBank sequence accession numbers |
|---|---|---|---|
| C parapsilosis (H59 isolate) | Kerman province, south east of Iran | HIV/AIDS patient | MT377825 |
| C parapsilosis (H40 isolate) | Kerman province, south east of Iran | HIV/AIDS patient | MT377826 |
| C albicans (H65 isolate) | Kerman province, south east of Iran | HIV/AIDS patient | MT377827 |
| C albicans (H6 isolate) | Kerman province, south east of Iran | HIV/AIDS patient | MT377828 |
| C glabrata (H60 isolate) | Kerman province, south east of Iran | HIV/AIDS patient | MT377829 |
| C kefyr (H41 isolate) | Kerman province, south east of Iran | HIV/AIDS patient | MT377830 |
| C tropicalis (S63 isolate) | Kerman province, south east of Iran | Non‐HIV person | MT377831 |
| C lusitaniae (S9 isolate) | Kerman province, south east of Iran | Non‐HIV person | MT377832 |
| C guilliermondii (S7 isolate) | Kerman province, south east of Iran | Non‐HIV person | MT377824 |
4. DISCUSSION
The purpose of this study was molecular identification of clinical Candida isolates by real‐time PCR‐HRMA and investigation of the genetic diversity of these species. High‐resolution melt analysis was able to identify among various clinical Candida species. 21 Real‐time PCR‐HRM could be considered as a quick, accurate, and inexpensive method for precise identification of yeast species such as Candida spp in clinical specimens. 13 , 22 Phenotypic assays, used for differentiation of Candida species, have also some limitations such as less accuracy, time‐consuming, and inability to identify some species. 10 Superiorities of HRMA over conventional methods for Candida identification were demonstrated in Nemcova et al's study. 16 Although some methods such as API‐ID32C, CANDIDA test 21, and pyrosequencing are not able to distinguish some Candida species, while these Candida species could be easily identified using HRMA. 16 In regard with that, due to its simplicity and rapidity, this method can be applied to predict the response of the treatment. 23
In the present study, two hundred and thirty two Candida isolates including 111 Candida isolates from the HIV/AIDS patients and 121 Candida isolates obtained from non‐HIV individuals were identified by real‐time PCR and high‐resolution melting curve analysis. Here, by considering the mean Tm ± 4 SD and the dMelt curve shape of eight Candida reference strains, six and eight different species in the HIV/AIDS patients and non‐HIV persons were distinguished, respectively. Nemcova et al 16 differentiated 23 out of 27 Candida species based on real‐time PCR amplification and HRMA of the ITS2 region with UNF1 and UNF2 primers. Mandviwala et al 24 used a relatively demanding analysis in MS Excel to identify 8 Candida species based on ITS1 and ITS2 sequences.
Arancia et al 21 distinguished five Candida species by HRMA analysis MP65 gene. In Somogyvari et al's study, 25 10 Candida spp were differentiated by analyzing the HRMA of the ITS2 sequence. In a study concluded by Asadzadeh et al, 26 real‐time PCR method based on melting point analysis with SYBR Green dye was introduced as a fast and reliable molecular method to perform accurate diagnosis and differentiation of C albicans and C dubliniensis. Alnuaimi et al 27 identified nine Candida spp using HRMA based on ITS1, 5.8S, and ITS2 sequences, in which they used eight Candida species as positive controls in each run. Decat et al 28 reported that the ITS2 sequence using five positive controls is a useful approach for discrimination of 17 Candida species.
In the present study, prevalent species included C albicans followed by C glabrata in the HIV/AIDS patients and C albicans followed by C glabrata and C krusei in non‐HIV persons. Similar to our results, C albicans was recognized as the most frequent species. 22 , 29 , 30 In contrast with the present study, C tropicalis was the second most common species. Similarly, the second most species was C glabrata. 13 In a study by Ziauddin Khan et al, the second most species were C parapsilosis, C tropicalis, and C glabrata. 30
According to the nucleotide changes in the ITS region and its importance in the molecular epidemiology of the disease, it seems that the phylogenetic study is essential. Also, these nucleotide variations would be more critical in the interpretation of PCR‐based molecular experimental results. Therefore, we used HRM analysis of partial amplification ITS region as well as specific primers along with comparing the nucleotide sequence with other reported records of Candida species.
The results of the phylogenetic analysis show that there are two main clades and six separate subclades. Accordingly, about 88.9% of the isolates were located in clade I and 11.10% of the studied isolates were in clade II. Subclades were numbered from I to VI, with subclade I consisting of six subclades and H40, H60, H6, H65, H40, H599, and S7 isolates. The second clade consisting isolate S9 was isolated from healthy individuals, which was C lusitaniae.
Some limitations of our study were the presence of HRM results with the confidence level lower than 50% and the closed melting Tm for some of species, for example, C albicans (87.385°C), C lusitaniae (87.84°C), and C krusei (87.04°C). To overcome these limitations, PCR assay with ITS 86 and ITS4 primers was performed for performing an accurate identification of these species. In this study, 42 of 232 Candida isolates were identified by PCR technique. Therefore, the error rate of HRM in this study was 18.10%. Moreover, other limitations in this study were the lack of cooperation by some of the HIV/AIDS patients for sampling, and the low number of samples selected for phylogenetic analysis due to budget constraints and rising dollar price in Iran.
5. CONCLUSION
Real‐time PCR followed by HRMA is a reliable, fast, and simple approach for performing an accurate identification of Candida species, especially in clinical samples. Accurate identification of Candida species is important due to antifungal susceptibility patterns that vary among these species, and a proper identification helps in the selection of antifungal drugs for prevention and treatment.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHORS’ CONTRIBUTIONS
PGHA and SS developed the study concept and design. EEN and MAM collected the data. SS analyzed and interpreted the data. SS wrote the article. PGHA and SS revised and edited the article. All authors read and approved the final article.
ETHICAL APPROVAL
The study was evaluated and approved by the Ethics Committee of the Kerman Medical University and Kerman Research Council (IR.KMU.REC.1396.2500).
ACKNOWLEDGMENT
This research was supported by a grant from the Deputy of Research and Technology of Kerman University of Medical Sciences (Grant No: 96000893).
Eghtedar Nejad E, Ghasemi Nejad Almani P, Mohammadi MA, Salari S. Molecular identification of Candida isolates by Real‐time PCR‐high‐resolution melting analysis and investigation of the genetic diversity of Candida species. J Clin Lab Anal. 2020;34:e23444 10.1002/jcla.23444
Contributor Information
Pooya Ghasemi Nejad Almani, Email: p_almani@kmu.ac.ir.
Samira Salari, Email: sa_salari@kmu.ac.ir.
REFERENCES
- 1. Zahabi ZF, Sharififar F, Almani PGN, Salari S. Antifungal activity of different fractions of Salvia rhytidea Benth as a valuable medicinal plant against various species of Candida in Kerman Province, southeast Iran. Gene Rep. 2020;1‐7. [Google Scholar]
- 2. Barati M, Mirkalantari S, Ansari S, Salari S, Fattahi A. Determination of antimicotic susceptibility pattern of Candida species isolated from patients with symptomatic candiduria. Res J Med Sci. 2019;24:1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salari S, Khosravi A, Mousavi S, Nikbakht‐Brojeni G. Mechanisms of resistance to fluconazole in Candida albicans clinical isolates from Iranian HIV‐infected patients with oropharyngeal candidiasis. J Mycol Med. 2016;26(1):35‐41. [DOI] [PubMed] [Google Scholar]
- 4. Li Y‐Y, Chen W‐Y, Li X, et al. Asymptomatic oral yeast carriage and antifungal susceptibility profile of HIV‐infected patients in Kunming, Yunnan Province of China. BMC Infect Dis. 2013;13(1):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pour AH, Salari S. Ghasemi Nejad Almani P. Oropharyngeal candidiasis in HIV/AIDS patients and non‐HIV subjects in the Southeast of Iran. Curr Med Mycol. 2018;4(4):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salari S, Ghasemi Nejad Almani P. Antifungal effects of Lactobacillus acidophilus and Lactobacillus plantarum against different oral Candida species isolated from HIV/ AIDS patients: an in vitro study. J Oral Microbiol. 2020;12(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh A, Verma R, Murari A, Agrawal A. Oral candidiasis: an overview. J Oral Maxillofac Pathol. 2014;18(Suppl 1):S81‐S85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Samaranayake L. Commensal oral Candida in Asian cohorts. Int J Oral Sci. 2009;1(1):2‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lourenço AG, Figueiredo LTM. Oral lesions in HIV infected individuals from Ribeirão Preto, Brazil. Med Oral Patol Oral Cir Bucal. 2008;1:281‐286. [PubMed] [Google Scholar]
- 10. Bakhshi T, Salari S, Naseri A, Esfandiarpour I, Mohammadi M, Almani P. Molecular identification of Candida species in patients with candidiasis in Birjand, Iran, using polymerase Chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) assay. J Isfahan Med Sch. 2016;33(359):1986‐1993. [Google Scholar]
- 11. Deorukhkar SC, Shahriar R. Identification of Candida Species: Conventional Methods in the Era of Molecular Diagnosis. Ann Microbiol Immunol. 2018;1(1):1‐6. [Google Scholar]
- 12. Thanyasrisung P, Kesakomol P, Pipattanagovit P, Youngnak‐Piboonratanakit P, Pitiphat W, Matangkasombut O. Oral Candida carriage and immune status in Thai human immunodeficiency virus‐infected individuals. J Med Microbiol. 2014;63(5):753‐759. [DOI] [PubMed] [Google Scholar]
- 13. Duyvejonck H, Cools P, Decruyenaere J, et al. Validation of high resolution melting analysis (HRM) of the amplified ITS2 region for the detection and identification of yeasts from clinical samples: comparison with culture and MALDI‐TOF based identification. PLoS One. 2015;10(8):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aslani N, Janbabaei G, Abastabar M, et al. Identification of uncommon oral yeasts from cancer patients by MALDI‐TOF mass spectrometry. BMC Infect Dis. 2018;18(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yazdanparast SA, Khodavaisy S, Fakhim H, et al. Molecular characterization of highly susceptible Candidaafricana from vulvovaginal candidiasis. Mycopathologia. 2015;180(5–6):317‐323. [DOI] [PubMed] [Google Scholar]
- 16. Nemcova E, Cernochova M, Ruzicka F, Malisova B, Freiberger T, Nemec P. Rapid identification of medically important Candida isolates using high resolution melting analysis. PLoS One. 2015;10(2):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High‐resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49(6):853‐860. [DOI] [PubMed] [Google Scholar]
- 18. Diezmann S, Cox CJ, Schönian G, Vilgalys RJ, Mitchell TG. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J Clin Microbiol. 2004;42(12):5624‐5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheon H, Yang H‐J, Choi M, Son J‐H. Effective demethylation of melanoma cells using terahertz radiation. Biomed Opt Express. 2019;10(10):4931‐4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gutzmer R, Mommert S, Küttler U, Werfel T, Kapp A. Rapid identification and differentiation of fungal DNA in dermatological specimens by LightCycler PCR. J Med Microbiol. 2004;53(12):1207‐1214. [DOI] [PubMed] [Google Scholar]
- 21. Arancia S, Sandini S, De Bernardis F, Fortini D. Rapid, simple, and low‐cost identification of Candida species using high‐resolution melting analysis. Diagn Microbiol Infect Dis. 2011;69(3):283‐285. [DOI] [PubMed] [Google Scholar]
- 22. Ninghui G, Bing W, Wei R, et al. Application of PCR and high‐resolution melting for rapid identification of yeasts routinely isolated in a clinical microbiology laboratory. Ann Clin Lab Sc. 2015;45(6):680‐685. [PubMed] [Google Scholar]
- 23. Reed GH, Kent JO, Wittwer CT. High‐resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics J. 2007;8(6):597‐608. [DOI] [PubMed] [Google Scholar]
- 24. Mandviwala T, Shinde R, Kalra A, Sobel JD, Akins RA. High‐throughput identification and quantification of Candida species using high resolution derivative melt analysis of panfungal amplicons. J Mol Diagn. 2010;12(1):91‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Somogyvari F, Horvath A, Serly J, Majoros H, Vagvolgyi C, Peto Z. Detection of invasive fungal pathogens by real‐time PCR and high‐resolution melting analysis. In Vivo. 2012;26(6):979‐983. [PubMed] [Google Scholar]
- 26. Asadzadeh M, Ahmad S, Al‐Sweih N, Khan Z. Rapid and accurate identification of Candida albicans and Candida dubliniensis by real‐time PCR and melting curve analysis. Med Princ Pract. 2018;27(6):543‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alnuaimi A, Wiesenfeld D, O'Brien‐Simpson N, Reynolds E, Peng B, McCullough M. The development and validation of a rapid genetic method for species identification and genotyping of medically important fungal pathogens using high‐resolution melting curve analysis. Mol Oral Microbiol. 2014;29(3):117‐130. [DOI] [PubMed] [Google Scholar]
- 28. Decat E, Van Mechelen E, Saerens B, et al. Rapid and accurate identification of isolates of Candida species by melting peak and melting curve analysis of the internally transcribed spacer region 2 fragment (ITS2‐MCA). Res Microbiol. 2013;164(2):110‐117. [DOI] [PubMed] [Google Scholar]
- 29. Mushi MF, Mtemisika CI, Bader O, et al. High oral carriage of non‐albicans Candida spp. among HIV‐infected individuals. Int J Infect Dis. 2016;49:185‐188. [DOI] [PubMed] [Google Scholar]
- 30. Khan Z, Mustafa AS, Alam FF. Real‐time LightCycler polymerase chain reaction and melting temperature analysis for identification of clinically important Candida spp. J Microbiol Immunol Infect. 2009;42(4):290‐295. [PubMed] [Google Scholar]


