Abstract
Somatic embryogenesis is the regeneration of embryos from the somatic cell via dedifferentiation and redifferentiation without the occurrence of fertilization. A complex network of genes regulates the somatic embryogenesis process. Especially, microRNAs (miRNAs) have emerged as key regulators by affecting phytohormone biosynthesis, transport and signal transduction pathways. miRNAs are small, non-coding small RNA regulatory molecules involved in various developmental processes including somatic embryogenesis. Several types of miRNAs such as miR156, miR157, miR 159, miR 160, miR165, miR166, miR167, miR390, miR393 and miR396 have been reported to intricate in regulating somatic embryogenesis via targeting the phytohormone signaling pathways. Here we review current research progress on the miRNA-mediated regulation involved in somatic embryogenesis via regulating auxin, ethylene, abscisic acid and cytokinin signaling pathways. Further, we also discussed the possible role of other phytohormone signaling pathways such as gibberellins, jasmonates, nitric oxide, polyamines and brassinosteroids. Finally, we conclude by discussing the expression of miRNAs and their targets involved in somatic embryogenesis and possible regulatory mechanisms cross talk with phytohormones during somatic embryogenesis.
Keywords: Somatic embryogenesis, miRNA, Target, Phytohormones
Introduction
Plant embryogenesis is the regeneration of embryo, either by zygotic embryogenesis (zygote to embryo), or by somatic embryogenesis (somatic cell to embryo). The zygotic embryogenesis occurs through fertilization of gametes; whereas somatic embryogenesis can be induced in vitro from the somatic cells of plants (Harada et al. 2010; Méndez-Hernández et al. 2019). However, the developmental and regulatory mechanisms are quite similar in both somatic and zygotic embryogenesis processes (Guan et al. 2016). Somatic embryogenesis can be induced in two different ways, such as indirect (via an intermediate callus stage) and direct (without intermediate callus stage) embryogenesis. Somatic embryogenesis is a widely used biotechnological tool for propagation, transformation and germplasm conservation (Loyola-Vargas and Ochoa-Alejo 2016). In addition, somatic embryogenesis (SE) is also an ideal model to conduct the research at physiological, cellular, and molecular levels (Nic-Can et al. 2015). However, few bottlenecks including poor responsiveness and somatic mutations during the SE process hampered the propagation of many plant species. (Awada et al. 2020). Though some woody plants are recalcitrant to SE, still it’s a promising technology for large scale propagation of important woody plants (Awada et al. 2020; Guan et al. 2016; Isah 2016).
Somatic embryogenesis is an apparent evidence of totipotency and induced by exogenous stimuli including stress (physical injury, low or high temperature, heavy metals, osmotic shock, drought or cold) (Ikeda-Iwai et al. 2003; Loyola-Vargas and Ochoa-Alejo 2016) or plant growth regulators (Wójcik et al. 2020). Auxin is the principle phytohormone for the induction of SE in plants (Wójcik et al. 2020). Other plant growth regulators including cytokinin, abscisic acid (ABA), gibberellin (GA), ethylene, jasmonates, polyamines and brassinosteroids also showed their efficacy in inducing SE in various plant species (Jiménez 2005; Yang and Zhang 2010). Somatic embryogenesis is regulated by the complex network of genes (Gulzar et al. 2020; Méndez-Hernández et al. 2019). The signal transduction genes such as somatic embryogenesis receptor-like kinase (SERK), calmodulin-like domain protein kinases (CPK) are essential for the induction of somatic embryogenesis (Davletova et al. 2001; Schmidt et al. 1997). In addition, numerous studies demonstrated the role of transcription factors encoding genes including LEAFY COTYLEDON (LEC1, LEC2, FUSCA3), ABAINSENSITIVE 3 (ABI3), AGAMOUS-LIKE 15 (AGL15), BABY BOOM (BBM), WUSCHEL (WUS) in SE process of different species (Boutilier et al. 2002; Horstman et al. 2017; Lotan et al. 1998; Stone et al. 2008; Su et al. 2009; Zheng and Perry 2013). Moreover, cell cycle regulation and cell wall development genes such as CEM6, SEPR, AGPs also play an important roles during the developmental stages of somatic embryogenesis (Joshi and Kumar 2013). Recent studies indicated that various microRNAs (miRNA) also regulate transcription factors and phytohormone metabolism during plant somatic embryogenesis at the post-transcriptional level (Siddiqui et al. 2019).
miRNAs are a class of endogenous non-coding small RNA molecules of 20–24 nt length and known to regulate gene expression at post-transcriptional levels through mRNA cleavage and/or translational repression of corresponding target genes (Wang et al. 2019). Since the first discovery of plant miRNAs in Arabidopsis (Reinhart et al. 2002), about 20,388 miRNA loci (MIRs) belonging to 5757 families, 1365 clusters, 1668 syntenic blocks and 141,327 predicted miRNA-target pairs in 88 species ranging from chlorophytes to angiosperms were recorded in PmiREN database (Guo et al. 2019). miRNAs play important roles in various biological processes including growth and development, metabolism, abiotic and biotic stress responses in plants (Guo et al. 2019; Wang et al. 2019). Moreover, their prominent role in embryogenesis was revealed by mutating miRNA biogenesis associated gene i.e. Dicer-Like 1 (DCL1). The DCL1 mutants of Arabidopsis plants showed impaired embryogenesis and development (Park et al. 2002). Further studies also revealed miRNAs roles in regulating the meristem development, cell proliferation and embryogenesis in plants (Long et al. 2018; Szczygiel-Sommer and Gaj 2019). In this review, we summarized the regulatory mechanisms of miRNAs during somatic embryogenesis of plants via targeting the phytohormone signaling pathways. Finally, we propose the future prospectives for dissecting functional roles of miRNAs in somatic embryogenesis.
miRNAs and target genes intricate in plant somatic embryogenesis
miRNAs were discovered in an invertebrate animal model Caenorhabditis elegans (Lee et al. 1993). However, 16 plant miRNAs and their functional role in developmental processes were first discovered in Arabidopsis (Reinhart et al. 2002). However, first Luo et al. (2006) reported the miRNAs that are associated with embryogenesis and post-embryonic development in embryogenic callus of rice. With the advent of miRNA identification methods, various miRNAs associated with SE have been identified in various plants including rice, maize, lily, coconut, Arabidopsis, cotton, citrus, longan, hybrid yellow popla, Japanese larch, and picea (Table 1).
Table 1.
List of miRNAs identified so far in various plants during somatic embryogenesis
| Different plant groups | Species | Developmental stage | Methods | References | |
|---|---|---|---|---|---|
| Angiosperms | Monocots | Rice (Oryza sativa) | undifferentiated calli, differentiated calli | Northern blot, NGS, qRT-PCR, | Luo et al. (2006) |
| Maize (Zea mays) | EC | NGS, Northern blot, NGS | Chavez-Hernandez et al. (2015) | ||
| Lily (Lilium pumilum) | NEC, EC, GE, TE, CE | NGS, qRT-PCR | Zhang et al. (2017), Yang et al. (2020) | ||
| Coconut (Cocos nucifera) | NEC, EC | Microarray, NGS | Sabana et al. (2020, 2018) | ||
| Dicots | Arabidopsis thaliana | EC, SE | NGS,qRT-PCR, Transformation | Szczygiel-Sommer and Gaj (2019), Szyrajew et al. (2017), Wójcik et al. (2017) | |
| Cotton (Gossypium hirsutum) | EC | NGS, qRT-PCR | Yang et al. (2013), Arora et al. (2020) | ||
| Orange (Citrus sinensis) | NEC, EC, GE, CE | NGS, qRT-PCR, Transformation | Long et al. (2018), Wu et al. (2011, 2015) | ||
| Longan (Dimocarpus longan) | NEC, EC, ICpEC, CpEC, GE, HE, TE, CE, ME | NGS, qRT-PCR, Transformation | Lin and Lai 2013, Li et al. 2015a, b), Xu et al. (2020), Zhang et al. (2020) | ||
| Hybrid yellow poplar (Liriodendron tulipifera) | pre-embryonic mass and SE at sequential developmental stages | NGS, microarray | Li et al. (2012) | ||
| Gymnosperms | Japanese Larch (Larix leptolepis) | EC, NEC | Microarray, NGS, qRT-PCR | Zhang et al. (2010), Liu et al. (2016a), Li et al. (2014) | |
| Picea balfouriana | EC | qRT-PCR | Li et al. (2017, 2019) | ||
Non-embryogenic callus (NEC), embryogenic callus (EC), incomplete pro-embryogenic culture (ICpEC), compact pro-embryogenic cultures (CpEC), globular embryo (GE), heart-shaped embryos (HE), torpedo shaped embryos (TE), cotyledonary embryos (CE), mature embryos (ME)
miRNAs and their targets expression in embryogenic callus
Various miRNAs are known to play a vital role in regulating the transition from callus into SE. miR156 is one of the most conserved miRNAs family in plants that targets Squamosa Promoter Binding Protein-Like (SPL) transcription factors (Wang et al. 2009). In Arabidopsis, the MIRNA genes and mature miRNA of miR156a-h were up-regulated during the SE process (Szyrajew et al. 2017). However, the expression pattern of miR156 is varied from monocots to eudicots plants. In citrus, the expression of miR156 is significantly higher in embryogenic callus (EC) than non-embryogenic callus (NEC) (Wu et al. 2011). In coconut, the expression pattern of miR156 is significantly higher in NEC than in EC. miRNA targeted SPL is involved in the development of plant leaves, flowers, fruit, juvenile-to-adult phase transition, stress response and secondary metabolism (Cui et al. 2014; Gou et al. 2011; Silva et al. 2014; Wang et al. 2009; Wu and Poethig 2006; Yu et al. 2015a, b). The expression of SPL transcription factors was principally reverse to that of miR156 expression in EC and NEC. The expression level of SPL2, -4, -5 were higher in NEC than in EC in citrus. miR156 could enhance the capability of SE induction from callus. The number of SEs formed in csi-miR156a overexpression lines and individual knock-down of the two target genes CsSPL3 and CsSPL14 was significantly higher than the controls (Long et al. 2018). The FUSCA3 (FUS3) is a B3 domain transcription factor that plays an important role in the induction of SE in plants (Liu et al. 2018). The expression of CsFUS3 in csi-miR156a overexpression lines is significantly up-regulated throughout the SE induction process. miR157a is another member of miR156 and involved in the regulation of cellular dedifferentiation and callus proliferation. The SPL10 transcription factor is the target of miR157a. The transcript level of GhmiR157a was found to be higher in the early stage of callus initiation and NEC; and lower transcript abundances in EC and SE development stage (Wang et al. 2018a). Moreover, callus proliferation rate in GhSPL10 overexpression lines is significantly increased (Wang et al. 2018b). These results indicated that miR156/157-SPLs might regulate the initial phases of SE induction.
miRNAs and phytohormone cross talk during somatic embryogenesis
Phytohormones such as auxin, cytokinin, ABA, gibberellin GA, ethylene and jasmonic acid play an essential role in inducing somatic embryogenesis of plants (Bhatia and Bera 2015; Jiménez 2005; Ruduś et al. 2001; Vondrákova et al. 2016). Among them, auxins are the most widely studied phytohormone for somatic embryogenesis induction (Nic-Can and Loyola-Vargas 2016; Wójcik et al. 2020). The regulatory networks of auxin are operated by the three dynamic processes such as auxin biosynthesis and inactivation, auxin polar transport and auxin signal transduction. Signal transduction pathway consists an array of transcriptional activation or repression of genes (Fukui and Hayashi 2018). Furthermore, several studies revealed the functional connections between miRNAs and phytohormones cross talk in the development processes of plant SE. Especially miR160, miR166, miR167, miR390, miR393 are involved in the regulation of auxin signaling during SE (Lin et al. 2015a, b; Wójcik and Gaj 2016; Wójcik et al. 2017). miR156/157 and miR159 are involved in the regulation of ethylene signaling (Wang et al. 2018a) and ABA signaling during SE, respectively (Li et al. 2013).
Ethylene signaling and miRNA-target genes
Ethylene plays important roles in SE induction and developmental processes (Wang et al. 2018a). miR157 is involved in the biosynthesis of ethylene, flavonoid and signaling of ethylene. The GhSPL10 is the target of GhmiR157a. The ethylene signaling-related genes (ETR1, CTR1, EBF1/2 and ERF1/2), ethylene biosynthesis gene (ACOs) and flavonoid synthesis genes (CHS, CHI, and DFR) were significantly activated in GhSPL10 overexpression lines (Wang et al. 2018a). Inhibition of ethylene biosynthesis by aminoethoxyvinylgycine treatment in GhSPL10 overexpression lines completely inhibited callus initiation, whereas an expression of ethylene biosynthesis genes led to higher callus proliferation in wild type (Wang et al. 2018a). Increased expression of flavonoid biosynthesis-related genes also observed by 1-aminocyclopropane carboxylic acid (ethylene biosynthesis precursor) treatment in wild type plants. miR157a negatively regulates its target SPL10 and resulted in callus induction by inducing ethylene responses, which further activate flavonoid biosynthesis to promote callus proliferation. It has revealed the miR157-SPL mediated regulation during somatic embryogenesis via modulating ethylene-mediated flavonoid biosynthesis.
ABA signaling and miRNA-target genes
ABA known to act as a signal substance in stress-induced somatic embryogenesis in various plants (Nishiwaki et al. 2000). ABA induces the accumulation of miR159 in Arabidopsis plants and miR159 targets the cleavage of MYB101 and MYB33 transcripts (Reyes and Chua 2007). The transcription factors MYB101 and MYB33 acts as positive regulators of ABA responses and are regulated by miR159 in ABA‐dependent manner (Millar and Gubler 2005; Reyes and Chua 2007; Tsuji et al. 2006). Expression analysis indicated that miR159 plays an important role in the SE. For instance, miR159 showed a high accumulation in E4, GE and CE stage, while MYB showed negative expression patterns (Li et al. 2013; Szyrajew et al. 2017; Wu et al. 2011; Zhang et al. 2012). The expression of miR159 in NEC is significantly higher than EC in larch, longan and citrus plants (Wu et al. 2011; Xu et al. 2020; Zhang et al. 2010). These suggested that miR159-MYB regulates somatic embryogenesis via modulating the ABA signaling pathways.
Auxin signaling and miRNA-target genes
miR160 and targets
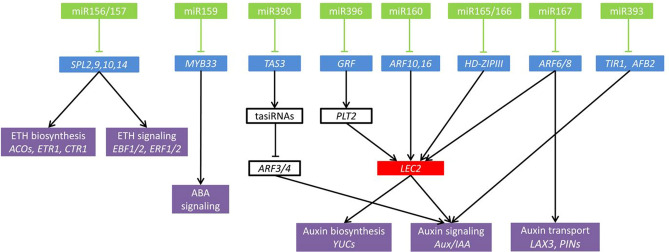
The auxin signal transduction pathways are also regulated by miRNAs. The AUXIN RESPONSE FACTORS (ARFs) such as ARF10, ARF16, ARF17 are the targets of miRNA160 (Liu et al. 2007; Mallory et al. 2005; Wang et al. 2005). miRNA160 is known to express in different developmental stages of somatic embryogenesis. miR160a of Longan plants was barely detectable during the early stages of SE, but highly expressed during the heart- and torpedo-shaped embryonic stages. Dlo-miR160a, -a∗ and -d∗ expression is significantly higher in torpedo-shaped embryos, globular embryos and cotyledonary embryos, respectively (Lin et al. 2015a, b). Loss-of- functional mutation of miR160a in Arabidopsis plants caused various embryonic developmental defects (Liu et al. 2010), suggesting the role of miR160 in embryo development. LEC2 is well known and an important regulator in inducing SE, via YUC-mediated auxin biosynthesis (Stone et al. 2008). A negative feedback regulation exists between ARF10/ARF16 and LEC2 during somatic embryogenesis. The relative expression of LEC2 was increased in miR160, mARF16 (carry the miR160-resistant form of ARF16) and arf10arf16 mutant lines; and expression of ARF10 and ARF16 was also increased in LEC2 overexpression lines (Wójcik et al. 2017). IAA-related indolic compound levels were significantly higher in the miR160b, miR160c insertion lines. Meanwhile, auxin biosynthesis genes (YUC2,YUC4, YUC10) and auxin-responsive genes (IAA1, IAA17 ACS4) were highly up-regulated in miR160b, miR160 and 35S:LEC2 lines (Stone et al. 2008; Wójcik et al. 2017). It has revealed that miR160-ARFs regulated SE induction through the LEC2-mediated auxin biosynthesis and signal transduction pathway.
miR165/166 and targets
In Arabidopsis, two copies of the MIR165 and six copies of the MIR166 genes produce the mature miR165 and miR166, varying by one nucleotide (Reinhart et al. 2002). The targets of miR166 belong to the class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIP III) family such as PHABULOSA (PHB) and PHAVOLUTA (PHV), genes that positively regulate the LEC2 (Liu et al. 2014; Wójcik et al. 2017). Various studies described the expression and role of miR166 in SE. The LaMIR166b transcript showed the highest level in mature somatic embryos and undetectable at the callus proliferation stage in Japanese Larch. The expression of PHB and PHV genes significantly increased at the early and late SE stages. During somatic embryogenesis in Japanese Larch, a positive feedback regulation has been found between PHB and PHV and LEC2 during somatic embryogenesis (Li et al. 2016). Accumulation of the PHB transcripts in the gain of-function phb1-d mutant and a STTM165/166 line (the target mimicry) led to the significant increase in LEC2 transcription; whereas overexpression of LEC2 significantly increased the level of the PHB and PHV transcripts during SE (Wójcik et al. 2017). PHB binds to the LEC2 promoter and activates its expression (Tang et al. 2012). It clearly demonstrated that miR165/166 regulates SE induction through the LEC2-mediated auxin biosynthesis and signal transduction pathway.
miR167 and targets
Two auxin response factors ARF6 and ARF8 are the cleavage targets of miRNA167. The overexpression lines of MIR167a (a predicted miR167 precursor gene) mimicked the arf6arf8 double mutants in Arabidopsis (Su et al. 2016). In the early stage of somatic embryogenesis, the expression of miR167 was dramatically decreased within 2 days during the culture of SE induction and was significantly higher in NEC than EC (Sabana et al. 2020; Wu et al. 2011; Yang et al. 2013). In the late stage of SE development, the expression of miR167 was dramatically increased. In citrus, the abundances of miR167 transcripts was increased continuously and reached the highest level at cotyledonary embryo (CE) stage (Wu et al. 2011), and a similar expression pattern was also reported in Japanese larch (Zhang et al. 2012). Overexpression of miR167 inhibited the SE formation (Su et al. 2016), and diminution of miR167 activity enhanced callogenesis and somatic embryogenesis (Arora et al. 2020). LEC2 is an important positive regulator in plant somatic embryogenesis, which promotes embryogenic induction in somatic tissues of Arabidopsis, via YUC-mediated auxin biosynthesis (Barbara et al. 2013). The relative expression of LEC2 gene was up regulated in miR167-mimic transformed callus of cotton. Besides auxin biosynthesis, the auxin distribution was regulated by miR167. In wild-type callus, the spatial distribution of auxin response signals was restricted to future SE initiation sites; in contrast, distribution of auxin response signals in embryonic callus of the 35S:mi167c transgenic lines were broader and more disorganized. The expression of auxin transporter genes LAX3 (auxin influx carrier), PIN1 and PIN2 (auxin efflux carrier) was also significantly increased in 35S-MIM167 callus in cotton (Su et al. 2016). These implied that the diminution of miR167 activity enhanced somatic embryogenesis via LEC2-YUC-meditaed auxin biosynthesis and LAX3-PIN-mediated auxin transport.
miR390 and targets
The role of miR390 in somatic embryogenesis of plants was first demonstrated in Longan. The trans-acting short-interfering RNAs (tasiRNAs) originated from TAS3 families via miR390 cleavage and targeted ARF3/-4 (Lin et al. 2015a, b). In longan, Pri-miR390 was highly expressed in friable-embryogenic callus (EC), and less expressed in incomplete compact pro-embryogenic cultures, while miR390 showed its lowest expression in EC and highest expression in torpedo-shaped embryos (TEs) (Lin and Lai 2013). The expression of miR390 in EC is significantly higher in EC compared to NEC stage in cotton (Yang et al. 2013). In Arabidopsis, the expression of miR390 is significantly higher in the early stages than the late stages (Szyrajew et al. 2017). The expression data analysis of miR390 in somatic embryogenesis indicated that miR390 is involved in the regulation of early stages of SE induction in plants. The DlTAS3 and DlARF4 exhibited lowest expressions in EC, and reached their peaks in the globular embryos stage, indicating their expression was inversely proportional to the expression of miR390, especially at the globular embryos to CEs stages. These observations suggesting that miR390 might involve in the SE induction via generating tasiARFs that repress ARFs transcripts.
miR393 and targets
miR393 is well known auxin-related miRNA and its targets were involved in the auxin signaling pathway. The F-box proteins TIR1, AFB1, AFB2, and AFB3 are receptors for auxin (Dharmasiri et al. 2005; Kepinski and Leyser 2005), are the targets of miR393 (Navarro et al. 2006). The expression of miR393 (including MIRNA gene and mature miRNA) is significantly higher at early stages of SE induction compared to later stages in Arabidopsis (Szyrajew et al. 2017; Wójcik and Gaj 2016).The expression of TIR1 and AFB2 at an early stage of SE induction was significantly reduced and increased significantly at later stages of SE. In contrast, the expression of AFB1 was gradually increased during somatic embryogenesis stages. The expression of miR393 was opposite to TIR1 and AFB2 expression at the early stage of SE induction. The frequency of embryogenic explants and the average number of SEs were significantly reduced in T-DNA insertion mutants (miR393a, miR393b, miR393a/miR393b) and overexpression lines (35S::MIR393a, 35S::MIR393b) (Wójcik and Gaj 2016), indicated that miR393 mediated regulation of SE. Explants treated with exogenous application of 5 μM of 2,4-D was found to be more effective for SE induction in Arabidopsis wild type plants (Col-0), whereas exogenous treatment of 1 μM of 2,4-D on miR393 mutant lines showed the highest embryogenic response. Over-expression of MIR393 exhibited a significantly reduced sensitivity to auxin treatment and their explants formed SE efficiently in the presence of 30 μM of 2,4-D. miR393 might influence the embryogenic potential of explant cells via modulating the auxin signaling pathway and through the modification of a cell’s sensitivity to auxin. miR393-mediated regulation of TIR1 and AFB2 during embryogenic transition induced through a modification of the explant sensitivity to auxin treatment.
miR396 and targets
miR396 is known to cleave the target of GROWTH-REGULATING FACTOR (GRF) transcription factors (Omidbakhshfard et al. 2015). In Arabidopsis, miR396 targets are GRF1, GRF4, GRF7, GRF8, and GRF9 (Jonesrhoades and Bartel 2004; Rodriguez et al. 2010). Further studies revealed the role of miR396 its role in somatic embryogenesis. The expression of miR396 was significantly increased during the induction of SE (Szczygiel-Sommer and Gaj 2019; Szyrajew et al. 2017). Whereas, the expression of GRFs (GRF1-9) was significantly decreased in SE. Decreased SE response and increased sensitivity to auxin treatment was observed in T-DNA insertion mutants (mir396a and mir396b), whereas, increased SE response and decreased sensitivity to auxin treatment was observed in 35S:MIR396b lines, compared to the WT plants. These indicated that miR396 may contribute to SE induction via an auxin-related pathway. Compared to the WT, the content of the IAA in mir396a and mir396b mutants was found to be 1.85- and 1.50-fold level increase, whereas the content of IAA was decreased by two-fold in the 35S:MIR396b lines. The expression of YUC1 and YUC2 was substantially up- and down-regulated in mir396 mutants and 35S:MIR396b expressing lines, during SE in Arabidopsis. PLETHORA genes (BBM and PLT2) are well-known candidates in triggering SE through the direct transcriptional regulation of the LEC1-ABI3-FUS3-LEC2 network that controls embryo identity (Horstman et al. 2017). Moreover, the expression of PLT1 and PLT2 was positively correlated with the expression of miR396. The expression of miR396 significantly reduced in ptl2 mutant lines and significantly increased in 35S:PLT:GRF lines. Its clearly demonstrated the miR396-mediated regulation of GRFs-PLT during somatic embryogenesis through a modification of the biosynthesis of auxin and also with exogenous sensitivity to auxin.
Cytokinin signaling and miRNA-target genes
Cytokinin signaling is also involved in the regulation of somatic embryogenesis (Ikeuchi et al. 2016). Cytokinins usually interacts with auxin to regulate callus initiation (Perianez-Rodriguez et al., 2014; Liu et al. 2016b). Cytokinin signaling type-A genes including Arabidopsis Response Regulator7 (ARR7) and ARR15 are negative regulators of cytokinin signaling (Buechel et al. 2010). Moreover, auxin response factor 10 (ARF10) down-regulates ARR15 via direct binding to the promoter region of ARR15 and modulates the cytokinin response (Liu et al. 2016b). The number of calli in Pro35S:miR160c and afr10afr16 double mutant lines are significantly lower than wild type. These results indicated that miR160 represses the callus initiation and formation by enhancing ARR15 expression.
Role of other phytohormones (Gibberellins, Jasmonates, Nitric oxide, Polyamines and Brassinosteroids) signaling and somatic embryogenesis
Gibberellins are also involved in the regulation of somatic embryogenesis in various plant species. In coconut, phytohormone gibberellin increases the formation of a number of calli that converts to somatic embryos (Montero-Córtes et al. 2010). In Medicago truncatula, gibberellin acts synergistically with abscisic acid and increases the induction of somatic embryogenesis (Nolan et al. 2014). However, till date the interaction between miRNA and gibberellins is mainly focused on flowering regulation and seed dormancy (Silva et al. 2019; Miao et al. 2019), and none of the reports have been published yet for elucidating the cross talk between miRNAs and gibberelins signaling pathways during somatic embryogenesis. In addition, jasmonates and nitric oxide can also regulate somatic embryogenesis through modulating expression of MYC2 and JAZ1 (Mira et al. 2016). miR319 regulates JA biosynthesis through targeting TCP transcription factors (Zhao et al. 2015; Schommer et al. 2008). Till date, no studies have been reported for the interaction between miRNA and jasmonates signaling for regulating somatic embryogenesis in plants. Polyamines are also involved in the regulation of somatic embryogenesis (Wu et al. 2009; Cheng et al. 2016; Zhu et al. 2018). Polyamines and its metabolites induce the somatic embryos in upland cotton (Gossypium hirsutum L.) (Cheng et al. 2015), and sugarcane (Reis et al. 2016). The exogenous spermidine treatment induces the miRNAs expression (Wang et al. 2018b) and miR396b can modulate ethylene-polyamine homeostasis through regulating ACC Oxidase gene expression (Zhang et al. 2016). The mechanism of the interaction between miRNAs and polyamines to regulate somatic embryogenesis in plants remains to be elucidated. Brassinosteroids control the cell division, elongation and differentiation and are essential for plant growth development. At present, there are few studies deciphered the cross talk between somatic embryogenesis and BR signaling pathway. The BRI1 (brassinosteroids receptor) can interact with SERK3. miRNA is involved in the regulation of brassinosteroids signaling pathway. For example, miR172 increases the sensitivity to BR by affecting the Bak1-mediated BR pathway, and promote seedling hypocotyl elongation and root growth in Arabidopsis thaliana (Kim et al., 2014). Osa-mir1848 regulated the synthesis of phytosterol and BR by splicing the target gene OsCYP51G3. The overexpression of Osa-mir1848 decreased the OsCYP51G3 expression resulted in some BR defect phenotypes, including plant dwarfism, erect leaves, semi-sterility of pollen grains, and short cell size (Xia et al., 2015). But the studies related to the interaction between miRNA and brassinolactone in regulating somatic embryogenesis in plants have not been reported yet.
Conclusion and future perspectives
As a negative regulatory factor, miRNAs play important roles in the development of plant SEs. There is an elementary understanding of the probable underlying mechanisms, such as regulation of auxin biosynthesis, transport and signaling transduction; ethylene biosynthesis and transduction (Fig. 1). However, there are few studies on the isolation and identification and elucidation of miRNA roles in the development stages of plant SE. The role of miRNAs in the development of embryos has not been comprehensively and systematically studied. The expression of miRNAs and their regulated target genes constitutes a relatively tight regulatory network, and the precise operation of this network ensures the normal operation of various physiological processes in plants. Therefore, to strengthen the research on the biological functions of miRNA during the process of plant embryo development will be helpful for further understanding of miRNA mediated embryo development via phtohormone signaling cross talk. In the process of plant embryo development, the mutual regulation of miRNAs and their downstream target genes are important factors in plant somatic embryogenesis.
Fig. 1.
A model depicting the miRNAs regulation via targeting phytohormone signaling pathways during somatic embryogenesis
Acknowledgements
This work was supported by the Central public-interest scientific institution basal research fund for the Chinese Academy of Tropical Agricultural Sciences (Nos. 1630152019002 and 1630152019001), and Hainan Provincial Natural Science Foundation of China (No. 319QN324).
Author contributions
LJ, RY, LZ, ZZ, and HC drafted the manuscript. LJ and RY collected the background information.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Arora S, Singh AK, Chaudhary B. Target-mimicry based miRNA167-diminution ameliorates cotton somatic embryogenesis via transcriptional biases of auxin signaling associated miRNAs and genes. Plant Cell Tissuse Org Cult. 2020;141:511–531. doi: 10.1007/s11240-020-01810-9. [DOI] [Google Scholar]
- Awada R, Verdier D, Froger S, Brulard E, de Faria Maraschin S, Etienne H, Breton D. An innovative automated active compound screening system allows high-throughput optimization of somatic embryogenesis in Coffea arabica. Sci Rep. 2020;10:810. doi: 10.1038/s41598-020-57800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbara W, Karolina J, PrzemyslAw G, Magdalena M, Katarzyna N, Gaj MD. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta. 2013;238:425–440. doi: 10.1007/s00425-013-1892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Bera T (2015) Somatic embryogenesis and organogenesis. In: Bhatia S, Sharma K, Dahiya R, Bera T (eds) Modern applications of plant biotechnology in pharmaceutical sciences. Academic Press, Salt Lake City, pp 214. 10.1016/B978-0-12-802221-4.00006-6
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechel S, Leibfried A, To JP, Zhao Z, Andersen SU, Kieber JJ, Lohmann JU. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur J Cell Biol. 2010;89:279–284. doi: 10.1016/j.ejcb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Chavez-Hernandez EC, Alejandri-Ramirez ND, Juarez-Gonzalez VT, Dinkova TD. Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front Plant Sci. 2015;6:1–14. doi: 10.3389/fpls.2015.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang F, Cheng X, Zhu Q, Sun Y, Zhu H, Sun J. Polyamine and its metabolite H2O2 play a key role in the conversion of embryogenic callus into somatic embryos in upland cotton (Gossypium hirsutum L.) Front Plant Sci. 2015;6:1063. doi: 10.3389/fpls.2015.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Zhu H, Tian W, Zhu S, Xiong X, Sun Y, Zhu Q, Sun J. De novo transcriptome analysis reveals insights into dynamic homeostasis regulation of somatic embryogenesis in upland cotton. Plant Mol Biol. 2016;92(3):279–292. doi: 10.1007/s11103-016-0511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Shan J, Shi M, Gao J, Lin H. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014;80:1108–1117. doi: 10.1111/tpj.12712. [DOI] [PubMed] [Google Scholar]
- Davletova S, Mészáros T, Miskolczi P, Oberschall A, Török K, Magyar Z, Dudits D, Deák M. Auxin and heat shock activation of a novel member of the calmodulin like domain protein kinase gene family in cultured alfalfa cells. J Exp Bot. 2001;52:215–221. doi: 10.1093/jexbot/52.355.215. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Fukui K, Hayashi K. Manipulation and sensing of auxin metabolism, transport and signaling. Plant Cell Physiol. 2018;59:1500–1510. doi: 10.1093/pcp/pcy076. [DOI] [PubMed] [Google Scholar]
- Gou J, De Felippes FF, Liu C, Weigel D, Wang J. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Li S, Fan X, Su Z. Application of somatic embryogenesis in woody plants. Front Plant Sci. 2016;7:938. doi: 10.3389/fpls.2016.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulzar B, Mujib A, Malik MQ, Sayeed R, Mamgain J, Ejaz B. Genes, proteins and other networks regulating somatic embryogenesis in plants. J Genet Eng Biotechnol. 2020;18(1):31. doi: 10.1186/s43141-020-00047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T, Cao X, Wei J, Li L, Yang X. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2019;48:1114–1121. doi: 10.1093/nar/gkz894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JJ, Belmonte MF, Kwong RW (2010) Plant embryogenesis (zygotic and somatic). In: Encyclopedia of life sciences. John Wiley & Sons, Chichester, pp 1–10. 10.1002/9780470015902.a0002042.pub2
- Horstman A, Li M, Heidmann I, Weemen M, Chen B, Muino JM, Angenent GC, Boutilier K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017;175:848–857. doi: 10.1104/pp.17.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H. Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J. 2003;34:107–114. doi: 10.1046/j.1365-313X.2003.01702.x. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K. Plant regeneration: cellular origins and molecular mechanisms. Development. 2016;143:1442–1451. doi: 10.1242/dev.134668. [DOI] [PubMed] [Google Scholar]
- Isah T. Induction of somatic embryogenesis in woody plants. Acta Physiol Plant. 2016;38:118. doi: 10.1007/s11738-016-2134-6. [DOI] [Google Scholar]
- Jiménez VM. Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul. 2005;47:91–110. doi: 10.1007/s10725-005-3478-x. [DOI] [Google Scholar]
- Jonesrhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Joshi R, Kumar P. Regulation of somatic embryogenesis in crops: A review. Agric Rev. 2013;34:1–20. [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim BH, Kwon Y, Lee B, Nam KH. Overexpression of miR172 suppresses the brassinosteroid signaling defects of bak1 in Arabidopsis. Biochem Biophys Res Commun. 2014;447(3):479–484. doi: 10.1016/j.bbrc.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Li T, Chen J, Qiu S, Zhang Y, Wang P, Yang L, Lu Y, Shi J. Deep sequencing and microarray hybridization identify conserved and species-specific microRNAs during somatic embryogenesis in hybrid yellow poplar. PLoS One. 2012;7:e43451. doi: 10.1371/journal.pone.0043451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang S, Han S, Wu T, Zhang J, Qi L. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tissue Org Cult. 2013;113:131–136. doi: 10.1007/s11240-012-0233-7. [DOI] [Google Scholar]
- Li W, Zhang S, Han S, Wu T, Zhang J, Qi L. The post-transcriptional regulation of LaSCL6 by miR171 during maintenance of embryogenic potential in Larix kaempferi (Lamb.) Carr. Tree Genet Genomes. 2014;10:223–229. doi: 10.1007/s11295-013-0668-y. [DOI] [Google Scholar]
- Li Z, Li S, Zhang L, Han S, Li W, Xu H, Yang W, Liu Y, Fan Y, Qi L. Over-expression of miR166a inhibits cotyledon formation in somatic embryos and promotes lateral root development in seedlings of Larix leptolepis. Plant Cell Tissue Org Cult. 2016;127:461–473. doi: 10.1007/s11240-016-1071-9. [DOI] [Google Scholar]
- Li Q, Deng C, Xia Y, Kong L, Zhang H, Zhang S, Wang J. Identification of novel miRNAs and miRNA expression profiling in embryogenic tissues of Picea balfouriana treated by 6-benzylaminopurine. PLoS One. 2017;12:1–17. doi: 10.1371/journal.pone.0176112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Deng C, Zhu T, Ling J, Zhang H, Kong L, Zhang S, Wang J, Chen X. Dynamics of physiological and miRNA changes after long-term proliferation in somatic embryogenesis of Picea balfouriana. Trees-Struct Funct. 2019;33:469–480. doi: 10.1007/s00468-018-1793-x. [DOI] [Google Scholar]
- Lin Y, Lai Z. Comparative analysis reveals dynamic changes in miRNAs and their targets andexpression during somatic embryogenesis in longan (Dimocarpus longan Lour.) Plos One. 2013;8:e60337. doi: 10.1371/journal.pone.0060337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lai Z, Tian Q, Lin L, Lai R, Yang M, Zhang D, Chen Y, Zhang Z. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front Plant Sci. 2015;6:956. doi: 10.3389/fpls.2015.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lin L, Lai R, Liu W, Chen Y, Zhang Z, Xu X, Lai Z. MicroRNA390-directed tas3 cleavage leads to the production of tasirna-arf3/4 during somatic embryogenesis in dimocarpus longan lour. Front Plant Sci. 2015;6:1119. doi: 10.3389/fpls.2075.07719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang J, Wang Y, Khanna K, Xie Z, Owen HA, Zhao D. The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 2010;62:416–428. doi: 10.1111/j.1365-313X.2010.04164.x. [DOI] [PubMed] [Google Scholar]
- Liu N, Tu L, Tang W, Gao W, Lindsey K, Zhang X. Small RNA and degradome profiling reveals a role for miRNAs and their targets in the developing fibers of Gossypium barbadense. Plant J. 2014;80:331–344. doi: 10.1111/tpj.12636. [DOI] [PubMed] [Google Scholar]
- Liu Y, Han S, Ding X, Li X, Zhang L, Li W, Xu H, Li Z, Qi L. Transcriptome analysis of mRNA and miRNA in somatic embryos of Larix leptolepis subjected to hydrogen treatment. Int J Mol Sci. 2016;17:1–14. doi: 10.3390/ijms17111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li J, Wang L, Li Q, Lu Q, Yu Y, Li S, Bai M, Hu Y, Xiang F. Repression of callus initiation by the miRNA-directed interaction of auxin-cytokinin in Arabidopsis thaliana. Plant J. 2016;87:391–402. doi: 10.1111/tpj.13211. [DOI] [PubMed] [Google Scholar]
- Liu Z, Ge X, Qiu W, Long J, Jia H, Yang W, Dutt M, Wu X, Guo W. Overexpression of the CsFUS3 gene encoding a B3 transcription factor promotes somatic embryogenesis in Citrus. Plant Sci. 2018;277:121–131. doi: 10.1016/j.plantsci.2018.10.015. [DOI] [PubMed] [Google Scholar]
- Long J, Liu C, Feng M, Liu Y, Wu X, Guo W. miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J Exp Bot. 2018;69:2979–2993. doi: 10.1093/jxb/ery132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Loyola-Vargas VM, Ochoa-Alejo N. Somatic embryogenesis: fundamental aspects and applications. Berlin: Springer; 2016. pp. 2–4. [Google Scholar]
- Luo Y, Zhou H, Li Y, Chen J, Yang J, Chen Y, Qu L. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006;580:5111–5116. doi: 10.1016/j.febslet.2006.08.046. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Hernández HA, Ledezma-Rodríguez M, Avilez-Montalvo RN, Juárez-Gómez YL, Skeete A, Avilez-Montalvo J, De-la-Peña C, Loyola-Vargas VM. Signaling overview of plant somatic embryogenesis. Front Plant Sci. 2019;10:1–15. doi: 10.3389/fpls.2019.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Wang Z, Zhang L, Yao J, Hua K, Liu X, Shi H, Zhu J. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat Commun. 2019;10(1):3822. doi: 10.1038/s41467-019-11830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MM, Wally OSD, Elhiti M, El-Shanshory A, Reddy DS, Hill RD, Stasolla C. Jasmonic acid is a downstream component in the modulation of somatic embryogenesis by Arabidopsis Class 2 phytoglobin. J Exp Bot. 2016;67(8):2231–2246. doi: 10.1093/jxb/erw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Córtes M, Sáenz L, Córdova I, Quiroz A, Verdeil J, Oropeza C. GA3 stimulates the formation and germination of somatic embryos and the expression of a KNOTTED-like homeobox gene of Cocos nucifera (L.) Plant Cell Rep. 2010;29(9):1049–1059. doi: 10.1007/s00299-010-0890-0. [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Nic-Can GI, Loyola-Vargas VM. The role of the auxins during somatic embryogenesis. Berlin: Springer International Publishing; 2016. pp. 171–182. [Google Scholar]
- Nic-Can GI, Galaz-Ávalos RM, De-la-Peña C, Alcazar-Magaña A, Wrobel K, Loyola-Vargas VM. Somatic embryogenesis: identified factors that lead to embryogenic repression. A case of species of the same genus. Plos One. 2015;10:e126414. doi: 10.1371/journal.pone.0126414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki M, Fujino K, Koda Y, Masuda K, Kikuta Y. Somatic embryogenesis induced by the simple application of abscisic acid to carrot (Daucus carota L.) seedlings in culture. Planta. 2000;211:756–759. doi: 10.1007/s004250000387. [DOI] [PubMed] [Google Scholar]
- Nolan KE, Song Y, Liao S, Saeed NA, Zhang X, Rose RJ. An unusual abscisic acid and gibberellic acid synergism increases somatic embryogenesis, facilitates its genetic analysis and improves transformation in Medicago truncatula. PLoS One. 2014;9(6):e99908. doi: 10.1371/journal.pone.0099908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Muellerroeber B. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol Plant. 2015;8:998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perianez-Rodriguez J, Manzano C, Moreno-Risueno MA. Post embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front Plant Sci. 2014;5:219. doi: 10.3389/fpls.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Gene Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RS, Vale EDM, Heringer AS, Santa-Catarina C, Silveira V. Putrescine induces somatic embryo development and proteomic changes in embryogenic callus of sugarcane. J Proteom. 2016;130:170–179. doi: 10.1016/j.jprot.2015.09.029. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Chua N. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruduś I, Kepczynski J, Kepczynska E. The influence of the jasmonates and abscisic acid on callus growth and somatic embryogenesis in Medicago sativa L. tissue culture. Acta Physiol Plant. 2001;23:103–107. doi: 10.1007/s11738-001-0029-6. [DOI] [Google Scholar]
- Sabana AA, Antony G, Rahul CU, Rajesh MK. In silico identification of microRNAs and their targets associated with coconut embryogenic calli. Agri Gene. 2018;7:59–65. doi: 10.1016/j.aggene.2018.01.002. [DOI] [Google Scholar]
- Sabana AA, Rajesh MK, Antony G. Dynamic changes in the expression pattern of miRNAs and associated target genes during coconut somatic embryogenesis. Planta. 2020;251:1–18. doi: 10.1007/s00425-020-03368-4. [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, Vries SCD. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1016/S1386-1425(01)00499-1. [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6(9):e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui ZH, Abbas ZK, Ansari MW, Khan MN. The role of miRNA in somatic embryogenesis. Genomics. 2019;111:1026–1033. doi: 10.1016/j.ygeno.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Silva GFFE, Silva EM, Silva Azevedo MD, Guivin MAC, Ramiro DA, Figueiredo CR, Carrer H, Peres LEP, Nogueira FTS. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014;78:604–618. doi: 10.1111/tpj.12493. [DOI] [PubMed] [Google Scholar]
- Silva GFF, Silva EM, Correa JPO, Vicente MH, Jiang N, Notini MM, Junior AC, De Jesus FA, Castilho P, Carrera E, López-Díaz I, Grotewold E, Peres LEP, Nogueira FTS. Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol. 2019;221(3):1328–1344. doi: 10.1111/nph.15492. [DOI] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh T, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. PNAS. 2008;105:3151–3156. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Zhao XY, Liu YB, Zhang CL, O Neill SD, Zhang XS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009;59:448–460. doi: 10.1111/j.1365-313X.2009.03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Liu YB, Zhou C, Li XM, Zhang XS. The microRNA167 controls somatic embryogenesis in Arabidopsis through regulating its target genes ARF6 and ARF8. Plant Cell Tissue Org Cult. 2016;124:405–417. doi: 10.1007/s11240-015-0903-3. [DOI] [Google Scholar]
- Szczygiel-Sommer A, Gaj MD. The miR396-GRF regulatory module controls the embryogenic response in arabidopsis via an auxin-related pathway. Int J Mol Sci. 2019;20:1–18. doi: 10.3390/ijms20205221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyrajew K, Bielewicz D, Dolata J, Wójcik AM, Nowak K, Szczygieł-Sommer A, Szweykowska-Kulinska Z, Jarmolowski A, Gaj MD. MicroRNAs are intensively regulated during induction of somatic embryogenesis in Arabidopsis. Front Plant Sci. 2017;8:1–16. doi: 10.3389/fpls.2017.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Bian S, Tang M, Lu Q, Li S, Liu X, Tian G, Nguyen V, Tsang EWT, Wang A, Rothstein SJ, Chen X, Cui Y. MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet. 2012;8:e1003091. doi: 10.1371/journal.pgen.1003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchitanaka M, Shimada Y, Nakazono M, Watanabe R, Nishizawa NK, Gomi K, Shimada A, Kitano H. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 2006;47:427–444. doi: 10.1111/j.1365-313X.2006.02795.x. [DOI] [PubMed] [Google Scholar]
- Vondrákova Z, Krajňákova J, Fisherová L, Vágner M, Eliášová K. Physiology and role of plant growth regulators in somatic embryogenesis. Seoul: National Institute of Forest Science; 2016. pp. 127–145. [Google Scholar]
- Wang J, Wang L, Mao Y, Cai W, Xue H, Chen X. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu N, Wang T, Li J, Wen T, Yang X, Lindsey K, Zhang X. The GhmiR157a-GhSPL10 regulatory module controls initial cellular dedifferentiation and callus proliferation in cotton by modulating ethylene-mediated flavonoid biosynthesis. J Exp Bot. 2018;69:1081–1093. doi: 10.1093/jxb/erx475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo S, Wang L, Wang L, He X, Shu S, Sun J, Lu N. Identification of microRNAs associated with the exogenous spermidine-mediated improvement of high-temperature tolerance in cucumber seedlings (Cucumis sativus L.) BMC Genom. 2018;19(1):285. doi: 10.1186/s12864-018-4678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mei J, Ren G. Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci. 2019;10:360. doi: 10.3389/fpls.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik AM, Gaj MD. miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta. 2016;244:231–243. doi: 10.1007/s00425-016-2505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik AM, Nodine MD, Gaj MD. miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front Plant Sci. 2017;8:1–17. doi: 10.3389/fpls.2017.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik AM, Wójcikowska B, Gaj MD. Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int J Mol Sci. 2020;21:1333. doi: 10.3390/ijms21041333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Dev. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang J, Liu J, Deng X. Involvement of polyamine biosynthesis in somatic embryogenesis of Valencia sweet orange (Citrus sinensis) induced by glycerol. J Plant Physiol. 2009;166(1):52–62. doi: 10.1016/j.jplph.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Wu XM, Liu MY, Ge XX, Xu Q, Guo WW. Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta. 2011;233:495–505. doi: 10.1007/s00425-010-1312-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Kou S, Liu Y, Fang Y, Xu Q, Guo W. Genome wide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA-and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol J. 2015;13:383–394. doi: 10.1111/pbi.12317. [DOI] [PubMed] [Google Scholar]
- Xia K, Ou X, Tang H, Wang R, Wu P, Jia Y, Wei X, Xu X, Kang S, Kim S, Zhang M. Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 2015;208(3):790–802. doi: 10.1111/nph.13513. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen X, Chen Y, Zhang Q, Su L, Chen X, Chen Y, Zhang Z, Lin Y, Lai Z. Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci Rep. 2020;10:4626. doi: 10.1038/s41598-020-60946-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang X. Regulation of somatic embryogenesis in higher plants. Crit Rev Plant Sci. 2010;29:36–57. doi: 10.1080/07352680903436291. [DOI] [Google Scholar]
- Yang X, Wang L, Yuan D, Keith L, Zhang X. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J Exp Bot. 2013;6:1521–1536. doi: 10.1093/jxb/ert013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang J, Wang J, Ren Y, Zhu Y, Sun H. Dynamic changes of miR166s at both the transcriptional and post-transcriptional levels during somatic embryogenesis in Lilium. Sci Hortic. 2020;261:1–8. doi: 10.1016/j.scienta.2019.108928. [DOI] [Google Scholar]
- Yu N, Niu Q, Ng K, Chua N. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015;83:673–685. doi: 10.1111/tpj.12919. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Wang LJ, Zhao B, Shan CM, Zhang YH, Chen DF, Chen XY. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol Plant. 2015;8:98–110. doi: 10.1016/j.molp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou J, Han S, Yang W, Li W, Wei H, Li X, Qi L. Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem Biophys Res Commun. 2010;398:355–360. doi: 10.1016/j.bbrc.2010.06.056. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang S, Han S, Wu T, Li X, Li W, Qi L. Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta. 2012;236:647–657. doi: 10.1007/s00425-012-1643-9. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang W, Wang M, Zhang H, Liu J. The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene-polyamine homeostasis. Plant Cell Physiol. 2016;57(9):1865–1878. doi: 10.1093/pcp/pcw108. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xue B, Gai M, Song S, Jia N, Sun H. Small RNA and transcriptome sequencing reveal a potential miRNA-mediated interaction network that functions during somatic embryogenesis in Lilium pumilum DC. fisch. Front Plant Sci. 2017;8:1–18. doi: 10.3389/fpls.2017.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QL, Su LY, Zhang ST, Xu XP, Chen XH, Li X, Jiang MQ, Huang SQ, Chen YK, Zhang ZH, Lai ZX, Lin YL. Analyses of microRNA166 gene structure, expression, and function during the early stage of somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol Biochem. 2020;147:205–214. doi: 10.1016/j.plaphy.2019.12.014. [DOI] [PubMed] [Google Scholar]
- Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot. 2015;66(15):4653–4667. doi: 10.1093/jxb/erv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Perry SE. AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 2013;161:2113–2127. doi: 10.1104/pp.113.216275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HG, Cheng WH, Tian WG, Li YJ, Liu F, Xue F, Zhu QH, Sun YQ, Sun J. iTRAQ-based comparative proteomic analysis provides insights into somatic embryogenesis in Gossypium hirsutum L. Plant Mol Biol. 2018;96(1–2):89–102. doi: 10.1007/s11103-017-0681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]