Abstract
Introduction
Lactate secreted by tumors is not just a byproduct, but rather an active modulator of immune cells. There are few studies aimed at investigating the true effect of lactate, which is normally confounded by pH. Such a knowledge gap needs to be addressed. Herein, we studied the immunomodulatory effects of lactate on dendritic cells (DCs) and macrophages (MΦs).
Methods
Bone marrow-derived innate immune cells were treated with 50 mM sodium lactate (sLA) and incubated for 2 days or 5 days at 37 °C. Controls included media, lipopolysaccharide (LPS), MCT inhibitors (α-cyano-4-hydroxycinnamic acid and AR-C15585). Flow cytometric analysis of immune phenotypes were performed by incubating cells with specific marker antibodies and viability dye. Differential expression analyses were conducted on R using limma-voom and adjusted p-values were generated using the Bejamini-Hochberg Procedure.
Results
Lactate exposure attenuated DC maturation through the downregulation of CD80 and MHCII expression under LPS stimulation. For MΦs, lactate exposure resulted in M2 polarization as evidenced by the reduction of M1 markers (CD38 and iNOS), and the increase in expression of CD163 and Arg1. We also revealed the role of monocarboxylate transporters (MCTs) in mediating lactate effect in MΦs. MCT4 inhibition significantly boosted lactate M2 polarization, while blocking of MCT1/2 failed to reverse the immunosuppressive effect of lactate, correlating with the result of gene expression that lactate increased MCT4 expression, but downregulated the expression of MCT1/2.
Conclusions
This research provides valuable insight on the influence of metabolic products on tumor immunity and will help to identify novel metabolic targets for augmenting cancer immunotherapies.
Electronic supplementary material
The online version of this article (10.1007/s12195-020-00652-x) contains supplementary material, which is available to authorized users.
Keywords: Lactate, Dendritic cells, Macrophages, Immune escape, Cancer, Immunosuppressive
Introduction
The field of cancer immunotherapy has recently been invigorated with the successful clinical trials of several cell-based approaches, including CAR T cell immunotherapy. Despite these improved outcomes in clinical trials showing response rates greater than 50% for liquid tumors, successful application of these approaches to solid tumors has been rare in the clinic.3,33 The tumor microenvironment (TME) of solid tumors has been directly implicated as the culprit behind the significant difference in clinical responses to cell-based immunotherapy between these two forms of cancers.39
It is now well established that the TME counters the efficacy of immune cells to detect and eliminate malignant cells by (i) physically preventing the migration of immune cells through the production of a dense, collagenous extracellular matrix (ECM), (ii) increased cell surface expression of inhibitory signaling molecules (e.g. programmed cell death protein 1 [PD-1]), (iii) recruitment of regulatory cell types (e.g. regulatory T cells and myeloid-derived suppressor cells) that cooperate with tumor cells to produce (iv) soluble anti-inflammatory mediators including cytokines (e.g. transforming growth factor-beta [TGF-β]) and tumor metabolic products.71,80 Cancer cells have a high metabolic demand which often leads to dependency on the glycolytic pathway for derivation of adenosine triphosphate (ATP). Some tumor cells perform glycolysis even in the presence of oxygen, a phenomenon known as the Warburg Effect.53 The major byproducts of glycolysis are lactate and H+ ions, which accumulate intratumorally due to the disorganized nature of tumor vasculature.14 Recent reports indicate that intratumoral lactic acid concentrations may get as high as 40 mM.76 Moreover, at extracellular concentrations lower than 40 mM, lactic acid can severely influence the phenotype and function of immune cells. For instance, Mendler et al. demonstrated that tumor lactic acidosis suppressed cytotoxic T lymphocyte function, including the complete inactivation of inflammatory cytokine production and impairment of lytic granule release, by preventing the phosphorylation of critical signaling proteins in the JNK, c-Jun and p38 pathways.45 This study along with others8,22 help to reveal the contributions of lactic acid to the derailment of adoptive T cell therapy for solid tumors. But its impacts aren’t limited to just tumor-infiltrating T cells. Macrophages (MΦs) and dendritic cells (DCs) are also susceptible to the immune modulatory effects of lactic acid.8,22
In the context of cancer, DCs are renowned for their role in immune surveillance and response to therapy. Dendritic cells are the ‘professional’ antigen-presenting cells of the immune system, as their primary function is to drive T cell selection and expansion following antigen uptake.6 Their interaction with adaptive immune cells is critical for inhibiting tumor development as they can effectively recognize danger-associated molecular patterns (DAMPs) and cross-present neo-antigens derived from tumor cells.72 In non-malignant tissues, MΦs typically assume a defensive role and once activated are efficient killers of microbes through phagocytosis and intracellular microbiocidal mechanisms.19 However, in cancerous tissue, MΦs seem to play a duplicitous role. Recent evidence suggests that the presence of MΦs in solid tumors fuels, rather than limits, tumor progression and negatively impacts various immunotherapies. In the tumor microenvironment MΦs, often called tumor-associated MΦs (TAMs), are shaped to promote immunosuppression, neovascularization, tumor metastasis, and poor responses to therapy.58 Unsurprisingly DCs derived from solid tumors also have impaired functionality including altered activation, impaired antigen presentation, and reduced capacity for pro-inflammatory cytokine production.40,52 Consequently, there is a lack of antitumor immunity.
The contribution of lactate to the resulting functional state of MΦs and DCs in tumors is still not clear. There are only a handful of studies that begin to isolate the effects of lactate on DC and MΦ phenotypes. Moreover, they often confound the effects of lactate with the accompanying acidity and hypoxia.9,12,27,56 For instance, a report by Bohn et al. indicated that tumor acidosis induced G protein–coupled receptor–dependent expression of the transcriptional repressor ICER in TAMs, which induced a non-inflammatory phenotype and promoted tumor growth. The Kreutz group demonstrated that tumor-derived lactic acid modulates DC maturation and antigen presentation.27 We recently investigated the effects of lactic acid derived from a biomaterial, poly(lactic-co-glycolic acid), on bone marrow-derived DC phenotypes and included lactate as a control in these studies. Interestingly, we found that lactate at high concentrations (> 50 mM), as well as polymer-derived lactic acid, can modulate the canonical NF-κB pathway by preventing the phosphorylation of TAK-1.1 However, our results also indicated that other pathways were involved in the polarization of DCs to an inactivated state.
Herein, we further expand upon those studies to identify the effects of lactate at a concentration relevant to the TME and, in an oxygenated, buffered environment, on both MΦ and DC phenotypes. Additionally, through probing the transcriptional profile, we aim to identify other key pathways modulated by lactate in these critical innate immune cells.
Materials and Methods
Experimental Animals
C57BL/6 and BALB/cByJ mice, ages 6–12 weeks, were purchased from Jackson Laboratory (Bar Harbor, ME). All animals were housed in specific pathogen-free environment in University of California, Davis facilities and used in accordance with detailed experimental protocols approved by University of California Institutional Animal Care and Use Committee (IACUC).
Generation of Bone Marrow-Derived Innate Immune Cells
Dendritic cells and MΦs were obtained from 6–12 weeks old, male and female, C57BL/6 mice in accordance with guidelines approved by the University of California, Davis, Animal Care and Use Committee (IACUC) using a modified 10-day protocol.43 Briefly, mice were euthanized by CO2 asphyxiation followed by cervical dislocation, and tibias and femurs were harvested for isolating bone marrow cells. The bone marrow cells were obtained by flushing the shaft of the long bones with a 25 g needle using RPMI medium with L-glutamine and 25 mM HEPES (Mediatech) containing 1% fetal bovine serum (Mediatech) and 1% penicillin/streptomycin (Hyclone) and mixed to make a homogeneous suspension. The suspension was then strained using 70 µm cell strainers (Becton Dickinson, NJ, United States), and cells were collected at 1800 rpm for 5 min. The red blood cells were lysed with ACK lysis buffer (Lonza), followed by centrifugation at 1800 rpm for 5 min to recover leukocytes. Leukocytes were then resuspended in DMEM/F-12 1:1 with L-glutamine (Cellgro), 10% fetal bovine serum (FBS, Corning), 1% sodium pyruvate (Lonza), 1% nonessential amino acids (Lonza), 1% penicillin/streptomycin (Hyclone) and 10 ng/mL GM-CSF (R&D Systems) (DC media) or 10% (v/v) L-929 supplement (MΦ media, prepared as previously described83) and plated on tissue culture flasks for 2 days to remove adherent cells. At two days, the nonadherent cells were transferred to low attachment plates and cultured in fresh DC or MΦ media for expansion of precursor cells. At six days, cells were transferred to tissue culture plates to allow for innate cell adhesion and proliferation. At ten days, the cells were used for the studies described below.
Lactate and MCT Inhibitor Treatment
To ensure that the pH of the media after addition of sodium lactate (sLA) were maintained, we measured the pH of media with or without 50 mM sLA using a pH meter. In the experiments aimed at evaluating the effect of sLA on immature (i) cells (in the absence of LPS), cells were treated with media (iDCs or iMΦs) or 50 mM (sLA) and incubated for 48 h or 120 h at 37 °C. LPS at concentrations of 1 or 0.1 µg/mL was used as stimulated controls for DCs and MΦs, respectively. To examine maturation resistance of sLA-treated DCs, cells were pretreated with sLA for 48 or 120 h. The cells were then washed with PBS to remove sLA, and subsequently cultured for additional 24 h with 1 µg/mL of LPS in culture media. Controls included in the resistance maturation experiment, were DCs cultured in media only and stimulated cells which were treated with 1 µg/mL of LPS for 24 h. The MCT inhibitors, AR-C155858 (ARC, Tocris) and α-cyano-4-hydroxycinnamic acid (CHCA, Aldrich) were reconstituted in DMSO to make 1 mM and 100 mM stocks, respectively. The stocks were diluted in media and co-incubated with sLA at concentrations of 10 µM for ARC or 1 mM for CHCA and the final concentration of DMSO used was 1%. 1% DMSO treatment was also used as a control in the experiment of MCT inhibition. For flow cytometric analysis of cell phenotypes, cells were incubated with antibodies against CD16/CD32 (Fcγ III/II Receptor) (clone 2.4G2, IgG2b, k); (BD Pharmingen) in 5% FBS for 20 min at 4 °C to block Fcγ receptors on DCs and MΦs. Following the incubation with Fc block, DCs were stained with CD80 (clone 16-10A1, IgG2, k) (BD Pharmingen), CD86 (clone GL1,IgG2a, k) (BD Pharmingen), I-A/I-E (clone M5/114.15.2, IgG2b, k) (BD Pharmingen), CD11c (clone HL3,IgG1, l2) (BD Pharmingen), and CD197 (clone 4B12, IgG2a, k) (Thermo Fisher Scientific) for 25 min at 4 °C in the dark. For MΦs, their surface markers were stained against APC-labelled anti-F4/80 (clone BM8, IgG2a, k) (Thermo Fisher Scientific), PerCP-eFluor710-labelled anti-CD38 (clone 90, IgG2a, k) (Thermo Fisher Scientific), super bright 600-labelled anti-CD163 (clone TNKUPJ, IgG2a, k) (Thermo Fisher Scientific), and/or PerCP-Cy5.5-labeled anti-CD197(CCR7) (clone 4B12, IgG2a, k) (Thermo Fisher Scientific). The intracellular staining of MΦs was performed through fixation and permeabilization using an intracellular fixation and permeabilization buffer set (Thermo Fisher Scientific) followed by a staining with PE-labelled anti-arginase1 (clone A1exF5, IgG2a, k) (Thermo Fisher Scientific), PE-Cy7-labelled anti-iNOS (clone CXNFT, IgG2a, k) (Thermo Fisher Scientific), and PerCP-eFluor710-labelled anti-RELM (clone DS8RELM, IgG1, k) (Thermo Fisher Scientific). UV Zombie dye (Biolegend) was used to examine the cell viability. Cells were then examined using an Attune Nxt Flow cytometer (Thermo Fisher Scientific) and at least 10,000 events were collected. Data analysis was performed using Attune software (Thermo Fisher Scientific). The cell populations were gating based on singlet then only live cells were selected using UV Zombie dye staining. Taking only live and singlet cells, the cells were chosen further according to the cell markers–CD11c for DCs, and F4/80 for MΦs. Dendritic cell or MΦ populations were then analyzed for other surface expressions. All the positive staining gates were set according to Fluorescence Minus One (FMO) controls (Fig. S1).
Cytokine and Chemokine Secretion
Cell culture supernatants were collected after respective incubations of sLA, MCT inhibitor, or control treatment, centrifuged to remove any cell debris, and stored at − 20 °C until analysis. The secretion of IL-12p70 (BD Biosciences), IL-10 (BD Biosciences), TGF-β (R&D Systems), CCL1 (R&D Systems), and CCL9 (R&D) were quantified using sandwich enzyme-linked immunosorbent assay (ELISA) kit (Becton Dickinson) according to manufacturer’s directions.
CD4+ T Cell Isolation and CFSE Staining
Mouse CD4+ T cells were purified from BALB/cByJ splenocyte suspensions by negative selection using CD4+ T cell isolation kit (Stemcell), following the manufacturer’s instructions. The relative purity as determined by flow cytometry was greater than 95%. T cells were subsequently stained with carboxyfluorescein succinimidyl ester (CFSE; Cell Trace Cell Proliferation Kit, Thermo Fisher Scientific) according to the manufacturer’s specifications.
Mixed Lymphocyte Reactions
For the mixed lymphocyte reaction studies, C57BL/6 DCs or MΦs (2.5 × 104 cells/well) were plated onto a 96-well tissue culture plate. Cells were co-cultured with sLA (50 mM) for 48 and 120 h. The C57BL/6 cells were then thoroughly washed with PBS, added with CFSE-stained BALB/cByJ CD4+ T cells (1.25 × 105 cells/well), and incubated at 37 °C for 72 h. After 72 h, nonadherent T cells were removed and stained with anti-CD4 (clone RM4-5, IgG2a, κ) (BD Pharmingen). Flow cytometry was then used to quantify T cell proliferation via CFSE dilution. More than 10,000 events were acquired for each sample, and data analysis was performed using Attune software (Thermo Fisher Scientific). Phorbol 12-myristate 13-acetate-(PMA; [81 ng/mL]) and ionomycin-(1.4 μg/mL) (Biolegend) stimulated cells were used as proliferation controls.
RNA Sequencing
Both DCs and MΦs were treated with media (control) or 50 mM sLA for 48 h at 37 °C. RNA extraction was performed by adding 1 mL of TRIzol Reagent (Thermo Fisher Scientific) to each well of 1 million cells. According to the manufacturer’s instructions, 200 µL of chloroform were added to each tube of 1 mL TRIzol. Following centrifugation, 400 µL of the top clear aqueous phase was taken from the samples and mixed with 400 µL of ethanol for precipitation. The RNA was further purified using an RNA Clean & Concentrator column (Zymo Research) including DNAse treatment in-column according to the manufacturer’s instructions. RNA quality was assessed using the LabChip GX Nucleic Acid Analyzer in the UC Davis DNA Core. Library preparation was performed by the DNA Core using the 3’-Tag-Seq (QuantSeq) Gene Expression Profiling protocol. The library was sequenced by the DNA Core via single-end RNA sequencing on the Illumina HiSeq 4000. Prior to analysis, genes with fewer than 2 counts per million reads were filtered out leaving 10856 DC genes and 11217 MΦ genes. Differential expression analyses were conducted on R using limma-voom and adjusted p-values were generated using the Bejamini-Hochberg Procedure. Before further analysis, genes with adjusted p-values > 0.05 and |log2FC| < 1 were removed, leaving 479 DC genes and 444 MΦ genes. Next, gene ontology (GO) enrichment analysis and KEGG analysis were performed on R using topGO and KEGGREST, respectively.
Statistical Analysis
Statistical analyses were performed using a one-way ANOVA for all experiments, unless otherwise noted. Statistical significance was determined by a one-way analysis of variance (ANOVA), followed by Dunnett’ s multiple-comparison test. In the experiments that evaluate the effect of sLA on immature cells (in the absence of LPS), the statistically significant differences between sLA-treated groups and iDCs were reported. In maturation resistance experiment, the statistically significant differences between sLA-treated group and LPS only-treated group were analyzed. For student’s t test, the means of each treatment group were compared. Differences were considered significant if p ≤ 0.05 using the Prism software (Version 8, GraphPad). All data are expressed as mean ± standard error (S.E.) of at least three independent experiments.
Results
Lactate Maintains iDC Phenotype but Downregulates CD11c
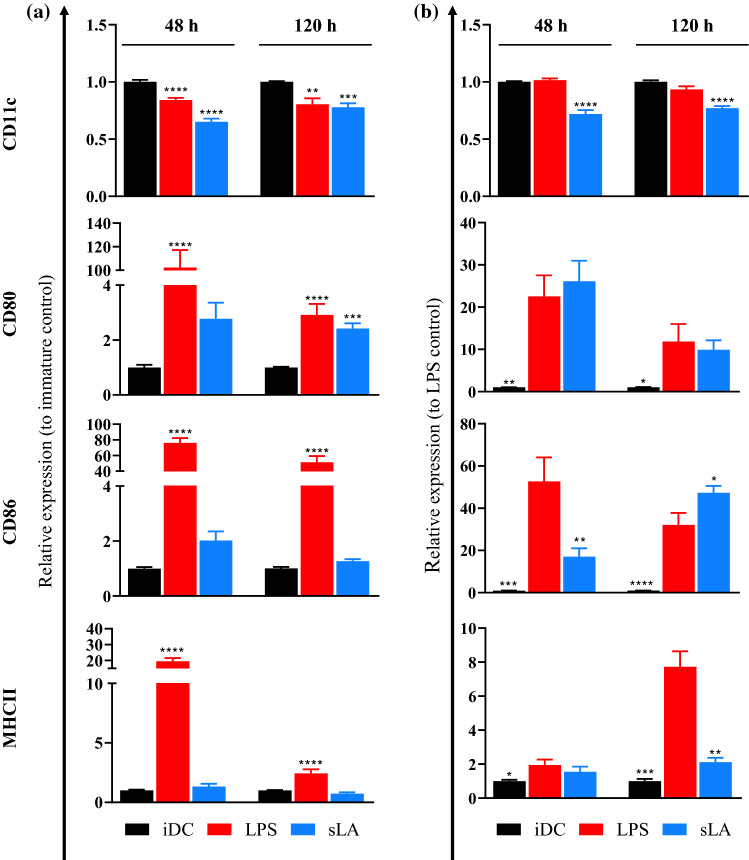
We first measured the pH of sLA-containing media incubated for 2 days and 5 days in an incubator. Due to the buffering capacity of the media, there were no significant changes in pH among samples either in the presence or absence of sLA (Table S1). To evaluate the immunogenic effect of sLA on DCs, we exposed DCs for 48 and 120 h with 50 mM sLA, which has been shown as a biologically-relevant concentration found in TME.17,29,75,76 The presence of specific cell surface markers and cytokine secretion have been used widely as one of the indicators to investigate the immune cell phenotypes. In this study, flow cytometry was used to assess the surface markers.18,42,49 The major histocompatibility complex class-II (MHCII) and costimulatory molecules, CD80 and CD86, were used as molecular markers to evaluate DC maturation status. These markers are necessary for DCs to activate T cells against target antigens and are generally used to measure the immunomodulatory effects of drugs on DCs. The change in expression level in inflammatory markers during cell activation dictates the phenotype of DCs.60 To differentiate DCs from other cells, CD11c was used as a DC marker. Bone marrow-derived DCs from C57BL/6 mice were cultured in 50 mM sLA for 48 and 120 h. Control groups for this study were iDCs and DCs treated with LPS, a common TLR-4 agonist. The expression of markers relative to iDC after sLA exposure was determined for 48 and 120 h (Figs. 1a and S2). At both time points, there was a significant downregulation of CD11c expression for sLA-treated cells compared to iDCs. At 48 h, CD80 expression for sLA-treated cells was comparable to iDCs. Interestingly, the expression became significantly upregulated after 120 h of incubation. Both CD86 and MHCII expression for sLA-treated cells were increased at 48 h and decreased at 120 h, respectively. The viability after sLA exposure is shown in Fig. S3. Soluble lactate treatment showed no cytotoxicity to DCs. These results suggest sLA (50 mM) treatment does not notably modulate the expression of any inflammatory markers for DCs but downregulates CD11c.
Figure 1.
Effects of sLA on immature and stimulated DC phenotype. DCs were treated with 50 mM sLA and incubated for 48 or 120 h (a). In resistance maturation experiment (b), DCs were pretreated with 50 mM sLA for 48 or 120 h. After sLA treatment, cells were cultured for an additional 24 h with 1 µg/mL of LPS in the absence of sLA (b). LPS only treatment at a concentration of 1 µg/mL was used as a stimulated control (a, b). Cells were then stained with fluorescently-tagged anti-CD11c, CD80, CD86, and MHCII. Flow cytometry was used to analyze the expression of the surface markers. Data is presented as the mean ± S.E. (n ≥ 3 independent experiments) and statistically significant differences were calculated in comparison with the immature cell control (a). In resistance maturation experiment, the statistically significant differences were calculated in comparison with LPS only treated group (b). The following symbols represent the indicated values: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 as determined by a one-way analysis of variance (ANOVA), followed by Dunnett’s multiple-comparison test.
Lactate Exposure Prevents Maturation of DCs
To further investigate the immunomodulatory effect of sLA on mature DCs, a resistance maturation experiment was performed as previously described by Allen et al.1 Dendritic cells were challenged with LPS after sLA treatment for 48 and 120 h. CD11c, CD80, CD86, and MHCII expression was quantified using flow cytometry. The expression of these molecules relative to LPS only group is shown in Figs. 1b and S4. At both incubation time points, lactate treatment induced a significant decrease in CD11c expression, relative to LPS control. Notably, the iDC control displayed very similar expression of CD11c to LPS control. Compared to the LPS control, CD80 expression for sLA-exposed cells was increased at 48 h and decreased at 120 h. At 48 h, CD86 expression for sLA-exposed cells was significantly lower than the LPS control. However, at 120 h, CD86 expression was significantly higher than the LPS control. Lactate-treated cells downregulated MHCII expression at 48 h, compared to the LPS control, and the decrease was further pronounced after 120 h incubation. Lipopolysaccharide and sLA treatment showed minimal cytotoxicity to DCs as shown by the viability result of DCs cultured with LPS after sLA pretreatment (Fig. S5).
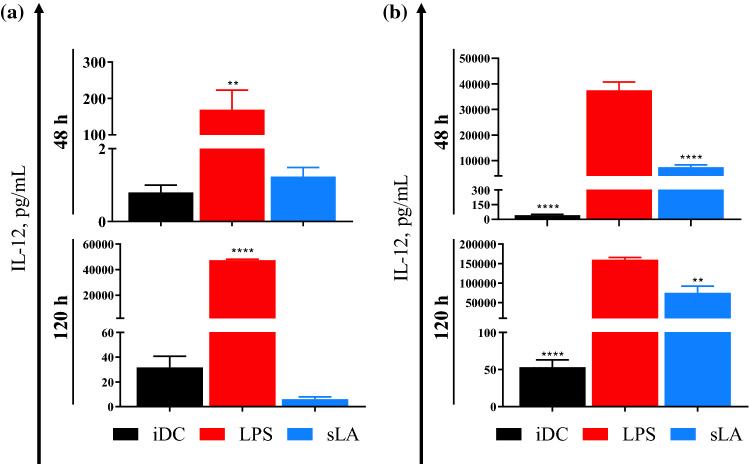
IL-12 Secretion from DCs is Inhibited, Even under LPS Stimulation
IL-12 is a pro-inflammatory cytokine that activates T cells through a Th1 response.30 The effect of sLA on IL-12 secretion from mature and iDCs was assessed and are depicted in Fig. 2. The results showed that LPS was able to activate IL-12 secretion as demonstrated by the strong secretion at both incubation time points. At 48 h, IL-12 secretion in sLA-treated DCs was higher than iDC control. However, at 120 h, the level of IL-12 was lower in the sLA-treated DCs compared to iDCs (Fig. 2a). In the maturation resistance experiments, there were substantial differences in IL-12 secretion between LPS-stimulated DCs and sLA-treated group at both incubation time points (Fig. 2b). Additionally, we evaluated IL-10 concentrations in culture media, but their levels were below the detection limit of the IL-10 ELISA kit (data not shown).
Figure 2.
sLA effects on IL-12 secretion from immature and stimulated DCs. Supernatants were collected from DCs co-cultured with 50 mM sLA after 48 and 120 h (a). For resistance maturation experiment, DCs were treated with 50 mM sLA for 48 and 120 h (b). After sLA incubation, cells were challenged with 1 µg/mL LPS for 24 h in the absence of sLA. The supernatants were collected after LPS challenge. LPS only treatment at a concentration of 1 µg/mL was used as a stimulated control (a, b). IL-12 levels were quantified using a sandwich ELISA. Data is presented as the mean ± S.E. (n ≥ 3 independent experiments) and statistically significant differences were calculated in comparison with the immature cell control. (a) In resistance maturation experiment, the statistically significant differences were calculated in comparison with LPS only treated group (b). The following symbols represent the indicated values: **p < 0.01 and ****p < 0.0001, as determined by a one-way analysis of variance (ANOVA), followed by Dunnett’s multiple-comparison test.
Macrophages are Polarized Towards M2 Phenotype in the Presence of Lactate
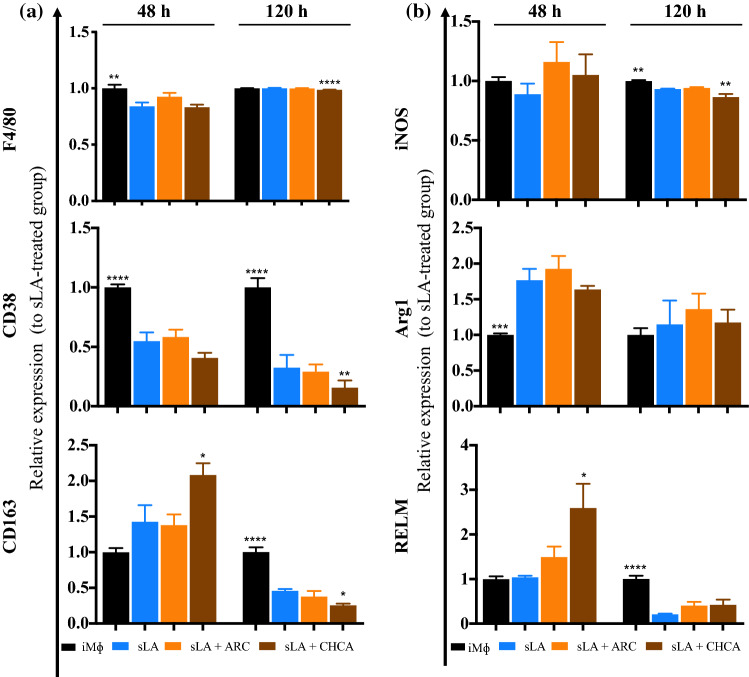
MΦs are plastic cells and can be polarized into two main phenotypes–classically (M1) and alternatively activated (M2) MΦs. In the context of cancer, M1 MΦs have the capacity to kill and remove tumor cells, on the other hand, M2 MΦs promote tumor growth and progression.2,26,35,41,48 CD38 and inducible nitric oxide synthase (iNOS) are known to be markers for M1 MΦs, while CD163, arginase 1 (Arg1), and resistin-like molecule (RELM) are considered M2 markers.2,26,35,41,48 To test the sLA effect on MΦ phenotype, we conducted experiments similar to those performed on DCs. MΦs were exposed to sLA at 50 mM for 48 and 120 h. Several surface and intracellular markers were investigated to determine MΦ polarization. The F4/80 marker was used identify MΦs and the expression of CD38, CD163, iNOS, Arg1, and RELM were also evaluated (Figs. 3 and S6). The expression of F4/80 and CD38 markers was upregulated in LPS-challenged MΦs, but the expression of CD163 marker was downregulated when compared to iMΦs. The results showed that the expression of F4/80 was reduced in sLA-treated MΦs at the 48 h time point, but was comparable to iMΦs after 120 h, respectively. A significant decrease in the expression of CD38 marker at both incubation time points was observed for sLA-treated MΦs, compared to iMΦ control. Contrastingly, the expression of CD163 marker was upregulated at both time points in sLA treatment, compared to iMΦ control. The results for the expression of intracellular markers are demonstrated in Figs. 3 and S6. Lipopolysaccharide induced the expression of iNOS and Arg1 markers while suppressing the expression of RELM at 48 h exposure. The expression of iNOS was downregulated after exposure to sLA for 48 h and 120 h. Additionally, sLA increased the expression of Arg1 at both incubation time points but had no effect on the RELM expression. The viability of MΦs cultured with sLA or LPS is shown in Fig. S7. Lipopolysaccharide showed minimal cytotoxicity to MΦs but no toxicity of sLA was observed. We also investigated chemokine secretions including CCL1, CCL9, and TGF-β in culture media after sLA treatment. The levels of CCL9 and TGF-β between sLA-treated MΦ and iMΦ were similar (Figs. S8–S9). CCL1 levels from culture media were undetectable (data not shown).
Figure 3.
MΦ phenotype after sLA exposure. MΦs were treated with 50 mM sLA and incubated for 48 or 120 h. LPS at a concentration of 0.01 µg/mL was used as a stimulating control. Cells were then stained with fluorescently-tagged anti-F4/80, CD38, CD163, iNOS, Arg1, and RELM. Flow cytometry was used to analyze the expression of the functional markers. Data is presented as the mean ± S.E. (n ≥ 3 independent experiments) and statistically significant differences were determined in comparison to the iMΦs. The following symbols represent the indicated values: *p < 0.05, ***p < 0.001, and ****p < 0.0001, as determined by a one-way analysis of variance (ANOVA), followed by Dunnett’s multiple-comparison test.
CCR7 Expression is Downregulated in Lactate
C-C chemokine receptor type 7 (CCR7) is an important molecule that is involved with the migration of antigen-presenting cells to adjacent lymph nodes for stimulation of T cells for appropriate immune responses.23 To study the effect of sLA on the migratory ability of DCs and MΦs, the expression of CCR7 were examined in DCs and MΦs. Bone marrow-derived DCs and MΦs were treated with 50 mM sLA for 48 and 120 h. The relative expression of CCR7 marker was normalized to the iDC or iMΦ population (Figs. 4 and S10). At 48 h, the level of CCR7 in cells exposed to sLA was significantly reduced on DCs and decreased in MΦs relative to the levels in immature cells. Similarly, at 120 h the expression of CCR7 remained downregulated in DCs and significantly decreased for MΦs compared to the levels of CCR7 in the corresponding controls.
Figure 4.
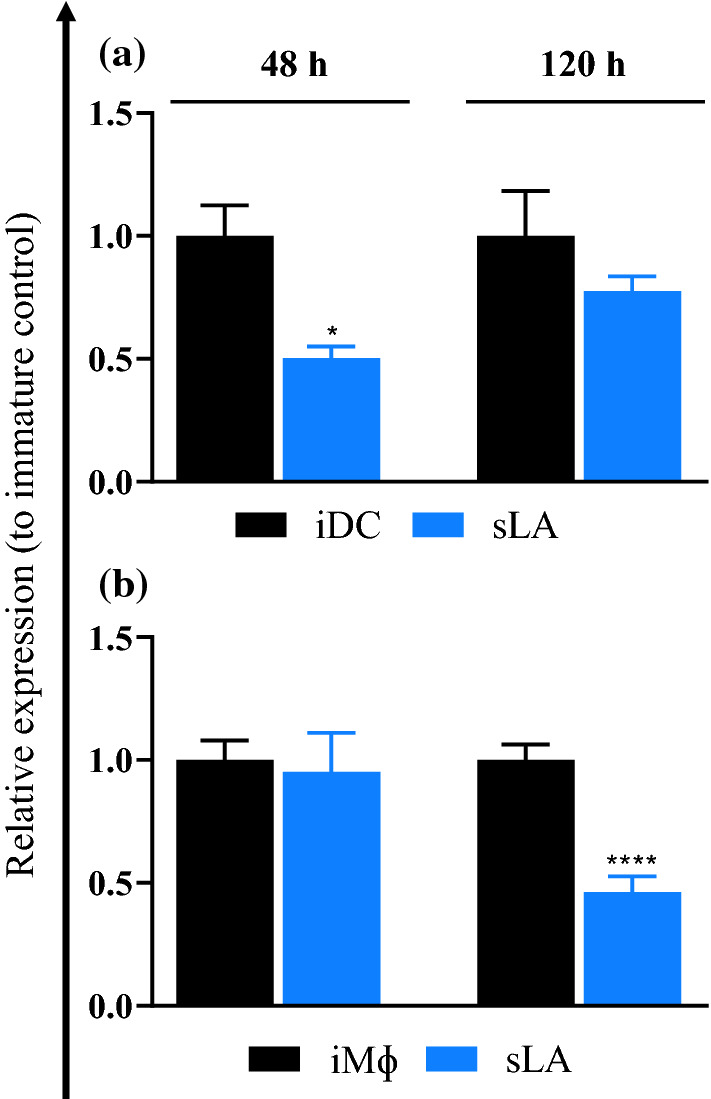
The effect of sLA exposure on CCR7 expression in DCs and MΦs. DCs (a) or MΦs (b) were treated with 50 mM sLA and incubated for 48 or 120 h. DCs and MΦs were then stained with fluorescently-tagged antibody for CCR7. Flow cytometry was used to analyze the expression of the surface marker. Data is presented as the mean ± S.E. (n = 2 [for DCs], and n = 3 [for MΦs] independent experiments). The following symbols represent the indicated values: *p < 0.05 and ****p < 0.0001 as determined by student’s t test.
Extended Lactate Exposure Results in Hyporesponsive T Cells to Allogeneic DC Challenge
Innate immune cells, such as DCs and MΦs, play a pivotal role in activating the adaptive immune system. In this regard, we wanted to test the allostimulatory capacity of DCs and MΦs following sLA exposure. To measure the difference in immunogenicity of sLA-treated DCs/MΦs from C57BL/6 mice, the proliferation of BALB/cByJ CD4+ T cells was quantified subsequent to co-culturing in a mixed lymphocyte reaction (MLR). T cells were stained and quantified with CFSE dye dilution and the percent proliferation in the co-cultured treatment group was normalized to the proliferation of iDCs/iMΦs plus CD4+ T cells group (iDCs/ iMΦs; Figs. 5 and S11). The control groups in this experiment included T cell only (negative control), T cells treated with PMA/Ionomycin (IONO) (positive control), and T cells co-cultured with iDCs or iMΦs. T cell only proliferation was expectedly lower in all time points than all other treatment groups for both DCs and MΦs. For PMA/IONO treatment group, all time points except 48 h, T cells exhibited a significantly higher proliferation rate compared to all other treatment groups. Likely an anomaly, the 48 h time point had a significant less T cell proliferation compared to the iDC control. At 48 h, sLA-treated DCs prompted less proliferation in allogeneic T cells than iDCs. At the 120 h time point, the proliferative response to sLA-treated DCs was significantly less. The relative proliferations at both time points for sLA-treated MΦs co-cultures were not dramatically altered. However, it should be noted that the proliferative response was of T cells exposed to sLA-treated macrophages was less than their respective iMΦ control.
Figure 5.

T cell proliferation for mixed lymphocyte reaction (MLR) between allogenic T cells and DCs or MΦs. DCs (a) or MΦs (b) were pretreated with 50 mM sLA for 48 or 120 h and then cultured with allogenic CD4+ T cells for 72 h in the absence of sLA. Cell proliferation was measured by CFSE dye dilution. Cell populations derived from T cells only and from T cells exposed to stimulatory cocktail of PMA and ionomycin were used as controls. Data present as the mean ± S.E. (n ≥ 3 independent experiments). Statistically significant differences were calculated in comparison with the immature cell control. The following symbols represent the indicated values: *p < 0.05, **p < 0.01, and ****p < 0.0001 as determined by a one-way analysis of variance (ANOVA), followed by Dunnett’s multiple-comparison test.
MCT4 Inhibitor Exacerbates the Effects of Lactate on Innate Immune Cells
The monocarboxylate transporters are membrane proteins that facilitate the transport of monocarboxylates such as pyruvate, LA, and ketone bodies across cell membrane. MCT comprises 14 members, of which MCT1, 2, and 4 have been extensively studied and characterized. MCT1/2 facilitate the transport of sLA into the cells, while MCT4 helps export sLA to the extracellular environment.7 We hypothesized that inhibition of MCTs would affect the response of innate immune cells to sLA. To block sLA shuttling, we treated MΦs with MCT inhibitors—along with 1% DMSO as a diluent. ARC and CHCA, generally regarded as inhibitors of MCT1/2 or MCT1/2/4, respectively, were used according to procedures described previously.13,20,47,59,69 We determined the viability of MΦs co-cultured with the MCT inhibitors and/or sLA (Fig. S13). There was no cytotoxicity of the inhibitors in the presence of sLA. After 48 h incubation with MCT inhibitors and/ or sLA, surface and intracellular markers including F4/80, CD38, CD163, iNOS, Arg1, and RELM were fluorescently labelled. Expression of all surface markers in the ARC and sLA co-treatment group were comparable to sLA-treated group (Figs. 6 and S12). Overall, ARC failed to modulate both extracellular and intracellular marker expression in comparison to the sLA-treated group. Treatment with CHCA diminished CD38 and this decrease was even more pronounced after 120 h. Compared to MΦs treated with sLA only, CD163 expression levels were higher in MΦ exposed to sLA and CHCA after 48 h, but lower after 120 h. In comparison with the sLA treatment group, iNOS level was upregulated and downregulated in CHCA treatment at 48 and 120 h post incubation, respectively. Arg1 level in MΦs co-treated with CHCA and sLA was not significantly different from sLA treatment group. At the 48 h time point, there was a dramatic increase in RELM expression in MΦs from CHCA and sLA co-treatment. However, at 120 h the RELM expression in MΦs exposed to both CHCA and sLA was comparable to that of sLA-treated MΦs. Interestingly, all groups that had sLA had reduced expression of RELM at the time, in comparison to the M0 macrophages. Overall, the treatment with ARC had little to no effect on the sLA-mediated responses. Conversely, the treatment with CHCA led to upregulation of M2 markers including CD163, RELM, and downregulation of M1 markers, CD38 and iNOS.
Figure 6.
MΦ phenotype after sLA exposure and MCT inhibitors. MΦs were co-treated with media (control), 50 mM sLA, ARC, and/or CHCA for 48 and 120 h. Cells were then stained with fluorescently-tagged antibodies for F4/80, CD38, CD163, iNOS, Arg1, and RELM. Flow cytometry was used to analyze the expression of these markers. Data is presented as the mean ± S.E. (n ≥ 3 independent experiments). The statistically significant differences were calculated in comparison with the sLA-treated group. The following symbols represent the indicated values: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 as determined by a one-way analysis of variance (ANOVA), followed by Dunnett’s multiple-comparison test.
Differential Expression of Genes in sLA-Treated Innate Immune Cells
To assess the potential underlying molecular mechanisms of lactate immunomodulation, RNA sequencing was performed on DCs treated with media (control) or sLA (50 mM) for 48 h. Any genes with less than 2 counts per million reads were discarded and 10856 DC genes were identified using differential gene expression. Raw p-values, adjusted p-values, average expression, and log2(fold change) (log2FC) values were generated for each gene. By setting a threshold of adjusted p-value < 0.05 and |log2FC| > 1, 479 significantly differentially expressed DC genes were found. Of these genes, 185 were upregulated (log2FC > 1) and 294 were downregulated (log2FC < − 1). Cutoffs for adjusted p-value and log2FC can be visualized in the volcano plot with all DC gene data (Fig. 7a). The highest upregulated genes for sLA-treated DCs were Fxyd2, Slco4a1, Serpini1, Lhfp, and Col27a1. The top downregulated genes identified were Ccna2, Clec12a, Adgre4, Cenpf, and Abcg3. G protein-coupled receptor genes with altered expression were also identified. Gprc5b was highly upregulated while Gpr34, Gpr183, Gpr35, and Gpr155 were downregulated. Several solute carrier family gene expression changes were also found. Upregulated genes in this family were Slco4a1, Slc6a9, Slc5a3, Slc6a12, Slc6a4, and Slc39a2. Downregulated genes were Slco2b1, Slc9a9, and Slc18a1. To further investigate the functions of these altered genes, GO enrichment analysis was performed on R using topGO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was done on R via KEGGREST. Any GO and KEGG terms with p-values < 0.05 associated with sLA-induced differential DC gene expression were identified (Fig. 7c). Gene ontology enrichment analysis yielded 18 significantly enriched biological process ontology terms, 6 significantly enriched molecular function ontology terms, and 9 significantly enriched cellular compartment ontology terms. The 4 GO: biological process ontology terms with the lowest p-values were “receptor-mediated endocytosis”, “phagocytosis, engulfment”, “innate immune response”, and “transmembrane receptor protein tyrosine kinase signaling pathway”. The highest GO: molecular function ontology terms were “protein homodimerization activity”, “protein C-terminus binding”, “magnesium ion binding”, and “Rab guanyl-nucleotide exchange factor activity”. The most significant GO: cellular compartment ontology terms were “nucleoplasm”, “anchored component of external side of plasma membrane”, “transcriptional repressor complex”, and “perinuclear region of cytoplasm”. KEGG pathway analysis revealed 19 significantly enriched pathways associated with the sLA-induced DC gene changes, with the top 4 pathways of “MicroRNAs in cancer”, “Parathyroid hormone synthesis, secretion and action”, “Signaling pathways regulating pluripotency of stem cells”, and “Mineral absorption”.
Figure 7.
Volcano plots were generated using the EnhancedVolcano package in R for 50 mM sLA treatment of (a) DCs and (b) MΦs. Data points represent differential gene expression comparisons between sLA treated and control conditions. Requirements for significantly differentially expressed genes were an adjusted p-value < 0.05 and a |log2FC| > 1. Data points colored grey are not significant, green have a |log2FC| > 1, blue have an adjusted p-value < 0.05, and red have both |log2FC| > 1 and adjusted p-value < 0.05. Differentially expressed genes of sLA-treated DCs were narrowed down from 10856 genes to 479 genes using log2FC and p-value cutoff requirements. Differentially expressed genes of sLA treated MΦs were also cut from 11217 genes down to 444 significant genes. (c, d) These significantly differentially expressed genes were then used to identify gene ontology (GO) and KEGG enrichment terms for DCs and MΦs. GO analysis was performed using topGO R package and KEGG analysis was performed in R using the KEGGREST package. Significant (p-value < 0.05) GO: Biological Process, GO: Cellular Compartment, GO: Molecular Function, and KEGG Pathway terms were plotted against their respective -log10(p-value) for both (C) DCs and (D) MΦs. Significance for differentially expressed genes was determined by Bejamini-Hochberg adjusted p-value < 0.05. The significance of GO terms was determined by p-value < 0.05 using a Kolmogorov-Smirnov test. KEGG terms were deemed significant on the basis of a Wilcoxon rank-sum test derived p-value < 0.05. Each treatment consisted of 5 biological replicates and 3 technical replicates.
RNA sequencing was also performed on MΦs treated with media (control) or sLA (50 mM) for 48hrs. 11217 MΦ genes with more than 2 counts per million reads were identified via differential gene expression analysis. Of these, 444 significantly altered genes were identified after sLA treatment based requirements of adjusted p-value < 0.05 and |log2(fold change)| > 1. Of these genes, 245 were upregulated and 199 were downregulated. These results can be visualized on a volcano plot shown in Fig. 7b. The highest upregulated genes were Slco4a1, Slc6a12, Fxyd2, Tal2, and Ly6a; and the highest downregulated genes were Myh15, Shisa9, Nptx1, Dcstamp, and Ocstamp. Expression of several G protein-coupled receptor genes was modified, with upregulation of Gpr141 and downregulation of Gpr34, Gpr171, Gpr157, and Gpr162. Many solute carrier family proteins were also significantly differentially expressed. There was high upregulation of genes Slco4a1, Slc6a12, Slc6a13, Slc5a3, Slc39a4, Slc2a6, and Slco3a1. Also, gene downregulation was observed for Slc40a1, Slc24a5, Slc24a3, Slc7a1, Slc26a11, and Slc6a8. Gene ontology enrichment analysis and KEGG pathway analysis were performed. Significant (p-value < 0.05) GO and KEGG terms were identified for sLA-induced differentially expressed MΦ genes (Fig. 7d) From the GO analysis, 16 significantly enriched biological process ontology terms, 4 significantly enriched cellular compartment ontology terms, and 4 significantly enriched molecular function ontology terms were found. The top 4 GO: biological process ontology terms were “innate immune response”, transmembrane transport”, “defense response to virus”, and “response to bacterium”. The 4 identified GO: cellular compartment ontology terms were “cytosol”, “nucleoplasm”, “synaptic vesicle”, and “fibrillar center”. The 4 identified GO: molecular function ontology terms were “identical protein binding”, “ATPase activity”, “sodium-independent organic anion transmembrane transporter activity”, and “GTPase activity”. From KEGG pathway analysis, 10 significantly enriched pathways were identified; the top 4 terms were “Mineral absorption”, “Pyrimidine metabolism”, “Hepatitis C”, and “RIG-I-like receptor signaling pathway”.
Discussion
Dendritic cells and MΦs are generally considered as prominent antigen-presenting cells having the ability to promote adaptive immunity response and present antigens, such as tumor-associated antigens, to T cells which subsequently suppress cancer progression and metastasis.6,46,54,55 Lactic acid secreted by tumors have been shown to help malignant cells avoid immune surveillance through the modulation of immune cells, including DCs, MΦs, and T cells.8,9,12,25,27,56 The effects posed by lactate exposure can be confounded by the accompanied acidic pH, as reported by Vermeulen et al.73 Evidently, the true effect of lactate on immune cells is a significant knowledge gap. Uncovering the sole effect of lactate on immunomodulation can further help develop successful immunotherapy and a better understanding of the behavior of lactate derived from biomaterials such as PLGA. Herein, we examined the effects of lactate on the immunogenicity of immune cells including DCs and MΦs.
Generally, CD11c and MHC–peptide complexes are expressed after DC differentiation from myeloid progenitor cells. Upon maturation, the expression of inflammatory markers in DCs, such as CD80, CD86, and MHCII is substantially increased.60 Once mature, they migrate to the lymph nodes where they interact with T cells and B cells to initiate the adaptive immune response.38 T cell activation primarily relies on the interaction between T-cell antigen receptors and MHC molecules on the surface of DC as well as the interaction through CD28 of T cells and stimulatory molecules on DC–CD80/CD86.15,74 In the present study, these inflammatory markers as well as a typical DC marker, CD11c, were used to investigate the immunomodulation of lactate in DCs. We observed the expression of CD80, CD86, and MHCII in sLA-treated cells to be comparable with iDC expression levels at 48 h. The results indicate that lactate (at cancer relevant concentrations) does not dramatically modulate the expression of inflammatory markers on DCs over a short period. However, we did observe a significant downregulation of CD11c in sLA-treated DCs and LPS-stimulated-DCs at both 48 and 120 h time points. Interestingly, a correlation between low CD11c expression and the risk of death and relapse in patients with gastric cancer has been demonstrated previously.78 The similarity in reduction of CD11c expression between lactate and LPS control may be explained by lactate interaction with TLR4 receptors reported by Samuvel et al..63 Further evidence to explain this phenomena was provided by the Decker group who showed that CD11c downregulation occurred after LPS activation.65
Lipopolysaccharide is a potent inflammatory mediator commonly used to induce maturation of innate immune cells, which after exposure to LPS, increase their expression of inflammatory molecules and secretion of stimulatory cytokines.37,68,77 To explore how sLA-treated DCs responded to LPS challenge, cells were incubated with LPS after sLA treatment and then immuno-stained for important maturation markers. We found that the effects of LPS on sLA-exposed DCs was time dependent. After 48 h incubation, 50 mM sLA failed to significantly deter LPS stimulation of the DCs. However, at the 120 h incubation time, CD11c, MHCII and CD80 expression were all downregulated compared to the LPS control. Additionally, after LPS challenge, the IL-12 levels for sLA-treated DCs were significantly lower than the LPS-only control at both time points. These results suggest IL-12 secretion from sLA-exposed DCs is inhibited, even under stimulation induced by LPS. These results are consistent with a similar maturation resistance experiment on the DCs previously performed by Allen et al.1 They reported that after 120 h of sLA exposure there was significantly lower expression of maturation markers (CD80, CD86, MHCII) in DCs. Consistently, we found that the effects of sLA exposure lessened the DC ability to mature. The increased CD86 expression after prolonged exposure to sLA may be a negative feedback mechanism, compensating the reduced expression of CD80 and MHCII.
As motioned earlier, MΦs can be divided into M1 and M2 phenotypes. M1 MΦs have antitumor capacity while M2 MΦs have protumoral functions.2,26,35,41,48 The phenotype of MΦs can be differentiate by the expression level of markers. CD38 has been designated as an inflammatory marker (M1) because it is selectively upregulated both in bone marrow-derived MΦs and in a murine model upon activation with inflammatory cytokines, such as LPS and IFN-γ or infectious disease models.2 Although the role of CD38 has not been extensively explored, it has been shown to be important during M1 activation and may regulate cellular processes such as cell adhesion, conversions of metabolic molecules, and Ca2+ signaling.2,41,48 M1 MΦs also upregulate inducible nitric oxide synthase (iNOS) that produces nitric oxide (NO) using L-arginine as a substrate.82 NO is an important pro-inflammatory mediator contributing to killing foreign microorganisms or cancer cells as well as normal cells. On the other hand, M2 MΦs express enzyme arginase 1 (Arg1), which catalyzes the hydrolysis of L-arginine to ornithine. Existing evidence showed that Arg1 was responsible for an inhibitory effect of TAMs or M2 MΦs on the activation of T cell responses and upregulation of wound healing process.4 Of common M2 markers, CD163 was studied extensively and was highly expressed in TAMs. It was found to be positively associated with tumor cell proliferation.64 Apart from Arg1 and CD163, resistin-like molecule (RELM) is also expressed in M2 MΦs, the function of which has been revealed as a regulator in restricting Th2 cell-mediated inflammation.50 In this regard, we used M1 markers (CD38 and iNOS) and M2 markers (CD163, Arg1, and RELM) to investigate the effect of sLA on MΦ polarization. Using these markers to investigate the role of sLA in MΦ phenotypes, our result indicates that sLA-activated MΦs are pushed toward M2 polarization through the downregulation of CD38 and upregulation of M2 markers, CD163 and RELM. In agreement with other studies published previously, LA secreted from cancer exerted immunosuppressive effect on MΦs.10,21,67
It is well established that CCR7 is essential for activation and mobilization of immune cells. In vitro studies have demonstrated that CCR7 was rapidly upregulated upon DC activation.66 A number of in vivo studies showed that CCR7-deficient mice had the impaired migration of DC from various primary organs including skin, intestinal lamina propria and lungs, to the draining lymph nodes.24,28,32,36,44,57,61,79 In the context of macrophages, CCR7 has been considered as one of the M1 markers and the role of CCR7 in recruitment/migration of M1 macrophages to the inflammation sites has also been revealed.81 Given, the importance of the CCR7, its expression in DCs and MΦs after sLA exposure was probed. sLA incubation resulted in the reduction of CCR7 expression in both DCs and MΦs, implying a diminished potential in migration to lymph nodes upon activation. Apart from migratory properties, the main function of antigen presenting cells is to activate T cells for initiating adaptive immune response.31,70 Antigen presenting cells migrate toward secondary lymphoid organs, where they stimulate T cells through receptors of T cells and molecules on APCs such as MHC, CD80 and CD8677, 78. We used a Mixed lymphocyte reaction (MLR), a standard in vitro assay widely used to determine the efficiency of antigen presenting cells in promoting T cell activation. MLR results provides evidence that particularly DCs after lactate exposure are unable to perform their primary function in activating T cells. Interestingly, MΦs maintained their capacity to stimulate T cells after lactate exposure. However, MΦs are poor stimulators of T cell responses in comparison to DCs.5 Allogeneic T cell hyporesponsiveness could be attributed to the low expression of stimulatory molecules, the low production of IL-12, and the M2 polarization after sLA exposure. Low expression of CCR7 together with impaired T cell activation suggested that tumor-resident innate immune cells could be trapped within the TME and therefore, likely to fail in prompting tumor-specific immunity at nearby lymph nodes. With respect to IL-12, several groups are now keenly investigating the introduction of IL-12 to solid tumors to boost cell-based immunotherapy responses.11,34,51
The mechanism of lactate uptake and export from DCs and MΦs has also been investigated previously.7 The SLC16A family of MCTs has been identified as a key transporter of lactate across the cell membrane MCT1 (Slc16a1 gene) and MCT2 (Slc16a7 gene) are responsible for uptake of lactate, while MCT3 (Slc16a8 gene) and MCT4 (Slc16a3 gene) can transport lactate out of the cell. To investigate the role of lactate shuttling in mediating innate immune phenotype, MΦs were treated with ARC for MCT1/2 inhibition or CHCA for MCT1/2/4 inhibition. We showed that the CHCA treatment boosts the suppressive effects of sLA in MΦs. However, the treatment with ARC had little or no effect on sLA-mediated responses, suggesting either that lactate-specific extracellular receptors may drive the observed suppressive effects of sLA, or that there may be other channels for sLA entry into the cell. Also, we found that prolonged exposure to DMSO, sLA and/or MCT inhibitors hindered the expression of CD163 and RELM, which was not consistent with our early results in just the presence of sLA (Fig. 6). This may be possibly due to the inhibiting effect of DMSO in the presence of lactate.16 Regarding gene expression, none of these SLC16A family genes were differentially expressed over the |log2FC| > 1 and adjusted p-value < 0.05 threshold. However, many of these genes were significantly differentially expressed, just with a less drastic fold change value. For instance, Slc16a1 expression was downregulated in sLA-treated MΦs (log2FC = − 0.4702, adjusted p-value = 0.0316) and no significant change was observed in sLA-treated DCs. Slc16a7 was also downregulated for DCs and MΦ s after sLA treatment (DC: log2FC = − 0.7523, adjusted p-value = 5.54E−05; MΦ: log2FC = − 0.3001, adjusted p-value = 6.82E−05). Expression of the Slc16a8 gene for the lactate exporter MCT3 was not significantly changed for DCs or MΦs. However, for the other lactate exporter MCT4, gene expression of Slc16a3 was significantly increased for both DCs and MΦs (DC: log2FC = 0.3094, adjusted p-value = 0.01251; MΦ: log2FC = 0.5742, adjusted p-value = 0.0085). The downregulation of MCT1/2 after lactate exposure helps explain why blocking this group of transporters had little effect on lactate-mediated response in macrophages. The results for gene expression also demonstrate that DCs and MΦs, upon exposure to high amounts of extracellular lactate, may alter their gene expression to reduce uptake (MCT1 and MCT2) and increase export (MCT4) in order to compensate for the environment. It was reported previously that CHCA treatment and MCT4 inhibition resulted in an intracellular accumulation of LA and decreased LA secretion. As such, CHCA treatment may contribute to the substantial adverse effects of sLA due to accumulation within the cell, correlating with studies reported by Stone et al.84 Other than the MCT proteins, several other solute carrier (SLC) family protein gene expression changes were also highlighted for both sLA-treated DCs and MΦs, with log2FC magnitude over 1 and adjusted p-value below 0.05. Overlapping upregulated SLC genes between DCs and MΦs were Slco4a1 (DC: log2FC = 3.5095, adjusted p-value = 1.21E−20; MΦ: log2FC = 7.4950, adjusted p-value = 1.31E−15), Slc6a12 (DC: log2FC = 1.6399, adjusted p-value = 1.43E−11; MΦ: log2FC = 5.9833, adjusted p-value = 4.70E−14), and Slc5a3 (DC: log2FC = 1.7867, adjusted p-value = 3.70E−15; MΦ: log2FC = 2.0597, adjusted p-value = 2.17E−18). Slco4a1 was the highest differentially expressed MΦ gene and second highest DC gene, signifying that this solute carrier may play an important role in response to lactate.
Previous work on immune cell response to LA/lactic acid highlighted associated G protein-coupled receptor pathways for proteins Gpr81 and Gpr132.10,62 Interestingly, the differential expression of both Gpr81 and Gpr132 genes were not deemed significant for both sLA-treated DCs and MΦs. Gpr132 expression was slightly increased but not significantly (DC: log2FC = 0.2894, adjusted p-value = 0.0992; MΦ: log2FC = 0.3573, adjusted p-value = 0.0559). Notably, the RNA sequencing data analysis identified several differentially expressed G protein-coupled receptor genes that fulfilled the cutoff requirement of |log2FC| > 1 and adjusted p-value < 0.05, implying that there may be other protein pathways involved in lactate signaling. For both DCs and MΦs, sLA treatment induced downregulation of Gpr34 (DC: log2FC = − 1.5973, adjusted p-value = 0.0023; MΦ: log2FC = − 1.4420, adjusted p-value = 1.38E−18). Differential expression of other solute carrier proteins and Gpr proteins apart from MCT proteins and Gpr81/Gpr132 could point to alternative or linked lactate pathways that have not been fully outlined in literature.
In conclusion, sLA, at a 50 mM concentration, does not modulate DC phenotype, but inhibits DC maturation in the presence of LPS, an inflammatory mediator. Moreover, sLA-treated DCs are poor stimulators for allogeneic T cells. Further, sLA promotes polarization towards an M2 phenotype. Our MCT inhibition studies indicate that lactate modulation of innate immune cells involves extracellular surface receptors. Intracellular accumulation of lactate can also have powerful effects, but the mechanisms involved are yet to be elucidated. Transcriptomic profiling provided some significant insight on the interactions of lactate with innate immune cells. Moreover, Gpr34 may be a pivotal molecule in lactate signaling and observed immune-modulatory effects in innate immune cells. Further analysis of the molecules identified in this study needs to be performed. However, the potential impact of deciphering lactate modulation is quite significant and powerful as we seek to advance the efficacy of cancer immune therapies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (Grants R35125012 and R01AI139399).
Author Contributions
RS contributed to the design and execution of experiments, analysis of data, and compilation of the manuscript. BT and HH contributed to the execution of experiments and analysis of data. NP contributed to the analysis of RNA sequencing data and discussion on gene expression. RA provided input on experimental protocols. JSL contributed to the design and execution of experiments, analysis of data, manuscript compilation and has primary responsibility for the content of the manuscript.
Conflict of interest
Rapeepat Sangsuwan, Bhasirie Thuamsang, Noah J. Pacifici, Hyunsoo Han, Svetlana Miakicheva, Riley Allen, and Jamal S. Lewis have no conflicts of interest to disclose.
Research Involving Human and Animal Rights
C57BL/6 and BALB/cByJ mice were purchased from Jackson Laboratories and were housed in specific pathogen-free environment conditions at the University of California, Davis TRACS facility and used according to the UC Davis Institutional Animal Care and Use Committee (IACUC).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Allen RP, Bolandparvaz A, Ma JA, Manickam VA, Lewis JS. Latent, immunosuppressive nature of poly(lactic-co-glycolic acid) microparticles. Acs Biomater. Sci. Eng. 2018;4:900–918. doi: 10.1021/acsbiomaterials.7b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amici SA, et al. CD38 is robustly induced in human macrophages and monocytes in inflammatory conditions. Front. Immunol. 2018;9:1593. doi: 10.3389/fimmu.2018.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annesley CE, Summers C, Ceppi F, Gardner RA. The evolution and future of CAR T cells for B-cell acute lymphoblastic leukemia. Clin. Pharmacol. Ther. 2018;103:591–598. doi: 10.1002/cpt.950. [DOI] [PubMed] [Google Scholar]
- 4.Arlauckas SP, et al. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics. 2018;8:5842. doi: 10.7150/thno.26888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banchereau, J. et al. Immunobiology of dendritic cells. Annu. Rev. Immunol.18, 767- + , 10.1146/annurev.immunol.18.1.767 (2000). [DOI] [PubMed]
- 6.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 7.Bosshart PD, Kalbermatter D, Bonetti S, Fotiadis D. Mechanistic basis of L-lactate transport in the SLC16 solute carrier family. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-10566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Bronte V. Macrophage response to lactic acid Tumor cells hijack macrophages via lactic acid. Immunol. Cell Biol. 2014;92:647–649. doi: 10.1038/icb.2014.67. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, et al. Gpr132 sensing of lactate mediates tumor–macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 2017;114:580–585. doi: 10.1073/pnas.1614035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi XW, et al. Significantly increased anti-tumor activity of carcinoembryonic antigen-specific chimeric antigen receptor T cells in combination with recombinant human IL-12. Cancer Med. 2019;8:4753–4765. doi: 10.1002/cam4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature513, 559– + , 10.1038/nature13490 (2014). [DOI] [PMC free article] [PubMed]
- 13.Colen CB, et al. Metabolic targeting of lactate efflux by malignant glioma inhibits invasiveness and induces necrosis: an in vivo study. Neoplasia. 2011;13:620–632. doi: 10.1593/neo.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbet C, Feron O. Tumour acidosis: from the passenger to the driver’s seat. Nat. Rev. Cancer. 2017;17:577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 15.Crespo HJ, Lau JT, Videira PA. Dendritic cells: a spot on sialic acid. Front. Immunol. 2013;4:491. doi: 10.3389/fimmu.2013.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng R, et al. Dimethyl sulfoxide suppresses mouse 4T1 breast cancer growth by modulating tumor-associated macrophage differentiation. J. Breast Cancer. 2014;17:25–32. doi: 10.4048/jbc.2014.17.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dienel GA. Brain lactate metabolism: the discoveries and the controversies. J. Cereb. Blood Flow Metab. 2012;32:1107–1138. doi: 10.1038/jcbfm.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elghetany MT. Surface marker abnormalities in myelodysplastic syndromes. Haematologica. 1998;83:1104–1115. [PubMed] [Google Scholar]
- 19.Elhelu MA. The role of macrophages in immunology. J. Natl Med. Assoc. 1983;75:314–317. [PMC free article] [PubMed] [Google Scholar]
- 20.Enerson BE, Drewes LR. Molecular features, regulation, and function of monocarboxylate transporters: implications for drug delivery. J. Pharm. Sci. 2003;92:1531–1544. doi: 10.1002/jps.10389. [DOI] [PubMed] [Google Scholar]
- 21.Errea, A. et al. Lactate inhibits the pro-inflammatory response and metabolic reprogramming in murine macrophages in a GPR81-independent manner. PLoS One11 (2016). [DOI] [PMC free article] [PubMed]
- 22.Fischer K, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 23.Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 24.Förster R, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 25.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genard G, Lucas S, Michiels C. Reprogramming of tumor-associated macrophages with anticancer therapies: radiotherapy versus chemo-and immunotherapies. Front. Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottfried E, et al. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood. 2006;107:2013–2021. doi: 10.1182/blood-2005-05-1795. [DOI] [PubMed] [Google Scholar]
- 28.Gunn MD, et al. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas, R. et al. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol.13 (2015). [DOI] [PMC free article] [PubMed]
- 30.Hamza T, Barnett JB, Li B. Interleukin 12 a key immunoregulatory cytokine in infection applications. Int. J. Mol. Sci. 2010;11:789–806. doi: 10.3390/ijms11030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He YT, Zhang Q-M, Kou QC, Tang B. In vitro generation of cytotoxic T lymphocyte response using dendritic cell immunotherapy in osteosarcoma. Oncol. Lett. 2016;12:1101–1106. doi: 10.3892/ol.2016.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hintzen G, et al. Induction of tolerance to innocuous inhaled antigen relies on a CCR7-dependent dendritic cell-mediated antigen transport to the bronchial lymph node. J. Immunol. 2006;177:7346–7354. doi: 10.4049/jimmunol.177.10.7346. [DOI] [PubMed] [Google Scholar]
- 33.Hou B, Tang Y, Li WH, Zeng QN, Chang DM. Efficiency of CAR-T therapy for treatment of solid tumor in clinical trials: a meta-analysis. Dis. Markers. 2019 doi: 10.1155/2019/3425291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang MTP, Fecek RJ, Qin TY, Storkus WJ, Wang YD. Single injection of IL-12 coacervate as an effective therapy against B16-F10 melanoma in mice. J. Control. Release. 2020;318:270–278. doi: 10.1016/j.jconrel.2019.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jablonski, K. A. et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS One10 (2015). [DOI] [PMC free article] [PubMed]
- 36.Jang MH, et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J. Immunol. 2006;176:803–810. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- 37.Jin P, et al. Molecular signatures of maturing dendritic cells: implications for testing the quality of dendritic cell therapies. J. Transl. Med. 2010;8:4. doi: 10.1186/1479-5876-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Mooney DJ. In vivo modulation of dendritic cells by engineered materials: towards new cancer vaccines. Nano today. 2011;6:466–477. doi: 10.1016/j.nantod.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiseleva, Y. Y., Shishkin, A. M., Ivanov, A. V., Kulinich, T. M. & Bozhenko, V. K. Car T-cell therapy of solid tumors: promising approaches to modulating antitumor activity of car T cells. Bulletin of Russian State Medical University, 5-12, 10.24075/brsmu.2019.066 (2019).
- 40.Kreutz M, et al. Tumor-derived lactic acid modulates dendritic cell activation and differentiation. Blood. 2004;104:147B. doi: 10.1182/blood-2005-05-1795. [DOI] [PubMed] [Google Scholar]
- 41.Lam JH, et al. Expression of CD38 on macrophages predicts improved prognosis in hepatocellular carcinoma. Front. Immunol. 2019;10:2093. doi: 10.3389/fimmu.2019.02093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanier L, et al. Correlation of functional properties of human lymphoid cell subsets and surface marker phenotypes using multiparameter analysis and flow cytometry. Immunol. Rev. 1983;74:143–160. doi: 10.1111/j.1600-065x.1983.tb01088.x. [DOI] [PubMed] [Google Scholar]
- 43.Lewis JS, Zaveri TD, Crooks CP, II, Keselowsky BG. Microparticle surface modifications targeting dendritic cells for non-activating applications. Biomaterials. 2012;33:7221–7232. doi: 10.1016/j.biomaterials.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martín-Fontecha A, et al. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 2003;198:615–621. doi: 10.1084/jem.20030448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendler AN, et al. Tumor lactic acidosis suppresses CTL function by inhibition of p38 and JNK/c-Jun activation. Int. J. Cancer. 2012;131:633–640. doi: 10.1002/ijc.26410. [DOI] [PubMed] [Google Scholar]
- 46.Mills CD, Lenz LL, Harris RA. A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res. 2016;76:513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miranda-Goncalves V, et al. Monocarboxylate transporters (MCTs) in gliomas: expression and exploitation as therapeutic targets. Neuro-oncology. 2013;15:172–188. doi: 10.1093/neuonc/nos298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morandi F, et al. CD38: a target for immunotherapeutic approaches in multiple myeloma. Front. Immunol. 2018;9:2722. doi: 10.3389/fimmu.2018.02722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriarty AT, Wiersema L, Snyder W, Kotylo PK, McCloskey DW. Immunophenotyping of cytologic specimens by flow cytometry. Diagn. Cytopathol. 1993;9:252–258. doi: 10.1002/dc.2840090303. [DOI] [PubMed] [Google Scholar]
- 50.Nair MG, et al. Alternatively activated macrophage-derived RELM-α is a negative regulator of type 2 inflammation in the lung. J. Exp. Med. 2009;206:937–952. doi: 10.1084/jem.20082048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakao, S. et al. Intratumoral expression of IL-7 and IL-12 using an oncolytic virus increases systemic sensitivity to immune checkpoint blockade. Sci. Transl. Med.12, 10.1126/scitranslmed.aax7992 (2020). [DOI] [PubMed]
- 52.Nasi A, Rethi B. Disarmed by density A glycolytic break for immunostimulatory dendritic cells? Oncoimmunology. 2013 doi: 10.4161/onci.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ngo H, Tortorella SM, Ververis K, Karagiannis TC. The Warburg effect: molecular aspects and therapeutic possibilities. Mol. Biol. Rep. 2015;42:825–834. doi: 10.1007/s11033-014-3764-7. [DOI] [PubMed] [Google Scholar]
- 54.Nikolic T, Roep B. Regulatory multitasking of tolerogenic dendritic cells–lessons taken from vitamin d3-treated tolerogenic dendritic cells. Front. Immunol. 2013;4:113. doi: 10.3389/fimmu.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nouri-Shirazi M, et al. Dendritic cells capture killed tumor cells and present their antigens to elicit tumor-specific immune responses. J. Immunol. 2000;165:3797–3803. doi: 10.4049/jimmunol.165.7.3797. [DOI] [PubMed] [Google Scholar]
- 56.Ohashi T, et al. M2-like macrophage polarization in high lactic acid-producing head and neck cancer. Cancer Sci. 2017;108:1128–1134. doi: 10.1111/cas.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohl L, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 58.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Ovens MJ, Davies AJ, Wilson MC, Murray CM, Halestrap AP. AR-C155858 is a potent inhibitor of monocarboxylate transporters MCT1 and MCT2 that binds to an intracellular site involving transmembrane helices 7-10. Biochem. J. 2010;425:523–530. doi: 10.1042/bj20091515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pearce EJ, Everts B. Dendritic cell metabolism. Nat. Rev. Immunol. 2015;15:18–29. doi: 10.1038/nri3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Randolph GJ, Angeli V, Swartz MA. Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat. Rev. Immunol. 2005;5:617–628. doi: 10.1038/nri1670. [DOI] [PubMed] [Google Scholar]
- 62.Ranganathan P, et al. GPR81, a cell-surface receptor for lactate, regulates intestinal homeostasis and protects mice from experimental colitis. J. Immunol. 2018;200:1781–1789. doi: 10.4049/jimmunol.1700604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samuvel DJ, Sundararaj KP, Nareika A, Lopes-Virella MF, Huang Y. Lactate boosts TLR4 signaling and NF-κB pathway-mediated gene transcription in macrophages via monocarboxylate transporters and MD-2 up-regulation. J. Immunol. 2009;182:2476–2484. doi: 10.4049/jimmunol.0802059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiraishi D, et al. CD163 is required for protumoral activation of macrophages in human and murine sarcoma. Cancer Res. 2018;78:3255–3266. doi: 10.1158/0008-5472.CAN-17-2011. [DOI] [PubMed] [Google Scholar]
- 65.Singh-Jasuja H, et al. The mouse dendritic cell marker CD11c is down-regulated upon cell activation through Toll-like receptor triggering. Immunobiology. 2013;218:28–39. doi: 10.1016/j.imbio.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 66.Sozzani S, et al. Cutting edge: differential regulation of chemokine receptors during dendritic cell maturation: a model for their trafficking properties. J. Immunol. 1998;161:1083–1086. [PubMed] [Google Scholar]
- 67.Stone SC, et al. Lactate secreted by cervical cancer cells modulates macrophage phenotype. J. Leukoc. Biol. 2019;105:1041–1054. doi: 10.1002/JLB.3A0718-274RR. [DOI] [PubMed] [Google Scholar]
- 68.Takashiba S, et al. Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor κB. Infect. Immun. 1999;67:5573–5578. doi: 10.1128/iai.67.11.5573-5578.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan Z, et al. The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J. Biol. Chem. 2015;290:46–55. doi: 10.1074/jbc.M114.603589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tourkova IL, Yurkovetsky ZR, Shurin MR, Shurin GV. Mechanisms of dendritic cell-induced T cell proliferation in the primary MLR assay. Immunol. Lett. 2001;78:75–82. doi: 10.1016/s0165-2478(01)00235-8. [DOI] [PubMed] [Google Scholar]
- 71.Umansky V. Immunosuppression in the tumor microenvironment: Where are we standing? Semin. Cancer Biol. 2012;22:273–274. doi: 10.1016/j.semcancer.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Veglia F, Gabrilovich DI. Dendritic cells in cancer: the role revisited. Curr. Opin. Immunol. 2017;45:43–51. doi: 10.1016/j.coi.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vermeulen M, et al. Acidosis improves uptake of antigens and MHC class I-restricted presentation by dendritic cells. J. Immunol. 2004;172:3196–3204. doi: 10.4049/jimmunol.172.5.3196. [DOI] [PubMed] [Google Scholar]
- 74.Villadangos JA, Schnorrer P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat. Rev. Immunol. 2007;7:543–555. doi: 10.1038/nri2103. [DOI] [PubMed] [Google Scholar]
- 75.Walenta, S. & Mueller-Klieser, (2017) W. F. in Semin. Radiat. Oncol. 267-274 (Elsevier). [DOI] [PubMed]
- 76.Walenta S, Schroeder T, Mueller-Klieser W. Lactate in solid malignant tumors: potential basis of a metabolic classification in clinical oncology. Curr. Med. Chem. 2004;11:2195–2204. doi: 10.2174/0929867043364711. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y-C, et al. Lipopolysaccharide-induced maturation of bone marrow-derived dendritic cells is regulated by notch signaling through the up-regulation of CXCR4. J. Biol. Chem. 2009;284:15993–16003. doi: 10.1074/jbc.M901144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, et al. High expression of CD11c indicates favorable prognosis in patients with gastric cancer. World J. Gastroenterol. W.J.G. 2015;21:9403. doi: 10.3748/wjg.v21.i31.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Worbs T, et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J. Exp. Med. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu AA, Drake V, Huang HS, Chiu SC, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology. 2015 doi: 10.1080/2162402x.2015.1016700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xuan W, Qu Q, Zheng B, Xiong S, Fan GH. The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. J. Leukoc. Biol. 2015;97:61–69. doi: 10.1189/jlb.1A0314-170R. [DOI] [PubMed] [Google Scholar]
- 82.Xue Q, Yan Y, Zhang R, Xiong H. Regulation of iNOS on immune cells and its role in diseases. Int. J. Mol. Sci. 2018;19:3805. doi: 10.3390/ijms19123805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaveri TD, Lewis JS, Dolgova NV, Clare-Salzler MJ, Keselowsky BG. Integrin-directed modulation of macrophage responses to biomaterials. Biomaterials. 2014;35:3504–3515. doi: 10.1016/j.biomaterials.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zorina ZA, Obozova TA. New data on the brain and cognitive abilities of birds. Zool. Z. 2011;90:784–802. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.