Abstract
Introduction
Bacteria and cancer cells share a common trait—both possess an electronegative surface that distinguishes them from healthy mammalian counterparts. This opens opportunities to repurpose antimicrobial peptides (AMPs), which are cationic amphiphiles that kill bacteria by disrupting their anionic cell envelope, into anticancer peptides (ACPs). To test this assertion, we investigate the mechanisms by which a pathogen-specific AMP, originally designed to kill bacterial Tuberculosis, potentiates the lytic destruction of drug-resistant cancers and synergistically enhances chemotherapeutic potency.
Materials and Methods
De novo peptide design, paired with cellular assays, elucidate structure-activity relationships (SAR) important to ACP potency and specificity. Using the sequence MAD1, microscopy, spectrophotometry and flow cytometry identify the peptide’s anticancer mechanisms, while parallel combinatorial screens define chemotherapeutic synergy in drug-resistant cell lines and patient derived ex vivo tumors.
Results
SAR investigations reveal spatial sequestration of amphiphilic regions increases ACP potency, but at the cost of specificity. Selecting MAD1 as a lead sequence, mechanistic studies identify that the peptide forms pore-like supramolecular assemblies within the plasma and nuclear membranes of cancer cells to potentiate death through lytic and apoptotic mechanisms. This diverse activity enables MAD1 to synergize broadly with chemotherapeutics, displaying remarkable combinatorial efficacy against drug-resistant ovarian carcinoma cells and patient-derived tumor spheroids.
Conclusions
We show that cancer-specific ACPs can be rationally engineered using nature’s AMP toolbox as templates. Selecting the antimicrobial peptide MAD1, we demonstrate the potential of this strategy to open a wealth of synthetic biotherapies that offer new, combinatorial opportunities against drug resistant tumors.
Electronic supplementary material
The online version of this article (10.1007/s12195-020-00626-z) contains supplementary material, which is available to authorized users.
Keywords: De novo design, Anticancer peptides, Supramolecular assembly, Nanostructures, Combinatorial therapy
Introduction
Chemotherapeutic resistance is a frequent cause of treatment failure in oncology, often preceding disease relapse and patient death.1,14 It is now common practice to select combinatorial cocktails of drugs with different mechanisms of action to prevent the emergence of resistant cell sub-populations. These regimens almost exclusively employ small molecule inhibitors that act on intracellular biochemical pathways,5,13 while compounds that target and disrupt the membranes of cancer cells are comparatively underexplored.3,33 Yet, there is accelerating clinical interest in adding tumor lytic agents to combinatorial therapies due to their ability to kill cancer cells through rapid, physical mechanisms that are not shared by conventional drugs.4,19,27
Anticancer peptides (ACPs) are one such class of potential therapeutics that exert their action by preferentially intercalating into and disrupting the plasma and mitochondrial membranes of cancer cells.19,43 These membrane-destabilizing, or membranolytic, effects can promote the intracellular diffusion and passive transport of otherwise poorly permeable small molecule drugs and biologics.42,45 More importantly, the rapid and non-stereospecific mechanisms of ACPs have led to the conclusion that evolutionary resistance towards these agents is probabilistically low.16,19 Taken together, these attributes suggest that ACPs could have significant potential to advance combinatorial strategies in precision medicine if properly exploited.
However, ACP discovery campaigns are constrained by the need for large empirical screens of complex natural product mixtures.18,39,46 These methods are costly, laborious and hindered by extensive med-chem optimization of lead candidates. As an alternative strategy, here we explore the potential to directly re-purpose nature’s toolbox of antimicrobial peptides to produce a robust ACP discovery pipeline. Antimicrobial peptides (AMPs) are a diverse family of natural, membrane-active antimicrobials that have evolved as vital weapons in the arms-race against pathogenic bacteria for a variety of lifeforms, from microorganisms to mammals (e.g., defensins).24,35 The bactericidal activity of cationic AMPs is initiated by electrostatic interactions with the highly anionic glycocalyx of bacteria, causing peptide-glycan binding that precedes interpolation into the cell envelope and self-assembly to form lytic pores.10,25 Similar to bacteria, cancer cells possess an increased electronegative surface charge compared to non-cancerous cells due to aberrant glycosylation and metabolism processes that yield sialic acid rich surface polysaccharides and overexpressed phosphatidylserine on the outer leaflet, respectively.17,22,29,36 These coincident phenotypes of bacteria and tumor cells encourage the use of AMPs for anticancer applications,19 and suggests we may be able to directly employ AMP templates, which have already been pre-optimized by evolution, to rationally design new ACPs. Here we test this assertion by investigating the ability of MAD1, a pathogen-specific AMP with potent activity towards bacterial Tuberculosis, to kill drug-resistant cancers and synergistically enhance the potency of chemotherapeutics.
Materials and Methods
Materials
1,2-ethanedithiol and diethyl ether were purchased from Acros Organics. 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP), piperidine, dimethyl sulfoxide spectrophotometer grade (DMSO), and thioanisole were purchased from Alfa Aesar. Vascular Cell Basal Medium, Rh VEGF, rg EGF, rg FGF basic, rg IGF-1, L-glutamine, heparin sulfate, hydrocortisone hemisuccinate, FBS, and ascorbic acid were purchased from ATCC. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were purchased from Avanti Polar Lipids. FITC Annexin V Apoptosis Detection Kit with PI was purchased from BioLegend. 0.2 μm nitrocellulose membranes were purchased from Bio-Rad. Cleaved Caspase-3 Rabbit Ab and β-Actin (8H10D10) Mouse mAb were purchased from Cell Signaling Technology. Fmoc-protected amino acids, oxyma, and Rink Amide ProTide Resin were purchased from CEM. Paraformaldehyde was purchased from Chem Cruz. 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), Fmoc-protected amino acids, N,N0-diisopropylcarbodiimide (DIC), O-(7-Azabenzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate (HATU), N,N-dimethylformamide (DMF), and Paclitaxel were purchased from Chem-Impex International, Inc. RPMI-1640, M199, trypsin and EDTA solution, Antibiotic-Antimycotic Solution, ultra-low attachment (ULA) plates, and 1× phosphate buffered saline (PBS) were purchased from Corning. Fetal bovine serum was purchased from HyClone. Hoechst 33342 trihydrochloride trihydrate dye was purchased from Invitrogen. Goat anti-mouse IgG (LC Specific) and Peroxidase AffiniPure Goat Anti-Rabbit IgG (H+L) were purchased from Jackson ImmunoResearch. Glycerol, tween 80, oadc supplement, MEM non-essential amino acid solution, d-(+)-Glucose, Insulin (Recombinant human), Transferrin Apo- (human plasma), hydrocortisone, OmniPur® Tween® 20 (Polyoxyethylene (20) Monolaurate), and sodium bicarbonate were purchased from Millipore-Sigma. N,N-Diisopropylethlamine (DIEA) and Doxorubicin HCl were purchased from Oakwood. 5% Blotto Immunoanalytical Grade (Non-Fat Dry Milk) was purchased from Rockland Immunochemicals. Cisplatin was purchased from Selleck Chemical S1166. MCDB-105 and FCCP (2-[2-[4-(trifluoromethoxy)phenyl]hydrazinylidene]-propanedinitrile) were purchased from Sigma-Aldrich. Cisplatin was purchased from Tocris Bioscience. Lysogeny broth, Mueller Hinton broth, 7H9 media, Nunc Lab-Tek Chambered Coverglass, sodium chloride (NaCl), hydrochloric acid (HCl), acetonitrile, trifluoroacetic acid (TFA), dichloromethane, dimethyl sulfoxide cell culture grade (DMSO), NHS-Fluorescein, 3500 MWCO Slide-A-Lyzer Dialysis Cassettes, Pierce ECL Western Blotting Substrate, SuperSignal West Dura Extended Duration Substrate, and formic acid LC/MS grade were purchased from ThermoFisher Scientific. Anisole was purchased from Tokyo Chemical Industry. F12 media, l-glutamine, sodium fluoride, sodium hydroxide, sodium phosphate monobasic, and sodium phosphate dibasic were purchased from VWR.
A549, OVCAR-3, T24, and NCI/ADR-RES cells were kindly provided by the laboratory of Dr. Joel P. Schneider (NCI, Frederick, MD). NL20 cell line was generously gifted by the laboratory of Dr. Matthew Taylor (Penn State University, College of Medicine, PA). Pseudomonas aeruginosa, Escherichia coli, Bacillus subtilis, methicillin-sensitive Staphylococcus aureus, methicillin-resistant Staphylococcus aureus, and Mycobacterium tuberculosis bacterial strains were generous gifts from the laboratories of Dr. Zissis Chroneos (Penn State, College of Medicine, Hershey, PA), Dr. Pak Kin Wong (Penn State, Biomedical Engineering, University Park, PA) and Dr. Kenneth Keiler (Penn State, Biochemistry and Molecular Biology, University Park, PA).
Analytical LCMS solvents were composed as follows: solvent A is 0.1% formic acid in water and solvent B is 9:1 acetonitrile and water with 0.1% formic acid. Preparative HPLC solvents consisted of solvent A (0.1% TFA in water) and solvent B (9:1 acetonitrile and water with 0.1% TFA).
Peptide Synthesis
Peptide synthesis reactions were performed as previously described,8 using Fmoc-based solid phase peptide synthesis with Oxyma/N,N′-Diisopropylcarbodiimide (DIC) activation on Rink Amide ProTide resin using a Liberty Blue Automated Microwave Peptide Synthesizer (CEM, Matthews, NC). To prepare fluorescently labeled peptides, MAD1 was further reacted on resin with 2 equivalents of NHS-Fluorescein (5/6-carboxyfluorescein succinimidyl ester) and 4 equivalents of diisopropylethalamine in DMF, and allowed to shake at room temperature for 4 h. Reaction progress was checked via Kaiser test to detect free amines.
Cleavage from the resin and de-protection was completed by stirring the crude with a trifluoroacetic acid:thioanisole:1,2-ethanedithiol:anisole (90:5:3:2) solution under a constant stream of argon. MAD1 was stirred at 30 °C for 3 h and DAP1/DAP2 were stirred for 2.5 h at room temperature. To precipitate the peptide from the deprotection cocktail, cold diethyl ether was added, followed by centrifugation at 5000 rpm for 8 min. After decanting the diethyl ether, the sample was lyophilized overnight to remove remaining trace amounts of organic solvent. Crude peptides were purified on a Phenomenex Semi-Prep Luna C18(2) column (Torrance, CA) by reverse-phase HPLC (Shimadzu, Columbia, MD). MAD1 was purified using a linear gradient of 0–25% solvent B over 25 min, followed by a linear gradient of 25–45% solvent B over 40 min. Purification of FITC-labeled MAD1 analogue was performed using a linear gradient of 0–12% solvent B over 6 min, followed by 12–32% solvent B over 20 min, and finally 32–38% solvent B over 12 min. For DAP1 peptide purification, a linear gradient of 0–42% solvent B over 38 min was used. For DAP2 peptide purification, a linear gradient of 0–10% solvent B was used over 10 min followed by a linear gradient of 10–25% solvent B over 30 min. After lyopholization to collect pure peptide, all compounds were characterized by LC-MS ESI (+) to confirm purity > 95% via MS peak integration (Supplementary Figs. 1–4).
Cell Cytotoxicity and Combinatorial Assays
All cells were cultured using standard culture conditions at 37 °C, 5% CO2, and 95% humidity. A549 (CCL-185), OVCAR-3 (HTB-161), T24 (HTB-4), and NCI/ADR-RES were cultured in RPMI-1640 supplemented with 10% v/v fetal bovine serum (FBS), 2mM L-glutamine (L-Gln), 10,000 units penicillin/mL, 10 mg/mL streptomycin, and 25 μg/mL amphotericin B. HUVEC (PCS-100-010) cells were cultured in Vascular Cell Basal Media (PCS-100-030) supplemented with an Endothelial Cell Growth Kit-VEGF (PCS-100-041) and 10 μg/mL gentamycin in culture flasks coated with 0.1% gelatin by incubating at 37 °C for 15 min. NL20 (CRL-2503) cells were cultured in Ham’s F12 supplemented with 1.5 g/L sodium bicarbonate, 2.7 g/L glucose, 2.0 mM l-glutamine, 0.1 mM nonessential amino acids, 0.005 mg/mL insulin, 10 ng/mL epidermal growth factor, 0.001 mg/mL transferrin, 500 ng/mL hydrocortisone and 4% fetal bovine serum, as advised by American Type Culture Collection (ATCC).
For cytotoxicity experiments, cells were first seeded in 96 well plates at 5 × 103 cells/well for A549, OVCAR-3, T24, and NCI/ADR-RES cells, and 10 × 103 cells/well for HUVEC and NL20 cells. After 24 h of incubation to allow the cells to adhere, the media was aspirated and 100 μL of treatments prepared in media with 5% FBS were added to each well; blank media and 20% DMSO served as negative and positive controls, respectively. Treatments were allowed to incubate for 48 h, followed by cell viability determination by MTT assay. In brief, wells were aspirated and replaced with 100 μL of 0.5 mg/mL thiazolyl blue tetrazolium bromide dissolved in media. The plates were allowed to incubate for 2–3 h and were then lysed with 100 μL of DMSO to dissolve the converted formazan product. Absorbance of the wells was measured via a microplate reader (Biotek, Winooski, VT) at 540 nm; percent viability was calculated by subtracting the positive control and normalizing to the negative control. To verify results from MTT asays, cell counting was used to as a secondary measure of cell death. Following MAD1 treatment, as described above, cells were fixed with 4% paraformaldehyde in PBS before staining the nuclei with Hoechst 33342. Five images were taken at each concentration using an Olympus IX73 microscope (Tokyo, Japan) will the well centered in the view field. Cell counting was performed by Matlab code modified from publicly available source at https://www.mathworks.com/help/images/ref/regionprops.html. Viability was calculated by comparing to an untreated control. Results were plotted in GraphPad Prism software with standard deviation reported.
For individual peptide cytotoxicity experiments, MAD1, DAP1, DAP2, AMP1, and AMP2 treatments were serially diluted to 100–0.001 μM concentrations. For individual chemotherapeutic experiments, doxorubicin, paclitaxel, and cisplatin were diluted 1:10 six times from 100 μM, 100 nM, and 100 μM, respectively. All treatments were prepared first as a stock solution in water and subsequently diluted into standard media with 5% FBS; paclitaxel and cisplatin stock solutions were first prepared in DMSO before diluting into media with a final DMSO concentration < 0.1%. All experiments were performed with n > 6.
For combinatorial cytotoxicity experiments, 96 well plates were set up in a checkerboard fashion such that peptide decreased in 1:10 concentration dilutions in one direction and chemotherapeutic decreased in 1:10 concentration dilutions in the orthogonal direction. 2× treatments were combined prior to cell administration for a final 1× concentration to prevent individual treatment effects. Experiments were performed in triplicate and heat maps were generated with MATLAB using logarithmic interpolation to obtain drug IC50 values at 20 μM MAD1.
Bacterial Cultures and Minimum Inhibitory Assays
For bacterial assays, bacteria were cultured according to the following established methods.37Bacillus subtilis (168) was cultured in lysogeny broth (LB). Escherichia coli (dh5-α), Pseudomonas aeruginosa (PAO-1), methicillin-resistant Staphylococcus aureus (MRSA; nrS72), and methicillin-sensitive Staphylococcus aureus (MSSA; usa300) were cultured in cation adjusted Mueller-Hinton broth (CAMHB). Mycobacterium tuberculosis (h37ra) was cultured in Middlebrook 7H9 supplemented with 0.05% (v/v) polysorbate 80, 0.5% (v/v) glycerol and 10% (v/v) oleic acid-albumin-dextrose-catalase (OADC). Each culture was grown in a shaking incubator at 37 °C at 200 rpm, according to guidelines provided by the Clinical and Laboratory Standards Institute (CLSI).
For MIC studies, peptide was first prepared in water at 320 μL (4×) and diluted 1:2 in appropriate treatment broth. Bacteria were prepared in appropriate media and diluted to an OD600 = 0.002. Next, 50 μL of treatment broth was added to each well in rows 2–8 of a 96 well plate, with the first three columns containing untreated broth as a negative control. In row 1, 100 μL of the 2× peptide in broth was added. Using a multi-channel pipette, 50 μL of the 2× peptide solution was diluted 1:2 in the subsequent row and repeated for all rows. 50 μL of bacteria was added to each well, incubated for 24 h, and read using visual evaluation. The MIC, or minimum inhibitory concentration, was determined by the lowest concentration with a significant reduction in bacteria pellet presence compared to the negative control.
Particle Formulation
Liposomes were prepared by extrusion techniques according to previous methods.8 Briefly, all liposomes were prepared in 2× concentration (5 mM). Healthy and cancer liposomal mimicking membranes were prepared with 72:23:5 and 45:24:30 phosphatidylcholine (POPC):phosphatidylethanolamine (DOPE):phosphatidylserine (POPS) lipids, respectively.34 The lipids dissolved in chloroform were mixed in proper molar proportions before being evaporated under a stream of argon. The lipid film was then dried completely by lyophilization overnight. Then, the lipids were rehydrated in 2× concentrated liposomal buffer (300 mM BTP, 100mM NaF, and pH 7.4) before 8 freeze/thaw cycles in liquid nitrogen and water bath 37 °C. The membranes were then extruded using an Avanti Mini Extruder (Alabaster, Alabama) with a 0.1 μM nuclepore polycarbonate membrane > 11 times. The extruded liposomes were then dialyzed in 3500 MWCO dialysis cassettes against water.
Circular Dichroism
Circular dichroism (CD) was performed using 20 mM sodium phosphate buffer (15.1 mM sodium phosphate dibasic heptahydrate, 0.49 mM sodium phosphate monobasic monohydrate, and pH 7.4) and a J-1500 Circular Dichroism Spectrometer (JASCO, Oklahoma City, OK). In a 1 mm path length quartz cell, 150 μL of membrane mimicking liposomes were combined with 150 μL of 200 μM peptide in buffer and exposed to a wavelength spectrum of 180 to 260 nm at 25 °C. Mean residue molar ellipticity [θ], measured in (10−1 deg cm2 dmol−1), was normalized using the following expression: where θmeas is measured ellipticity (mdeg), l is light path length (cm), c is molar concentration (mol/L), and r is number of amino acid residues.
Parallel optical density experiments were conducted measuring the absorbance at 600 nm (OD600) to track the kinetics and specificity of membrane disruption. Measurements were taken every 30 seconds for the first 10 min, followed by measurements every minute until 15 min.
Scanning Electron Microscopy
SEM was conducted on OVCAR-3 cells after first being seeded onto uncoated glass coverslips in a 12 well plate at 3 × 104 cells/well and allowed to adhere overnight. Next, the cells were treated with 10 μM of peptide in 5% FBS media for 1 or 4 h, and a negative control in peptide-free media. The cells were then washed with fresh serum free media and fixed with cold 5% glutaraldehyde in 0.1 mM sodium cacodylate and left overnight at 4 °C. The cells were subsequently rinsed three times with 0.1 mM sodium cacodylate before being subjected to an ascending ethanol dehydration series (25, 50, 70, 85, 90, 95, 100%). The cells were stored in 100% ethanol before transferring to a Leica EM CPD300 Critical Point Dryer (Buffalo Grove, Illinois). Dried samples were mounted onto metal SEM stubs using double sided carbon tape, and sputter coated with Au/Pd 60:40 using a Bal-tec SCD-050 Sputter Coater (Capovani Brothers Inc., Scotia, New York). Finally, samples were imaged using a Zeiss SIGMA VP-FESEM (Thornwood, New York) using secondary electron imaging. Experiments were performed in duplicate.
Confocal Fluorescent Microscopy
OVCAR-3 cells were seeded at 3 × 104 cells/well in a chambered coverslip and allowed to adhere for 24 h. Cells were treated at the peptide IC50 as determined by preliminary experimentation in serum free media for 1, 4, and 10 h. Serum free media was used as a negative control. After treatment incubation periods, cells were washed with cold PBS and fixed with 4% paraformaldehyde for 20 min at room temperature. After washing with cold PBS, 2 µg/mL Hoechst in PBS was added and left to incubate for 20 min. Slides were imaged on an Olympus Fluoview 1000 Confocal Microscope (Olympus, Shinjuku, Tokyo, Japan) with a PlanApo 60×/1.4 oil objective lens using 405 and 488 nm single photon lasers for DAPI and FITC, respectively.
Western Blotting
For western blots, OVCAR-3 cells were first seeded at 2.5 × 105 cells/well in a 6 well plate and allowed to adhere overnight. Cells were then treated in 5% FBS media with 1 × IC50 and 2 × IC50, with untreated controls of media alone. Following 2 or 48 h of treatment, the whole-cell protein lysates were prepared in RIPA buffer (50 mM Tris NaOH at pH 7.4, 150 mM NaCl, 1% IGEPAL-CA630 [octylphenoxy poly(ethyleneoxy)ethanol], 0.5% sodium deoxycholate, 0.1% SDS, 2mM EDTA, protease/phosphatase inhibitors) by first scraping the bottom of the well and subsequently briefly sonicated. The lysates were centrifuged to clear cellular debris, separated by 12% SDS-PAGE, and transferred to a 0.2 µm Nitrocellulose Membrane using the Bio-Rad Trans-Blot Turbo transfer machine (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s protocols. The blot was incubated in 5% Immunoanalytical Grade Non-fat Dry Milk for 1 h, followed by incubation with antibodies against cleaved caspase-3 (1:1000) or actin (1:1000) at 4 °C for 16 h. The blots were then incubated with 1:2000 dilution in 1× BSA of anti-mouse or anti-rabbit antibodies for 1 h before washing with TBST three times for 15 min and developing using Pierce ECL system according to the manufacturer’s protocol.
Flow Cytometry
OVCAR-3 cells were first seeded at 2 × 105 cells/well in 12 well plates. The cells were then allowed to adhere overnight. Treatments were conducted at 1 × IC50 of MAD1 at 2 and 24 h in culture media adjusted to 5% serum. Wells were left in 5% serum media for negative and compensation controls. Cells were then trypsinized, washed in cold PBS, and centrifuged at 3000 rpm at 15 °C for 5 min. After the supernatant was aspirated, the cells were resuspended in 100 μL of Annexin V Binding Buffer. In the dark, 5 μL of FITC Annexin V and 10 μL of Propidium Iodine solution were added to the cells, with compensation controls of a negative control with no staining, a FITC Annexin V only control, and a PI only control. The cells were then vortexed gently to combine and allowed to incubate for 15 min at room temperature. 400 μL of Annexin V Binding Buffer was then added to each tube and stored on ice prior to analyzing by flow cytometry.
Flow cytometry was conducted on a LSRFortessa Flow Cytometer (BD Biosciences, Franklin Lakes, New Jersey) using 488 and 532 nm lasers for FITC and PE-Texas Red signals to detect Annexin V and PI, respectively. Compensation was completed for 10,000 cells, and experiments were conducted over 20,000 cells.
Tetramethylrhodamine, Ethyl Ester (TMRE) Flow Cytometry
OVCAR-3 cells were treated with the indicated concentrations of MAD1 peptide for 48 h or the positive control FCCP (2-[2-[4-(trifluoromethoxy)phenyl]hydrazinylidene]-propanedinitrile) for 5 min (5 μM). Cells were incubated for 30 min with 200 nM TMRE, washed twice with PBS, and analyzed for fluorescence by flow cytometry on a BD FACSCanto 10 (BD Biosciences, Franklin Lakes, New Jersey).
Spheroid Culturing
OVCAR-3 cells were cultured using RPMI 1640 supplemented with 5% FBS and 1% penicillin-streptomycin. EOC patient-derived cells were cultured in 1:1 MCDB-105: M199 supplemented with 10% FBS and 1% penicillin-streptomycin. EOC patient-derived cells were provided by Dr. Katherine M. Aird under an Institutional Review Board-approved protocol. EOC patient-derived cells were collected from patient ascites fluid after paracentesis procedures. Cells were routinely tested for mycoplasma as previously described.44 OVCAR-3 and patient-derived cells (EOC15, EOC17, and EOC19) were seeded at 1 × 105 cells/well in ultra-low attachment (ULA) plates. 24 h after seeding, spheroids were treated alone or in combination with 2 µM cisplatin and/or 4 µM MAD1 peptide for 48 h. After 48 h, 10× images were taken of the spheroids (Nikon Elements Ts2, Nis Elements F 4.51.00). Spheroid length and width was calculated using ImageJ. Spheroid volume was calculated using the following equation . Experiments were performed in triplicate at least two independent times.
Statistical Methods
Cell viability experiments were completed n > 6 in two independent experiments and analyzed using GraphPad Prism 8 to fit curves and calculate IC50 values using inhibitor vs. normalized response non-linear regression model. IC50 values are reported as mean values ± standard deviation. For bacterial MIC experiments, values are reported as the average of 3 replicates. Pore size frequency distribution was completed by ImageJ FIJI on 4 independent cells and n = 30 pores from SEM images. UV–Vis OD600 and CD spectra are means of 5 and 3 replicates, respectively and reported as mean ± standard deviation. Individual UV–Vis and CD spectra, as well as fluorescent confocal and TEM micrographs, shown are representative plots from three individual experiments. CD data is with a variance in ellipticity of 0.2 millidegrees at each wavelength. Relative fluorescence data calculated from fluorescent confocal images represents the average of 10 independent measurements ± standard deviation. For spheroid volume analysis, a Kruskal-Wallis test and Dunn’s multiple comparisons test was performed using GraphPad Prism 8.
Results and Discussion
Design and Activity of De Novo ACPs
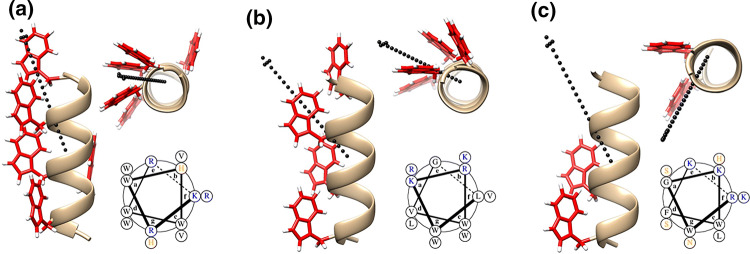
To begin our design of new ACPs, we turned to the model antimicrobial peptide MAD1 (myco-membrane associated disruption 1 sequence). This sequence, recently characterized by our lab,38 preferentially disrupts mycobacterial membranes to elicit anti-Tuberculosis specific activity. Key to MAD1’s bactericidal action is its ability to adopt a facially amphiphilic alpha helix that is defined by a hydrophobic, tryptophan-rich surface (Fig. 1a). Intermolecular zippering of tryptophan side chains enables supramolecular assembly of MAD1 peptides to form a lytic nanostructure within the Tuberculosis pathogen cell wall. Given its antimicrobial importance, we first investigate the role of this tryptophan-rich helical surface on the potential anticancer activity of MAD1. To do this we designed two ACP analogues, DAP1 and DAP2 (de novo designed anticancer peptide), in which both sequences exhibit variations in hydrophobic amino acid packing and spatial display relative to the MAD1 template. For DAP1, the cationic and hydrophobic faces of the helix are spatially segregated with respect to each other (see helical wheel in Fig. 1b, bottom), resulting in a more uniform angular alignment of tryptophan residues in the axial direction. To accommodate this dense packing of hydrophobic groups, the number and spacing of lysine and arginine residues was altered for the DAP1 scaffold. Collectively this design leads to an increase in sequence hydrophobicity (defined by logD value40) and a decrease in helical hydrophobic moment (a vector-based measure of helix amphiphilicity32) of DAP1 compared to MAD1 (Table 1). Conversely, DAP2 is designed such that tryptophan residues are axially isolated to only the N-terminal half of the peptide, leading to a decrease in the overall hydrophobic surface area relative to MAD1 (Fig. 1c). This results in a significant decrease in hydrophobicity, as defined by a reduction of its logD value, and corollary increase in hydrophobic moment for DAP2 relative
Figure 1.
Rational design of de novo anticancer peptides. Minimized model of (a) MAD1, (b) DAP1, (c) DAP2 peptide helices. For all panels, Left: helical profile; Top right: axial view (red = tryptophan residues). Vector of the hydrophobic moment shown as dotted black arrow. Bottom right: helical wheel (black = hydrophobic, blue = basic, orange = polar residues).
Table 1.
Sequence and physiochemical properties of de novo designed ACPs.
| Peptide | Sequencea | Sequence length | Formal chargeb | logD | Hydrophobic moment |
|---|---|---|---|---|---|
| MAD1 | KRWHWWRRHWVVW-NH2 | 13 | + 7 | − 3.41 | 14.828 |
| DAP1 | LWKRWVGVWRKWL-NH2 | 13 | + 5 | − 3.23 | 13.879 |
| DAP2 | RWGKWFKKNSHLS-NH2 | 13 | + 5 | − 9.37 | 15.250 |
| AMP1 | WKWLKKWIK-NH2 | 9 | + 4 | − 6.46 | 13.449 |
| AMP2 | KRWWKWWRR-NH2 | 9 | + 5 | − 6.90 | 15.084 |
aAll peptides are prepared with amidated C-terminus
bFormal charge includes N-terminal amine. Histidine considered partially protonated for purposes of formal charge calculation
to MAD1. To further deconvolute the effects of logD and hydrophobic moment on ACP potency in later studies, we also include two additional tryptophan-rich antimicrobial peptides, AMP1 and AMP2.31,38 AMP1 exhibits a similar spatial distribution of tryptophan residues, and thus an analogous hydrophobic moment, as DAP1 (Supplementary Fig. 5a, Table 1) but is significantly more hydrophilic (logD = − 6.46). AMP2, on the other hand, is defined by facially segregated amphiphilic regimes (see helical wheel in Supplementary Fig. 5b) that impart it with a comparable hydrophobic moment as DAP2, while the logD value remains consistent with AMP1. Together, these peptides allow us to systematically evaluate how changes in sequence logD and hydrophobic moment influence their cytotoxic effects, with the expectation that anticancer potency will follow DAP1 > MAD1 > AMP1 > AMP2 > DAP2.
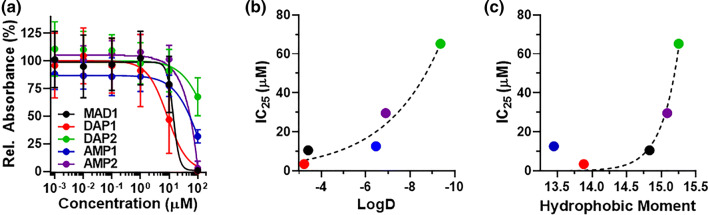
To test this assertion, we measured the cytotoxicity of each sequence against a panel of human cancer cell lines, as well as non-cancerous controls (Fig. 2a, Table 2 and Supplementary Fig. 6). As predicted, DAP1 was the most potent ACP of the five sequences, with IC50 values that ranged from 8.6 to 15.3 μM, dependent on cell line. MAD1 displayed preferential activity towards the ovarian carcinoma cell line OVCAR-3 (IC50 = 14.2 μM, Fig. 2a), moderate activity against A549 (lung carcinoma) and NCI/ADR-RES (drug resistant ovarian cancer) cells, and was inactive towards T24 (bladder cancer). Given that these results are derived from MTT experiments, which measures mitochondrial activity and thus subject to artifacts from metabolic quiescence, we performed a secondary nuclei-counting assay (Supplementary Fig. 7). These results support the claim that MAD1 mediates toxicity by killing, or inhibiting the growth, of OVCAR-3 cells (IC50 = 3.3 μM) and is not solely a result of changes in cellular metabolism. Finally, consistent with our expectations, AMP1 and AMP2 were less effective compared to DAP1 and MAD1 (IC50 ≈ 50 μM, Fig. 2a) in OVCAR-3 cells, while DAP2 was inactive towards all four of the tumor cell lines tested.
Figure 2.
ACP cytotoxicity and structure-activity relationships. (a) Cytotoxicity curves for MAD1, DAP1, DAP2, AMP1, and AMP2 peptides against OVCAR-3 ovarian carcinoma cells, shown as percent relative MTT absorbance. Curves for all four cancer cell lines tested are shown in Supplementary Fig. 6. Relative change in OVCAR-3 IC25 value as a function of (b) sequence hydrophobicity (logD) and (c) helix facial amphiphilicity (hydrophobic moment). Exponential fit (dashed line, GraphPad Prism) included to guide the eye.
Table 2.
Peptide cytotoxicity towards cancerous and non-cancerous cell lines.
| Peptide | Cancerous (IC50, μM) | Healthy (IC50, μM) | Therapeutic indexa | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A549 | OVCAR-3 | NCI/ADR-RES | T24 | HUVEC | NL20 | A549 | OVCAR-3 | NCI/ADR-RES | T24 | |
| MAD1 | 36.2 | 14.2 | 85.6 | > 100 | > 100 | 76.9 | 2.8 | 7.0 | 1.2 | – |
| DAP1 | 11.9 | 8.6 | 14.0 | 15.3 | 19.5 | 19.0 | 1.6 | 2.3 | 1.4 | 1.3 |
| DAP2 | > 100 | > 100 | > 100 | > 100 | > 100 | > 100 | – | – | – | – |
| AMP1 | – | 51.1 | – | – | – | – | – | – | – | – |
| AMP2 | – | 53.5 | – | – | – | – | – | – | – | – |
aTherapeutic index of each peptide for indicated cell type relative to HUVEC controls (IC50-HUVEC/IC50-indicated cell)
Interestingly, although DAP1 was the most potent ACP of the five peptides, it showed nearly equal toxicity against the two non-cancerous cell lines tested: human umbilical vein endothelial cells (HUVEC) and human lung epithelium (NL20). Accordingly, this defines therapeutic indices,
a quantitative measure of relative drug safety, for DAP1 of ≤ 2.3 (Table 2). MAD1 on the other hand was well tolerated by both control cell lines and, as a result, generally possessed superior therapeutic indices compared to DAP1. Notably, MAD1 displayed an index of 7 towards OVCAR-3 cells, suggesting a distinct selectivity of the peptide towards ovarian cancers; an assertion we later confirm in ex vivo studies. This specificity may be a result of variations in glycan composition between the tested cell lines. Ovarian cancer cells, for example, are defined by highly sialylated N-glycans displayed at the cell surface.7 Bladder cancer cells, on the other hand, are characterized by increased expression of neutral mannose glycans and β 1,6-branched oligosaccharides.30 Given that many cationic ACPs preferentially engage cancer cells via binding to negatively charged sialic acid residues,22 this suggests MAD1-sensitive versus -insensitive cell lines may be distinguished by their glycan manifold.
Thus far, our cytotoxicity data supports the claim that AMPs can be re-designed to afford new ACP candidates. Given the shared mechanisms of action between these two classes of peptides, the reverse should also be true: ACPs can function as AMPs. To test this assertion, we measured the antimicrobial activity of MAD1, DAP1, and DAP2 against a polymicrobial panel (Table 3). In line with our hypothesis, MAD1 and DAP1 both displayed bactericidal activity, with DAP1 generally showing more potent broad-spectrum effects relative to the MAD1 peptide. Conversely, DAP2 is inactive towards all six of the bacterial lines tested.
Table 3.
Minimum inhibitory concentration (MIC) of de novo ACPs.
| Bacteria | MIC (μM) | |||
|---|---|---|---|---|
| MAD1 | DAP1 | DAP2 | ||
| Gram Positive | B. subtilis (168) | 0.2 | < 0.6 | > 80 |
| MS S. aureus | > 80a | 7 | > 80 | |
| MR S. aureus | > 80a | 5 | > 80 | |
| Gram Negative | P. aeruginosa | 50a | 20 | > 80 |
| E. coli | 25 | 10 | > 80 | |
| Mycobacteria | M. tuberculosis | 2.5a | 20 | > 80 |
aData reported in Ref. 38
ACP Structure-Activity Relationships and Ovarian Cancer Specificity
To better understand the physiochemical drivers of ACP toxicity in tumor cells we further examined how anticancer potency of these peptides is influenced by their hydrophobicity (logD, Fig. 2b) and facial amphiphilicity (hydrophobic moment, Fig. 2c). For these analyses we utilized cytotoxicity data from the OVCAR-3 cell line and plotted the appropriate IC25 value of each sequence. Here, IC25 was chosen as DAP2 did not achieve an IC50 at the concentrations tested (see green data in Fig. 2a). Results demonstrate a clear correlation between peptide toxicity and both sequence logD (Fig. 2b) and hydrophobic moment (Fig. 2c); with changes in the latter causing a particularly sharp shift in ACP potency. This suggests there may exist an amphiphilicity threshold that differentiates active from inactive sequences. In our studies, peptide helices with a hydrophobic moment < 15 were sufficiently amphiphilic to cause potent oncolytic activity (IC25 < 15 µM). Taken together, this suggests that the cytotoxic potential of a given ACP is largely dependent on a combination of its hydrophobicity and amphiphilicity, or lipophilicity. This further validates the claim of shared molecular mechanisms between ACPs and AMPs, as it is well established that lipophilicity is a key driver of AMP bacteriolytic activity.19,41
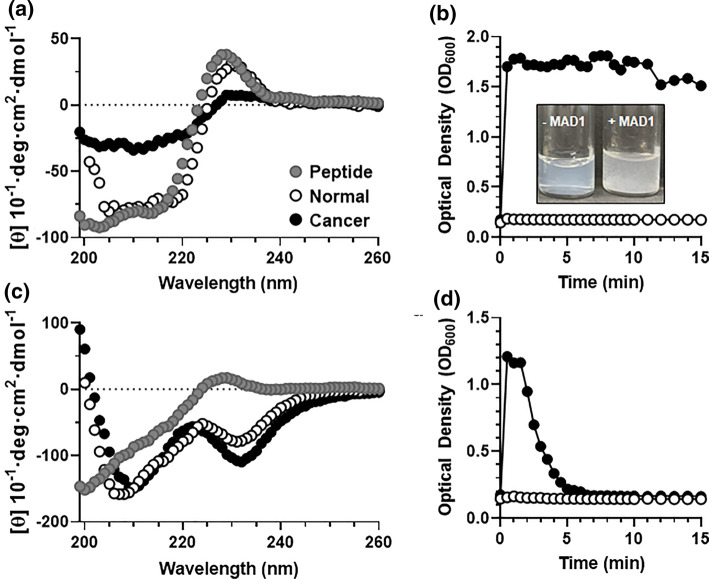
Our cell-based studies collectively demonstrate that MAD1 preferentially targets and disrupts cancer cell membranes over that of normal, healthy counterparts. To mechanistically investigate this, circular dichroism (CD) spectroscopy was performed to monitor changes in peptide secondary structure in the absence and presence of normal or cancerous liposomal membrane analogues. CD spectra shown in Fig. 3a demonstrate that MAD1 adopts an α-helical conformation in aqueous solutions, with characteristic minima in ellipticity at 204 and 216 nm. An exciton band at 228 nm is indicative of interactions between aromatic indole chromophores as a result of intermolecular tryptophan pairing between peptides.12 When mixed with liposomes mimicking the composition of normal mammalian cell membranes (72:23:5 POPC:DOPE:POPS),34 MAD1 largely maintains its α-helical structure. A slight red-shift (+ 2 nm) in both the Trp-Trp ellipticity maximum (230 nm) and α-helical minima (206 and 218 nm) suggests MAD1 benignly adsorbs to the surface of normal membranes.28 However, when mixed with cancer membranes (45:24:30 POPC:DOPE:POPS),34 a dramatic absorption flatting of MAD1’s CD signal is observed (Fig. 3a, black data). These changes can be attributed to the organization of peptide chromophores during their ordered interpolation within regions of high lipid density.28 This indicates that MAD1 is able to rapidly assemble within the cancer cell liposomal membrane to cause significant membrane destabilization and subsequent lysis, which is further corroborated by an increase in solution optical density (Fig. 3b) and the generation of macroscopic lipid fragments (Fig. 3b inset and Supplementary Fig. 8). Conversely, MAD1 did not lyse the healthy liposomal counterparts.
Figure 3.
Tumor membrane-templated ACP assembly. (a) Circular dichroism spectra of the MAD1 peptide in aqueous solution (●), or in the presence of normal (○) and cancer cell (●) membrane models. (b) Relative disruption of normal (○) and cancer (●) model membranes by the MAD1 peptide, as determined from optical density measurements (OD600). Inset: image of cancer membrane liposomal solution before (-MAD1) and after (+MAD1) treatment with the peptide. Formation of large flocculates provides visual confirmation of liposome disruption. (c) Circular dichroism spectra and (d) optical density measurements of DAP1 in the absence or presence of model membranes (● = no membrane, ○ = normal membrane, ● = cancer membrane).
DAP1, on the other hand, evolves from an ensemble of random coil conformations to a CD spectrum indicative of β-sheet rich and tryptophan stacked conformations in the presence of both normal and cancer cell membranes, as indicated by ellipticity minima at 212 and 230 nm, respectively (Fig. 3c).6 Parallel optical density measurements demonstrate that DAP1 exhibits a transient interaction with cancer cell membranes, leading to a temporary increase in optical density (Fig. 3d). Yet, DAP1 does not completely lyse cancer liposomes, as evidenced by rapid restoration of optical density back to baseline (Fig. 3d) and the absence of large lipid flocculates in solution (Supplementary Fig. 8). This temporary turbidity change may be explained by transitory adsorption of the peptide to the lipid surface, or integration of the sequence into the bilayer to form stable peptide-lipid complexes without causing membrane lysis (Fig. 3d). Interestingly, DAP1 showed little interaction affinity with non-cancerous liposomes, despite eliciting potent toxicity towards healthy cell lines tested in our in vitro assays (Table 2). This suggests these highly simplified liposomal membrane models may be missing other factors that influence ACP activity and specificity, including sphingolipids, cholesterol and surface glycans; the latter particularly influential on ACP potency.22
Collectively, our biophysical data suggests that MAD1 preferentially interpolates into the membranes of cancer cells to elicit selective oncolytic activity. The supramolecular assembly of DAP1, conversely, appears to poorly discriminate healthy from cancerous membranes (Fig. 3c), providing further mechanistic insight into the differential cell cytotoxicity profiles of the two peptides (Table 2).
MAD1 Mechanism of Action
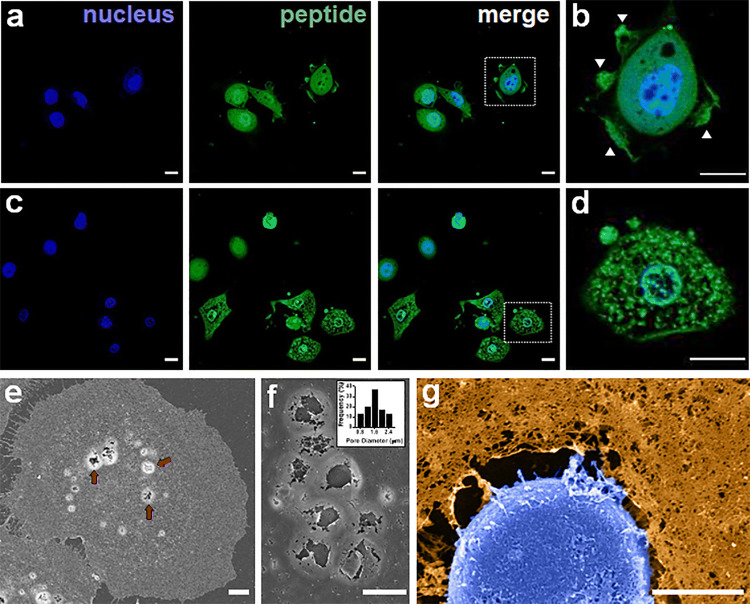
Recent studies suggest that many ACPs exploit diverse physical (lytic) and biochemical (apoptotic) modes of toxicity, which operate in either an independent or coincident fashion depending on the cell type.11,21,43 This is exemplified by our current study, in which DAP1 displayed similar potency across all cell lines tested, whereas MAD1 demonstrated a strong preference for OVCAR-3 ovarian carcinoma cells (Table 2). Thus, DAP1 may employ a cell-type agnostic mechanism, while MAD1 potentially elicits a combination of necrosis and apoptosis that is particularly potent towards ovarian carcinomas. To investigate this, we monitored the time-dependent subcellular localization of a fluorescently-labeled MAD1 analogue in OVCAR-3 cells (Fig. 4a). Results show that, shortly after addition, MAD1 decorates the surfaces of treated ovarian cancer cells. Interestingly, exposure of OVCAR-3 to MAD1 evoked a ruffled morphology of the cellular surface, particularly at the migratory leading edges (Fig. 4b). This is complemented by the appearance of micron-sized defects in the plasma membrane. After this initial surface binding, MAD1 translocates to the cytoplasm (Fig. 4c) and subsequently localizes to the nuclear envelope (Fig. 4d, Supplementary Fig. 9). To confirm these observations, as well as directly visualize MAD1-induced damage to the plasma and nuclear membranes, we performed scanning electron microscopy. SEM imaging at 1 h after peptide addition confirms that MAD1 rapidly assembles within the membranes of cancer cells to form micron-sized pores (Fig. 4e, see untreated control in Supplementary Fig. 10), with an average diameter of ~1.5 μm (Fig. 4f). This is followed later by peptide-mediated degradation of the nuclear envelope (Fig. 4g, Supplementary Fig. 11), as well as significant morphologic changes to membrane nano-topography (Supplementary Fig. 12). Although ACP-mediated damage to plasma and mitochondrial membranes has been observed previously, to our knowledge MAD1 is the first example of an amphipathic peptide with the capacity to preferentially bind and disrupt the nuclear envelope.
Figure 4.
MAD1 integration into cancer cells and subcellular trafficking. (a) Confocal micrographs of OVCAR-3 ovarian carcinoma cells treated with 14 μM of fluorescein-labeled MAD1 for 1 h. (b) Magnification of boxed cell in merged image of panel a. Membrane ruffling marked by white arrows. (c) Micrographs of OVCAR-3 cells following a 10 h incubation with MAD1. (d) Magnification of boxed cell in merged image of panel c demonstrating peptide localization to the nuclear envelope (see Supplementary Fig. 9 for 3D z-stacks). Scale bars for panels a-d = 15 μm. (e) SEM image of membrane-templated MAD1 pores following a 1 h treatment of OVCAR-3 cells with MAD1. Peptide-induced surface pores highlighted by red arrows. (f) Magnification of membrane pores formed by MAD1. Inset: Histogram of pore diameter. (g) Magnified SEM micrograph of an OVCAR-3 cell treated for 4 h with MAD1 (full image can be found in Supplementary Fig. 11). Cell membrane (orange) and nucleus (blue) have been false-colored to aid visualization. Scale bar for panels (e)–(g) = 5 μm.
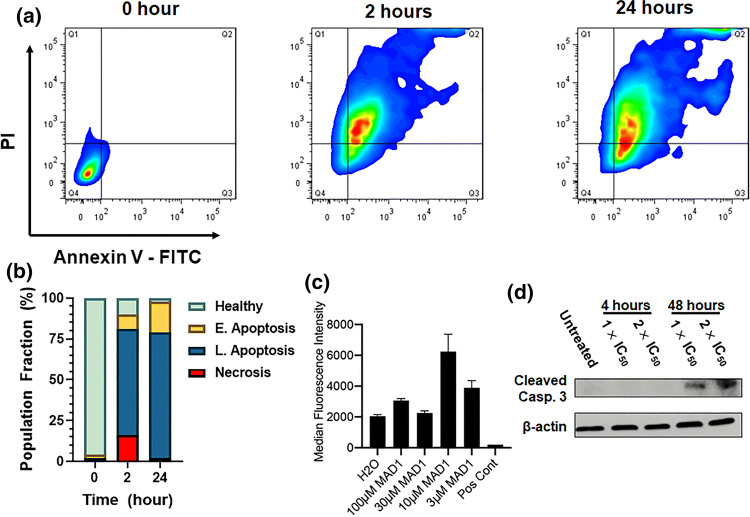
Taken together, this suggests that insertion and supramolecular assembly of MAD1 within the membranes of cancer cells alters bilayer tension and destabilizes the protective nuclear enclosure. Accordingly, this may lead to the induction of apoptotic signaling cascades after initial necrotic priming of cancer cells by the peptide. To test this, we performed annexin V-PI flow cytometry assays following treatment of OVCAR-3 cells at the IC50 of MAD1 for 2 and 24 h (Fig. 5a). Results show that, although MAD1 causes considerable cell necrosis (16% of population) at early time points (2 h), the majority of cells (65%) adopt a late apoptotic phenotype (Fig. 5b). After 24 h cells predominantly occupy early (19%) and late (77%) apoptotic stages. This rapid induction of apoptosis may be caused by mitochondrial depolarization, which can activate caspase cascades within 30 min. To investigate this, we utilized a TMRE-staining flow cytometry assay to evaluate mitochondrial transmembrane potential in OVCAR-3 cells after a 48 h incubation with the peptide. Here, intact mitochondria take up the TMRE dye and brightly fluoresce, while depolarized or inactive mitochondria fail to sequester the TMRE fluorophore. Results in Fig. 5c and Supplementary Fig. 13 show that OVCAR-3 mitochondrial integrity is generally unchanged in the presence of MAD1, and at certain peptide concentrations appears to increase. This indicates that MAD1 does not affect mitochondrial activity to potentiate intrinsic apoptotic signaling in treated cancer cells. Yet, parallel western blot experiments confirm the presence of cleaved caspase 3, a common effector of both intrinsic and extrinsic apoptotic pathways, in MAD1-treated OVCAR-3 cells (Fig. 5d). Taken together, this indicates that MAD1’s early lytic effects are complemented later by the induction of extrinsic apoptotic pathways. Given that the peptide assembles at the surfaces of cancer cells (Fig. 4), the most likely explanation for this is that membrane-templated supramolecular assembly of MAD1 leads to ligation of extrinsic FAS, TNF or TRAIL death receptors.9
Figure 5.
Mechanism of antitumor action by MAD1. (a) Representative plots from flow cytometric PI/Annexin V-FITC apoptosis assays of OVCAR-3 cells treated with 14 μM of MAD1 for 2 or 24 h. (b) Quadrant quantification of flow cytometry data defining the healthy cell population from necrotic cells, or those in early and late apoptosis, as a function of incubation time with the peptide. (c) Quantification of TMRE fluorescence for OVCAR-3 cells treated with varying concentrations of MAD1. Water or FCCP were included as a negative and positive control, respectively. (d) Western blot analysis of cleaved caspase 3 from OVCAR-3 cells after incubation with medium alone (untreated), or MAD1 at 1× and 2× its IC50 for 4 and 48 h.
However, our western blot data, which demonstrates that MAD1-mediated activation of caspase 3 does not occur until 48 h, contradicts the annexin V-PI assays which show that the peptide induces apoptosis within 2 h (Figs. 5a and 5b). One explanation for these incongruous results in that early MAD1-mediated poration of the cancer cell plasma membrane allows the cytosolic diffusion of the otherwise cell-impermeant annexin-V probe, which can then bind its phosphatidylserine lipid target on the inner leaflet. An alternative hypothesis is that membrane interpolation of MAD1 rearranges lipids in the asymmetric bilayer and physically translocates phosphatidylserine to the outer leaflet, where it then binds the annexin-V protein. In both scenarios MAD1 treated cells would stain positive for apoptosis at these early time points, but ultimately experience an oncolytic mechanism. Similar results have been observed for other AMPs tested for antitumor activity, which rapidly induce late apoptotic and necrotic phenotypes in treated cancer cells.15,20,23,26
Anticancer Synergy
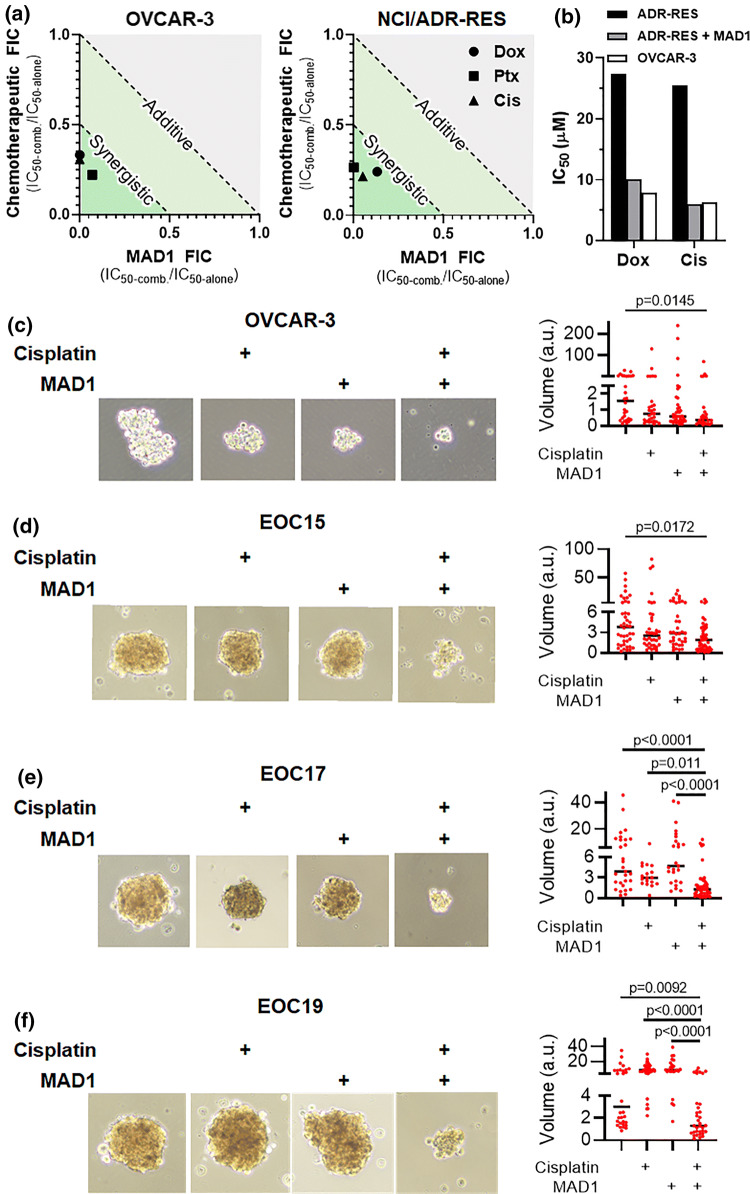
Given MAD1’s selective action against ovarian cancer cells, and its co-induction of physical (lysis) and biochemical (extrinsic apoptosis) death pathways, we next tested its potential to synergistically enhance the potency of three chemotherapeutics commonly used to treat ovarian carcinomas: Doxorubicin (Dox), Paclitaxel (Ptx) and Cisplatin (Cis). Remarkably, in both drug-
refractory (OVCAR-3) and multidrug resistant (NCI/ADR-RES) ovarian cancer lines, MAD1 potently synergizes with all three therapeutics tested (Fig. 6a, Supplementray Table 1 and Supplementary Fig. 14). As a notable example, co-treatment of multidrug resistant NCI/ADR-RES cells in the presence of 20 μM MAD1 restored the potency of both Dox and Cis to a level equivalent to the more sensitive OVCAR-3 cell line (Fig. 6b, analysis derived from combinatorial heat maps shown in Supplementary Fig. 14). Conversely, Paclitaxel showed only a modest improvement in potency when combined with 20 μM MAD1 (Supplementary Fig. 15).
Figure 6.
Chemotherapeutic synergy. (a) Isobolograms of MAD1 and Doxorubicin (Dox), Paclitaxel (Ptx) or Cisplatin (Cis) combinatorial synergy in OVCAR-3 (left) and NCI/ADR-RES (right) cells. Fractional inhibitory concentration (FIC) < 1 and < 0.5 represent additive and synergistic effects, respectively. (b) Comparison of Dox and Cis IC50 towards NCI/ADR-RES as either a monotherapy (ADR-RES, black) or in combination with 20 μM MAD1 (ADR-RES + MAD1, grey). Activity of each drug as a monotherapy in pre-resistant OVCAR-3 (OVCAR-3, white) cells shown for comparison. (c–f) Indicated ovarian cancer cell line or patient-derived ovarian carcinoma cells were cultured in ultra-low attachment conditions and treated for 48 h with 2 μM cisplatin or 4 μM MAD1 alone and in combination. Data represent volume in arbitrary units (a.u.) and median.
Clinically, late-stage epithelial ovarian cancer (EOC) is characterized by dissemination of the disease into the peritoneal cavity to form tumor spheroids, which is then typically treated with platinum-based chemotherapy (e.g., Cisplatin).2 In order to mimic these conditions ex vivo, ultra-low attachment culture plates were used to induce the formation of 3D tumor spheroids using OVCAR-3 cells, as well as three patient-derived lines (EOC15, 17, 19), which were then treated alone or in combination with Cisplatin and/or MAD1 (Figs. 6c to 6f). Remarkably, all three patient-derived tumor cell spheroids showed minimal reduction in size when treated with either Cisplatin or MAD1 alone, while the combination produced potent and synergistic anticancer effects. These ex vivo studies confirm that MAD1 synergizes with platinum-based drugs to decrease ovarian cancer cell size, even in advanced patient-derived carcinomas that are often refractory to the standard of care.
Conclusions
Although AMPs have been extensively studied and developed as treatments for infectious diseases, their translation towards anticancer applications has been underexplored. Here, we demonstrate general design principles that can be used to repurpose AMP scaffolds into cancer-specific host defense peptides. This approach eliminates the need for large empirical screens and, hence, allows for a robust and rapid ACP discovery pipeline. Unique to this strategy is that it can develop anticancer agents that do not target a resistance-susceptible biochemical pathway, but instead act by physically disrupting key structural elements, including the plasma and nuclear membranes, that are difficult for cancer cells to mutationally alter. As an exemplary application, we show that the Tuberculosis-specific host defense peptide, MAD1, can be repurposed as a potent ovarian cancer targeting ACP. Surprisingly, this peptide displays an unusual specificity for human ovarian carcinomas and synergistically enhances the activity of clinical therapies towards drug-refractory and -resistant tumors. This is particularly impactful as many ovarian cancer patients are diagnosed at an advanced stage, and either relapse or succumb to the disease as a result of chemotherapeutic resistance. This suggests that ACPs engineered from AMP templates, which synergistically exploit lytic and apoptotic anticancer mechanisms, may be a unique and potentially transformative addition to combinatorial therapies against drug-resistant gynecological cancers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the laboratory of Dr. Joel Schneider at the National Cancer Institute for providing cancer cell lines. We also thank Dr. Matthew Taylor (Penn State University, College of Medicine, Hershey, PA) for donating the NL20 cell line. We thank the laboratories of Dr. Zissis Chroneos (Penn State University, College of Medicine, Hershey, PA), Dr. Pak Kin Wong (Penn State University, PA), and Dr. Kenneth Keiler (Penn State University, PA) for sharing the bacterial strains employed in these studies. We acknowledge and thank the Penn State Microscopy and Cytometry Facility—University Park, PA for assistance with confocal and electron microscopy. We also acknowledge the Penn State X-Ray Crystallography Facility—University Park, PA for use of the CD spectrophotometer. Funding for this research was provided by the Penn State Institute of Energy and the Environment Human Health and the Environment Seed Grant (S.H.M.), NIH:R00CA194309 (K.M.A.), NIH:F31CA236372 (E.S.D.), Penn State College of Engineering Grant (M.R.A.) and Penn State Graduate Research Fellowship (A.W.S.).
Conflict of interest
Authors Matthew R. Aronson, Erika S. Dahl, Jacob A. Halle, Andrew W. Simonson, Rose A. Gogal, Adam B. Glick, Katherine M. Aird and Scott H. Medina declare they have no conflict of interest.
Research Involving Human Rights
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer. 2003;3:502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed N, Stenvers K. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front. Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan C-X, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 4.Al-Benna S, Shai Y, Jacobsen F, Steinstraesser L. Oncolytic activities of host defense peptides. Int. J. Mol. Sci. 2011;12:8027–8051. doi: 10.3390/ijms12118027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012;30:679. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 6.Andrushchenko VV, Vogel HJ, Prenner EJ. Solvent-dependent structure of two tryptophan-rich antimicrobial peptides and their analogs studied by FTIR and CD spectroscopy. Biochim. Biophys. Acta - Biomembr. 2006;1758:1596–1608. doi: 10.1016/j.bbamem.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Anugraham M, Jacob F, Nixdorf S, Everest-Dass AV, Heinzelmann-Schwarz V, Packer NH. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: Glycan structures reflect gene expression and DNA methylation status. Mol. Cell. Proteomics. 2014;13:2213–2232. doi: 10.1074/mcp.M113.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronson MR, Simonson AW, Orchard LM, Llinás M, Medina SH. Lipopeptisomes: Anticancer peptide-assembled particles for fusolytic oncotherapy. Acta Biomater. 2018;80:269–277. doi: 10.1016/j.actbio.2018.09.025. [DOI] [PubMed] [Google Scholar]
- 9.Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals. 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Ding H, Feng P, Lin H, Chou K-C. iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget. 2016;7:16895. doi: 10.18632/oncotarget.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochran AG, Skelton NJ, Starovasnik MA. Tryptophan zippers: stable, monomeric β-hairpins. Proc. Natl. Acad. Sci. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeStefano CB, Liu SV. Combinatorial immunotherapy and chemotherapy. In: Patel SP, Kurzrock R, editors. Early Phase Cancer Immunotherapy. Cham: Springer; 2018. pp. 199–218. [Google Scholar]
- 14.Eckstein N. Platinum resistance in breast and ovarian cancer cell lines. J. Exp. Clin. Cancer Res. 2011;30:91. doi: 10.1186/1756-9966-30-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emelianova AA, Kuzmin DV, Panteleev PV, Sorokin M, Buzdin AA, Ovchinnikova TV. Anticancer activity of the goat antimicrobial peptide ChMAP-28. Front. Pharmacol. 2018;9:1501. doi: 10.3389/fphar.2018.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felício MR, Silva ON, Gonçalves S, Santos NC, Franco OL. Peptides with dual antimicrobial and anticancer activities. Front. Chem. 2017;5:5. doi: 10.3389/fchem.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 18.Gajski G, Garaj-Vrhovac V. Melittin: a lytic peptide with anticancer properties. Environ. Toxicol. Pharmacol. 2013;36:697–705. doi: 10.1016/j.etap.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Gaspar D, Veiga AS, Castanho MARB. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013;4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou L, Zhao X, Wang P, Ning Q, Meng M, Liu C. Antitumor activity of antimicrobial peptides containing CisoDGRC in CD13 negative breast cancer cells. PLoS ONE. 2013;8:e53491. doi: 10.1371/journal.pone.0053491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Wang X, Wang H, Liu Y, Chen Y. Studies on mechanism of action of anticancer peptides by modulation of hydrophobicity within a defined structural framework. Mol. Cancer Ther. 2011;10:416–426. doi: 10.1158/1535-7163.MCT-10-0811. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa K, Medina SH, Schneider JP, Klar AJSS. Glycan alteration imparts cellular resistance to a membrane-lytic anticancer peptide. Cell Chem. Biol. 2017;24:149–158. doi: 10.1016/j.chembiol.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim I-W, Lee JH, Kwon Y-N, Yun E-Y, Nam S-H, Ahn M-Y, Kang D-C, Hwang JS. Anticancer activity of a synthetic peptide derived from harmoniasin, an antibacterial peptide from the ladybug Harmonia axyridis. Int. J. Oncol. 2013;43:622–628. doi: 10.3892/ijo.2013.1973. [DOI] [PubMed] [Google Scholar]
- 24.Kościuczuk EM, Lisowski P, Jarczak J, Strzałkowska N, Jóźwik A, Horbańczuk J, Krzyżewski J, Zwierzchowski L, Bagnicka E. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012;39:10957–10970. doi: 10.1007/s11033-012-1997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar P, Kizhakkedathu JN, Straus SK. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules. 2018;8:4. doi: 10.3390/biom8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda K, Fukuda T, Isogai H, Okumura K, Krstic-Demonacos M, Isogai E. Antimicrobial peptide FF/CAP18 induces apoptotic cell death in HCT116 colon cancer cells via changes in the metabolic profile. Int. J. Oncol. 2015;46:1516–1526. doi: 10.3892/ijo.2015.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawler SE, Speranza M-C, Cho C-F, Chiocca EA. Oncolytic viruses in cancer treatment: a review. JAMA Oncol. 2017;3:841–849. doi: 10.1001/jamaoncol.2016.2064. [DOI] [PubMed] [Google Scholar]
- 28.Manzo G, Scorciapino MA, Wadhwani P, Bürck J, Pietro Montaldo N, Pintus M, Sanna R, Casu M, Giuliani A, Pirri G, Luca V, Ulrich AS, Rinaldi AC. Enhanced amphiphilic profile of a short β-stranded peptide improves its antimicrobial activity. PLoS ONE. 2015;10:1–18. doi: 10.1371/journal.pone.0116379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina SH, Schneider JP. Cancer cell surface induced peptide folding allows intracellular translocation of drug. J. Control. Release. 2015;209:317–326. doi: 10.1016/j.jconrel.2015.05.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Przybyło M, Hoja-Lukowicz D, Lityńska A, Laidler P. Different glycosylation of cadherins from human bladder non-malignant and cancer cell lines. Cancer Cell Int. 2002;2:6. doi: 10.1186/1475-2867-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramón-García S, Mikut R, Ng C, Ruden S, Volkmer R, Reischl M, Hilpert K, Thompson CJ. Targeting mycobacterium tuberculosis and other microbial pathogens using improved synthetic antibacterial peptides. Antimicrob. Agents Chemother. 2013;57:2295–2303. doi: 10.1128/AAC.00175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reißer S, Strandberg E, Steinbrecher T, Ulrich AS. 3D hydrophobic moment vectors as a tool to characterize the surface polarity of amphiphilic peptides. Biophys. J. 2014;106:2385–2394. doi: 10.1016/j.bpj.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riedl S, Zweytick D, Lohner K. Membrane-active host defense peptides–challenges and perspectives for the development of novel anticancer drugs. Chem. Phys. Lipids. 2011;164:766–781. doi: 10.1016/j.chemphyslip.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivel T, Ramseyer C, Yesylevskyy S. The asymmetry of plasma membranes and their cholesterol content influence the uptake of cisplatin. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-41903-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo M-D, Won H-S, Kim J-H, Mishig-Ochir T, Lee B-J. Antimicrobial peptides for therapeutic applications: a review. Molecules. 2012;17:12276–12286. doi: 10.3390/molecules171012276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma B, Kanwar SS. Phosphatidylserine: a cancer cell targeting biomarker. Semin. Cancer Biol. 2018;52:17–25. doi: 10.1016/j.semcancer.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Simonson AW, Lawanprasert A, Goralski TDP, Keiler KC, Medina SH. Bioresponsive peptide-polysaccharide nanogels—a versatile delivery system to augment the utility of bioactive cargo. Nanomedicine. 2019;17:391–400. doi: 10.1016/j.nano.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonson AW, Mongia AS, Aronson MR, Alumasa JN, Chan DC, Bolotsky A, Ebrahimi A, Proctor EA, Keiler KC, Medina SH. Pathogen-specific de novo antimicrobials engineered through membrane porin biomimicry. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12267992.v1. [DOI] [Google Scholar]
- 39.Suarez-Jimenez G-M, Burgos-Hernandez A, Ezquerra-Brauer J-M. Bioactive peptides and depsipeptides with anticancer potential: sources from marine animals. Mar. Drugs. 2012;10:963–986. doi: 10.3390/md10050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao P, Wang R, Lai L. Calculating partition coefficients of peptides by the addition method. J. Mol. Model. 1999;5:189–195. [Google Scholar]
- 41.Teixeira V, Feio MJ, Bastos M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012;51:149–177. doi: 10.1016/j.plipres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Thundimadathil J. Cancer treatment using peptides: current therapies and future prospects. J. Amino Acids. 2012;2012:967347. doi: 10.1155/2012/967347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyagi A, Kapoor P, Kumar R, Chaudhary K, Gautam A, Raghava GPS. In silico models for designing and discovering novel anticancer peptides. Sci. Rep. 2013;3:2984. doi: 10.1038/srep02984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uphoff CC, Drexler HG. Detection of mycoplasma contaminations. In: Helgason CD, Miller CL, editors. Basic Cell Culture Protocols. Totowa, NJ: Humana Press; 2005. pp. 13–23. [Google Scholar]
- 45.Zhao J, Huang Y, Liu D, Chen Y. Two hits are better than one: synergistic anticancer activity of α-helical peptides and doxorubicin/epirubicin. Oncotarget. 2015;6:1769. doi: 10.18632/oncotarget.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng L-H, Wang Y-J, Sheng J, Wang F, Zheng Y, Lin X-K, Sun M. Antitumor peptides from marine organisms. Mar. Drugs. 2011;9:1840–1859. doi: 10.3390/md9101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.