Highlights
-
•
This meta-analysis is a comprehensively studies of microRNA in MM.
-
•
MicroRNA clusters helped to distinguish MM and healthy people.
-
•
MicroRNA is a potential noninvasive biomarker in early diagnosis of MM.
Keywords: Multiple myeloma, MicroRNAs, Diagnosis, Meta-analysis
Abbreviations: MM, Multiple myeloma; microRNA, miRNA; QUADAS-2, Quality Assessment of Diagnostic Accuracy Study 2; SE, Sensitivity; SP, Specificity; PLR, Positive likelihood ratio; NLR, Negative likelihood ratio; DOR, Diagnostic odds ratio; AUC, Area under the curve; CI, confidence interval; MGUS, Monoclonal gammopathy of undetermined significance; PCL, Plasma cell leukemia
Abstract
Background
Multiple myeloma (MM) is the second incurable hematological malignancy. In recent years, due to the rise of microRNA (miRNA), many scholars have participated in the study of its value in the diagnosis of MM, and have obtained good but inconsistent results. Therefore, in order to determine the role of miRNA in the early diagnosis of MM, we performed this meta-analysis.
Methods
We searched for related studies including PubMed, Web of Science, EMBASE, Cochrane Library, China National Knowledge Infrastructure (CNKI) and Wanfang Database as of July 20, 2020 to conduct this meta-analysis. To improve the accuracy, the quality assessment of Diagnostic Accuracy Study 2 (QUADAS-2) was used. We also applied random effects models to summarize sensitivity and specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and area under the curve (AUC) to measure diagnostic values, and subgroup analysis used to discover potential sources of heterogeneity.
Results
We finally collected 32 studies from 15 articles that included a total of 2053 MM patients and 1118 healthy controls in this meta-analysis. The overall sensitivity, specificity, PLR, NLR, DOR and AUC were 0.81, 0.85, 5.5, 0.22, 25 and 0.90, respectively. Subgroup analysis shows that the down-regulation of microRNA clusters with larger samples size of plasma type could carry out a better diagnostic accuracy of MM patients. In addition, publication bias was not found.
Conclusions
Circulating miRNA could be a potential non-invasive biomarker for early diagnosis of MM. However, multi-center, more rigorous, and larger-scale studies are needed to verify our conclusions.
1. Introduction
Multiple myeloma (MM) is a malignant proliferative disease of plasma cells mediated by B lymphocytes, characterized by excessive proliferation of abnormal plasma cells in the bone marrow, bone damage, and immune deficiency[1]. The disease is still the second incurable hematological malignancy, accounting for about 1% of all types of human cancers and 13% of all hematological malignancies. It is more common in middle-aged and elderly people over 40 ages[2], and the survival time of the patient ranges from several months to several years[3], and the incidence of MM has gradually increased in recent years[4], [5]. The onset of MM is a gradual evolutionary process, from the initial monoclonal gammopathy of undetermined significance (MGUS) to smoldering MM, intramedullary MM, and finally to non-bone marrow MM / plasma cell leukemia (PCL)[6], [7]. Despite the advent of multiple targeted new drugs such as immunomodulatory drugs (IMiDs)[8] and proteasome inhibitors (PI)[9], which have greatly improved the quality of life of patients with MM, some patients have been affected by cytogenetic abnormalities and changes in the bone marrow microenvironment, and malignant transformation of plasma cells has occurred, causing MGUS to progress to MM[10]. As the traditional gold standard for diagnosing MM, bone marrow biopsy may not be accepted by all patients, because of its invasive injury that causes pain for patients, and the condition may have reached an advanced stage. There is an urgent need to find a more sensitive, convenient and non-invasive biomarker for early clinical diagnosis of MM.
MicroRNAs, a class of small non-coding RNAs (comprising 18 to 22 nucleotides), which regulates gene expression, affects a variety of cell biological processes and linked to cancer development[11], [12]. Circulating miRNA have been put forward as attractive diagnostic tools[13], [14] due to its strong specificity, high accuracy and stability[15]. During the formation and evolution of MM cells, many miRNAs related to the pathogenesis of MM were abnormally expressed, suggesting that miRNAs may also play an important role in the occurrence and development of MM[16]. Studies have found that in the bone marrow tissues of patients with MM, the expression of miRNA-181a was increased, and the expression of miRNA-373 was decreased. The expressions of the two were negatively correlated and jointly participate in the occurrence and development of tumors, and proved that the expression levels of these two miRNAs were related to the age, clinical stage, degree of differentiation, and lymph node metastasis of MM patients[17]. Therefore, the study of miRNA expression profiles and gene regulatory networks related to MM can help to understand the mechanism of MM and which has opened up new possibilities for improving the early diagnosis and treatment of MM.
With the rise of miRNAs in recent years, many studies have shown that the accuracy of circulating miRNAs in the early diagnosis of MM is satisfactory but inconsistent[[18], [19], [20]]. This may be caused by factors such as different detection technologies, platforms, standards, and insufficient clinical samples. Therefore, we carried out this meta-analysis to evaluate the worth of circulating miRNA in the early diagnosis of MM patients.
2. Materials and methods
2.1. Search strategy and literature selection
Two investigators independently searched the PubMed, Web of Science, EMBASE, Cochrane Library, China National Knowledge Infrastructure (CNKI) and Wanfang database without language restrictions. The search MeSH terms were used as follows: “multiple myeloma” and “ miRNA ” or “ microRNA ”. The searches were confined to publications involving human subjects, and the last search was conducted on 20/07/2020.
2.2. Inclusion and exclusion criteria
The inclusion criteria: (a) All studies involved newly diagnosed multiple myeloma patients and healthy controls; (b) The obtained miRNA is derived from serum or plasma specimens; (c) The literature contained relevant statistics such as sensitivity, specificity, and AUC value.
The exclusion criteria: (a) Duplicated information; (b) The literature were letters, comments, case reports or reviews; (c) The microRNA obtained is not derived from peripheral circulating blood, such as animal experiments, cell lines or biopsy; (d) Lack of sufficient data.
2.3. Data collection and study assessment
Two researchers separately extracted the following data from the full text and corresponding supplementary information of eligible articles: the name of the first author, country/region, year of publication, miRNA type (single or cluster), gene expression (up-regulation or down-regulation), sample size (number of patients with MM / healthy people), type of specimen (serum or plasma), as well as relevant statistical data and methodological quality information. The quality of included studies were assessed independently by two investigators using diagnostic accuracy studies-2 (QUADAS-2) criteria[22]. If two investigators disagreed, consulted the third investigator (WTZ) and reached a consensus.
2.4. Statistical analysis
All statistical analyses were done by Review Manager 5.2 and version 13.0 of STATA. The Higgins’s inconsistency index (I2) statistic was used to assess the heterogeneity between these studies. If the I2 value was over 50%, it considered that there was obvious heterogeneity, and then the random effects model was conducted. We also combined sensitivity, specificity, diagnostic odds ratio (DOR), positive likelihood ratio (PLR), negative likelihood ratio (NLR) and the area under the curve (AUC), and the corresponding 95% CI was calculated the overall and subgroup analysis. In addition, subgroup analysis was performed to explore the heterogeneity. At last, we used Deeks’ funnel plot analysis to explore the potential publication bias.
3. Results
3.1. Article screening flowchart
Two investigators independently searched databases, including PubMed, Web of Science, Embase, Cochrane Library, Chinese National Knowledge Infrastructure (CNKI) and Wan-fang databases, a total of 1670 articles. After excluding irrelevant, duplicates, reviews, letters, case report, and not human studies, 67 articles were left. We read these articles carefully to assess eligibility and screening based on inclusion and exclusion criteria, 15 articles (32 studies) were finally included in this meta-analysis. (Fig. 1)
Fig. 1.
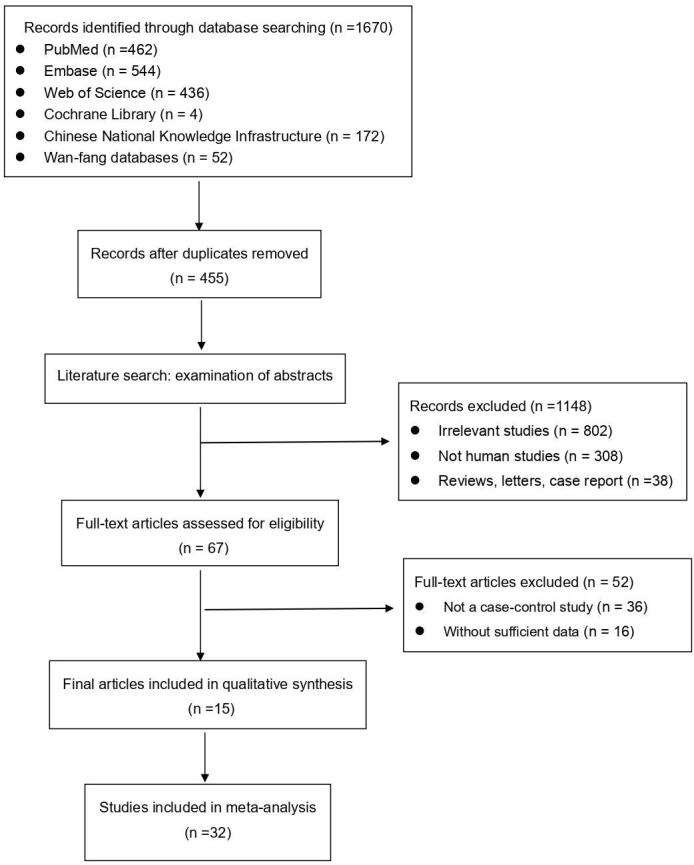
Flow chart of the meta-analysis.
3.2. Basic characteristics and quality assessment of included literature
The main characteristics of the 32 studies were shown in Table 1, ranging from 2012 to 2020. Refer to the QUADAS-2 tool to evaluate the quality of all the included literature. The evaluation results show that the overall quality of the studies included in this meta-analysis was high. (Fig. 2)
Table 1.
Characteristics of the included studies.
| Author | Year | Country | microRNAs | Regulation mode | Sample size | Speci-men | Diagnostic power | |||
|---|---|---|---|---|---|---|---|---|---|---|
| MM | Healthy | Sen (%) | Spe (%) | AUC | ||||||
| Single miRNA | ||||||||||
| Kubiczkova, L.[23] | 2014 | Czech Republic | let-7d | Downregulated | 103 | 30 | Serum | 0.641 | 0.867 | 0.804 |
| Kubiczkova, L.[23] | 2014 | Czech Republic | let-7e | Downregulated | 103 | 30 | Serum | 0.888 | 0.633 | 0.829 |
| Li, J.[24] | 2020 | China | miR-15a-5p | Upregulated | 23 | 18 | Serum | 0.870 | 0.610 | 0.804 |
| Li, F.[25] | 2015 | China | miR-16–1 | Downregulated | 90 | 19 | Plasma | 1.000 | 0.730 | 0.864 |
| Hao, M.[26] | 2015 | China | miR-19a | Upregulated | 108 | 56 | Serum | 0.773 | 0.897 | 0.910 |
| Qiu, X. Y.[27] | 2013 | China | miR-20a | Downregulated | 40 | 20 | Plasma | 0.63 | 0.85 | 0.74 |
| Sevcikova, S.[28] | 2013 | Czech Republic | miR-29a | Upregulated | 91 | 30 | Serum | 0.880 | 0.700 | 0.832 |
| Xu, Y. N.[29] | 2017 | China | miR-29a | Upregulated | 40 | 20 | Serum | 0.815 | 0.722 | 0.763 |
| Kubiczkova, L.[23] | 2014 | Czech Republic | miR-34a | Upregulated | 103 | 30 | Serum | 0.777 | 0.700 | 0.790 |
| Yoshizawa, S.[19] | 2012 | Japan | miR-92a | Downregulated | 62 | 133 | Plasma | 0.919 | 0.991 | 0.981 |
| Hao, M.[26] | 2015 | China | miR-92a | Upregulated | 108 | 56 | Serum | 0.724 | 0.869 | 0.830 |
| Jiang, Y.[30] | 2018 | China | miR-125b-5p | Upregulated | 35 | 20 | Plasma | 0.860 | 0.960 | 0.954 |
| Kubiczkova, L.[23] | 2014 | Czech Republic | miR-130a | Downregulated | 103 | 30 | Serum | 0.575 | 0.900 | 0.722 |
| Li, J.[24] | 2020 | China | miR-134-5p | Upregulated | 23 | 18 | Serum | 0.870 | 0.667 | 0.812 |
| Hao, M.[26] | 2015 | China | miR-135b-5p | Upregulated | 108 | 56 | Serum | 0.667 | 0.833 | 0.810 |
| Nidhi Gupta.[21] | 2019 | Germany | miR-143 | Upregulated | 30 | 30 | Serum | 0.767 | 0.767 | 0.854 |
| Nidhi Gupta.[21] | 2019 | Germany | miR-144 | Upregulated | 30 | 30 | Serum | 0.733 | 0.733 | 0.784 |
| Xie, L. L. [31] | 2018 | China | miR-148a | Upregulated | 50 | 30 | Serum | 0.760 | 0.700 | 0.791 |
| Xu, Y. N.[29] | 2017 | China | miR-155 | Downregulated | 40 | 20 | Serum | 0.800 | 0.722 | 0.862 |
| Nidhi Gupta.[21] | 2019 | Germany | miR-199 | Upregulated | 30 | 30 | Serum | 0.800 | 0.800 | 0.90 |
| Nidhi Gupta.[21] | 2019 | Germany | miR-203 | Upregulated | 30 | 30 | Serum | 0.833 | 0.833 | 0.930 |
| Jiang, Y.[30] | 2018 | China | miR-490-3p | Upregulated | 35 | 20 | Plasma | 0.600 | 0.850 | 0.866 |
| Cai, L.[32] | 2019 | China | miR-497 | Downregulated | 63 | 50 | Serum | 0.860 | 0.960 | 0.933 |
| Jones, C. I.[33] | 2012 | UK | miR-720 | Upregulated | 24 | 13 | Serum | 0.872 | 0.923 | 0.911 |
| Kubiczkova, L.[23] | 2014 | Czech Republic | miR-744 | Downregulated | 103 | 30 | Serum | 0.728 | 0.667 | 0.715 |
| Jones, C. I.[33] | 2012 | UK | miR-1308 | Downregulated | 24 | 13 | Serum | 0.821 | 0.923 | 0.892 |
| Hao, M.[26] | 2015 | China | miR-4254 | Upregulated | 108 | 56 | Serum | 0.793 | 0.985 | 0.920 |
| Shen, X.[34] | 2017 | China | miR-4449 | Upregulated | 71 | 64 | Serum | 0.789 | 0.913 | 0.885 |
| miRNA cluster | ||||||||||
| Hao, M.[26] | 2015 | China | miR-19a + miR-4254 | Upregulated | 108 | 56 | Serum | 0.917 | 0.905 | 0.950 |
| Liu, B.[35] | 2015 | China | miR-21/miR-199b-5p | Upregulated | 24 | 30 | Serum | 0.960 | 1.000 | 0.990 |
| Xu, Y. N.[29] | 2017 | China | miR-29a/miR-155 | Upregulated | 40 | 20 | Serum | 0.808 | 0.833 | 0.874 |
| Kubiczkova, L.[23] | 2014 | Czech Republic | miR-34a + let-7e | Upregulated | 103 | 30 | Serum | 0.806 | 0.867 | 0.898 |
Fig. 2.
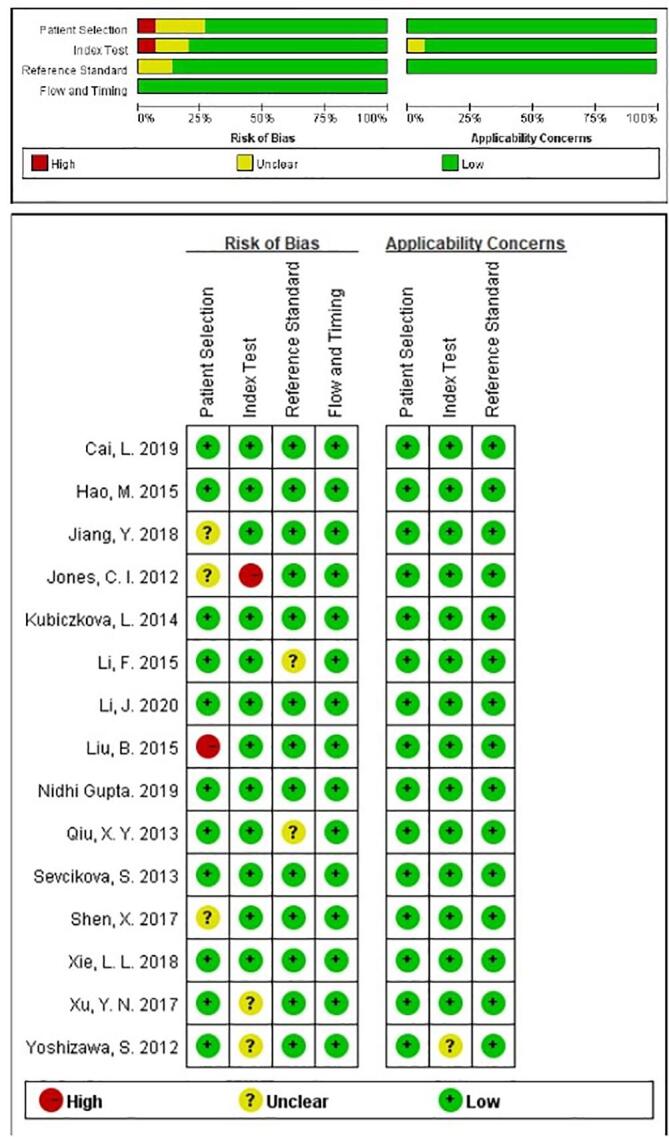
Overall methodological quality assessments of included articles based on QUADAS-2 tool.
3.3. Diagnostic accuracy of circulating microRNAs in MM patients from healthy controls
We included in the pooled analysis a total of 32 studies involving 3,171 participants (2,053 MM and 1,118 healthy controls). Significant heterogeneity was found in our meta-analysis, as demonstrated by the results (I2 = 78.75% for sensitivity and I2 = 72.02% for specificity, respectively). Thus, the random-effect model was used in our meta-analysis. Overall, the pooled sensitivity was 0.81 (95% CI: 0.77–0.85), specificity was 0.85 (95% CI: 0.82–0.89), PLR was 5.5 (95% CI: 4.1–7.5), NLR was 0.22 (95% CI: 0.18–0.27), DOR was 25 (95% CI: 16–39), and AUC was 0.90 (95% CI: 0.87–0.92). (Fig. 3) The above results suggest that miRNA can be served as an adjuvant tool in differentiating MM patients from healthy controls.
Fig. 3.
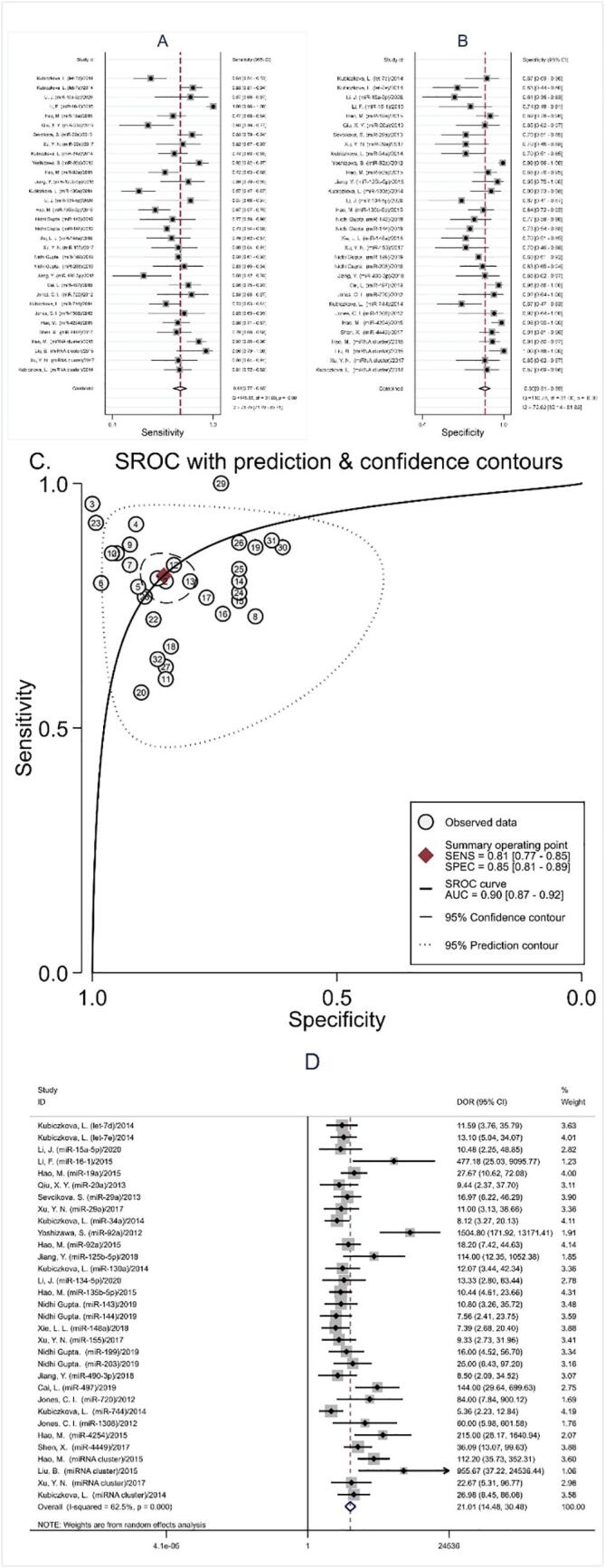
Diagnostic value of microRNAs in MM patients from healthy controls in all studies. (A) Sensitivity; (B) Specificity; (C) AUC; (D) DOR.
3.4. Diagnostic value of miRNA cluster in MM patients
There were 4 studies focused on miRNA clusters. As shown in Fig. 4, the pooled sensitivity was 0.88 (95% CI: 0.78–0.94), specificity was 0.92 (95% CI: 0.82–0.96), NLR was 0.13 (95% CI: 0.07–0.26), PLR was 10.4 (95% CI: 4.4–24.8), DOR was 80 (95% CI: 19–336), and AUC was 0.96 (95% CI: 0.93–0.97). The results manifested that microRNAs cluster had a relatively high diagnostic accuracy in early diagnosis of MM patients.
Fig. 4.
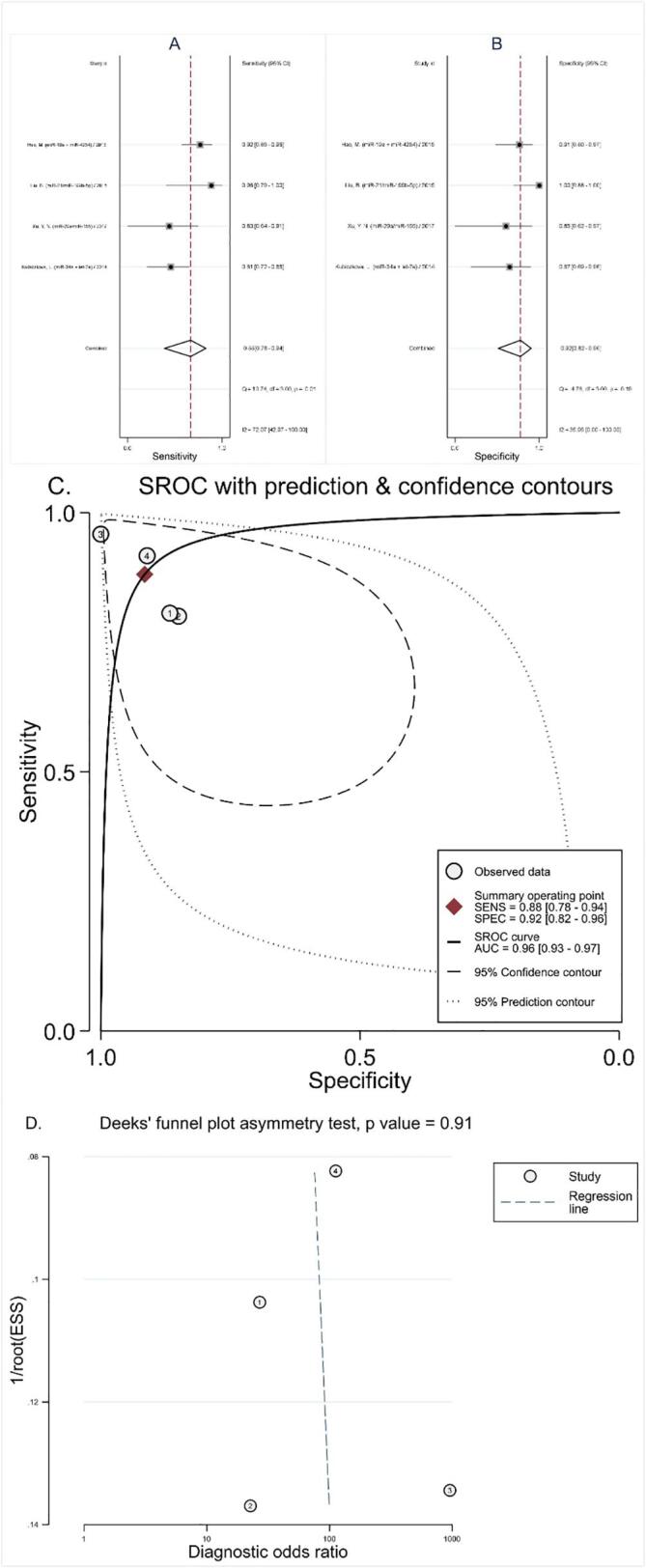
Diagnostic value of miRNA cluster in diagnosing MM patients from healthy controls. (A) Sensitivity; (B) Specificity; (C) AUC; (D) Funnel plot.
3.5. Subgroup analysis
Since the above methods proved that there was heterogeneity between the included study results, further subgroup analysis was performed according to specimen types, regulation mode, miRNAs profiling, sample size and ethnicity. The relevant statistic for each subgroup analysis were demonstrated in Table 2. It showed that using miRNA clusters to diagnose MM had more advantages than single miRNA: sensitivity (0.88 vs. 0.80), specificity (0.92 and 0.84) and AUC (0.96 and 0.89). Furthermore, down-regulated miRNAs have better diagnostic accuracy than up-regulated miRNAs: sensitivity (0.83 vs. 0.80), specificity (0.87 vs. 0.84) and AUC (0.92 vs. 0.88). In addition, compared with serum, the sensitivity, specificity and AUC of miRNA testing in plasma were higher, 0.89 vs. 0.80, 0.92 vs. 0.84, and 0.96 vs. 0.88, respectively. Moreover, the larger sample size showed higher accuracy: sensitivity (0.82 vs. 0.80), specificity (0.88 vs. 0.81) and AUC (0.92 vs. 0.87). Finally, the analysis based on ethnicity demonstrated miRNA yield a better diagnosis accuracy in the Asian race populations than European.
Table 2.
Summary estimates of diagnostic power and their 95% confidence intervals.
| Subgrupo | Se (95% CI) | Sp(95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) |
|---|---|---|---|---|---|---|
| miRNAs profiling | ||||||
| Single miRNA | 0.80 [0.76–0.84] | 0.84 [0.79–0.88] | 5.1 [3.7–6.9] | 0.24 [0.19–0.29] | 21 [14–33] | 0.89 [0.86–0.91] |
| Multiple miRNAs | 0.88 [0.78–0.94] | 0.92 [0.82–0.96] | 10.4 [4.4–24.8] | 0.13 [0.07–0.26] | 80 [19–336] | 0.96 [0.93–0.97] |
| Regulation modo | ||||||
| Upregulated | 0.80 [0.77–0.83] | 0.84 [0.79–0.88] | 5.2 [3.8–6.9] | 0.23 [0.19–0.28] | 22 [14–34] | 0.88 [0.85–0.91] |
| Downregulated | 0.83 [0.70–0.91] | 0.87 [0.76–0.94] | 6.5 [3.2–13.1] | 0.20 [0.11–0.36] | 33 [11–97] | 0.92 [0.89–0.94] |
| Sample size | ||||||
| ≥100 | 0.82 [0.75–0.87] | 0.88 [0.82–0.92] | 6.7 [4.2–10.7] | 0.21 [0.15–0.29] | 32 [17–64] | 0.92 [0.89–0.94] |
| <100 | 0.80 [0.75–0.84] | 0.81 [0.75–0.86] | 4.2 [3.1–5.9] | 0.25 [0.19–0.32] | 17 [10–29] | 0.87 [0.84–0.90] |
| Specimen types | ||||||
| Serum | 0.80 [0.76–0.83] | 0.84 [0.79–0.88] | 4.9 [3.7–6.4] | 0.24 [0.20–0.29] | 20 [14–30] | 0.88 [0.85–0.90] |
| Plasma | 0.89 [0.60–0.98] | 0.92 [0.78–0.98] | 11.7 [3.9–35.6] | 0.12 [0.03–0.52] | 98 [14–675] | 0.96 [0.94–0.98] |
| Etnia | ||||||
| Asian | 0.83 [0.77–0.88] | 0.88 [0.82–0.93] | 7.2 [4.5–11.6] | 0.19 [0.14–0.27] | 38 [19–74] | 0.92 [0.89–0.94] |
| European | 0.78 [0.72–0.83] | 0.78 [0.73–0.83] | 3.6 [2.9–4.5] | 0.28 [0.23–0.35] | 13 [9–18] | 0.85 [0.81–0.88] |
Se: sensitivity, Sp specificity, PLR: positive likelihood ratios, NLR: negative likelihood ratios, DOR: diagnostic odds ratio, AUC: area under the curve, CI: confidence interval.
3.6. Publication bias
Stata 13.0 was used to evaluate publication bias and draw a Deeks funnel chart. As demonstrated in Fig. 5, Fig. 4, the pooled Deeks test result of all studies was P = 0.12, the pooled Deeks’ test result of microRNA clusters was P = 0.91, which indicated that no found publication bias in this meta-analysis.
Fig. 5.
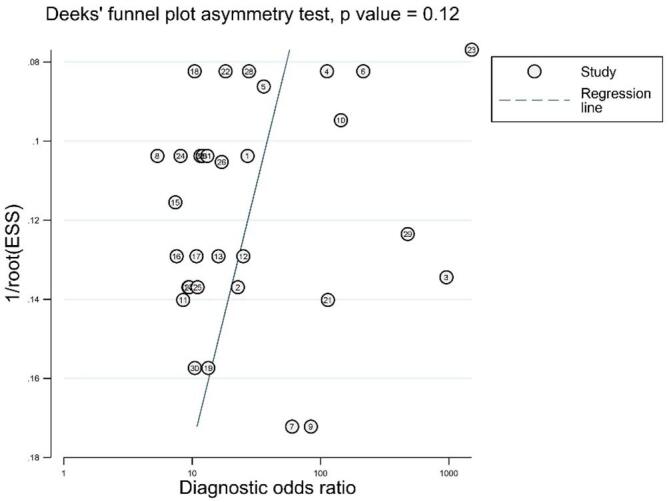
Deeks’ linear regression test of funnel plot asymmetry.
4. Discussion
As a common malignant tumor in clinical practice, MM has always been a hot spot for scholars. The emergence of targeted new drugs has greatly improved the quality of life of MM patients. However, for the early diagnosis of MM, that is, after the patient has bone pain, clinicians obviously cannot make a timely and accurate diagnosis based on traditional imaging examinations, which leads to the development of the patient's condition and delays the treatment[36]. Bone marrow biopsy is the traditional gold standard for diagnosis, but it is an invasive test that is very painful for patients. Therefore, we need a more accurate and advanced noninvasive biomarker for the early diagnosis of MM. According to the latest research, miRNA had strong specificity and sensitivity in diagnosing the occurrence and metastasis of malignant tumors[13], [37], and can be used as a new method for early diagnosis and monitoring of tumors. Several studies had analyzed the accuracy of circulating miRNAs in diagnosing MM. For instance, Cai[32] and Nidhi[21] et al. pointed out that the sensitivity and specificity of miR-497 and miR-199 in diagnosing MM were 86.0% and 96.0%, 80.0% and 80.0%, respectively. According to reports by Yoshizawa[19], the sensitivity and specificity of miRNA-92a were 91.9% and 99.1%, respectively, and its diagnostic accuracy was satisfactory. However, Hao et al.[26] found that the sensitivity and specificity of miRNA-92a were low, 72.4% and 86.9%, respectively. It can be seen that even the same miRNA has different accuracy due to different specimens, sample sizes and detection techniques. In addition, it had been shown that miRNA clusters (miR-21, miR-199b-5p) can separate MM patients from healthy peoples with a high sensitivity and specificity (96.0% and 100%)[35], which were more reliable than the results of a single miRNA.
Therefore, we carried out this meta-analysis to evaluate the value of circulating miRNAs in the early diagnosis of MM. The overall results of showed that the pooled sensitivity, specificity, PLR, NLR, and DOR of circulating miRNA detection in the diagnosis of MM were 0.81 (95% CI: 0.77–0.85), 0.85 (95% CI: 0.82–0.89), 5.5 (95% CI: 4.1–7.5), 0.22 (95% CI: 0.18–0.27), 25 (95% CI: 16–39), and 0.90 (95% CI: 0.87–0.92), respectively. AUC is a measure of the diagnostic accuracy test method: the closer the AUC is to 1, the greater the diagnostic value[38]. In addition, the pooled DOR showed that the probability of correctly diagnosing MM individuals was 25 times higher than the false negative diagnosis of healthy individuals. All of the results proved that circulating miRNAs showed relatively high accuracy in the diagnosis of MM.
In this meta-analysis, subgroup analysis was performed to find probable sources of heterogeneity. We found that down-regulation of microRNA clusters with larger samples size of plasma type could carry out a better diagnostic accuracy of MM patients. The expression of a single miRNA in serum or plasma lacks specificity in cancer detection, and it might fluctuate in other diseases[37]. However, miRNA clusters with complex molecular mechanisms can reflect the occurrence and development of tumors in many ways, forming a more stable and reliable network diagnostic structure[39]. According to reports, more proteins could be retained in plasma for co-isolation of miRNAs[40], and it had more clinical applications, which was consistent with our results. Our research also found that there was a difference between the down-regulation and the up-regulation of miRNA, this was consistent with Hui’s research results[41], this difference may be due to the number of miRNAs analyzed, the type and size of samples contained, the design statistical methods, and the different platforms used for microarray technology. Finally, the diagnostic value of miRNA for Asian races was higher than that for European. This may be related to the morbidity and sensitivity of race, and more multi-center genetic studies are needed to explain the reasons.
To the best of our knowledge, this meta-analysis was the latest, most comprehensive, objective and credible on the diagnostic value of miRNA in multiple myeloma. However, there were still some limitations. Although a comprehensive search strategy is used in our literature search, some valuable research may be lost. In addition, individual studies included relatively few patients, which limited the strength of our meta-analysis conclusions. Finally, in recent years, the microRNA has been widely used as a potential biomarker in different clinical settings, however, the diagnosis of MM in clinic using only circulating miRNAs still needs to be solved, and even other auxiliary examinations are needed.
5. Conclusion
In short, with the gradual deepening of research on circulating miRNAs, it had shown great advantages in the early diagnosis of MM. This method not only had high sensitivity and strong specificity, but also had non-invasive and no radiation risks. It is worth continuing to optimize its practicality. In the future, multi-center, more rigorous, and high-quality case-control studies are still needed in clinical practice to improve the efficacy of circulating miRNA in the early diagnosis of MM.
6. Ethics Committee
The data comes from literature search and does not require any ethics committee approval and patient informed consent.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Fairfield H., Falank C., Avery L., Reagan M.R. Multiple myeloma in the marrow: pathogenesis and treatments. Ann. N. Y. Acad. Sci. 2016;1364(1):32–51. doi: 10.1111/nyas.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander D.D., Mink P.J., Adami H.O., Cole P., Mandel J.S., Oken M.M., Trichopoulos D. Multiple myeloma: a review of the epidemiologic literature. Int. J. Cancer. 2007;120(Suppl 12):40–61. doi: 10.1002/ijc.22718. [DOI] [PubMed] [Google Scholar]
- 3.Fonseca R., Abouzaid S., Bonafede M., Cai Q., Parikh K., Cosler L., Richardson P. Trends in overall survival and costs of multiple myeloma, 2000–2014. Leukemia. 2017;31(9):1915–1921. doi: 10.1038/leu.2016.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong J., Lee J.H. Recent advances in multiple myeloma: a Korean perspective. Korean J Intern Med. 2016;31(5):820–834. doi: 10.3904/kjim.2015.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyle R.A., Larson D.R., Therneau T.M., Dispenzieri A., Kumar S., Cerhan J.R., Rajkumar S.V. Long-Term Follow-up of Monoclonal Gammopathy of Undetermined Significance. The New England journal of medicine. 2018;378(3):241–249. doi: 10.1056/NEJMoa1709974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kyle R.A., Rajkumar S.V. Monoclonal Gammopathy of Undetermined Significance and Smoldering Multiple Myeloma. Hematol. Oncol. Clin. North Am. 2007;21(6):1093–1113. doi: 10.1016/j.hoc.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Landgren O., Kyle R.A., Pfeiffer R.M., Katzmann J.A., Caporaso N.E., Hayes R.B., Dispenzieri A., Kumar S., Clark R.J., Baris D. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113(22):5412–5417. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raza S., Safyan R.A., Lentzsch S. Immunomodulatory Drugs (IMiDs) in Multiple Myeloma. Curr. Cancer Drug Targets. 2017;17(9):846–857. doi: 10.2174/1568009617666170214104426. [DOI] [PubMed] [Google Scholar]
- 9.Hungria V.T.D.M., Crusoé E.D.Q., Bittencourt R.I., Maiolino A., Magalhães R.J.P., Sobrinho J.D.N., Pinto J.V., Fortes R.C., Moreira E.D.S., Tanaka P.Y. New proteasome inhibitors in the treatment of multiple myeloma. Hematol Transfus Cell Ther. 2019;41(1):76–83. doi: 10.1016/j.htct.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhodapkar M.V. MGUS to myeloma: a mysterious gammopathy of underexplored significance. Blood. 2016;128(23):2599–2606. doi: 10.1182/blood-2016-09-692954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felekkis K., Touvana E., Stefanou C., Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236–240. [PMC free article] [PubMed] [Google Scholar]
- 12.Schwerk J., Savan R. Translating the Untranslated. Region. 2015;195(7):2963–2971. doi: 10.4049/jimmunol.1500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu B., Ju S., Chu H., Shen X., Zhang Y., Luo X., Cong H. The potential function of microRNAs as biomarkers and therapeutic targets in multiple myeloma. Oncology letters. 2018;15(5):6094–6106. doi: 10.3892/ol.2018.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Peng R., Wang J., Qin Z., Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clinical epigenetics. 2018;10(1):59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handa H., Murakami Y., Ishihara R., Kimura-Masuda K., Masuda Y. The Role and Function of microRNA in the Pathogenesis of Multiple Myeloma. Cancers. 2019;11(11):1738. doi: 10.3390/cancers11111738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan R., Liu N., Yang J., Peng J., Liu L., Guo X. The expression and role of miR-181a in multiple myeloma. Medicine. 2018;97(35):e12081. doi: 10.1097/MD.0000000000012081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao M., Zang M., Zhao L., Deng S., Xu Y., Qi F., An G., Qin Y., Sui W., Li F. Serum high expression of miR-214 and miR-135b as novel predictor for myeloma bone disease development and prognosis. Oncotarget. 2016;7(15):19589–19600. doi: 10.18632/oncotarget.7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshizawa S., Ohyashiki J.H., Ohyashiki M., Umezu T., Suzuki K., Inagaki A., Iida S., Ohyashiki K. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood cancer journal. 2012;2(1):e53. doi: 10.1038/bcj.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J., Zhang M., Wang C. Circulating miRNAs as diagnostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Journal of clinical laboratory analysis. 2020;34(6):e23233. doi: 10.1002/jcla.23233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta N., Kumar R., Seth T., Garg B., Sati H.C., Sharma A. Clinical significance of circulatory microRNA-203 in serum as novel potential diagnostic marker for multiple myeloma. J. Cancer Res. Clin. Oncol. 2019;145(6):1601–1611. doi: 10.1007/s00432-019-02896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M., Sterne J.A., Bossuyt P.M. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 23.Kubiczkova-Besse L., Sedlarikova L., Kryukov F., Radova L., Nekvindova J., Nemec P., Drandi D., Caltagirone S., Omedè P., Krejci M. Circulating miR-130a in multiple myeloma and extramedullary myeloma patients. Blood. 2014;124(21) [Google Scholar]
- 24.Li J., Zhang M., Wang C. Circulating miRNAs as diagnostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. J. Clin. Lab. Anal. 2020;34(6) doi: 10.1002/jcla.23233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F., Xu Y., Deng S.H., Li Z.J., Zou D.H., Yi S.H., Sui W.W., Hao M., Qiu L.G. MicroRNA-15a/16-1 cluster located at chromosome 13q14 is down-regulated but displays different expression pattern and prognostic significance in multiple myeloma. Oncotarget. 2015;6(35):38270–38282. doi: 10.18632/oncotarget.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao M., Zang M., Wendlandt E., Xu Y., An G., Gong D., Li F., Qi F., Zhang Y., Yang Y. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int. J. Cancer. 2015;136(8):1835–1844. doi: 10.1002/ijc.29199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiaoyan Q., Zhao M., Wu S., Wenjun Y., Jiaren X., Xu J., Li J., Lijuan C. Circulating microrna 483–5p in multiple myeloma as a novel biomarker for diagnosis and predicting survival. Blood. 2013;122(21) [Google Scholar]
- 28.Sevcikova S., Kubiczkova L., Sedlarikova L., Slaby O., Hajek R. Serum miR-29a as a marker of multiple myeloma. Leukemia & lymphoma. 2013;54(1):189–191. doi: 10.3109/10428194.2012.704030. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y.N., Xiao C.R., Huang Y.D., Lu Q.Y. Circulating Serum MicroRNA as Diagnostic Biomarkers for Multiple Myeloma. Zhongguo shi yan xue ye xue za zhi. 2017;25(2):471–475. doi: 10.7534/j.issn.1009-2137.2017.02.029. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y., Luan Y., Chang H., Chen G. The diagnostic and prognostic value of plasma microRNA-125b-5p in patients with multiple myeloma. Oncology letters. 2018;16(3):4001–4007. doi: 10.3892/ol.2018.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie L.L., Shi X.Y., Cong H. Expression and clinical significance of serum microRNA-148a in patients with multiple myeloma. [J]. Jiangsu Medical. 2018;44(10):1143–1147. doi: 10.19460/j.cnki.0253-3685.2018.10.014. [DOI] [Google Scholar]
- 32.Cai L., Ping Z. The Diagnostic and Prognostic Value of Plasma MicroRNA497 in Patients with Multiple Myeloma. [J] Inner Mongolia Med. 2019;51(8):903–906. doi: 10.16096/J.cnki.nmgyxzz.2019.51.08.003. [DOI] [Google Scholar]
- 33.Jones C.I., Zabolotskaya M.V., King A.J., Stewart H.J., Horne G.A., Chevassut T.J., Newbury S.F. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br. J. Cancer. 2012;107(12):1987–1996. doi: 10.1038/bjc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen X., Ye Y., Qi J., Shi W., Wu X., Ni H., Cong H., Ju S. Identification of a novel microRNA, miR-4449, as a potential blood based marker in multiple myeloma. Clin. Chem. Lab. Med. 2017;55(5):748–754. doi: 10.1515/cclm-2015-1108. [DOI] [PubMed] [Google Scholar]
- 35.Bo Liu, Long Li Shi, Yu You Jian. Studies on the roles of miRNA expression forecast in the multiple myeloma. Chinese Journal of Laboratory Diagnosis. 2015;(2):259–261. [Google Scholar]
- 36.Bergstrom D.J., Kotb R., Louzada M.L., Sutherland H.J., Tavoularis S., Venner C.P. Consensus Guidelines on the Diagnosis of Multiple Myeloma and Related Disorders: Recommendations of the Myeloma Canada Research Network Consensus Guideline Consortium. Clinical lymphoma, myeloma & leukemia. 2020;20(7):e352–e367. doi: 10.1016/j.clml.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Condrat C.E., Thompson D.C., Barbu M.G., Bugnar O.L., Boboc A., Cretoiu D., Suciu N., Cretoiu S.M., Voinea S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells. 2020;9(2) doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swets J.A. Measuring the Accuracy of Diagnostic Systems. Science (New York, NY) 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 39.Gao S.-S., Wang Y.-J., Zhang G.-X., Zhang W.-T. Potential diagnostic value of miRNAs in peripheral blood for osteosarcoma: A meta-analysis. Journal of Bone. Oncology. 2020;23 doi: 10.1016/j.jbo.2020.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei H., Pu K., Liu X.G., Li B.X., Zhang H.S., Wang H., Wang H., Sun W.M., Wang Y.P. The diagnostic value of circulating microRNAs as a biomarker for gastric cancer: A meta-analysis. Oncol. Rep. 2019;41(1):87–102. doi: 10.3892/or.2018.6782. [DOI] [PMC free article] [PubMed] [Google Scholar]


