Abstract
Cilia or flagella of eukaryotes are small micro-hair like structures that are indispensable to single-cell motility and play an important role in mammalian biological processes. Cilia or flagella are composed of nine doublet microtubules surrounding a pair of singlet microtubules called the central pair (CP). Together, this arrangement forms a canonical and highly conserved 9+2 axonemal structure. The CP, which is a unique structure exclusive to motile cilia, is a pair of structurally dimorphic singlet microtubules decorated with numerous associated proteins. Mutations of CP-associated proteins cause several different physical symptoms termed as ciliopathies. Thus, it is crucial to understand the architecture of the CP. However, the protein composition of the CP was poorly understood. This was because the traditional method of identification of CP proteins was mostly limited by available Chlamydomonas mutants of CP proteins. Recently, more CP protein candidates were presented based on mass spectrometry results, but most of these proteins were not validated. In this study, we re-evaluated the CP proteins by conducting a similar comprehensive CP proteome analysis comparing the mass spectrometry results of the axoneme sample prepared from Chlamydomonas strains with and without CP complex. We identified a similar set of CP protein candidates and additional new 11 CP protein candidates. Furthermore, by using Chlamydomonas strains lacking specific CP sub-structures, we present a more complete model of localization for these CP proteins. This work has established a new foundation for understanding the function of the CP complex in future studies.
Keywords: Cilia, Flagella, Central Pair, Mass Spectrometry, Electron Microscopy
Significance
Cilia and flagella are highly organized structures of eukaryotes which propel the cell motions or generate liquid flows around the cells. The central pair complex is located at the center of cilia and flagella. It is known to be essential for the regulation of beating of cilia and flagella, but its protein composition and architecture were poorly understood. By exploiting comprehensive mass spectrometry and several mutant strains of Chlamydomonas, we identified novel central pair proteins and mapped these proteins to the structure. Our model can be used as a foundation to understand the functions of the central pair complex.
Introduction
Cilia and flagella are common terms used to describe the same hair-like structure of eukaryotic cells and will therefore be used interchangeably in this paper. It is known that defective cilia are implicated in a variety of different human diseases, from developmental disorders to metabolic syndromes [1]. However not all cilia are alike. Primary cilia are non-motile and are commonly reported as sensory receptors [2]. Motile cilia, on the other hand, show a beating motion at high frequencies driven by motor protein dyneins [3]. This rudimentary motion is the driving force for a plethora of multi-level systems from single cell movement to mammalian organ function and maintenance [1]. Motile cilia also present in our respiratory system that beat together in order to clear mucus build up and infectious agents [4].
Cilia-related diseases, otherwise known as ciliopathies, such as primary ciliary dyskinesia (PCD), are derived from the impairment of motile cilia [5,6]. Patients who suffer from PCD often experience a wide spectrum of symptoms ranging from male infertility to an increased susceptibility to respiratory infections [7]. Failure to recognize or diagnose PCD early on can often be lethal later in life [4]. A common practice used to diagnose PCD is a cross-section analysis of patient nasal epithelium cilia using transmission electron microscopy (TEM). However due to the diversity of PCD mutations, many different defective proteins can lead to similar malformations [7]. Additionally, not all mutations produce visible differences at standard TEM resolution while still inducing PCD-like symptoms. The largest obstacle to our understanding of cilia-related defects is our limited comprehension of the proteins that comprise cilia.
Cilia are highly complex structures composed of different compartments. Motile cilia consist of nine doublet microtubules (DMTs) surrounding a pair of singlet microtubules called the central pair (CP) [8]. This specific arrangement defines the “9+2” structure of the axoneme (Fig. 1A). There exists axonemal dyneins (outer dynein arm, ODA, and inner dynein arm, IDA) attached to DMTs which are responsible for the beating of cilia. Radial spoke (RS) complexes extend from DMTs toward the CP. Intraflagellar transport (IFT) driven by IFT dyneins and IFT kinesins takes place on the DMTs [9]. This arrangement of the axoneme structure is highly conserved in all eukaryotes with motile cilia, suggesting that there exists a similar set of proteins and processes. Thus, we can study the axoneme composition using model organisms like a green algae Chlamydomonas reinhardtii
Figure 1.

Structures of the axoneme and the CP complex. (A and B) Schematic diagrams of the axoneme (A) and the CP complex structure (B) viewed from the base of flagella. The axoneme structure consists of nine DMTs radially surrounding the CP complex. The DMTs are decorated with ODA, IDA and RS complexes. IFT trains are transported at the space between the membrane and the DMTs. The CP consists of two structurally dimorphic singlets termed C1 and C2 and are connected by the bridge. Several distinct sub-structures bind around the singlets with a repeating pattern along the axis of the axoneme. Diagonal link is also known to connect the C2 with the C1b region. The model of the CP structure is adopted from [15]. (C) Cryo-ET of purified WT Chlamydomonas axoneme. Cross section (left) and longitudinal section (right) of 3D volume of demembranated WT axoneme are shown. Our purified WT axoneme retained both singlet microtubules of the CP complex with protruding sub-structures after demembranation step as shown in red arrowheads. Yellow dashed line indicates the section in the right panel. Scale bar represents 50 nm.
The presence of the CP distinguishes motile cilia from its immotile counterpart, primary cilia. The CP has a diverse array of functions including the regulation of local Ca2+ concentration, ATP/ADP concentration and axonemal dynein activities through mechanical interactions with the RS [10–13]. The CP is a huge protein complex composed of a pair of structurally and functionally dimorphic singlet microtubules named C1 and C2 and many other associated proteins [14]. The CP has a variety of sub-structures such as C1a, C1b, C1c and C1d on the C1 singlet, or C2a, C2b and C2c on the C2 singlet that were characterized by traditional cross-sectional electron microscopy (EM). C1 and C2 microtubules are connected by two structures called the “bridge” and the “diagonal link”. With recent higher resolution cryo-electron tomography (cryo-ET) structures, more details of these sub-structures have been characterized allowing the C1a to be sub-classified as C1a/e, the C1b as C1b/f, the C2a as C2a/e and the C2c as C2c/d (Fig. 1B) [15]. Through this manuscript, we follow the newer nomenclature of the CP sub-structures as in Figure 1B. These sub-structures bind with 16- or 32-nm repeating units along the axonemal axis [15]. Despite their unique existence in motile cilia and their importance to motility, the proteins that comprise the CP have been poorly understood. Traditionally, 22 proteins (apart from α- and β-tubulins) have been characterized as components of the CP complex (Table 1). For example, kinesin-like protein 1 (KLP1) is a phosphoprotein that localizes at the C2 microtubule around C2c/d region [25]. Mutations in several known CP proteins such as Hydin located at the C2b and FAP221 (PCDP1) located at the C1d have been previously shown to cause ciliopathic symptoms [10,16]. However, it was generally believed that there should be more unidentified proteins inside the CP complex. Due to random insertions of transgenes into the Chlamydomonas reinhardtii genome, previous characterization of CP proteins largely relied on phenotype-based screening of CP protein mutants [32,33]. This approach, however, remains inefficient and biased towards proteins which produce visible phenotypes. Recently, Zhao, L., et al. used a more comprehensive approach taking advantage of relative quantitative mass spectrometry (MS) comparing Chlamydomonas strains with and without intact CP [30]. By doing so, 44 proteins were proposed to be new CP protein candidates in addition to the traditionally known CP proteins. Though some of these proteins, FAP47, FAP76, FAP99, FAP246 and DPY30, were confirmed to locate at the CP by immunofluorescence study, the other candidates remain to be tested. Furthermore, the localizations of these new CP protein candidates in CP sub-structures were not clear.
Table 1.
Summary of MS results of traditionally known CP proteins
| Names | Uniprot ID | WT Exclusive unique peptide counta (Quantitative values after normalizationa) |
pf15
Exclusive unique peptide counta (Quantitative values after normalizationa) |
pf15/WT ratio (%) (Quantitative values were used) |
p-values (WT vs pf15) |
Localizations | References |
|---|---|---|---|---|---|---|---|
| Hydin | A8HQ52 | 68, 87, 27 (68, 75, 53) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.00059 | C2b | [16, 17] |
| FAP42 (C1b-350) | A8J614 | 63, 59, 36 (68, 50, 69) | 2, 2, 2 (2, 2, 2) | 3.6 | 0.00062 | C1b/f | [18] |
| PF6 | Q9ATK5 | 53, 65, 32 (65, 72, 82) | 4, 3, 5 (5, 3, 5) | 6.0 | 0.00016 | C1a/e | [19, 20] |
| CPC1 | Q6J4H1 | 55, 57, 30 (76, 66, 74) | 6, 7, 4 (7, 8, 4) | 8.8 | <0.00010 | C1b/f | [19] |
| FAP54 | A8J666 | 46, 61, 27 (44, 48, 49) | 0, 0, 1 (0, 0, 1) | 0.7 | <0.00010 | C1d | [10, 21] |
| FAP46 | A8ICS9 | 40, 50, 35 (52, 44, 78) | 1, 0, 2 (1, 0, 3) | 2.5 | 0.0052 | C1d | [10, 21] |
| FAP74 | D4P3R7 | 32, 42, 15 (34, 32, 33) | 0, 0, 1 (0, 0, 1) | 1.0 | <0.00010 | C1d | [10, 21] |
| FAP69 (C1b-135) | A8IF19 | 16, 24, 11 (17, 23, 26) | 1, 2, 2 (1, 2, 2) | 8.3 | 0.0023 | C1b/f | [18] |
| PF16 | A8J0A5 | 17, 19, 16 (34, 69, 74) | 1, 1, 1 (1, 1, 1) | 1.9 | 0.010 | C1 C1a-e-c complex | [22] [23] |
| HSP70b | A8JEU4 | 14, 22, 6 (17, 18, 10) | 5, 5, 2 (7, 6, 2) | 33 | 0.028 | C1b/f | [18, 24] |
| KLP1 | A8I9T2 | 15, 16, 3 (17, 13, 7) | 1, 2, 1 (1, 2, 2) | 15 | 0.025 | C2c/d | [25, 26] |
| FAP101 | A8I345 | 13, 16, 14 (17, 20, 30) | 0, 0, 1 (0, 0, 1) | 1.6 | 0.0055 | C1a/e | [19] |
| Enolaseb | A8JH98 | 17, 12, 9 (23, 31, 17) | 7, 9, 6 (9, 11, 8) | 39 | 0.017 | C1b/f | [18] |
| FAP221 (Pcdp1) | A8J6X7 | 7, 9, 7 (6, 5, 12) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.015 | C1d | [10, 21] |
| FAP114 (C1a-32) | Q45QX5 | 7, 7, 5 (13, 8, 16) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0081 | C1a/e | [19] |
| FAP119 (C1a-34) | Q45QX4 | 7, 7, 3 (8, 7, 5) | 0, 0, 1 (0, 0, 1) | 5.1 | 0.0034 | C1a/e | [19] |
| FAP297 (WDR93) | A8HQE0 | 4, 8, 2 (6, 6, 3) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0040 | C1d | [21] |
| PF20 | A8ITB4 | 3, 7, 2 (3, 6, 3) | 1, 0, 0 (1, 0, 0) | 9.9 | 0.028 | C1-C2 bridge | [27] |
| PP1ab | Q9XGU3 | 2, 2, 1 (2, 2, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | <0.00010 | C1 | [28] |
| FAP174b | A8I439 | 1, 2, 4 (2, 4, 13) | 3, 2, 4 (5, 2, 8) | 79 | 0.75 | C2 C1b/f | [29] [30] |
| Calmodulinb | A8IDP6 | 1, 3, 3 (1, 2, 7) | 4, 4, 4 (6, 5, 5) | 160 | 0.34 | C1a/e | [19, 31] |
| FAP227 (C1a-18) | Q45QX6 | n.d. | n.d. | — | — | C1a/e | [19] |
a Three values from biological triplicate for both exclusive unique peptide counts and quantitative values are shown.
b These proteins are shared with other compartments of the axoneme.
Here, we re-examined the CP protein components by a similar proteomic approach and obtained mostly the same result but with 11 additional CP protein candidates. Furthermore, we localized these newer CP protein candidates to certain CP sub-structures by using several different Chlamydomonas mutant strains of CP sub-structures. Our results have established a new foundation for understanding the CP architecture.
Materials and Methods
Strains and culture condition
Chlamydomonas reinhardtii strains used in this study are as follows: CC-124 (Wild Type, WT), CC-1033 (pf15, CP-less mutant) [34], CC-5148 (cpc1, C1b/f-less mutant) [14], CC-1034 (pf16, mutant with an unstable C1 structure) [14,22,35] and CC-1029 (pf6, C1a/e-less mutant) [15,36]. The cells were purchased from the Chlamydomonas resource center. Cells were grown first on a Tris-acetatephosphate (TAP) [37] solid plate containing 1.5% agar and then cultured in TAP liquid media under shaking or stirring conditions under a 12 hr light and dark cycle. For flagella purification, each Chlamydomonas strain was cultured in 1 L liquid TAP media with stirring until the OD600 reached 0.5–0.6.
Chlamydomonas flagella isolation and purification of microtubule fraction
The cells were harvested by low-speed centrifugation (700 g for 7 min at 4°C), and flagella were removed from the cell bodies by pH shock [38]. Cell bodies were removed by low speed centrifugation (1,800 g for 5 min at 4°C) in HMDS (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM DTT, 4% sucrose containing 10 μg/ml aprotinin and 5 μg/ml leupeptin) and flagella were collected by higher-speed centrifugation (4,696 g for 40 min at 4°C). Isolated flagella were resuspended in HMDEKP buffer (30 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM potassium acetate, 0.5% polyethylene glycol, (MW 20,000) containing 10 μM paclitaxel, 1 mM Phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin and 5 μg/ml leupeptin). Paclitaxel, PMSF, leupeptin and aprotinin were added to the buffer throughout the purification after this step. Flagella were demembraned by incubating with HMDEKP buffer containing 1.5% NP40 for 30 min on ice. For cryo-electron microscopy (cryo-EM), sonication was performed for better splitting of the axoneme after NP40 treatment. Chlamydomonas axonemes were then spun down by a table top centrifuge (7,800 g for 10 min at 4°C). To split the bundle of axoneme, the samples were incubated with ADP (final concentration of 1 mM) for 10 min at room temperature to activate dyneins and then incubated with 0.1 mM ATP for 10 min at room temperature to induce doublet sliding. The samples were then spun down (16,000 g for 10 min at 4°C). Protease was not added for splitting. After this, Chlamydomonas microtubule fraction was incubated twice with HMDEKP buffer containing 0.6 M NaCl for 30 min on ice, spun down (16,000 g for 10 min at 4°C) and resuspended in HMDEKP buffer. Purification process was performed three times for each strain for biological triplicates.
Cross-sectional EM
For cross-sectional EM, the samples (200–300 μg/ml) were fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) and washed three times with washing buffer (0.1 M sodium cacodylate buffer). Samples were post fixed with 1% aqueous OsO4 and 1.5% aqueous potassium ferrocyanide for 2 h and washed three times with washing buffer. Specimens were dehydrated in a graded ethanol-dH2O from 30, 50, 70, 80, 90 to 100% ethanol. The samples were infiltrated with a graded Epon-ethanol series (1:1, 3:1), embedded in 100% Epon and then polymerized in an oven at 60°C for 48 hr. Ultrathin sections (90 to 100-nm thickness) were prepared from the polymerized blocks with a Diatome diamond knife using a Leica Microsystems EM UC7 ultramicrotome, transferred onto 200-mesh copper grids and stained with 4% uranyl acetate for 6 min and Reynold’s lead for 5 min. The TEM grids were imaged by a FEI Tecnai G2 Spirit 120 kV TEM equipped with a Gatan Ultrascan 4000 CCD Camera Model 895.
Cryo-EM observation
3.5 μl of sonicated microtubule fraction sample (~4 mg/ml) purified from WT Chlamydomonas was applied to glow-discharged holey carbon grids (Quantifoil R2/2), blotted and vitrified in liquid ethane using the Vitrobot Mark IV (Thermo Fisher Scientific). Micrographs were obtained at 59 kx nominal magnification on the direct electron detector Falcon II with the Titan Krios (Thermo Fisher Scientific) using a total dose of ~28 electrons/Å2 and 7 frames (calibrated pixel size of 1.375 Å/pixel). The defocus range was set to between –1.2 and –3.8 μm.
Cryo-ET
A demembranated WT Chlamydomonas flagella sample (~500 μg/ml, 3.5 μl) was applied to glow-discharged holey carbon grids (Quantifoil R2/2), blotted and vitrified in liquid ethane using the Vitrobot Mark IV (Thermo Fisher Scientific). Vitrified grids were then loaded onto Titan Krios (Thermo Fisher Scientific) equipped with Falcon II direct electron detector (Thermo Fisher Scientific) operating at 300 kV. Tilt series were obtained using Tomography software (Thermo Fisher Scientific) in the range of –40 to +60 degrees with a 4-degree increment at nominal magnification of 29 kx so that the total dose is limited to ~100 electrons/Å2. The defocus was set to –5 μm.
MS analysis
4x Laemmli buffer (#1610747, Bio-Rad) was added to the microtubule fraction samples in HMDEKP buffer so that it will be 1x, and 25–30 μg protein was loaded onto the SDS-PAGE gel. To detect all proteins in the sample, electrophoresis was terminated before the proteins entered the separation gel. A single band containing all proteins in the sample was then cut out from the gel and subjected to in-gel digestion [39]. Resultant ~2 μg peptides, which is a maximum protein amount can be loaded, were chromatographically separated on a Dionex Ultimate 3000 UHPLC. First, peptides were loaded onto a Thermo Acclaim Pepmap (Thermo, 75 μm ID×2 cm with 3 μm C18 beads) precolumn and then onto an Acclaim Pepmap Easyspray (Thermo, 75 μm×25 cm with 2 μm C18 beads) analytical column and separated with a flow rate of 200 nl/min with a gradient of 2–35% solvent (acetonitrile containing 0.1% formic acid) over 2 hours. Peptides of charge 2+ or higher were recorded using a Thermo Orbitrap Fusion mass spectrometer operating at 120,000 resolution (FWHM in MS1, 15,000 for MS/MS). The data was searched against Chlamydomonas reinhardtii protein dataset from UniProt (https://www.uniprot.org/).
Data analysis
MS data was analyzed using Scaffold_4.8.4 (Proteome Software Inc.). Total spectrum counts of each protein was divided by total spectrum of the whole sample for normalization to obtain quantitative values. Proteins with mean values of exclusive unique peptide counts of 2 or more in the WT MS results were used for further analysis. To identify CP protein candidates, Student’s t-test was applied to pf15 and WT MS results using biological triplicates. Proteins exhibiting a minimum four-fold change and a statistical significance threshold (p<0.05) in pf15 or proteins which were completely missing in pf15 were categorized as CP candidates. For statistical analysis using several mutant strain MS results, one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test was performed using GraphPad Prism 8.
Results and Discussion
Purification of the axoneme fraction retains the CP proteins with a minimal amount of unrelated proteins
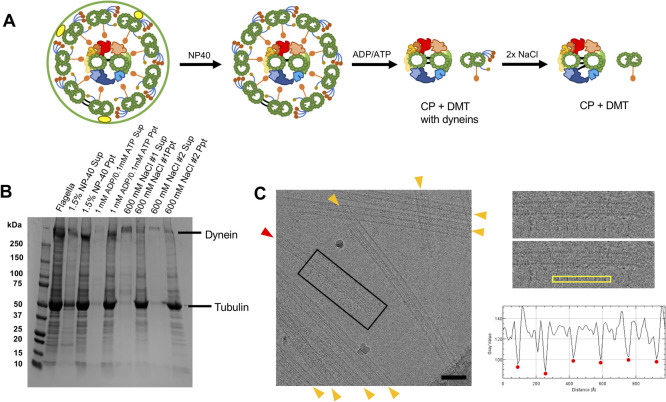
Previous approaches targeting each CP protein one by one presented 22 proteins with clear evidences [10,14,16,17,21,22,34,36,40] (Table 1). A recent study comparing MS results from the Chlamydomonas axoneme with and without a whole CP complex presented more candidates of CP proteins but most of them were not validated [30]. Here, we aimed to re-evaluate the CP protein composition by similar MS analysis. Due to the sensitive nature of MS, peptide detection tends to have an unfavorable preference for large and abundant proteins. Previous proteomic analysis of whole Chlamydomonas flagella showed the presence of an abundant amount of proteins from membrane and matrix fractions and large proteins such as dynein heavy chains [30,41]. Therefore, we tried to obtain samples which contain CP proteins with less contaminant proteins. To achieve this, the microtubule fraction was purified from WT Chlamydomonas flagella by sequential purification following the deflagellation by pH shock. First, proteins from the membrane and matrix fraction were removed by NP-40 treatment. Flagella and demembranated axoneme samples were verified by cross-sectional EM and cryo-ET. The axoneme containing two singlet microtubules belonging to the CP with protruding sub-structures were observed (Fig. 1C and Supplementary Fig. S1A). Demembranated axonemes were then treated with ADP/ATP and with 0.6 M NaCl twice to extract axonemal dyneins (Fig. 2A). From SDS-PAGE, significant amounts of proteins were removed in the final extract leaving the tubulin band which is a main component of the CP and DMT (Fig. 2B). Though we tried to remove the RS complexes by dialysis against a low salt buffer [42] or a KI treatment [31], it was not possible to remove Chlamydomonas RS complexes while maintaining the integrity of the microtubule structure. Thus, the RS complexes remained in our sample. It was previously reported that a high salt treatment destabilizes the CP structure, especially the C2 microtubule [14]. Cross-sectional EM result of our NaCl treated sample were consistent with this (Supplementary Fig. S1B and S1E). Nevertheless, twice salt treated sample was also imaged using cryo-EM and the remaining singlet microtubules from the CP were confirmed to retain the repeating sub-structures (Fig. 2C and Supplementary Fig. S2).
Figure 2.
Preparation of microtubule fraction for MS. (A) A schematic diagram of sequential purification of the axoneme. Flagella were demembranated using a detergent NP-40 following the isolation from Chlamydomonas cells. Demembranated axonemes were incubated with ADP and ATP to induce splitting of the DMTs and the CP. The samples were then treated with 0.6 M NaCl twice to shed large protein complexes such as dyneins. Note that illustrations here show protein compositions rather than the actual structures. (B) SDS-PAGE gel demonstrating protein shedding after sequential purification. The signal of the dynein heavy chain band (>500 kDa) was decreased significantly after NaCl treatments. In contrast, the tubulin band which is a main component of the CP and DMTs showed little change after sequential purification. (C) A typical cryo-EM image of purified sample showing the presence of singlet microtubule from the CP. In our cryo-electron micrographs of our purified microtubule fraction, both DMTs (orange arrowheads) and singlet microtubule from the CP (red arrowhead) with characteristic protruding sub-structures were observed (see also Supplementary Fig. S2). Boxed area of the micrograph is shown in the right panel. The plot profile of yellow box area was generated by ImageJ and the distances between the peaks (red dots) were measured. The averaged distance between the protrusions was 16.7 nm which is consistent with the known repeating unit of the CP [15]. There were more numbers of DMTs compared with singlets from the CP reflecting the stoichiometry inside the axoneme. Scale bar represents 100 nm.
To detect all proteins in the sample, the purified microtubule fraction was subjected to SDS-PAGE, and proteins were cut out as a single band and analyzed by MS (see details for Materials and Methods). In our MS results, almost all (21 out of 22) known CP proteins such as Hydin, CPC1, PF6, FAP69, PF16, KLP1 and FAP221 (PCDP1) were detected by our MS (Table 1). The only known CP protein which we failed to detect was FAP227 (C1a-18) [19]. Since the size of FAP227 is small (18 kDa), it is unfavorable for detection by MS. Detected traditionally known CP proteins were previously shown to localize at different CP sub-structures (Table 1). Together with the EM results, we concluded that our purification method retained the CP proteins and was usable for proteomic analysis.
Known microtubule inner proteins (MIPs) like Rib43a and Rib72 [43,44] and RS proteins were also detected since these structures are tightly associated with DMTs (Supplementary Table S1). Proteins from other axonemal components such as IFT complex proteins, IFT dyneins, IFT kinesins, axonemal dyneins and dynein regulatory complex were still detected due to the high sensitivity of MS though we tried to reduce them as much as possible.
Re-evaluation of CP proteins by comparative proteomic analysis
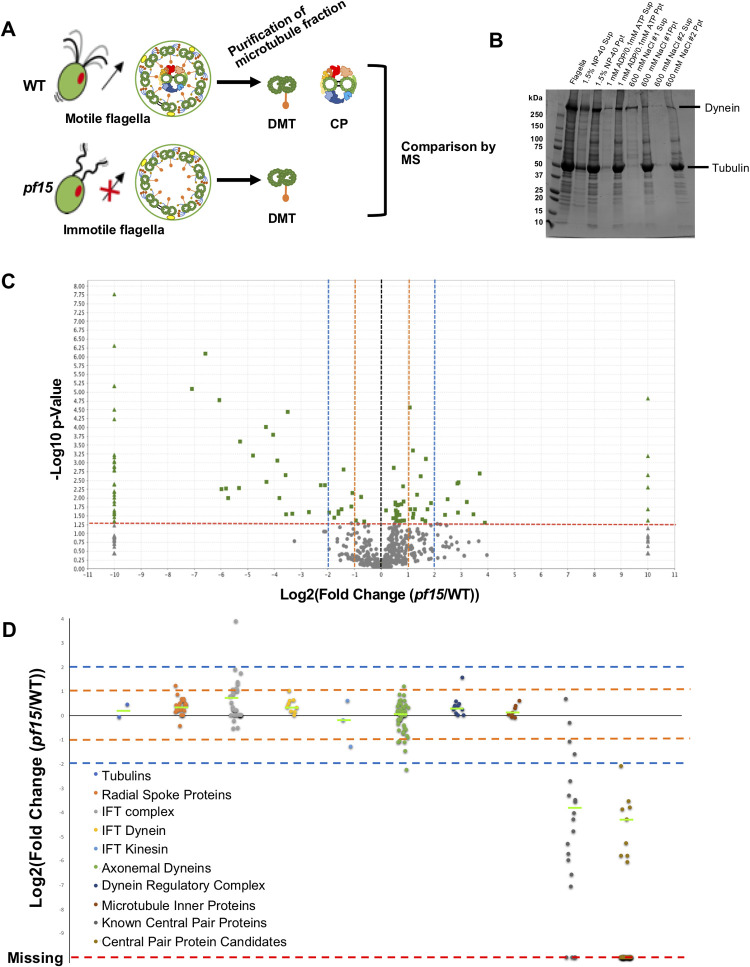
Next, we tried to identify the CP proteins by MS. Since it is not possible to sub-fractionate CPs from DMTs as they are both microtubule-based structures, we used a comparative proteomic approach using a specific Chlamydomonas mutant lacking a whole CP complex similar to Zhao, L., et al., (2019) [30]. Chlamydomonas mutant strain pf15 contains a mutation in a gene encoding the p80 subunit of microtubule severing enzyme Katanin [34]. The resulting effects prevent the entire CP complex from assembling and lead to paralyzed flagella while leaving other components like DMTs intact (Fig. 3A). pf15 strain was chosen in this study instead of pf18 used in [30] since the genetic background of the p15 strain has been well-characterized. An identical purification process was used for pf15 mutant flagella and WT (Fig. 3A and B). The MS analysis was performed for the microtubule fraction of pf15, and normalized MS results of pf15 and WT were compared (see details for Materials and Methods). In the MS result of pf15, there were many proteins significantly reduced or completely missing when compared with the WT (Fig. 3C, left side). Along with traditionally known CP proteins listed in Table 1, there were proteins totally missing or significantly reduced in pf15. These proteins were possible candidates of the CP proteins.
Figure 3.
Identification of new CP proteins by MS. (A) Schematic diagrams of the axoneme structures from WT and pf17_071-g015.tif flagella. Obtained microtubule fractions from WT and pf15 were analyzed by MS and the results were compared. (B) SDS-PAGE result of sequential purification of microtubule fraction from pf15 flagella showing similar pattern with that of WT flagella. (C) A volcano plot comparing WT and pf15 MS results. Changes in a protein abundance between WT (n=3) and pf15 (n=3) results were plotted. A dashed red line indicates the significance threshold of p<0.05 and proteins meet this criterion are shown in green. Triangle dots represent completely missing proteins in either WT or pf15 result. Two- and four-fold changes are shown by the orange and blue dashed lines, respectively. There were more proteins completely missing in pf15 results while many others showed more than two-fold decrease in pf15 results. (D) Plot of fold changes of proteins categorized into different groups. Proteins identified by MS were arranged in groups (Tubulins; RS proteins; IFT complex proteins; IFT dynein; IFT kinesin; axonemal dyneins; dynein regulatory complex; MIP candidates and known CP proteins) and fold change between WT and pf15 results of each protein was plotted. Two- and four-fold changes are shown by the orange and blue dashed lines, respectively. Green lines indicate the median value for each category. Statistical significance compared with tubulin result was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test. Among these classes, only known CP protein class were significantly reduced with a p-value of 0.00050. Fold changes of our CP protein candidates are also shown at the rightmost column. Red line represents proteins that were completely missing in pf15. Proteins included in each class are listed in Tables 1 and 2, and Supplementary Table S4.
There were also proteins detected in higher amounts in pf15 or only detected in pf15 (Fig. 3C, right side). The numbers of these proteins were less than the proteins that were only detected in WT. In our MS, some IFT complex proteins (IFT22, IFT56, IFT57, IFT74 and BBS1) were detected in higher amounts in pf15 (Supplementary Table S2). Previous studies have shown that the IFT complex proteins accumulate in the CP-less Chlamydomonas mutant axonemes, possibly trapped in the empty space where CP complex is supposed to be [45]. Therefore, these increased amounts of IFT proteins are probably due to the accumulation of the IFT particles. Other increased proteins might also be trapped inside the axoneme similarly. There were also several proteins only detected in pf15 result, but the amounts of detected peptides of these proteins were generally low, and thus these proteins might be due to contamination (Supplementary Table S2).
To clearly see the differences between WT and pf15, we performed a comparison based on protein categories which include tubulins, RS proteins, IFT complex, IFT dynein, IFT kinesin, axonemal dyneins, dynein regulatory complex proteins, MIPs and traditionally known CP proteins (Fig. 3D). Since values were normalized, α- and β-tubulins were detected at the same level between WT and pf15 results and served as a control. As clusters, only traditionally known CP proteins were significantly decreased in pf15, thus validating our strategy. Other classes like IFT dynein, IFT kinesin, RS proteins, axonemal dyneins, dynein regulatory complex proteins and MIPs did not show any significant decrease as groups. From these results, we concluded that our method is valid to identify CP proteins.
When we focused on each protein, 18 out of the 20 traditionally known CP proteins detected in WT were completely missing or significantly decreased in pf15 (Table 1). The exceptions were Calmodulin and FAP174 which did not show significant decrease and fell into the region between two-fold decrease and two-fold increase (between two orange lines in Fig. 3D). Calmodulin is shared with the RS complex [31], and thus its decrease is masked by signals from the remaining RS in our purification. FAP174 also did not show significant decrease, and this could mean that FAP174 is also present in other axonemal structures (discussed later). Other CP proteins like enolase and HSP70, which are also known to be shared with other axonemal components, showed significant decrease but fell into the region between two- to four-fold decreases in pf15 (between lower orange and blue lines in Fig. 3D). This area also contained proteins such as DHC9, p38 and KAP from other categories. Almost all the proteins from other complexes did not show decreases of four-fold or larger except DHC8 (below the lower blue line in Fig. 3D). Thus, proteins which were decreased significantly (p<0.05) with a 4-fold decrease or greater in pf15, or completely missing in pf15, were categorized as new CP protein candidates. With this criteria, 37 proteins, such as FAP7, FAP47, FAP65 and FAP70 were identified as CP candidates (Fig. 3D and Table 2). DPY30 was originally not included in this criteria, but it is included in Table 2 since it is a confirmed CP protein in [30].
Table 2.
MS results of new CP proteins and their localizations inside the CP complex
| Names | Uniprot ID | WT exclusive unique peptide counta (quantitative valuea) |
pf15
exclusive unique peptide counta (quantitative valuea) |
pf15/WT ratio (%) (quantitative values were used) |
p-values (WT vs pf15) |
Localizations |
|---|---|---|---|---|---|---|
| CHLREDRAFT_150638 | A8J566 | 1, 5, 0 (1, 4, 0) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.23 | C1a/e (this study) |
| CHLREDRAFT_170023 | A8IMQ8 | 3, 9, 0 (3, 6, 0) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.17 | C1b/f (this study) |
| CHLREDRAFT_177061 | A8J9A4 | 7, 7, 2 (6, 5 , 3) | 0, 0, 1 (0, 0, 1) | 7.1 | 0.0098 | C1b/f (this study) |
| DPY30b, c | A8J1X7 | 1, 2, 1 (1, 1, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0041 | C1a/e ([30] and this study) |
| EF-3 | A8ISZ1 | 4, 2, 1 (4, 1, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.047 | Not assigned |
| FAP7d | A8IVW2 | 14, 16, 7 (26, 33, 28) | 1, 0, 1 (1, 0, 1) | 2.6 | 0.00025 | C1a/e (this study) |
| FAP47b | A8IPW8 | 22, 35, 15 (23, 27, 26) | 0, 0, 0 (0, 0, 0) | 0.0 | <0.00010 | C2b (this study) |
| FAP65d | A8JFU2 | 13, 23, 10 (12, 19, 16) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0016 | C2a, c, d, e and Bridge (this study) |
| FAP70d | A8I7W0 | 12, 22, 11 (14, 26, 23) | 0, 1, 0 (0, 1, 0) | 33 | 0.0053 | C2a, c, d, e and Bridge (this study) |
| FAP75d | A8HYW3 | 13, 19, 8 (13, 16, 15) | 0, 1, 1 (0, 1, 1) | 5.0 | <0.00010 | C2a, c, d, e and Bridge (this study) |
| FAP76b | A8J128 | 24, 26, 14 (26, 23, 26) | 0 , 1, 0 (0, 1, 0) | 1.5 | <0.00010 | C1a-e-c complex ([23] and this study) |
| FAP81d | A8IPC1 | 23, 27, 12 (24, 24, 23) | 0, 0, 0 (0, 0, 0) | 0.0 | <0.00010 | C1a-e-c complex ([23] and this study) |
| FAP92d | A8HR45 | 28, 30, 17 (29, 23, 36) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0017 | C1a-e-c complex ([23] and this study) |
| FAP99b | A8IUG5 | 9, 13, 1 (10, 9, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.060 | C1d (this study) |
| FAP105d | A8IKV8 | 3, 5, 0 (3, 4, 0) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.12 | C1a-e-c complex ([23] and this study) |
| FAP108d | A8IPA9 | 2, 3, 1 (2, 2, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.00090 | C1a-e-c complex ([23] and this study) |
| FAP123d | A8IEJ6 | 4, 3, 0 (4, 2, 0) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.16 | C1a-e-c complex (this study) |
| FAP125d | A8IY87 | 14, 18, 9 (15, 18, 16) | 0, 0, 0 (0, 0, 0) | 0.0 | <0.00010 | C1c (this study) |
| FAP147d | A8IT32 | 7, 10, 3 (6, 7, 7) | 0, 0, 0 (0, 0, 0) | 0.0 | <0.00010 | C2a, c, d, e and Bridge (this study) |
| FAP171d | A8IUF4 | 4, 9, 2 (4 , 7, 3) | 1, 0, 0 (1, 0, 0) | 8.4 | 0.029 | C2a, c, d, e and Bridge (this study) |
| FAP173 | A8JAF7 | 3, 3, 1 (4, 2, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.012 | Not assigned |
| FAP194d | A8J5U4 | 9, 13, 4 (10, 10, 7) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0013 | C2a, c, d, e and Bridge (this study) |
| FAP199 | A8J1E6 | 1, 2, 3 (1, 1, 7) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.19 | Not assigned |
| FAP209 | A8J100 | 6, 8, 3 (6, 5, 5) | 0, 1, 0 (0, 1, 0) | 6.7 | 0.00086 | C1c (this study) |
| FAP216d | A8JGM3 | 12 , 16, 4 (13, 13, 7) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0064 | C1a-e-c complex [23] C1a/e (this study) |
| FAP219 | A8J9I0 | 5, 7, 1 (5, 5, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.022 | C1c (this study) |
| FAP225d | A8HNF2 | 14, 19, 4 (14, 22, 7) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.034 | C2a, c, d, e and Bridge (this study) |
| FAP239d | A8J319 | 0, 5, 2 (0, 3, 3) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.12 | C2a, c, d, e and Bridge (this study) |
| FAP244e | A8IZG0 | 12, 14, 5 (14, 10, 12) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.00069 | tether complex of I1 dynein of IDA [46, 47] C2a, c, d, e and Bridge (this study) |
| FAP246b | A8HNZ7 | 7, 6, 3 (7, 6, 5) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.00096 | C1b/f ([30] & this study) |
| FAP266d | A8JB69 | 4, 5, 2 (4, 3 , 3) | 1, 1, 0 (1, 1, 0) | 23 | 0.0043 | C2a, c, d, e and Bridge (this study) |
| FAP279 | A8HWC6 | 5, 7, 1 (6, 4, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.029 | C1a-e-c complex (this study) |
| FAP289d | A8JCZ9 | 8, 12, 4 (9, 9, 8) | 0, 0, 0 (0, 0, 0) | 0.0 | <0.00010 | C1a-e-c complex (this study) |
| FAP312d | A8IUV6 | 2, 5, 1 (2, 3, 2) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0066 | C2 [30] |
| FAP348d | A8JBI2 | 2, 3, 2 (2, 2, 3) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.0095 | C1a/e (this study) |
| FAP412d | A8JGL8 | 6, 9, 0 (6, 6, 0) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.12 | C1a-e-c complex [23] |
| MOT17d | A8J798 | 3, 4, 3 (3, 2, 7) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.044 | C1c (this study) |
| Phosphoglycerate mutasee | A8HVU5 | 4, 10, 0 (4, 7, 0) | 0, 0, 0 (0, 0, 0) | 0.0 | 0.15 | Membrane+matrix fraction [38] C1b/f? (this study) |
a Three values from biological triplicate for both exclusive unique peptide counts and quantitative values
are shown.
b CP proteins which were confirmed by [30].
c Proteins that has 2 or more of mean values of detected exclusive unique peptides in WT are listed, except DPY30 which was confirmed as a CP protein in [30].
d CP candidates shared with [30].
e CP proteins shared with other axonemal compartments.
A recent proteomic study used CP-less mutant pf18 instead of pf15 to identify CP proteins [30]. Only demembranated whole axonemes were analyzed by quantitative MS in [30] instead of purified microtubule fractions. Despite the differences in strains and methods used, 26 out of 37 identified proteins in our work were shared with Zhao, L., et al., (2019) [30] making these proteins strong candidates for the CP proteins (din Table 2). One of these shared CP protein candidates was FAP125. FAP125 is a kinesin-like protein which was previously proposed to be part of the CP complex without direct evidence [48]. Our study presented further evidence showing that FAP125 is a CP-associated kinesin in addition to KLP-1.
Eleven CP candidate proteins were uniquely identified in our study while 19 (including DPY30) were unique to Zhao, L., et al. (2019) [30] (Table 2 and Supplementary Table S3). These differences might be due to contaminated proteins from remaining axonemal components in Zhao, L., et al.’s purification method since only demembranated samples were used in the study. Alternatively, it is possible that some of the CP proteins fell off in our purification method. NAP was shown to be a component of the C1a/e region by immunoprecipitation by Zhao, L., et al., [30], but we failed to detect NAP in our MS. This could be due to the weak association of NAP to the C1a/e region. It is also possible that differences are due to different strains used. Nevertheless, it is noteworthy that while using different strains and methods, many common proteins were identified as new CP protein candidates. Our work can be therefore be used complementarily to understand the architecture of the CP complex.
One of the proteins identified only in our result was phosphoglycerate mutase which was detected in WT but completely missing in pf15 (Table 2). Phosphoglycerate mutase was previously shown to be present in the membrane and matrix fraction and play roles in ATP production together with enolase [18]. Interestingly, enolase which is involved in the same ATP synthesis pathway as phosphoglycerate mutase is known to be a component of the CP as well as being present in the membrane and matrix fractions. Since these enzymes work together, it is likely that phosphoglycerate mutase is also integrated into the CP complex to facilitate the reaction.
In our criteria, FAP244 was also identified as a CP protein. FAP244 was previously shown to be a component of the tether complex of I1 dynein of the IDA by cryo-ET studies [46,47]. In our NaCl treated conditions, FAP244 was detected in WT with high abundancies but not detected in any of the pf15 triplicate (Table 2). FAP244 was also decreased in the pf18 strain replicates by Zhao, L., et al., (XP_001694394.1 in Supplementary Table S1 in [30]) but it was not proposed as a CP protein. It is likely that the FAP244 portion from axonemal dyneins was masking the decrease of FAP244 from the CP complex in the previous study. Based on these results, FAP244 is probably shared with the CP complex and the I1 dynein. FAP43 was previously shown to have a weak sequence similarity with FAP244 and to have a redundant function with FAP244 in I1 dynein [46,47]. In our MS results, FAP43 was not decreased in pf15 triplicate (Supplementary Table S1). FAP44, which makes up the tether complex of I1 dynein together with FAP43 and FAP244, also did not show any reduction in pf15 (Supplementary Table S1). Therefore, FAP244 in the CP complex might have a different and independent function from I1 dynein.
Localizing CP protein candidates into sub-structures of the CP complex
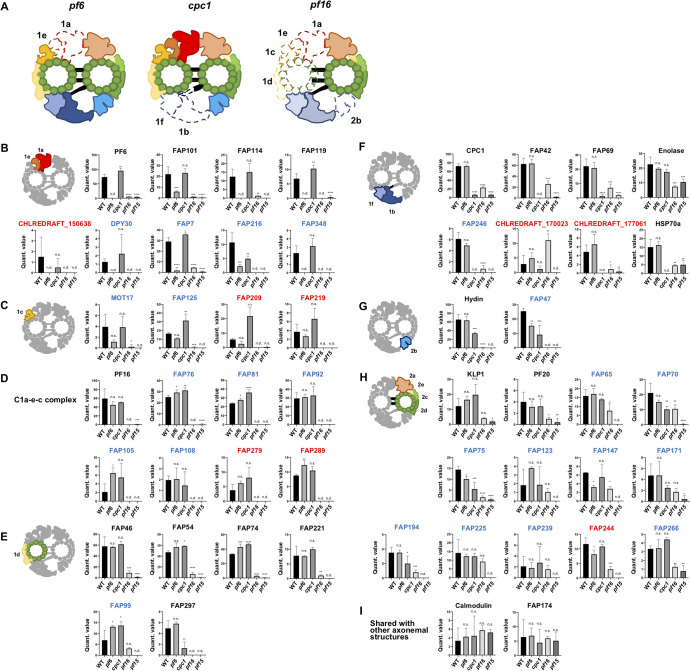
There are several Chlamydomonas mutant strains corresponding to specific CP sub-structures. The pf16 strain is one of such strains with an unstable C1 microtubule [14,22,35]. In the pf16 axoneme, the C1 microtubule can be assembled. However, the whole C1 structure is reported to be lost after demembranation or NaCl treatment due to the missing of PF16 protein which localizes to the C1 microtubule [14]. Using this feature, Zhao, L., et al., assigned their CP candidates into the C1 or C2 microtubule by comparing MS results of pf16 with that of WT [30]. In this study, we further aimed to identify the localizations of these CP protein candidates around the CP complex. To achieve this, we used different kinds of Chlamydomonas mutants lacking specific CP sub-substructures, pf6 (C1a/e mutant), cpc1 (C1b/f mutant) as well as pf16 (Fig. 4A). In our purification condition, the majority of flagella purified from pf16 strain had only one of the CP microtubules, presumably the C2 microtubule (Supplementary Fig. S1C and E) consistent with previous reports. Following the same sample preparations as WT, there were remaining structures observed at the center of the axoneme by cross-sectional EM (Supplementary Fig. S1D) and purified microtubule fractions from different CP mutants were analyzed by MS (Supplementary Fig. S3). The normalized MS results from five different strains were compared, and MS detection profiles for each protein of interest were generated (Fig. 4B-I and Supplementary Fig. S4). Traditionally known CP proteins, which localize at the same area, shared similar MS profiles which allowed us to assign newer CP candidates to certain CP sub-structures.
Figure 4.
MS analyses using Chlamydomonas mutant strains lacking CP sub-structures. (A) Schematic diagrams of CP compositions from mutants lacking sub-structures of the CP. Sub-structures of the CP which are missing in pf17_071-g016.tif strains are shown in dashed lines. pf6 strain is missing the C1a/e structure (formerly the C1a), cpc1 strain lacks the C1b/f structure (formerly the C1b) while pf16 strain has an unstable C1 structure. In the pf16 model, The C1b/f region is in transparent since this region can remain attached to the C2 microtubule with the diagonal link [14]. Note that these diagrams show the protein compositions in the samples rather than the actual structures. (B-I) MS profiles of CP proteins and their possible localizations. Detected levels of proteins were compared among strains (WT, pf6, cpc1, pf16 and pf15). Mean values of normalized quantitative values of each CP protein are shown (error bars represent SD for biological triplicate). Known CP proteins that have been localized to specific sub-complexes showed similar MS profiles. These proteins were used as references to assign newer CP candidates to certain sub-structures, such as the C1a/e area (B), the C1c area (C), the C1a-e-c complex (D), the C1d region (E), the C1b/f area (F), the C2b area (G), the C2a, c, d, e & bridge area (H), and proteins shared with other axonemal structures (I). Known CP proteins are labelled in black, CP candidates shared with [30] are labelled in blue, and CP candidates obtained only in our work are in red. One-way ANOVA followed by Dunnett’s multiple comparisons test comparing with WT values (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; n.s., not significant; n.d., not detected). Plots not shown here are presented in Supplementary Figure S4.
C1a/e proteins
CP proteins like PF6, FAP101, FAP114 (C1a-32) and FAP119 (C1a-34), which were shown to be located at the C1a/e sub-structure [19,36], were detected both in WT and cpc1 strains since they retain the C1a/e structure. These proteins were either not detected or detected in low amounts in pf6, pf15 and pf16 because of the lack of the C1a/e region (Fig. 4B). To verify this statistically, we performed one-way ANOVA test and these known C1a/e proteins were all significantly decreased in pf6, pf15 and pf16 but not in cpc1 (Fig. 4B). DPY30 which was proposed to be located at the C1a/e region by immunoprecipitation [30] also met this standard, further verifying our strategy. Among new CP candidates, FAP7, FAP348 and CHLREDRAFT_150638 showed the same pattern by one-way ANOVA test and were likely to be present in the C1a/e region as well. FAP348 was previously proposed to be located somewhere in the C1 complex [30]. Based on our MS results, FAP348 is likely to be located at C1a/e region. FAP7 was shared with Zhao, L., et al.,’s CP candidates but it was not assigned to any region of the CP complex. Our results assigned FAP7 to the C1a/e region. The new CP protein CHLREDRAFT_150638 only identified in our result is likely a component of C1a/e region. After we submitted this manuscript, FAP216 was localized around the base of C1a/e structure by cryo-ET [23]. Though FAP216 showed similar MS pattern with other C1a/e proteins, it was also decreased in cpc1. This could mean that some part of FAP216 is reaching to the cpc1 region or interacting with proteins that compose it.
C1a-e-c complex proteins
Through cryo-ET and immunoprecipitation using PF16 protein as a bait, proteins like FAP76, FAP81, FAP92, FAP105, FAP108, FAP216, FAP412 and MOT17 (FAP305) were proposed to form a super-complex inside the CP structure [23]. In our MS results, most of these proteins (PF16, FAP76, FAP81, FAP92, FAP105 and FAP108) showed similar MS profiles being highly detected in WT, pf6 and cpc1 strains but not detected or detected only very little amount in pf16 and pf15 strains (Fig. 4D). In terms of the location, some of these proteins like PF16 and FAP92 are at the C1a/e region [23], but they might be stably tethered to the C1 microtubule due to their interaction even with the apparent loss of the structure in pf6. In our MS analysis, FAP279 and FAP289 also showed similar MS profiles and are likely components of the C1a-e-c super-complex (Fig. 4D). FAP279 is a leucine-rich repeat-containing protein that was not assigned as a CP protein before. A homologue of FAP279 is also present in humans (LRRC72). FAP289 was not previously assigned to certain CP sub-structure.
C1d proteins
There are several CP proteins known to localize at the C1d area. FAP46, FAP54, FAP74, FAP221 (PCDP1) are such proteins [10]. These proteins were proposed to form a complex located at the C1d region. In our MS profile, these proteins show a similar trend being significantly reduced only in pf15 and pf16 strains since they lack this region (Fig. 4E). The difference with known C1a-c-e complex proteins was that these C1d proteins were still detected in pf16. FAP99 showed a similar MS profile as these C1d proteins and was therefore assigned to this area (Fig. 4E). FAP99 was previously confirmed to be an authentic CP protein and was proposed to be located at the C1 microtubule [30]. We were then able to further narrow down its localization.
FAP297 (WDR93) was also proposed to be a component of the C1d complex [21] but older work failed to detect this protein [10]. Unlike the other C1d proteins, FAP297 showed a significant reduction in the cpc1 strain lacking the C1b/f region (Fig. 4A). This could mean that FAP297 is located at the interface between the C1d and the C1f and interacting with proteins from the C1f. Zhao, L., et al., also failed to detect FAP297 by immunoprecipitation using FAP46 as a bait, though all other known C1d proteins were detected [30]. This further supports our idea that FAP297 is located away from other known C1d proteins.
C1c proteins
Some proteins like MOT17, FAP125, FAP209 and FAP219 showed a significant reduction only in pf15 and pf16 strains by one-way ANOVA tests. However, these proteins were slightly but not significantly reduced in pf6 (Fig. 4C). These MS patterns were somewhat in between that of known C1a/e proteins (significant reduction in pf6 result) and C1d proteins (WT level detection in pf6 result). Hence, these proteins are probably at the interface of the C1a/e and the C1d, namely the C1c area. One of these proteins, MOT17 was shown to interact with known C1a/e proteins by immunoprecipitation which is the neighboring region of the C1c [30]. FAP125 was previously proposed to be somewhere in the C1 microtubule, but its localization to a specific sub-structure was not achieved. Our results located FAP125 into specific sub-structure of the C1 microtubule. For FAP209 and 219, there was no localization information in the axoneme at all.
C1b/f proteins
Traditionally known proteins belonging to the C1b/f sub-complex like CPC1, FAP42 (C1b-350) and FAP69 (C1b-135) also share similar patterns distinct with significant decrease in cpc1, pf15 and pf16 but with modest decreases in pf16 (Fig. 4F). At first, we were puzzled with this result of only a modest decrease in pf16 since pf16 is generally assumed to have an unstable, and therefore missing C1 to which the C1b/f region is also attached. Therefore, we looked into the article characterizing the pf16 mutant and realized that the C1b/f part remains with the C2 microtubule due to the diagonal link connecting these structures even though other sub-structures like the C1a, c, d and e, and C2b were missing [14]. This was also mentioned by the authors but has been overlooked in recent articles. Thus, we concluded that the C1b/f proteins are partially present in the pf16 structure in our purification condition (Fig. 4A). Enolase and HSP70A showed different MS patterns from other known C1b/f proteins (Fig. 4F), but these proteins were previously shown to be present both in CP complex and in other compartments of the axoneme. The differences represent the presence of these proteins in other compartments of the axoneme [18,24]. Therefore, the MS profile shared with CPC1, FAP42 and FAP69 was used as a standard for C1b/f proteins. FAP246 was previously shown to interact with other C1b/f proteins by immunoprecipitation [30]. Our assignment is consistent with this. Among our new CP candidates, CHLREDRAFT_170023 and CHLREDRAFT_177061 showed C1b/f-like profile and are likely to be located at this region (Fig. 4F). CHLREDRAFT_170023 and CHLREDRAFT_177061 were not proposed to be localized to the CP complex before.
C2b proteins
Hydin is the only protein known to be associated with the C2b region [16,17]. Based on previous cross-sectional EM results, this region solubilizes before the C1b/f region in pf16 CP structure [14]. Consistent with this, Hydin was missing in pf16 MS (Fig. 4G). Hydin was also decreased in cpc1 compared with WT. From previous studies, the C2b projection is in close proximity to the C1b sub-structure [15]. It is possible that the interaction between neighboring sub-structures C2b and C1b are tighter than previously assumed. In our MS profile, FAP47 showed a similar trend with Hydin (Fig. 4G). In recent MS results, FAP47 also showed an elution pattern similar to Hydin [30]. Taken together, FAP47 is located at C2b region. In Zhao, L., et al., (2019) [30], FAP49, 72, 154 and 416 were also identified as CP proteins and proposed to form a complex with FAP47 based on their immunoprecipitation results [30]. In our MS result, FAP49, 72 and 154 shared peptide sequences and therefore we were not able to conclude if these proteins are decreased in pf15 (Supplementary Table S3). FAP416 was not detected in our MS.
Proteins localized at C2a, c, d, e and bridge
KLP1 is known to localize at the C2c/d area [25]. KLP1 was detected in most strains but in a very small amounts in the pf15 strain (Fig. 4H). This is consistent with the results of previous cross-section EM showing that the C2c/d region is stably bound to the C2 microtubule in pf16 [14] (Fig. 4A). Interestingly, PF20 protein which is known to be localized at the “bridge” that connects the C1 and C2 microtubules [27] also showed an MS profile similar with that of KLP1 being detected slightly more in pf16 strain compared with pf15 result (Fig. 4H). By immunogold labeling EM in previous studies, gold particles were found to be bound to only one of the CP singlets, presumably the C2 microtubule [27]. Our results also suggest that PF20 is associated more tightly to the C2 microtubule. Proteins like FAP65, 70, 75, 123, 147, 171, 194, 225, 239, 244 and 266 showed similar MS profiles with KLP1 and PF20 (Fig. 4H) and therefore are likely to be present at the C2a, c, d, e and the bridge region. The C2a/e area is also included since this part is known to be stably attached to the C2 microtubule in pf16 strain [14]. Previous cryo-ET studies have shown that there are protein structures inside the C2 microtubule tubulin lattice similar to the MIPs in the DMTs [15]. Some of these proteins might correspond to these inner proteins of the C2 microtubule. FAP65, 70, 75, 147, 171 and 239 were previously proposed to be somewhere at the C2 microtubule [30] and we were able to further narrow down the localizations of these proteins. For FAP123, 194, 225 and 266, there was no previous information about their localization inside the CP complex. FAP244 is probably present in both the tether complex of I1 dynein and this region of the CP as mentioned earlier.
Other CP protein candidates
In our results, some of the proteins showed MS profiles which were not readily assigned to certain classes. Calmodulin, a traditionally known CP protein, did not show a significant decrease in either of the strains used in our study (Fig. 4I). As mentioned, Calmodulin is known to be shared with the RS which was left in our sample preparation. Thus, the decrease of Calmodulin can be masked by the signal from the RS.
Similarly, FAP174, which is a traditionally known CP protein, did not show significant decrease in either of our strains by one-way ANOVA test (Fig. 4I). FAP174 might be shared with other compartments of the axoneme similar to Calmodulin. In the previous study, FAP174 was shown to be reduced in CP-less mutant but the signals were still detected by Western blots [29]. The remaining FAP174 might correspond to the portion from other axonemal components. Previously, FAP174 was immunoprecipitated by FAP246 with other traditionally known C1b/f proteins [30]. In our MS profile, though it was not significant, FAP174 showed a slight decrease in cpc1 which lacks the C1b/f region. Based on these results, FAP174 is probably located at the C1b/f area.
There were some remaining CP candidates which we were unable to assign to a certain CP sub-structure (Supplementary Fig. S4), including FAP312 and 412 which were shared with [30]. FAP312 was proposed to be a C2 protein [30] and FAP412 was recently proposed to be a part of C1a-e-c super-complex [23]. However, due to low detection of these proteins in our MS, we could not confidently assign these proteins to a certain CP sub-structure. A known C1 protein, PP1a was detected only in WT (Supplementary Fig. S4) and therefore we were not able to assign it to a certain sub-structure of the CP. Though it was not clear by our MS profile (Supplementary Fig. S4), phosphoglycerate mutase might be a part of the C1b/f region since this protein is known to work with enolase which was described to localize to this sub-structure. The other CP candidates like EF-3, FAP173 and FAP199 were not assigned to certain sub-structures. These proteins were, however, detected in only small amounts and could be false positives of MS and require further testing.
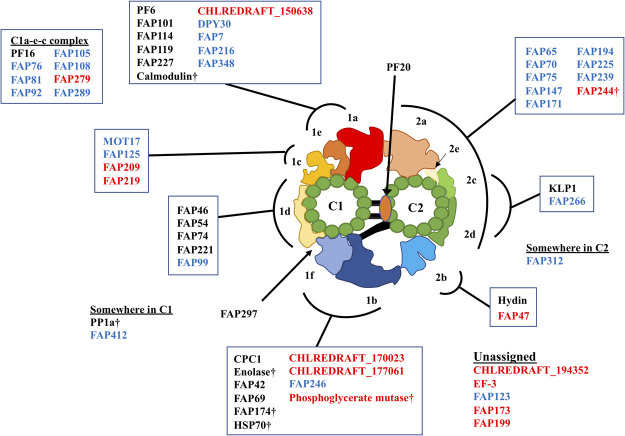
Model of CP protein localization and insights into functions in the flagella
From our MS profile, we were able to build a more complete model of the localizations of the CP proteins (Fig. 5 and Table 2). Our model of localization also gave some insights into the regulation of flagellar motility. FAP125 is a kinesin-like protein recently identified as a CP protein [30] and our work localized it to the C1c area by our study. The presence of FAP125 at the C1c area is interesting since KLP-1, another known CP kinesin, is located at the C2c/d which is at the opposite side of the CP complex. KLP-1 was proposed to work as a conformational switch in the CP [25], and thus symmetrical binding of two CP-kinesins onto separate singlet microtubules might play a role in waveform switching or planar waveform in a coordinated way. Further functional analysis of FAP125 in future work will reveal this point. Like this example, our model of the localizations of CP proteins can be used to understand how each CP protein is organized and is working as a complex.
Figure 5.
Model of localizations of CP proteins. Proteins are mapped to CP sub-structures based on our MS profiles. Traditionally known CP proteins are shown in black, CP candidates shared with [30] are in blue and proteins identified only in our results are in red. Daggers denote the proteins possibly shared with other axonemal structures.
Very recently, CP proteins at the C1a-e-c regions were shown to form a super-complex across the CP sub-structures [23]. Our MS results also have shown that the components of this super-complex share very similar MS profiles. Other than this region, FAP147 showed a very similar MS profile with FAP244. FAP266’s MS profile was especially similar with KLP1. Therefore, these protein pairs might also be forming sub-complexes inside the CP structure (Fig. 5). Biochemical experiments and structural analysis of the CP complex in a higher resolution would reveal this point in future studies.
Conclusion
PCD is a rare but prevalent congenital disease that derives from the impairment of motile cilia. An insufficient understanding of the protein composition of axonemal complexes directly affects the success and efficiency of clinical diagnosis of a wide variety of ciliopathies. By using comprehensive MS analysis of Chlamydomonas strains, we have identified more CP proteins and localized them to specific sub-structures of the CP which allows for more informed interpretation of whole exome sequence data and cross-sectional analysis. Through this method, we circumvent traditional means of protein identification and localization and provide a more comprehensive insight into the entire making of the CP complex. Such proteomic approaches that exploit mutant strains could also be applicable for other uninvestigated areas of the axoneme. Our assignments of these CP proteins into each CP sub-structure will serve as primers for future modeling of CP proteins when a high-resolution structure of the CP complex is obtained by cryo-EM.
A preliminary version of this work, doi: 10.1101/739383, was deposited in the bioRxiv on August 19, 2019.
Acknowledgements
We thank Dr. Kaustuv Basu and Ms. Jeannie Mui at the Facility for Electron Microscopy Research of McGill University for help in preparation of cross-section EM microscope operation and data collection. We are indebted to Mr. Lorne Taylor and Ms. Amy Ho Yee Wong from MUHC for help with MS. This work was supported by JSPS KAKENHI Grant Numbers JP19K23726 and JP20K15733, and the Foundation for Nara Institute of Science and Technology (R2290001) to MI. KHB was supported by grants from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-04954), Canada Institute of Health Research (PJT-156354) and the Canada Institute for Advanced Research Arzieli Global Scholars Program and McGill University.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
MI and KHB conceived the project and designed the experiments. DD and MI performed culture of the cells, purification of microtubule fractions from flagella for MS analysis with the help of KP and RR. DD performed cross-sectional EM, and MI performed cryo-EM and cryo-ET observations with the aid of KHB. DD and MI analyzed the results. All authors were involved in the manuscript writing process.
Data Availability
The mass spectrometry is deposited in Dryad (https://doi.org/10.5061/dryad.x0k6djhg6).
Supplementary Material
References
- [1].Waters A. M. & Beales P. L.. Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. 26, 1039–1056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wheway G., Nazlamova L. & Hancock J. T.. Signaling through the Primary Cilium. Front. Cell Dev. Biol. 6, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lindemann C. B. & Lesich K. A.. Flagellar and ciliary beating: the proven and the possible. J. Cell Sci. 123, 519–528 (2010). [DOI] [PubMed] [Google Scholar]
- [4].Tilley A. E., Walters M. S., Shaykhiev R. & Crystal R. G.. Cilia dysfunction in lung disease. Annu. Rev. Physiol. 77, 379–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Praveen K., Davis E. E. & Katsanis N.. Unique among ciliopathies: primary ciliary dyskinesia, a motile cilia disorder. F1000Prime Rep. 7, 36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McKenzie, C. W., Preston, C. C., Finn, R., Eyster, K. M., Faustino, R. S. & Lee, L. Strain-specific differences in brain gene expression in a hydrocephalic mouse model with motile cilia dysfunction. Sci. Rep. 8: 13370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Popatia R., Haver K. & Casey A.. Primary Ciliary Dyskinesia: An Update on New Diagnostic Modalities and Review of the Literature. Pediatr. Allergy Immunol. Pulmonol. 27, 51–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sloboda R. D. & Rosenbaum J. L.. Making sense of cilia and flagella. J. Cell Biol. 179, 575–582 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stepanek L. & Pigino G.. Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352, 721–724 (2016). [DOI] [PubMed] [Google Scholar]
- [10].DiPetrillo C. G. & Smith E. F.. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J. Cell Biol. 189, 601–612 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kikushima K. Central pair apparatus enhances outer-arm dynein activities through regulation of inner-arm dyneins. Cell Motil. Cytoskeleton 66, 272–280 (2009). [DOI] [PubMed] [Google Scholar]
- [12].Oda T., Yanagisawa H., Yagi T. & Kikkawa M.. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J. Cell Biol. 204, 807–819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang H. & Mitchell D. R.. Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J. Cell Sci. 117, 4179–4188 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mitchell D. R. & Sale W. S.. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144, 293–304 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Carbajal-González B. I., Heuser T., Fu X., Lin J., Smith B. W., Mitchell D. R., et al. . Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton 70, 101–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lechtreck K. F., Delmotte P., Robinson M. L., Sanderson M. J. & Witman G. B.. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180, 633–643 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lechtreck K. F. & Witman G. B.. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176, 473–482 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mitchell, B. F. Pedersen L. B., Feely M., Rosenbaum J. L. & Mitchell D. R.. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell 16, 4509–4518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wargo M. J., Dymek E. E. & Smith E. F.. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J. Cell Sci. 118, 4655–4665 (2005). [DOI] [PubMed] [Google Scholar]
- [20].Rupp G., O’Toole E. & Porter M. E.. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 12, 739–751 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brown J. M., DiPetrillo C. G., Smith E. F. & Witman G. B.. A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell Sci. 125, 3904–3913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Smith E. F. & Lefebvre P. A.. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 132, 359–370 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fu G., Zhao L., Dymenk E., Hou Y., Song K., Phan N., et al. . Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. J. Cell Biol. 218, 4236–4251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bloch M. A. & Johnson K. A.. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 108, 3541–3545 (1995). [DOI] [PubMed] [Google Scholar]
- [25].Yokoyama R., O’toole E., Ghosh S. & Mitchell D. R.. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA 101, 17398–17403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bernstein M., Beech P. L., Katz S. G. & Rosenbaum J. L.. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125, 1313–1326 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Smith E. F. & Lefebvre P. A.. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol. Biol. Cell 8, 455–467 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang P., Fox L., Colbran R. J. & Sale W. S.. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J. Cell Sci. 113, 91–102 (2000). [DOI] [PubMed] [Google Scholar]
- [29].Rao V. G., Sarafdar R. B., Chowdhury T. S., Sivadas P., Yang P., Dongre P. M., et al. . Myc-binding protein orthologue interacts with AKAP240 in the central pair apparatus of the Chlamydomonas flagella. BMC Cell Biol. 17, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao L., Hou Y., Picariello T., Craige B. & Witman G. B.. Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol. 218, 2051–2070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang P., Diener D. R., Yang C., Kohno T., Pazour G. J., Dienes J. M., et al. . Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119, 1165–1174 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li X., Zhang R., Patena W., Gang S. S., Blum S. R., Ivanova N., et al. . An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Teves M. E., Nagarkatti-Gude D. R., Zhang Z. & Strauss J. F.. 3rd Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton 73, 3–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dymek E. E., Lefebvre P. A. & Smith E. F.. PF15p is the Chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot. Cell 3, 870–879 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dutcher S. K., Huang B. & Luck D. J.. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98, 229–236 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Goduti D. J. & Smith E. F.. Analyses of functional domains within the PF6 protein of the central apparatus reveal a role for PF6 sub-complex members in regulating flagellar beat frequency. Cytoskeleton 69, 179–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gorman D. S. & Levine R. P.. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54, 1665–1669 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Craige B., Brown J. M. & Witman G. B.. Isolation of Chlamydomonas flagella. Curr. Protoc. Cell Biol. 50, 3.41.1‐3.41.9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shevchenko A., Tomas H., Havlis J., Olsen J. V. & Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- [40].Johnson K. A., Haas M. A. & Rosenbaum J. L.. Localization of a kinesin-related protein to the central pair apparatus of the Chlamydomonas reinhardtii flagellum. J. Cell Sci. 107, 1551–1556 (1994). [DOI] [PubMed] [Google Scholar]
- [41].Pazour G. J., Agrin N., Leszyk J. & Witman G. B.. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ichikawa M., Liu D., Kastritis P. L., Basu K., Hsu T. C., Yang S., et al. . Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat. Commun. 8, 15035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ichikawa M., Khalifa A. A. Z., Kubo S., Dai D., Basu K., Maghrebi M. A. F., et al. . Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins. Proc. Natl. Acad. Sci. USA 116, 19930–19938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ma M., Stoyanova M., Rademacher G., Dutcher S. K., Brown A. & Zhang R.. Structure of the Decorated Ciliary Doublet Microtubule. Cell 179, 909–922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lechtreck K. F., Gould T. J. & Witman G. B.. Flagellar central pair assembly in Chlamydomonas reinhardtii. Cilia 2, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fu G., Wang Q., Phan N., Urbanska P., Joachimiak E., Lin J., et al. . The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol. Biol. Cell 29, 1048–1059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kubo T., Hou Y., Cochran D. A., Witman G. B. & Oda T.. A microtubule-dynein tethering complex regulates the axonemal inner dynein f (I1). Mol. Biol. Cell 29, 1060–1074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cole, D. G. The Chlamydomonas Sourcebook Second Edition (Academic Press, London, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry is deposited in Dryad (https://doi.org/10.5061/dryad.x0k6djhg6).