Abstract
Genome-wide copy number surveys associated chromosome 11q11 with obesity. As this is an olfactory receptor-rich region, we hypothesize that genetic variation in olfactory receptor genes might be implicated in the pathogenesis of obesity. Multiplex Amplicon Quantification analysis was applied to screen for copy number variants at chromosome 11q11 in 627 patients with obesity and 330 healthy-weight individuals. A ± 80 kb deletion with an internally 1.3 kb retained segment was identified, covering the three olfactory receptor genes OR4C11, OR4P4, and OR4S2. A significant increase in copy number loss(es) was perceived in our patient cohort (MAF = 27%; p = 0.02). Gene expression profiling in metabolic relevant tissues was performed to evaluate the functional impact of the obesity susceptible locus. All three 11q11 genes were present in visceral and subcutaneous adipose tissue while no expression was perceived in the liver. These results support the ‘metabolic system’ hypothesis and imply that gene disruption of OR4C11, OR4P4, and OR4S2 will negatively influence energy metabolism, ultimately leading to fat accumulation and obesity. Our study thus demonstrates a role for structural variation within olfactory receptor-rich regions in complex diseases and defines the 11q11 deletion as a risk factor for obesity.
Keywords: Obesity, Olfactory receptor, Copy number variant, Chromosome 11q11, Expression profiling
Highlights
-
•
Olfactory receptor-rich 11q11 region increases the predisposition for obesity.
-
•
Fine-mapping reveals OR4C11, OR4P4 and OR4S2 as risk factors for obesity.
-
•
Adipose tissue expression of 11q11 genes indicates their metabolic relevance.
-
•
New hypothesis in obesity pathogenesis exposed for three olfactory receptor genes.
1. Introduction
Obesity is a complex heterogeneous disorder in which body fat has accumulated as a result of the chronic imbalance in energy homeostasis. Although excessive food intake and lack of physical activity are perceived as the major contributors, heritability studies revealed that 40–70% of the interindividual variability in body mass index (BMI) is attributed to genetic factors [[1], [2], [3]]. Extensive search for genes involved in body weight regulation led to the recognition of the leptin-melanocortin signaling pathway as a key regulator of food intake and energy expenditure [4]. Mutations in a number of genes from this pathway have been proven to be responsible for early-onset monogenic obesity [5]. Nevertheless, obesity in most individuals has a complex etiology and involves the interaction of multiple genes and environmental factors. The introduction of genome-wide association studies (GWASs) resulted in the identification of >500 genomic loci that account for 16–40% of BMI variability [6]. As these findings only partially explain the heritability estimates for BMI, it warrants the need to examine other forms of genetic variation.
Copy number variants (CNVs) have been predicted to play a significant role in the genetic susceptibility of human disease [[7], [8], [9]]. They are defined as DNA segments ranging in size from 1 kb to several Mb and present themselves as variable copy numbers across individuals. To explore the contribution of CNVs to obesity, genome-wide surveys have been performed in patient populations (Table 1). In 2009, Sha et al.[10] associated a 194 kb copy number variable region (CNVR) at chromosome 10q11.22 with BMI. The region spans four genes of which the neuropeptide Y4 receptor (NPY4R) is acknowledged as an important regulatory gene in food intake. Statistical analysis revealed that 1.6% of the estimated BMI variation could be explained by the CNVR. This was later supported by the study of Aerts et al. (2016), which showed an essential role for genetic and structural NPY4R variation in the pathogenesis of obesity[11]. Furthermore, a meta-analysis by the GIANT Consortium revealed a 10 kb and a 45 kb CNV upstream of neuronal growth regulator 1 (NEGR1). Both deleted regions affect non-overlapping conserved elements present at the locus [12]. NEGR1 is expressed in the brain and hypothalamus, which are implicated in central nervous system processes of body weight regulation. Although its function in the pathogenesis of obesity still has to be determined, the 1p31.1 deletion has been established as a CNVR regulating energy balance [13,14]. In 2010, Bochukova et al. identified a few rare CNVs with a size range of 220 kb to 1.7 MB at chromosome 16p11.2. Different genes were reported for the deleted regions but always contained Src homology 2B adaptor protein 1 (SH2B1)[15]. The gene is part of the leptin-melanocortin pathway and involved in leptin and insulin signaling. Subjects carrying the 16p11.2 deletion exhibit hyperphagia and severe insulin resistance. In 2011, a genome-wide CNV analysis associated a 80 kb deletion at chromosome 11q11 with early-onset extreme obesity [16]. The CNVR represents a locus enclosing the three olfactory receptor genes olfactory receptor family 4 subfamily P member 4 (OR4P4), subfamily S member 2 (OR4S2) and subfamily C member 6 (OR4C6). While the phenotypic diversity in olfactory receptors (ORs) by copy number variability is well-known [[17], [18], [19]], Jarick et al. (2011) were the first to link the CNVR 11q11 with early-onset extreme obesity.
Table 1.
Overview of the most important BMI-associated copy number variants.
| Locus | Position (Mb) | Size (kb) | Overlap Genes | Regulatory pathways |
|---|---|---|---|---|
| 1p31.1 | Chr1: 72,541,074-72,583,749 | 42.7 | NEGR1 | Feeding behavior Locomotory behavior Neuron project development |
| 10q11.22 | Chr10: 46,943,377-47,136,996 | 193.6 | SYT15, GPRIN2, NPY4R, LOC728643 | Pancreatic polypeptide receptor signaling GPCR signaling |
| 11q11 | Chr11: 55,374,020-55,453,589 | 79.6 | OR4P4, OR4S2, OR4C6 | Olfactory signaling pathway GPCR signaling |
| 16p11.2 | Chr16: 28,823,927-29,043,875 | 220.0 | ATXN2L, TUFM, SH2B1, ATP2A1, RABEP2, CD19, NFATC2IP, SPNS1, LAT | Intracellular signal transduction |
Genes responsible for association of CNV with obesity pathogenesis are underlined.
Chromosomal locations shown in genome build GRCh37/hg19.
ORs are generally known for their function in odor recognition. They interact with odorants in the nasal cavity to initiate a neural cascade resulting in the perception of smell [20]. In this way, odor signals can act as a sensor of the metabolic state with the intention of influencing appetite and satiety in humans [21]. The interindividual differences perceived in food intake behavior could be explained by genetic variation in OR genes [22,23]. These findings support the idea for a possible link between chemosensation and obesity. Nevertheless, deep sequencing revealed that these G-protein coupled receptors are also expressed in non-olfactory tissues where they exert diverse functions beyond chemosensation [[24], [25], [26], [27]]. A recent study by Wu et al. (2017) determined the possible function of an ectopically expressed OR in cellular energy metabolism and obesity. They discovered that mouse olfactory receptor 544 (Olfr544 - human homologue OR52K1) is highly expressed in the two major metabolic tissues; activation of the receptor stimulates lipolysis in adipocytes and induces fatty acid oxidation and ketogenesis in the liver. Another study by Giusepponi et al. (2018) analyzed gene expression of olfactory receptor family 6 subfamily C member 3 (OR6C3) in human adipose tissue samples of various body weight. They observed significantly lower OR6C3 expression in subjects with obesity compared to normal-weight individuals. Both studies imply an antiobesogenic effect of OR genes where disruption of their function will result in body fat accumulation.
Although it is not known whether OR genes can have a direct influence on appetite control or energy homeostasis, the OR-rich 11q11 CNV has previously been recognized as an interesting region for obesity [16,30]. Further comprehensive investigation of this CNV might offer novel insights into the genetic architecture of obesity (missing heritability) and can help in revealing new disease mechanisms. In this respect, our objective was to examine the association of the 11q11 CNV candidate region in a Caucasian population of children and adults with obesity. Structural variation screening and expression profiling in metabolic relevant tissues were performed to determine susceptibility for disease.
2. Materials and methods
2.1. Structural variation screening of the olfactory receptor-rich 11q11 region
2.1.1. Study population
A total of 627 patients with obesity (324 children and 303 adults) and 330 normal-weight adults were included for structural variation screening in our study (Table 2). The pediatric patient cohort consists of unrelated children and adolescents (age ≥ 12 years) with obesity that were recruited at the Obesity Clinic for Children from the Antwerp University Hospital (Antwerp) and Jessa Hospital (Hasselt) in Belgium. The adult patient cohort consists of unrelated adults (age ≥ 18 years) with obesity that were recruited at the Obesity Clinic from the Antwerp University Hospital (Antwerp) in Belgium. Patients with mutations in the Melanocortin-4 receptor gene, the most common cause of monogenic obesity, have been excluded from the screening samples. The control population includes normal-weight adults of Caucasian origin recruited among employees from the Antwerp University Hospital and the University of Antwerp as well as among couples seeking prenatal counselling at the Centre of Medical Genetics (due to increased triple test or high maternal age). Couples seeking prenatal genetic counselling because of familial disease history were excluded. All subjects gave their written informed consent and parental permission was provided in case children participated. The study protocol was approved by the local ethics committee (Medical Ethics committee UAntwerp - registration number A04 21) and performed according to the Declaration of Helsinki.
Table 2.
Population characteristics.
| Patients with obesity |
Healthy-weight adults | ||
|---|---|---|---|
| Children | Adults | ||
| N | 324 | 303 | 330 |
| Male (n) | 140 | 81 | 136 |
| Female (n) | 184 | 222 | 194 |
| Age (years) | 12 ± 0.23 | 41 ± 0.74 | 34 ± 0.32 |
| Weight (kg) | 72.3 ± 2.26 | 113.9 ± 1.13 | 65.8 ± 0.54 |
| Height (m) | 1.49 ± 0.018 | 1.69 ± 0.005 | 1.73 ± 0.005 |
| BMI (kg/m2) | 31.57 ± 0.40 | 39.87 ± 0.32 | 21.85 ± 0.10 |
| BMI Z-score | 2.67 ± 0.03 | N.A. | N.A. |
Mean value ± standard error of the mean is shown for all parameters, except N and gender distribution (absolute numbers). N.A.: not applicable.
2.1.2. Anthropometry
Weight was measured on a digital scale to the nearest 0.01–0.2 kg whereas height was measured to the nearest 0.1–0.5 cm, respectively for children and adults. The BMI was calculated for all individuals as weight (in kg) divided by height (in m) squared. For adults, BMI cut-off values were applied as defined by the World Health Organization [31]. Only patients with a BMI ≥ 30 were included while controls were excluded in case 18.5 < BMI ≥ 25. Children with obesity were identified by the use of the Flemish Growth Charts 2004 [32,33]. Percentile lines that cross a BMI of 30 kg/m2 at 18 years of age on the Flemish age- and sex-specific BMI growth curves were used as cut-off values for the diagnosis of obesity. BMI Z-scores were calculated based on data depicted from the Flemish Growth Charts 2004.
2.1.3. CNV analysis by multiplex amplicon quantification
Copy number changes in the genomic region of interest were detected by the use of Multiplex Amplicon Quantification (MAQ), which has been recognized as a valuable diagnostic tool with an assay performance approaching 100% [34]. The technique involves the simultaneous amplification of several fluorescently labelled target and reference amplicons, followed by capillary electrophoresis and fragment analysis.
Genomic DNA was extracted from blood samples for all patient and control samples. The target region was set at chr11:55,300,000-55,700,000 (genome build GRCh37/hg19) and includes the previously reported OR-rich CNVR of Jarick et al. (2011) as well as ten additional OR genes. Primer pairs were designed with the MAQ primer design tool and are available upon request (Supplementary Fig. 1). MAQ-assays were performed with a total of 50 ng input DNA and following the manufactured protocol (Agilent Technologies, Antwerp, Belgium). Two negative control samples were included in each experiment for accurate normalization.
The resulting MAQ-PCR products were analyzed by capillary electrophoresis on an ABI Prism Genetic Analyzer 3130xl (Applied Biosystems Inc., Foster City, CA, USA). Generated raw data were sized relative to the GS500 ROX internal-lane size standard and target amplicons were scored using Genemarker software V2.6.4 (SoftGenetics LLC., Oakwood, PA, USA). This software computes and visualizes the dosage quotient (DQ) by comparing the intensities of the target and reference amplicons in the test individual with those in the experimental control. A DQ of 0.25–0.75 was considered indicative of a deletion while a DQ of 1.25–1.75 was indicative of a duplication.
2.1.4. Characterization of the olfactory receptor-rich 11q11 deletion
OR genes involved in the 11q11 deletion were confirmed by Sanger sequencing (Genbank accession nos. AB065774, AB065775, and BK004390). Gene-specific primers were designed using Primer3 software and are available upon request (genome build GRCh37/hg19). Polymerase chain reactions (PCR) were carried out under standard conditions followed by direct sequencing of the purified PCR product on an ABI Prism Genetic Analyzer 3130xl (Applied Biosystems Inc., Foster City, CA, USA). Sequences were analyzed using CLC DNA workbench (CLC Bio, Aarhus, Denmark).
2.2. Olfactory receptor expression profiling of 11q11 genes in metabolic relevant tissues
Qualitative gene expression of relevant 11q11 ORs was investigated in adult human liver and adipose tissue from three biological replicates. Total RNA from cells and tissues of specimens affected by obesity was isolated using the Quick-RNA™ Microprep kit (Zymo Research, Irvine, CA, USA) for liver and RNeasy® Lipid Tissue kit (Qiagen, Hilden, Germany) for visceral and subcutaneous adipose tissue. cDNA was synthesized with 0.5–3 μg input RNA using SuperScript® III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA). Amplification was performed by a touchdown PCR protocol under standard conditions. The presence of OR expression in both tissues was examined by visualization after gel electrophoresis. The housekeeping gene actin beta (ACTB) was included as internal control. Gene-specific cDNA primers were designed using Primer3 software and are available upon request (genome build GRCh37/hg19).
Supplementary, expression in the olfactory epithelium and metabolic relevant tissues was assessed by browsing the Genotype-Tissue Expression (GTEx) Portal and available liver transcriptome data.
2.3. Statistics
The power of the current study was estimated using the genetic power calculator [35]. Assuming a prevalence of the disease of 14% and a disease allele frequency of 25%, the current study design with 627 cases and 330 controls holds 80% power to detect a disease allele with a genotype relative risk of 1.31 under an additive model, at a significance level of 0.05.
The genotypic probability of disease between patients with obesity and lean adults was statistically evaluated by the Cochran-Armitage trend test. The effect of genotype on BMI was assessed by simple linear regression in case BMI was treated as continuous variable while a Chi-square Test was applied when the different BMI categories (mild – moderate – extreme obesity) were used for analysis. Significance level was set at p = 0.05. All statistical analyses were performed using Rstudio.
3. Results
3.1. Structural variation screening of the olfactory receptor-rich 11q11 region
3.1.1. Common deletion at chromosome 11 associated with obesity
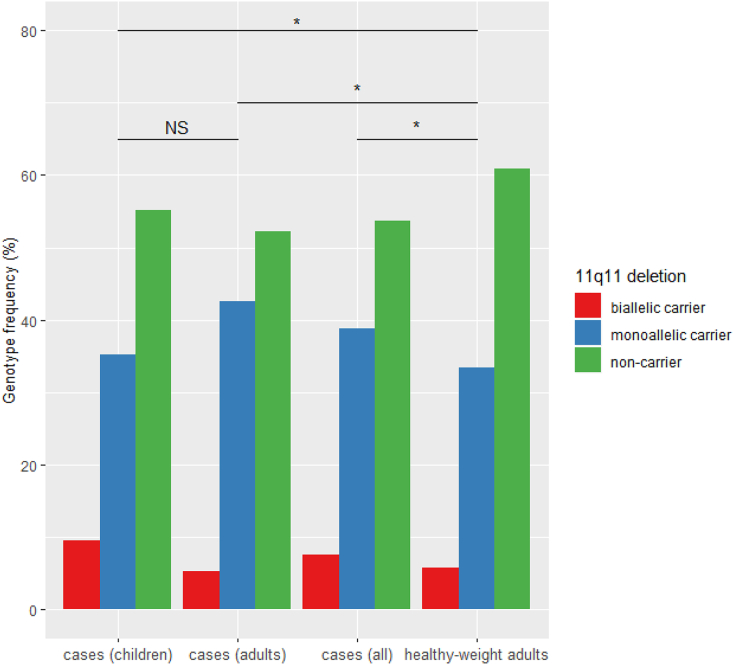
Nine hundred fifty-seven individuals were screened for copy number changes in the 11q11 region. A ± 80 kb deletion with an internally 1.3 kb retained segment was identified. We observed 42.57% heterozygotes and 5.28% homozygotes for the deletion in the adult patient cohort. For the children and adolescents with obesity, heterozygous and homozygous deletion carriers were observed in respectively 35.19% and 9.57%. Natural variation at CNVR 11q11 was considered in controls and identified 33.33% as carriers of heterozygous deletions and 5.76% as carriers of homozygous deletions (Fig. 1). No duplications in the OR-rich region were perceived among subjects with obesity nor in individuals with a healthy weight.
Fig. 1.
Genotypic probability of disease between patients with obesity and adults with healthy weight.
Genotype frequencies for copy number variable region 11q11 are displayed for each phenotype. The genetic risk estimate for obesity was statistically evaluated. Significance levels are presented on the figure as NS: p > 0.05 and *: ≤ 0.05.
Significant differences in genotype frequency (Table 3) were perceived for age-matched patients with obesity versus controls (p = 0.041, odds ratio = 1.25 [0.97–1.62]) as well as for not age-matched patients with obesity versus controls (p = 0.028, odds ratio = 1.29 [1.00–1.59]). These consistent findings allowed us to take both patient groups together, which resulted in an even higher significance level and effect size (p = 0.018). The calculated odds ratio of 1.27 [1.02–1.59] indicates an increased prevalence of the deletion in our patient cohort (MAF = 26.87%) compared to our control population (MAF = 22.42%). No significant difference between both patient groups (children versus adults) could be assigned in respect to the 11q11 deletion frequency (p = 0.41, odds ratio = 1.03 [0.80–1.32]); neither a correlation between BMI and genotype could be recognized when BMI was treated as a continuous variable (p = 0.19), nor when the different BMI categories were assumed (p = 0.07).
Table 3.
Characteristics of the 11q11 CNVR related to the different studied cohorts.
| Patients with obesity | Healthy-weight adults | MAFobese | MAFlean | odds ratio | p-value | |
|---|---|---|---|---|---|---|
| Age-matched cases versus controls | 303 | 330 | 26.57 | 22.42 | 1.25 [0.98–1.62] | 0.041 |
| Not age-matched cases versus controls | 324 | 330 | 27.16 | 22.42 | 1.29 [1.00–1.66] | 0.028 |
| All cases versus controls | 627 | 330 | 26.87 | 22.42 | 1.27 [1.02–1.59] | 0.018 |
3.1.2. Fine-mapping results in characterization of the involved OR genes
Our detailed MAQ design led to fine-mapping of the 11q11 CNV (Supplementary Fig. 1); absence of MAQ probes 5–9 and 12–13 was perceived while probes 1–4, 10–11 and 14–16 were present in CNV-carriers. This corresponded to a minimal size deletion of ±80 kb spanning chr11:55,368,225-55,448,559 and a maximum size deletion of ±148 kb spanning chr11:55,356,922-55,504,384. Internally, a retained segment was discovered with ±1.3 kb as minimal size spanning chr11:55,432,491-55,433,765 and ± 16 kb as maximal size spanning chr11:55,419,331-55,435,229 (Table 4).
Table 4.
Genomic features of 11q11 CNVR.
| Minimal deleted region | Maximal deleted region | Minimum size (kb) | Maximum size (kb) |
|---|---|---|---|
| Chr11:55,368,225-55,448,559 OR4C11 OR4P4 OR4S2 |
Chr11:55,356,922-55,504,384 OR4C11 OR4P4 OR4S2 |
80.3 |
147.5 |
| Minimal internal retained region | Maximal internal retained region | Minimum size (kb) | Maximum size (kb) |
| Chr11:55,432,491-55,433,765 OR4C6 |
Chr11:55,419,331-55,435,229 OR4C6 |
1.3 |
15.9 |
Chromosomal locations shown in genome build GRCh37/hg19.
In-depth analysis showed that OR4C11, OR4P4 and OR4S2 were deleted in 11q11 carriers while OR4C6 was positioned in the internally retained segment. Genotype-specific Sanger sequencing of the four different genes confirmed CNV screening results. Our findings further indicated that the nine additional studied OR genes, lying on the long arm of chromosome 11, were not part of the deletion.
3.2. Olfactory receptor expression profiling of 11q11 genes in metabolic relevant tissues
The expression of genes positioned in the deletion was further examined in metabolic relevant tissues. Although qualitative gene expression and available GTEx and transcriptome data did not allow the detection of transcripts coding for OR4C11 (NM_001004700), OR4S2 (NM_001004059) and OR4P4 (NM_001004124) in the liver, expression profiling in visceral and subcutaneous adipose tissue indicated the presence of these genes in body fat. Gene expression of 11q11 OR genes in the olfactory epithelium could not be demonstrated based on expression databases and literature.
4. Discussion
The present study explored whether structural variation within the olfactory receptor-rich 11q11 candidate region is implicated in the pathogenesis of obesity. An increased risk for childhood and adult obesity was perceived in case an individual carries at least one copy of the deleted allele (p = 0.018). Additionally, a high copy number loss frequency (MAFobese = 26.87%) and low odds ratio (1.27; 95% CI = 1.02–1.59) were observed, assuming that the 11q11 CNVR will have a rather small effect size. Although this is true for monogenic disease forms, common CNVs have already been associated with susceptibility for complex diseases [[36], [37], [38]]. We therefore conclude that the 11q11 deletion can be recognized as a risk factor for complex obesity.
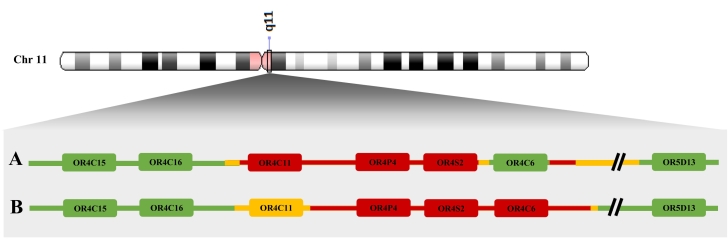
Fine-mapping of the 11q11 CNVR revealed a minimal size deletion of ±80 kb enclosing the three olfactory receptor genes OR4C11, OR4P4 and OR4S2. An internally retained ±1.3 kb segment was discovered that encompasses OR4C6. A difference in size and chromosomal location was perceived with the initial 11q11 CNV study by Jarick et al. (2011). However, we believe that our results identified the same CNVR (Fig. 2). The observed dissimilarity can be ascribed to the used technique for CNV calling. The initial 11q11 CNV loss study applied a GWAS approach using the Affymetrix Genome-Wide Human CNV Array 6.0 while we designed a MAQ assay specifically for the 11q11 region. High-density SNP array experiments (1.8 million probes) provide a cost-effective way for CNV discovery as they can easily detect the total number of copies in a genomic region. Nonetheless, precise breakpoint identification as well as predicting CNV length is rather restricted by the coverage of probes on the microarray [39]. A wide variation in probe distribution along the genome is noticed [40]; some regions are densely located (e.g. known copy number polymorphic sites) while others lack probes (e.g. centromeres). Another drawback is the genomic architecture of the target region. Genomic regions such as segmental duplications, tandem repeats, and complex CNV areas are hard to call by SNP arrays due to limited sensitivity [41]. Both reasons can give an explanation for the perceived differences in the deleted region as it is located around the centromere of chromosome 11 encompassing multiple genes of the hypervariable OR family. Consequently, Jarick et al. (2011) detected a CNVR containing OR4P4, OR4S2 and OR4C6. The use of a more targeted CNV screening approach by our research group leads to the discovery that OR4C11 is also involved in the 11q11 deletion while OR4C6 is still present in deletion carriers. These findings result from a well-thought-out probe design. The strength of using a MAQ assay for CNV screening is that you can specifically design probes in the region of interest. In this way, we were able to find primer pairs within three genes of the OR-rich 11q11 region that were not covered by SNP array probes (Supplementary Fig. 1 & Supplementary Fig. 2). Probes directly positioned up- and downstream of the gene (gene boundaries) were chosen in case no primer pairs were found within the gene itself. This made it possible to discover the involvement of OR4C11 in the 11q11 deletion, a gene that was not identified in the initial CNV loss study by Jarick et al. (2011). However, this gene is positioned in the deletion when considering the maximal deleted region identified by Jarick et al. (2011). Fine-mapping of the 11q11 deletion by our research group makes it possible to hypothesize that genetic variation in OR4C11, OR4P4 and OR4S2 increases the risk for obesity while OR4C6 does not contribute to the observed disease susceptibility resulting from the deletion.
Fig. 2.
Detected copy number variable region in current and initial 11q11 copy number variant loss study.
Comparison of the 11q11 deletion discovered in (A) our study by multiplex amplicon quantification and (B) the study of Jarick et al. (2011) by the Affymetrix Genome-Wide Human CNV Array 6.0. Red coloring represents the minimal deleted region while yellow coloring signifies the maximal deleted region. Green coloring indicates genes that are not positioned in the deletion. Differences in size and chromosomal location are observed. In the current study, ± 80 kb spanning chr11:55,368,225-55,448,559 was identified as minimal region and a ± 147 kb deletion spanning chr11:55,356,922-55,504,384 as maximal region. An internally retained segment was present with ±1.3 kb as minimal size spanning chr11:55,432,491-55,433,765 and ± 16 kb as maximal size spanning chr11:55,419,331-55,435,229. In contrast, Jarick et al. (2011) identified a minimum size deletion of ±80 kb spanning chr11:55,374,020-55,453,589 and a maximum size deletion of ±97 kb spanning chr11: 55,363,328-55,460,696. Both copy number variable regions cover the olfactory receptor genes OR4P4 and OR4S2; inconsistencies are perceived for genes OR4C11 and OR4C6.
HgLiftOver was applied to convert the deleted region of the initial 11q11 CNV loss study from genome build GRCh36/hg18 to GRCh37/hg19.
The same CNVR on chromosome 11 was identified by a study extensively investigating copy number changes in OR-rich regions [42]. They noticed that the CNV was only present as a deletion across geographically diverse populations (MAF = 36%). Carriers of the deletion showed absence of OR genes OR4C11, OR4P2 and OR4S2 while OR4C6 was present in all individuals. Additionally, Young et al. (2008) examined the formation mechanism of the identified 11q11 CNV. The mutational process underlying this copy number change was inferred by the presence of alternative structural alleles around the rearrangement breakpoints. Different formation mechanisms were considered including non-allelic homologous recombination (NAHR), non-homologous end-joining (NHEJ), shrinking or expansion of variable number of tandem repeats (VNTRs) and mobile element insertions (MEI). The most convenient mechanism for OR-containing regions is NAHR since ORs typically occur as highly homologous tandem repeat sequences in the genome. However, Young et al. (2008) characterized the region as a complex CNV area resulting from a combination of multiple deletion and inversion events. They proposed that the presence of highly similar L1 repeats, flanking the 11q11 region, initiate improper pairing of the repeats with the corresponding region. The outcome is the creation of a loop structure wherein the different deletions and inversions could have occurred. Final analysis of the deletion/inversion breakpoints implicated NHEJ as the formation mechanism of this complex CNV area. This is in line with a paper by Mills et al. (2011) who associated specific formation mechanisms with structural variation chromosomal positioning, size and type. They observed (i) high abundance of VNTR near centromeres while NAHR was clustered near telomeres, (ii) occurrence of small structural variants as a result of VNTR or MEI (in presence of Alu or L1 repeats) while NHEJ- and NAHR-based mechanisms were perceived across a wide size range, and (iii) NHEJ as dominating deletion mechanism while MEI was seen as main insertion process [43]. Specific for the identified rearranged area, we infer the presence of NHEJ, VNTR and MEI as the 11q11 region is a complex CNVR consisting of multiple small and large deletion and insertion events near the centromere.
Assessment of the functional impact of gene disrupting CNVs by Mills et al. (2011) noticed a significant enrichment for genes involved in cell defense and sensory perception. Accordingly, genetic variation affecting sensory acuity and perception could explain phenotypic differences observed in the sense of smell [22]. Recent evidence showed that orexigenic agouti-related protein-expressing neurons are regulated by energy status and sensory perception. Copy number loss of ORs could thus result in partial or total insensitivity of the corresponding odorant, for which might be compensated by an increase in food intake [21]. This ‘olfactory system’ hypothesis proposes that disruption of OR genes could influence eating behavior, ultimately leading to hyperphagia and obesity [23,[44], [45], [46], [47]]. An alternative hypothesis has been suggested since the discovery of ectopic OR expression in metabolic relevant tissues. Research has demonstrated that ORs could have a protective role against fat accumulation in the liver and adipose tissue [28,29]. This ‘metabolic system’ hypothesis implies that gene disruption of ORs will negatively influence energy metabolism with obesity as a consequence. Although we were not able to confirm nor reject the “olfactory system” hypothesis with our study design, we could exclude the presence of 11q11 OR gene expression in the liver. In addition, gene expression of OR4C11, OR4P4 and OR4S2 was detected in adipose tissue. This is the first time that expression of these genes has been studied in relation to obesity and gives us the possibility of postulating about the underlying mechanism. Based on our results, the ‘metabolic system’ hypothesis is most likely the responsible mechanism for the increased disease susceptibility perceived in 11q11 deletion carriers. This implies that the responsible OR genes regulate energy metabolism in visceral and subcutaneous adipose tissue. The 11q11 region could thus be recognized as an obesity susceptible locus in which copy number loss(es) will result in a disturbed energy balance with fat accumulation as outcome. Future research will be necessary to elucidate the involvement of the three OR genes in body weight regulation. On one hand, the possible association of obesity with olfactory dysfunction (“olfactory system” hypothesis) will need to be excluded. However, this is not easy as the primary tissue of interest for expression profiling is the olfactory epithelium. On the other hand, the results of the present study need to be confirmed in vitro (e.g. adipocyte cell line) or in vivo (e.g. mouse model). This will make it possible to depict the responsible gene for the increased disease susceptibility or determine whether this is the result of a synergetic effect. Additionally, it will extend our knowledge of the physiological functions of the 11q11 OR in energy metabolism.
5. Conclusion
Our findings indicated an increased prevalence of the OR-rich 11q11 deletion in patients with obesity. Although its effect size is rather small, the CNVR will substantially contribute to the missing heritability of complex obesity. Accordingly, the OR genes encompassing the CNVR (OR4C11, OR4S2, OR4P4) are identified as risk factors for this complex disease. Extensive fine-mapping further revealed a more complex CNV area than originally discovered by Jarick et al. (2011) and exposed the responsible mechanism for CNV formation. Expression profiling demonstrated that all three genes encompassing the 11q11 deletion were expressed in adipose tissue and direct towards the ‘metabolic system’ hypothesis. To our knowledge, we are the first research group to indicate a functional role of the OR-rich 11q11 region in energy homeostasis and susceptibility to obesity (both in children and adults).
Funding
This research was supported by an Interuniversity Attraction Pole Project (Phase VII project 43, BELSPO) and a Research Fund of the University of Antwerp (GOA project: FFB180348/36572). A subcohort of the adult obese population was recruited as part of the European Commission projects HEPADIP (Hepatic and adipose tissue and functions in the metabolic syndrome; Contract LSHM-CT-2005-018734) and RESOLVE (A systems biology approach to RESOLVE the molecular pathology of two hallmarks of patients with metabolic syndrome and its co-morbidities; hypertriglyceridemia and low HDL-cholesterol; contract FP7-305707).
Authors contribution
SD designed the study, carried out experiments, analyzed data and wrote manuscript. SH carried out experiments. WVH contributed to the research design and manuscript revisions. AV, GM, KVH, SV and LVG recruited and clinically screened subjects. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declared no conflict of interest.
Acknowledgements
The research team would like to acknowledge the individuals and organizations who participated in this study. A power calculation was performed by Erik Fransen, Department of Medical Genetics, University of Antwerp, Antwerp, Belgium. Liver transcriptome data was made available thanks to University Lille, Inserm, CHU-Lille, Institut Pasteur de Lille, U1011-EGID, Lille, France.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymgmr.2020.100656.
Appendix A. Supplementary data
References
- 1.Stunkard A.J., Foch T.T., Hrubec Z. A twin study of human obesity. JAMA. 1986;256(1):51–54. 3712713 Epub 1986/07/04. [PubMed] [Google Scholar]
- 2.Stunkard AJ, Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, Schull WJ. An adoption study of human obesity. N Engl J Med. 1986;314(4):193–198. doi: 10.1056/NEJM198601233140401. 3941707 Epub 1986/01/23. [DOI] [PubMed] [Google Scholar]
- 3.Coady SA, Jaquish CE, Fabsitz RR, Larson MG, Cupples LA, Myers RH. Genetic variability of adult body mass index: a longitudinal assessment in framingham families. Obes Res. 2002;10(7):675–681. doi: 10.1038/oby.2002.91. 12105290 Epub 2002/07/10. [DOI] [PubMed] [Google Scholar]
- 4.Beckers S., Zegers D., Van Gaal L.F., Van Hul W. The role of the leptin-melanocortin signalling pathway in the control of food intake. Crit. Rev. Eukaryot. Gene Expr. 2009;19(4):267–287. doi: 10.1615/critreveukargeneexpr.v19.i4.20. 19817705 [DOI] [PubMed] [Google Scholar]
- 5.Zegers D., Van Hul W., Van Gaal L.F., Beckers S. Monogenic and complex forms of obesity: insights from genetics reveal the leptin-melanocortin signaling pathway as a common player. Crit. Rev. Eukaryot. Gene Expr. 2012;22(4):325–343. doi: 10.1615/critreveukargeneexpr.v22.i4.60. 23272802 Epub 2013/01/01. [DOI] [PubMed] [Google Scholar]
- 6.Diels S., Vanden Berghe W. Insights into the multifactorial causation of obesity by integrated genetic and epigenetic analysis; Obes Rev: 2020. Van Hul W.32170999 Epub 2020/03/15. [DOI] [PubMed] [Google Scholar]
- 7.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. 17122850 PMC2669898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollox E.J., Hoh B.P. Human gene copy number variation and infectious disease. Hum. Genet. 2014;133(10):1217–1233. doi: 10.1007/s00439-014-1457-x. 25110110 [DOI] [PubMed] [Google Scholar]
- 9.Jensen M., Kooy R.F., Simon T.J., Reyniers E., Girirajan S., Tassone F. A higher rare CNV burden in the genetic background potentially contributes to intellectual disability phenotypes in 22q11.2 deletion syndrome. Eur. J. MEd. Genet. 2018;61(4):209–212. doi: 10.1016/j.ejmg.2017.11.016. 29191496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha B.Y., Yang T.L., Zhao L.J., Chen X.D., Guo Y., Chen Y. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J. Hum. Genet. 2009;54 doi: 10.1038/jhg.2009.10. (4):199–202. Epub 2009/02/21. PubMed PMID: 19229253; PubMed Central PMCID: PMCPMC2733232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerts E, Beckers S, Zegers D, Van Hoorenbeeck K, Massa G, Verrijken A. CNV analysis and mutation screening indicate an important role for the NPY4R gene in human obesity. Obesity (Silver Spring). 2016;24(4):970–976. doi: 10.1002/oby.21435. 26921218 Epub 2016/02/28. [DOI] [PubMed] [Google Scholar]
- 12.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. 19079261 PMC2695662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renstrom F., Payne F., Nordstrom A., Brito E.C., Rolandsson O., Hallmans G. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum. Mol. Genet. 2009;18(8) doi: 10.1093/hmg/ddp041. 1489–96. Epub 2009/01/24. PubMed PMID: 19164386; PubMed Central PMCID: PMCPMC2664142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Bradfield J.P., Li M., Wang K., Zhang H., Kim C.E. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity (Silver Spring) 2009;17(12) doi: 10.1038/oby.2009.159. 2254–7. Epub 2009/05/30. PubMed PMID: 19478790; PubMed Central PMCID: PMCPMC2860782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463(7281) doi: 10.1038/nature08689. 666–70. Epub 2009/12/08. PubMed PMID: 19966786; PubMed Central PMCID: PMCPMC3108883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarick I., Vogel C.I., Scherag S., Schafer H., Hebebrand J., Hinney A. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum. Mol. Genet. 2011;20(4) doi: 10.1093/hmg/ddq518. 840–52. Epub 2010/12/07. PubMed PMID: 21131291; PubMed Central PMCID: PMCPMC3024044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasin-Brumshtein Y, Lancet D, Olender T. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 2009;25(4):178–184. doi: 10.1016/j.tig.2009.02.002. 19303166 Epub 2009/03/24. [DOI] [PubMed] [Google Scholar]
- 18.Zozulya S., Echeverri F., Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2(6) doi: 10.1186/gb-2001-2-6-research0018. 11423007 RESEARCH0018. PMC33394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasin Y., Olender T., Khen M., Gonzaga-Jauregui C., Kim P.M., Urban A.E. High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genet. 2008;4(11) doi: 10.1371/journal.pgen.1000249. e1000249. Epub 2008/11/08. PubMed PMID: 18989455; PubMed Central PMCID: PMCPMC2570968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young J.M., Trask B.J. The sense of smell: genomics of vertebrate odorant receptors. Hum. Mol. Genet. 2002;11(10):1153–1160. doi: 10.1093/hmg/11.10.1153. 12015274 [DOI] [PubMed] [Google Scholar]
- 21.Rebolledo-Solleiro D., Solleiro-Villavicencio H., Velasco M., Roldan-Roldan G. Obesity, metabolic syndrome and olfactory perception. Rev. Neurol. 2020;70(2) doi: 10.33588/rn.7002.2019204. 31930471 53–66. Epub 2020/01/14. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449(7161):468–472. doi: 10.1038/nature06162. 17873857 Epub 2007/09/18. [DOI] [PubMed] [Google Scholar]
- 23.Choquette AC, Bouchard L, Drapeau V, Lemieux S, Tremblay A, Bouchard C. Association between olfactory receptor genes, eating behavior traits and adiposity: results from the Quebec Family Study. Physiol Behav. 2012;105(3):772–776. doi: 10.1016/j.physbeh.2011.10.015. 22044667 Epub 2011/11/03. [DOI] [PubMed] [Google Scholar]
- 24.Griffin C.A., Kafadar K.A., Pavlath G.K. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell. 2009;17(5):649–661. doi: 10.1016/j.devcel.2009.09.004. 19922870 PMC2780437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flegel C., Manteniotis S., Osthold S., Hatt H., Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0055368. e55368. Epub 2013/02/14. PubMed PMID: 23405139; PubMed Central PMCID: PMCPMC3566163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang A.J., Ortega F.E., Riegler J., Madison D.V., Krasnow M.A. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature. 2015;527(7577):240–244. doi: 10.1038/nature15721. 26560302 PMC4765808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z., Zhao H., Fu N., Chen L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell. Physiol. 2018;233(3):2104–2115. doi: 10.1002/jcp.25929. 28338216 [DOI] [PubMed] [Google Scholar]
- 28.Giusepponi ME, Kern M, Chakaroun R, Wohland T, Kovacs P, Dietrich A. Gene expression profiling in adipose tissue of Sprague Dawley rats identifies olfactory receptor 984 as a potential obesity treatment target. Biochem Biophys Res Commun. 2018;505(3):801–806. doi: 10.1016/j.bbrc.2018.09.137. 30297106 Epub 2018/10/10. [DOI] [PubMed] [Google Scholar]
- 29.Wu C., Hwang S.H., Jia Y., Choi J., Kim Y.J., Choi D. Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J. Clin. Invest. 2017;127(11):4118–4123. doi: 10.1172/JCI89344. 28990936 PMC5663348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D., Li Z., Wang H., Yang M., Liang L., Fu J. Interactions between obesity-related copy number variants and dietary behaviors in childhood obesity. Nutrients. 2015;7 doi: 10.3390/nu7043054. (4):3054–66. Epub 2015/04/29. PubMed PMID: 25912042; PubMed Central PMCID: PMCPMC4425189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.WHO Fact sheet 2018 [30/03/20]. Available from. https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight
- 32.Roelants M, Hauspie R, Hoppenbrouwers K. References for growth and pubertal development from birth to 21 years in Flanders, Belgium. Annals of human biology. 2009;36(6):680–694. doi: 10.3109/03014460903049074. 19919503 Epub 2009/11/19. [DOI] [PubMed] [Google Scholar]
- 33.Hauspie R., Roelants M. Flemish Growth Charts 2004 2004 [22 July 2013]. Available from. http://www.vub.ac.be/groeicurven
- 34.Kumps C., Van Roy N., Heyrman L., Goossens D., Del-Favero J., Noguera R. Multiplex amplicon quantification (MAQ), a fast and efficient method for the simultaneous detection of copy number alterations in neuroblastoma. BMC Genomics. 2010;11:298. doi: 10.1186/1471-2164-11-298. 20459859 PMC2879279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. 12499305 Epub 2002/12/25. [DOI] [PubMed] [Google Scholar]
- 36.Hollox E.J., Huffmeier U., Zeeuwen P.L., Palla R., Lascorz J., Rodijk-Olthuis D. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40(1) doi: 10.1038/ng.2007.48. 23–5. Epub 2007/12/07. PubMed PMID: 18059266; PubMed Central PMCID: PMCPMC2447885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poniah P., Mohd Zain S., Abdul Razack A.H., Kuppusamy S., Karuppayah S., Sian Eng H. Genome-wide copy number analysis reveals candidate gene loci that confer susceptibility to high-grade prostate cancer. Urol. Oncol. 2017;35(9) doi: 10.1016/j.urolonc.2017.04.017. 28527622 545 e1- e11. Epub 2017/05/22. [DOI] [PubMed] [Google Scholar]
- 38.Sul JH, Service SK, Huang AY, Ramensky V, Hwang SG, Teshiba TM. Contribution of common and rare variants to bipolar disorder susceptibility in extended pedigrees from population isolates. Transl Psychiatry. 2020;10(1):74. doi: 10.1038/s41398-020-0758-1. 32094344 Epub 2020/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W., Olivier M. Current analysis platforms and methods for detecting copy number variation. Physiol. Genomics. 2013;45(1):1–16. doi: 10.1152/physiolgenomics.00082.2012. 23132758 PMC3544484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin C.F., Naj A.C., Wang L.S. Analyzing copy number variation using SNP Array data: protocols for calling CNV and association tests. Curr Protoc Hum Genet. 2014;79:1–27. doi: 10.1002/0471142905.hg0127s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z.F., Ruivenkamp C., Staaf J., Zhu H., Barbaro M., Petillo D. Detection of submicroscopic constitutional chromosome aberrations in clinical diagnostics: a validation of the practical performance of different array platforms. Eur. J. Hum. Genet. 2008;16(7):786–792. doi: 10.1038/ejhg.2008.14. 18285835 [DOI] [PubMed] [Google Scholar]
- 42.Young J.M., Endicott R.M., Parghi S.S., Walker M., Kidd J.M., Trask B.J. Extensive copy-number variation of the human olfactory receptor gene family. Am. J. Hum. Genet. 2008;83 doi: 10.1016/j.ajhg.2008.07.005. (2):228–42. Epub 2008/08/05. PubMed PMID: 18674749; PubMed Central PMCID: PMCPMC2495065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332) doi: 10.1038/nature09708. 59–65. Epub 2011/02/05. PubMed PMID: 21293372; PubMed Central PMCID: PMCPMC3077050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariman E.C., Szklarczyk R., Bouwman F.G., Aller E.E., van Baak M.A., Wang P. Olfactory receptor genes cooperate with protocadherin genes in human extreme obesity. Genes Nutr. 2015;10(4) doi: 10.1007/s12263-015-0465-3. 465. Epub 2015/05/07. PubMed PMID: 25943692; PubMed Central PMCID: PMCPMC4420755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel Z.M., DelGaudio J.M., Wise S.K. Higher Body Mass Index Is Associated with Subjective Olfactory Dysfunction. Behav. Neurol. 2015;2015 doi: 10.1155/2015/675635. 675635. Epub 2015/07/23. PubMed PMID: 26199458; PubMed Central PMCID: PMCPMC4496469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pastor A., Fernandez-Aranda F., Fito M., Jimenez-Murcia S., Botella C., Fernandez-Real J.M. A Lower Olfactory Capacity Is Related to Higher Circulating Concentrations of Endocannabinoid 2-Arachidonoylglycerol and Higher Body Mass Index in Women. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0148734. e0148734. Epub 2016/02/06. PubMed PMID: 26849214; PubMed Central PMCID: PMCPMC4746072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simchen U, Koebnick C, Hoyer S, Issanchou S, Zunft HJ. Odour and taste sensitivity is associated with body weight and extent of misreporting of body weight. Eur J Clin Nutr. 2006;60(6):698–705. doi: 10.1038/sj.ejcn.1602371. 16435003 Epub 2006/01/26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.