Abstract
Background and Aims
While nuclear DNA content variation and its phenotypic consequences have been well described for animals, vascular plants and macroalgae, much less about this topic is known regarding unicellular algae and protists in general. The dearth of data is especially pronounced when it comes to intraspecific genome size variation. This study attempts to investigate the extent of intraspecific variability in genome size and its adaptive consequences in a microalgal species.
Methods
Propidium iodide flow cytometry was used to estimate the absolute genome size of 131 strains (isolates) of the golden-brown alga Synura petersenii (Chrysophyceae, Stramenopiles), identified by identical internal transcribed spacer (ITS) rDNA barcodes. Cell size, growth rate and genomic GC content were further assessed on a sub-set of strains. Geographic location of 67 sampling sites across the Northern hemisphere was used to extract climatic database data and to evaluate the ecogeographical distribution of genome size diversity.
Key Results
Genome size ranged continuously from 0.97 to 2.02 pg of DNA across the investigated strains. The genome size was positively associated with cell size and negatively associated with growth rate. Bioclim variables were not correlated with genome size variation. No clear trends in the geographical distribution of strains of a particular genome size were detected, and strains of different genome size occasionally coexisted at the same locality. Genomic GC content was significantly associated only with genome size via a quadratic relationship.
Conclusions
Genome size variability in S. petersenii was probably triggered by an evolutionary mechanism operating via gradual changes in genome size accompanied by changes in genomic GC content, such as, for example, proliferation of transposable elements. The variation was reflected in cell size and relative growth rate, possibly with adaptive consequences.
Keywords: Intraspecific DNA content variation, genome size, flow cytometry, golden-brown algae, Synura petersenii, GC content, biovolume, growth rate, environmental conditions, ITS
INTRODUCTION
The nuclear genome constitutes an essential cell component. While the quality of nuclear DNA (expressed by nucleotide sequences or presence of specific alleles) has been intensively in the focus of biologists for more than a half of century, its quantity per cell has received far less attention. Nonetheless, genomes contain orders of magnitude more DNA than required to sustain the basic cell functioning (a phenomenon called the ‘C-value enigma’; Gregory, 2001a) and this may suggest that even the overall quantity of nuclear DNA has an adaptive potential (Mirsky and Ris, 1951; Thomas, 1971; Cavalier-Smith, 2005). Our knowledge of the genome size variation and its evolutionary consequences mainly comes from plant and animal studies (e.g. Leitch et al., 1998; Gregory, 2005; Beaulieu et al., 2008; Liedtke et al., 2018; Trávníček et al., 2019). Much less of this topic is known from single-celled eukaryotes, despite the fact that protist genome size ranges >28 600-fold compared with 6600-fold variation among plants and animals (Veldhuis et al., 1997; Gregory, 2005; Keeling and Slamovits, 2005; Pellicer and Leitch, 2020).
There are several evolutionary mechanisms responsible for variation of the amount of nuclear DNA . The genome size can either increase or decrease by chromosomal aberrations (aneuploidy), non-homologous recombination and changes in the relative genome-wide frequency of insertions to deletions (Devos et al., 2002; Roux et al., 2003; Lynch and Conery, 2003; Wu et al., 2018). Conversely, only an increase in genome size is possible via higher activity of transposable elements and, more abruptly, by whole-genome doubling (polyploidization; Soltis and Soltis, 1999; Kidwell, 2002; Cavalier-Smith, 2005; Sun et al., 2012). Recent or past hybridization events between closely related but separate species can also contribute to differentiation of the amount of nuclear DNA (Baack et al., 2005).
Variation in nuclear DNA content is usually accompanied by phenotypic consequences. The genome size directly affects the size of the nucleus (Sparrow and Evans, 1961; Baetcke et al., 1967; Bennett, 1972; Gregory, 2001b; Zubáčová et al., 2008) and through it also fundamentally relates to the cell size (Cavalier-Smith and Beaton, 1999; Gregory, 2001a, b; Cavalier-Smith, 2005). The genome size–cell size correlation, also known as the karyoplasmic ratio, has been observed across the eukaryotic tree of life, including many protist lineages (Wilson, 1925; Bennett, 1972; Suzuki et al., 1982; Shuter et al., 1983; Veldhuis et al., 1997; LaJeunesse et al., 2005; Connolly et al., 2008; von Dassow et al., 2008). Although the exact cause of this relationship is still debated, evolutionary changes in cell size may either cause or be caused by changes in genome size (Gregory, 2001a). The cell size is a particularly important trait in single-celled organisms as it inversely correlates with metabolic rate and growth rate, and directly correlates with generation time (Van’t Hof and Sparrow, 1963; Bennett, 1972; Shuter et al., 1983; Gregory, 2001a; Cavalier-Smith, 2005). Changes in cell size may also be reflected in protist ecology, for example by altering the grazing pressure, efficiency of nutrient acquisition and/or light harvesting (Garcia-Pichel, 1994; Finkel et al., 2001; Smetacek et al., 2004; Irwin et al., 2006). Therefore, genome size variation could be (via cell size) subject to natural selection in populations of protists (Cavalier-Smith, 2005).
Another understudied genomic parameter with possibly adaptive nature is the relative genome-wide frequency of AT to GC base pairs, often expressed as a %GC content (Bennett and Leitch, 2011; Šmarda and Bureš, 2012). The higher genomic GC content is sometimes associated with extreme climatic conditions such as pronounced cold, drought or temperature fluctuations, probably due to the increased thermal stability of the DNA double helix (Šmarda and Bureš, 2012; Šmarda et al., 2014; Trávníček et al., 2019). Additionally, the GC content variation may be linked to changes in genome size, since the genomic nucleobase composition might be altered by high activity of transposable elements (TEs) or chromosomal aberrations (Wichman et al., 1993; Armbrust, 2004; Derelle et al., 2006). However, only little is known about the evolutionary impact of GC content variation and, what is known, comes almost exclusively from studies on prokaryotes, plants and animals (Goodsell and Dickerson, 1994; Hildebrand et al., 2010; Šmarda et al., 2014; Mugal et al., 2015; Veleba et al., 2017; Trávníček et al., 2019).
Both genome size and GC content may be estimated using flow cytometry (FCM). This technique is based on measuring the properties of fluorescent-stained particles (e.g. cells or isolated nuclei) in a stream of fluid and allows rapid and precise nuclear DNA analysis (Doležel et al., 2007). While FCM has found a wide spectrum of applications in genomic surveys on plants and animals (Dionisio Pires et al., 2004; Kron et al., 2007; Galbraith, 2012; Pellicer and Leitch, 2014), its potential has not yet been explored in depth in protist studies (but see Figueroa et al., 2010), and robust methodological protocols allowing work with diverse protist material are missing.
The dearth of data is especially pronounced when it comes to intraspecific genome size variation in unicellular eukaryotes. This can be attributed to analysing only a single strain per species in most studies (Veldhuis et al., 1997; Mazalová et al., 2011) and, possibly, to the fact that in contrast to, for example, plant studies, best-practice protocols preventing false reports based on methodological and instrumental errors (Greilhuber, 2005) are not routinely applied.
To study the evolution of genome size and its phenotypic and physiological consequences, we chose the golden-brown algal species Synura petersenii (Chrysophyceae, Stramenopiles) as our model system. In general, protist species have a short generation time and huge population size, which allows them to respond quickly to environmental change (Lynch and Conery, 2003; Foissner, 2007; Ribeiro et al., 2013). Synura petersenii is an autotrophic flagellate with assumed worldwide distribution. It creates colonies of cells covered by siliceous scales with species-specific ornamentation and a characteristic pronounced central keel (Kristiansen and Preisig, 2007). The species has recently undergone thorough taxonomic revision supported by molecular markers and morphometric analysis of its siliceous scales, which revealed 15 separate species in the formerly recognized S. petersenii species complex (Wee et al., 2001; Boo et al., 2010; Kynčlová et al., 2010; Škaloud et al., 2012, 2014; Jo et al., 2016). One of these taxonomically revised species, S. petersenii sensu stricto (s.s.) is used as a model species in this study. During our pilot FCM analysis, we detected intraspecific genome size variation among the strains of S. petersenii. The general aims of the study are to prove the existence of intraspecific genome size variation in S. petersenii and investigate in detail, for the first time, the extent of intraspecific variability in genome size in a microalgal species. Additionally, we ask the following questions. (1) Is the variability in DNA content linked to GC content variation? (2) Are there any phenotypic and physiological consequences of varying genome size? (3) Is genome size variation among strains reflected in their ecogeographical distribution?
MATERIALS AND METHODS
Origin, cultivation and identification of the investigated strains
Altogether, 131 isolates of the species Synura petersenii were obtained from various freshwater localities across the Northern hemisphere. Sampling details are listed in Supplementary data Table S1. To establish new cultures, water samples were taken using a 25 µm mesh plankton net and single Synura colonies were captured by micro-pipetting and transferred into separate culture wells filled with WC medium (Guillard and Lorenzen, 1972). All cultures were maintained at 17 °C (cooling box Pol-Eko Aparatura Sp.J., model ST 1, Wodzisław Śląski, Poland) with 24 h light mode under illumination of 30 µmol m–2 s–1 (TLD 18 W/33 fluorescent lamps, Philips, Amsterdam, the Netherlands).
All strains were identified based on their internal transcribed spacer sequence of nuclear ribosomal DNA (ITS1, 5.8S and ITS2 rDNA) since this is the most variable of the commonly used molecular markers in this group (Jo et al., 2016). For this purpose, genomic DNA was extracted from a centrifuged pellet of cells by InstaGene Matrix (Bio-Rad, USA) and the resulting supernatant was directly used as a PCR template. The amplifications were performed using the universal primer ITS4 (White, 1990) and a lineage-specific primer Kn1.1 (Wee et al., 2001). The PCRs were carried out in a total volume of 20 µL with a PCR mix containing 0.2 µL of MyTaqHS DNA polymerase (Bioline), 4 µL of MyTaqHS buffer (Bioline), 0.4 µL of each primer, 14 µL of double-distilled water and 1 µL of template DNA (not quantified). The amplifications were performed in Eppendorf Mastercycler ep Gradient 5341 (Eppendorf GmbH, Hamburg, Germany) using the following program: 1 min of denaturation at 95 °C; followed by 35 cycles of denaturation at 95 °C (15 s), annealing at 52 °C (30 s) and elongation at 72 °C (40 s), concluded with a final extension at 72 °C (7 min) and held at 10 °C. The PCR products were sized on a 1 % agarose gel and then purified using AMPure XP magnetic beads (Agencourt). The purified DNA templates were sequenced by the Sanger Sequencing method at Macrogen, Inc. (Seoul, Korea, http://dna.macrogen.com). Finally, the obtained sequences were identified using BLAST in the National Center for Biotechnology Information (NCBI) Search database and our personal ITS database created during previous studies (Škaloud et al., 2012, 2014). The strains with ITS rDNA sequence identical to that of S. petersenii were transferred into Erlenmeyer flasks with 30 mL of WC medium and kept for longer cultivation. The collection was further supplemented with five strains from previous studies (Kynčlová et al., 2010; Kim et al., 2019).
DNA content estimation
To estimate the nuclear genome size of our strains, we employed propidium iodide FCM. Approximately 2 weeks before the planned FCM analyses, cultures were inoculated into fresh medium. For sample preparation, 1 mL of well-grown culture was centrifuged (5500 rpm for 5 min) and the superfluous medium was removed by pipetting. Subsequently, 350 µL of ice-cold nuclei isolation buffer Otto I (0.1 m citric acid, 0.5 % Tween-20; Otto, 1990) was added to the algal pellet, causing the release of the sample nuclei. The resulting suspension was thoroughly shaken and kept on ice. Solanum pseudocapsicum (2C = 2.59 pg; Temsch et al., 2010) was used as an internal standard. To release nuclei of the standard, an approx. 20 mg piece of fresh leaf tissue was chopped with a razor blade in a plastic Petri dish with 250 µL of ice-cold Otto I buffer. Both suspensions (with algal and standard nuclei) were thoroughly mixed and filtered through a 42 µm nylon mesh into a special 3.5 mL cuvette for direct use with the flow cytometer. Following a 20 min incubation at room temperature, the sample was mixed with 1 mL of staining solution consisting of Otto II buffer (0.4 m Na2HPO4·12H2O; Otto, 1990), 50 μg mL−1 propidium iodide, 50 μg mL−1 RNase IIA and 2 μL mL−1 β-mercaptoethanol. The stained sample was immediately analysed using a Partec CyFlow SL cytometer (Partec GmbH, Münster, Germany) equipped with a green solid-state laser (Cobolt Samba, 532 nm, 100 mW). Measurements on each sample were taken for up to 5000 particles, and the resulting FCM histograms analysed using FloMax ver. 2.4d (Partec, Münster, Germany). Since there is no knowledge of the ploidy level in the genus Synura (Olefeld et al., 2018), we identified the first sample peak on the FCM histogram as G1 (vegetative cells) and the second peak as G2 (dividing cells). The absolute nuclear DNA amount (C-value) was calculated as sample G1 peak mean fluorescence/standard G1 peak mean fluorescence × standard 2C DNA content (according to Doležel, 2005).
In the case of low quality measurement, i.e. G1 sample coefficient of variation (CV) >5 %, the sample preparation and analysis was repeated. To minimize the effect of random instrumental shift, each S. petersenii strain was analysed at least three times on separate days. Whenever the three independent genome size estimates differed by >3 %, the most outlying measurement was discarded and a new measurement conducted, until this condition was fulfilled. In order to corroborate genome size differences among strains, simultaneous analysis of multiple selected strains (i.e. A64, D55 and G61) was performed. We also tested for strain genome size stability during its cultivation. Following inoculation into a fresh medium, three strains exhibiting the highest variation among repeated measurements (i.e. F19, G16 and H11) were analysed once a week for the period of 6–7 weeks. Two other strains (961 and S63.B3) were then re-analysed 2 years following the first measurements.
GC content estimation
To assess variation in genomic GC content, we analysed 38 strains of S. petersenii using FCM with the AT-selective dye DAPI (4',6-diamidino-2-phenylindole) and compared the results with propidium iodide FCM. The strains for GC content estimation, cell size measurements and growth rate analysis (see below) were selected representatively across the whole range of genome size diversity; however, a different set of strains was used for each assessment (Supplementary data Table S2). This was due to unavailability of some strains at the time of particular assessments (e.g. the cultured strains did not survive, provided limited biomass or a contamination occurred); replacement strains with similar genome size were then randomly selected. We employed the same sample preparation as for propidium iodide FCM, except that the staining solution consisted of 1 mL of Otto II buffer, 4 µg mL−1 DAPI and 2 μL mL−1 β-mercaptoethanol. Samples were immediately analysed using a Partec PA II flow cytometer (Partec GmbH, Münster, Germany) equipped with a 488 nm UV LED as a source of excitation light. Analyses were run up to 5000 particles and the resulting FCM histograms were analysed using FloMax ver. 2.4d (Partec, Münster, Germany). Computation of the base content was done according to Šmarda et al. (2008) via a publicly available Excel spreadsheet (http://sci.muni.cz/botany/systemgr/download/Festuca/ATGCFlow.xls).
Cell size measurements
After 2 weeks of cultivation in fresh medium, the cell size of 39 selected S. petersenii strains was analysed by imaging FCM using Benchtop B3 Series FlowCAM (Fluid Imaging Technologies, Yarmouth, ME, USA). The FlowCAM settings were AutoImage mode, 50 μm flow cell, ×20 objective and flow rate 0.020 mL min–1. The mean biovolume of 100 cells per strain was calculated on manually selected images using VisualSpreadsheet® Particle Analysis Software ver. 4.11.12 as the volume of a sphere of the area-based radius measured automatically by the FlowCAM for each cell.
Growth rate
Test of growth rate was performed on eight replicate cultures of each of the 31 selected S. petersenii strains. The chlorophyll fluorescence, i.e. the effective quantum yield of photochemical energy conversion in photosystem II (ΦPSII), of cultures starting with the same initial concentration was measured daily at the same hour over 15 d using a PAM 2500 fluorometer (Heinz Walz GmbH, Effeltrich, Germany). The variable ΦPSII is a relative parameter calculated as (FM′ − F)/FM′, where F is the steady-state fluorescence in the light-adapted state and FM′ is the maximum fluorescence in the light-adapted state measured after the application of a saturation pulse (Roháček and Barták, 1999). The growth rate was subsequently derived as an inverse value of the median time at which the population density reaches half the carrying capacity, i.e. the inflection point (t-mid value–1) in R software ver. 3.4.3 (R Development Core Team, 2017) using the package growthcurver ver. 0.3.0 (Sprouffske and Wagner, 2016).
Ecogeographical patterns
Geographical distribution of the genome size diversity in Synura was visualized on a map using ArcGIS 10.0 (ESRI, Redlands, CA, USA). In order to assess putative ecogeographical trends in the distribution of genome size diversity well beyond any obvious spatial patterns, we also tested for associations between the genome size of Synura and database-derived climatic variables. We used ArcGIS to extract climate data from 19 Bioclim variables of the WorldClim database ver. 2 (http://worldclim.org/; Fick and Hijmans, 2017) downloaded in the highest available resolution (30 arc seconds). Climatic conditions may affect aquatic microalgae e.g. via temperature-regulated onset and duration of their seasonal blooms or precipitation frequently being associated with input of nutrients into aquatic ecosystems (Baek et al., 2009). Each of 67 sampling sites was assigned values of the climatic variables and geographic latitude, included as an additional ecogeographically relevant parameter. When multiple strains were collected at a site, we only retained those with genome size estimates differing by at least 5 % (i.e. an arbitrarily selected threshold to prevent pseudoreplication of data) and such strains were then treated as independent observations. If two or more strains with a similar genome size (<5 % difference) originated from the same site, all except one randomly selected strain were excluded from the dataset. The resulting dataset consisting of 82 strains was analysed using a redundancy analysis (RDA) implemented in Canoco 5 (Lepš and Šmilauer, 2014); the genome size of Synura was used as an explanatory variable. All response variables were standardized prior to the RDA and statistical significance was tested using a Monte Carlo test with 999 permutations. The RDA was then also used to test the effect of GC content.
Statistical analysis
Unless stated otherwise, statistical data analysis was conducted in R. Separate linear regression models were applied to test whether the variation in cell size and growth rate can be explained by genome size (log transformed). These analyses were conducted on a sub-set of 39 and 31 strains, respectively, for which data were available. Both models were also re-run using the GC content as an explanatory variable.
We then employed a regression model to assess the relationship between GC content (response variable) and genome size (explanatory variable, log transformed). Due to previous reports of a quadratic relationship between the two genomic parameters in plants (e.g. Šmarda et al., 2014), we used manual Akaike information criterion (AIC)-based forward selection with the function ‘addterm’ from the R package MASS ver. 7.3-50 (Venables and Ripley, 2002) to test whether incorporating the logarithm of genome size either in linear (approx. log.GS) or quadratic form [approx. log.GS + I(log.GS2)] will significantly improve the model performance.
RESULTS
Intraspecific variability in genome size
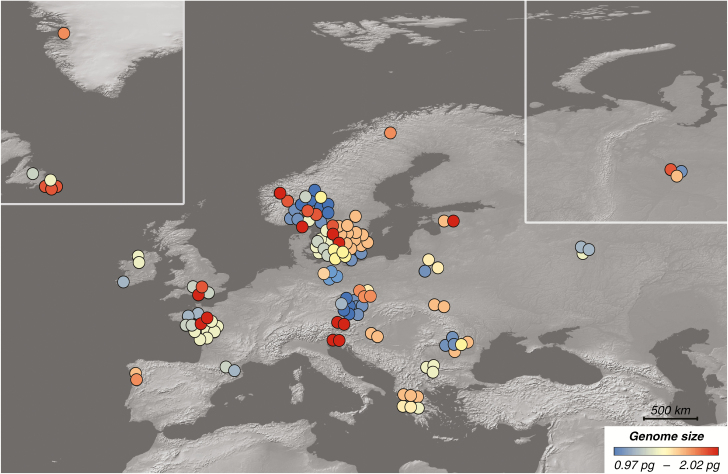
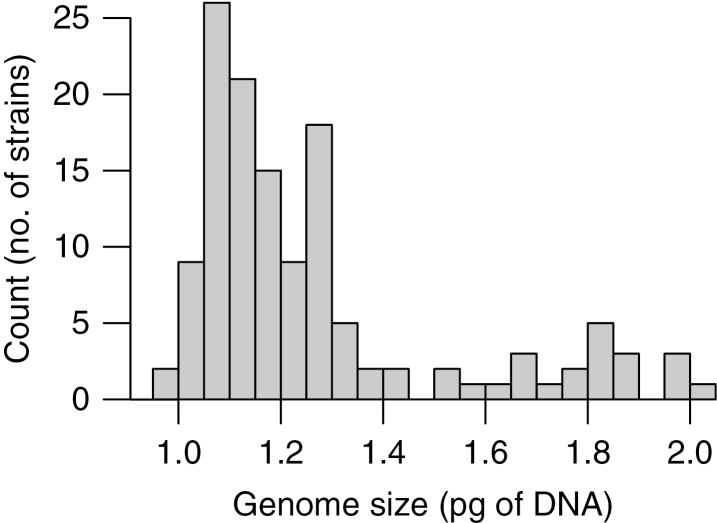
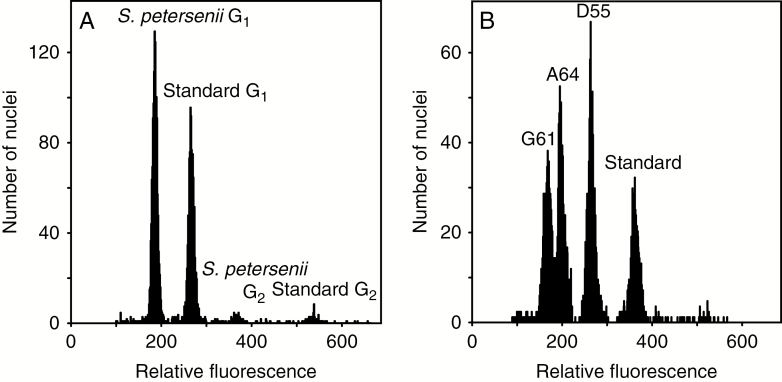
We successfully determined the absolute nuclear DNA amount in 131 strains of S. petersenii with identical ITS rDNA sequence from 67 localities across the Northern hemisphere (Fig. 1). The sampled S. petersenii strains exhibited a 2.1-fold variation in their genome size, ranging from 0.971 to 2.022 pg of DNA (Supplementary data Table S2). The frequency distribution of the genome size values was conspicuously positively skewed (median = 1.170 pg, mean = 1.296 pg; Fig. 2). Sufficient precision of flow cytometric measurements was ensured by relatively low CVs for both sample and standard G1 nuclei peaks (mean CV = 3.29 and 2.25 %, respectively); see Fig. 3A for a representative analysis. The intraspecific variability in genome size was also verified in a simultaneous analysis of three S. petersenii strains with contrasting nuclear DNA amounts that resulted in three clearly differentiated peaks in the flow cytometric histogram (Fig. 3B).
Fig. 1.
Distribution of 131 strains of Synura petersenii under study and their estimated genome size. Symbol colour refers to different genome size categories.
Fig. 2.
Frequency distribution of genome size categories among investigated Synura petersenii strains.
Fig. 3.
Flow cytometric histograms showing relative fluorescence of propidium iodide-stained nuclei of Synura petersenii samples and Solanum pseudocapsicum (Standard). (A) A representative analysis of a single S. petersenii strain with G1 and G2 phase nuclei apparent for both the analysed sample and standard. (B) A simultaneous analysis of three strains with distinct genome size (G61, A64 and D55) to confirm the existing differences.
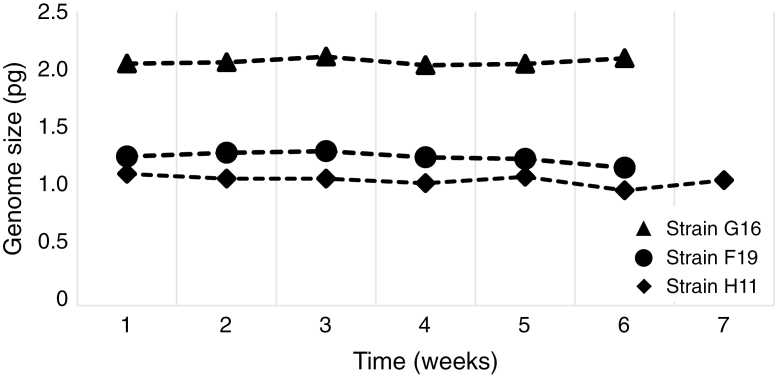
To corroborate the longer term stability of genome size differences among strains in cultivation, two selected strains were re-analysed 2 years following the first measurements: strain 961 (1.071 and 1.077 pg of DNA, respectively) and strain S63.B3 (1.516 and 1.492 pg of DNA, respectively). In both cases, the deviation between repeated estimates fell within the limits of instrumental precision (i.e. mean CV). Similarly, three other strains investigated for genome size stability via regular weekly measurements (F19, G16 and H11) did not show any substantial deviations from the original estimates (Fig. 4).
Fig. 4.
Temporal stability of the genome size of three selected Synura petersenii strains.
Phenotypic consequences and ecogeographical distribution of genome size diversity
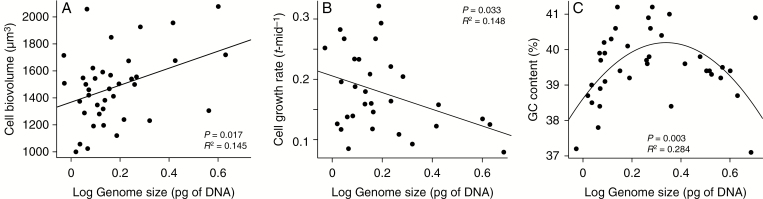
Genome size was significantly associated with both cell size (F1,37 = 6.25, P = 0.017, R2 = 0.145; Fig. 5A) and growth rate (F1,29 = 5.03, P = 0.033, R2 = 0.148; Fig. 5B); an increase in genome size led to an increase in cell size and a decrease in growth rate. However, the two observed relationships were not affected by a putative strong correlation between the cell size and growth rate (t23 = –1.32, P = 0.201, r = –0.265).
Fig. 5.
Three genomic and phenotypic traits associated with intraspecific genome size diversity in Synura petersenii: cell size (A), relative growth rate (B) and genomic GC content (C). Model predictions are depicted using lines in (A and B) and the curve of the quadratic function in (C).
Synura strains with smaller or larger genome size did not display any apparent spatial trends in their geographic distribution (Fig. 1). This was supported by the lack of a significant association between spatial distribution of genome size diversity and climatic variables or latitude in the RDA (P = 0.505; 999 permutations). The first, constrained RDA axis explained only 1.0 % of overall variation, whereas the second, unconstrained axis explained 41.6 % of variation (Supplementary data Fig. S1).
Diversity in genomic GC content
The other genomic parameter, GC content, varied from 37.1 to 41.2 %, with a mean value of 39.5 % (Supplementary data Table S2). The GC content had no significant effect on either cell size (F1,15 = 0.29, P = 0.598, R2 = 0.019) or growth rate (F1,11 = 1.96, P = 0.189, R2 = 0.151). No significant association between GC content diversity and climatic variables or latitude was detected in RDA (P = 0.890; 999 permutations). The first, constrained RDA axis explained only 0.8 % of the overall variation, whereas the second, unconstrained axis explained 42.2 % of the variation (Supplementary data Fig. S1). On the other hand, a significant quadratic relationship was detected between GC content and genome size (F2,35 = 6.95, P = 0.003, R2 = 0.284; Fig. 5C). The appropriateness of including the predictor in a quadratic form was corroborated using manual forward selection, as such a model significantly outperformed both the linear relationship (F1,36 = 0.11, P = 0.743, R2 = 0.003) and the null model without any predictors.
DISCUSSION
Genome size variability and its evolutionary sources
Currently, there is an apparent dearth of genome size estimates from protists, especially regarding the degree of intraspecific genome size variation. This might be due to the fact that the use of FCM, an efficient and widely applied technique of nuclear DNA content estimation (Doležel et al., 2007), is often methodologically challenging in protists as a result of difficulties in obtaining sufficient amounts of biomass, protoplast extraction or the presence of a wide variety of pigments and secondary metabolites interfering with fluorescent staining (Veldhuis et al., 1997; Kapraun, 2007; Mazalová et al., 2011; Poulíčková et al., 2014). Considering the above, we adopted an FCM protocol with several steps improving the robustness of our estimates that included, inferring each genome size estimate from mean value of three analyses on different days, simultaneous analysis of strains with different genome size to confirm the existing differences (multiple peaks in an FCM histogram) and re-analysing strains after a period of time to account for genome size stability under cultivation. In our study, we successfully estimated genome size of >130 strains belonging to a single microalgal species and, to our knowledge, this is the most comprehensive intraspecific genome size screening conducted on protists so far. We revealed considerable variability in genome size of Synura petersenii, ranging 2-fold across the analysed strains, from 0.97 to 2.02 pg of DNA. Our estimates (median value = 1.17 pg) are not consistent with an earlier estimate of S. petersenii genome size of 0.78 pg made by Olefeld et al. (2018). However, the published data belonged to the strain WA18K-A (with other designation CCMP 2892) that in the taxonomic revision of the S. petersenii species complex was assigned to a different species, S. heteropora (Škaloud et al., 2014). Therefore, we present the first genome size data for S. petersenii s.s.
There are several possible scenarios for what could be the source of genome size diversity observed among S. petersenii strains. First, owing to the robust FCM protocol, consistent methodology and generally high precision of our analyses, we are convinced that the error of measurement has not substantially contributed to the genome size variation. Taking into account the 2-fold difference between the lowest and highest genome size estimates, alternating life cycle stages or whole-genome doubling (polyploidization) events would seem to be likely explanations. However, none of these mechanisms can be the sole source of the diversity observed in Synura as there were no discrete genome size categories that would reflect the inherent ploidy shifts (Fig. 2). Another argument against the alternating life cycle stages is that strains re-analysed after weeks (or 2 years) exhibited more or less stable genome size estimates (Fig. 4). This contrasts with the genome size differentiation that emerged within a long-term cultivated strain of Thalassiosira weissflogii belonging to diatoms, a more intensively studied group of Stramenopiles, implying the ability to rapidly change the amount of DNA (von Dassow et al., 2008), possibly in the context of sexual reproduction or (theoretically) local adaptation. On the other hand, genome size appeared to be stable in cultivation of another diatom, Ditylum brightwellii, where intraspecific variation among strains was also previously detected (Koester et al., 2010). While we are unable to exclude the possibility that strains at both extreme ends of the observed genome size continuum are in fact different ploidy cytotypes or distinct stages of the S. petersenii life cycle, other evolutionary mechanisms operating with more gradual increases or decreases in genome size were most probably involved.
Alternative explanations may be provided by proliferation of TEs, unequal frequency of insertions to deletions and multiplication of larger genomic segments or even whole chromosomes (supernumerary chromosomes or aneuploidy; Jones et al., 2008; Šmarda and Bureš, 2010; Ruiz-Ruano et al., 2011; Stelzer et al., 2019). For example, genome size diversity in the diatom T. weissflogii was attributed to polyploidization, aneuploidization and gene duplications (von Dassow et al., 2008). A recent chromosome doubling was also detected in the diatom T. pseudonana (Armbrust, 2004). Unfortunately, despite a considerable effort, we did not succeed with karyotyping of S. petersenii strains and neither chromosome counts nor complete genomic sequences are available for any representative of the class Chrysophyceae (including the genus Synura). It is thus unclear whether prompt karyotype evolution or chromosomal aberrations could be responsible for the observed intraspecific genome size variation. Genome size changes caused by chromosomal aberrations or increased TE activity may be accompanied by significant alterations of genomic GC content (i.e. the relative proportion of GC base pairs; Wichman et al., 1993; Armbrust, 2004; Derelle et al., 2006). Interestingly, we found a significant relationship between the genome size of Synura strains and their genomic GC content, which had a quadratic nature and predicted highest GC content in medium-sized genomes (Fig. 5C). A similar quadratic relationship between the two variables was previously documented in monocot plants (Veselý et al., 2012; Šmarda et al., 2014), where it was explained by involvement of GC-rich long terminal repeat (LTR) retrotransposons in genome size expansion in combination with a mechanism responsible for decreasing GC content in large genomes (e.g. lower energetic cost of synthesis of dATPs and dTTPs leading to their misincorporation into the newly synthesized DNA as a mutational bias toward an AT-rich genome; Rocha and Danchin, 2002; Grover and Wendel, 2010). It is worth emphasizing that in contrast to a 207-fold genome size variation among monocot plants in the dataset analysed by Šmarda et al. (2014), we were able to detect the significant quadratic relationship with GC content on a very fine scale of 2-fold genome size variation. This might suggest that the observed GC content variation is a mere by-product of the mechanism governing genome size evolution in S. petersenii, which is in line with the absence of any biological or environmental correlates of GC content diversity in our dataset.
Intraspecific variability vs. cryptic diversity
As a general rule in multicellular organisms, individuals belonging to the same species share a constant nuclear DNA content (Swift, 1950). Nonetheless, intraspecific genome size variation manifested either via multiple ploidy cytotypes or at a homoploid level is occasionally observed among both plants and animals (Jeffery et al., 2016; Kolář et al., 2017; Stelzer et al., 2019). There is also some evidence of intraspecific genome size variation among microalgae coming from diatoms (von Dassow et al., 2008; Koester et al., 2010), desmids (Poulíčková et al., 2014) and haptophytes (Medlin et al., 1996; Veldhuis et al., 1997; Read et al., 2013). Nonetheless, the frequency of this phenomenon and its prevalence across various groups of protists is still poorly documented, and the evolutionary mechanisms involved are only exceptionally addressed. Theoretically, there are two mutually non-exclusive evolutionary scenarios that would result in intraspecific genome size variation. First, the mechanisms of genome size change might act recurrently with high enough frequency to compensate for only a transient character of induced changes (e.g. via aneuploidy or presence of supernumerary chromosomes). Secondly, intraspecific genome size variation could be maintained in populations when it is coupled with a reproductive barrier that prevents crosses between conspecific individuals with different genome sizes. The reproductive barrier in the latter scenario could either directly result from the mechanism inducing genome size differences (e.g. polyploidization) or arise independently (e.g. spatial or temporal isolation or a specific mate recognition systems).
A unique insight into mechanisms maintaining intraspecific genome size variation was recently documented in one species of rotifer (Stelzer et al., 2019). The variation, apparent already at the within-population level, was possibly due to independently segregating large genomic elements present in males. Regardless of the genome size difference, individuals were able to interbreed and produce viable offspring, stressing their identity to one species (Stelzer et al., 2019). However, under natural conditions, the species relies predominantly on asexual reproduction mediated by parthenogenetic females. Similar to rotifers, S. petersenii also reproduces mainly clonally (by a cell division), though sexual reproduction has been documented (Sandgren and Flanagin, 1986). Despite our great efforts, we were unable to experimentally interbreed S. petersenii strains. Neither crosses between contrasting genome size categories nor those performed between strains with similar-sized genomes were successful, possibly suggesting inadequate conditions for mating. It thus remains unclear whether the different genome size categories of S. petersenii strains are coupled with a reproductive barrier or not. Another similarity between our studied S. petersenii populations and the case study on rotifers is that many strains of different genome size categories co-occurred contemporarily at the same locality. We detected the common presence of two or more strains differing in their genome size (up to 1.8-fold difference) in 14 out of 67 localities (21 %). However, the actual rate may be even higher as our sampling strategy was primarily focused at between-locality comparisons and there seem to be no clear trends in geographical distribution of genome size categories. It is likely that the prevalence of clonal reproduction contributes to the maintenance of strains of different genome size categories and their coexistence in S. petersenii populations. Synura petersenii is a colonial species and it is generally unknown whether the colonies are composed of genetically identical cells or may combine multiple genotypes (strains). Since the cultures for this study were established from one colony of cells each and always had uniform genome size, we hypothesize that strains of different genome size coexist at a locality in well-separated colonies.
Our results cannot rule out the scenario that various genome size categories in S. petersenii are coupled with reproductive barriers and thus reflect cryptic diversity within the taxon. Were this the case, it would mean that the nuclear ITS rDNA region is not always a sufficient molecular marker to separate microalgal species, even though this marker (and rDNA in general) is widely used as a barcode for species identification in many algal studies (e.g. Helms et al., 2001; Connell, 2002; von Dassow et al., 2008; Whittaker et al., 2012; Jo et al., 2016). Genome size estimation using FCM might then serve as a useful tool for identifying potential cryptic diversity in protists. Such an approach has already been applied in some diatoms and harmful dinoflagellates (Figueroa et al., 2010; Koester et al., 2010).
Adaptive role of genome size variation
An important aspect of intraspecific genome size diversity and its evolutionary maintenance is its putative adaptive potential, i.e. whether strains with a certain genome size have a fitness (dis-)advantage in some environmental or evolutionary context. Among S. petersenii strains, an increase in genome size resulted in a significant increase in cell size and a significant decrease in relative growth rate. The genome size–cell size correlation has been previously documented in many other protists (LaJeunesse et al., 2005; Connolly et al., 2008; von Dassow et al., 2008; Poulíčková et al., 2014; Olefeld et al., 2018), though in our study the relationship was not as tight as presumed, explaining 14.5 % of the overall variation (Fig. 5A). There are two likely explanations for this discrepancy. First, in the other studies, the relationship was tested across different species, thus with a much broader range of both genome sizes and cell sizes, increasing the chance of finding a general trend. Secondly, the cells of Synura lack a cell wall and flexibly adjust their volume under varying temperature, nutrient composition, etc. (Němcová et al., 2010; Řezáčová-Škaloudová et al., 2010; Pichrtová and Němcová, 2011), which could have contributed to residual model variance.
The genome size was further associated with the relative growth rate of particular strains (14.8 % of overall variation explained), leading to up to a 3-fold difference in growth rates between strains from the lowest and highest genome size categories (Fig. 5B). As a result, strains with larger genomes could not grow and divide as quickly as their counterparts with a smaller genome size, a feature that should be reflected in their relative fitness at least under specific environmental conditions. In aquatic micro-organisms, rapid population growth is a key factor for successful colonization of a new site and effective monopolization of local resources (i.e. the monopolization hypothesis; De Meester et al., 2002). Once a population is well established and possibly also locally adapted, the existence of a large bank of resting propagules (in this case Synura cysts) provides a powerful buffer against newly invading genotypes. Under this scenario, S. petersenii strains with larger genomes should be inferior colonizers of new sites, possibly sometimes outcompeted at the existing localities by other strains with smaller genomes. This is in line with the frequency distribution of genome size categories across S. petersenii populations, which was strongly skewed towards smaller genomes (Fig. 2). The strains with larger genomes could then be maintained in populations either due to their recurrent in situ origin from smaller genome progenitors or because of other compensatory adaptive traits that were not included in our study, possibly stemming from their larger cell size, e.g. more efficient nutrient uptake and/or photosynthesis (Finkel et al., 2001; Irwin et al., 2006).
The question is whether the identified phenotypic consequences of genome size variation could also have translated into contrasting ecogeographical distributions of Synura strains.
Based on our results, this does not seem to be the case. It was already suggested by the lack of any clear geographical trends in distribution of strains from particular genome size categories (Fig. 1) and was further strengthened by the occasional co-occurrence of strains with different genome size at the sampled localities. The RDA on a dataset consisting of 19 database-derived climatic variables and geographical latitude characterizing the Synura sampling sites provided a more comprehensive assessment (Supplementary data Fig. S1). The non-significant effect of genome size in the RDA indicated that the current spatial distribution of different genome size categories in Synura is not a result of large-scale environmental filtering. To our knowledge, our study was the first attempt to evaluate intraspecific genome size variation in protists in an ecogeographical context.
Conclusions
Genome size variation and its evolutionary consequences are highly understudied among protists, particularly on the intraspecific level, with nearly no data available. We present the most comprehensive intraspecific genome size screening conducted to date, revealing a gradient of continuous genome size variation among S. petersenii strains. Even though we were unable to identify the main evolutionary mechanism responsible for genome size variation in this species, it probably operates via gradual changes in genome size which are accompanied by changes in genomic GC content. We hypothesize that proliferation of TEs and multiplication of larger genomic segments or even whole chromosomes are the most likely scenarios. Interestingly, the genome size variability was reflected in cell size and relative growth rate but not in distinct ecogeographical distribution of strains. Occasionally, we even detected strains with different genome size coexisting at the same locality. Whether these strains are associated with reproductive barriers (suggesting cryptic diversity within S. petersenii) remained unresolved, though prevailing clonal reproduction of the species could substantially contribute to the maintenance of local genome size diversity even in their absence.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: collection details for Synura petersenii strains used in this study. Table S2: genomic and physiological parameters recorded on investigated strains of Synura petersenii. Figure S1: redundancy analyses testing associations between genomic parameters and climatic conditions across the collection sites.
ACKNOWLEDGEMENTS
We would like to thank to E. Gusev (Papanin’s Institute for Biology of Inland Waters, Russian Academy of Sciences) for providing us with cultures.
FUNDING
This work was supported by the Charles University [GAUK, project no. 1304317]. Part of the work was carried out with the support of RECETOX Research Infrastructure [ID LM2015051, MEYS CR, 2016–2019].
LITERATURE CITED
- Armbrust EV, Berges JA, Bowler C, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- Baack EJ, Whitney KD, Rieseberg LH. 2005. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytologist 167: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Shimode S, Kim H, Han M-S, Kikuchi T. 2009. Strong bottom-up effects on phytoplankton community caused by a rainfall during spring and summer in Sagami Bay, Japan. Journal of Marine Systems 75: 253–264. [Google Scholar]
- Baetcke KP, Sparrow AH, Nauman CH, Schwemmer SS. 1967. The relationship of DNA content to nuclear and chromosome volumes and to radiosensitivity (LD50). Proceedings of the National Academy of Sciences, USA 58: 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist 179: 975–986. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences 181: 109–135. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. 2011. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Annals of Botany 107: 467–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boo SM, Kim HS, Shin W, et al. 2010. Complex phylogeographic patterns in the freshwater alga Synura provide new insights into ubiquity vs. endemism in microbial eukaryotes. Molecular Ecology 19: 4328–4338. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany 95: 147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Beaton MJ. 1999. The skeletal function of non-genic nuclear DNA: new evidence from ancient cell chimaeras. Genetica 106: 3–13. [DOI] [PubMed] [Google Scholar]
- Connell L. 2002. Rapid identification of marine algae (Raphidophyceae) using three-primer PCR amplification of nuclear internal transcribed spacer (ITS) regions from fresh and archived material. Phycologia 41: 15–21. [Google Scholar]
- Connolly JA, Oliver MJ, Beaulieu JM, Knight CA, Tomanek L, Moline MA. 2008. Correlated evolution of genome size and cell volume in diatoms (Bacillariophyceae). Journal of Phycology 44: 124–131. [DOI] [PubMed] [Google Scholar]
- von Dassow P, Petersen TW, Chepurnov VA, Armbrust EV. 2008. Inter- and intraspecific relationships between nuclear DNA content and cell size in selected members of the centric diatom genus Thalassiosira (Bacillariophyceae). Journal of Phycology 44: 335–349. [DOI] [PubMed] [Google Scholar]
- De Meester L, Gómez A, Okamura B, Schwenk K. 2002. The monopolization hypothesis and the dispersal–gene flow paradox in aquatic organisms. Acta Oecologica 23: 121–135. [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proceedings of the National Academy of Sciences, USA 103: 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio Pires LM, Jonker RR, Van Donk E, Laanbroek HJ. 2004. Selective grazing by adults and larvae of the zebra mussel (Dreissena polymorpha): application of flow cytometry to natural seston. Freshwater Biology 49: 116–126. [Google Scholar]
- Doležel J. 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany 95: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. 2007. Flow cytometry with plant cells. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA. [Google Scholar]
- Fick SE, Hijmans RJ. 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302–4315. [Google Scholar]
- Figueroa RI, Garcés E, Bravo I. 2010. The use of flow cytometry for species identification and life-cycle studies in dinoflagellates. Deep-Sea Research Part II: Topical Studies in Oceanography 57: 301–307. [Google Scholar]
- Finkel ZV, Platt T, Sathyendranath S, et al. 2001. Light absorption and size scaling of light-limited metabolism in marine diatoms. Limnology and Oceanography 46: 86–94. [Google Scholar]
- Foissner W. 2007. Protist diversity and distribution: some basic considerations. Dordrecht: Springer, 1–8. [Google Scholar]
- Galbraith DW. 2012. Flow cytometry and fluorescence-activated cell sorting in plants: the past, present, and future. Biomédica 30: 65. [Google Scholar]
- Garcia-Pichel F. 1994. A model for internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreens. Limnology and Oceanography 39: 1704–1717. [Google Scholar]
- Goodsell DS, Dickerson RE. 1994. Bending and curvature calculations in B-DNA. Nucleic Acids Research 22: 5497–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2001a Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews of the Cambridge Philosophical Society 76: 65–101. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2001b The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells, Molecules & Diseases 27: 830–843. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2005. Genome size evolution in animals. In: Gregory TR, ed. The evolution of the genome. Elsevier Academic Press, 3–87. [Google Scholar]
- Greilhuber J. 2005. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany 95: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover CE, Wendel JF. 2010. Recent insights into mechanisms of genome size change in plants. Journal of Botany 2010: 1–8. [Google Scholar]
- Guillard RRL, Lorenzen CJ. 1972. Yellow-green algae with chlorophyllide C. Journal of Phycology 8: 10–14. [Google Scholar]
- Helms G, Friedl T, Rambold G, Mayrhofer H. 2001. Identification of photobionts from the lichen family Physciaceae using algal-specific ITS rDNA sequencing. The Lichenologist 33: 73–86. [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A. 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genetics 6: e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin AJ, Finkel ZV, Schofield OME, Falkowski PG. 2006. Scaling-up from nutrient physiology to the size-structure of phytoplankton communities. Journal of Plankton Research 28: 459–471. [Google Scholar]
- Jeffery NW, Hultgren K, Chak STC, Gregory TR, Rubenstein DR. 2016. Patterns of genome size variation in snapping shrimp. Genome 59: 393–402. [DOI] [PubMed] [Google Scholar]
- Jo BY, Kim JI, Škaloud P, Siver PA, Shin W. 2016. Multigene phylogeny of Synura (Synurophyceae) and descriptions of four new species based on morphological and DNA evidence. European Journal of Phycology 51: 413–430. [Google Scholar]
- Jones RN, Viegas W, Houben A. 2008. A century of B chromosomes in plants: so what? Annals of Botany 101: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF. 2007. Nuclear DNA content estimates in green algal lineages: chlorophyta and streptophyta. Annals of Botany 99: 677–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Slamovits CH. 2005. Causes and effects of nuclear genome reduction. Current Opinion in Genetics & Development 15: 601–608. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63. [DOI] [PubMed] [Google Scholar]
- Kim JI, Shin H, Škaloud P, et al. 2019. Comparative plastid genomics of Synurophyceae: inverted repeat dynamics and gene content variation. BMC Evolutionary Biology 19: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester JA, Swalwell JE, von Dassow P, Armbrust EV. 2010. Genome size differentiates co-occurring populations of the planktonic diatom Ditylum brightwellii (Bacillariophyta). BMC Evolutionary Biology 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolář F, Čertner M, Suda J, Schönswetter P, Husband BC. 2017. Mixed-ploidy species: progress and opportunities in polyploid research. Trends in Plant Science 22: 1041–1055. [DOI] [PubMed] [Google Scholar]
- Kristiansen J, Preisig HR. 2007. Chrysophyte and haptophyte algae. In: Büdel B, Gärtner G, Krienitz L, Preisig HR, Schagerl M, eds. Süβwasserflora von Mitteleuropa. Freswater Flora of Central Europe. Berlin: Springer, 118. [Google Scholar]
- Kron P, Suda J, Husband BC. 2007. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology, Evolution, and Systematics 38: 847–876. [Google Scholar]
- Kynčlová A, Škaloud P, Škaloudová M. 2010. Unveiling hidden diversity in the Synura petersenii species complex (Synurophyceae, Heterokontophyta). Nova Hedwigia 136: 283–298. [Google Scholar]
- LaJeunesse TC, Lambert G, Andersen RA, Coffroth MA, Galbraith DW. 2005. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. Journal of Phycology 41: 880–886. [Google Scholar]
- Leitch I, Chase MW, Bennett MD. 1998. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany 82: 85–94. [Google Scholar]
- Lepš J, Šmilauer P. 2014. Multivariate analysis of ecological data using Canoco 5, 2nd edn Cambridge: Cambridge University Press. [Google Scholar]
- Liedtke HC, Gower DJ, Wilkinson M, Gomez-Mestre I. 2018. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nature Ecology & Evolution 2: 1792–1799. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302: 1401–1404. [DOI] [PubMed] [Google Scholar]
- Mazalová P, Šarhanová P, Ondřej V, Poulíčková A. 2011. Quantification of DNA content in freshwater microalgae using flow cytometry: a modified protocol for selected green microalgae. Fottea 11: 317–328. [Google Scholar]
- Medlin LK, Barker GLA, Campbell L, et al. 1996. Genetic characterisation of Emiliania huxleyi (Haptophyta). Journal of Marine Systems 9: 13–31. [Google Scholar]
- Mirsky AE, Ris H. 1951. The composition and structure of isolated chromosomes. Journal of General Physiology 34: 475–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugal CF, Arndt PF, Holm L, Ellegren H. 2015. Evolutionary consequences of DNA methylation on the GC content in vertebrate genomes. G3 5: 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Němcová Y, Neustupa J, Kviderová J, Řezáčová-Škaloudová M. 2010. Morphological plasticity of silica scales of Synura echinulata (Synurophyceae) in crossed gradients of light and temperature – a geometric morphometric approach. Nova Hedwigia 136: 21–32. [Google Scholar]
- Olefeld JL, Majda S, Albach DC, Marks S, Boenigk J. 2018. Genome size of chrysophytes varies with cell size and nutritional mode. Organisms Diversity & Evolution 18: 163–173. [Google Scholar]
- Otto F. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods in Cell Biology 33: 105–110. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Leitch IJ. 2014. The application of flow cytometry for estimating genome size and ploidy level in plants. Methods in Molecular Biology 1115: 279–307. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Leitch IJ. 2020. The plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226: 301–305. [DOI] [PubMed] [Google Scholar]
- Pichrtová M, Němcová Y. 2011. Effect of temperature on size and shape of silica scales in Synura petersenii and Mallomonas tonsurata (Stramenopiles). Hydrobiologia 673: 1–11. [Google Scholar]
- Poulíčková A, Mazalová P, Vašut RJ, Šarhanová P, Neustupa J, Škaloud P. 2014. DNA content variation and its significance in the evolution of the genus Micrasterias (desmidiales, streptophyta). PLoS One 9 e86247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Read BA, Kegel J, Klute MJ, et al. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499: 209–213. [DOI] [PubMed] [Google Scholar]
- Řezáčová-Škaloudová M, Neustupa J, Němcová Y. 2010. Effect of temperature on the variability of silicate structures in Mallomonas kalinae and Synura curtispina (Synurophyceae). Nova Hedwigia 136: 55–69. [Google Scholar]
- Ribeiro S, Berge T, Lundholm N, Ellegaard M. 2013. Hundred years of environmental change and phytoplankton ecophysiological variability archived in coastal sediments. PLoS One 8: e61184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. 2002. Base composition bias might result from competition for metabolic resources. Trends in Genetics 18: 291–294. [DOI] [PubMed] [Google Scholar]
- Roháček K, Barták M. 1999. Technique of the modulated chlorophyll fluorescence: basic concepts, useful parameters, and some applications. Photosynthetica 37: 339–363. [Google Scholar]
- Roux N, Toloza A, Radecki Z, Zapata-Arias FJ, Dolezel J. 2003. Rapid detection of aneuploidy in Musa using flow cytometry. Plant Cell Reports 21: 483–490. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ruano FJ, Ruiz-Estévez M, Rodríguez-Pérez J, López-Pino JL, Cabrero J, Camacho JP. 2011. DNA amount of X and B chromosomes in the grasshoppers Eyprepocnemis plorans and Locusta migratoria. Cytogenetic and Genome Research 134: 120–126. [DOI] [PubMed] [Google Scholar]
- Sandgren CD, Flanagin J. 1986. Heterothallic sexuality and density dependent encystment in the Chrysophycean alga Synura petersenii Korsh. Journal of Phycology 22: 206–216. [Google Scholar]
- Shuter BJ, Thomas JE, Taylor WD, Zimmerman AM. 1983. Phenotypic correlates of genomic DNA content in unicellular eukaryotes and other cells. The American Naturalist 122: 26–44. [Google Scholar]
- Škaloud P, Kynčlová A, Benada O, Kofroňová O, Škaloudová M. 2012. Toward a revision of the genus Synura, section Petersenianae (Synurophyceae, Heterokontophyta): morphological characterization of six pseudo-cryptic species. Phycologia 51: 303–329. [Google Scholar]
- Škaloud P, Škaloudová M, Procházková A, Němcová Y. 2014. Morphological delineation and distribution patterns of four newly described species within the Synura petersenii species complex (Chrysophyceae, Stramenopiles). European Journal of Phycology 49: 213–229. [Google Scholar]
- Šmarda P, Bureš P. 2010. Understanding intraspecific variation in genome size in plants. Preslia 82: 41–61. [Google Scholar]
- Šmarda P, Bureš P. 2012. The variation of base composition in plant genomes. In: Wendel J, Greilhuber J, Dolezel J, Leitch I, eds. Plant genome diversity, Vol. 1. Vienna: Springer Vienna, 209–235. [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. 2008. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany 101: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, et al. 2014. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proceedings of the National Academy of Sciences, USA 111: E4096–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetacek V, Assmy P, Henjes J. 2004. The role of grazing in structuring Southern Ocean pelagic ecosystems and biogeochemical cycles. Antarctic Science 16: 541–558. [Google Scholar]
- Soltis DE, Soltis PS. 1999. Polyploidy: recurrent formation and genome evolution. Trends in Ecology & Evolution 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Sparrow AH, Evans HJ. 1961. Nuclear factors affecting radiosensitivity. I. The influence of nuclear size and structure, chromosome complement, and DNA content. Brookhaven Symposia in Biology 14: 76–100. [PubMed] [Google Scholar]
- Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer CP, Pichler M, Stadler P, Hatheuer A, Riss S. 2019. Within-population genome size variation is mediated by multiple genomic elements that segregate independently during meiosis. Genome Biology and Evolution 11: 3424–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, López Arriaza JR, Mueller RL. 2012. Slow DNA loss in the gigantic genomes of salamanders. Genome Biology and Evolution 4: 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nishibayashi S, Kuroiwa T, Kanbe T, Tanaka K. 1982. Variance of ploidy in Candida albicans. Journal of Bacteriology 152: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift H. 1950. The constancy of desoxyribose nucleic acid in plant nuclei. Proceedings of the National Academy of Sciences, USA 36: 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temsch EM, Greilhuber J, Krisai R. 2010. Genome size in liverworts. Preslia 82: 63–80. [Google Scholar]
- Thomas CA., Jr 1971. The genetic organization of chromosomes. Annual Review of Genetics 5: 237–256. [DOI] [PubMed] [Google Scholar]
- Trávníček P, Čertner M, Ponert J, Chumová Z, Jersáková J, Suda J. 2019. Diversity in genome size and GC content shows adaptive potential in orchids and is closely linked to partial endoreplication, plant life-history traits and climatic conditions. New Phytologist 224: 1642–1656. [DOI] [PubMed] [Google Scholar]
- Van’t Hof J, Sparrow AH. 1963. A relationship between DNA content, nuclear volume, and minimum mitotic cycle time. Proceedings of the National Academy of Sciences, USA 49: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis MJW, Cucci TL, Sieracki ME. 1997. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. Journal of Phycology 33: 527–541. [Google Scholar]
- Veleba A, Šmarda P, Zedek F, Horová L, Šmerda J, Bureš P. 2017. Evolution of genome size and genomic GC content in carnivorous holokinetics (Droseraceae). Annals of Botany 119: 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S. New York: Springer. [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T. 2012. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany 109: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee JL, Fasone LD, Sattler A, Starks WW, Hurley DL. 2001. ITS/5.8S DNA sequence variation in 15 isolates of Synura petersenii Korshikov (Synurophyceae). Nova Hedwigia 122: 245–258. [Google Scholar]
- White TJ. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, ed. PCR protocols: a guide to methods and applications. San Diego: Academic Press, 315–322. [Google Scholar]
- Whittaker KA, Rignanese DR, Olson RJ, Rynearson TA. 2012. Molecular subdivision of the marine diatom Thalassiosira rotula in relation to geographic distribution, genome size, and physiology. BMC Evolutionary Biology 12: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, Van Den Bussche RA, Hamilton MJ, Baker RJ. 1993. Transposable elements and the evolution of genome organization in mammals. Dordrecht: Springer, 149–157. [DOI] [PubMed] [Google Scholar]
- Wilson EB. 1925. The karyoplasmic ratio. In: The cell in development and heredity. New York: The Macmillan Company, 727–733. [Google Scholar]
- Wu Y, Sun Y, Sun S, et al. 2018. Aneuploidization under segmental allotetraploidy in rice and its phenotypic manifestation. Theoretical and Applied Genetics 131: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubáčová Z, Cimbůrek Z, Tachezy J. 2008. Comparative analysis of trichomonad genome sizes and karyotypes. Molecular and Biochemical Parasitology 161: 49–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.