Abstract
To investigate the U2AF1 gene mutation site, mutation load and co-mutations genes in patients with myelodysplastic syndrome (MDS) and their effects on prognosis. Gene mutation detection by next-generation sequence and related clinical data of 234 MDS patients were retrospectively collected and analyzed for the relationship between the clinical characteristics, treatment efficacy and prognosis of U2AF1 gene mutation. Among the 234 MDS patients, the U2AF1 gene mutation rate was 21.7% (51 cases), and the median variant allele frequency was 39.5%. Compared with the wild type, the U2AF1 mutant had a higher incidence of chromosome 8 aberration, and was positively correlated with the occurrence of ASXL1, RUNX1, SETBP1 gene mutation, negatively correlated with SF3B1, NPM1 genes mutation (p < 0.05). The most common mutation site of U2AF1 was S34F (32 cases), while U2AF1 Q157P site mutations had a higher incidence of chromosome 7 abnormalities (p = 0.003). The U2AF1 gene mutation more frequently coincided with signal pathway related gene mutations (p = 0.043) with a trend of shortened overall survival. Among patients with U2AF1 gene mutations, those with ASXL1 mutations were prone to develop into acute myeloid leukemia, those with RUNX1 mutations had an increased risk of relapse, and those with TET2 mutations had higher 1-year survival rate. Compared with the patient group of lower mutation load (VAF ≤ 40%), the group with higher mutation load of U2AF1 (VAF > 40%) had a significantly lower 1-year survival rate (46.1% and 80.5%, p = 0.027). The criteria of U2AF1 VAF > 40% is an independent indicator for poor prognosis of MDS patients. VAF > 40% of U2AF1 is an independent factor of short OS in MDS patients. MDS patients with a mutation in the Q157P site of U2AF1 and a higher U2AF1 mutation load suggests poor prognosis, and co-mutated genes in U2AF1 can affect disease progression and prognosis.
Subject terms: Medical research, Biomarkers, Prognostic markers
Introduction
Myelodysplastic syndrome (MDS) is a group of diseases with myeloid ineffective hematopoiesis, morbid hematopoiesis and high-risk transformation into acute myeloid leukemia (AML). It is characterized by malignant clonal hematopoietic stem/progenitor cell disease. About 70–90% MDS patients has one or more gene mutations1–3. A number of studies have confirmed that gene mutations play important role in the occurrence, development, treatment and prognosis in MDS patients2–6. New molecular techniques, however, such as next-generation sequencing (NGS) identifying important genetic alterations, have paved the path for new drug development targeting those specific gene mutations, and an array of new gene mutation-targeted agents are now available for MDS and AML treatment7. As a RNA shearing gene mutation, U2AF1 is an early driving gene of MDS8. Currently, the effect of U2AF1 on the MDS prognosis is not clear. Some studies show that U2AF1 mutation has no effect on prognosis9–11, other studies show that U2AF1 gene mutation with poor prognosis12–15. Studies show that U2AF1 mutation sites may affect the prognosis. Q157 site mutation has worse prognosis than S34 mutation14,16,17. Gradual accumulation of mutant genes can promote the occurrence and progress of MDS. Therefore, the prognosis of MDS may be related to the accumulation of different co-mutation gene combinations18. In this study, gene mutation detection by NGS and related clinical data from 234 MDS patients had been analyzed to explore U2AF1 mutation sites, mutation load and co-mutation genes of MDS and its clinical significance.
Result
Clinical characteristics
Among the 234 MDS patients, 142 were male (60.6%) and 92 were female (39.4%) with the median age of 55 (17–86) years old. The median number of bone marrow blasts was 6.0 (0–18.8) % at the diagnosis. The median peripheral white blood cell count (WBC) was 2.5 (0.02–20.3) × 109/L; the median neutrophil (n) count was 0.95 (0–17.7) × 109/L; the median hemoglobin concentration (Hgb) was 71 (24–145) g/L. The median platelet count (PLT) was 49 (3–745) × 109/L. There were 32 (13.6%) patients relapsed, 36 (15.3%) developed to AML. The overall response rate (ORR) of 212 patients was 30.7% (65), and 31.6% (48) among the 134 patients with Decitabine (DEC) treatment.
Mutant gene detection
Among the 234 patients, there were 347 mutated genes, 72.2% (169/234 patients) had at least one mutated gene, with a median mutants of 2 (1–6), while 38.0% (89/234 patients) had two or more mutated genes. 21.7% (51 patients) had U2AF1 mutation with total of 52 mutation sites detected. Among those with U2AF1 mutation, 84.3% (43 patients) had exon 2 mutation, 62.7% (32 patients) had U2AF1 mutated protein sequence showing the 34th serine was substituted by phenylalanine (S34F rs371769427 , hereinafter referred as S34F), and 21.6% (11 cases) had the 34th serine was substituted by tyrosine (S34Y). 13.7% (7 cases) had the 157th glutamate was substituted by proline (Q157P). In one case, both S34F and Q157P mutations were detected at the same time. The median allele frequency (mVAF) of 51 mutation sites was 39.5% (1.38% ~ 49.02%), all of which were heterozygous mutations. Among 51 patients with U2AF1 mutation, 39 cases (76.5%) had co-mutated genes. The common co-mutated genes were ASXL1 (33.3%, 17 cases), TET2 (31.4%, 16 cases), RUNX1 (21.6%, 11 cases) and SETBP1 (11.8%, 6 cases).
Clinical characteristics and prognosis of U2AF1 mutation
Compared with the non-mutated U2AF1 group, the frequency of co-mutation between U2AF1 gene and signal pathway associated genes of was higher in the U2AF1 mutated patient group (22.9% and 10.6%, p = 0.026). The incidence of abnormal chromosome 8 (22.4% and 10.3%, p = 0.063) and chromosome 20 (14.6% and 7.4%, p = 0.161) were higher in U2AF1 mutation group, but the difference was not statistically significant. U2AF1 gene mutation was positively correlated with ASXL1, RUNX1 and SETBP1 gene mutations, but was negatively correlated with SF3B1 gene mutation (p < 0.05). However, there were no significant differences in recurrence rate, risk of AML transformation, treatment response to DEC and median OS (mOS: 628 days and 790 days, p = 0.765) between the U2AF1 mutation and U2AF1 non-mutated groups;
Clinical characteristics and prognostic significance of different U2AF1 mutation sites
In MDS patients with U2AF1 mutation, due to the cases of MDS-RS (0 case), MDS (5q -) (2 cases) and MDS-U (1 case) were so few, so only 48 cases of MDS-SLD/MLD/EB1/EB2 with U2AF1 mutation were included to analyze the mutation site, mutation load and co mutation gene of U2AF1.
Compared to S34F and S34Y mutations, MDS patients with U2AF1 Q157P mutation site had a significant high incidence of chromosome 7 abnormalities (p = 0.01), older age, relatively high risk groups with IPSS-R risk stratification, lower hemoglobin concentration, higher incidence of complex karyotypes. OS was shorter (mOS of S34F, S34Y and Q15P: 628, 804 and 284 days, respectively, p = 0.391). However, the difference was not statistically significant (p > 0.05) compared to S34F and S34Y mutations. The co-mutation rates of S34F, S34Y and Q157P amino acid mutation sites were 71.9%, 66.7% and 100%, respectively. The co-mutation frequency of two co-mutation genes for S34Y was higher than that of S34F and S34Y mutation sites. There was no significant difference for the type and proportion of co-mutation genes between S34F and S34Y loci (Table 1).
Table 1.
U2AF1 mutation: protein sequence and clinical characteristics.
| Total | U2AF1mutation protein sequence [case (%)] | Total X2/Z | Total p value | ||
|---|---|---|---|---|---|
| S24F | S34Y | Q157P | |||
| 32 | 9 | 6 | |||
| Median VAF | 0.4127(0.0613~0.4902) | 0.397(0.0138~0.4808) | 0.37215(0.0401~0.4196) | 0.535 | |
| Age (year) | 55(21~77) | 53(33~69) | 67.5(43~83) | 0.088 | |
| Gender (male) | 23(71.9) | 8(88.9) | 2(33.3) | 4.854 | 0.091 |
| MDS subtype 1 | 1.154 | 0.655 | |||
| MDS-SLD and -MLD | 12(37.5) | 5(55.6) | 2(33.3) | ||
| MDS-EB1 + EB2 | 20(62.5) | 4(44.4) | 4(66.7) | ||
| MDS subtype 2 | 9.350 | 0.097 | |||
| MDS-SLD | 3(9.4) | 1(11.1) | 2(33.3) | ||
| MDS-MLD | 9(28.1) | 4(44.4) | 0(0) | ||
| MDS-EB1 | 7(21.9) | 0(0) | 3(50) | ||
| MDS-EB2 | 13(40.6) | 4(44.4) | 1(16.7) | ||
| IPSS-R group | 1.291 | 0.633 | |||
| Relatively low risk group | 7(21.9) | 2(22.2) | 0(0) | ||
| Relatively high risk group | 25(78.1) | 7(77.8) | 6(100) | ||
| Laboratory Results | |||||
| BM blast at diagnosis (%) | 6.2(0.4~18.4) | 4.8(0.4~14) | 1.25(1~2.5) | 0.752 | |
| N (109/L) | 0.77(0.06~8.63) | 0.6(0.03~8.1) | 1.16(0.32~6.5) | 0.603 | |
| PLT (109/L) | 52(7~459) | 38(29~394) | 50(14~118) | 0.86 | |
| HGB (g/L) | 73.5(28~131) | 102(45~145) | 61.5(49~87) | 0.245 | |
| Chromosomal abnormality | 13(40.6) | 6(66.7) | 4(66.7) | 2.706 | 0.323 |
| Chromosome 7 abnormality | 2(6.3) | 0(0) | 4(66.7) | 11.659 | 0.002 |
| Chromosome 8 abnormality | 6(18.8) | 4(44.4) | 0(0) | 3.922 | 0.102 |
| Chromosome 20 abnormality | 5(15.6) | 2(22.2) | 0(0) | 1.172 | 0.574 |
| Complex karyotype (≥ 3) | 1(3.1) | 0(0) | 1(16.7) | 2.577 | 0.275 |
| DEC treatment response | 0.913 | 0.738 | |||
| Non-ORR | 13(65) | 3(60) | 5(83.3) | ||
| ORR | 7(35) | 2(40) | 1(16.7) | ||
| Total | |||||
| Relapse | 4(12.5) | 0(0) | 0(0) | 1.028 | 0.750 |
| Transformed to AL | 4(12.5) | 1(11.1) | 0(0) | 0.538 | 1.000 |
| Median OS (day) | |||||
| Co-mutation gene | |||||
| TET2 mutation | 8(25) | 3(33.3) | 4(66.7) | 3.849 | 0.134 |
| ASXL1 mutation | 13(40.6) | 3(33.3) | 1(16.7) | 1.153 | 0.660 |
| RUNX1 mutation | 8(25) | 1(11.1) | 1(16.7) | 0.713 | 0.860 |
| TP53 mutation | 1(3.1) | 0(0) | 1(16.7) | 2.577 | 0.275 |
| Epigenetic gene (+) | 18(56.3) | 4(44.4) | 4(66.7) | 0.806 | 0.744 |
| Signal pathway gene (+) | 6(18.8) | 1(11.1) | 4(66.7) | 6.074 | 0.040 |
| Other pathway gene (+) | 10(31.3) | 2(22.2) | 3(50) | 1.34 | 0.450 |
| Number of co-mutation genes | 8.396 | 0.160 | |||
| 0 | 9(28.1) | 3(33.3) | 0(0) | ||
| 1 | 11(34.4) | 2(22.2) | 1(16.7) | ||
| 2 | 2(6.3) | 2(22.2) | 3(50) | ||
| ≧ 3 | 10(31.3) | 2(22.2) | 2(33.3) | ||
Clinical characteristics and prognosis of co-mutation of U2AF1, ASXL1, TET2 and RUNX1
-
A.
Compared to U2AF1 + ASXL1-, patients with U2AF1 + ASXL1 + had a higher risk of progress to AML (29.4% and 0.0%, p = 0.04). There was a increasing recurrent rate (17.6% and 2.9%, p = 0.102). Between these two groups, there was no significant difference in IPSS-R grouping, age, gender and the response to DEC treatment and prognosis.
-
B.
Compared to U2AF1 + TET2- patients, the mutation load of U2AF1 was lower in 16 cases of U2AF1 + TET2 + (mVAF: 41.3% and 35.4%, p = 0.034), the one year OS rate had trend of increase (one year OS: 60.3% and 66.7%, p = 0.109), and the ORR rate was higher in the group of DEC treatment (23.8% and 45.4%, p = 0.252) with no statistically significance. Among the 9 cases with mutation of U2AF1 S34F site mutation with TET2 + , 8 patients survived, one case lost to follow-up, suggesting that U2AF1 patients with S34F co-mutation with TET2 may have a better prognosis.
-
C.
Compared to U2AF1 + RUNX1-, the recurrence rate is high (30.0% and 2.6%, p = 0.025). However, there were no significant differences in MDS subtype distribution, IPSS-R group, age, gender, response to DEC treatment, disease progression and prognosis between the two groups.
Clinical characteristics and prognosis of mutation load of U2AF1 gene.
According to mVAF (39.5%) of U2AF1 mutation, the patients were divided into two groups: the group with relatively low mutation load (VAF ≤ 40%) and the group with relatively high mutation load (VAF > 40%). The incidence of chromosome 7 abnormalities was lower in the group with higher U2AF1 mutation load (0% and 23.1%, p = 0.025), and the 1-year survival rate was significantly lower (46.1% and 80.5%, p = 0.027) (Fig. 1). There was no significant difference in age, gender, response to DEC treatment, disease progress, and recurrence between the two groups.
Figure 1.
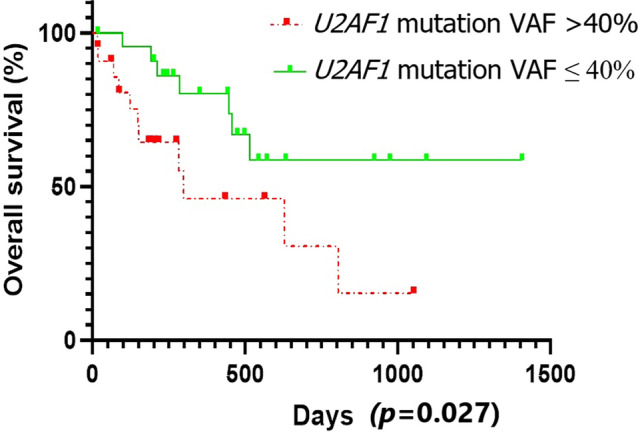
Compared with the group with relatively low mutation load (VAF ≤ 40%), the group with relatively high mutation load (VAF > 40%) has a significantly lower OS (46.1% and 80.5%, p = 0.027).
Gene mutation and prognosis
Among the 234 MDS patients, the median OS was 286 days (2–1406 days), with 90 (38.5%) patients died and 45 (19.2%) patients lost of follow-up. Univariate analysis results showed different factors were all correlated with a shorter OS: AML transformation (mOS: 447 days and 802 days, p = 0.028), chromosome 7 abnormality (-7, del(7q)) (mOS: 400 days and 830 days, p < 0.001), chromosome 20 abnormality (del(20q)) (mOS: 412 days and 802 days, p = 0.047), EZH2 mutation (mOS: 256 days and 791 days, p = 0.005), TP53 mutation (mOS: 497 days and 7 97 days, p = 0.023), CBL mutation (mOS: 158 days and 791 days, p = 0.038), RUNX1 VAF > 40% (mOS: 432 days and 909 days, p = 0.034), U2AF1 VAF > 40% (one-year OS rate: 46.1% and 80.5%, p = 0.027). Meanwhile, the age, proportion of bone marrow blasts, IPSS-R stratification and MDS subtype were included in the multivariate COX analysis. The results showed that VAF of U2AF1 > 40% [HR = 3.261, 95% CI (1.654–6.430), p = 0.001], chromosome 7 abnormality [HR = 3.885, 95% CI (1.527–5.148), p < 0.001], EZH2 mutation [HR = 8.523, 95% CI (2.558–28.395), p < 0.001], TP53 mutation [HR = 4.532, 95% CI (1.885–10.895), p = 0.001], CBL mutation [HR = 4.286, 95% CI (1.294–14.193), p = 0.017] were independent adverse factors of OS (Table 2).
Table 2.
MDS patients multivariate analysis.
| Factors | HR | 95%CI | p value |
|---|---|---|---|
| Chromosome 7 abnormality | 3.885 | 2.117~7.131 | < 0.001 |
| EZH2 mutation | 8.523 | 2.558~28.395 | < 0.001 |
| TP53 mutation | 4.532 | 1.885~10.895 | 0.001 |
| CBL mutation | 4.286 | 1.294~14.193 | 0.017 |
| U2AF1 VAF > 40% | 3.261 | 1.654~6.430 | 0.001 |
Discussion
U2AF1 gene is an RNA shearing related gene, which is the early mutation gene of MDS8. The mutation rate of U2AF1 gene in MDS patients is about 5% to 20%9,14,16,19. Studies have shown that in MDS patients, the mutation rate of U2AF1 in Asians is higher than that in Caucasians, and there is a significant correlation between U2AF1 mutation and chromosome 8 abnormality (trisomy 8) in Asians9. In this study, the mutation rate of U2AF1 in 234 MDS patients was 21.7%, higher than that in previous studies9,17. Compared with those without U2AF1 mutation, the incidence of abnormal chromosome 8 and chromosome 20 in U2AF1 mutant increased, but the difference was not significant; Among the 51 patients with U2AF1 mutation, the mutation sites were all with S34 and Q157, and the mutation rate of S34F had the highest rate, similar to that in previous studies9,17. In other studies, Q157 mutation rate was the highest in patients with primary myelofibrosis and AML17,20. In addition, previous studies9,14,17 demonstrated that U2AF1 mutant is prone to anemia and thrombocytopenia, and S34F mutant of U2AF121 can have reduced erythropoiesis and abnormal granulo-monocyte differentiation via regulating the downstream target gene H2AFY and STRAP to form abnormal splicing. This study showed that the number of neutrophils in U2AF1 mutant is low, but there is no significant difference among the three mutation sites. Due to the sample size, further verification is needed.
Recent studies have shown that specific mutations are closely related to cytogenetic abnormalities and may promote the occurrence and development of leukemia through unique pathogenesis. Kim et al.6 showed the S34Y site mutation of U2AF1 was significantly correlated to chromosome 8 abnormality. The S34F site mutation was correlated to chromosome 20 abnormality, and the Q157 mutation had a high incidence of chromosome 7 abnormality, but there was no statistical difference.
In this study we observed at the first time that compared with S34F and S34Y mutants, Q157P mutation had significantly higher incidence of chromosome 7 abnormalities (p = 0.002), higher co-mutation rate of signaling pathway related genes (p = 0.004). There were also higher incidence of co-mutation with TP53, higher risk of AML transformation, and shorter overall survival time in the Q157P mutation group. However, there was no significant difference between the two groups, which was probably related to the small sample size. Zhang et al.22 have shown that U2AF1 mutation can affect the expression of p53 signaling pathway and MAPK signaling pathway proteins by down-regulating the transcription of related genes. Therefore, we speculate that Q157P mutation of U2AF1 may lead to genetic instability and increase the incidence of some specific pathogenic genes by down-regulating the transcription of related genes, thus affecting the progress and prognosis of the disease.
Li et al.18 showed that the clonal patterns with initial mutations (ASXL1, DNMT3A and TET2) promoted the occurrence of MDS, while the some additional driver mutations (SF3B1, U2AF1 or RUNX1) played roles to keep the basic disease features, or give rise to different phenotypes (BCOR, EZH2 or TP53) in individual patients. These mutations were identified as last events for MDS development to be AML. Last mutations can exist at MDS diagnosis, or emerge at AML transformation, and involve a small group of genes.
Single-allele CEBPA mutations and diverse TP53 mutations were checked as the most common last event mutations. Considering the necessity of last event mutations and limited gene involvement in AML transformations, it is possible to validate a small group of last events involved mutations to develop some new strategies to block MDS progression. It is suggested that the occurrence and development of MDS is the process of gene mutation accumulation, and the different forms of co-mutation gene may have different effect for disease prognosis. At present, most of the researches focus on U2AF1 mutation status, mutation sites, clinical characteristics and prognostic impact of the combined abnormalities of cytogenetics, and less on the clinical characteristics and prognostic impact of U2AF1 and other co-mutation genes. In this study, the clinical characteristics and prognosis of patients with U2AF1 mutation co-mutated with ASXL1, TET2 and RUNX1 mutations were compared with those without related gene mutations, respectively, in order to elucidate the role of co-mutation genes in patients with U2AF1 gene mutation. The results demonstrated that U2AF1 mutation was positively correlated with ASXL1, RUNX1 and SETBP1, and negatively correlated with SF3B1 (p < 0.05), consistent with previous studies9,14,16,17. Different mutation sites may have fixed co-mutation patterns. In this study, we found that the co-mutation ratio of Q157P and signal pathway related genes was significantly higher than that of other sites. In addition, the incidence of TP53 co-mutation at Q15P was higher than that at S34, which was consistent with the study by Tefferi et al.17. This study also showed that the co-mutation rate of Q157 with TP53, ASXL1, IDH1, SETBP1 was higher than that of S34. Hou et al.23 showed that in MDS patients with U2AF1 mutation, those with RUNX1 mutation were independent adverse factors affecting hematopoietic stem cell transplantation, suggesting that the type of co-mutation gene may have an impact on prognosis. In this study, patients with U2AF1 mutation had a higher recurrence rate than those without RUNX1 mutation. We considered that it might be related to the higher mutation load of U2AF1 in this study. At the same time, we found that compared with those without ASXL1 mutation, those with ASXL1 mutation had a higher mutation load of U2AF1 (p = 0.674), and the proportion of bone marrow blasts was higher (p = 0.145), which was more distributed in patients with MDS-EB (p = 0.092). The risk of transformation to AML is higher (p = 0.004) and the recurrence rate is higher (p = 0.121). However, the mutation load of U2AF1 in TET2 mutation group is lower than that in TET2 non-mutation group (41.3% and 35.4%, p = 0.034), and the one-year OS rate increases (60.3% and 66.7%, p = 0.109). The ORR rate with DEC treatment is higher (33.3% and 30.0%), and the difference is not statistically significant. This study found that the mutation load of U2AF1 in different co-mutation gene combinations may affect the mutation load of U2AF1. The mutation load may have certain effect on disease progression and prognosis. In this study, the number of cases of U2AF1 and TET2, ASXL1, RUNX1 co -mutation were 16, 17 and 10, respectively, with a small sample size. Therefore, the clinical data and survival analysis were not conducted according to the mutation load of the co-mutation genes. The sample size will be further expanded in the future to study the impact of each co-mutation gene load in MDS patients with U2AF1 mutation.
Currently, the effect of U2AF1 mutation on prognosis is not clear yet. Some studies showed that U2AF1 mutation has no effect on prognosis 9–11), and others show that the prognosis of patients with U2AF1 mutation is poor 12–15. Recent studies have shown that there are differences in prognosis among different U2AF1 mutation sites. The prognosis of patients with U2AF1 Q157 mutant is worse than that of S3414,16,17. In this study, the OS of Q157P has a shorter OS, compared with other loci (S34F, S34Y and mOS of Q157P: 628 days, 804 days and 284 days, p = 0.418), but the difference is not significant. Further validation is needed with a large sample study. In addition, some studies have shown that the mutation load of TET2 and TP53 has an effect on the overall survival time24–26. At present, few studies have reported that the mutation load of U2AF1 has an effect on the prognosis. In this study, we found that compared with the group with relatively low mutation load of U2AF1 (VAF ≤ 40%), the group with relatively high mutation load of U2AF1 (VAF > 40%) has a significantly lower one-year survival rate (49.1% and 80.5%, p = 0.027). There was no significant difference in age, gender, response to DEC treatment, disease progression and recurrence between the two groups. The poor prognosis may be related to the higher incidence of RUNX1 mutation (31.8% and 11.5%, p = 0.152). In previous studies, the patients with RUNX1 co-mutation are more likely to progress to AML with poor prognosis27,28. In this study, multivariate analysis confirmed for the first time that VAF > 40% of U2AF1 was an independent impact factor of MDS patients with a shorter OS. It has further verified the influence of U2AF1 mutation load on disease progression and prognosis when U2AF1 co-mutated with TET2, ASXL1 and RUNX1. However, more studies are needed to explore the relationship between co-mutation gene and mutation load, as well as its role in disease progression and prognosis.
In summary, our results show that VAF > 40% of U2AF1 is an independent factor of short OS in MDS patients, and the OS of MDS patients with Q157 mutation of U2AF1 was shortened, but the difference was not significant, which needs to be further verified in the future.
Materials and methods
Patient cohort
From January 2016 to January 2020, data from 234 MDS patients were collected retrospectively. Informed consent was obtained from all patients or legal guardians and the study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University and based on the ethical principles for medical research involving human subjects of the Helsinki Declaration. Diagnosis was done according to the standard diagnosis with MICM criteria based on the 2016 WHO MDS diagnosis and classification criteria29. Inclusion criteria:
De novo MDS patients diagnosed according to the WHO (2016) MDS diagnosis and classification criteria;
Tested with NGS at the first diagnosis;
Older than 16 years old.
According to the MDS diagnosis criteria, 17 Cases (7.3%) patients diagnosed with MDS with single-lineage dysplasia (MDS-SLD), 52 cases (22.2%) patients diagnosed with MDS with multi-lineage dysplasia (MDS-MLD), 51 cases (21.8%) diagnosed with MDS with excess blasts subtype 1 (MDS-EB1), 98 cases (41.9%) diagnosed with MDS with excess blasts subtype 2 (MDS-EB2), 10 cases (4.3%) MDS with ring sideroblasts (MDS-RS), 4 cases (1.7%) MDS with isolated del (5q) and 2 cases (0.9%) with MDS unclassifiable (MDS-U), respectively. According to the revised international prognostic scoring system (IPSS-R), 205 MDS patients with chromosomal karyotype were stratified by IPSS-R. There were 54 cases in the intermediate-risk group (26.3%), and 70 cases in the high-risk group (34.2%) and 43 cases in the very-high risk group (23.9%). According to the prognosis score system, MDS patients were divided into relatively low-risk group (IPSS-R score ≤ 3.5) and relatively high-risk group (IPSS-R score > 3.5), with 47 cases (22.9%) and 158 cases (77.1%), respectively.
Next generation sequence
PCR was used to amplify and sequence genes. Gene mutation detection was done with standard NGS technology on a Illumina MiSeq System (Illumina, San Diego, CA) high-throughput sequencing platform with Amplicon panel targeting complete coding exons and their adjacent splice junctions from 22 genes was designed using Illumina Design Studio software v6.0 (https://designstudio.illumina.com/Home/SelectAssay) . Details of the variant calling, filtering, and annotation are described in our recently published reports2,30,31. Analyses were conducted of the relevant mutations of 22 related genes, including FLT3-ITD, NPM1, KIT, CEBPA, DNMT3A, IDH1, IDH2, TET2, EZH2, RUNX1, ASXL1, PHF6, TP53, SF3B1, SRSF2, U2AF1, ZRSR2, NRAS, CBL, SETBP1, ETV6, and JAK2. For the gene length longer than 150 bp, such as FLT3-ITD, alternative RT-PCR method was used for analysis. The pathogenic mutation sites were determined according to the COSMIC database and literature reports 30, 31).
Treatment methods
Of the 234 MDS patients, 212 were treated with different regimen. 67 were treated with demethylation agent (HMA), 85 were treated with combination of HMA and CAG regimen, 60 patients were treated with other treatment groups (including supportive treatment and chemotherapy alone). Total of 152 patients were treated with HMA, of which 13 were treated with DEC and Azacytidine (AZA), 5 with AZA alone, 134 with DEC alone. Among the 134 patients treated with DEC, there were 60 patients with low dose (6-8 mg/m2/d) and 74 patients with high dose (12-15 mg/m2/d) of DEC. In addition, according to whether they were treated with chemotherapy, 58 patients were classified into the DEC treatment alone group and 76 patients into the DEC + other agents’ chemotherapy group.
Treatment and efficacy criteria
Based on the efficacy criteria of the International Working Group on myelodysplastic syndrome (IWG), the ORR was defined as the sum of complete remission (CR), complete remission of bone marrow (MCR), partial remission (PR) and hematological improvement (HI) after 2–6 courses of drug treatment.
Follow up
In this study, the medical records and telephone follow-up methods were used for both hospitalized and clinic visited patients. The starting point of follow-up was the date of diagnosis of MDS, and the end point of follow-up was either of the deadlines of follow-up (2020.2.1), the time of death or the time of loss of follow-up (LOF). As of the follow-up date, the mOS of 234 MDS patients was 189 (2–1406) days, including 99 survivals (42.3%), 90 deaths (38.5%) and 45 LOF (19.2%). Overall survival (OS) is defined as the time from start of randomization to death for any reason. If the patient is still alive by the end of the follow-up date, then the end time of total survival period is the end time of follow-up (2020.2.1).
Statistical analysis
SPSS 26.0 software was used for statistical analysis, and chi square test or Fisher exact probability methods were used for counting data, non-parameter test was used for measurement and grade data. Spearman correlation analysis was used for non-normal distribution data. Logistic regression analysis was used for multivariate analysis. Kaplan–Meier method was used for survival analysis. Log rank test was used for comparison of differences between groups. Cox regression analysis was used for significant indicators in survival analysis. P < 0.05 was considered as statistically significance.
Ethics approval and consent to participate
Informed consent was obtained from all patients or legal guardians and the study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University and based on the ethical principles for medical research involving human subjects of the Helsinki Declaration.
Consent for publication
All authors agreed to publish.
Acknowledgements
We thank Dr. Yafei Li and Dr. Runqing Lu of The First Affiliated Hospital of Zhengzhou University for their help in laboratory testing and patient clinical information collection.
Abbreviations
- AML
Acute myeloid leukemia
- AZA
Azacytidine
- CR
Complete remission
- DEC
Decitabine
- HI
Hematologic improvement
- HMA
Hypomethylating agent
- IPSS-R
Revised international prognostic scoring system
- IWG
International Working Group
- MCR
Complete remission of bone marrow
- MDS
Myelodysplastic syndrome
- MDS (5q-)
MDS with isolated del (5q)
- MDS-EB1
MDS with excess blasts subtype 1
- MDS-EB2
MDS with excess blasts subtype 2
- MDS-MLD
MDS with multi-lineage dysplasia
- MDS-RS
MDS with ring sideroblasts
- MDS-SLD
MDS with single-lineage dysplasia
- MDS-U
MDS unclassifiable
- mOS
Median overall survival
- mVAF
Median variant allele frequency
- NGS
Next-generation sequence
- ORR
Overall response rate
- OS
Overall survival
- PR
Partial remission
- VAF
Variant allele frequency
- WBC
White blood cell count
Author contributions
R.G. and J.Y. designed and directed the study, and wrote the manuscript. H.W. and Y.G. contributed for data collection and analysis. Z.D. T.L. contributed for laboratory testing. X.X., D.W. Z. J. and R.G. contributed for clinical data and patients’ treatment. All authors contributed to drafting and revising the article and agree to be accountable for all aspects of the work. R.G. and J.Y. approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (81,070,445); Henan Natural Science Foundation of China (182,300,410,301); Henan Medical Science and Technology Research Project (201,403,029), Key Scientific Research Project of Henan Provincial Education Department (20A320062). The funding bodies did not participate in study design, in data collection, analysis, and interpretation, and in writing the manuscript.
Data availability
Data and material will be available upon corresponding author approval. All data sets generated/analyzed for this study are included in the manuscript and the additional files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Haiqiong Wang and Yongbo Guo.
Contributor Information
Jifeng Yu, Email: yujifengzzu@163.com.
Rong Guo, Email: fccguor@zzu.edu.cn.
References
- 1.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu J, Li Y, Li T, Li Y, Xing H, Sun H, et al. Gene mutational analysis by NGS and its clinical significance in patients with myelodysplastic syndrome and acute myeloid leukemia. Exp. Hematol. Oncol. 2020;9:2. doi: 10.1186/s40164-019-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, Li Y, Zhang D, Wan D, Jiang Z. Clinical implications of recurrent gene mutations in acute myeloid leukemia. Exp. Hematol. Oncol. 2020;9:4. doi: 10.1186/s40164-020-00161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bejar R. Implications of molecular genetic diversity in myelodysplastic syndromes. Curr. Opin. Hematol. 2017;24(2):73–78. doi: 10.1097/MOH.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill H, Leung AY, Kwong YL. Molecular and cellular mechanisms of myelodysplastic syndrome: Implications on targeted therapy. Int. J. Mol. Sci. 2016;17(4):440. doi: 10.3390/ijms17040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627; quiz 99. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Jiang PYZ, Sun H, Zhang X, Jiang Z, Li Y, et al. Advances in targeted therapy for acute myeloid leukemia. Biomark. Res. 2020;8:17. doi: 10.1186/s40364-020-00196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SY, Kim K, Hwang B, Im K, Park SN, Kim JA, et al. The high frequency of the U2AF1 S34Y mutation and its association with isolated trisomy 8 in myelodysplastic syndrome in Asians, but not in Caucasians. Leuk Res. 2017;61:96–103. doi: 10.1016/j.leukres.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Hong JY, Seo JY, Kim SH, Jung HA, Park S, Kim K, et al. Mutations in the spliceosomal machinery genes SRSF2, U2AF1, and ZRSR2 and response to decitabine in myelodysplastic syndrome. Anticancer Res. 2015;35(5):3081–3089. [PubMed] [Google Scholar]
- 11.Hwang SM, Kim SY, Kim JA, Park HS, Park SN, Im K, et al. Short telomere length and its correlation with gene mutations in myelodysplastic syndrome. J. Hematol. Oncol. 2016;9(1):62. doi: 10.1186/s13045-016-0287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung SH, Kim YJ, Yim SH, Kim HJ, Kwon YR, Hur EH, et al. Somatic mutations predict outcomes of hypomethylating therapy in patients with myelodysplastic syndrome. Oncotarget. 2016;7(34):55264–55275. doi: 10.18632/oncotarget.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Song L, Xu L, Chang C, Xu F, Wu D, et al. Genetic landscape of recurrent ASXL1, U2AF1, SF3B1, SRSF2, and EZH2 mutations in 304 Chinese patients with myelodysplastic syndromes. Tumour Biol. 2016;37(4):4633–4640. doi: 10.1007/s13277-015-4305-2. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Liu J, Jia Y, Wang J, Xu Z, Qin T, et al. Clinical features and biological implications of different U2AF1 mutation types in myelodysplastic syndromes. Genes Chromosom. Cancer. 2018;57(2):80–88. doi: 10.1002/gcc.22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Zhang N, Wu X, Zheng X, Ling Y, Gong Y. Prognostic value of U2AF1 mutant in patients with de novo myelodysplastic syndromes: a meta-analysis. Ann. Hematol. 2019;98(12):2629–2639. doi: 10.1007/s00277-019-03843-3. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Zou D, Yang S, Ouyang G, Mu Q. Prognostic significance of U2AF1 mutations in myelodysplastic syndromes: A meta-analysis. J. Int. Med. Res. 300060519891013 (2019). [DOI] [PMC free article] [PubMed]
- 17.Tefferi A, Mudireddy M, Finke CM, Nicolosi M, Lasho TL, Hanson CA, et al. U2AF1 mutation variants in myelodysplastic syndromes and their clinical correlates. Am J Hematol. 2018;93(6):E146–E148. doi: 10.1002/ajh.25084. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Xu F, Wu LY, Zhao YS, Guo J, He Q, et al. A genetic development route analysis on MDS subset carrying initial epigenetic gene mutations. Sci. Rep. 2020;10(1):826. doi: 10.1038/s41598-019-55540-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganguly BB, Kadam NN. Mutations of myelodysplastic syndromes (MDS): An update. Mutat. Res. Rev. Mutat. Res. 2016;769:47–62. doi: 10.1016/j.mrrev.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Zhao TY, Guan LX, Zheng WS, Wang ML, Cheng LC, Hu YL, et al. Clinical characteristics and prognosis of U2AF1 mutation in patients with acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(1):7–11. doi: 10.19746/j.cnki.issn.1009-2137.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Yip BH, Steeples V, Repapi E, Armstrong RN, Llorian M, Roy S, et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J. Clin. Invest. 2017;127(9):3557. doi: 10.1172/JCI96202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Zhao H, Wu K, Peng Y, Han X, Zhang H, et al. Knockdown of spliceosome U2AF1 significantly inhibits the development of human erythroid cells. J. Cell Mol. Med. 2019;23(8):5076–5086. doi: 10.1111/jcmm.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou C, Jiang SH, Shen HJ, Ding ZX, Miao M, Chen SN, et al. Clinical features and prognostic analysis of myelodysplastic syndromes patients with U2AF1 mutation. Zhonghua Xue Ye Xue Za Zhi. 2020;41(1):72–75. doi: 10.3760/cma.j.issn.0253-2727.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montalban-Bravo G, Kanagal-Shamanna R, Benton CB, Class CA, Chien KS, Sasaki K, et al. Genomic context and TP53 allele frequency define clinical outcomes in TP53-mutated myelodysplastic syndromes. Blood Adv. 2020;4(3):482–495. doi: 10.1182/bloodadvances.2019001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang L, Luo Y, Zhu S, Wang L, Ma L, Zhang H, et al. Mutation status and burden can improve prognostic prediction of patients with lower-risk myelodysplastic syndromes. Cancer Sci. 2020;111(2):580–591. doi: 10.1111/cas.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallman DA, Padron E. Integrating mutation variant allele frequency into clinical practice in myeloid malignancies. Hematol. Oncol. Stem Cell Ther. 2016;9(3):89–95. doi: 10.1016/j.hemonc.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Tsai SC, Shih LY, Liang ST, Huang YJ, Kuo MC, Huang CF, et al. Biological activities of RUNX1 mutants predict secondary acute leukemia transformation from chronic myelomonocytic leukemia and myelodysplastic syndromes. Clin Cancer Res. 2015;21(15):3541–3551. doi: 10.1158/1078-0432.CCR-14-2203. [DOI] [PubMed] [Google Scholar]
- 28.Chen CY, Lin LI, Tang JL, Ko BS, Tsay W, Chou WC, et al. RUNX1 gene mutation in primary myelodysplastic syndrome–the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br. J. Haematol. 2007;139(3):405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 29.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 30.Liu S. Expert consensus on the application of next-generation sequencing in hematological neoplasms. Zhonghua Xue Ye Xue Za Zhi. 2018;39:881–886. doi: 10.3760/cma.j.issn.0253-2727.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and material will be available upon corresponding author approval. All data sets generated/analyzed for this study are included in the manuscript and the additional files.


