Abstract
Leaf rolling is an important mechanism to mitigate the effects of moisture stress in several plant species. In the present study, a set of 92 wheat recombinant inbred lines derived from the cross between NI5439 × HD2012 were used to identify QTLs associated with leaf rolling under moisture stress condition. Linkage map was constructed using Axiom 35 K Breeder’s SNP Array and microsatellite (SSR) markers. A linkage map with 3661 markers comprising 3589 SNP and 72 SSR markers spanning 22,275.01 cM in length across 21 wheat chromosomes was constructed. QTL analysis for leaf rolling trait under moisture stress condition revealed 12 QTLs on chromosomes 1B, 2A, 2B, 2D, 3A, 4A, 4B, 5D, and 6B. A stable QTL Qlr.nhv-5D.2 was identified on 5D chromosome flanked by SNP marker interval AX-94892575–AX-95124447 (5D:338665301–5D:410952987). Genetic and physical map integration in the confidence intervals of Qlr.nhv-5D.2 revealed 14 putative candidate genes for drought tolerance which was narrowed down to six genes based on in-silico analysis. Comparative study of leaf rolling genes in rice viz., NRL1, OsZHD1, Roc5, and OsHB3 on wheat genome revealed five genes on chromosome 5D. Out of the identified genes, TraesCS5D02G253100 falls exactly in the QTL Qlr.nhv-5D.2 interval and showed 96.9% identity with OsZHD1. Two genes similar to OsHB3 viz. TraesCS5D02G052300 and TraesCS5D02G385300 exhibiting 85.6% and 91.8% identity; one gene TraesCS5D02G320600 having 83.9% identity with Roc5 gene; and one gene TraesCS5D02G102600 showing 100% identity with NRL1 gene were also identified, however, these genes are located outside Qlr.nhv-5D.2 interval. Hence, TraesCS5D02G253100 could be the best potential candidate gene for leaf rolling and can be utilized for improving drought tolerance in wheat.
Subject terms: Genetics, Molecular biology, Plant sciences
Introduction
Bread wheat (Triticum aestivum L.) is one of the world's major staple food crops accounting for 20% of calories consumed globally1. Global wheat production amounts to 771 million tonnes (mt) of grains from an area of 218 million hectares (mha)2. Drought is among one of the major stress factors affecting wheat yields, especially in rain fed agriculture. As much as 21 percent reduction in wheat yield has been observed at approximately 40% water reduction3. Almost 50% of cultivation in the developing countries occurs in rain fed system4. Besides, the ongoing climate change crisis is making the problem far worse with infrequent and erratic precipitation leading to the expansion of drought prone areas. As much as 50% productivity reduction in wheat is observed in many countries in drought years leading to significant per capita food production decline5–7. Breeding drought tolerant cultivars is the best strategy for mitigating the effects of water deficit conditions. Drought tolerance is a complex trait involving several morphological, agronomical, physiological, and biochemical components. Drought stress is the result of an imbalance between evapo-transpiration flux and water intake from soil8. Moisture stress conditions result in physiological dehydration at the cellular level9. Response of plants to moisture stress at cellular level is complex and involves several factors including signaling molecules, transcription factors, hormones, and secondary metabolites7. Hence, estimation of drought tolerance is a difficult task. From the breeders’ point of view, drought tolerance is the ability of plants to survive moisture stress conditions with minimum reduction in yield. Response of plants to drought stress is broadly classified into two categories: avoidance and tolerance. Avoidance mechanisms include morphological and physiological adjustments such as reduced stomatal number and conductance, increased leaf thickness and decreased leaf area, increased root system and leaf rolling to reduce evapo-transpiration and thus enabling plants to escape moisture stress7, 10, 11. Tolerance mechanisms, on the other hand, maintain cellular turgor pressure by biochemical modifications through osmotic adjustments12. Leaf rolling is an escape mechanism evolved by plants to increase water use efficiency under moisture deficit conditions. In wheat, leaf lamina rolls transversally along the mid-rib to form a cylinder under severe moisture stress and/or high temperature and unrolls when stress is relieved13. This enables the plant to conserve water by decreasing transpiration and reducing leaf temperature. Wheat genotypes showing this trait exhibit better stomatal regulation, higher water use efficiency (WUE), and photosynthetic efficiency under stress conditions14.
Leaf rolling is a complex quantitative trait that is controlled by multiple genes following non-Mendelian inheritance15. Leaf rolling is a typical response related to mechanism of reducing moisture stress in field crops which is well studied in rice16, 17 and maize15, 18. Five mutants in maize19–22 and 17 mutants in rice with leaf rolling traits22 have been characterized. Additionally, more than 70 genes/QTLs associated with leaf rolling have been mapped in rice23. In contrast, studies regarding leaf rolling and identification of associated genomic regions or genes in wheat are scarcely reported. A study on RIL (recombinant inbred lines) population developed by crossing durum wheat (T. turgidum ssp. durum) with wild emmer wheat (T. turgidum ssp. dicoccoides) identified 11 significant QTLs associated with flag leaf rolling24. In another related species Secale cereale (Rye), four stable QTLs associated with leaf rolling were reported on chromosomes 3R, 5R, and 7R25 utilizing DArT markers. Wheat is a polyploid species and identifying genes with minor effects like those affecting drought tolerance is a daunting task. Quantitative trait loci (QTL) mapping using biparental populations is one of the commonly used approaches to analyze complex traits to identify genomic regions associated with complex traits such as drought tolerance. Successful QTL mapping requires high density of markers for precise QTL detection and accurate identification of candidate genes for complex traits. Earlier, SSR and other DNA based markers have been utilized for QTL mapping. However, with the advent of single nucleotide polymorphism (SNPs) genotyping platforms and availability of whole genome sequence of bread wheat26 it has become possible to develop high density molecular maps with greater precision.
In the present study leaf rolling trait was investigated in a mapping population comprising of RILs derived from the cross NI5439 x HD2012. Array based SNP markers along with SSR markers were used for generating a high-density linkage map. Polymorphic SNP and SSR markers were utilized for QTL mapping of leaf rolling. The objectives of this study were: (1) to create a genetic linkage map utilizing the SNP markers derived using 35 K SNP Array; (2) to identify QTLs controlling leaf rolling trait; (3) to infer potential candidate gene(s) responsible for leaf rolling under moisture stress and (4) comparison of leaf rolling genes predicted in wheat with cloned leaf rolling genes in rice.
Results
Genetic linkage mapping
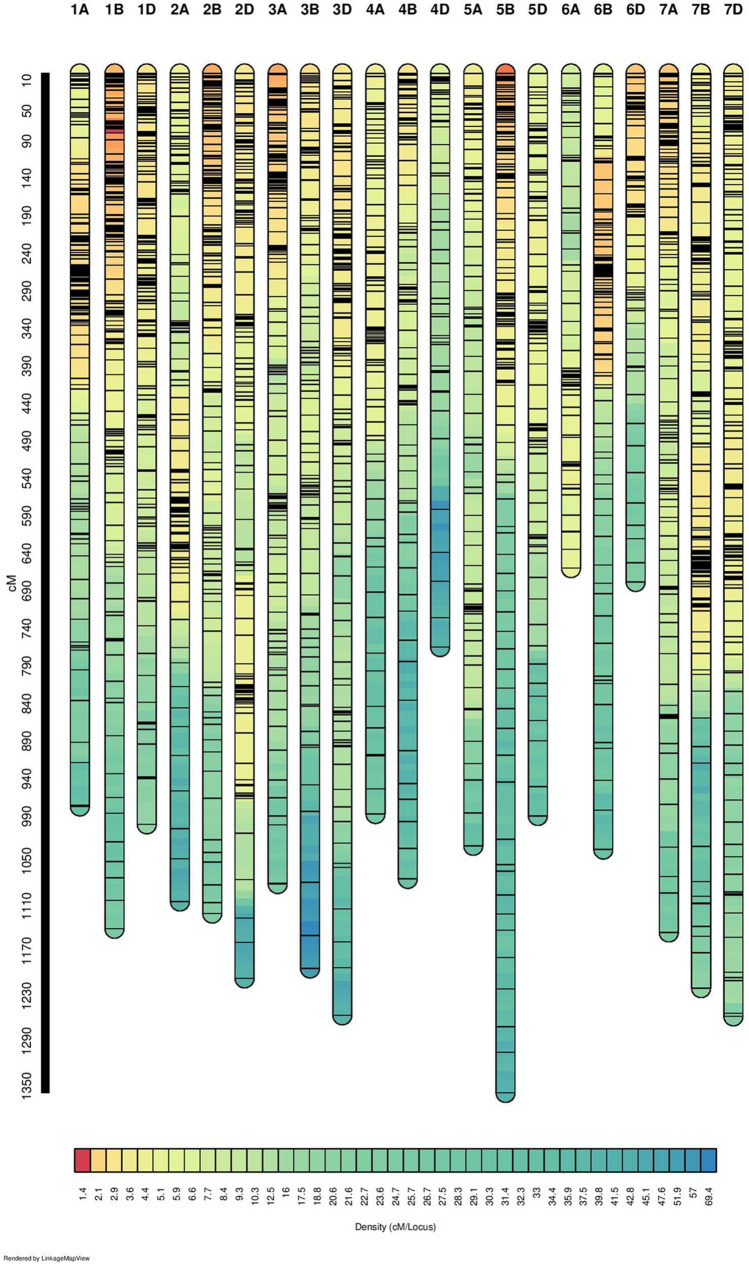
The genetic linkage map was constructed using a mapping population of 92 RILs derived from the cross between NI5439 and HD2012. A total of 3661 markers comprising of 3589 SNP and 72 SSR markers were utilized for construction of linkage map (Fig. 1, Supplementary Figure S1). The linkage map spanned 22,275.01 cM in length distributed across the 21 wheat chromosomes (Table 1). Probe set ID, marker allele for each genotype, affymetrix SNP ID, reference chromosome, physical position (bp) and sequence details for the filtered 3589 SNPs are given in Supplementary data sheet S1. Average distance between two linked markers ranged from 4.27 cM (1B) to 14.43 cM (4D) with an overall marker interval of 6.08 cM. Marker density ranged from 0.07 cM per marker (4D) to 0.23 cM per marker (1B) with an average of 0.17 cM per marker. Genome wise length analysis of map revealed that A, B, and D genomes spanned 6982.80 cM (31.34%), 8139.80 cM (36.54%), and 7152.42 cM (32.10%), respectively. The number of markers varied from 53 in 4D chromosome to 267 in 1B chromosome. The shortest chromosome was 6A which harbored 96 markers with a genetic length of 659.43 cM. The longest chromosome was 5B which had 220 markers with a genetic length of 1359.04 cM. A genome and D genome had 0.16 cM marker density with 1103 and 1125 total markers respectively, as compared to B genome which has marker density of 0.18 cM per marker with 1433 markers.
Figure 1.
Linkage map constructed from SNP and SSR genotyping in a recombinant inbred population derived from a cross between NI5439 and HD2012.
Table 1.
Marker statistics of linkage map constructed from recombinant inbred lines derived from NI5439/HD2012.
| Chr | No. of markers | Chromsome length (cM) | %Marker | Average marker interval (resolution) | Marker density |
|---|---|---|---|---|---|
| 1A | 192 | 977.08 | 5.24 | 5.09 | 0.20 |
| 1B | 267 | 1140.19 | 7.29 | 4.27 | 0.23 |
| 1D | 182 | 1001.05 | 4.97 | 5.50 | 0.18 |
| 2A | 164 | 1103.75 | 4.48 | 6.73 | 0.15 |
| 2B | 237 | 1119.90 | 6.47 | 4.73 | 0.21 |
| 2D | 228 | 1206.40 | 6.23 | 5.29 | 0.19 |
| 3A | 205 | 1080.63 | 5.60 | 5.27 | 0.19 |
| 3B | 150 | 1193.09 | 4.10 | 7.95 | 0.13 |
| 3D | 191 | 1255.56 | 5.22 | 6.57 | 0.15 |
| 4A | 128 | 987.20 | 3.50 | 7.71 | 0.13 |
| 4B | 126 | 1073.67 | 3.44 | 8.52 | 0.12 |
| 4D | 53 | 764.73 | 1.45 | 14.43 | 0.07 |
| 5A | 120 | 1029.56 | 3.28 | 8.58 | 0.12 |
| 5B | 220 | 1359.04 | 6.01 | 6.18 | 0.16 |
| 5D | 128 | 989.77 | 3.50 | 7.73 | 0.13 |
| 6A | 96 | 659.43 | 2.62 | 6.87 | 0.15 |
| 6B | 209 | 1034.56 | 5.71 | 4.95 | 0.20 |
| 6D | 137 | 677.88 | 3.74 | 4.95 | 0.20 |
| 7A | 198 | 1145.15 | 5.41 | 5.78 | 0.17 |
| 7B | 224 | 1219.35 | 6.12 | 5.44 | 0.18 |
| 7D | 206 | 1257.04 | 5.63 | 6.10 | 0.16 |
| A genome | 1103 | 6982.80 | 30.13 | 6.33 | 0.16 |
| B genome | 1433 | 8139.80 | 39.14 | 5.68 | 0.18 |
| D genome | 1125 | 7152.42 | 30.73 | 6.35 | 0.16 |
| Total | 3661 | 22,275.01 | 6.08 | 0.17 |
Phenotypic evaluation
The NI5439 × HD2012 derived RIL population showed significant variation for leaf rolling. The descriptive statistical parameters and frequency distribution over all the three environments for leaf rolling is shown in Supplementary Table S1 and Supplementary Figure S2. The mapping population was characterized by average mean value of lines varying from 2.00 to 5.00. Coefficient of variation (CV) ranged from 26.11 to 32.20 with an average CV of 27.82. Standard deviation (SD) ranged from 0.63 to 0.80 with average SD of 0.68. Positive correlation was observed within the replicates of each environment and within each environment for leaf rolling trait (Supplementary Tables S2–S3). Analysis of variance showed significant differences among genotypes of the RIL population in all 3 years (Supplementary Table S4). RILs showed extreme values for leaf rolling than that of parents across all the environments signifying transgressive segregation. This indicates polygenic inheritance of the trait and suggests that alleles with positive effects are distributed among the parents. Mean heritability of 0.94 was observed for leaf rolling trait (Supplementary Table S4).
QTL identification for leaf rolling
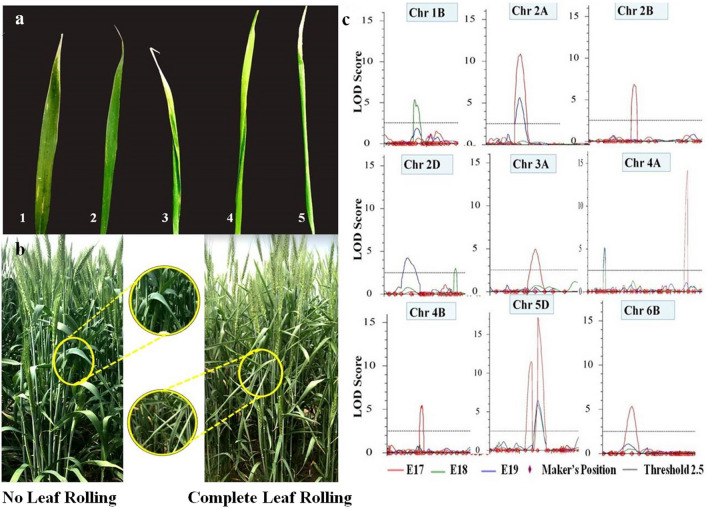
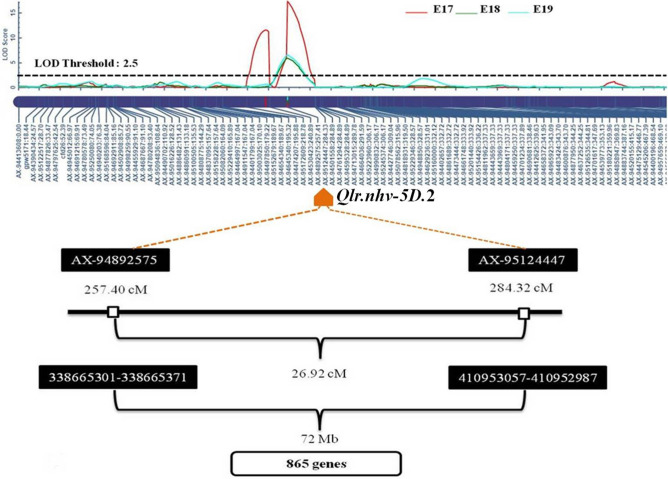
For the leaf rolling trait, a total of 12 QTLs were detected under drought stress condition during all 3 years (Fig. 2c; Table 2). LOD scores of these QTLs ranged from 3.01 to 17.19. Phenotypic variance explained (%) varied from 4.06 to 20.10%. QTLs identified were associated with chromosomes 1B, 2A, 2B, 2D, 3A, 4A, 4B, 5D and 6B. Eight QTLs were identified during 2017–2018 (E17), whereas, four QTLs were detected during 2018–2019 (E18) and 2019–2020 (E19), with all QTLs showing LOD score greater than 3. On 5D chromosome, a stable QTL Qlr.nhv-5D.2 was detected consistently throughout all the environments (Fig. 3). Qlr.nhv-5D.2 was flanked by markers AX-94892575 and AX-95124447 (5D:338665301–5D:410952987). In E17, the QTL Qlr.nhv-5D.2 showed LOD score of 17.19 explaining phenotypic variance of 20.10%. In E18, the QTL Qlr.nhv-5D.2 showed LOD score 5.84 explained 16.08% of phenotypic variance. While, in E19, the QTL Qlr.nhv-5D.2 showed LOD score 6.45 and explained 14.25% of phenotypic variance. Another QTL Qlr.nhv-5D.1 was also detected on 5D chromosome only in E17. On chromosome 2A, QTL Qlr.nhv-2A was detected in two environments (E17 and E19). Qlr.nhv-2A was flanked by markers AX-94942225–AX-95254393 having LOD score 10.85 and PVE 12.03% in E17, whereas in E19 Qlr.nhv-2A explained LOD score 5.64 and PVE 12.13%. On chromosome 4A, QTL Qlr.nhv-4A.2 was detected in E18 and E19. Qlr.nhv-4A.2 was flanked by markers AX-95212081–AX-94470023 having LOD score 5.13 and PVE 13.43% in E18 and LOD score of 5.06 and PVE 11.38% in E19. In E17, another QTL Qlr.nhv-4A.1 flanked by markers AX-94739181–AX-94500554 (LOD score 14.18 and PVE 15.26%) was also identified. On chromosome 2D, two distinct QTLs were detected in E18 and E19. Qlr.nhv-2D.1 was flanked by markers AX-94743285–AX-94823535 having LOD score 3.01 and PVE 7.44%. On the other hand, Qlr.nhv-2D.2 was flanked by markers AX-94661194–AX-94603691 having LOD score 4.21 and PVE 15.80%. On rest of the chromosomes viz., 1B (Qlr.nhv-1B), 2B (Qlr.nhv-2B), 3A (Qlr.nhv-3A), 4B (Qlr.nhv-4B), 6B (Qlr.nhv-6B) QTLs were detected only during a single year.
Figure 2.
(a) Scoring of leaf rolling trait ranging from 1 (no leaf rolling) to 5 (complete leaf rolling); (b) RILs of NI5439 × HD2012 mapping population showing difference in leaf rolling from no rolling of the leaf to complete rolling; (c) graphical illustration of QTLs detected in three environments for leaf rolling. Red, green and blue represent the three environments E17, E18 and E19.
Table 2.
QTLs for leaf rolling in RIL mapping population of NI5439 and HD2012.
| Chr | QTL | Env | Pos | Flanking markers | LOD | PVE(%) | Add |
|---|---|---|---|---|---|---|---|
| 1B | Qlr.nhv-1B | E18 | 172 | AX-95149749-gwm153 | 5.32 | 15.35 | − 0.26 |
| 2A | Qlr.nhv-2A | E17 | 681 | AX-94942225–AX-95254393 | 10.85 | 12.03 | − 0.32 |
| E19 | 679 | AX-94942225–AX-95254393 | 5.64 | 12.13 | − 0.27 | ||
| 2B | Qlr.nhv-2B | E17 | 686 | AX-94711931–AX-95260437 | 6.86 | 6.49 | − 0.23 |
| 2D | Qlr.nhv-2D.1 | E18 | 430 | AX-94743285–AX-94823535 | 3.01 | 7.44 | − 0.18 |
| Qlr.nhv-2D.2 | E19 | 283 | AX-94661194–AX-94603691 | 4.21 | 15.80 | − 0.31 | |
| 3A | Qlr.nhv-3A | E17 | 823 | AX-94598770–AX-94844071 | 4.95 | 4.06 | 0.18 |
| 4A | Qlr.nhv-4A.1 | E17 | 482 | AX-94739181–AX-94500554 | 14.18 | 15.26 | 0.36 |
| Qlr.nhv-4A.2 | E18 | 80 | AX-95212081–AX-94470023 | 5.13 | 13.43 | 0.24 | |
| E19 | 83 | AX-95212081–AX-94470023 | 5.06 | 11.38 | 0.26 | ||
| 4B | Qlr.nhv-4B | E17 | 262 | AX-94993235–AX-94508980 | 5.44 | 5.43 | − 0.21 |
| 5D | Qlr.nhv-5D.1 | E17 | 237 | AX-95172609–AX-94530423 | 11.47 | 14.01 | 0.35 |
| Qlr.nhv-5D.2 | E17 | 258 | AX-94892575–AX-95124447 | 17.19 | 20.10 | − 0.41 | |
| E18 | 258 | AX-94892575–AX-95124447 | 5.84 | 16.08 | − 0.26 | ||
| E19 | 258 | AX-94892575–AX-95124447 | 6.45 | 14.25 | − 0.29 | ||
| 6B | Qlr.nhv-6B | E17 | 142 | AX-94500240–AX-94980566 | 5.31 | 4.47 | − 0.19 |
Figure 3.
Stable QTL (Qlr.nhv-5D.2) for leaf rolling on 5D chromosome during E17 (red), E18 (green) and E19 (blue). Genetic and physical map location of Qlr.nhv-5D.2 flanked by markers AX-94892575- AX-95124447.
Candidate gene identification and in-silico gene expression
A stable QTL for leaf rolling Qlr.nhv-5D.2 was located on 5D chromosome, flanked by markers AX-94892575 and AX-95124447. Putative gene(s) for leaf rolling was predicted in this marker interval (Fig. 3). Out of 865 genes present in the interval, 14 protein coding genes related to drought tolerance in T. aestivum were identified viz., Chlorophyll a/b binding protein, Potassium transporter, HVA22-like protein, Glycosyltransferase, Patatin, Histone proteins: Histone H2A and Histone H4 variant TH011, Peroxidase gene, Dirigent protein, Calcium-transporting ATPase, Auxin efflux carrier component, MADS box protein, 3-ketoacyl-CoA synthase, and heat shock protein 90 (Table 3). Gene ontology (GO) term analysis was carried out for these 14 genes and data is furnished in the Supplementary Table S5. Gene expression of these predicted 14 potential candidate genes under stress and non stress treatments was analyzed in-silico using the wheat expression browser (Supplementary Figure S3). Out of the 14 genes, TraesCS5D02G238300, TraesCS5D02G268000, TraesCS5D02G240900 and TraesCS5D02G249800 were expressed equally throughout all treatments indicating that they are not dependent on the stress/no stress stimulus for their expression. Low to no expression was observed for genes, TraesCS5D02G293300 and TraesCS5D02G294500 in all three treatments. The genes, TraesCS5D02G248600, TraesCS5D02G252700, TraesCS5D02G308500 and TraesCS5D02G316200 showed different levels of expression in control treatment but showed low to no expression in the other two treatments. The two genes TraesCS5D02G240200 and TraesCS5D02G256400 were up-regulated in drought stress treatment as compared to both the control treatments. TraesCS5D02G284100 showed similar expression in no stress control and drought stress and displayed slightly lesser expression in the PEG control. While TraesCS5D02G309500 showed almost similar expression in no stress control and drought stress and slightly higher expression in the PEG control. Based on the in-silico gene expression analysis, six genes exhibited differential expression in no stress control and drought stress treatments. Out of these six genes, four genes viz., TraesCS5D02G248600, TraesCS5D02G252700, TraesCS5D02G308500 and TraesCS5D02G316200 showed down-regulation and two genes viz., TraesCS5D02G240200 and TraesCS5D02G256400 showed up-regulation under drought stress. Hence out of the 14 genes these six may be the best candidate genes.
Table 3.
Candidate gene(s) identified related to drought stress in the marker interval region of QTL Qlr.nhv-5D.2.
| S. no. | Gene stable ID | Gene description | Source | Gene bp start | Gene bp end |
|---|---|---|---|---|---|
| 1 | TraesCS5D02G238300 | Chlorophyll a-b binding protein, chloroplastic | UniProtKB/TrEMBL;Acc:W5FYI0 | 346636682 | 346638071 |
| 2 | TraesCS5D02G240200 | Potassium transporter | UniProtKB/TrEMBL;Acc:A0A1D6S5W8 | 348479747 | 348485739 |
| 3 | TraesCS5D02G240900 | HVA22-like protein | UniProtKB/TrEMBL;Acc:A0A096UT86 | 349934068 | 349935316 |
| 4 | TraesCS5D02G248600 | Glycosyltransferase | UniProtKB/TrEMBL;Acc:A0A1D5ZMZ8 | 355607639 | 355609171 |
| 5 | TraesCS5D02G249800 | Patatin | UniProtKB/TrEMBL;Acc:W5FVZ6 | 356200779 | 356204674 |
| 6 | TraesCS5D02G252700 | Histone H2A | UniProtKB/TrEMBL;Acc:A0A1D5ZZ26 | 358798314 | 358798998 |
| 7 | TraesCS5D02G256400 | Peroxidase | UniProtKB/TrEMBL;Acc:A0A1D5ZID1 | 362661987 | 362665323 |
| 8 | TraesCS5D02G268000 | Heat shock protein 90 | UniProtKB/TrEMBL;Acc:Q0Q0I7 | 371469091 | 371472885 |
| 9 | TraesCS5D02G284100 | Calcium-transporting ATPase | UniProtKB/TrEMBL;Acc:W5FZP2 | 384852906 | 384863954 |
| 10 | TraesCS5D02G293300 | Auxin efflux carrier component | UniProtKB/TrEMBL;Acc:A0A1D5ZW76 | 390504143 | 390506578 |
| 11 | TraesCS5D02G294500 | MADS box protein | UniProtKB/TrEMBL;Acc:Q718F3 | 391729992 | 391737307 |
| 12 | TraesCS5D02G308500 | Histone H4 variant TH011 | UniProtKB/Swiss-Prot;Acc:P62785 | 405334206 | 405334791 |
| 13 | TraesCS5D02G309500 | Dirigent protein | UniProtKB/TrEMBL;Acc:A0A1D5ZQ97 | 406461906 | 406462620 |
| 14 | TraesCS5D02G316200 | 3-Ketoacyl-CoA synthase | UniProtKB/TrEMBL;Acc:A0A1D5ZJN8 | 410200495 | 410202372 |
Homology modeling of leaf rolling genes in wheat and rice
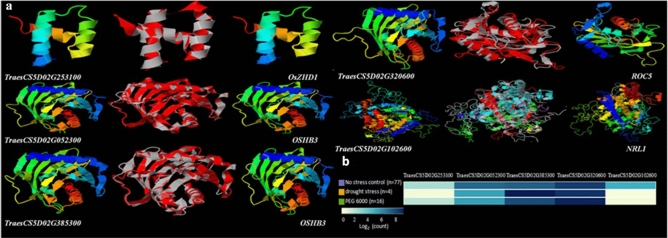
In maize and rice, extensive studies have been undertaken for leaf rolling as a component trait of drought tolerance. We compared the protein sequences of leaf rolling genes with the 5D chromosome of wheat reference genome to identify putative genes in wheat. In case of maize, three reported genes viz., Lbl1 (leafbladeless1), Rld1 (Rolled leaf1), ZmOCL5 (Outer Cell Layer5) were used for comparison. However, none of them showed more than 80% identity in the prescribed QTL interval. In case of rice, 17 genes related to leaf rolling viz., YABBY1, COW1 (CONSTITUTIVELY WILTED1)/NAL7 (NARROW LEAF7), OsHB1, ADL1 (ADAXIALIZED LEAF1), SSL1 (SHALLOT-LIKE1)/RL9, NRL1 (NARROW AND ROLLED LEAF1), ACL1 (Abaxially Curled Leaf1), ACL2, LC2, Roc5 (Rice outermost cell-specific gene5), SRL1 (SEMI-ROLLED LEAF1), RL14, OsZHD1, REL1 (Rolled and Erect Leaf1), REL2, LRRK1, OsHB3 were utilized for comparison. We identified five genes in wheat showing more than 80% identity with four reported genes in rice viz., NRL1, OsZHD1, Roc5, and OsHB3 (Table 4). Out of these, one gene (TraesCS5D02G253100) falls exactly in the QTL interval (338665301–410952987) and is showing 96.9% identity with OsZHD1. Two genes similar to OsHB3 were identified viz. TraesCS5D02G052300 and TraesCS5D02G385300 exhibiting 85.6% and 91.8% identity, respectively. One gene TraesCS5D02G320600 was having 83.9% identity with Roc5 gene. Lastly, TraesCS5D02G102600 showed 100% identity with NRL1 gene. The last four genes did not fall in the exact QTL interval. Nevertheless, they are reported on 5D chromosome near to the interval. The 3D protein structure of these five wheat genes was predicted and superimposed on the respective protein from rice (Fig. 4A). The 3D protein structure of all the five proteins was found to be significantly similar to the corresponding rice protein. The physiochemical properties of these proteins were also predicted, and data is furnished in Supplementary Table S6.
Table 4.
Homology modeling of Leaf rolling genes in Wheat and Rice.
| S. no | Rice genes | Gene hit | Genomic location | Query % | E-val | Identity (%) | Subcellular location | P value | Raw score |
|---|---|---|---|---|---|---|---|---|---|
| 1 | OsZHD1 | TraesCS5D02G253100 | 5D: 359474816–359475706 | 60.2 | 3.60E−38 | 96.9 | Nucleus | 0.00E+00 | 168.00 |
| 2 | OsHB3 | TraesCS5D02G052300 | 5D: 50483246–50488859 | 75.4 | 0 | 85.6 | Nucleus | 0.00E+00 | 620.69 |
| 3 | TraesCS5D02G385300 | 5D: 454414059–454419771 | 48.4 | 0 | 91.8 | Chloroplast | 0.00E+00 | 550.73 | |
| 4 | ROC5 | TraesCS5D02G320600 | 5D:412737994–412744775 | 75.8 | 8.20E−79 | 83.9 | Nucleus | 1.52E−12 | 359.98 |
| 5 | NRL1 | TraesCS5D02G102600 | 5D:115947470–115952140 | 80.8 | 3.00E−30 | 100.0 | Chloroplast outer membrane | 1.64 E -08 | 1134.16 |
Figure 4.
(a) 3D protein structures of the predicted leaf rolling genes in wheat (left) along with the protein structure of corresponding gene in rice (right) and their superimposed (middle) 3D structure in red and grey color. Red color represents wheat protein whereas grey represents rice protein. (b) Expression profiles of predicted leaf rolling wheat genes in three different conditions (no stress control, drought stress and PEG 6000). The dark and light intensity of the blue color represents the higher and lower relative abundance of the transcript.
Discussion
Leaf rolling is an important drought tolerance mechanism having polygenic control with additive effects which has been reported in various crops like wheat, rice, maize, and sorghum27. In a study on genetics of leaf rolling, involvement of polygenes with additive effects was reported in wheat27. Another study in tetraploid wheat T. turgidum ssp. dicoccoides (AABB, 2n = 4x = 28) reported eleven QTLs associated with leaf rolling, located on chromosomes 1A, 2A, 2B, 4B, 5A, 5B, 6A, 6B, 7A, and 7B24. Five of these QTLs significantly co-localized with QTLs associated with plant productivity. In rye (RR, 2n = 14), a distant relative of wheat, four stable QTLs for flag leaf rolling located on chromosomes 3R, 5R, and 7R were reported25. However, only few studies on leaf rolling is reported in hexaploid wheat. Leaf rolling was found out to be a trait showing non-Mendelian inheritance. Frequency distribution of leaf rolling in the mapping population showed a skewed normal distribution with large number of RILs showing lesser values of leaf rolling score under drought stress. The trait displayed high heritability and significant positive correlation within the replicates of each environment and within each environment which shows its stability. Mapping population showed significant genotypic variation. Transgressive segregation was evident since RILs showed extreme values for leaf rolling than that of parents across all the environments. All the above points suggest polygenic inheritance of leaf rolling trait. In order to dissect complex polygenic traits like yield a reductionist approach is successful where the complex trait is partitioned into several component traits28. Drought tolerance is also a complex trait whose inheritance is hard to follow. Hence, we have studied a component trait viz., leaf rolling for quantifying drought tolerance. Bi-parental RIL populations are best suited for studying polygenic traits which when combined with genome-wide molecular markers make pin pointing QTLs more efficient in a polyploid crop like wheat. In the current study, 35 K Axiom Wheat Breeder’s SNP Array and microsatellite markers were utilized for mapping of QTLs for flag leaf rolling. A genetic map was constructed using 3661 markers distributed across 21 wheat chromosomes. The 35 K wheat array provided sufficient number of polymorphic markers to construct genetic map and to identify potential putative QTLs. QTL mapping in the RIL population identified 12 QTLs for leaf rolling trait under drought condition in three consecutive years. These QTLs were associated with chromosomes 1B, 2A, 2B, 2D, 3A, 4A, 4B, 5D and 6B. Interestingly, a stable QTL Qlr.nhv-5D.2 was identified on 5D chromosome with a common marker interval (AX-94892575–AX-95124447) indicating that the putative genes related to leaf rolling/drought tolerance are residing in this interval.
In the physical interval 5D:338665301–5D:410952987, 14 putative genes related to drought tolerance were identified. In-silico gene expression analysis showed varied expression of these genes under three treatments viz., no stress control, drought stress and PEG6000. Four genes viz., TraesCS5D02G238300, TraesCS5D02G268000, TraesCS5D02G240900 and TraesCS5D02G249800 displayed equal expression across treatments which ruled out their possibility to be candidate genes. The lack of expression of two genes viz., TraesCS5D02G293300 and TraesCS5D02G294500 in all three treatments also ruled out their possibility to be candidate genes. Other two genes TraesCS5D02G284100 and TraesCS5D02G309500 can also be ruled out since they showed similar expression in no stress control and drought stress while slight variation under PEG control. Hence, out of 14 genes only six are qualified to be the potential candidate genes since they showed differential expression under drought and no stress conditions. Out of these six genes, two genes TraesCS5D02G240200 and TraesCS5D02G256400 are up-regulated under drought stress. The first gene, a Potassium transporter gene (TraesCS5D02G240200) is reported to be a positive regulator (OsHAK1-High Affinity Potassium Transporter) of drought tolerance in rice29. OsHAK1 overexpression plants were shown to be yielding 35% more than wild type plants under water stress conditions. The second gene TraesCS5D02G256400 is the Peroxidase gene. Under abiotic stress the principal defensive mechanism at cell level exhibited by plants is an increase in the reactive oxygen species (ROS) viz., hydroxyl radical, singlet oxygen, superoxide and hydrogen peroxide (H2O2)30. Excessive production of ROS has its side effects leading to protein degradation, enzyme inhibition, DNA and RNA damage, cellular damage and finally cell death. This makes ROS detoxification important for protecting plant cell from the toxic impact of ROS. It is achieved by enzymatic and non-enzymatic antioxidant elements. One of the enzymatic antioxidants is peroxidase. In a study involving wild relatives of wheat, the expression of peroxidases was found to be higher under drought stress31. These evidences prove that both the genes are actively involved in drought tolerance and positively regulates the plant’s response to drought stress.
The four genes viz., TraesCS5D02G248600, TraesCS5D02G252700, TraesCS5D02G308500 and TraesCS5D02G316200 were shown to be down-regulated under drought stress and PEG treatments. The gene TraesCS5D02G248600 is shown to have Glycosyltransferase activity. Uridine diphosphate (UDP)-glycosyltransferases (UGTs) are group of enzymes which are involved in transferring sugar moieties onto an array of small molecules and regulate various metabolic pathways32. Studies in Arabidopsis showed that glycosyltransferase genes viz., UGT79B2 and UGT79B332 and AtUGT76C233 were induced by abiotic stresses like cold, salt and drought stresses. The two genes TraesCS5D02G252700 and TraesCS5D02G308500 identified in our study were histone related genes, Histone H2A and Histone H4 variant TH011, respectively. This clearly points to the chromatin changes occurring under drought stress conditions34. An abiotic stress responsive H2A variant gene TaH2A.7 was reported in wheat which enhanced drought tolerance and promotes stomatal closure when overexpressed in Arabidopsis while it had no effect on the response to saline, osmotic and oxidative stresses35. Histone acetylation is reported to be correlated with drought stress and ABA responses in plants34. In rice, induction of HAT (Histone acetyltransferase) genes enhanced acetylation of histone molecules including H4 under drought stress conditions36. TraesCS5D02G316200 is having 3-ketoacyl-CoA synthase activity. 3-ketoacyl-CoA synthase (KCS) genes are involved in the biosynthesis of cuticular wax which forms the first protective layer of plants37. Hence, cuticular wax plays a major role in abiotic and biotic stress tolerance. It is reported that the total amount of wax per unit leaf area increased by 80% in plants under water stress than control. In wheat, glaucous genotypes have a significantly higher yield than non-glaucous in normal and moderate drought environments38. A similar positive correlation has been observed in barley39. OsWSL1 is one of the 3-ketoacyl-CoA synthase (KCS) genes in rice which is related to drought tolerance since Oswsl1 mutant exhibited drought sensitivity40. AtCER6 is another KCS gene characterized in Arabidopsis which when transformed in tomato (transgenic) lines showed significant increase in water use efficiency (WUE) and enhanced drought tolerance as compared to wild type control41. Even though these genes are associated with drought tolerance, none of them are directly linked to leaf rolling. Hence we have compared reported leaf rolling genes in rice and maize with the wheat genome sequence.
Rolled leaf trait is well studied in rice where it is one of the breeding objectives for genetic improvement. This trait provides additional benefits like reduced water loss by transpiration, thus imparting drought tolerance. In rice, around 70 genes/QTLs for rolled leaf trait have been reported22, 23. Some of the cloned genes are YABBY142, COW1/NAL743, 44, OsHB1, OsHB3 and OsHB545, ADL146, SSL1/RL947, NRL148, ACL1 and ACL249, LC250, Roc551, SRL152, RL1453, OsZHD1 and OsZHD254, REL155, REL256, OsARVL457, LRRK158. We have utilized 17 of these genes for comparative studies. Comparative homology modeling and the 3D protein structure superimposition of leaf rolling rice genes with wheat revealed five important genes in 5D chromosome of wheat genome. These five genes exhibited more than 80% identity with four reported genes in rice viz., NRL1, OsZHD1, Roc5, and OsHB3. Out of the five genes, only TraesCS5D02G253100 (5D: 359474816–359475706) falls exactly within the QTL interval region of Qlr.nhv-5D.2 (5D:338665301-5D:410952987). TraesCS5D02G253100 has 96.9% structure similarity to OsZHD1, which is zinc finger homeodomain class homeobox transcription factor (TF). Studies in most of the cloned leaf rolling genes in rice, the involvement of bulliform cells in regulation of rolling has been confirmed. For instance, the expression of the genes viz., Roc5 and OsZHD1 regulate the number, size and/or arrangement of bulliform cells leading to adaxial/abaxial rolling of leaf15 while OsHB3 controlled leaf rolling by affecting leaf polarity46. Having close structural similarity to OsZHD1, TraesCS5D02G253100 might be responsible for increasing the no. of bulliform cells leading to abadixal leaf rolling in wheat54. Similarly, other identified genes viz. TraesCS5D02G052300, TraesCS5D02G385300, TraesCS5D02G320600 and TraesCS5D02G102600 might have the same function as of their respective genes in rice OsHB3, ROC5 and NRL1. In-silico expression profiling patterns of the five genes were predicted under drought stress as compared to no stress control and PEG treatment (Fig. 4b). Expression of TraesCS5D02G253100 was reduced under drought stress as compared to both the controls. While the expression of TraesCS5D02G052300 decreased under drought stress and PEG treatment. Under drought stress expression of TraesCS5D02G385300 was higher than the no stress control. The gene TraesCS5D02G102600 was not at all expressed under drought stress and PEG treatment. Hence, there is differential expression of these four genes under drought and no stress conditions. Whereas, in case of TraesCS5D02G320600 expression was similar in all the treatments. Since TraesCS5D02G253100 (Zn finger homeodomain class homeobox TF) shows > 95% structure similarity to OsZHD1, falls exactly in the QTL interval Qlr.nhv-5D.2 and shows differential expression under drought stress and no stress condition, it could be the best candidate gene for leaf rolling which needs further functional validation across diverse genotypes.
Conclusion
In the present study, a linkage map of RIL population from the cross NI5439 × HD2012 spanning 22,275.01 cM using 3589 SNPs and 72 SSR was constructed. This linkage map was utilized to identify QTLs for leaf rolling under moisture stress condition. Twelve QTLs were detected on chromosomes viz., 1B, 2A, 2B, 2D, 3A, 4A, 4B, 5D and 6B. A stable QTL Qlr.nhv-5D.2 was identified on 5D chromosome. Marker assisted breeding can be employed for transfer of this QTL to improve drought tolerance in wheat cultivars. Six best putative candidate genes associated with drought tolerance were identified within the QTL interval on 5D. Moreover, five genes in wheat showing more than 80% identity with four reported leaf rolling genes in rice viz., NRL1, OsZHD1, Roc5, and OsHB3 were also identified. Out of these, TraesCS5D02G253100 coding for Zn finger homeodomain class homeobox transcription factor located within the QTL interval Qlr.nhv-5D.2 could be the best candidate gene for leaf rolling. Further investigation can validate the exact gene for map-based cloning, understanding the molecular basis of leaf rolling and utilization for improvement of drought tolerance in wheat.
Material and methods
Plant material
Mapping population consisted of 92 F2:16 RILs developed from cross between genotypes NI5439 and HD2012 by single spike descent method59. NI5439 is a variety released for cultivation in the peninsular region of India with parentage REMP 80/3* NP 710. HD2012 is a variety developed from the cross HD1467/HB208.
Experimental design and phenotyping
The experiment was conducted during 2017–2018, 2018–2019 and 2019–2020 growing seasons at Indian Agriculture Research Institute, New Delhi (28.6377° N, 77.1571° E). Ninety-two RILs along with parental lines were analyzed in a randomized complete block design (RCBD) in two replications. The experimental unit consisted of a four-row plot of 1 m length and 25 cm row to row distance. The RILs were given irrigation after 21 days of sowing in order to avoid any crop failure, after which no irrigation was provided. Phenotyping of flag leaf rolling was visually scored after the flowering stage on the scale from 1 (no leaf rolling) to 5 (complete leaf rolling) (Fig. 2a,b). The three environments i.e. 2017–2018, 2018–2019 and 2019–2020 hereafter will be mentioned as E17, E18 and E19, respectively.
Statistical analysis
Descriptive statistical parameters analysis and correlation studies were conducted using Statistica 13.0 software package (Stat-Soft, Inc., USA, https://www.statsoft.com). ANOVA and Heritability of the trait was calculated using the AOV functionality of QTLIcimapping version 4.2. The following model was used in ANOVA (1) and estimation of heritability per mean (2).
| 1 |
where the number of genotypes is equal to g, and the number of blocks is equal to r. Assuming yjk is the oberservation of the ith genotype in the kth block
| 2 |
For QTL mapping Best linear unbiased estimates (BLUE) were calculated using QTL IciMapping software v4.260.
Genotyping and linkage map construction
High quality genomic DNA of parental genotypes and RILs was isolated using CTAB method61. Qualitative and quantitative check was done using 0.8% agarose gel and Nanodrop spectrophotometer. For SSR markers, PCR was performed in reaction volume of 10 μl comprising 1 μl of each primer (5 pmol/μl), 2 μl of genomic DNA (25 ng) and 2 μl of nuclease free water added to 4 μl of reaction mix (Go Green Taq Promega) in 96-well PCR plates with thermal seal in Eppendorf thermal cycler with a thermal profile of initial denaturation step of 94.0 °C for 4 min, followed by 45 cycles of 94.0 °C for 1 min (denaturation), 50–60 °C (annealing temperature depending on primer) for 1 min, 72.0 °C for 1 min (primer extension) and a final extension of 72.0 °C for 10 min and storage at 4.0 °C. The SSRs found to be polymorphic between parents were used for genotyping of 92 RILs. The amplified PCR products of SSR markers were subsequently resolved on 3.5% MetaPhorTM (Lonza) gel in1X TBE buffer. The gel stained in ethidium bromide was visualized under UV-trans-illuminator in a gel documentation system (Syngene G: Box Gel Documentation System).
SNP Genotyping was conducted using Axiom 35 K Breeder’s SNP Array62 by Imperial Life Sciences (Gurgaon, India). Genotyping calls for 35,143 SNP markers were obtained, which were filtered in a series of steps. Only those SNPs which showed polymorphism between two parents were utilized for linkage map construction. Out of 7601 polymorphic SNP markers, SNPs lacking any chromosome ID and position were removed. Polymorphic markers having more than 10 missing values for the RILs were also filtered out. For construction of genetic map, only the polymorphic markers with minor allele frequency of more than 0.3 were considered. These markers were tested for significant segregation distortion using Chi-square test. In addition to the selected 4287 SNP markers, 139 polymorphic SSR markers were also used in development of linkage map.
Before linkage map construction, BIN tool algorithm implemented in QTL IciMapping software v4.260 was used for binning of markers having identical segregation patterns. A total of 3661 (3589 SNP + 72 SSR) markers were finally utilized for linkage map construction. Twenty one linkage groups (LGs) were determined based on LOD (logarithm of odds) threshold value greater than 3. The ordering of 3661 markers distributed over 21 chromosomes was performed using SER (SERiation) algorithm implemented in QTL IciMapping software v4.263. Kosambi mapping function was used for conversion of recombination frequencies between markers to centiMorgan. The R package LinkageMapView was utilized to graphically display linkage map64.
QTL mapping
QTL mapping was performed using BIP functionality in QTL IciMapping v 4.2 (https://www.isbreeding.net/software/)60 employing the Inclusive Composite Interval Mapping of Additive function (ICIM-ADD). The following parameters were used: scanning step was set at 1.00 cM, p value was set at 0.001, minimum 2.50 LOD threshold was set to declare significant QTLs. QTLs detected for leaf rolling were designated according to standard nomenclature65.
Candidate gene prediction
QTL intervals obtained in linkage map were further studied for prediction of candidate gene(s) associated with the respective QTLs using wheat sequence (International Wheat Genome Sequencing Consortium, 2018) available at Ensembl Plants (https://plants.ensembl.org/Triticum_aestivum/Info/Index). Marker intervals were mapped on for their physical locations and sequence between the interval was retrieved using BLAST, homology was selected on basis of E value = 1E−100 and 100% identity. The number and kind of genes present in the sequence were obtained using BioMart tool.
Homology modeling
Amino acid sequence of already reported genes for leaf rolling in rice and maize were retrieved using RAP-DB (Rice Annotation Project Database; https://rapdb.dna.affrc.go.jp/) and MaizeGDB (Maize Genetics and Genomics database; https://www.maizegdb.org/). Putative wheat genes responsible for leaf rolling present within the QTL interval region were identified by BLASTP search using these sequences against the fully annotated latest release of reference genome (International Wheat Genome Sequencing Consortium, 2018) using Ensembl Plants (https://plants.ensembl.org/Triticum_aestivum/Info/Index). Sequences having E value below zero and identity greater than 80% were selected. The identified wheat genes were then analyzed on ProtParam tool of Expasy website (https://web.expasy.org/protparam/) for the various physiochemical properties. BUSCA (https://busca.biocomp.unibo.it/) was used for predicting the subcellular localization of predicted leaf rolling wheat genes. Protein 3D structure of the wheat genes was predicted using Swiss-Model Server (https://swissmodel.expasy.org/). In order to check significant similarity between the predicted genes and their corresponding genes in rice, the predicted genes were superimposed based on statistical and probability scores and root-square deviation (RMSD) using FATCAT tool (flexible structure alignment by changing fragment pairs allowing twists)66. To perform the expression profiling patterns of the identified leaf rolling genes under drought stress RNA-seq expression data was accessed through Wheat expression Browser (https://www.wheat-expression.com/) powered by expression Visualization and Integration Platform (expVIP)67, 68.
Supplementary information
Acknowledgements
The authors acknowledge ICAR sponsored project “Incentivizing research in agriculture” for funding and ICAR-Indian Agricultural Research Institute, New Delhi for providing infrastructure facilities.
Author contributions
S.K., N.M. and V. conceived and designed the experiments; A.V. performed the experiments; A.V., M.N and P.A. analyzed the data and prepared figures; M.N. and A.V. wrote the paper. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Aakriti Verma, M. Niranjana, S. K. Jha and Niharika Mallick.
Supplementary information
is available for this paper at 10.1038/s41598-020-75703-4.
References
- 1.Qaseem MF, Qureshi R, Shaheen H. Effects of pre-Anthesis drought, heat and their Combination on the growth, yield and physiology of diverse wheat (Triticum aestivum L.) genotypes varying in sensitivity to heat and drought stress. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-43477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FAOSTAT. https://www.fao.org/faostat/en/.
- 3.Daryanto S, Wang L, Jacinthe PA. Global synthesis of drought effects on maize and wheat production. PLoS One. 2016;11:e0156362. doi: 10.1371/journal.pone.0156362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta P, Balyan H, Gahlaut V. QTL analysis for drought tolerance in wheat: Present status and future possibilities. Agronomy. 2017;7:5. doi: 10.3390/agronomy7010005. [DOI] [Google Scholar]
- 5.Wheaton E, Kulshreshtha S, Wittrock V, Koshida G. Dry times: Hard lessons from the Canadian drought of 2001 and 2002. Can. Geogr. 2008;52:241–262. doi: 10.1111/j.1541-0064.2008.00211.x. [DOI] [Google Scholar]
- 6.Epule ET, Peng C, Lepage L, Chen Z. The causes, effects and challenges of sahelian droughts: A critical review. Reg. Environ. Change. 2014;14:145–156. doi: 10.1007/s10113-013-0473-z. [DOI] [Google Scholar]
- 7.Lamaoui M, Jemo M, Datla R, Bekkaoui F. Heat and drought stresses in crops and approaches for their mitigation. Front. Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipiec J, Doussan C, Nosalewicz A, Kondracka K. Effect of drought and heat stresses on plant growth and yield: A review. Int. Agrophys. 2013;27:463–477. doi: 10.2478/intag-2013-0017. [DOI] [Google Scholar]
- 9.Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005;16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sicher RC, Timlin D, Bailey B. Responses of growth and primary metabolism of water-stressed barley roots to rehydration. J. Plant Physiol. 2012;169:686–695. doi: 10.1016/j.jplph.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Goufo P, et al. Cowpea (Vigna unguiculata L. Walp.) metabolomics: Osmoprotection as a physiological strategy for drought stress resistance and improved yield. Front. Plant Sci. 2017;8:586. doi: 10.3389/fpls.2017.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blum A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant. Cell Environ. 2017;40:4–10. doi: 10.1111/pce.12800. [DOI] [PubMed] [Google Scholar]
- 13.Sirault XRR, Condon AG, Wood JT, Farquhar GD, Rebetzke GJ. ‘Rolled-upness’: Phenotyping leaf rolling in cereals using computer vision and functional data analysis approaches. Plant Methods. 2015;11(1):1–11. doi: 10.1186/s13007-015-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sariyeva G, Kenjebaeva S, Lichtenthaler HK. Photosynthesis performance of non-rolling and flag leaf rolling wheat genotypes during temperature stress. Acta Bot. Hung. 2009;51:185–194. doi: 10.1556/ABot.51.2009.1-2.18. [DOI] [Google Scholar]
- 15.Gao L, et al. Fine mapping and candidate gene analysis of a QTL associated with leaf rolling index on chromosome 4 of maize (Zea mays L.) Theor. Appl. Genet. 2019;132:3047–3062. doi: 10.1007/s00122-019-03405-1. [DOI] [PubMed] [Google Scholar]
- 16.Duan L, et al. Novel digital features discriminate between drought resistant and drought sensitive rice under controlled and field conditions. Front. Plant Sci. 2018;9:492. doi: 10.3389/fpls.2018.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cal AJ, et al. Leaf morphology, rather than plant water status, underlies genetic variation of rice leaf rolling under drought. Plant Cell Environ. 2019;42:1532–1544. doi: 10.1111/pce.13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baret F, et al. Leaf-rolling in maize crops: From leaf scoring to canopy-level measurements for phenotyping. J. Exp. Bot. 2018;69(10):2705–2716. doi: 10.1093/jxb/ery071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson JM, Lane B, Freeling M. Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development. 2002;129:4581–4589. doi: 10.1242/dev.129.19.4581. [DOI] [PubMed] [Google Scholar]
- 20.Xu, D. Gene mapping for roll-leaf-above-ear and the other plant-type and ear traits in maize (Zea mays L.). Dissertation, Southwest University.(2008)
- 21.Meng Q, Yuan J, Chen Y, Zhao W. Discovery and characteristics of a novel maize mutant with rolled leaves. J. Maize Sci. 2011;19(3):79–81. [Google Scholar]
- 22.Liu X, et al. Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 2016;67:2139–2150. doi: 10.1093/jxb/erw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiang Z, et al. Joint mapping and allele mining of the rolled leaf trait in rice (Oryza sativa L) PLoS One. 2016;11:e0158246. doi: 10.1371/journal.pone.0158246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peleg Z, et al. Genomic dissection of drought resistance in durum wheat × wild emmer wheat recombinant inbreed line population. Plant Cell Environ. 2009;32:758–779. doi: 10.1111/j.1365-3040.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- 25.Myşków B, Góralska M, Lenarczyk N, Czyczyło-Mysza I, Stojałowski S. Putative candidate genes responsible for leaf rolling in rye (Secale cereale L.) BMC Genet. 2018;19:57. doi: 10.1186/s12863-018-0665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appels R, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:6403. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 27.Sirault X. R. R., Condon, A. G., Rebetzke, G. J., & Farquhar G. D. Genetic analysis of leaf rolling in wheat. (2008). https://ses.library.usyd.edu.au/handle/2123/3213. [DOI] [PMC free article] [PubMed]
- 28.Brinton J, Uauy C. A reductionist approach to dissecting grain weight and yield in wheat. Special Issue: Barley and wheat biology. J. Integr. Plant Biol. 2019;61(3):337–358. doi: 10.1111/jipb.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G, et al. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 2017;8:1885. doi: 10.3389/fpls.2017.01885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hossain MS, Dietz KJ. Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front. Plant Sci. 2016;7:548. doi: 10.3389/fpls.2016.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehrabad Pour-Benab S, Fabriki-Ourang S, Mehrabi AA. Expression of dehydrin and antioxidant genes and enzymatic antioxidant defense under drought stress in wild relatives of wheat. Biotechnol. Biotechnol. Equip. 2019;33:1063–1073. doi: 10.1080/13102818.2019.1638300. [DOI] [Google Scholar]
- 32.Li P, et al. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017;89:85–103. doi: 10.1111/tpj.13324. [DOI] [PubMed] [Google Scholar]
- 33.Li YJ, Wang B, Dong RR, Hou BK. AtUGT76C2, an Arabidopsis cytokinin glycosyltransferase is involved in drought stress adaptation. Plant Sci. 2015;236:157–167. doi: 10.1016/j.plantsci.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Kim JM, Sasaki T, Ueda M, Sako K, Seki M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015;6:114. doi: 10.3389/fpls.2015.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu W, Li Y, Cheng Z, Xia G, Wang M. A wheat histone variant gene TaH2A.7 enhances drought tolerance and promotes stomatal closure in Arabidopsis. Plant Cell Rep. 2016;35:1853–1862. doi: 10.1007/s00299-016-1999-6. [DOI] [PubMed] [Google Scholar]
- 36.Fang H, Liu X, Thorn G, Duan J, Tian L. Expression analysis of histone acetyltransferases in rice under drought stress. Biochem. Biophys. Res. Commun. 2014;443:400–405. doi: 10.1016/j.bbrc.2013.11.102. [DOI] [PubMed] [Google Scholar]
- 37.Xue D, Zhang X, Lu X, Chen G, Chen ZH. Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 2017;8:621. doi: 10.3389/fpls.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson DA, Richards RA, Turner NC. Yield, water relations, gas exchange, and surface reflectances of near-isogenic wheat lines differing in glaucousness 1. Crop Sci. 1983;23:318–325. doi: 10.2135/cropsci1983.0011183X002300020033x. [DOI] [Google Scholar]
- 39.González A, Ayerbe L. Effect of terminal water stress on leaf epicuticular wax load, residual transpiration and grain yield in barley. Euphytica. 2010;172:341–349. doi: 10.1007/s10681-009-0027-0. [DOI] [Google Scholar]
- 40.Yu D, et al. Wax Crystal-Sparse Leaf1 encodes a-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Springer. 2008;228:675–685. doi: 10.1007/s00425-008-0770-9. [DOI] [PubMed] [Google Scholar]
- 41.Karaba, A. Improvement of water use efficiency in rice and tomato using Arabidopsis wax biosynthetic genes and transcription factors. PhD thesis Wageningen University (2007). https://library.wur.nl/WebQuery/wurpubs/fulltext/121877.
- 42.Dai M, et al. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 2007;144:121–133. doi: 10.1104/pp.107.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo YM, et al. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007;65:125–136. doi: 10.1007/s11103-007-9203-6. [DOI] [PubMed] [Google Scholar]
- 44.Fujino K, et al. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008;279:499–507. doi: 10.1007/s00438-008-0328-3. [DOI] [PubMed] [Google Scholar]
- 45.Itoh JI, Hibara KI, Sato Y, Nagato Y. Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008;147:1960–1975. doi: 10.1104/pp.108.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hibara K, et al. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 2009;334:345–354. doi: 10.1016/j.ydbio.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 47.Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. 2009;21:719–735. doi: 10.1105/tpc.108.061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, et al. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010;73:283–292. doi: 10.1007/s11103-010-9614-7. [DOI] [PubMed] [Google Scholar]
- 49.Li L, et al. Overexpression of ACL1 (abaxially curled leaf 1) increased Bulliform cells and induced Abaxial curling of leaf blades in rice. Mol. Plant. 2010;3:807–817. doi: 10.1093/mp/ssq022. [DOI] [PubMed] [Google Scholar]
- 50.Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 2010;20:935–947. doi: 10.1038/cr.2010.109. [DOI] [PubMed] [Google Scholar]
- 51.Zou L, et al. Leaf rolling controlled by the homeodomain leucine zipper class IV gene roc5 in rice. Plant Physiol. 2011;156:1589–1602. doi: 10.1104/pp.111.176016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang JJ, Zhang GH, Qian Q, Xue HW. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012;159:1488–1500. doi: 10.1104/pp.112.199968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang L, et al. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 2012;10:524–532. doi: 10.1111/j.1467-7652.2012.00679.x. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, et al. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta. 2014;239:803–816. doi: 10.1007/s00425-013-2009-7. [DOI] [PubMed] [Google Scholar]
- 55.Chen Q, et al. Characterization of rolled and erect leaf 1 in regulating leave morphology in rice. J. Exp. Bot. 2015;66:6047–6058. doi: 10.1093/jxb/erv319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang SQ, et al. REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice. 2016;9:37. doi: 10.1186/s12284-016-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, et al. Characterization and fine mapping of the rice gene OsARVL4 regulating leaf morphology and leaf vein development. Plant Growth Regul. 2016;78:345–356. doi: 10.1007/s10725-015-0097-z. [DOI] [Google Scholar]
- 58.Zhou Y, et al. LRRK1, a receptor-like cytoplasmic kinase, regulates leaf rolling through modulating bulliform cell development in rice. Mol. Breed. 2018;38:48. doi: 10.1007/s11032-018-0811-4. [DOI] [Google Scholar]
- 59.Tomar RS, et al. Development of mapping populations and their characterization for drought tolerance in wheat. Indian J. Genet. Plant Breed. 2012;72:195–207. [Google Scholar]
- 60.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3:269–283. doi: 10.1016/j.cj.2015.01.001. [DOI] [Google Scholar]
- 61.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen AM, et al. Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum) Plant Biotechnol. J. 2017;15:390–401. doi: 10.1111/pbi.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buetow KH, Chakravarti A. Multipoint gene mapping using seriation. I. General methods. Am. J. Hum. Genet. 1987;41:20. [PMC free article] [PubMed] [Google Scholar]
- 64.Ouellette LA, Reid RW, Blanchard SG, Brouwer CR. LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics. 2018;34(2):306–307. doi: 10.1093/bioinformatics/btx576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McIntosh R. A., Yamazaki Y., Dubcovsky J., Rogers J.,Morris C., Appels R. & Xia X. C. Catalogue of gene symbols for wheat. In 12th International Wheat Genetics Symposium, 8–13 September,Yokohama, Japan (2013). https://wheat.pw.usda.gov/GG2/Triticum/wgc/2013/.
- 66.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 67.Kumar A, et al. Genome-wide identification, characterization, and expression profiling of SPX gene family in wheat. Int. J. Biol. Macromol. 2019;140:17–32. doi: 10.1016/j.ijbiomac.2019.08.105. [DOI] [PubMed] [Google Scholar]
- 68.Borrill P, Ramirez-Gonzalez R, Uauy C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016;170:2172–2186. doi: 10.1104/pp.15.01667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.