Abstract
SARS-CoV-2 infection shows a wide-ranging clinical severity, requiring prognostic markers. We focused on S100B, a calcium-binding protein present in biological fluids, being a reliable biomarker in disorders having inflammatory processes as common basis and RAGE as main receptor. Since Covid-19 is characterized by a potent inflammatory response also involving RAGE, we tested if S100B serum levels were related to disease severity. Serum samples (n = 74) were collected from hospitalized SARS-CoV-2 positive patients admitted to Covid center. Illness severity was established by admission clinical criteria and Covid risk score. Treatment protocols followed WHO guidelines available at the time. Circulating S100B was determined by ELISA assay. Statistical analysis used Pearson’s χ2 test, t-Test, and ANOVA, ANCOVA, Linear Regression. S100B was detected in serum from Covid-19 patients, significantly correlating with disease severity as shown both by the level of intensity of care (p < 0.006) as well by the value of Covid score (Multiple R-squared: 0.3751); the correlation between Covid-Score and S100B was 0.61 (p < 0.01). S100B concentration was associated with inflammation markers (Ferritin, C-Reactive Protein, Procalcitonin), and organ damage markers (Alanine Aminotransferase, Creatinine). Serum S100B plays a role in Covid-19 and can represent a marker of clinical severity in Sars-CoV-2 infected patients.
Subject terms: Biomarkers, Diseases, Medical research
Introduction
Evaluation of Covid-19 severity and possible outcomes is limited by clinical heterogeneity and lack of specific markers1–3. Several laboratory parameters are considered in clinical practice, but the identification of novel indicators in blood specimens is a key issue for understanding the underlying biological mechanisms and improving prognostic accuracy4–9.As a candidate marker we focused on the S100B protein, which is regarded to be involved in inflammatory processes as a Danger-Associated Molecular Patterns (DAMP) molecule10,11. S100B is a small acidic calcium-binding protein, originally isolated in the nervous system, where it is concentrated in astrocytes, being also present in oligodendrocytes, Schwann cells, enteric glial cells, and some neuron subpopulations12,13. It is also present in definite non-neural cell types, including dendritic cells, certain lymphocyte subpopulations, chondrocytes, Langerhans cells, melanocytes, adrenal medulla satellite cells, Leydig cells, skeletal muscle satellite cells, and adipocytes, which intriguingly constitute a site of concentration for the protein comparable to the nervous tissue10. S100B can be detected in biological fluids (cerebrospinal fluid, peripheral blood, urine, saliva, feces) in particular conditions10,12,14. Besides, the protein has been shown to be actively released and interact with target cells through the multiligand transmembrane immunoglobulin-like Receptor for Advanced Glycation Endproducts (RAGE) which is able to initiate an intracellular signaling cascade15 and was associated to several pathological conditions, reasonably referred to inflammatory processes12. Based on these findings, S100B is considered a reliable biomarker for a variety of neural and non-neural pathological conditions, even displaying a predictive role14–17.
Indeed, S100B appears to share similar characteristics to DAMPs molecules, including the interaction with RAGE and, once released in the microenvironment, the active participation to the inflammatory tissue reaction to damage10,18. Interestingly, the S100B-RAGE axes has been shown to participate in pulmonary inflammatory processes19,20.
Thus, in the light of the notion that SARS CoV-2 infection can induce a severe acute respiratory syndrome with a complex pattern of clinical manifestations characterized by a potent inflammatory response18,21–23 we tested the possibility that S100B could be present in detectable amounts in serum of Covid-19 patients, as well as the possible relationship between severity of the disease and increase in S100B serum levels.
Results
S100B is present in serum of Covid patients, correlating with disease severity
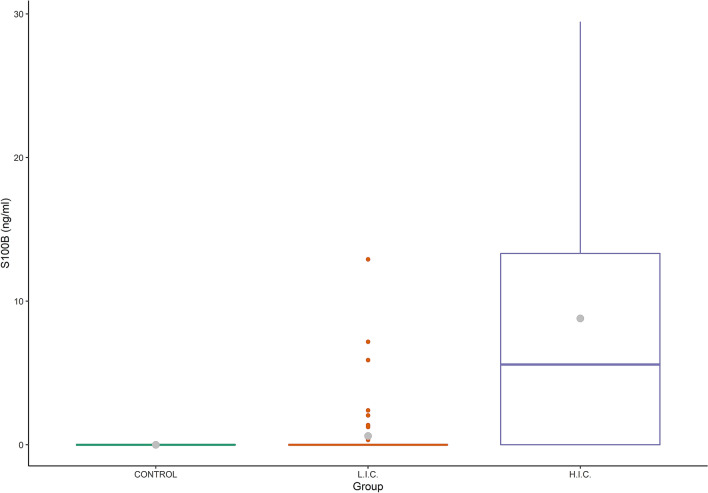
S100B was detected at concentrations over the LOD in the serum of 19 patients out of 74 (25.7%), ranging from 0.25 to 29.46 ng/mL. Results obtained by non-parametric Wilcox test showed a positive significant association (p < 0.001) between S100B serum concentrations and the severity of the disease as measured based on HIC or LIC wards where the Covid-19 patients were hospitalized. S100B levels showed a higher mean value in HIC than in LIC (8.80 ± 10.24 and 0.62 ± 2.10 ng/mL, respectively; p < 0.006), as reported in Table 1 and Fig. 1.
Table 1.
Overview of patients included in the study and their serum data.
| Participants | All (n = 74) | HICa (n = 19) | LICb (n = 55) | p value |
|---|---|---|---|---|
| Characteristics | ||||
| Median age (IQR)—years | 66 (32–89) | 63 (35–85) | 66 (32–89) | 0.66 |
| Female – number (%) | 25 (49) | 9 (64) | 16 (43) | |
| Period from hospitalization to blood sample collection—days (SD) | 18.0 ± 18.0 | 19.5 ± 17.8 | 17.6 ± 18.3 | 0.72 |
| Period from blood sample collection to hospital discharge—days (SD) | 13.2 ± 11.5 | 14.0 ± 11.8 | 13.0 ± 11.5 | 0.79 |
| Serum data | ||||
| S100B—ng/mL (SD) | 2.39 ± 6.04 | 8.80 ± 10.24 | 0.62 ± 2.10 | 0.006 |
| White Blood Cell count per mm3 (SD) | 7.21 ± 3.01 | 7.78 ± 4.56 | 7.05 ± 2.45 | 0.55 |
| Lymphocyte count per mm3 (SD) | 1.49 ± 0.73 | 1.41 ± 0.85 | 1.51 ± 0.69 | 0.67 |
| Alanine Aminotransferase—IU/L (SD) | 29.8 ± 26.2 | 39.4 ± 43.0 | 27.2 ± 18.9 | 0.28 |
| Creatinine—mg/dL (SD) | 0.96 ± 0.57 | 0.89 ± 0.45 | 0.98 ± 0.60 | 0.52 |
| d-Dimer—ng/mL (SD) | 583 ± 514 | 810 ± 585 | 520 ± 479 | 0.08 |
| Prothrombin—seconds (SD) | 13.7 ± 2.0 | 13.9 ± 1.9 | 13.6 ± 2.1 | 0.62 |
| Ferritin—mg/L (SD) | 782 ± 914 | 1212 ± 1387 | 663 ± 705 | 0.16 |
| Procalcitonin—ng/mL (SD) | 334 ± 583 | 586 ± 1192 | 265 ± 197 | 0.30 |
| C Reactive Protein—mg/dL (SD) | 6.00 ± 7.60 | 6.27 ± 8.34 | 5.92 ± 7.46 | 0.88 |
aHigh Intensity Care ward.
bLow Intensity Care ward.
Figure 1.
Detection of S100B in Covid patients and controls. Box plot showing the distribution of S100B in Covid (n = 74) patients with different clinical severity of disease. Also data from controls (n = 5 healthy individuals: negative for SARS-CoV-2 detection by PCR and negative by serologic test) are included. Grey dot: mean value, Line: median value; HIC: High Intensity Care; LIC: Low Intensity Care.
No statistically significant differences were observed for other variable and in particular among the groups for age, gender and number of days of hospitalization to the date of blood sampling. The Wilcox test showed no statistically significant differences between S100B levels and gender.
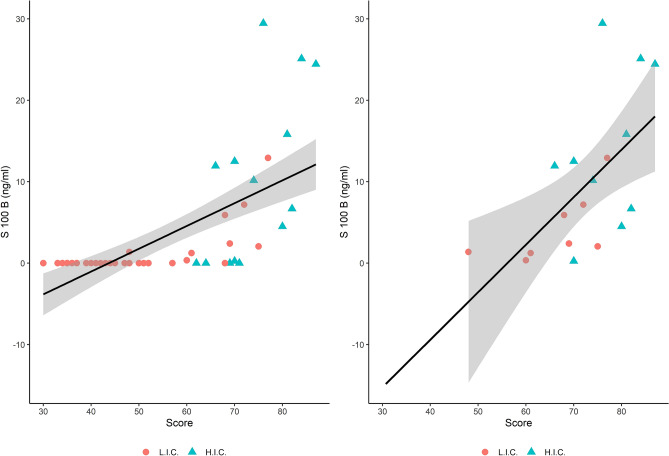
Finally, the number of patients positive for serum S100B was significantly higher in HIC (p < 0.01). This result was confirmed by both Welch-ANOVA model (p < 0.01) and Games-Howell test (p < 0.01). The incremental trend of S100B vs clinical severity measured by Covid-score was statistically significant. Figure 2 shows the distribution of S100B concentration and Covid Score in all patients (A) and the subgroup of patient where S100B was detected in the serum (B). Most patients admitted in LIC ward show a presence of S100B below the LOD, even if with different values of Covid Score (A). Additional information is reported in supplementary material (Figs. S1 and S2). When considering only those samples over the LOD, a hypothetical trendline was extrapolated (B), suggesting a theorical distribution. Both trendlines show significant correlations between S100B and the score for evaluating clinical risk (multiple R-squared 0.4369 and 0.3751, respectively), supporting a putative role of S100B as a marker for clinical severity.
Figure 2.
Relationship between Covid-Score and concentration of S100B. The scatterplot shows a positive correlation between S100B concentration (ng/mL) and clinical severity of the disease as represented by Covid-score. Analysis considering all samples (A) or only those with S100B detected in serum (B), from both HIC and LIC wards. Most of the samples with a concentration of S100B below the LOD belongs to the group hospitalized in the LIC wards (A). When considering only patients with S100B over the LOD and from both wards (B), the regression equation was Y = 0.584X−32.778. Figure A is reported as a comparison respect to figure B to highlight the distribution of S100B levels below the LOD and the independent linear regression curves are reported in supplementary materials (S1). The hospitalization ward is indicated for each patient (Red Dots: Low Intensity Care (LIC); Blue Triangles: High Intensive Care (HIC). The linear regression lines and their confidence intervals (95%) are showed by the gray areas. The correlation between Covid-Score and S100B is equal to 0.66 (p < 0.001) (A) and 0.61 (p < 0.01) (B).
S100B correlates with several blood markers
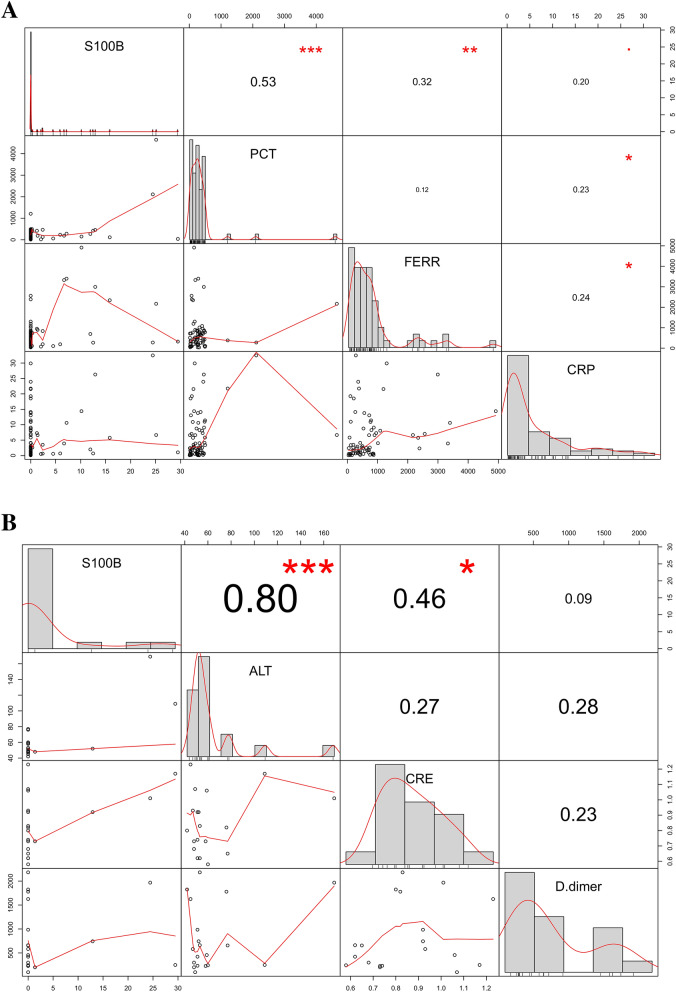
A positive and significant correlation was observed between S100B and Ferritin concentrations (p < 0.01, 74 Observations), as well as for other parameters (Table 2), and in particular PCT (p < 0.01), d-Dimer (p < 0.05) and CRP (p < 0.1). The ANCOVA model displayed the marginal effects of the CRP and the hospitalization ward (HIC as corner point) variables on the S100B levels, showing a positive and statistically significant relation (p < 0.05 and p < 0.01, respectively). Similar results were found for PCT (p < 0.01) and ALT (p < 0.01). In order to estimate the marginal effects of the measured parameters, a stepwise selection procedure was carried out, showing ALT and PCT variables as the best predictors of the S100B levels. A further linear regression was used to verify the single and interactive effects of CRP, ALT, PCT variables, showing a significant interaction with ALT and PCT (p < 0.001) and a correlation with CRP (Fig. 3). The whole of the results suggests that both S100B presence and levels correlate with the severity of the disease, the trend of CRP and Ferritin values and inflammatory status, as well as with other key parameters of Covid-19 severity, such as PCT, d-Dimer, ALT. Moreover, we observed a significant correlation between S100B and CRE levels (p < 0.01) in the subgroup of patients with high levels of ALT (> 40 IU/L, n = 19)18, thus suggesting the possibility of an independent association with a liver and/or kidney damage.
Table 2.
Correlation matrix: Pearson correlation coefficients and relative p values.
| S100Ba | WBCb | LYMc | ALTd | CREe | d-Dimerf | PTg | FERRh | PCTi | CRPj | agek | days beforel | days afterm | Covid scoren | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 74 | n = 74 | n = 74 | n = 74 | n = 74 | n = 74 | n = 74 | n = 69 | n = 74 | n = 74 | n = 74 | n = 74 | n = 71 | n = 56 | |
| S100Ba | 0.05 | 0.03 | 0.36 | 0.1 | 0.21 | − 0.06 | 0.32 | 0.53 | 0.2 | − 0.14 | − 0.07 | − 0.03 | 0.66 | |
| n = 74 | 0.7 | 0.805 | 0.002** | 0.402 | 0.073+ | 0.599 | 0.007** | 0.001*** | 0.082+ | 0.245 | 0.557 | 0.83 | 0.001*** | |
| WBCb | 0.05 | − | 0.13 | − 0.09 | 0.22 | 0.12 | 0.15 | 0.3 | 0.14 | 0.07 | 0.06 | − 0.12 | 0.07 | − 0.08 |
| n = 74 | 0.7 | − | 0.266 | 0.441 | 0.06+ | 0.324 | 0.188 | 0.011* | 0.223 | 0.542 | 0.595 | 0.303 | 0.54 | 0.579 |
| LYMc | 0.03 | 0.13 | − | − 0.17 | − 0.04 | − 0.21 | − 0.1 | − 0.15 | 0.04 | − 0.23 | − 0.2 | − 0.11 | − 0.28 | − 0.004 |
| n = 74 | 0.805 | 0.266 | − | 0.138 | 0.717 | 0.067+ | 0.385 | 0.209 | 0.723 | 0.046* | 0.096+ | 0.331 | 0.02* | 0.974 |
| ALTd | 0.36 | − 0.09 | − 0.17 | − | − 0.11 | 0.32 | − 0.09 | − 0.02 | 0.12 | 0.31 | − 0.23 | − 0.13 | 0.03 | 0.11 |
| n = 74 | 0.002** | 0.441 | 0.138 | − | 0.347 | 0.006** | 0.452 | 0.866 | 0.299 | 0.008** | 0.048* | 0.279 | 0.84 | 0.432 |
| CREe | 0.1 | 0.22 | − 0.04 | − 0.11 | − | − 0.03 | 0.21 | − 0.13 | 0.29 | − 0.05 | 0.2 | 0.02 | 0.03 | − 0.09 |
| n = 74 | 0.402 | 0.06+ | 0.717 | 0.347 | − | 0.822 | 0.073+ | 0.276 | 0.013* | 0.701 | 0.09+ | 0.895 | 0.83 | 0.506 |
| d− Dimerf | 0.21 | 0.12 | − 0.21 | 0.32 | − 0.03 | − | 0.08 | 0.32 | 0.15 | 0.26 | − 0.01 | − 0.15 | 0.02 | 0.21 |
| n = 74 | 0.073+ | 0.324 | 0.067+ | 0.006** | 0.822 | − | 0.509 | 0.008** | 0.197 | 0.026* | 0.921 | 0.19 | 0.89 | 0.117 |
| PTg | − 0.06 | 0.15 | − 0.1 | − 0.09 | 0.21 | 0.08 | − | 0.05 | − 0.03 | − 0.07 | 0.03 | 0.23 | 0.23 | − 0.07 |
| n = 74 | 0.599 | 0.188 | 0.385 | 0.452 | 0.073+ | 0.509 | − | 0.678 | 0.791 | 0.539 | 0.805 | 0.049* | 0.05* | 0.612 |
| FERRh | 0.32 | 0.3 | − 0.15 | − 0.02 | − 0.13 | 0.32 | 0.05 | – | 0.12 | 0.24 | 0.09 | − 0.1 | − 0.04 | 0.38 |
| n = 69 | 0.007** | 0.011* | 0.209 | 0.866 | 0.276 | 0.008** | 0.678 | – | 0.333 | 0.043* | 0.447 | 0.435 | 0.76 | 0.005*** |
| PCTi | 0.53 | 0.14 | 0.04 | 0.12 | 0.29 | 0.15 | − 0.03 | 0.12 | – | 0.23 | − 0.05 | − 0.16 | 0.02 | 0.28 |
| n = 74 | 0.000*** | 0.223 | 0.723 | 0.299 | 0.013* | 0.197 | 0.791 | 0.333 | – | 0.048* | 0.693 | 0.177 | 0.89 | 0.04* |
| CRPj | 0.2 | 0.07 | − 0.23 | 0.31 | − 0.05 | 0.26 | − 0.07 | 0.24 | 0.23 | – | 0.07 | − 0.05 | 0.15 | 0.04 |
| n = 74 | 0.082+ | 0.542 | 0.046* | 0.008*** | 0.701 | 0.026* | 0.539 | 0.043* | 0.048* | – | 0.538 | 0.661 | 0.2 | 0.766 |
| agek | − 0.14 | 0.06 | − 0.2 | − 0.23 | 0.2 | − 0.01 | 0.03 | 0.09 | − 0.05 | 0.07 | – | 0.26 | 0.24 | − 0.06 |
| n = 74 | 0.245 | 0.595 | 0.096+ | 0.048 | 0.09+ | 0.921 | 0.805 | 0.447 | 0.693 | 0.538 | – | 0.026* | 0.05+ | 0.63 |
| days beforel | − 0.07 | − 0.12 | − 0.11 | − 0.13 | 0.02 | − 0.15 | 0.23 | − 0.1 | − 0.16 | − 0.05 | 0.26 | – | 0.75 | 0.11 |
| n = 74 | 0.557 | 0.303 | − 0.2 | 0.279 | 0.895 | 0.19 | 0.049* | 0.435 | 0.177 | 0.661 | 0.026* | – | 0.001*** | 0.422 |
| days afterm | − 0.03 | 0.07 | 0.096 | 0.03 | 0.03 | 0.02 | 0.23 | − 0.04 | 0.02 | 0.15 | 0.24 | 0.75 | – | 0.05 |
| n = 71 | 0.83 | 0.54 | 0.02* | 0.84 | 0.83 | 0.89 | 0.05+ | 0.76 | 0.89 | 0.2 | 0.05+ | 0.001*** | – | 0.72 |
| Covid scoren | 0.66 | − 0.08 | − 0.004 | 0.11 | − 0.09 | 0.21 | − 0.07 | 0.38 | 0.28 | 0.04 | − 0.06 | 0.11 | 0.05 | – |
| n = 56 | 0.001*** | 0.579 | 0.974 | 0.432 | 0.506 | 0.117 | 0.612 | 0.005*** | 0.04* | 0.766 | 0.63 | 0.422 | 0.72 | – |
Statistically significant results are highlighted in bold.
n number of patients.
aS100B protein, ng/mL.
bWhite Blood Cell count per mm3.
cLymphocyte count per mm3.
dAlanine Aminotransferase—IU/L.
eCreatinine—mg/dL.
fd-Dimer—ng/mL.
gProthrombin—seconds.
hFerritin—mg/L.
iProcalcitonin—ng/mL.
jC Reactive Protein—mg/dL.
kPatients’ age—years.
lPeriod from hospitalization to blood sample collection—days.
mPeriod from blood sample collection to hospital discharge—days.
nCovid score—%.
+p < 0.1; *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 3.
Correlation of S100B with other blood markers. (A) Correlation of S100B versus inflammatory markers. S100B significantly correlates with PCT, FERR, CRP. Scatterplots of pairwise variable are shown. Values in the middle of boxes are referred to the Pearson Correlation. PCT: Procalcitonin; FERR: Ferritin, CRP: C-Reactive Protein. Red Stars and dots are referred to the p-values (***p < 0.0001; **p < 0.001; *p < 0.01; +p < 0.1). (B) Correlation of S100B versus Organ Damage markers. S100B significantly correlates with ALT and CRE (subgroup of patients showing ALT > 40 IU/L). Scatterplots of pairwise variable are shown. Values in the middle of boxes are referred to the Pearson Correlation. ALT: Alanine Aminotransferase; CRE: Creatinine; d-Dimer: d-Dimer. Values in the middle of boxes are referred to the Pearson Correlation. Red Stars and dots are referred to the p values (***p < 0.0001; **p < 0.001; *p < 0.01; +p < 0.1).
Discussion
In this study, we investigated the levels of circulating S100B in serum of patients affected by SARS-CoV-2 virus at various stages of the disease. S100B serum concentrations resulted correlated with the severity of the disease, as indicated by clinical/laboratory parameters. The major part of data at present available on the S100B protein as a biomarker and pathogenic factor deals with disorders primarily related to the nervous system whereas disorders primarily related to other systems having been essentially disregarded12: thus, present results appear to enlarge the field of investigation on this protein and its potentials in Covid-19 and, more in general, infectious diseases. Indeed, the discrete distribution of S100B in definite extra-neural cell types offers the basis for a functional/pathogenic role of the protein in extra-neural tissues, which at present has not been exhaustively investigated.
This study was performed during the epidemic peak, with the advantage of collecting homogeneous data from the same outbreak but with all the restrictions present while managing the emergency, including the limits in sample size and in the number of additional clinical or laboratory parameters to be assessed. However, the acquired data were stressed by an accurate statistical analysis, strongly reporting a role for S100B in Covid-19.
The increased levels of S100B are reasonably related to inflammatory processes, as also supported by its significant correlation with CRP, which is known to be a recognized inflammatory hallmark24. S100B is known to participate in inflammatory processes25,26 which are also known to be raised during SARS-CoV-2 disease19,21. Interestingly, in this respect we observed that S100B levels are correlated with indicators of distress involving non-neural districts, such as ALT, d-Dimer, PCT, suggesting, in this case, a wider and systemic valence for S100B as a putative biomarker.
The source of increased serum S100B in SARS-CoV-2 patients remains to be identified. Since information indicating a prevalent involvement of the nervous system in pathogenic processes of SARS-CoV-2 at present is lacking27, it seems unlikely that in this case the protein is primarily released from this tissue, which at present is regarded to be the natural source of the protein in biological fluids in the major part of pathological conditions already known. Among the cell types which are known to express and putatively release the protein, adipocytes, dendritic and lymphoid cell types28–31 appear to be putative sources for serum S100B in this disease. Adipocytes are known to secrete inflammatory cytokines which play a recognized role in crosstalk with the immune system, which at present are known to be especially relevant in processes leading to obesity32,33. Since the role(s) of molecules secreted by this intriguing cell type, including S100B, is at present largely unknown, this finding might add a novel element deserving interest. In the case of dendritic and lymphoid cell types, their role in inflammatory processes is widely known34,35, so that the mechanistic involvement of S100B in their function would merely increase the breadth of knowledge in this respect. It is interesting to note that, under pulmonary inflammation, S100B has been reported to be upregulated in bronchiolar epithelial cells and airway dendritic cells36,37. Moreover, as shown in alveolar cell types, S100B can stimulate the secretion of pro-inflammatory cytokines, that are commonly involved in lung inflammation, following a similar process suspected to be present also in Covid-1919–22,36,37.
Additional studies will be needed in order to define the source of serum S100B in patients affected by SARS-CoV-2, but the finding of increased serum levels of the protein, correlated with the gravity of the disease and inflammatory processes, offers a novel biomarker potentially useful to monitor the disease. In addition, in the light of growing evidence candidating S100B as an active factor in inflammatory processes12, the present findings may even propose the protein as a therapeutic target to counteract the potent inflammatory processes characterizing this infectious disease.
In conclusion, increased serum levels of S100B correlate with the severity of Covid-19 and inflammatory processes, offering a novel biomarker potentially beneficial in monitoring the disease course and prognosis. In the light of growing evidence candidating S100B as an active factor in inflammatory processes driven by DAMP and RAGE, the present findings propose this protein as a severity marker and its cellular pathways as candidate targets to unravel pathogenetic mechanisms and counteract the potent inflammatory processes characterizing SARS-CoV-2 infection.
Methods
Dataset
74 serum samples from patients with confirmed SARS-CoV-2 infection hospitalized in an academic Covid hospital in Rome, Italy, were collected during the epidemic pick (from January 29th to May 6th, 2020). The inclusion criteria were Covid-19 diagnosis and age (≥ 18 y.o.), while exclusion criteria were concomitant or pre-existing neurological disorders, cardiovascular diseases, diabetes and cancer. According to WHO clinical criteria at admission time, patients were hospitalized in High (HIC) and Low Intensity Care (LIC) wards, respectively. Severity of Covid-19 at the time of blood sampling was quantified using a Covid-score, attributing a value ranging from 0 to 100%4–7,9. Main variables incorporated in the Covid-score included: older age, male sex, comorbidities, respiratory rate, oxygenation, radiographic severity, higher neutrophils, higher CRP and lower albumin at presentation, predicted critical care admission and mortality; in particular: age > 50, male, oxygen saturation < 93%, radiological severity score > 3, neutrophil count > 8.0 × 109/L, CRP > 40 mg/L, albumin < 34 g/L, creatinine > 100 μmol/L, comorbidity and chronic lung disease, ALT > 40 IU/L; Creatinine > 100 μmol/L; D-dimer > 0.5 μg/L; Prothrombin-time > 16 s; Ferritin > 300 μg/L; Procalcitonin > 0.1 ng/mL.
Namely, patients recovered in HIC (n = 16) presented: severe pneumonia (fever or suspected respiratory infection, plus one of the following: respiratory rate > 30 breaths/min; severe respiratory distress or SpO2 ≤ 93% on room air); and LIC (n = 58) comprehended a group spanning from uncomplicated disease to pneumonia but without signs of severe pneumonia. In both groups, all patients received treatment in accordance with the guidelines available at the time of the study1. Time of hospitalization ranged from 7 to 85 days (average: 29.97). Table 1 shows patients’ characteristics together with data obtained on serum samples. Patients were balanced with respect to gender (female = 49%), their age ranging from 32 to 89 (median: 66.0). Serum samples from healthy subjects (n = 5) negative for SARS-CoV-2 detection by PCR and negative by serologic test were used as controls for the ELISA test.
Ethics
The project protocol involved the rapid recruitment of patient-participants during the pandemic pick and no additional project-related procedures (we used only material from routine venipunctures). Anonymity was assured by the hospital privacy protocol and written consent obtained by each of recruited participants. The study was approved by the Sant’Andrea University Hospital Ethical Committee/Institutional Review Board (N. 5773/2020).
Laboratory Tests
The serum S100B concentrations were measured by adapting the enzyme-linked immunosorbent assay (ELISA) kit following manufacturer protocols (S100B ELISA Kit, Abcam, England). Samples resulting below the detection limit of the test (LOD, equal to 0.1 ng/mL) were considered negative and assigned a value of 0 ng/mL. From the same serum, a panel of consolidated blood markers was also assessed, including White Blood Cell count (WBC), Lymphocytes (LYM), Alanine Aminotransferase (ALT), Creatinine (CRE), d-Dimer, Prothrombin (PT), Ferritin (FERR), Procalcitonin (PCT), C Reactive Protein (CRP).
Statistical analysis
Continuous variables were S100B serum concentration (ng/mL), WBC (count per mm3), LYM (count per mm3), ALT (IU/L), CRE (mg/dL), d-Dimer (ng/mL), PT (seconds), FERR (ng/mL), PCT (pg/mL), CRP (mg/dL), period from hospitalization to blood sample collection (days), period from sample collection to hospital discharge (days), severity of the disease (Covid score), age (years). Categorical variables were gender (male = 0, female = 1), S100B protein in detectable amount (below LOD = 0 and over LOD = 1), hospitalization ward (LIC = 0 and HIC = 1). Relations between continuous variables were compared by Pearson’s correlation. Pearson’s χ2 test was used to compare the frequencies of the categorical variables and non parametric test Wilcox or Kruskal were used to compare the two groups under the non-normality assumption. Linear models (ANOVA, ANCOVA, and Linear Regression) were performed to evaluate the effect of variables observed on concentration of S100B. Whenever the test of homogeneity of variance was not satisfied, the ANOVA results were substituted by those from Welch ANOVA. Tukey test were performed to evaluate the difference of means between groups. If equal variance assumption is violated during the ANOVA process, pairwise comparison was based on the Games-Howell statistics. Stepwise regression was performed to obtain the model with the best predictors. All analyses were considered statistically significant at a p value of less than 0.05, if not differently indicated. All data were analyzed using the R environment for statistical computing (Version 4.0.1).
Supplementary information
Acknowledgements
The authors thank Dr. Gianluca Gianfranceschi for the technical support and Tiziana Zilli for the library support; The study was partially supported by Nando-Elsa Peretti Foundation Project (grant assigned to F.M.; NaEPF 2019-041), by Fondazione per la Ricerca Scientifica Termale grant: “Development of innovative strategies for thermal water treatments: nanotechnologies & perspectives for hygiene” (CUP H81I18000070008) and by the Foundation for Biology and Regenerative Medicine, Tissue Engineering and Signaling T.E.S (PPP).
Author contributions
A.A., V.R.S. and F.M. designed the experiments. A.A., E.S., and M.S. collected clinical data. E.S., G.S., G.G., F.V. and M.S. performed laboratory analysis. L.M.M., M.O. and F.V. analysed data. A.A., M.O., G.D.S., R.D.L., F.R., P.P.P., M.V., V.R.S. and F.M. interpreted the results. M.O., M.V., V.R.S. and F.M. wrote the Article. M.V., V.R.S. and F.M. edited and revised the Article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Vincenzo Romano Spica and Fabrizio Michetti.
Supplementary information
is available for this paper at 10.1038/s41598-020-75618-0.
References
- 1.WHO. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected: interim guidance, 13 March 2020 [Online]. Available from: https://apps.who.int/iris/handle/10665/331446. Accessed 24 March 2002.
- 2.Guan W, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao, D. et al. A comparative study on the clinical features of Coronavirus 2019 (COVID-19) pneumonia with other pneumonias. Clin. Infect. Dis. 2020 Mar 12. 10.1093/cid/ciaa247 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 4.Galloway JB, Norton S, Barker RD, Brookes A, Carey I, et al. A clinical risk score to identify patients with COVID-19 at high risk of critical care admission or death: an observational cohort study. J. Infect. 2020;81:282–288. doi: 10.1016/j.jinf.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao L, et al. Development and validation of the HNC-LL score for predicting the severity of coronavirus disease 2019. EBioMedicine. 2020;57:102880. doi: 10.1016/j.ebiom.2020.102880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji, D. et al. Prediction for progression risk in patients with COVID-19 pneumonia: The CALL Score. Clin. Infect. Dis. 2020 Apr 9. 10.1093/cid/ciaa414 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 7.Bhargava, A. et al. Predictors for Severe COVID-19 Infection. Clin. Infect. Dis. 2020 May 30. 10.1093/cid/ciaa674 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 8.Liu R, et al. The value of urine biochemical parameters in the prediction of the severity of coronavirus disease 2019. Clin Chem Lab Med. 2020;58:1121–1124. doi: 10.1515/cclm-2020-0220. [DOI] [PubMed] [Google Scholar]
- 9.Wynants L, et al. Prediction models for diagnosis and prognosis of Covid-19 infection: systematic review and critical appraisal. Br Med J. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 11.Yang D, Han Z, Oppenheim JJ. Alarmins and immunity. Immunol Rev. 2017;280:41–56. doi: 10.1111/imr.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michetti F, et al. The S100B story: from biomarker to active factor in neural injury. J Neurochem. 2019;148:168–187. doi: 10.1111/jnc.14574. [DOI] [PubMed] [Google Scholar]
- 13.Lippi G, Cervellin G. Protein S100B: from cancer diagnostics to the evaluation of mild traumatic brain injury. Clin Chem Lab Med. 2016;54:703–705. doi: 10.1515/cclm-2016-0144. [DOI] [PubMed] [Google Scholar]
- 14.Michetti F, Massaro A, Russo G, Rigon G. The S-100 antigen in cerebrospinal fluid as a possible index of cell injury in the nervous system. J Neurol Sci. 1980;44:259–263. doi: 10.1016/0022-510X(80)90133-1. [DOI] [PubMed] [Google Scholar]
- 15.Kato K, Kimura S, Semba R, Suzuki F, Nakajima T. Increase in S-100 protein levels in blood plasma by epinephrine. J Biochem. 1983;94:1009–1011. doi: 10.1093/oxfordjournals.jbchem.a134397. [DOI] [PubMed] [Google Scholar]
- 16.Gazzolo D, et al. Increased urinary S100B protein as an early indicator of intraventricular hemorrhage in preterm infants: correlation with the grade of hemorrhage. Clin Chem. 2001;47:1836–1838. doi: 10.1093/clinchem/47.10.1836. [DOI] [PubMed] [Google Scholar]
- 17.Di Liddo R, et al. S100B as a new fecal biomarker of inflammatory bowel diseases. Eur Rev Med Pharmacol Sci. 2020;24:323–332. doi: 10.26355/eurrev_202001_19929. [DOI] [PubMed] [Google Scholar]
- 18.Cheng C, et al. Expression profiling of endogenous secretory receptor for advanced glycation end products in human organs. Mod Pathol. 2005;18:1385–1396. doi: 10.1038/modpathol.3800450. [DOI] [PubMed] [Google Scholar]
- 19.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rojas A, Gonzalez I, Morales MA. SARS-CoV-2-mediated inflammatory response in lungs: should we look at RAGE? Inflamm Res. 2020;69:641–643. doi: 10.1007/s00011-020-01353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J, et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prati D, et al. Updated definitions of healthy ranges for serum alanine aminotransferase levels. Ann Intern Med. 2002;137:1–10. doi: 10.7326/0003-4819-137-1-200207020-00006. [DOI] [PubMed] [Google Scholar]
- 24.Sproston NR, Ashworth JJ. Role of C-reactive protein at sites of inflammation and infection. Front Immunol. 2018;9:754. doi: 10.3389/fimmu.2018.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorci G, et al. S100B Protein, a damage-associated molecular pattern protein in the brain and heart, and beyond. Cardiovasc Psychiatry Neurol. 2010;2010:656481. doi: 10.1155/2010/656481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, et al. S100B regulates inflammatory response during osteoarthritis via fibroblast growth factor receptor 1 signalling. Mol Med Rep. 2018;18:4855–4864. doi: 10.3892/mmr.2018.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frontera, J. et al. Global consortium study of neurological dysfunction in COVID-19 (GCS-NeuroCOVID): study design and rationale. Neurocrit Care 2020 May 22. 10.1007/s12028-020-00995-3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 28.Michetti F, Dell’Anna E, Tiberio G, Cocchia D. Immunochemical and immunocytochemical study of S-100 protein in rat adipocytes. Brain Res. 1983;262:352–356. doi: 10.1016/0006-8993(83)91032-6. [DOI] [PubMed] [Google Scholar]
- 29.Cocchia, D., Tiberio, G., Santarelli, R. & Michetti, F. S-100 protein in “follicular dendritic” cells or rat lymphoid organs. An immunochemical and immunocytochemical study. Cell Tissue Res. 1983;230:95–103. [DOI] [PubMed]
- 30.Steiner J, et al. Human CD8(+) T cells and NK cells express and secrete S100B upon stimulation. Brain Behav Immun. 2011;25:1233–1241. doi: 10.1016/j.bbi.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 31.Miki Y, et al. Morphologic, flow cytometric, functional, and molecular analyses of S100B positive lymphocytes, unique cytotoxic lymphocytes containing S100B protein. Eur J Haematol. 2013;90:99–110. doi: 10.1111/ejh.12036. [DOI] [PubMed] [Google Scholar]
- 32.Engin AB. Adipocyte-macrophage cross-talk in obesity. Adv Exp Med Biol. 2017;960:327–343. doi: 10.1007/978-3-319-48382-5_14. [DOI] [PubMed] [Google Scholar]
- 33.Maurizi G, Della Guardia L, Maurizi A, Poloni A. Adipocytes properties and crosstalk with immune system in obesity-related inflammation. J Cell Physiol. 2018;233:88–97. doi: 10.1002/jcp.25855. [DOI] [PubMed] [Google Scholar]
- 34.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian C, Cao X. Dendritic cells in the regulation of immunity and inflammation. Semin Immunol. 2018;35:3–11. doi: 10.1016/j.smim.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Sorci G, et al. The danger signal S100B integrates pathogen- and danger-sensing pathways to restrain inflammation. PLoS Pathog. 2011;7:e1001315. doi: 10.1371/journal.ppat.1001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piazza O, et al. S100B induces the release of pro-inflammatory cytokines in alveolar type I-like cells. Int J Immunopathol Pharmacol. 2013;26:383–391. doi: 10.1177/039463201302600211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.