Heterozygous missense mutations in COPA underlie constitutive interferon signaling through STING, thereby defining a novel type I interferonopathy and emphasizing the importance of the ER–Golgi axis in interferon homeostasis. Based on these results, three patients have been treated with interferon signaling (JAK1) inhibitors.
Abstract
Heterozygous missense mutations in coatomer protein subunit α, COPA, cause a syndrome overlapping clinically with type I IFN-mediated disease due to gain-of-function in STING, a key adaptor of IFN signaling. Recently, increased levels of IFN-stimulated genes (ISGs) were described in COPA syndrome. However, the link between COPA mutations and IFN signaling is unknown. We observed elevated levels of ISGs and IFN-α in blood of symptomatic COPA patients. In vitro, both overexpression of mutant COPA and silencing of COPA induced STING-dependent IFN signaling. We detected an interaction between COPA and STING, and mutant COPA was associated with an accumulation of ER-resident STING at the Golgi. Given the known role of the coatomer protein complex I, we speculate that loss of COPA function leads to enhanced type I IFN signaling due to a failure of Golgi-to-ER STING retrieval. These data highlight the importance of the ER–Golgi axis in the control of autoinflammation and inform therapeutic strategies in COPA syndrome.
Graphical Abstract
Introduction
Type I IFNs are critical mediators of the antiviral immune response induced upon detection of pathogens, principally viruses, through the sensing of their nucleic acids by innate receptors (Hartmann, 2017). At the same time, a tight regulation of the IFN signaling pathway is required to prevent overactivation of the innate immune system, inducing tissue damage and pathology. The study of the type I interferonopathies, Mendelian diseases characterized by chronic up-regulation of IFN and IFN-stimulated genes (ISGs), has led to the definition of innate immune sensing pathways affected in these diseases and thereby the homeostatic mechanisms necessary to avoid autoinflammation (Uggenti et al., 2019). These include the degradation, modification, and compartmentalization of self-nucleic acids to avoid their aberrant detection (Crow et al., 2006a, 2006b; Rice et al., 2012; Dhir et al., 2018; Mackenzie et al., 2017), and the regulation of key components of the IFN signaling pathway (Rice et al., 2014; Zhang et al., 2015; Meuwissen et al., 2016). Part of the viral infection detection system, the sensor of cytoplasmic DNA cyclic GMP-AMP synthase (cGAS) signals through the production of cyclic GMP-AMP (cGAMP) to activate the ER protein stimulator of IFN genes (STING) and IFN induction through TBK1 activation and IRF3 nuclear translocation. Recent observations have highlighted the ER–Golgi axis as important in this context, with translocation of STING from the ER to the ER–Golgi intermediate compartment (ERGIC) and the Golgi central to the transmission and termination of cytoplasmic DNA signaling to IFN (Ogawa et al., 2018; Gonugunta et al., 2017; Gui et al., 2019). Indeed, heterozygous gain-of-function mutations in STING induce constitutive translocation of STING to the ERGIC/Golgi, causing a well-characterized type I interferonopathy, STING-associated vasculopathy with onset in infancy (SAVI; Liu et al., 2014; Jeremiah et al., 2014; Dobbs et al., 2015; Mukai et al., 2016). The precise mechanisms involved in the regulation of such trafficking, and the termination of STING signaling, remain unclear.
In 2015, heterozygous missense mutations in COPA, encoding coatomer protein subunit α (COPA), were described to cause an inflammatory syndrome demonstrating lung, renal, and joint involvement (Watkin et al., 2015). Of note, this disease is characterized by variable expression and a remarkably high frequency of clinical nonpenetrance (Watkin et al., 2015; Vece et al., 2016; Jensson et al., 2017; Volpi et al., 2018; Noorelahi et al., 2018; Tsui et al., 2018; Taveira-DaSilva et al., 2019; Patwardhan and Spencer, 2019; Boulisfane-El Khalifi et al., 2019; Frémond et al., 2020; Krutzke et al., 2019). COPA constitutes part of the coatomer protein complex I (COPI), playing a role in the retrograde transport of cargo proteins between the Golgi and the ER and the movement of vesicles within the Golgi (Brandizzi and Barlowe, 2013; Arakel and Schwappach, 2018; Lavieu et al., 2013). Evidence of ER stress, and priming of a helper T cell 17 response, was reported in the context of mutant COPA protein (Watkin et al., 2015), and an increased frequency of autoantibodies led to the suggestion that COPA syndrome was an autoimmune disease. More recently, Volpi et al. (2018) described an increased expression of ISGs in circulating blood cells of five patients with COPA syndrome, consistent with an autoinflammatory state related to type I IFN. The link between mutations in COPA and IFN induction is unknown. However, the suggested clinical similarities between COPA syndrome and SAVI (Volpi et al., 2018), the importance of trafficking for STING signaling, and the involvement of COPA in the ER–Golgi axis led us to hypothesize that COPA might regulate the function of STING through intracellular transport.
Herein we provide extended evidence of up-regulation of the type I IFN pathway in symptomatic patients carrying heterozygous mutations in COPA, and document the pathological overlap between COPA syndrome and SAVI, thereby supporting the assignation of COPA syndrome to the type I interferonopathy grouping. We then define the link between COPA dysfunction and IFN signaling to be STING dependent, and demonstrate a physical, likely indirect, interaction between COPA and STING. Finally, we show that mutant COPA is associated with an accumulation of STING in the Golgi compartment, leading us to hypothesize that COPA has a role in the retrograde trafficking of STING from the Golgi to the ER. These data highlight the importance of the ER–Golgi axis in type I IFN induction, and emphasize the potential of therapeutic strategies targeting this axis in COPA syndrome.
Results and discussion
COPA syndrome is characterized by persistent up-regulation of type I IFN signaling and demonstrates pathological overlap with SAVI
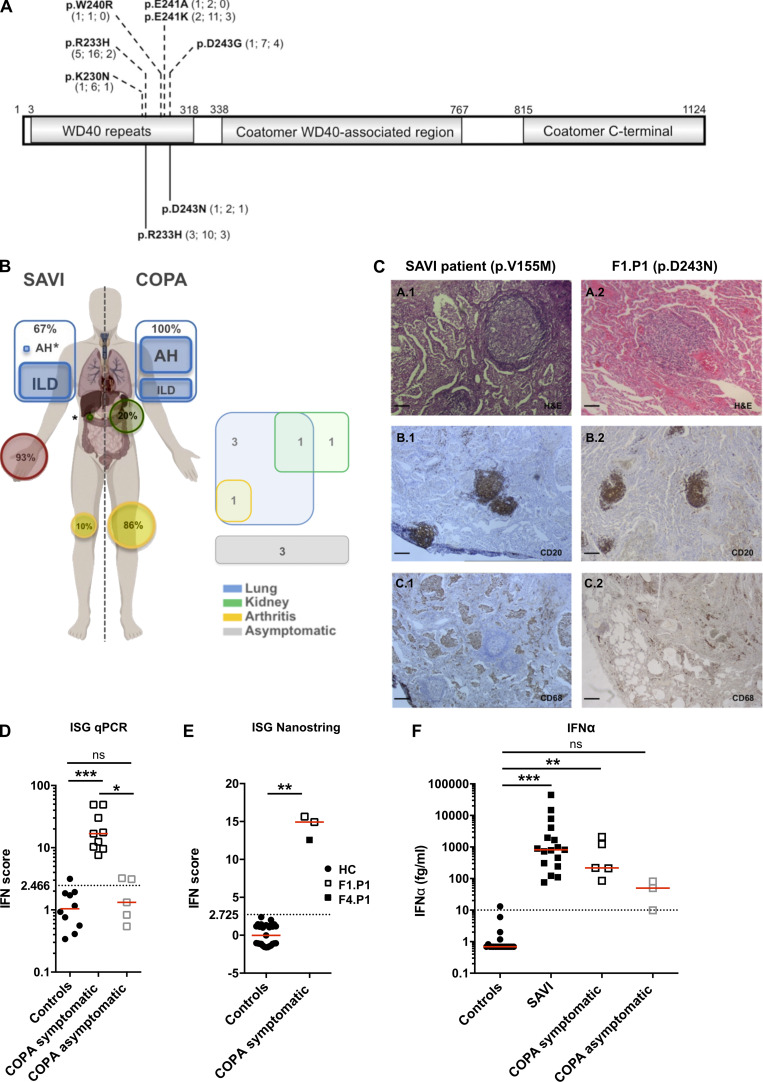
We studied nine individuals, comprising six symptomatic patients and three asymptomatic relatives from four unrelated families, each harboring a pathogenic heterozygous missense substitution in COPA, i.e., either p.R233H or p.D243N (Fig. 1 A, Table S1, and Fig. S1 A). The p.R233H substitution has been previously reported in several families (Watkin et al., 2015; Volpi et al., 2018; Taveira-DaSilva et al., 2019; Krutzke et al., 2019), while a substitution of the aspartate at position 243 has been described in one family, in that case to a glycine (p.D243G; Watkin et al., 2015; Fig. 1 A). There were six females and three males, with a median age of 33 yr (range, 13–55 yr), and a median age at disease onset (symptomatic patients) of 2.5 yr (range, 1–50 yr). COPA syndrome is characterized by clinical nonpenetrance, and three of the nine mutation carriers in this study were asymptomatic (Fig. 1 B; Watkin et al., 2015). Five of six symptomatic patients demonstrated clinical and/or histopathological lung involvement (Fig. 1, B and C; Table S1; and Fig. S1 B). Renal disease was observed in two patients, while one had arthritis and two reported arthralgia. Other features suggestive of autoinflammation included myositis and macrophage activation syndrome. Positive autoantibody titers were frequently detected.
Figure 1.
Heterozygous mutations in COPA, comparison of COPA syndrome and SAVI, and constitutive activation of the type I IFN pathway in patients. (A) Schematic representation of COPA adapted from Watkin et al. (2015) highlighting the WD40 repeat domain, the coatomer WD40-associated region, and the coatomer C-terminal region. Previously reported mutations (Watkin et al., 2015; Noorelahi et al., 2018; Patwardhan and Spencer, 2019) are shown above (p.K230N, p.R233H, p.W240R, p.E241K, p.E241A, and p.D243G). The mutations carried by the patients in this study are shown below (previously reported p.R233H and newly described p.D243N). Numbers in brackets refer to the number of families identified, mutation carriers, and asymptomatic individuals, respectively. (B) Comparison of the clinical phenotypes of SAVI and COPA syndrome according to published data (left, and see Table S2), and features present in the patients of this study (right). Lung, kidney, skin, and joint involvement are represented in blue, green, red, and yellow, respectively. AH, alveolar hemorrhage; asterisks represent a single patient reported in the literature (Volpi et al., 2019; Tang et al., 2020). (C) Comparison of histopathological features of the lung of a patient with SAVI carrying a p.V155M mutation in STING (Jeremiah et al., 2014; left) and F1.P1 (right). H&E staining showing lymphoid follicles (A.1 and A.2). Immunohistochemical staining identified a majority of CD20+ B cells within the B cell follicles (B.1 and B.2), and macrophage (CD68+) alveolar infiltration in the SAVI patient (C.1), and the presence of rare macrophages (CD68+ cells) within the alveoli of F1.P1 (C.2). Original magnification: ×40 (A; scale bars, 100 µm), ×10 (B and C; scale bars, 400 µm). (D) IFN scores calculated from the median fold change in relative quantification values for a set of six ISGs (Rice et al., 2013; IFI27, IFI44L, IFIT1, ISG15, RSAD2, SIGLEC1; normal <2.466) recorded in patients, as compared with 10 HCs. Red lines indicate median values. Median values in symptomatic patients (n = 9 samples from five individuals; 16.87; IQR, 9.953–39.73) were significantly higher than in HCs (1.045; IQR, 0.5225–1.870; ***, P= 0.0002), whereas median values in asymptomatic carriers (n = 5 samples from three individuals; 1.319; IQR, 0.6865– 3.179) were comparable to HCs (by Kruskal–Wallis test). One asymptomatic carrier (F2.P2) displayed a mildly positive IFN signature on two occasions (IFN scores above the dotted line). (E) IFN scores calculated from the median fold change in relative quantification values for a set of 24 ISGs (see Materials and methods and Table S4, normal <2.725) recorded in the peripheral blood of F1.P1 (sampled twice, depicted as open squares) and F4.P1 (depicted as a black square), as compared with 27 HCs (depicted as circles). Red lines indicate median values. Data were statistically analyzed using the Mann–Whitney test (**, P < 0.01). (F) Concentrations of IFN-α protein assessed by ultra-sensitive digital ELISA (Rodero et al., 2017) in plasma or serum from HCs (n = 20, <10 fg/ml), and patients with mutations in TMEM173 encoding STING (n = 17 samples from nine SAVI patients) or COPA (n = 5 samples from five symptomatic patients and n = 3 samples from two asymptomatic carriers). Red lines indicate median values of 817.8 fg/ml (IQR, 275–3,039), 217.7 fg/ml (IQR, 148–1,658), and 49.61 fg/ml (IQR, 9.809–81), respectively, in SAVI patients, COPA symptomatic patients, and asymptomatic carriers. Data were statistically analyzed using the Kruskal–Wallis test (**, P < 0.01; ***, P < 0.0001; ns, not significant).
Figure S1.
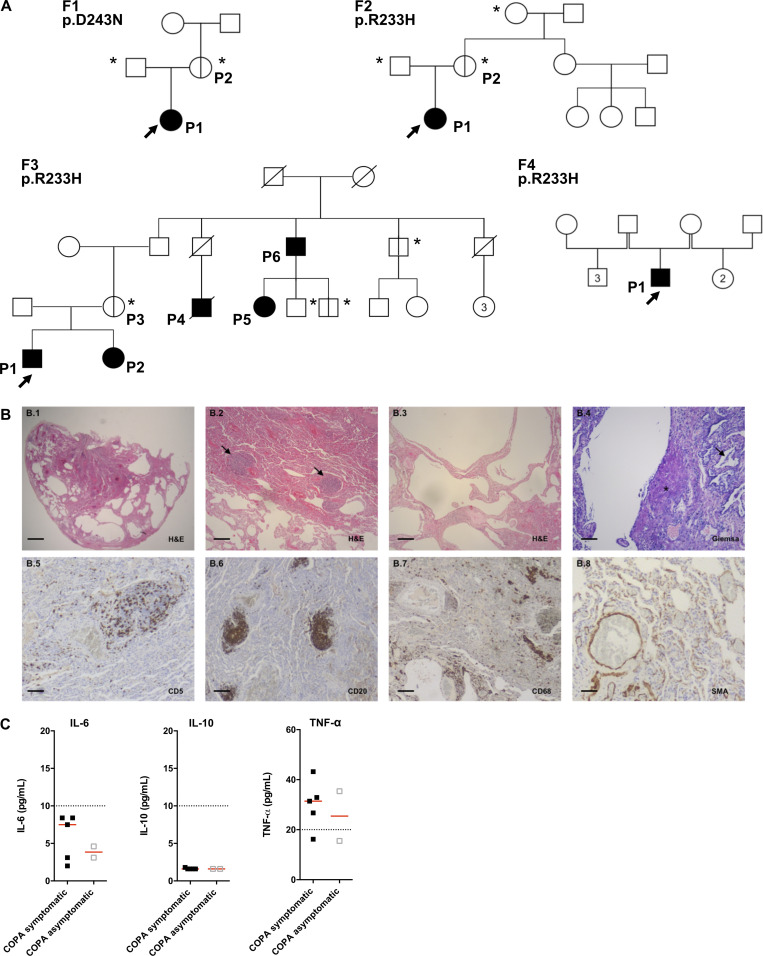
Pedigrees of the patients, lung pathology of F1.P1, and concentration of NF-κB–related cytokines observed in the patients. (A) Pedigrees of the four families (F) in this study. Circles (females) and squares (males) blackened and clear with vertical lines indicate respectively symptomatic and asymptomatic carriers of the annotated heterozygous mutation in COPA. Diagonal bars indicate deceased individuals. Arrows and asterisks indicate, respectively, index cases and asymptomatic individuals screened for the mutation. Numbers inside the symbols indicate the number of individuals of the same gender. (B) Histopathological analysis of the lung biopsy of F1.P1. (B.1–B.4) H&E and Giemsa staining showing subpleural emphysema with local interstitial thickening of the remaining interalveolar septa (B.1 and B.2), lymphoid follicles (arrows; B.3), and mildly cellular fibroblastic foci (star), as well as mild macrophagic alveolitis (arrow; B.4, B.5, B.6, and B.7). Immunohistochemical staining identified a majority of CD20+ B cells within the B cell follicles, scattered CD5+ T cells in the interstitium, and the presence of rare macrophages (CD68+ cells) within the alveoli. (B.8) Anti-smooth muscle actin (SMA) staining demonstrating normal staining of vessels and absence of myofibroblastic proliferation in the interstitium. Original magnification: ×4 (B.1; scale bar, 1 mm), ×10 (B.2, B.3, B.6, and B.7; scale bars, 400 µm), ×40 (B.4, B.5, and B.8; scale bars, 100 µm). (C) Concentrations of IL-6 protein (normal < 10 pg /ml), IL-10 protein (normal < 10 pg /ml), and TNF-α protein (normal < 20 pg/ml) measured in the plasma of COPA patients (n = 5 samples from five symptomatic patients and n = 2 samples from two asymptomatic carriers). The dotted lines indicate the normal values. Red lines depict median values.
An overlap between the clinical features of COPA syndrome and those observed in the well-defined type I interferonopathy SAVI (Liu et al., 2014; Jeremiah et al., 2014; Melki et al., 2017), which can also associate interstitial lung disease, joint involvement (Liu et al., 2014; Clarke et al., 2020), and hemorrhagic alveolitis (Tang et al., 2020; summarized in Fig. 1 B and Table S2), suggested the possibility of shared pathological determinants. Lung biopsy tissue from patient F1.P1 (COPA p.D243N), and from a patient carrying a p.V155M STING-activating mutation (Jeremiah et al., 2014), both demonstrated emphysema, lymphoid follicles, and macrophagic alveolitis (Fig. 1 C and Fig. S1 B), consistent with previous reports of interstitial lung disease in SAVI (Table S3). The lymphoid infiltrate was characterized by follicular CD20+ B cells within the nodules and lung-infiltrating CD5+ T lymphocytes, confirmed by immunohistochemical staining (Fig. S1 B).
The type I interferonopathies are characterized by chronic up-regulation of IFN signaling, revealed by increased expression of ISGs (Liu et al., 2014; Jeremiah et al., 2014; Rice et al., 2013, 2017; de Jesus et al., 2020), referred to as an “IFN signature,” and high levels of IFN-α protein in peripheral blood (Rodero et al., 2017). We recorded a marked and persistent IFN signature, within the range seen in SAVI patients, in the six symptomatic COPA patients on every occasion tested (one to six time points over a period of up to 3 yr), assessed by quantitative reverse transcription PCR (RT-qPCR) of six ISGs or the expression of 24 ISGs measured on a NanoString platform (Fig. 1, D and E). In contrast, asymptomatic mutation carriers demonstrated no, or a minimally elevated, IFN signature. Ultra-sensitive digital ELISA (Rodero et al., 2017) revealed raised concentrations of IFN-α in plasma from all symptomatic individuals (median of 217.7 fg/ml; interquartile range [IQR], 148–1,658), with mildly increased levels in the two asymptomatic carriers tested (median of 49.61 fg/ml; IQR, 9.809–81) compared with healthy controls (HCs; Fig. 1 F). In contrast, the concentration of other cytokines was normal (IL-6, IL-10) or minimally elevated (TNF-α) in the plasma from patients (Fig. S1 C).
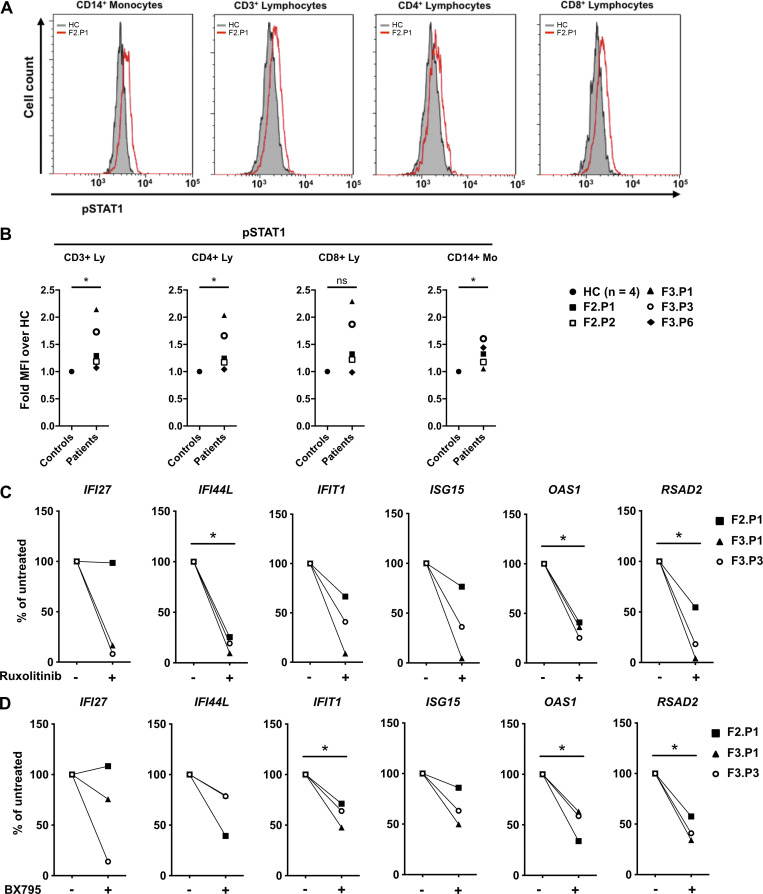
Upon induction of expression through STING activation and phosphorylation of TBK1 and IRF3, type I IFNs trigger the expression of ISGs following binding to their unique receptor IFNAR, subsequently activating the kinases JAK1 and Tyk2 and the transcription factors STAT1/2. Ex vivo flow cytometry assays demonstrated increased levels of phosphorylated STAT1 in T lymphocytes and monocytes from COPA patients compared with controls (Fig. 2, A and B), confirming IFN signaling through STAT1. We previously demonstrated that, in vitro, ruxolitinib, a JAK1/2 inhibitor, and BX795, a TBK1/IKKE inhibitor, reduced ISG expression in peripheral blood mononuclear cells (PBMCs) from SAVI patients (Frémond et al., 2016, 2017). Similarly, in vitro exposure to ruxolitinib or BX795 reduced constitutively increased ISG expression in PBMCs from two symptomatic patients with COPA syndrome and an asymptomatic mutation carrier with a (minimally) positive IFN signature (Fig. 2, C and D). Ruxolitinib has been successfully used in the treatment of certain monogenic type I interferonopathies (Frémond et al., 2016; Briand et al., 2019; Sanchez et al., 2018). Indeed, given these data, JAK1/2 inhibitors are being tried in three patients in our study, including F2.P1, who has been receiving ruxolitinib for >1 yr, with improvement of her severe lung phenotype (Frémond et al., 2020).
Figure 2.
Type I IFN is responsible for the constitutive induction of ISGs in COPA syndrome. (A) Flow cytometry analysis of phosphorylated STAT1 (pSTAT1) in monocytes, CD3+, CD4+, and CD8+ lymphocytes from F2.P1 compared with a HC. (B) pSTAT1 measured as in A in CD3+, CD4+, and CD8+ lymphocytes (Ly) and CD14+ monocytes (Mo) from COPA patients compared with a HC sampled the same day, and expressed as fold mean fluorescence intensity (MFI) of over that of the HC. Blood from F3.P1 and F3.P3 was processed during the same experiment and compared with one HC. Data were statistically analyzed using the Mann–Whitney test (*, P < 0.05; ns, not significant). (C and D) RT-qPCR gene expression analysis in PBMCs from three patients (F2.P1, F3.P1, and F3.P3), after overnight culture in the absence of treatment or presence of ruxolitinib (C) or BX795 (D). Results are expressed as percentage of the baseline elevated levels and show that IFIT1, IFI27, IFI44L, ISG15, OAS1, and RSAD2 levels decreased after in vitro treatment (except IFI27 in F2.P1) for both drugs. Data were statistically analyzed using a one-sample t test (*, P < 0.05).
The above data characterize COPA syndrome as a novel monogenic type I interferonopathy, sharing clinical and histopathological features with SAVI. The high degree of clinical nonpenetrance, around 30%, in reported families segregating a pathogenic mutation in COPA, is striking and suggests that additional factors are required to perturb IFN signaling and clinical disease to manifest. Importantly, IFN signaling levels in COPA patients may enable the distinction between symptomatic and asymptomatic status, thereby providing a tool for clinical investigation and treatment decision making. Of note, while nonpenetrance is a feature of other type I interferonopathy genotypes (e.g., due to gain-of-function of IFIH1; Rice et al., 2014), our experience to date has been that such asymptomatic individuals almost invariably demonstrate a persistent up-regulation of ISG transcripts irrespective of clinical status (Rice et al., 2020). The reasons for this difference remain unclear.
COPA mutants induce type I IFN production and signaling through STING in a dominant-negative manner
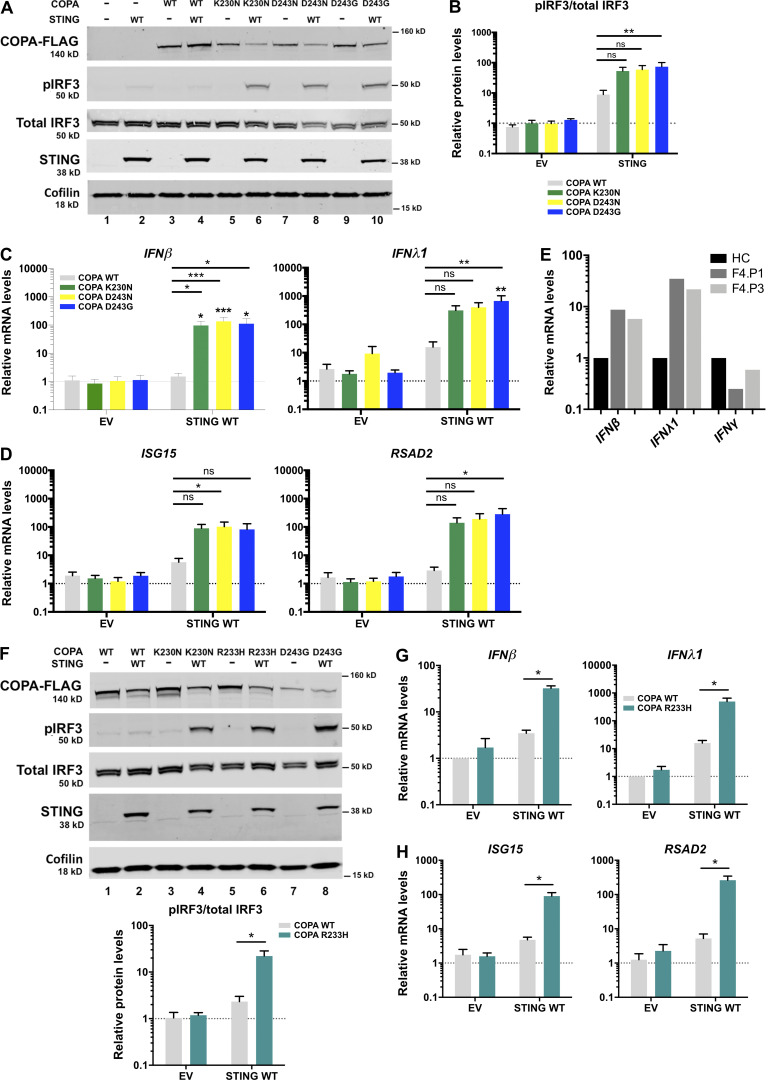
To decipher the constitutive activation of type I IFN recorded above, we transiently transfected human embryonic kidney (HEK) 293T cells, expressing endogenous COPA but not STING (Watkin et al., 2015; Burdette et al., 2011), with WT STING, and with either WT COPA or a COPA construct carrying a p.K230N, p.R233H, p.D243G, or p.D243N pathogenic mutation (with the p.K230N substitution having been described previously in another family; Watkin et al., 2015). Enhanced phosphorylation of IRF3 was observed in cells cotransfected with STING and mutant COPA, as opposed to cells cotransfected with WT COPA (Fig. 3, A, B, and F). Activation of the type I IFN pathway was dependent on STING, since no phosphorylation of IRF3 was detected when any of the mutant COPA plasmids were cotransfected with an empty vector (EV). Consistently, we recorded a significant increase in the expression of IFNβ transcripts in cells coexpressing any of the four mutant COPA plasmids and STING (Fig. 3, C and G). DNA stimulation can induce the production of IFNλ1, a type III IFN with similar activities to type I IFN, through a Ku70-STING complex in human primary macrophages and cell lines including HEK293 (Sui et al., 2017). As for IFNβ, we recorded increased expression of IFNλ1 in HEK293T cells cotransfected with mutant COPA and STING, as compared with EV (Fig. 3, C and G). Of note, mRNA expression of both IFNβ and IFNλ1, but not of type II IFN (IFNγ), was also higher in PBMCs from one symptomatic patient (F4.P1) and his asymptomatic mother (F4.P3), compared with HCs (Fig. 3 E). Finally, consistent with the positive IFN signature recorded in patients, and the induction of IRF3 phosphorylation and IFNβ transcription in HEK293T cells, the expression of the ISGs ISG15 and RSAD2 was enhanced in cells cotransfected with STING and mutant COPA, but not with WT COPA (Fig. 3, D and H).
Figure 3.
COPA mutants induce IFN production and signaling through STING. (A) Western blot analysis of FLAG, IRF3 phosphorylated (pIRF3) at Ser396, total IRF3, STING, and Cofilin in whole-cell lysates of HEK293T cells cotransfected with WT STING and WT COPA or mutant COPA plasmids (K230N, D243N, D243G) or corresponding control (EV or −) plasmids for 48 h. Representative results from four independent experiments. (B) Quantification of phosphorylated IRF3 (at position Ser396) relative to total IRF3 expressed as fold over EV signal as observed in A. Mean ± SEM of four independent experiments, analyzed with the Kruskal–Wallis test (**, P= 0.0069). (C and D) mRNA expression analysis assessed by RT-qPCR of IFNβ, IFNλ1 (C), and two ISGs (ISG15, RSAD2; D) in HEK293T cotransfected with EV or WT STING and WT COPA or mutant COPA plasmids. Means ± SEM of five independent experiments, statistically analyzed using two-way ANOVA and the Dunnett’s multiple comparisons test (statistics above black lines), with results above each bar indicating the comparison of expression in cells cotransfected with WT STING and each of the three COPA mutant plasmids compared with the cells cotransfected with EV and the corresponding COPA mutant plasmid by two-way ANOVA with Sidak’s multiple comparisons test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant). (E) RT-qPCR gene expression analysis of IFNβ (type I IFN), IFNλ1 (type III IFN), and IFNγ (type II IFN) in PBMCs from one symptomatic patient (F4.P1) and his asymptomatic mother (F4.P3), and one HC, after overnight culture in one experiment. (F) Western blot analysis of HEK293T cotransfected with WT STING and WT COPA or mutant COPA plasmids (K230N, R233H, D243G) or EV (−) as in A. Representative results for three experiments. Results were quantified for R233H mutant compared with WT and averaged below. Data were statistically analyzed using two-way ANOVA with Sidak’s test for multiple comparison (*, P < 0.05). (G and H) mRNA expression analysis assessed by RT-qPCR of IFNβ, IFNλ1 (G), and two ISGs (ISG15, RSAD2; H) in HEK293T cotransfected with EV or WT STING and WT COPA or COPA R233H plasmids. Means ± SEM of three independent experiments, statistically analyzed using two-way ANOVA and Sidak’s multiple comparisons test (*, P < 0.05).
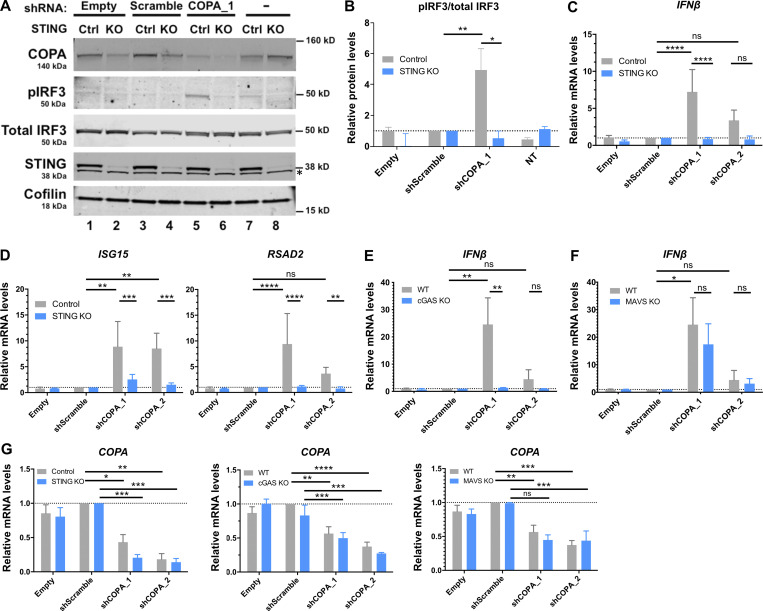
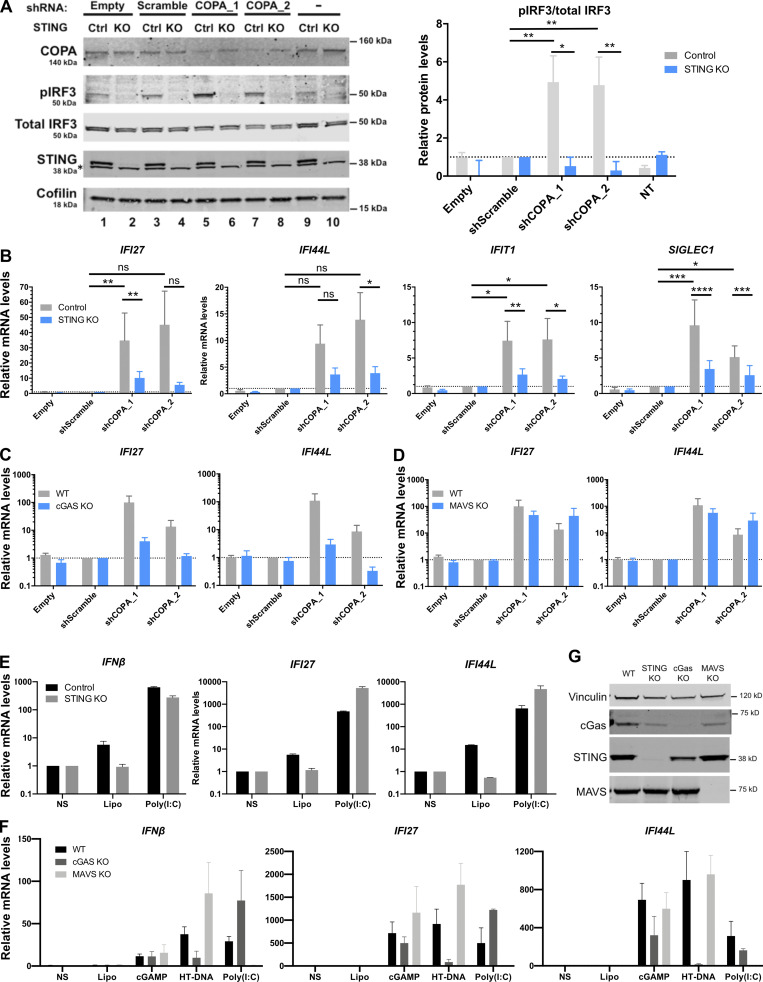
To determine whether heterozygous substitutions in COPA confer an effect through a gain-of-function or dominant-negative mechanism, we silenced COPA in cells competent for STING signaling to IFN, i.e., the THP-1 monocytic cell line. shRNA knockdown of COPA was sufficient to induce IRF3 phosphorylation and increased IFNβ and ISG expression (Fig. 4, A–D and G; and Fig. S2, A and B). This suggested that the IFN pathway is activated by loss of COPA, and that COPA syndrome–associated substitutions act as dominant-negative mutations. Sensing of intracellular DNA induces the synthesis of cGAMP by cGAS, thereby activating STING to signal through the ER–Golgi axis and induce IFN production. IFN signaling conferred by COPA down-regulation was abrogated in both STING and cGAS KO THP-1 cells (Fig. 4, A–E and G; and Fig. S2, A–C). In contrast, IFN signaling was not dependent on mitochondrial antiviral-signaling protein (MAVS), an adaptor molecule essential for cytoplasmic RNA sensing (Fig. 4, F and G; and Fig. S2 D). KO THP-1 cell lines were functionally validated by assessing IFNβ and ISG (IFI27 and IFI44L) expression after stimulation with agonists of the RNA (i.e., poly(I:C)) and DNA (i.e., herring testes DNA [HT-DNA] and cGAMP) pathways (Fig. S2, E–G; Cerboni et al., 2017; Hertzog et al., 2020 Preprint). As expected, STING and cGAS KO cells were normally responsive to RNA ligand poly(I:C), contrary to MAVS KO cells. Furthermore, MAVS and cGAS KO THP-1 cells responded normally to cGAMP stimulation while MAVS KO cells were normally responsive to cytoplasmic HT-DNA, in contrast to cGAS KO THP-1 cells.
Figure 4.
Loss of COPA induces STING-dependent type I IFN signaling. (A) Western blot analysis of IRF3 phosphorylated at Ser396, total IRF3, COPA, STING, and Cofilin in whole-cell lysates of control (Ctrl) or STING KO THP-1 cells transduced with an EV, a scrambled shRNA or an shRNA targeting COPA (shCOPA_1), or non-transduced (NT; left). Data are representative of three independent experiments. Asterisk indicates unspecific band. Results for a second shRNA targeting COPA (shCOPA_2) are shown in Fig. S2, A and B. (B) Quantification of phosphorylated IRF3 (at position Ser396) relative to total IRF3 expressed as fold over scrambled shRNA signal observed in A. Means ± SEM of three independent experiments were statistically analyzed using one-way ANOVA and two-way ANOVA with Sidak’s multiple comparisons test for comparison of results with shScramble vs. shCOPA_1 (**, P = 0.0026) and results in control vs. STING KO THP-1 cells for shCOPA_1 (*, P = 0.0104), respectively. (C and D) mRNA expression analysis assessed by RT-qPCR of IFNβ (C) and two of six tested ISGS (ISG15, RSAD2, [D]; and see Fig. S2 B) in control or STING KO THP-1 cells transduced with an EV, a scrambled shRNA, two different shRNAs targeting COPA (shCOPA_1 and shCOPA_2), or nontransduced (NT). Means ± SEM of three independent experiments, statistically analyzed using one-way ANOVA and two-way ANOVA with Sidak’s multiple comparisons test for comparison of results with shScramble vs. each shCOPA, and results in control vs. STING KO THP-1 cells for each shCOPA, respectively. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (E and F) mRNA expression analysis assessed by RT-qPCR of IFNβ in WT or THP-1 cells null for cGAS (E) or MAVS (F) transduced with an EV, a scrambled shRNA, or two different shRNAs targeting COPA (shCOPA_1 and shCOPA_2). Means ± SEM of three independent experiments, statistically analyzed using one-way ANOVA and two-way ANOVA with Sidak’s multiple comparisons test for comparison of results with shScramble vs. each shCOPA, and results in WT vs. cGAS KO THP-1 cells for each shCOPA, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001). (G) mRNA expression of COPA assessed by RT-qPCR in THP-1 cells control and null for STING (left), or WT and null for cGAS (middle) or MAVS (right) transduced with an EV, a scrambled shRNA, or shCOPA_1 and shCOPA_2. Mean ± SEM of three independent experiments were statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for WT and control THP-1 cells) or two-way ANOVA (WT or control vs. THP-1 cells null for STING, cGAS, or MAVS, for each shCOPA) with Sidak’s multiple comparisons test: *, P <0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
Figure S2.
Knockdown of COPA induces STING-dependent, MAVS-independent type I IFN signaling in THP-1 cells. (A) Left: Western blot analysis of IRF3 phosphorylated at Ser396, total IRF3, COPA, STING, and Cofilin in whole-cell lysates of control or STING KO THP-1 cells transduced with an EV, a scrambled shRNA, or two different shRNAs targeting COPA (shCOPA_1 and shCOPA_2, respectively lanes 5 and 6, and lanes 7 and 8), or nontransduced (NT). Data are representative of three independent experiments. Asterisk indicates unspecific band. Right: Quantification of phosphorylated IRF3 (at position Ser396) relative to total IRF3 expressed as fold over scrambled shRNA relative signal, statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for control THP-1 cells; **, P = 0.0026 for shCOPA_1 and **, P = 0.004 for shCOPA_2) or two-way ANOVA (control vs. STING KO THP-1 cells for each shCOPA; *, P = 0.0104 for shCOPA_1 and **, P = 0.0094 for shCOPA_2) with Sidak’s multiple comparisons test (mean ± SEM). (B) mRNA expression analysis assessed by RT-qPCR of four out of six tested ISGS (IFI27, IFI44L, IFIT1, and SIGLEC1; see other results in Fig. 4 D) in control or STING KO THP-1 cells transduced with an EV, a scrambled shRNA, or shCOPA_1 and shCOPA_2. Mean ± SEM of three independent experiments were statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for control THP-1 cells) or two-way ANOVA (control vs. STING KO THP-1 cells for each shCOPA) with Sidak’s multiple comparisons test (*, P <0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant). (C and D) mRNA expression analysis assessed by RT-qPCR of two ISGs (IFI27, IFI44L; D) in WT or THP-1 cells null for cGAS or MAVS transduced with an EV, a scrambled shRNA, or shCOPA_1 and shCOPA_2. Mean ± SEM of three independent experiments were statistically analyzed in one-way ANOVA (shScramble vs. shCOPA for WT THP-1 cells) or two-way ANOVA (WT vs. MAVS KO THP-1 cells for each shCOPA) with Sidak’s multiple comparisons test (ns, nonsignificant). (E and F) mRNA expression analysis assessed by RT-qPCR of IFNβ and two ISGS (IFI27, IFI44L) in control or THP-1 cells null for STING, cGAS, or MAVS and nonstimulated (NS) or stimulated with 1 µg/ml poly(I:C), 1 µg/ml 2′3′cGAMP, or 0.25 µg/ml HT-DNA for 24 h combined with Lipofectamine (Lipo). Mean ± SEM of two (poly[I:C]) to three (cGAMP, HT-DNA) independent experiments. (G) Western Blot analysis of cGAS, STING, and MAVS expression in THP-1 WT, STING, cGAS, or MAVS KO, and loading control Vinculin. Representative of two independent experiments. Ctrl, control.
These data indicate that heterozygous mutations in COPA act through a dominant-negative mechanism, with loss of COPA function leading to STING-dependent IFN induction. This is in agreement with results published by Watkin et al. (2015) showing that mutant COPA acts as a dominant-negative in respect to ER homeostasis. In addition, the above in vitro data suggest both a primary role for the (mutant) COPA–STING interaction in driving disease in COPA syndrome and variable cGAS-dependency. Specifically, overexpressed STING activated IFN signaling in response to COPA mutants in HEK293T cells that lack cGAS expression (Sun et al., 2013), while IFN induction following COPA knockdown in THP-1 cells was dependent on cGAS. The latter observation suggests a tonic level of cGAMP production by cGAS is present in THP-1 cells, perhaps driven by mitochondrial DNA or nuclear DNA released into the cytosol as a result of DNA damage (Mackenzie et al., 2017; Härtlova et al., 2015; Kim et al., 2019), or activation of nuclear cGAS by chromatin (Lahaye et al., 2018; Gentili et al., 2019), and is required to activate STING. In contrast, the ability of COPA mutants to activate IFN signaling in the absence of cGAS upon overexpression of STING in HEK293T cells suggests an effect that overwhelms any requirement for cGAMP, perhaps most likely STING oligomerization due to overexpression (Ergun et al., 2019).
COPA mutants disrupt the physical interaction with STING
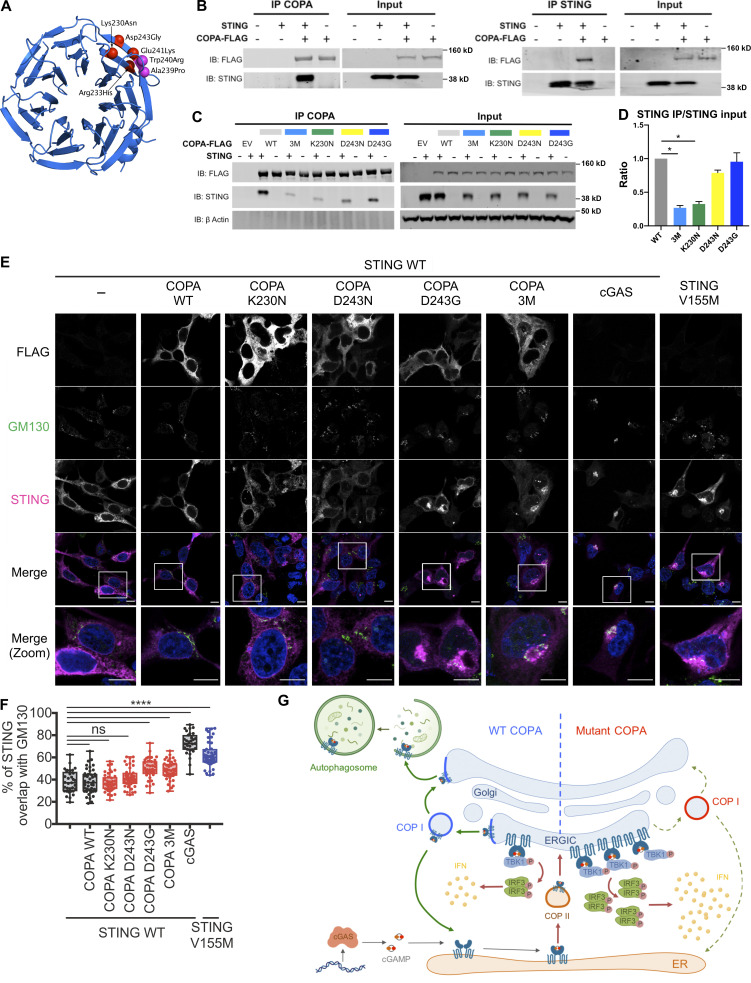
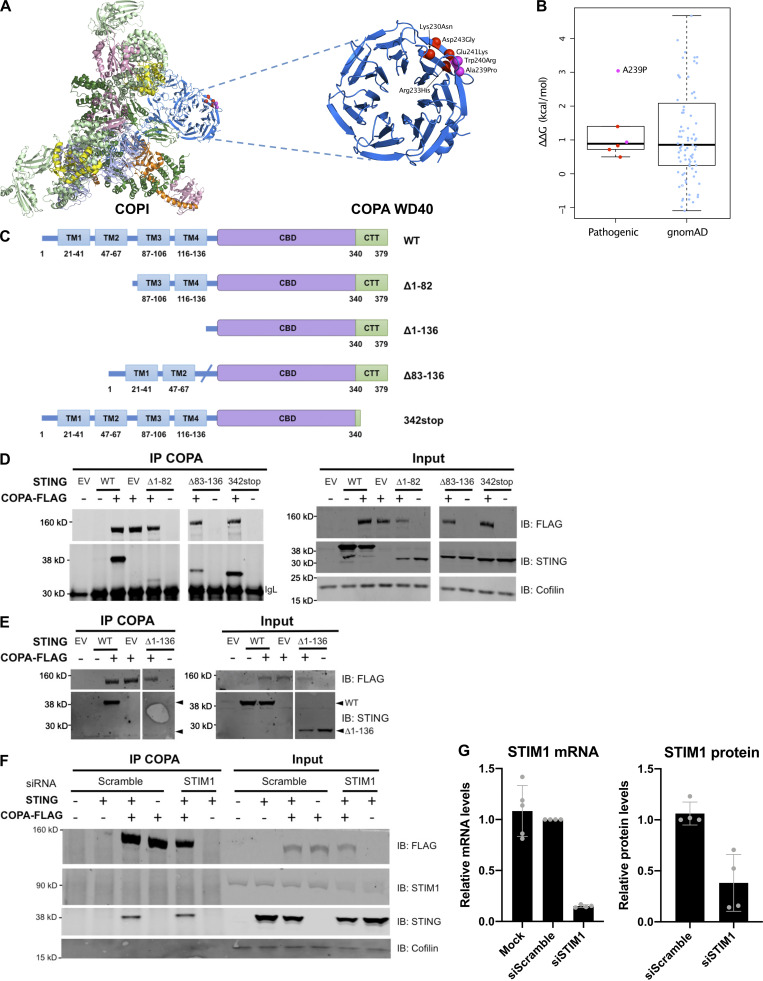
COPA syndrome–associated mutations are restricted to the WD40 domain of the protein, involved in the recognition of dilysine motifs (KKxx and KxKxx) on cargo proteins and their packaging into COPI-coated vesicles (Brandizzi and Barlowe, 2013; Letourneur et al., 1994; Ma and Goldberg, 2013; Fig. 1 A). Examination of the crystal structure of the human COPA WD-repeat domain revealed that four recurrently substituted amino acids are located close to each other in a pair of anti-parallel β-strands (Fig. 5 A and Fig. S3 A). A further previously reported mutation (p.W240R; Noorelahi et al., 2018), and an unpublished mutation annotated as “likely pathogenic” in ClinVar (p.A239P), mapped to the same region. Five of these six mutated residues are located on the protein surface. Inspection of the electron microscopy structure of the COPI coat leaf (Dodonova et al., 2017) shows that this region of COPA is also located on the surface of the COPI complex, and does not participate in any interactions within the leaf (Fig. S3 A). Using molecular modeling, we found that the predicted effects of disease-associated mutations on protein stability were similar to those of annotated variants recorded in control individuals on gnomAD v2.1 (Karczewski et al., 2019; Fig. S3 B), with only the unreported mutation (p.A239P) predicted to moderately destabilize protein folding (ΔΔG >1.5 kcal/mol). These data suggest that disease-causing mutations in COPA are more likely to perturb an intermolecular interaction, rather than destabilize the protein. Indeed, normal levels of COPA protein have been previously observed in patient material (Watkin et al., 2015), and two mutations associated with COPA syndrome (p.E241K and p.K230N) have been shown to confer impaired binding to proteins carrying COPI cargo dilysine motifs (Watkin et al., 2015).
Figure 5.
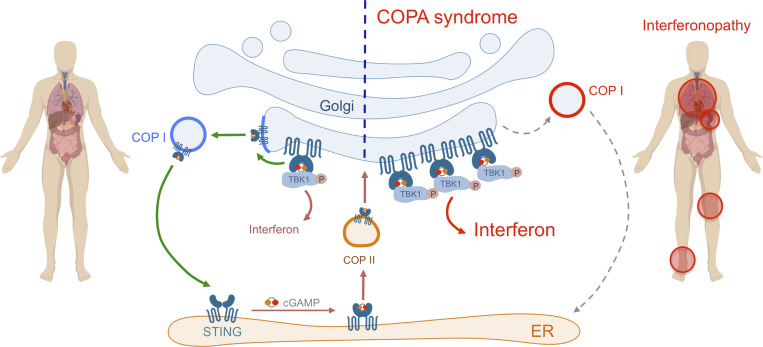
Interaction of COPA and STING and STING localization. (A) Location of pathogenic missense mutations within the structure of the COPA WD-repeat domain (PDB ID: 6PBG). The four mutations studied in this report are shown in red, while two further previously identified mutations are indicated in magenta. Location of the pathogenic missense mutations within the structure of the COPI coat leaf is given in Fig. S3 B. (B) Western blot analysis of FLAG and STING in proteins IP with an antibody against FLAG (IP COPA, left) or STING (right), and in whole-cell lysates (input) of HEK293T cells cotransfected with EV (−) or WT STING and WT COPA plasmids. Data are representative of three independent experiments. (C) Western blot analysis of FLAG, STING, and β-actin in proteins IP with an antibody against FLAG (IP COPA) and whole-cell lysates (input) of HEK293T cells cotransfected with EV (EV or −) or WT STING and WT COPA, or with individual mutant COPA plasmids (K230N, D243N, D243G) or with a plasmid carrying three substitutions (3M: K230N/R233H/D243N). Data are representative of three independent experiments. (D) Quantification of STING protein levels co-immunoprecipitated (STING IP) with COPA-FLAG compared with the signal recorded in the input, as observed in C. Mean ± SEM of three independent experiments were statistically analyzed using Kruskal–Wallis test (*, P < 0.05). (E) Representative images of STING and COPA localization in HEK293FT cells transfected with WT, mutant COPA (3M carrying three substitutions: K230N/R233H/D243N), and STING plasmids or corresponding control plasmids (−), and transfected with cGAS or control plasmid the next day. Cells were fixed and stained for nuclear DNA (DAPI; blue), COPA (FLAG; gray), Golgi (GM130; green), and STING (magenta). “Merge” row shows an overlay of DAPI, GM130, and STING signals, GM130 and STING costaining being represented in white. “Merge (zoom)” depicts an enlargement of the square above. Images are representative of three independent experiments. Scale bar, 10 µm. (F) Quantification of the ratio of STING signal localized to the GM130/Golgi compartment over total STING signal in images as in A, for at least 10 cells per condition per experiment (one-way ANOVA with Dunnett’s correction: ****, P < 0.0001). ns, not significant. (G) Left: DNA from pathogens and damaged cells induces the production of cGAMP by cGAS. Upon binding of cGAMP, STING translocates, in a COPII-dependent process, to the ERGIC and Golgi, where it triggers TBK1 and IRF3 phosphorylation and subsequent IFN production. We hypothesize that COPA, within the COPI complex, plays a role in STING trafficking after signaling, taking STING back to the ER, or on to endolysosomes/the autophagy pathway (Gonugunta et al., 2017), and thereby leading to resolution of IFN signaling. Our data indicate that STING may be a cargo of COPI through COPA. Right: When COPA is mutated in the cargo-binding WD40 domain, STING can no longer be sorted into COPI vesicles, and so is retained in the ERGIC/Golgi in an active state, leading to continued IFN production. COP, coatomer protein; IB, immunoblot.
Figure S3.
COPI complex and physical interaction of COPA and STING. (A) Location of pathogenic COPA mutations within the structure of the COPI coat leaf (PDB ID: 5NZR). The four mutations from this study are shown in red, and two previously reported mutations are shown in magenta. (B) Predicted effects of pathogenic mutations and putatively benign substitutions present in the gnomAD database on protein stability, calculated with FoldX. (C) Schematic representation of WT STING and truncated STING plasmids (i.e., Δ1-82, Δ83-136, Δ1-136, and 342stop). (D) Western blot analysis of FLAG, STING, and Cofilin in proteins IP with an antibody against FLAG (IP COPA), and whole-cell lysates (input) of HEK293T cells cotransfected with EV (EV or −), or WT COPA and WT STING, or truncated STING plasmids, i.e., Δ1-82, Δ83-136, and 342stop (data representative of three independent experiments). (E) Western blot analysis as in F of proteins IP with an antibody against FLAG (IP COPA), and whole-cell lysates (input) of HEK293T cells cotransfected with EV (EV or −), or WT COPA and WT STING, STING Δ1-136 (data representative of two independent experiments). (F) Western blot analysis of FLAG, STING, STIM1, and Cofilin in proteins IP with an antibody against FLAG and in whole-cell lysates (input) of HEK293T cells cotransfected with EV (−) or WT STING and WT COPA plasmids and with a scrambled siRNA or an siRNA pool targeting STIM1. Results are representative of three independent experiments. (G) Quantification of STIM1 silencing assessed by mRNA (left) and protein (right) levels measured, respectively, by RT-qPCR and Western blot 72 h after transfection. Error bars represent SDs. Results are representative of three independent experiments. CTT, C-terminal tail.
Activation of ER-resident STING requires trafficking to the Golgi (Burdette et al., 2011; Dobbs et al., 2015), with resolution of signaling involving further trafficking to endolysosomes and degradation (Gonugunta et al., 2017). As part of COPI, COPA participates in the transport of proteins from the Golgi back to the ER, and within the Golgi compartment itself (Brandizzi and Barlowe, 2013; Popoff et al., 2011). Thus, we hypothesized that COPA might be involved in the sorting and recruitment of STING to COPI-coated vesicles at the Golgi, and that mutations in COPA may result in altered STING trafficking and sustained activation at the Golgi. To explore this possibility, we performed reciprocal coimmunoprecipitation (coIP) in HEK293T cells cotransfected with COPA and STING, observing a consistent interaction between WT COPA and WT STING (Fig. 5 B). We recorded a weaker interaction when the COPA construct carried the p.K230N mutation (Fig. 5, C and D). This was apparently not the case when the COPA plasmid carried a mutation at position 243 (i.e., p.D243N or p.D243G). To investigate whether combined substitutions of different amino acids in the WD40 domain could have additive effects, we also generated a COPA construct, referred to as 3M, comprising three disease-causing COPA substitutions located in the mutation hotspot (i.e., p.K230N, p.R233H, and p.D243N). In this assay, we observed a decreased interaction of COPA with STING similar to that seen with the p.K230N substitution alone (Fig. 5, C and D). Thus, while every mutation that we tested resulted in enhanced IFN signaling ex vivo and in vitro, we noted differential effects on STING binding of certain mutants, perhaps explained by a bias of our overexpression system favoring or masking the impact of the mutation.
To gain insight into the details of the above interaction, we investigated the domains of STING involved in the interaction with COPA. STING is an ER membrane protein containing four transmembrane (TM) domains and N- and C-terminal portions facing the cytosol (Ouyang et al., 2012; Shang et al., 2019). We performed coIP assays using STING constructs lacking TM1 and TM2 (deletion 1–82), TM3 and TM4 (deletion 83–136), all four TM domains (deletion 1–136), or the cytoplasmic C-terminal tail (codon-stop at position 342; Fig. S3 C). All truncations, except the 1–136 deletion, localized to the ER (data not shown). The interaction of COPA with 342-STOP STING was conserved, suggesting that the physical association between COPA and STING does not require the TBK1- and IRF3-binding C-terminal tail of STING (Fig. S3 D). Consistently, although some binding was retained, there appeared to be a weaker interaction in the absence of STING TM1 and TM2, or TM3 and TM4, and we could not detect any interaction in the absence of all four TMs (Fig. S3, D and E).
STING does not have a typical C-terminal dilysine motif, suggesting an indirect interaction and implicating an adaptor protein enabling binding to the COPI complex. Given that COPA is cytosolic (Waters et al., 1991), this would be consistent with a dependency of the COPA and STING interaction on the TM domains of STING. We considered that one possible candidate might be the calcium sensor STIM1 (stromal interaction molecule 1), which does possess a dilysine motif (Lavieu et al., 2010), and was recently shown to act as a retention factor for STING at the ER membrane (Srikanth et al., 2019). However, knockdown of STIM1 did not affect the interaction between COPA and STING (Fig. S3, F and G).
COPA mutants result in abnormal STING localization
Finally, we wanted to investigate whether COPA dysfunction could lead to STING signaling by affecting STING trafficking. Gain-of-function heterozygous mutations in STING lead to constitutive activation and translocation of STING to the ERGIC/Golgi independent of an interaction with its ligand, cGAMP (Dobbs et al., 2015; Jeremiah et al., 2014; Cerboni et al., 2017). In HEK293FT cells, we noted increased localization of WT STING to a GM130-positive Golgi location when cells were cotransfected with p.D243G and the 3M triple mutant COPA, equivalent to the SAVI mutant STING V155M, and to WT STING cotransfected with cGAS to induce STING activation by cGAMP. This was not the case with WT COPA, or when WT STING was transfected alone (Fig. 5, E and F), suggesting that loss of COPA function led to STING accumulation at the Golgi and activation of the IFN pathway.
COPA (α-COP) is one of seven subunits (α, β, β′, δ, ε, γ, and ζ-COP) of the COPI complex, described to mediate the retrograde transport of proteins between the Golgi and the ER (Brandizzi and Barlowe, 2013; Arakel and Schwappach, 2018), and vesicle movement within the Golgi per se (Popoff et al., 2011; Park et al., 2015). Protein cargoes can be recognized by COPA and the β′-COP subunits through well-characterized dilysine motifs, thus allowing for their packaging into COPI-coated vesicles (Letourneur et al., 1994; Ma and Goldberg, 2013). COPI also enables the retrieval of escaped ER-resident luminal proteins through the recruitment of K/HDEL receptors binding to the K/HDEL signal of these proteins (Brandizzi and Barlowe, 2013). On the other side of the ER–Golgi interface, coatomer complex II (COPII) mediates anterograde, ER to Golgi, transport of proteins to be further modified in the Golgi and/or secreted (Brandizzi and Barlowe, 2013; Fig. 5 G).
DNA recognition by cGAS leads to production of the second messenger cGAMP, which in turn binds to STING. After activation and conformational changes, STING moves to the ERGIC and Golgi (Dobbs et al., 2015) to induce type I IFN transcription through phosphorylation of TBK1 and IRF3 (Liu et al., 2015), a process in which COPII has been implicated (Gui et al., 2019). In contrast, SAVI-associated gain-of-function STING mutants traffic to the ERGIC in the absence of cGAMP stimulation (Jeremiah et al., 2014; Dobbs et al., 2015). We observed a mutation-dependent physical interaction between COPA and STING, and enrichment of STING in the Golgi compartment in the context of mutated COPA. One possible explanation for these data is that STING is actively trafficked back to the ER from the ERGIC/Golgi by COPI, and that such trafficking is reduced due to COPA loss-of-function, leading to sustained signaling (Fig. 5 G). It is also possible that COPA contributes to the attenuation of IFN signaling through the movement of STING through the Golgi compartment (Pellett et al., 2013). COPA, β′-COP, and β-COP have additionally been implicated in autophagy and the maturation of autophagosomes necessary for content degradation, through the maintenance of early endosome function (Razi et al., 2009). Although we did not detect an accumulation of STING when COPA is mutated, we also did not observe the expected degradation of STING seen when it signals (Gonugunta et al., 2017), leaving open the question of disrupted STING degradation in this context. Of note, STING trafficking and signaling can be disrupted by inhibiting the function of ARF1, a coactivator of COPI (Dobbs et al., 2015; Gui et al., 2019), through the de-myristoylation of ARF1 GTPase by the Shigella effector protein IpaJ, by ARF1 GTPase activity inhibition upon treatment with brefeldin A (Dobbs et al., 2015), or by siRNA knock-down of ARF1 (Gui et al., 2019). Given our data, these observations may reflect a function of ARF1 in mediating IFN signaling unrelated to COPA/COPI (Sztul et al., 2019), or of COPA unrelated to COPI/ARF1.
Summarizing, we describe STING-dependent constitutive induction of type I IFN due to heterozygous dominant-negative missense mutations in COPA. These data indicate a role for COPA in STING trafficking and signaling necessary for the maintenance of cellular homeostasis, and highlight the ER–Golgi axis as a potential therapeutic target in certain autoinflammatory states.
Materials and methods
Patients
Patients were recruited from the Pediatric Rheumatology Department, Royal Hospital for Sick Children (Edinburgh, UK), the Pediatric Pneumology Department, Trousseau Hospital (Assistance Publique–Hôpitaux de Paris, Paris, France), the Pneumology Department, Cochin Hospital (Assistance Publique–Hôpitaux de Paris, Paris, France) and the Pediatric Rheumatology unit, University Hospital of Lille (Lille, France). Data relating to four patients (F2.P1, F2.P2, F3.P5, and F3.P6) have been previously reported (Boulisfane-El Khalifi et al., 2019; Frémond et al., 2020). We recorded clinical and laboratory data, including lung histology when performed, as well as information relating to IFN status. The latter included the expression of ISGs in peripheral blood, measurement of IFN-α in serum or plasma using an ultra-sensitive digital ELISA assay (Rodero et al., 2017), and STAT1 phosphorylation in lymphocytes and monocytes extracted from whole blood. Measurement of cytokines other than IFN-α was performed using ELISA. The study was approved by the Comité de protection des personnes Ile de France II and the French advisory committee on data processing in medical research (ID-RCB: 2014-A01017-40). Consent of the parents and/or patients, depending on age, was obtained for conducting the experiments.
Histological analyses
Formalin-fixed paraffin-embedded lung biopsy slides of patient 1 from family 1 (F1.P1) were analyzed in comparison to equivalent samples from a patient carrying a TMEM173-activating mutation (Jeremiah et al., 2014). Sections underwent H&E, Giemsa, and immunohistochemical staining, using antibodies for CD5, CD20, CD68, and smooth-muscle actin.
IFN score (Rice et al., 2013)
Total RNA was extracted from whole blood with a PAXgene (PreAnalytix) RNA isolation kit. RNA concentration was assessed with a spectrophotometer (FLUOstar Omega, Labtech). RT-qPCR analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) and cDNA derived from 40 ng total RNA. Using TaqMan probes for IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs00356631_g1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1), and SIGLEC1 (Hs00988063_m1), the relative abundance of each target transcript was normalized to the expression level of HPRT1 (Hs03929096_g1) and 18S (Hs999999001_s1), and assessed with the Applied Biosystems StepOne software v2.1 and DataAssist software v3.01. For each of the six probes, individual (patient and control) data were expressed relative to a single calibrator. The median fold change of the six genes compared with the median of 29 previously collected HCs was used to create an IFN score for each individual, with an abnormal score being defined as >2 SDs above the mean of the control group, i.e., 2.466.
For NanoString ISG analysis, total RNA was similarly extracted from whole blood with a PAXgene (PreAnalytix) RNA isolation kit. Analysis of 24 genes and 3 housekeeping genes was conducted using the NanoString customer designed CodeSets according to the manufacturer’s recommendations (NanoString Technologies). Agilent Tapestation was used to assess the quality of the RNA. 100 ng of total RNA was loaded for each sample. Data were processed with nSolver software (NanoString Technologies). The data were normalized relative to the internal positive and negative calibrators, the three reference probes, and the control samples. The median of the 24 probes for each of 27 HC samples was calculated. The mean NanoString score of the 27 HCs +2 SD of the mean was calculated. Scores above this value (>2.724) were designated as positive. The list of probes used in NanoString ISG analysis is supplied in Table S4.
Quantification of IFN-α protein by Simoa assay
The Simoa IFN-α assay (Rodero et al., 2017) was developed using a Quanterix Homebrew Simoa assay according to the manufacturer’s instructions, and using two autoantibodies specific for IFN-α isolated and cloned from two APS1/autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients as described (Meyer et al., 2016; Rodero et al., 2017). The 8H1 antibody clone was used as a capture antibody after coating on paramagnetic beads (0.3 mg/ml), and the 12H5 was biotinylated (biotin/antibody ratio = 30/1) and used as the detector. Recombinant IFNα17/αI (PBL Assay Science) was used to produce a standard curve after cross-reactivity testing. The limit of detection was calculated as the mean value of all blank runs +3 SD and was 0.23 fg/ml.
Structural modeling
The effects of missense mutations on the stability of the COPA WD-repeat domain structure (PDB ID: 6PBG) were modeled with FoldX (Guerois et al., 2002) using default parameters. The presented ΔΔG values were calculated as the average of 10 FoldX replicates. The FoldX “RepairPDB” function was run before modeling the mutations. Protein structures were visualized with PyMol.
Cell culture
PBMCs were isolated by Ficoll-Paque density gradient (Lymphoprep, Proteogenix) from the blood of patients and healthy donors. Fresh or cryopreserved PBMCs were used for the assays. Control PBMCs were obtained from the Etablissement Français du Sang blood bank. PBMCs were cultured at 37°C in 5% CO2 in RPMI 1640 GlutaMax medium (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (GIBCO). PBMCs were treated with ruxolitinib 1 µM or BX795 2 µM. HEK 293T and 293FT cells (ATCC) were grown in 6-, 12-, or 96-well plates at 37°C in 5% CO2 in DMEM (GIBCO) supplemented with 10% (vol/vol) fetal bovine serum (GIBCO). Control and STING KO THP-1 monocytic cell lines were previously generated in the Manel laboratory (Cerboni et al., 2017). WT and cGAS KO THP-1 cells were from InvivoGen (THP1 Dual). MAVS KO THP-1 cell lines were recently generated in the Rehwinkel laboratory (Hertzog et al., 2020 Preprint). All THP-1 cell lines were cultured at 37°C in 5% CO2 in RPMI (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (GIBCO). THP-1 cells were stimulated for 24 h with 1 µg/ml poly(I:C) (High Molecular Weight; InvivoGen), 1 µg/ml 2′3′cGAMP (InvivoGen), or 0.25 µg/ml HT-DNA (Sigma-Aldrich) combined with Lipofectamine 2000 (Invitrogen), following the manufacturer’s instructions.
Plasmids
pCMV6-Entry vector encoding Myc-DKK-tagged human WT COPA (Origene) was used as the parental vector for mutagenesis. Mutant plasmids of COPA were generated via site-directed mutagenesis using the Q5 kit (E0554S; New England Biolabs) according to the manufacturer’s instructions. NEB 5-α Competent Escherichia coli was transformed with the newly synthesized plasmid DNA, and colonies were screened for the presence of the desired variants (primers are shown in Table S5). The p.V155M STING mutant was previously produced in the Crow laboratory, from the pMSCV-hygro+ plasmid carrying the WT TMEM173 gene (Addgene). pTRIP-SFFV-mTagBFP-2A, pTRIP-SFFV-mTagBFP2-2A STING WT, and pTRIP-SFFV-mTagBFP2-2A STING V155M were previously described (Cerboni et al., 2017). STING with deletions 1–82, 83–136, 1–136, and 342stop were obtained by overlapping PCR mutagenesis and then cloned in pTRIP-SFFV-tagBFP-2A, resulting in plasmids pTRIP-SFFV-mTagBFP2-2A STING del 1–82, del 83–136, and 342stop. In all final constructs, the DNA fragments originating from the PCR and encompassing the restriction sites used for cloning were fully verified by sequencing. Mouse cGAS was codon-optimized, synthetized, and cloned in pVAX1 (R-D Biotech and Invitrogen), resulting in plasmid pVAX1-cGAS.
Cell transfection
At 70% confluency, HEK293T cells were transiently cotransfected with 100 ng (96-well plates assays) or 1,000 ng (12-well plates assays) of total DNA plasmid using TransIT-293 (Mirus, MIR2700). The plasmids employed were either pCMV6-Myc-DKK encoding WT and mutant COPA, pMSCV-hygro(+) encoding WT or mutant TMEM173/STING variant, or pTRIP-SFFV-mtagBFP-2A encoding WT and truncated TMEM173 (see above), and corresponding EVs as controls. For STIM1 silencing, HEK293T cells were transiently cotransfected with 2,500 ng (6-well plates assays) of total DNA plasmid together with a siRNA targeting STIM1 (reference L-011785-00-0005 from Dharmacon) using TransIT-X2 (Mirus, MIR6000). The plasmids employed were pCMV6-Myc-DKK encoding WT COPA, pMSCV-hygro(+) encoding WT TMEM173/STING, and corresponding EVs as controls.
coIP
48 h after transfection, HEK293T cells were harvested in 0.5% CHAPS buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA) for FLAG IP or radioimmunoprecipitation assay (Thermo Fisher Scientific) lysis buffer for STING IP. Extracts were incubated overnight with 20 µg/ml of anti-FLAG M2 (Sigma-Aldrich; Table S6) or 5 µg/ml anti-STING (R&D Systems; Table S6) antibody in the presence of Protein G magnetic dynabeads (Invitrogen) and the resulting complexes washed with PBS (pH 7.4), 0.02% Tween 20, denatured, and eluted according to the manufacturer’s instructions.
Knockdown of gene expression by short hairpin RNA
Lentiviral constructs containing shRNA targeting COPA (TRCN0000065268, TRCN0000065269, and TRCN0000244426 for shCOPA_1 in WT, cGAS KO, and MAVS KO THP-1) were constructed by ligating annealed oligonucleotides into pLKO.1 (TRC cloning vector, a gift from D. Root, Broad Institute, Cambridge, MA; Addgene plasmid 10878), according to the RNAi consortium protocol (https://portals.broadinstitute.org/gpp/public/). pLKO.1 EV was from Open Biosystems and pLKO.1 control shRNA (nontarget, SHC016) from Sigma-Aldrich. Lentiviral vectors carrying these constructs were produced by calcium phosphate transfection of 293FT cells with shRNA constructs in combination with packaging vectors psPAX2, a gift from D. Trono (École polytechnique fédérale de Lausanne, Lausanne, Switzerland; Addgene plasmid 12260), and envelope pCMV-VSV-G (Addgene plasmid 8454). Medium of 70% confluent 293FT in 75-cm2 flasks was changed 2 h before transfection. Calcium phosphate precipitates were prepared by mixing 12.5 µg shRNA vectors with 12.5 µg psPAX2 and 5 µg pCMV-VSV-G in water for a final volume of 875 µl. 125 µl 2 M CaCl2 and 1 ml Hepes-buffered saline 2× (50 mM Hepes, 10 mM KCl, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.05) were sequentially added dropwise in slowly vortexed solution. Solutions were incubated at room temperature for 20 min and mixed gently with 293FT supernatant. Medium was replaced by 7 ml of culture medium 24 h later. Supernatants were collected, centrifuged at 1,700 rpm for 5 min, and 0.4-µm–filtered. 500,000 THP1 cells were transduced with 0.5 ml lentiviral vectors, 8 µg/ml polybrene (Millipore), and 10 mM Hepes (Invitrogen) in 12-well plates, and medium was replaced 24 h later. Cells were collected for analysis 4 d after transduction.
Western blot analysis
For whole cell lysate analysis, proteins were extracted from THP-1 and transfected HEK293T cells using lysis buffer (radioimmunoprecipitation assay, 1% protease inhibitor, 1% phosphatase inhibitor). Bolt LDS Sample Buffer (4×; Novex Life Technologies) and Bolt Sample Reducing agent (10×; Novex Life Technologies) were added to protein lysates, samples resolved on 4–12% Bis-Tris Plus gels (Invitrogen), and then transferred to nitrocellulose membrane (Invitrogen). Where protein phosphorylation status was investigated, membranes were blocked in LI-COR buffer and primary phospho-antibodies incubated for 48 h in the blocking solution. Otherwise, membranes were blocked with 5% nonfat milk in TBS and primary antibodies incubated overnight in blocking buffer supplemented with 0.1% Tween. A list of antibodies used in this study is supplied in Table S6. Membranes were washed and incubated with appropriate anti-mouse or anti-rabbit secondary antibodies for 45 min at room temperature (LI-COR). Signal was detected using the Odyssey CLx System (LI-COR). For STING Δ1-136 revelation in coIP assays, Quick Western Blot kit (LI-COR) was used to label STING antibody and avoid anti-FLAG light chain signal, according to the manufacturer’s instructions. Comparative signal analyses were performed using Image Studio Lite (LI-COR).
RT-qPCR quantification of gene expression
Total RNA was extracted using the RNAqueous-Micro Kit (Ambio), and reverse transcription performed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Levels of cDNA were quantified by RT-qPCR using Taqman Gene Expression Assay (Applied Biosystems) and normalized to the expression level of HPRT1. A list of the probes used in this study is supplied in Table S7.
Confocal microscopy
HEK293FT cells were plated on fibronectin (10 µg/ml; Sigma-Aldrich)–coated coverslips. The next day, cells were cotransfected with COPA and STING plasmids (1.5 µl of TransIT-293 Transfection Reagent [Mirus, MIR2704] for 0.25 µg of each plasmid in OptiMEM [Thermo Fisher Scientific, 51985042]) or corresponding control plasmids (pTRIP-SFFV-mTagBFP-2A or pCMV6-Entry). The next day, cells were transfected with cGAS plasmid (pVAX1-cGAS) or control plasmid (pVAX1; 1.5 µl of TransIT-293 Transfection Reagent for 0.5 µg of plasmid). 20 h later, cells were washed twice with PBS (GIBCO) and fixed on the coverslip by adding 500 µl of 4% PFA in PBS buffer for 30 min at room temperature. Coverslips were washed twice with PBS, and incubated with 500 ml of fresh PBS-glycine buffer (375 mg glycine in 50 ml of PBS) for 10 min at room temperature. After two washes in PBS, coverslips were incubated with 500 ml of staining buffer consisting of PBS, 0.2% BSA (Euromedex), 0.05% Saponin (Sigma-Aldrich, S4521), and 10% goat serum (Sigma-Aldrich, G9023) for 30 min at room temperature. For STING, GM130, and COPA visualization, coverslips were incubated with primary antibodies against STING, GM130, and FLAG-COPA (rabbit IgG anti-GM130 at 1:200, Abcam, ab52649; or rabbit IgG anti-GM130 at 1/100, Invitrogen, PA5-85643; mouse IgG2b anti-STING at 1/400, R&D Systems, MAB7169; mouse IgG1 anti-FLAG M2 at 1/1,500, Sigma-Aldrich, F3165) diluted in staining buffer for 1 h at room temperature in the dark. Coverslips were washed four times with staining buffer and incubated with secondary antibodies diluted in staining buffer (goat anti-rabbit IgG–Alexa 546 at 1:400, Invitrogen, A-11010; goat anti-mouse IgG2b–Alexa 647 at 1:400, Life Technologies, A-21242; goat anti-mouse IgG1–Alexa 488 at 1:400, Thermo Fisher Scientific, A-21121) for 1 h in the dark. Coverslips were washed five times with staining buffer, rinsed in water, and mounted on slides with DAPI Fluoromount-G (SouthernBiotech, 0100–20) and dried for 20 min at 37°C. Z-stacks of images were acquired with a Leica DmI8 inverted microscope equipped with an SP8 confocal unit using a 63× (1.4 NA) objective.
Image analysis
Analysis was performed on one selected focal plane per cell in a z-stack. Images were processed and analyzed using ImageJ Fiji. For STING/GM130 colocalization analysis, a region of interest corresponding to the cytoplasm of each cell was drawn manually, and a mask of the Golgi was obtained using a threshold on the GM130 channel. STING intensities were measured in both regions. The percentage of colocalization was defined as the percentage of STING summed pixel intensities contained inside the Golgi region divided by STING summed pixel intensities in the total cytoplasm region.
STAT phosphorylation ex vivo assay staining
Whole blood or ficolled PBMCs were fixed using Beckman Coulter PerFix Expose Fixation Buffer (10 min at room temperature) and then permeabilized using Beckman Coulter PerFix Expose Permeabilizing Buffer (5 min at 37°C). Cells were subsequently stained with PE-anti-STAT1 pY701, and cell surface markers (APC-anti-CD3, BV421-anti-CD8a, Alexa Fluor 750–anti-CD14) for 1 h at room temperature protected from light. Flow cytometry analysis was performed on a Beckman Coulter Gallios flow cytometer, and results were analyzed using Kaluza software v1.3.
Statistics
Analyses were performed with PRISM software (v6 for Macintosh, GraphPad Inc.) as indicated in the figure legends. A P value <0.05 was considered significant.
Online supplemental material
Fig. S1 shows the pedigrees of patients, lung pathology of patient F1.P1, and concentration of NF-κB–related cytokines in patients. Fig. S2 documents further characterization of IFN signaling upon down-regulation of COPA with a second shRNA in THP-1 WT, STING control, cGAS KO, and MAVS KO and shows functional characterization of KO cell lines. Fig. S3 presents data relating to the physical interaction of COPA and STING, as well as the effect of STIM1 silencing on the COPA–STING interaction. Table S1 shows patient characteristics. Table S2 shows the comparison of COPA patients from this study to those in the literature and to SAVI patients. Table S3 summarizes the pulmonary histopathological features of COPA patients compared with SAVI patients. Table S4, Table S5, Table S6, and Table S7 describe the NanoString probes, cloning primers, antibodies, and RT-qPCR primers used in this study, respectively.
Supplementary Material
shows characteristics of the patients.
displays a comparison of the COPA patients in this study with those in the literature and with SAVI patients.
shows pulmonary histopathological features of COPA patients in comparison to SAVI patients.
lists probes used for NanoString ISG analysis.
shows primers used for site-directed mutagenesis of COPA in this study.
is a list of antibodies used in this study.
is a list of primers used in this study for RT-qPCR.
Acknowledgments
We thank Immunoqure AG for provision of antibodies for the Simoa assay.
M.-L. Frémond received a grant from the Institut National de la Santé et de la Recherche Médicale (000427993) and acknowledges La Fondation Square. Y.J. Crow acknowledges the European Research Council (GA309449 and 786142-E-T1IFNs), and a state subsidy managed by the Agence Nationale de la Recherche under the “Investments for the Future” program (ANR-10-IAHU-01). Y.J. Crow and D. Duffy acknowledge the Agence Nationale de la Recherche (grant CE17001002). J.A. Marsh is supported by a Medical Research Council Career Development Award (MR/M02122X/1) and is a Lister Institute of Preventive Medicine Research Prize Fellow. N. Manel was supported by LABEX DCBIOL (ANR-10-IDEX-0001-02 PSL* and ANR-11-LABX-0043), ANR-17-CE15-0025-01, ANR-18-CE92-0022-01, Fondation BMS, Agence Nationale de Recherches sur le Sida et les Hépatites Virales (ECTZ71745), Sidaction (VIH2016126002), and Ile-de-France Emergence. M. Le Bihan received a doctoral fellowship from Ile-de-France ARDoc. The project was supported by MSDAVENIR (Devo-Decode Project).
Author contributions: Conceptualization: A. Lepelley, C. Uggenti, N. Manel, Y.J. Crow, and M.-L. Frémond; Methodology: A. Lepelley, M.J. Martin-Niclós, M. Le Bihan, C. Uggenti, G.I. Rice, D. Duffy, J. Hertzog, J. Rehwinkel, N. Manel, Y.J. Crow, and M.-L. Frémond; Formal analysis: J.A. Marsh; Investigation: A. Lepelley, M.J. Martin-Niclós, M. Le Bihan, C. Uggenti, G.I. Rice, V. Bondet, J. Hertzog, E. Carter, M. Depp, T.J. Molina, L. Seabra, L. Chatenoud, S. Chhun, and M.L. Frémond; Writing (original draft): A. Lepelley, Y.J. Crow, and M.-L. Frémond; Writing (review and editing): A. Lepelley, M. Le Bihan, J.A. Marsh, J. Hertzog, J. Rehwinkel, K.J. Mackenzie, B. Neven, N. Nathan, N. Manel, Y.J. Crow, and M.-L. Frémond; Funding acquisition: M.L. Frémond, N. Manel, and Y.J. Crow; Resources: J. Hertzog, J. Rehwinkel, S. Amselem, S. Boulisfane-El Khalifi, M. Brennan, A. Coulomb l’Hermine, M. Legendre, K.J. Mackenzie, J. Marey, C. McDougall, K.J. McKenzie, B. Neven, C. Thumerelle, M. Wislez, and N. Nathan; Supervision: A. Lepelley, J.A. Marsh, J. Rehwinkel, N. Nathan, N. Manel, Y.J. Crow, and M.-L. Frémond.
References
- Arakel E.C., and Schwappach B.. 2018. Formation of COPI-coated vesicles at a glance. J. Cell Sci. 131 jcs209890 10.1242/jcs.209890 [DOI] [PubMed] [Google Scholar]
- Boulisfane-El Khalifi S., Viel S., Lahoche A., Frémond M.-L., Lopez J., Lombard C., Dubos F., Reumaux H., Gnemmi V., Legendre M., et al. 2019. COPA syndrome as a cause of lupus nephritis. Kidney Int. Rep. 4:1187–1189. 10.1016/j.ekir.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F., and Barlowe C.. 2013. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14:382–392. 10.1038/nrm3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand C., Frémond M.-L., Bessis D., Carbasse A., Rice G.I., Bondet V., Duffy D., Chatenoud L., Blanche S., Crow Y.J., et al. 2019. Efficacy of JAK1/2 inhibition in the treatment of chilblain lupus due to TREX1 deficiency. Ann. Rheum. Dis. 78:431–433. 10.1136/annrheumdis-2018-214037 [DOI] [PubMed] [Google Scholar]
- Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., and Vance R.E.. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 478:515–518. 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni S., Jeremiah N., Gentili M., Gehrmann U., Conrad C., Stolzenberg M.-C., Picard C., Neven B., Fischer A., Amigorena S., et al. 2017. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214:1769–1785. 10.1084/jem.20161674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S.L.N., Robertson L., Rice G.I., Seabra L., Hilliard T.N., Crow Y.J., and Ramanan A.V.. 2020. Type 1 interferonopathy presenting as juvenile idiopathic arthritis with interstitial lung disease: report of a new phenotype. Pediatr. Rheumatol. Online J. 18:37 10.1186/s12969-020-00425-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C., et al. 2006a Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 38:917–920. 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. 2006b Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38:910–916. 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- de Jesus A.A., Hou Y., Brooks S., Malle L., Biancotto A., Huang Y., Calvo K.R., Marrero B., Moir S., Oler A.J., et al. 2020. Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J. Clin. Invest. 130:1669–1682. 10.1172/JCI129301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A., Dhir S., Borowski L.S., Jimenez L., Teitell M., Rötig A., Crow Y.J., Rice G.I., Duffy D., Tamby C., et al. 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 560:238–242. 10.1038/s41586-018-0363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs N., Burnaevskiy N., Chen D., Gonugunta V.K., Alto N.M., and Yan N.. 2015. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe. 18:157–168. 10.1016/j.chom.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodonova S.O., Aderhold P., Kopp J., Ganeva I., Röhling S., Hagen W.J.H., Sinning I., Wieland F., and Briggs J.A.G.. 2017. 9Å structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments. eLife. 6 e26691 10.7554/eLife.26691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun S.L., Fernandez D., Weiss T.M., and Li L.. 2019. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell. 178:290–301.e10. 10.1016/j.cell.2019.05.036 [DOI] [PubMed] [Google Scholar]
- Frémond M.-L., Rodero M.P., Jeremiah N., Belot A., Jeziorski E., Duffy D., Bessis D., Cros G., Rice G.I., Charbit B., et al. 2016. Efficacy of the Janus kinase 1/2 inhibitor ruxolitinib in the treatment of vasculopathy associated with TMEM173-activating mutations in 3 children. J. Allergy Clin. Immunol. 138:1752–1755. 10.1016/j.jaci.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Frémond M.-L., Uggenti C., Van Eyck L., Melki I., Bondet V., Kitabayashi N., Hertel C., Hayday A., Neven B., Rose Y., et al. 2017. Brief Report: Blockade of TANK-Binding Kinase 1/IKKɛ Inhibits Mutant Stimulator of Interferon Genes (STING)-Mediated Inflammatory Responses in Human Peripheral Blood Mononuclear Cells. Arthritis Rheumatol. 69:1495–1501. 10.1002/art.40122 [DOI] [PubMed] [Google Scholar]
- Frémond M.-L., Legendre M., Fayon M., Clement A., Filhol-Blin E., Richard N., Berdah L., Roullaud S., Rice G.I., Bondet V., et al. 2020. Use of ruxolitinib in COPA syndrome manifesting as life-threatening alveolar haemorrhage. Thorax. 75:92–95. 10.1136/thoraxjnl-2019-213892 [DOI] [PubMed] [Google Scholar]
- Gentili M., Lahaye X., Nadalin F., Nader G.P.F., Puig Lombardi E., Herve S., De Silva N.S., Rookhuizen D.C., Zueva E., Goudot C., et al. 2019. The N-Terminal Domain of cGAS Determines Preferential Association with Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Rep. 26:2377–2393.e13. 10.1016/j.celrep.2019.01.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonugunta V.K., Sakai T., Pokatayev V., Yang K., Wu J., Dobbs N., and Yan N.. 2017. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Rep. 21:3234–3242. 10.1016/j.celrep.2017.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R., Nielsen J.E., and Serrano L.. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320:369–387. 10.1016/S0022-2836(02)00442-4 [DOI] [PubMed] [Google Scholar]
- Gui X., Yang H., Li T., Tan X., Shi P., Li M., Du F., and Chen Z.J.. 2019. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 567:262–266. 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., et al. 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 42:332–343. 10.1016/j.immuni.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Hartmann G. 2017. Nucleic Acid Immunity. Adv. Immunol. 133:121–169. 10.1016/bs.ai.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog J., Rigby R.E., Roll S., Cursi C., Chauveau L., Davenne T., and Rehwinkel J.. 2020. Varicella-Zoster Virus ORF9 Is an Antagonist of the DNA Sensor cGAS. bioRxiv 10.1101/2020.02.11.943415 (Preprint posted February 11, 2020) [DOI] [PMC free article] [PubMed]
- Jensson B.O., Hansdottir S., Arnadottir G.A., Sulem G., Kristjansson R.P., Oddsson A., Benonisdottir S., Jonsson H., Helgason A., Saemundsdottir J., et al. 2017. COPA syndrome in an Icelandic family caused by a recurrent missense mutation in COPA. BMC Med. Genet. 18:129 10.1186/s12881-017-0490-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeremiah N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.-C., Goudin N., Frémond M.-L., Nitschke P., Molina T.J., et al. 2014. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124:5516–5520. 10.1172/JCI79100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The Genome Aggregation Database Consortium . 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210 (Preprint posted August 13, 2019) [DOI]
- Kim J., Gupta R., Blanco L.P., Yang S., Shteinfer-Kuzmine A., Wang K., Zhu J., Yoon H.E., Wang X., Kerkhofs M., et al. 2019. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 366:1531–1536. 10.1126/science.aav4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzke S., Rietschel C., and Horneff G.. 2019. Baricitinib in therapy of COPA syndrome in a 15-year-old girl. Eur. J. Rheumatol. 1–4:1–4. 10.5152/eurjrheum.2019.18177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X., Gentili M., Silvin A., Conrad C., Picard L., Jouve M., Zueva E., Maurin M., Nadalin F., Knott G.J., et al. 2018. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell. 175:488–501.e22. 10.1016/j.cell.2018.08.062 [DOI] [PubMed] [Google Scholar]
- Lavieu G., Orci L., Shi L., Geiling M., Ravazzola M., Wieland F., Cosson P., and Rothman J.E.. 2010. Induction of cortical endoplasmic reticulum by dimerization of a coatomer-binding peptide anchored to endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 107:6876–6881. 10.1073/pnas.1002536107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G., Zheng H., and Rothman J.E.. 2013. Stapled Golgi cisternae remain in place as cargo passes through the stack. eLife. 2 e00558 10.7554/eLife.00558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F., Gaynor E.C., Hennecke S., Démollière C., Duden R., Emr S.D., Riezman H., and Cosson P.. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 79:1199–1207. 10.1016/0092-8674(94)90011-6 [DOI] [PubMed] [Google Scholar]
- Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Sanchez G.A.M., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S., et al. 2014. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371:507–518. 10.1056/NEJMoa1312625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., et al. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347 aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Ma W., and Goldberg J.. 2013. Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 32:926–937. 10.1038/emboj.2013.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K.J., Carroll P., Martin C.-A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 548:461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki I., Rose Y., Uggenti C., Van Eyck L., Frémond M.-L., Kitabayashi N., Rice G.I., Jenkinson E.M., Boulai A., Jeremiah N., et al. 2017. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol. 140:543–552.e5. 10.1016/j.jaci.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Meuwissen M.E.C., Schot R., Buta S., Oudesluijs G., Tinschert S., Speer S.D., Li Z., van Unen L., Heijsman D., Goldmann T., et al. 2016. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 213:1163–1174. 10.1084/jem.20151529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Woodward M., Hertel C., Vlaicu P., Haque Y., Kärner J., Macagno A., Onuoha S.C., Fishman D., Peterson H., et al. ; APECED patient collaborative . 2016. AIRE-Deficient Patients Harbor Unique High-Affinity Disease-Ameliorating Autoantibodies. Cell. 166:582–595. 10.1016/j.cell.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T., Barber G.N., Arai H., and Taguchi T.. 2016. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7:11932 10.1038/ncomms11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorelahi R., Perez G., and Otero H.J.. 2018. Imaging findings of Copa syndrome in a 12-year-old boy. Pediatr. Radiol. 48:279–282. 10.1007/s00247-017-3961-3 [DOI] [PubMed] [Google Scholar]
- Ogawa E., Mukai K., Saito K., Arai H., and Taguchi T.. 2018. The binding of TBK1 to STING requires exocytic membrane traffic from the ER. Biochem. Biophys. Res. Commun. 503:138–145. 10.1016/j.bbrc.2018.05.199 [DOI] [PubMed] [Google Scholar]
- Ouyang S., Song X., Wang Y., Ru H., Shaw N., Jiang Y., Niu F., Zhu Y., Qiu W., Parvatiyar K., et al. 2012. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 36:1073–1086. 10.1016/j.immuni.2012.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., Yang J.-S., Schmider A.B., Soberman R.J., and Hsu V.W.. 2015. Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42. Nature. 521:529–532. 10.1038/nature14457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A., and Spencer C.H.. 2019. An unprecedented COPA gene mutation in two patients in the same family: comparative clinical analysis of newly reported patients with other known COPA gene mutations. Pediatr. Rheumatol. Online J. 17:59 10.1186/s12969-019-0359-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P.A., Dietrich F., Bewersdorf J., Rothman J.E., and Lavieu G.. 2013. Inter-Golgi transport mediated by COPI-containing vesicles carrying small cargoes. eLife. 2 e01296 10.7554/eLife.01296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff V., Adolf F., Brügger B., and Wieland F.. 2011. COPI budding within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3 a005231 10.1101/cshperspect.a005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M., Chan E.Y.W., and Tooze S.A.. 2009. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185:305–321. 10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Kasher P.R., Forte G.M.A., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A., et al. 2012. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44:1243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Forte G.M.A., Szynkiewicz M., Chase D.S., Aeby A., Abdel-Hamid M.S., Ackroyd S., Allcock R., Bailey K.M., Balottin U., et al. 2013. Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 12:1159–1169. 10.1016/S1474-4422(13)70258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Del Toro Duany Y., Jenkinson E.M., Forte G.M., Anderson B.H., Ariaudo G., Bader-Meunier B., Baildam E.M., Battini R., Beresford M.W., et al. 2014. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 46:503–509. 10.1038/ng.2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Melki I., Frémond M.-L., Briggs T.A., Rodero M.P., Kitabayashi N., Oojageer A., Bader-Meunier B., Belot A., Bodemer C., et al. 2017. Assessment of type I interferon signaling in pediatric inflammatory disease. J. Clin. Immunol. 37:123–132. 10.1007/s10875-016-0359-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Park S., Gavazzi F., Adang L.A., Ayuk L.A., Van Eyck L., Seabra L., Barrea C., Battini R., Belot A., et al. 2020. Genetic and phenotypic spectrum associated with IFIH1 gain-of-function. Hum. Mutat. 41:837–849. 10.1002/humu.23975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., Decalf J., Bondet V., Hunt D., Rice G.I., Werneke S., McGlasson S.L., Alyanakian M.-A., Bader-Meunier B., Barnerias C., et al. 2017. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J. Exp. Med. 214:1547–1555. 10.1084/jem.20161451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez G.A.M., Reinhardt A., Ramsey S., Wittkowski H., Hashkes P.J., Berkun Y., Schalm S., Murias S., Dare J.A., Brown D., et al. 2018. JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J. Clin. Invest. 128:3041–3052. 10.1172/JCI98814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G., Zhang C., Chen Z.J., Bai X.-C., and Zhang X.. 2019. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature. 567:389–393. 10.1038/s41586-019-0998-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Woo J.S., Wu B., El-Sherbiny Y.M., Leung J., Chupradit K., Rice L., Seo G.J., Calmettes G., Ramakrishna C., et al. 2019. The Ca2+ sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 20:152–162. 10.1038/s41590-018-0287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Zhou M., Imamichi H., Jiao X., Sherman B.T., Lane H.C., and Imamichi T.. 2017. STING is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci. Signal. 10 eaah5054 10.1126/scisignal.aah5054 [DOI] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E., Chen P.-W., Casanova J.E., Cherfils J., Dacks J.B., Lambright D.G., Lee F.S., Randazzo P.A., Santy L.C., Schürmann A., et al. 2019. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell. 30:1249–1271. 10.1091/mbc.E18-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Xu H., Zhou C., Peng Y., Liu H., Liu J., Li H., Yang H., and Zhao S.. 2020. STING-Associated Vasculopathy with Onset in Infancy in Three Children with New Clinical Aspect and Unsatisfactory Therapeutic Responses to Tofacitinib. J. Clin. Immunol. 40:114–122. 10.1007/s10875-019-00690-9 [DOI] [PubMed] [Google Scholar]
- Taveira-DaSilva A.M., Markello T.C., Kleiner D.E., Jones A.M., Groden C., Macnamara E., Yokoyama T., Gahl W.A., Gochuico B.R., and Moss J.. 2019. Expanding the phenotype of COPA syndrome: a kindred with typical and atypical features. J. Med. Genet. 56:778–782. 10.1136/jmedgenet-2018-105560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui J.L., Estrada O.A., Deng Z., Wang K.M., Law C.S., Elicker B.M., Jones K.D., Dell S.D., Gudmundsson G., Hansdottir S., et al. 2018. Analysis of pulmonary features and treatment approaches in the COPA syndrome. ERJ Open Res. 4 00017-2018 10.1183/23120541.00017-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggenti C., Lepelley A., and Crow Y.J.. 2019. Self-Awareness: Nucleic Acid-Driven Inflammation and the Type I Interferonopathies. Annu. Rev. Immunol. 37:247–267. 10.1146/annurev-immunol-042718-041257 [DOI] [PubMed] [Google Scholar]
- Vece T.J., Watkin L.B., Nicholas S., Canter D., Braun M.C., Guillerman R.P., Eldin K.W., Bertolet G., McKinley S., de Guzman M., et al. 2016. Copa syndrome: a novel autosomal dominant immune dysregulatory disease. J. Clin. Immunol. 36:377–387. 10.1007/s10875-016-0271-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi S., Tsui J., Mariani M., Pastorino C., Caorsi R., Sacco O., Ravelli A., Shum A.K., Gattorno M., and Picco P.. 2018. Type I interferon pathway activation in COPA syndrome. Clin. Immunol. 187:33–36. 10.1016/j.clim.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Volpi S., Insalaco A., Caorsi R., Santori E., Messia V., Sacco O., Terheggen-Lagro S., Cardinale F., Scarselli A., Pastorino C., et al. 2019. Efficacy and adverse events during janus kinase inhibitor treatment of SAVI syndrome. J. Clin. Immunol. 39:476–485. 10.1007/s10875-019-00645-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.G., Serafini T., and Rothman J.E.. 1991. ‘Coatomer’: a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 349:248–251. 10.1038/349248a0 [DOI] [PubMed] [Google Scholar]
- Watkin L.B., Jessen B., Wiszniewski W., Vece T.J., Jan M., Sha Y., Thamsen M., Santos-Cortez R.L.P., Lee K., Gambin T., et al. ; Baylor-Hopkins Center for Mendelian Genomics . 2015. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat. Genet. 47:654–660. 10.1038/ng.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., et al. 2015. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 517:89–93. 10.1038/nature13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows characteristics of the patients.
displays a comparison of the COPA patients in this study with those in the literature and with SAVI patients.
shows pulmonary histopathological features of COPA patients in comparison to SAVI patients.
lists probes used for NanoString ISG analysis.
shows primers used for site-directed mutagenesis of COPA in this study.
is a list of antibodies used in this study.
is a list of primers used in this study for RT-qPCR.