By elevating levels of the GATA2 transcription factor in hematopoietic progenitor cells, a transcriptional enhancer establishes multilineage differentiation potential. During embryogenesis, deleting the enhancer upregulates innate immune machinery and deconstructs multilineage potential to favor monocytic differentiation.
Abstract
Stem and progenitor cell fate transitions constitute key decision points in organismal development that enable access to a developmental path or actively preclude others. Using the hematopoietic system, we analyzed the relative importance of cell fate–promoting mechanisms versus negating fate-suppressing mechanisms to engineer progenitor cells with multilineage differentiation potential. Deletion of the murine Gata2−77 enhancer, with a human equivalent that causes leukemia, downregulates the transcription factor GATA2 and blocks progenitor differentiation into erythrocytes, megakaryocytes, basophils, and granulocytes, but not macrophages. Using multiomics and single-cell analyses, we demonstrated that the enhancer orchestrates a balance between pro- and anti-fate circuitry in single cells. By increasing GATA2 expression, the enhancer instigates a fate-promoting mechanism while abrogating an innate immunity–linked, fate-suppressing mechanism. During embryogenesis, the suppressing mechanism dominated in enhancer mutant progenitors, thus yielding progenitors with a predominant monocytic differentiation potential. Coordinating fate-promoting and -suppressing circuits therefore averts deconstruction of a multifate system into a monopotent system and maintains critical progenitor heterogeneity and functionality.
Introduction
Stem and progenitor cell fate transitions are critical determinants of development, physiological homeostasis, and adaptive responses to life-threatening stresses. In principle, engineering a particular fate potential into a cell with developmental plasticity can be accomplished by instigating fate-promoting or negating fate-suppressing mechanisms. Whether these operationally distinct mechanisms are commonly mutually exclusive or interlinked is unclear. It is instructive to consider this problem in the context of hematopoietic stem and progenitor cells (HSPCs) that generate the diverse cells comprising blood. Major progress has been made in identifying the heterogeneous HSPC populations and the intrinsic transcriptional networks and microenvironment-based mechanisms that control their functional transitions (Haas et al., 2018; Weissman, 2016). These studies have forged principles that guide a wide swath of basic and translational research and extend well beyond the hematopoietic system.
A plethora of macromolecules (proteins, RNAs, and metabolites) control hematopoiesis and generate HSPC heterogeneity (Haas et al., 2018; Orkin and Zon, 2008; Rossi et al., 2012). A major challenge has been to establish a global perspective of how factors, signals, and pathways functionally intersect or operate independently to sustain HSPC pools and accommodate massive demands to generate diverse progeny. Conventional strategies involve loss-of-function or gain-of-function analyses while measuring mechanistic (e.g., impact on a transcriptome) and biological (e.g., impact on a cell state transition) consequences. Transcriptomes of tens of thousands of mRNAs, which might or might not be highly concordant with proteomes, often weigh heavily in functional assessments. Proteomic advances permit quantification of several thousand proteins in a mammalian cell, far short of comprehensive coverage (Richards et al., 2015). Although single-cell transcriptomic (Watcham et al., 2019) and proteomic (Palii et al., 2019) analyses have utility for deconvoluting mechanisms emerging from cell population data, when deployed alone, these approaches may have intrinsic limitations. Amalgamating transcriptomic and proteomic data can surmount limitations to mechanistic discovery, including those involving master regulators that instigate complex networks to establish and/or maintain cellular states.
Transcription factors, such as the master hematopoietic regulator GATA2 (Tsai and Orkin, 1997; Tsai et al., 1994), establish genetic networks that promote HSPC proliferation, survival, and differentiation (Katsumura et al., 2017). Mutations of murine Gata2 (de Pater et al., 2013; Ling et al., 2004; Rodrigues et al., 2005; Tsai et al., 1994) or +9.5 and −77 enhancers (+9.5 kb downstream and −77 kb upstream of the Gata2 transcription start site; Gao et al., 2013; Grass et al., 2006; Johnson et al., 2012; Johnson et al., 2015; Mehta et al., 2017; Soukup et al., 2019) abrogate HSPC genesis and function. Human GATA2 coding (Dickinson et al., 2011; Hahn et al., 2011; Hsu et al., 2011; Ostergaard et al., 2011) or +9.5 enhancer (Hsu et al., 2013; Johnson et al., 2012) mutations yield immunodeficiency and predisposition to develop myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML; Churpek and Bresnick, 2019; Dickinson et al., 2014; McReynolds et al., 2018; Spinner et al., 2014). In poor-prognosis 3q21;q26 AML, an inversion translocates −77 next to MECOM (encoding the transcription factor EVI1) to generate a super-enhancer, increasing EVI1 and reducing GATA2 to cause leukemia (Gröschel et al., 2014; Katayama et al., 2017; Yamazaki et al., 2014).
As an erythroid and megakaryocytic fate-promoting mechanism, GATA2 increases GATA1 expression (Mehta et al., 2017), which upregulates its own coregulator friend of GATA1 (FOG1; Crispino et al., 1999). As a fate-suppressing mechanism, GATA1 antagonizes PU.1, thereby blocking myelopoiesis (Nerlov et al., 2000; Zhang et al., 2000); however, whether this occurs in single cells is unclear (Hoppe et al., 2016). Solitary and integrated fate-promoting and -suppressing mechanisms provide the architectural framework for building complex developmental and biological processes.
Prior to GATA1 expression, GATA2 must meet unique challenges to know when and where to induce HSPC cell genesis and function (Churpek and Bresnick, 2019; Katsumura et al., 2017). Although GATA2 and PU.1 cooperatively stimulate progenitor differentiation into mast cells (Walsh et al., 2002), the converging mechanisms that endow GATA2 (and other master regulatory transcription factors) with fate-promoting versus -suppressing activities are undefined. We describe a mechanism in which an enhancer (−77) promotes GATA2 expression, inducing a fate-promoting mechanism and concomitantly abrogating a fate-suppressing mechanism. −77−/− progenitors mounted a response to sustain the fate-suppressing mechanism involving innate immune machinery. The consequence of this genetic aberration is deconstruction of an integrated multipotent differentiation system into a predominantly unipotent system. Furthermore, the innovative multiomics resource of WT and enhancer mutant primary progenitor cells, involving quantitative proteomics, single-cell transcriptomics, and population transcriptomics, linked to GATA2 rescue, will catalyze many additional discoveries beyond those described herein.
Results
Discovering progenitor cell fate mechanisms using multiomics
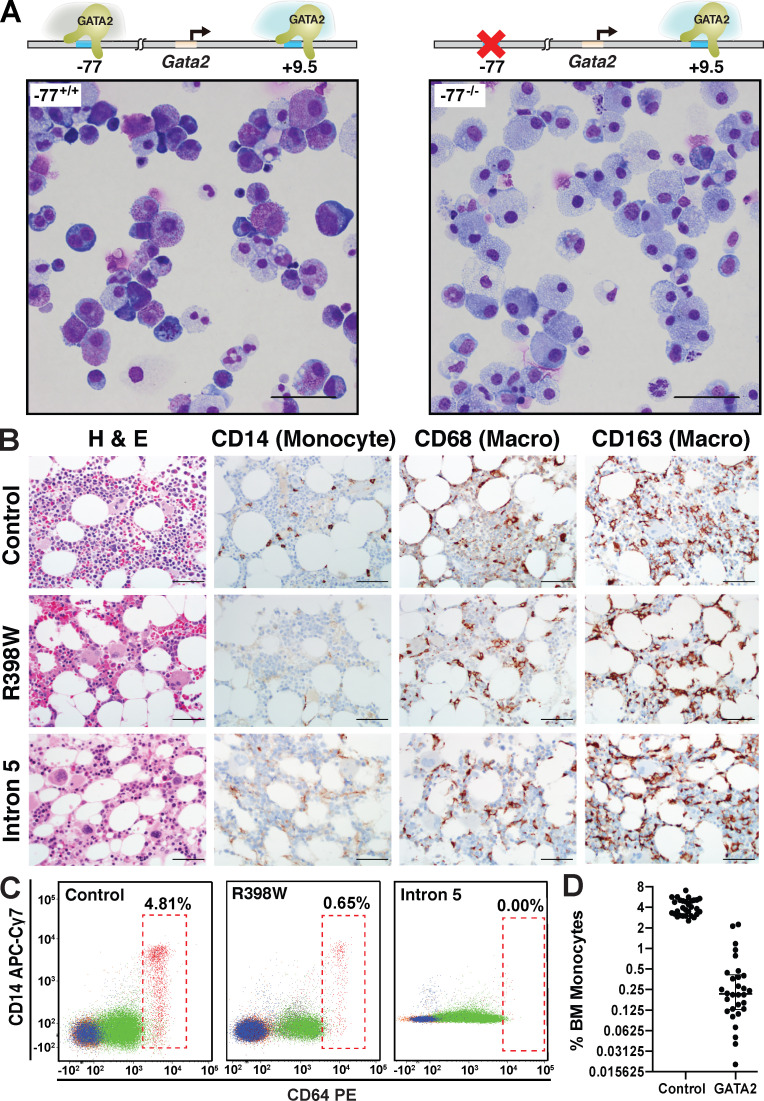
Hematopoietic progenitors from murine fetal liver, WT for the Gata2 −77 enhancer (−77+/+), can undergo erythroid, megakaryocytic, granulocytic, and monocytic differentiation ex vivo. By contrast, progenitors with a −77 homozygous deletion (−77−/−) exhibit a predominant monocytic cell fate and generate abundant macrophages (Fig. 1 A; Johnson et al., 2015). Human GATA2 deficiency syndrome, resulting from GATA2 coding (R398W) or intron 5 (human equivalent of murine +9.5 enhancer) mutations, is characterized by bone marrow hypocellularity, dysplastic megakaryocytes, and monocytopenia (Fig. 1 B; Calvo et al., 2011). Flow cytometry confirmed monocyte depletion from marrow aspirates (Fig. 1, C and D). Despite the monocytopenia, bone marrow macrophages were abundant in heterozygous R398W or intron 5 mutant patients (Fig. 1 B), consistent with the capacity of GATA2-deficient murine progenitors to generate macrophages ex vivo.
Figure 1.
Gata2 and GATA2 enhancer and coding mutations deconstruct a multifate program. (A) Representative Giemsa staining of dissociated colonies from −77+/+ and −77−/− E14.5 fetal liver cells cultured for 8 d in M3434 complete methylcellulose media. −77+/+ and −77−/− alleles are depicted. Scale bars = 50 µm. (B) Immunohistochemical (IHC) detection of CD68+ bone marrow macrophages from patients with GATA2 coding or intron 5 heterozygous mutations. IHC of bone marrow from a healthy control subject, a 17-yr-old female with GATA2 deficiency and germline GATA2 mutation (c.1192C>T; p. R398W) with early dysplastic changes and normal bone marrow karyotype, and a 29-yr-old female with GATA2 deficiency and germline mutation in intron 5 (c.1017+572C>T) diagnosed with MDS with abnormal bone marrow karyotype involving trisomy 1q. Sections were stained with H&E or CD68 or CD163 antibody to detect bone marrow macrophages and CD14 antibody to detect monocytes. Scale bars = 50 µm. (C) Flow cytometric analysis of marrow aspirates from control subject and patient samples described in B using CD14 versus CD64 to detect monocytes and monocytic precursors (RBCs in hatched boxes). Percentages are of total marrow cells. (D) Quantitation of bone marrow (BM) monocytes from 30 healthy volunteer control subjects and 30 patients with GATA2 mutations presenting with bone marrow failure, pre-MDS, or overt MDS. Individual data points are graphed on a log2 scale with median values and interquartile ranges demarcated. P = 2.99e−23 using unpaired two-tailed Student’s t tests. APC, allophycocyanin; Cy7, cyanine 7.
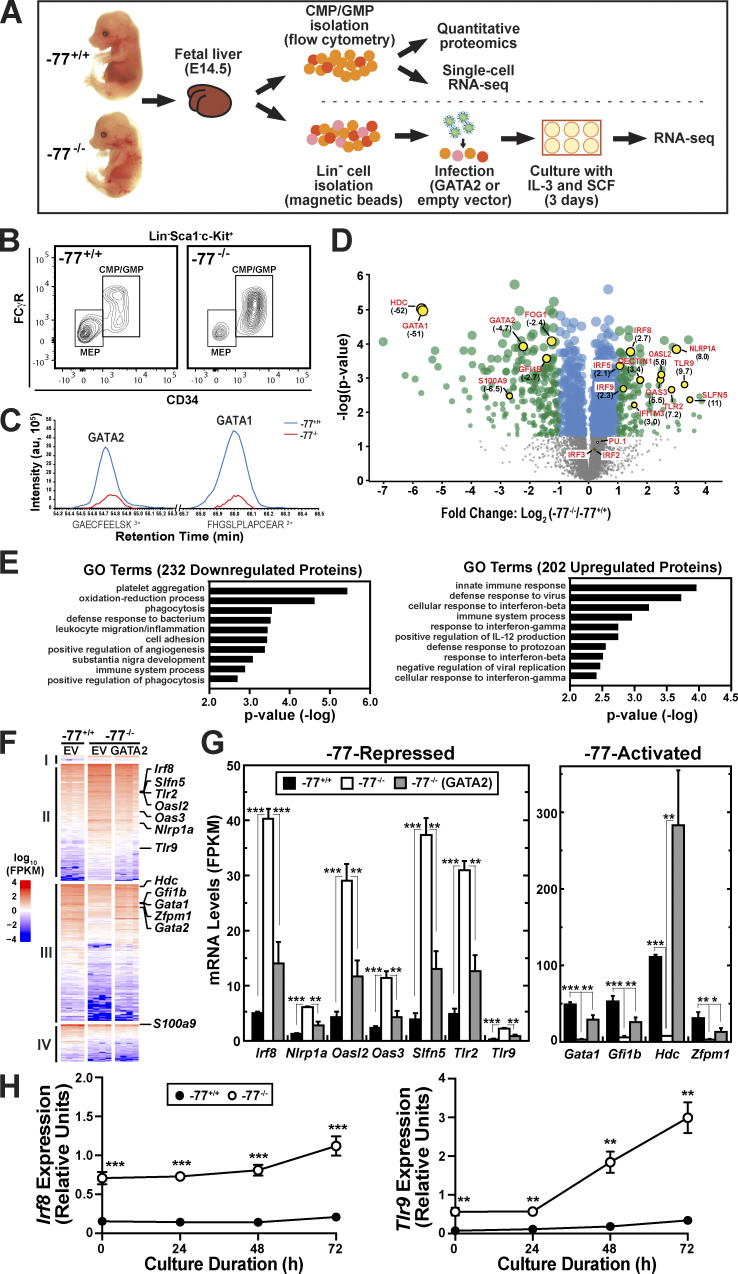
To address the contribution of cell fate–promoting versus –suppressing mechanisms to the activity of a cell fate–regulatory enhancer, we isolated a lineage-negative (Lin−)Sca1−c-Kit+CD34+ myeloid progenitor population (common myeloid progenitor [CMP] and granulocyte-monocyte progenitor [GMP]) from fetal liver of embryonic day 14.5 (E14.5) −77+/+ and −77−/− mouse embryos (Fig. 2, A and B). The CMP/GMP pool is a complex mixture of progenitors with diverse transcriptional profiles (Olsson et al., 2016; Paul et al., 2015). Previously, we showed that −77+/+ fetal liver CMPs possessed erythroid and myeloid colony-forming capacity ex vivo, whereas GMPs only generated myeloid cells. −77−/− CMPs and GMPs were largely restricted toward macrophage production (Johnson et al., 2015). Herein, we analyzed the molecular changes resulting from loss of the −77 enhancer, which underlie the restricted cell fate potential. Quantitative proteomics was conducted to discover the −77-regulated protein ensemble endowing progenitors with multifate potential (Fig. 2, C and D). GATA2 was 4.7-fold lower in −77−/− progenitors. GATA2 directly activates Gata1 transcription in progenitors (Mehta et al., 2017), and although essential GATA1 functions are manifested principally in committed megakaryocytes, erythrocyte and mast cell precursors, and their developing progeny (Fujiwara et al., 1996; Pevny et al., 1995; Pevny et al., 1991; Tsang et al., 1998), GATA1 protein was detected in progenitors. GATA1 was 51-fold lower in −77−/− progenitors (Fig. 2, C and D), and the GATA1-induced gene/protein FOG1 (Crispino et al., 1999) was 2.4-fold lower (Fig. 2 D). GATA2 directly activates Hdc and Gfi1b transcription (Gao et al., 2013; Katsumura et al., 2016; Katsumura et al., 2014; Mehta et al., 2017), and respective proteins were 52- and 2.7-fold lower in −77−/− progenitors.
Figure 2.
Elucidating cell fate mechanisms through multiomics and genetic rescue of transcriptomic aberrations in enhancer mutant progenitors. (A) Schematic representation of experimental workflow for the multiomic analyses. (B) Representative flow plot of the CMP/GMP pool (Lin−Sca1−cKit+CD34+) isolated for proteomic and scRNA-seq analyses. The CMP:GMP ratio was 1:2 in both −77+/+ and −77−/− fetal livers. The megakaryocyte-erythrocyte–restricted progenitor (MEP) population was excluded because the −77 deletion strongly reduces fetal liver MEPs. (C) Quantitative proteomic analysis performed by MS. Flow-sorted cells were pooled into replicates of 5–6 × 106 cells. Three pools of −77+/+ (n = 17 from 11 litters) and four pools of −77−/− (n = 13 from seven litters) were analyzed. The plot shows reduced recovery of representative peptides of GATA2 and GATA1 in −77−/− samples. (D) Volcano plot depicting 202 upregulated and 232 downregulated proteins in −77−/− CMP/GMP pool (q < 0.05). Select upregulated IFN targets and downregulated proteins are highlighted. Fold change relative to −77+/+ is shown in parentheses. See also Table S1. (E) Top categories from Gene Ontology (GO) analysis of down- and upregulated proteins using DAVID Bioinformatics Resources (https://david.ncifcrf.gov). See also Fig. S1. (F) Population RNA-seq analysis. Heatmap of the 3,161 differentially expressed (DE) genes (fold change ≥2 and adjusted P value <0.05) from comparing −77−/− (n = 4) and −77+/+ (n = 4) Lin− fetal liver cells infected with empty pMSCV vector (EV) and cultured for 3 d. Infection of −77−/− with a GATA2 expression vector (n = 4) parses the DE genes into four categories: (I) no rescue of upregulated genes (n = 51), (II) rescue of upregulated genes (n = 1,251), (III) rescue of downregulated genes (n = 1,463) and (IV) no rescue of downregulated genes (n = 396). See also Fig. S2. (G) Comparison of mRNA levels for select DE genes mined from the RNA-seq data. See also Table S2. (H) Quantitation of mRNA levels in −77+/+ (n = 5 from three litters) and −77−/− (n = 7 from three litters) Lin− fetal liver cells cultured for 72 h. In all graphs, error bars represent mean ± SEM. Statistics were calculated using unpaired two-tailed Student’s t test; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. FPKM, fragments per kilobase of transcript per million mapped reads.
Although GATA2 is not known to be a key regulator of genes/proteins mediating innate or adaptive immune processes, innate immune machinery was upregulated, including IFN signaling pathway components (Schneider et al., 2014; Table S1). IFN-inducible transcription factors, termed IFN regulatory factors (IRFs), were upregulated in −77−/− progenitors, including IRF5 (2.1-fold), IRF8 (2.7-fold), and IRF9 (2.3-fold). Deletion of murine Irf8 and biallelic human IRF8 mutations that cause severe monocytopenia and primary immunodeficiency disease (Bigley et al., 2018; Hambleton et al., 2011; Kurotaki et al., 2013; Yáñez et al., 2015) revealed IRF8 to be an essential monocytic differentiation determinant. PU.1 upregulates IRF8 expression (Schönheit et al., 2013), and IRF8 and PU.1 function collectively to control myeloid and inflammatory genes through composite binding sites (Marecki et al., 2001; Meraro et al., 2002). However, expression of Spi1 encoding PU.1 is not altered in −77−/− CMPs or GMPs (Johnson et al., 2015), and PU.1 levels were unaltered in the CMP/GMP pool (Fig. 2 D).
Other upregulated IFN-inducible innate immune components included the pattern recognition receptors TLR9 (9.7-fold) and TLR2 (7.2-fold; Fig. 2 D); the negative regulator of TLR signaling IRAK3 (IRAK-M; Kobayashi et al., 2002) was downregulated 2.8-fold. Other IFN-inducible proteins upregulated in −77−/− progenitors were oligoadenylate synthase-like protein 2 (OASL2; 5.6-fold), nucleotide-binding domain, leucine-rich repeat protein 1A (NLRP1A; 8.0-fold), the non-TLR pattern recognition receptor dectin-1 (3.4-fold), and the IFNγ target ISG15 (3.4-fold).
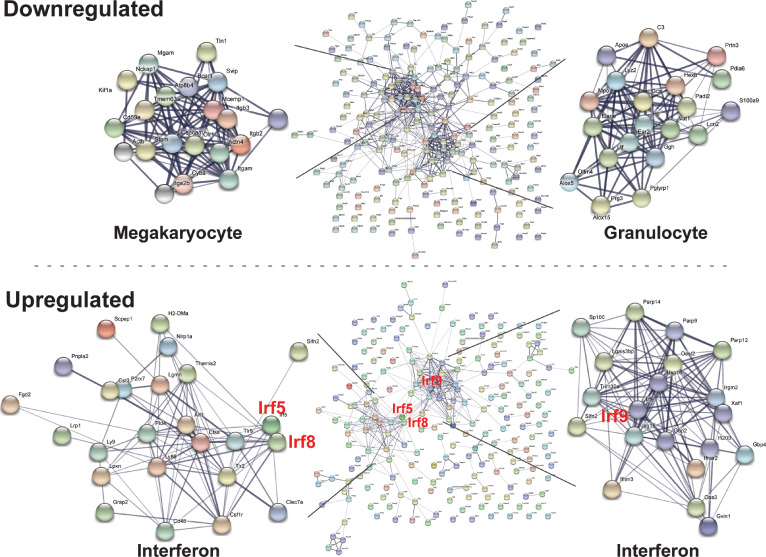
We used gene ontology and STRING network prediction tools to infer global consequences of proteome alterations. The 202 upregulated proteins highlighted a spectrum of immune and inflammatory mechanisms, whereas the 232 downregulated proteins were linked to functional processes in megakaryocyte and granulocyte biology (Fig. 2 E). This illustrated the overt loss of megakaryocyte and granulocyte biology–linked proteins, indicative of abrogated cell fate programs, with acquisition of IFN response proteins (Fig. S1).
Figure S1.
Network analysis reveals loss of erythroid, megakaryocyte, and granulocyte proteins and induction of innate immune proteins in progenitor cells. Functional relationships between up- or downregulated cohorts of proteins identified in our proteomic analysis of the −77−/− CMP/GMP pool were evaluated by using STRING (https://string-db.org). Relationships reveal loss of erythroid/megakaryocyte and granulocyte proteins and induction of inflammatory proteins in progenitor cells.
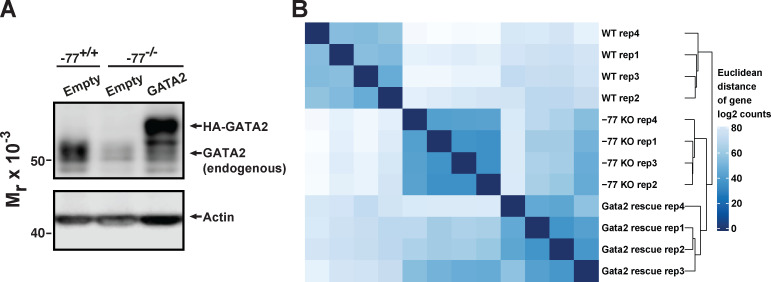
To test if the aberrant −77−/− progenitor proteome reflects GATA2 downregulation and not genes topologically associated with the −77 enhancer, we used our previously described genetic rescue system in primary −77−/− progenitor cells (Katsumura et al., 2018). Lin− progenitors from −77+/+ or −77−/− E14.5 fetal liver (Fig. 2 A; McIver et al., 2018) were infected with GATA2-expressing or control retroviruses and cultured for 3 d. Under conditions in which GATA2 is expressed at near-physiological levels (Fig. S2 A), transcriptomes (four replicates of each condition; Fig. S2 B) were elucidated with RNA sequencing (RNA-seq). Of the 3,161 differentially expressed genes between −77+/+ and −77−/− progenitors, GATA2 expression rescued or partially rescued the majority (86%; 2,714) of the alterations (Fig. 2 F). Rescue was defined as genes that were up- or downregulated in the GATA2 rescue system and in −77+/+ versus −77−/− progenitors.
Figure S2.
Reproducibility of biological replicates from the population RNA-seq analysis. (A) Representative Western blot of GATA2 expression in samples used for transcriptomic analysis. Lin− fetal liver progenitors were infected with retrovirus for expression of hemagglutinin (HA)-tagged GATA2 or the empty vector MSCV-PIG. The Western blot was probed with an antibody to detect both endogenous and expressed GATA2. (B) RNA-seq read counts for all genes were transformed to the log2 scale by using the DESeq2 rlog function and employed for calculating Euclidean distance. Hierarchical clustering of all the 12 RNA-seq datasets was performed on the basis of Euclidean distance and denoted to the right of the heatmap.
Many −77-repressed and -induced proteins (Fig. 2 D) were also regulated at the mRNA level (Fig. 2, F and G). Using criteria for protein and mRNA changes of twofold or greater and false discovery rate <0.05, we analyzed the 92 and 57 proteins downregulated and upregulated, respectively, in −77−/− progenitors. This analysis revealed 58 targets, 67% of which were regulated in a qualitatively indistinguishable manner at the mRNA level (Table S2). The rescue analysis demonstrated that GATA2 loss caused the IFN response and many of the gene expression alterations in −77−/− progenitors. Thus, the ectopic innate immune machinery induction in −77−/− progenitors was also detected in cultured −77−/− progenitors.
To further test if innate immune machinery induction is sustained when progenitors are segregated from niche components, progenitors were cultured for up to 3 d. At all times tested, Irf8 and Tlr9 mRNA levels in −77−/− exceeded those of −77+/+ progenitors (Fig. 2 H). Because the GATA2-reversible, aberrant −77−/− progenitor transcriptome is maintained upon progenitor propagation without a heterocellular microenvironment, these results support a cell-intrinsic transcriptome/proteome dysregulation in GATA2-downregulated progenitors.
Enhancer-instigated developmental circuits in single progenitor cells
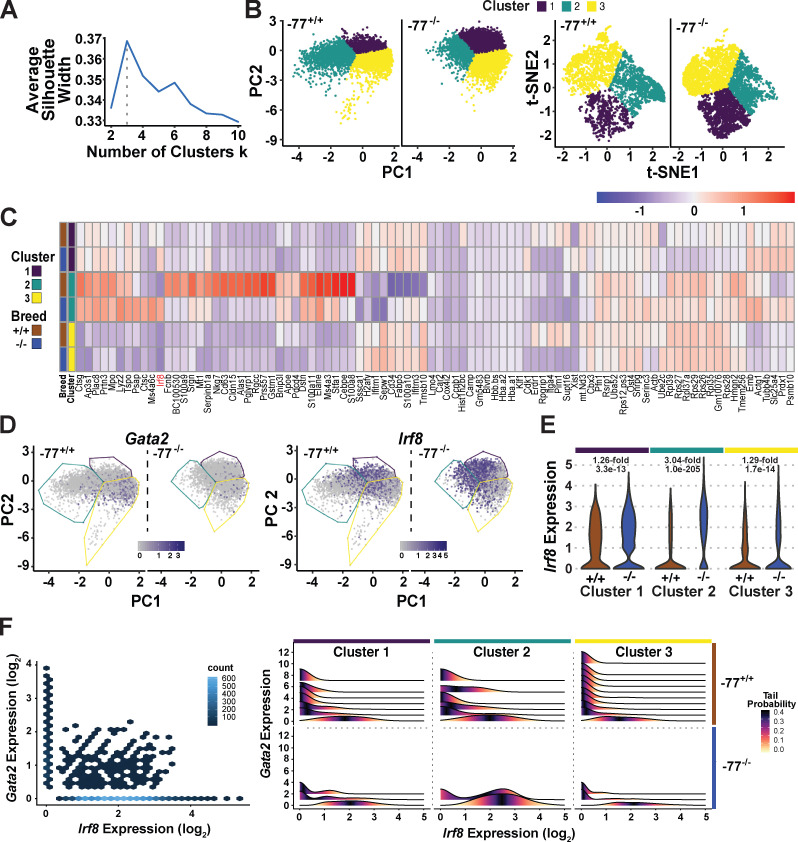
On the basis of proteomic and transcriptomic analyses of progenitor populations (Fig. 2), we considered the relationship between −77-regulated GATA2 expression and the GATA2-dependent transcriptome and proteome in single progenitors. Single-cell RNA sequencing (scRNA-seq) of Lin−Sca1−c-Kit+CD34+ cells was conducted with the 10x Genomics platform and analyzed using multiple dimensional reduction approaches and tools (principal component analysis [PCA], t-distributed stochastic neighbor embedding [t-SNE]; van der Maaten and Hinton, 2008; and Uniform Manifold Approximation and Projection; Becht et al., 2019), which yielded qualitatively similar conclusions. scRNA-seq data derived from PCA was subjected to k-means clustering. Maximizing average silhouette width, calculated as a function of the number of clusters, revealed three clusters to be optimal (Fig. 3, A and B). t-SNE analyses, which leverage nonlinear dimensional reduction, also revealed three or four clusters to be optimal (Fig. 3 B and Fig. S3 A). On the basis of cohorts of genes enriched in each cluster, the transcriptome of cluster 2 cells is characterized by an innate immune response, whereas enriched genes of cluster 3 cells highlight transcriptional and translational processes linked to red cell production (Fig. S3 B). A comparison of differential gene expression between −77+/+ and −77−/− progenitors included prominent myeloid gene expression in cluster 2, which was reduced in −77−/− progenitors (Fig. 3 C). A subset of cells in all clusters expressed Gata2, and Gata2 expression was uniformly lower in −77−/− progenitors (Fig. 3 D). Irf8 was expressed in a subset of cells in all clusters, highly upregulated in cluster 2 in mutants (3.04-fold increase in expression per cell; P = 1.0e−205), and upregulated significantly but to a lesser extent in clusters 1 (1.26-fold increase; P = 3.3e−13) and 3 (1.29-fold increase; P = 1.7e−14; Fig. 3 E).
Figure 3.
Enhancer-dependent genetic network in single progenitor cells: anticorrelative Gata2 and Irf8 expression. (A) Selection of optimal numbers of clusters by maximizing the average silhouette width in k-means clustering after dimension reduction with PCA. See also Fig. S3. (B) Comparison of cell clusters revealed by linear and nonlinear dimension reduction methods, PCA, and t-SNE. PCA was used for subsequent clustering. (C) Heatmap of median expression of selected DE genes across clusters. The genes displayed include DE genes of each cluster with adjusted P value <0.05 and fold change within the top 1% of the cluster. (D) Cluster-specific distribution of Gata2 and Irf8 expression overlaid on the PCA map. Blue and gray colors indicate high and low expression levels, respectively. (E) Violin plots of cluster-specific Irf8 expression. Fold increases (in −77+/+ versus −77−/−) and P values are indicated for each cluster. (F) Hex and ridgeline plots of Gata2 and Irf8 coexpression. The hex plot compares coexpression in the entire population. In the ridgeline plots, cells were stratified by Gata2 expression within each cluster. The Irf8 expression distribution within each Gata2 stratum is colored so that the darkest value represents the median Irf8 distribution within each stratum. PC, principal component.
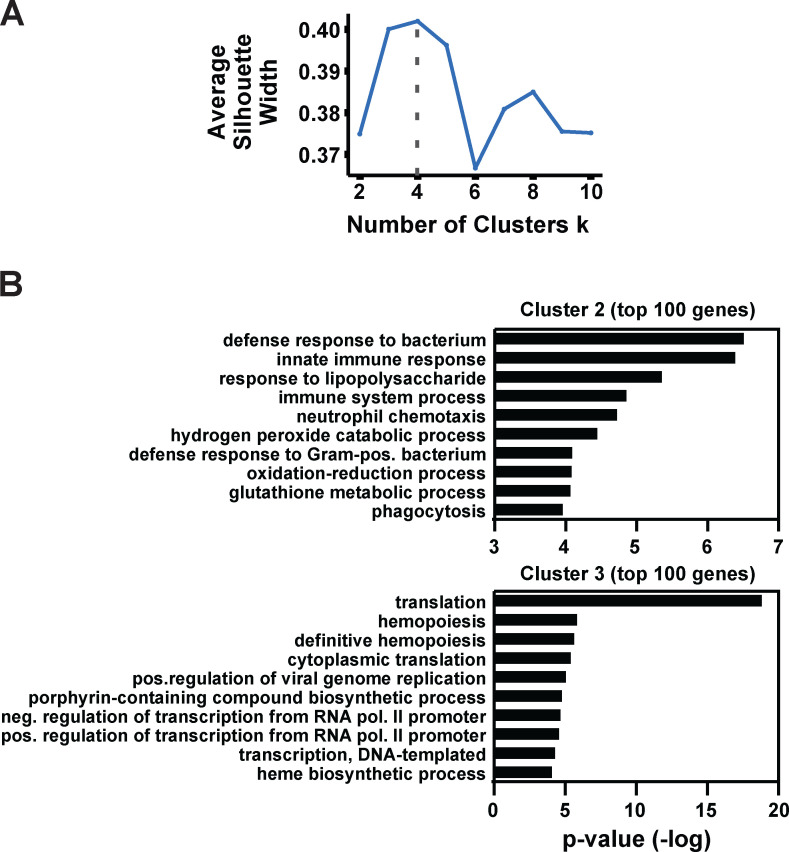
Figure S3.
Cluster number optimization and cluster-specific features derived from gene expression patterns. (A) Selection of optimal numbers of clusters by maximizing the average silhouette width in k-means clustering after dimension reduction with t-SNE. (B) Top categories obtained from GO analysis of the 100 most enriched genes for PCA clusters 2 and 3 using DAVID Bioinformatics Resources (https://david.ncifcrf.gov). Genes enriched in cluster 1 did not parse into specific categories. The enriched genes in each cluster were determined using the MAST function of Seurat’s FindAllMarkers.
Because Gata2 and Irf8 were expressed in a subset of cells in all clusters, and because −77 deletion downregulated Gata2 and upregulated Irf8 transcription, we tested whether this opposing expression pattern occurs in distinct, common, or both cohorts of cells. In the heterogeneous population, most cells lacked coexpression of Gata2 and Irf8 (Fig. 3 F, hex plot). When single cells were parsed on the basis of Gata2 expression levels, and when expression of Irf8, relative to Gata2, was compared by cluster (Fig. 3 F), cluster 1 cells exhibited a broad range of Gata2 expression, and Irf8 expression was detected only in cells with a low level of or no Gata2. −77 deletion greatly reduced Gata2 expression, and only cells with the lowest Gata2 levels expressed Irf8. Cluster 1 and 2 cells shared a common range of Gata2 expression, and Irf8 expression was detected in cells with little to no Gata2. −77 deletion nearly abrogated Gata2 expression, and Irf8 expression emerged prominently, with the highest level detected in cluster 2 cells. Cluster 3 cells also exhibited a broad range of Gata2 expression, with Irf8 detectable only in cells expressing little to no Gata2. The −77 deletion abrogated Gata2 and upregulated Irf8 identically to the other clusters. This analysis identified cells expressing both Gata2 and Irf8, with one high and the other low. Taken together with IRF8 being a vital monocytic differentiation determinant and our discovery that GATA2 restoration in −77−/− progenitors rescued Irf8 expression, these results support a model in which −77 loss downregulates GATA2, corrupting the transcriptome and proteome. Under these conditions, Irf8 expression increases and IRF8 enables or drives the predominant monocytic differentiation.
Genetic construction and deconstruction of progenitor heterogeneity and developmental trajectories
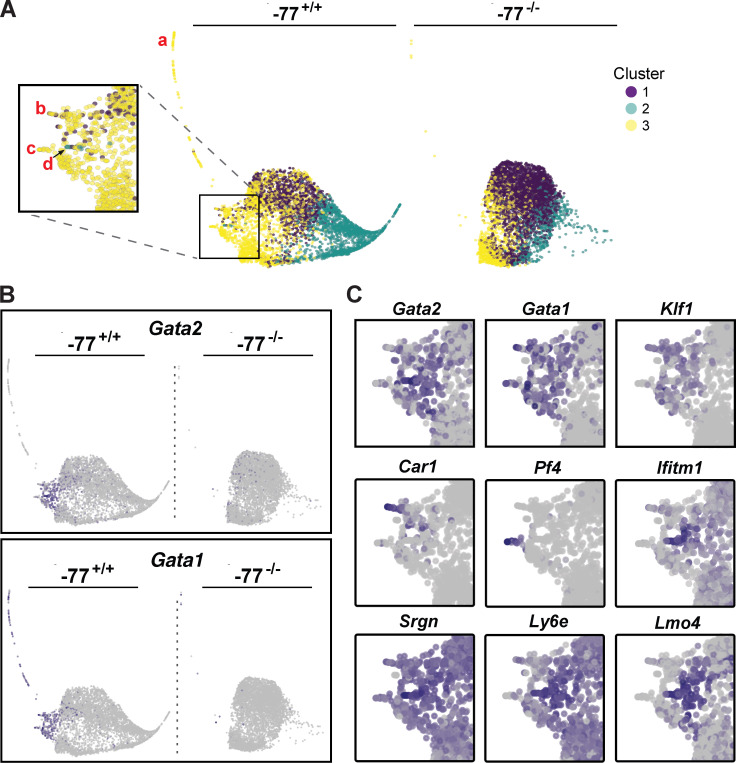
To further dissect how an enhancer deletion deconstructs a multifate system to yield a predominant solitary fate, the progenitor scRNA-seq data were analyzed with the pseudotime trajectory tool SPRING (Weinreb et al., 2018). By segregating cells with disparate transcriptomes, pseudotime trajectory analysis identifies potential developmental paths. It was unclear, however, if this analysis would unveil trajectories when applied to a more restricted progenitor population. SPRING trajectory plots were color coded to illustrate the relationship of the trajectories (Fig. 4 A) with the previously established PCA-defined clusters (Fig. 3 B). One prominent trajectory (Fig. 4 A, trajectory a) and additional, less pronounced trajectories (Fig. 4 A, boxed inset, trajectories b–d) extrude from cluster 3 in the −77+/+ plot. Cells expressing the highest Gata2 levels composed the smaller trajectories (Fig. 4, B and C). Gata1 expression overlapped extensively with that of Gata2 in the short leftward extrusion (Fig. 4, B and C) but was also detected throughout trajectory a that lacked Gata2. Restricted expression of multiple erythroid-specific genes (e.g., Klf1, Hba-a1, Alas2, Slc4a1), as with Gata1, which is expressed in erythroid cells and additional cell types, provides evidence that this extended trajectory reflects progressive erythroid differentiation. These cells compose just 2% of the total −77+/+ population and may represent erythroid-primed cells within the CMP population. Within the inset, trajectory b cells of cluster 3 expresses erythroid markers Klf1 and Car1 (Fig. 4 C), whereas the megakaryocytic gene Pf4 is restricted to trajectory c. A distinct subset of cells, trajectory d, expressed basophil markers Lmo4, Ifitm1, Ly6e, and Srgn (Tusi et al., 2018). Because all of these trajectories are absent or greatly diminished in −77−/− progenitors, the enhancer deletion abrogated erythroid, megakaryocytic, and basophil developmental trajectories.
Figure 4.
Enhancer-dependent developmental trajectories. Developmental trajectories were established by SPRING analysis (Tusi et al., 2018; Weinreb et al., 2018) using 4,980 −77+/+ or −77−/− progenitors. (A) SPRING analysis identified lineage trajectories that are nearly absent in −77−/− progenitors. The PCA-derived cluster designation of each cell (Fig. 2) was mapped onto the SPRING trajectory plots. Prominent trajectories of cells from cluster 2 or 3 were absent in −77−/− samples. Red letters mark specific developmental trajectories: erythroid (a, b), megakaryocyte (c), and basophil (d). (B) Global distribution of Gata2- and Gata1-expressing cells. Both genes are abundantly expressed in cluster 3. (C) Gene expression patterns define lineage trajectories for megakaryocytes (Pf4), basophils (Ifitm1, Srgn, Ly6e, and Lmo4), and erythroid cells (Gata1, Klf1, and Car1). Expression of each gene is shown for the boxed area of A.
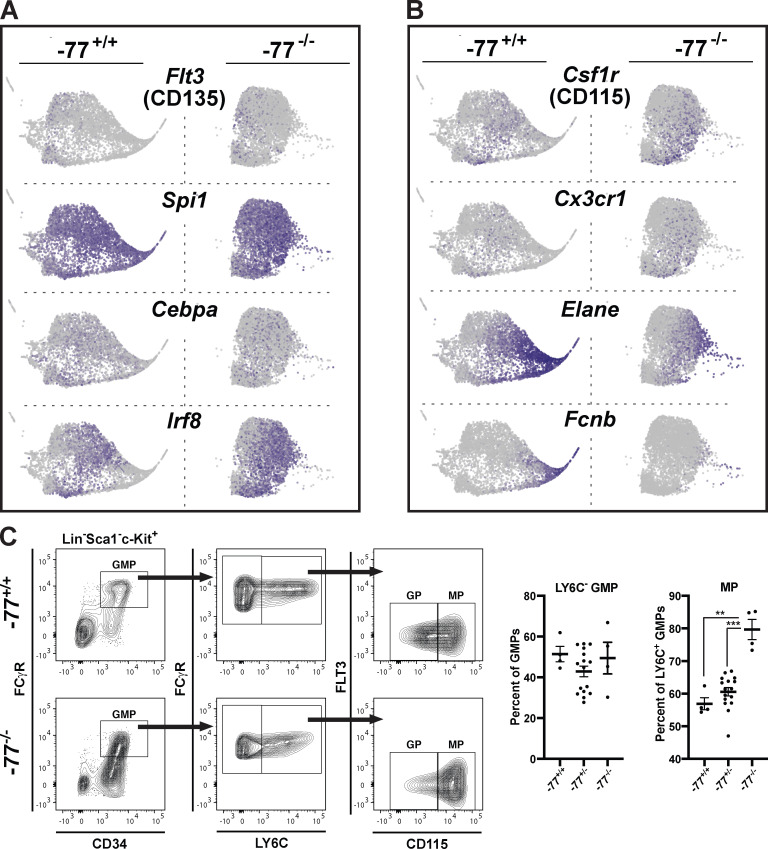
Cluster 1 cells comprise the central mass of the SPRING plot and express myeloid transcripts, including Flt3, Spi1, Cebpa, and Irf8 (Fig. 5 A). Expression of the monocyte progenitor markers Csf1r (CD115) and Cx3cr1 in the central mass was restricted to a cell cohort that was expanded in the −77−/− samples (Fig. 5 B). A rightward extrusion comprising cluster 2 cells was enriched in neutrophil transcripts (e.g., Elane and Fcnb; Fig. 5 B), with the left-to-right directionality characterized by attributes associated with progressive neutrophil development. Although the −77 deletion had little to no impact on Spi1- and Cepba-expressing cells in the central mass, it abrogated the neutrophil developmental trajectory. Expansion of GMP-derived monocyte progenitors and loss of granulocyte progenitors was confirmed by flow cytometry. Bipotential Ly6C− GMPs were unaffected by the −77 deletion (Fig. 5 C).
Figure 5.
Loss of neutrophils but retention of monocyte progenitors in −77−/− progenitors. (A) Expression of myeloid regulatory genes in cluster 1. In our proteomic analysis, FLT3, a marker for monocyte–dendritic cell precursors, was elevated 2.4-fold in −77−/− progenitors, whereas PU.1 (Spi1) and CEBPα were unchanged. (B) −77 enhancer deletion abrogates a granulocyte trajectory (Elane and Fcnb) in cluster 2 but promotes monocyte progenitors (Csf1r, Cx3cr1). CSF1R was elevated 2.6-fold in −77−/− progenitors. (C) Flow cytometric analysis of granulocyte and monocyte progenitors within the GMP pool. Quantitation of Ly6C− GMPs and monocyte progenitors. −77+/+ (n = 4; two litters), −77+/− (n = 16; three litters), −77−/− (n = 4; two litters). Error bars represent mean ± SEM. Statistics were calculated using unpaired two-tailed Student’s t test; **, P ≤ 0.01; ***, P ≤ 0.001.
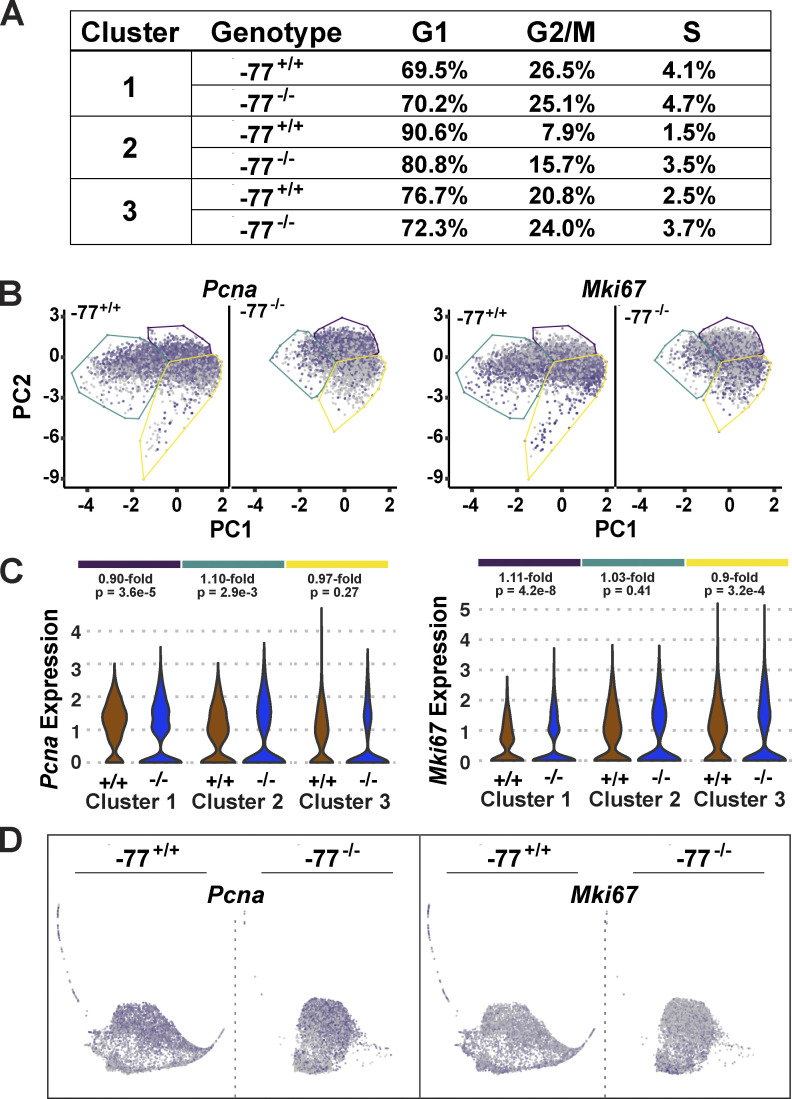
In the −77−/− progenitors, Irf8 expression was a common attribute of many cells within the central mass, consistent with Irf8 upregulation as an early step in acquisition of the predominant monocytic fate program. Thus, −77 and GATA2 endow myeloid progenitors with erythroid-, megakaryocyte-, basophil-, and neutrophil-primed transcriptomes, and this multilineage transcriptomic heterogeneity suggests a mechanism underlying the diverse differentiation potentials of the heterotypic progenitor population. Accordingly, −77 deletion attenuates transcriptomic heterogeneity, thus deconstructing progenitor multipotentiality to yield a predominant monocytic fate. This deconstruction of the multifate system occurs without gross changes in progenitor cell cycle status, though 2.0- and 2.3-fold increases in G2/M- and S-phase cells were detected in a small percentage of cluster 2 cells (Fig. S4 A). These alterations were not associated with changes in genes expressed specifically in proliferating cells (Pcna and Mki67). Pcna and Mki67 were expressed broadly in cells within all clusters, and −77 deletion had little to no impact on their expression (Fig. S4, B–D). By coordinating fate-promoting and -suppressing circuits, this enhancer mechanism generates progenitor functional heterogeneity to accommodate physiological requirements.
Figure S4.
Cell fate deconstruction does not impact progenitor proliferative status. (A) Cell cycle status was determined for each cluster using the scran function cyclone. (B) Comparison of the distribution of Pcna- and Mki67-expressing cells. Red and gray colors indicate high and low expression levels, respectively. (C) Violin plots depicting cluster-specific Pcna and Mki67 expression. Fold increases (in −77+/+ versus −77−/−) and P values are indicated for each cluster. (D) Developmental trajectories were established by SPRING analysis.
Coordinating fate-promoting and -suppressing circuits to generate multipotency: Mechanistic considerations
How does an enhancer deletion trigger progenitors to mount an ectopic innate immune response? IFN signaling is implicated in diverse HSPC functions in physiology and pathology (Baldridge et al., 2010; de Bruin et al., 2014; Essers et al., 2009; Li et al., 2014; Zoumbos et al., 1985), and its regulation is controlled at multiple levels (Schneider et al., 2014). During fetal liver hematopoiesis, GATA2-mediated suppression of IFN signaling may balance IFN-activated and -suppressed processes, thus minimizing the emergence of dysregulated signaling and deleterious consequences. GATA2 downregulation would disrupt this defensive mechanism, causing ectopically high IFN signaling. Alternatively, GATA2 downregulation might desensitize IFN signaling components, resulting in subphysiological signaling insufficient to support progenitor functions. Elevated IFN signaling may constitute an attempt to restore physiological IFN-dependent signaling outputs. Finally, GATA2 downregulation might upregulate IFN signaling components independent of IFN receptor signaling.
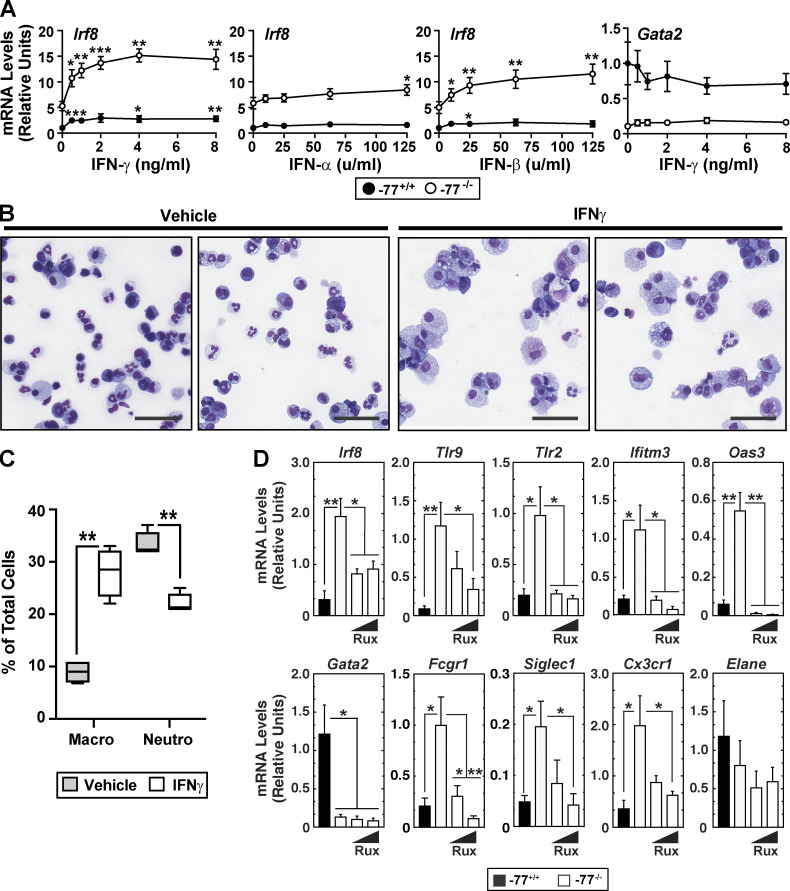
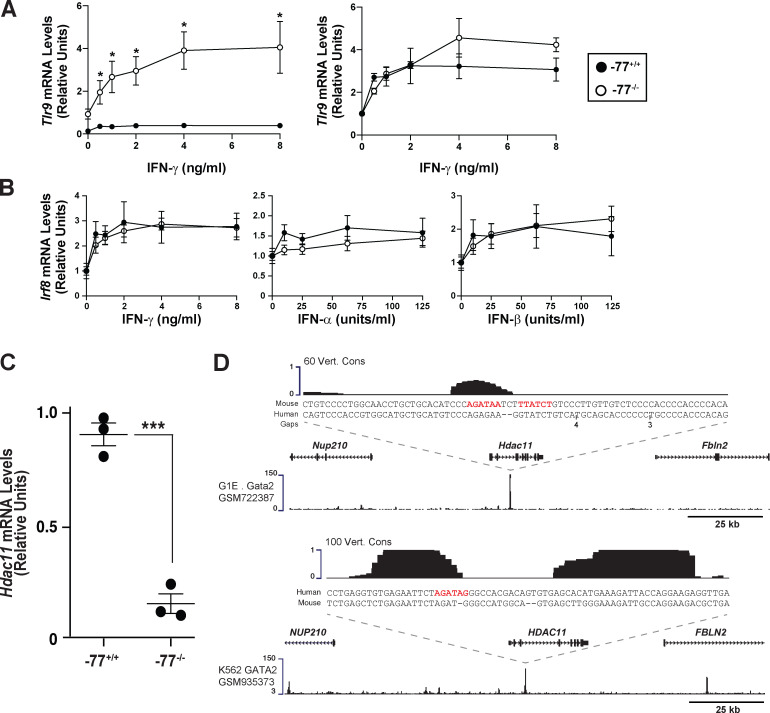
To address these potential mechanisms, we asked whether −77−/− progenitors with upregulated IFN signaling components are responsive to exogenous IFN or if the preinduced state reflects maximal pathway activity and lack of competence to respond further. Because gene ontology analysis, gene set enrichment analysis, and individual gene attributes did not reveal that upregulated components conform to a strict type I (α and β) or type II (γ) IFN signature, we analyzed the responsiveness of −77+/+ and −77−/− cells to α, β, or γ IFN. IFNγ increased Irf8 expression 2.8-fold (P = 0.007) and 2.9-fold (P = 0.005) in −77+/+ and −77−/− cells, respectively (Fig. 6 A). Similarly, IFNγ increased Tlr9 expression 2.9-fold (P = 0.012) and 4.4-fold (P = 0.049) in −77+/+ and −77−/− cells, respectively (Fig. S5 A). IFNα and IFNβ were less effective inducers than IFNγ in −77+/+ cells and elicited similar responses in −77+/+ and −77−/− cells (Fig. 6 A). Normalization of the data revealed that −77+/+ and −77−/− progenitors had a comparable sensitivity to IFNγ-mediated induction of Irf8 and Tlr9 expression (Fig. S5, A and B). Thus, the preinduced Irf8 and Tlr9 state of −77−/− progenitors did not preclude or impact the IFN dose-dependent transcriptional response. IFNγ did not affect Gata2 expression in −77+/+ or −77−/− progenitors (Fig. 6 A, right). Because IFN induced a hyperphysiological expression response in −77−/− cells, in which multifate potential was deconstructed into a singular fate, these results support a model in which −77 and GATA2 suppress IFN signaling, thus averting the emergence of dysregulated signaling with consequences deleterious for cellular differentiation.
Figure 6.
Mechanisms underlying the ectopic innate immune response in GATA2-deficient progenitors. (A) Responsiveness of Irf8 and Gata2 to IFN treatment in −77−/− versus −77+/+ Lin− cells. Lin− E15.5 fetal liver cells were cultured for 24 h with type I (IFNα or IFNβ) or type II (IFNγ) IFN, and RNA was quantitated by quantitative RT-PCR (qRT-PCR). n = 3–7 biological replicates from −77−/− and −77+/+ littermates were analyzed for each condition. Mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001, by two-tailed, paired Student’s t test for comparison of expression for each IFN concentration relative to untreated cells. See also Fig. S5. (B) Representative Giemsa staining of dissociated colonies from −77+/+ and −77−/− E14.5 fetal liver cells grown for 8 d in M3434 complete methylcellulose media supplemented with 20 ng/ml IFNγ or vehicle (PBS, 0.01% BSA). Scale bars = 50 µm. (C) Quantitation of macrophages and neutrophils as a percentage of Giemsa-stained cells recovered from colonies. n = 4 biological replicates for each condition. (D) JAK1/2 inhibition by ruxolitinib (Rux) suppresses IFNγ and monocytic target gene expression in −77−/− progenitors. n = 4 biological replicates. In all graphs, error bars represent mean ± SEM. Statistics were calculated using unpaired two-tailed Student’s t test; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Figure S5.
GATA2 directly induces Hdac11, encoding a suppressor of IFN signaling. (A) Responsiveness of Tlr9 to IFNγ treatment in −77−/− versus −77+/+ Lin- fetal liver cells. Cells were cultured for 24 h in the presence of IFNγ, and RNA was quantitated by qRT-PCR. n = 4 biological replicates from −77−/− and −77+/+ littermates were analyzed for each condition. Mean ± SEM; *, P ≤ 0.05 by two-tailed, paired Student’s t test for comparison of expression for each IFN concentration relative to untreated cells. (B) Comparable sensitivity of −77+/+ and −77−/− progenitors to IFNγ-dependent transcriptional regulation. The means for Irf8 expression in untreated −77+/+ and −77−/− samples have been normalized to 1, with treated conditions set relative to the untreated value. n = 3–7 biological replicates. (C) Hdac11 mRNA expression in −77+/+ or −77−/− progenitors from lineage-depleted E14.5 fetal livers cultured for 3 d. n = 3 biological replicates. Error bars represent mean ± SEM. Statistics were calculated using unpaired two-tailed Student’s t test; ***, P ≤ 0.001. (D) GATA2 occupancy at Hdac11 was mined from existing mouse (GATA1-null G1E cells; upper profile) and human (K562 cells; lower profile) chromatin immunoprecipitation–sequencing datasets.
Because IFN signaling components are upregulated in −77−/− progenitors with skewed, predominant monocytic differentiation, and since IFNγ induced Irf8 and Tlr9 expression in −77+/+ progenitors, we asked if IFNγ suffices to skew differentiation. Cells were treated with 20 ng/ml IFNγ when plated for colony formation. 8 d later, cells were analyzed by Wright-Giemsa staining (Fig. 6 B). Consistent with the prior report that IFNγ favors monocytic over granulocytic differentiation (de Bruin et al., 2012), IFNγ increased monocytic and decreased granulocytic progeny by 3.1-fold (P = 0.0004) and 1.5-fold (P = 0.0004), respectively (Fig. 6 C).
IFN induces Irf8 and Tlr9 expression (Schneider et al., 2014), both being ectopically high in −77−/− progenitors. In dendritic cells, TLR9 signaling requires IRF8 for NF-κB activation (Tsujimura et al., 2004). Thus, in certain contexts, IRF8 and TLR9 mechanisms are functionally intertwined. In other contexts, IFNγ and TLR signaling synergistically control cell function (Hu and Ivashkiv, 2009).
Is IFN signaling responsible for ectopic innate immune machinery upregulation in −77−/− progenitors? IFNγ dimerizes and binds a heterodimeric receptor of IFNGR1 and IFNGR2 subunits, which recruits Janus kinases (JAK1 and JAK2), leading to phosphorylation and STAT1 transcription factor activation (Stark and Darnell, 2012). If increased IFNγ signaling causes elevated Irf8 and Tlr9 expression, blocking JAK1/2 should attenuate or abrogate aberrant gene expression. If Irf8 and Tlr9 upregulation does not involve canonical IFNγ signaling, JAK1/2 inhibition should not impact expression. After treatment with the JAK1/2 inhibitor ruxolitinib for 48 h, the elevated expression of Irf8, Tlr9, and other IFN-inducible genes in −77−/− progenitors was attenuated to a level resembling that of −77+/+ progenitors (Fig. 6 D). Ruxolitinib did not significantly affect the low Gata2 expression in −77−/− versus −77+/+ progenitors.
Because the Irf8 expression level dictates monocytic versus granulocytic fate (Bigley et al., 2018; Giladi et al., 2018; Hambleton et al., 2011; Kurotaki et al., 2013; Yáñez and Goodridge, 2016; Yáñez et al., 2015), and because ruxolitinib downregulated Irf8 expression, we reasoned that ruxolitinib would attenuate the preferential monocytic fate potential. Ruxolitinib treatment reduced expression of monocytic genes Fcgr1, Siglec1, and Cx3cr1 to levels comparable with those of WT cells, whereas expression of the granulocytic gene Elane was unaffected (Fig. 6 D). Thus, the GATA2 deficiency–instigated fate-deconstructing mechanism requires JAK1/2 signaling to elevate expression of IFN response and monocytic genes.
The histone deacetylase HDAC11 is a suppressor of type I and type II IFNs. T cells from Hdac11-knockout mice display elevated IFNγ levels, which increase IFN signaling (Woods et al., 2017), and HDAC11 suppresses type I IFN signaling via limiting receptor deposition in the plasma membrane (Cao et al., 2019). Fatty acylation of SHMT2 promotes its localization to endosomes/lysosomes, where association with BRISC (BRCC36 isopeptidase complex) induces deubiquitination and stabilization of IFNαR1. HDAC11-mediated defatty-acylation of SHMT2 impairs endosome/lysosome localization, leading to reduced receptor recycling (Cao et al., 2019). Consistent with this mechanism, upregulated IFN signaling was associated with a 5.6-fold reduction in Hdac11 mRNA in primary −77−/− progenitors (Fig. S5 C). Chromatin immunoprecipitation sequencing revealed GATA2 occupancy at mouse and human Hdac11 loci, and GATA2 motifs (WGATAR) reside at the occupancy site (Fig. S5 D), suggesting direct regulation.
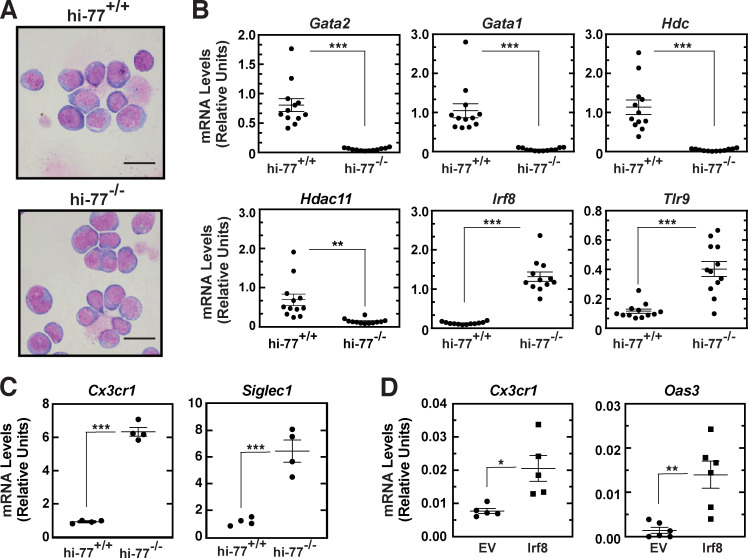
To further test whether the gene expression differences in −77−/− versus −77+/+ primary fetal liver progenitors are stable when progenitors are removed from an in vivo environment, we used estrogen-regulated HoxB8 (Wang et al., 2006) to immortalize the progenitors (Fig. 7 A). HoxB8-immortalized (hi) −77−/− cells retained lower expression of Gata2 and its target genes Gata1, Hdc, and Hdac11 versus hi−77+/+. However, innate immune (Irf8, Tlr9; Fig. 7 B) and monocytic (Cx3cr1 and Siglec1; Fig. 7 C) gene expression were higher in hi−77−/− progenitors. Expressing exogenous IRF8 in hi−77+/+ cells increased levels of both monocytic (Cx3cr1) and innate immunity (Oas3) mRNAs (Fig. 7 D), analogous to the high Irf8, Cs3cr1, and Oas3 expression in hi−77−/− cells. Thus, the −77−/− phenotype is stable ex vivo, and, on the basis of known monocytic differentiation activity of IRF8 in mouse (Kurotaki et al., 2013; Yáñez et al., 2015) and humans (Bigley et al., 2018; Hambleton et al., 2011) and our gain-of-function analysis, elevated IRF8 contributes to the phenotype.
Figure 7.
Differential expression of innate immunity and monocytic genes is retained in immortalized −77−/− progenitors. (A) Representative Giemsa staining of HoxB8-immortalized (hi) fetal liver progenitor cells. Scale bars = 20 µm. (B) qRT-PCR analysis of mRNA expression in hi−77+/+ and hi−77−/− progenitors. Each point depicts the value from an independently derived hi line. n = 12. (C) qRT-PCR analysis of mRNA expression in hi−77+/+ and hi−77−/− clones. n = 4 biological replicates. (D) qRT-PCR analysis of mRNA expression in clonal hi−77+/+ cells transiently expressing IRF8. In all graphs, error bars represent mean ± SEM. Statistics were calculated using unpaired two-tailed Student’s t test; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Discussion
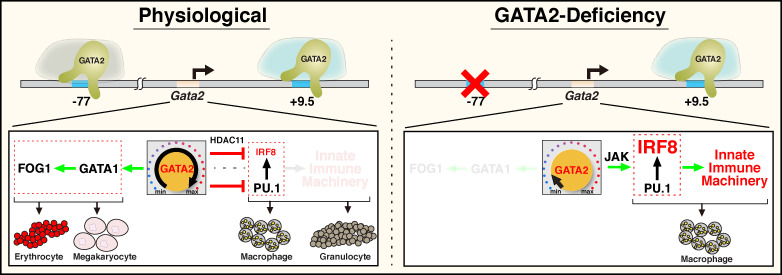
Stem and progenitor cell activity to efficiently generate diverse progeny requires mechanisms that enable, propel, or restrict cell fate transitions to achieve specific developmental outputs. For multipotent cells, it is instructive to consider if the distinct fate potentials are acquired concomitantly, via independent steps, with each dedicated to an individual fate, or via a hybrid mechanism. Using hematopoietic progenitors and an enhancer mutant allele of a gene encoding the master regulator GATA2, we demonstrated that GATA2 primes the progenitor genome to generate a transcriptome and proteome that endow erythroid, megakaryocytic, granulocytic, and monocytic fates. The enhancer deletion renders GATA2 limiting, corrupting the GATA2-dependent transcriptome and proteome that confer multifate potential, leading to a predominant monocytic fate. Because restoring GATA2 reverses the transcriptomic aberrations, this powerful system was used to elucidate mechanisms that construct and deconstruct multifate systems. The −77 enhancer builds the multifate system by establishing a fate-promoting circuit and concomitantly negating a fate-suppressive circuit. The fate-promoting circuit requires the downstream target GATA1 with its coregulator FOG1 to establish erythroid- and megakaryocyte-primed transcriptomes (Fig. 8). The fate-suppressing circuit revealed a link between GATA2 and innate immune machinery, IFN signaling pathway components and sensors of pathogen constituents and activities, which defend against pathogen intruders. The analysis therefore unveiled mechanistic underpinnings of a multicell fate system construction process.
Figure 8.
Coordinating cell fate–promoting and –suppressing circuitry to establish a multifate progenitor system. The model depicts a physiological mechanism in which GATA2 induces a fate-promoting circuit involving another transcription factor, GATA1, and its coregulator FOG1 and opposes an IRF8-dependent fate-suppressing circuit. Because PU.1 commonly occupies chromatin and functions with IRF8 (Mancino et al., 2015), repression might involve the reported GATA2–PU.1 antagonism (Walsh et al., 2002) or a PU.1-independent mechanism involving HDAC11-mediated suppression of IFNγ signaling. PU.1 levels are constant in −77+/+ and −77−/− progenitors (Fig. 2 D). −77 enhancer deletion abrogates circuits promoting erythroid and megakaryocytic fates, and −77−/− cells mount an ectopic response in which the IRF8-dependent fate-suppressive circuit prevails, skewing multilineage potential into a predominant monocytic program.
Foundational insights into circuitry underlying fate mechanisms have emerged from a transdifferentiation system in which pre–B cells acquire the capacity to form macrophages (Bussmann et al., 2009). Pre–B cells are heterogeneous vis-à-vis their potential to transdifferentiate. Single cells that generate macrophages rapidly are more refractory to reprogramming into induced pluripotent cells than are cells exhibiting slow transdifferentiation (Francesconi et al., 2019). The levels of a single protein, c-Myc, determine rapid transdifferentiation/low reprogramming (low c-Myc) efficiency versus slow transdifferentiation/high reprogramming (high c-Myc) efficiency states. Analogous to c-Myc, variable GATA2 levels control circuits that alter fate output. Gata2 +9.5−/− aorta gonad mesonephros is quantitatively defective in its capacity to generate HSPCs, and +9.5+/− embryos exhibit intermediate phenotypes in aorta gonad mesonephros and fetal liver hematopoiesis (Gao et al., 2013; Johnson et al., 2012). Gata2+/− embryos and adults also exhibit intermediate phenotypes (Ling et al., 2004; Rodrigues et al., 2005). Contrasting with these quantitative differences, GATA2 levels dictate establishment of a multifate system with qualitatively distinct fate outputs by induction of a fate-promoting circuit concomitant with negating a fate-suppressing circuit (Fig. 8).
The deconstruction of a multifate system, with emergence of a dominant fate-suppressing circuit, was unpredictable on the basis of transcriptional or developmental paradigms. Why would this circuit consist of innate immune machinery including IFN signaling components and pattern recognition receptors? In GATA2 deficiency syndrome, mechanisms that trigger MDS and AML are enigmatic (Churpek and Bresnick, 2019). GATA2 loss creates a disease predisposition, and we proposed that GATA2-low cells are vulnerable to genetic and/or environmental insults that launch HSPCs on a pathogenic path (Soukup et al., 2019). By instructing an efficient monocytic program, IRF8 induction in GATA2-deficient cells during embryogenesis may consume progenitors vulnerable to genetic or environmental insults. Pathogen infection of progenitors can elevate NLRP1A, an inflammasome component that senses pathogen enzymatic activity (Martinon et al., 2002) and triggers cell death via pyroptosis to clear pathogen-harboring progenitors (Masters et al., 2012); Nlrp1a is upregulated in −77−/− progenitors. Alternatively, IFN response gene expression may confer pathogen resistance and preserve progenitors, analogous to viral resistance of human embryonic stem cells and differentiated neural stem cell progeny (Wu et al., 2018). By extrapolation, innate immune machinery upregulation in −77−/− progenitors may ensure integrity of the progenitor pool. Pattern recognition receptors, which confer pathogen resistance (Ronald and Beutler, 2010), would provide sensors for progenitors to respond to bacterial (and Mycobacterium, a common pathogen in GATA2 deficiency syndrome; Dickinson et al., 2014; Spinner et al., 2014), viral, and fungal pathogens to evade deleterious consequences of infection. Without the upregulated innate immune machinery safety net, infection-induced stress might constitute a pathogenic trigger for an otherwise silent GATA2 mutation. Our results revealed some differences from IFN actions on embryonic stem cells (Wu et al., 2018). Embryonic stem cells with upregulated IFN components exhibit an attenuated IFNγ response, and oligoadenylate synthase family members, important determinants of pathogen immunity, were not upregulated (Wu et al., 2018). By contrast, −77−/− progenitors retain normal IFNγ responsiveness, and oligoadenylate synthase family members were upregulated. Given direct GATA2 activation of HDAC11 transcription described herein (Fig. S5 and Fig. 7) and established links between HDAC11 and reduced IFN signaling (Cao et al., 2019; Woods et al., 2017), it is attractive to propose that the fate-suppressing circuit involves a GATA2–HDAC11–innate immune axis.
In summary, we elucidated a mechanism in which an enhancer constructs a multifate system via coordinating fate-promoting and -suppressing circuits. The enhancer deletion instigates an ectopic innate immune response that deconstructs the multifate system, which unveils new dimensions to GATA factor, hematopoiesis, and immune mechanisms. Further dissecting the system will almost certainly yield additional principles governing broadly operational fate-regulatory circuits and transformative insights into innate immune machinery function in multipotent cells in physiology and pathology. In addition, the innovative multiomics resource of WT and enhancer mutant primary progenitor cells, involving quantitative proteomics, scRNA-seq, and population RNA-seq coupled with GATA2 genetic rescue, will uniquely enable diverse molecular/cellular discoveries beyond those described herein.
Materials and methods
Contact for reagent and resource sharing
A detailed list of reagents and resources is provided in Table S3. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, E.H. Bresnick.
Human subjects
Patients with germline GATA2 mutations were enrolled in clinical protocols approved by the institutional review board at the National Institute of Allergy and Infectious Diseases (ClinicalTrials.gov identifier, NCT01905826) and in accordance with the Declaration of Helsinki. Patient-specific information is provided in Fig. 1. Bone marrow biopsies were performed with informed consent.
Mice
Hematopoietic progenitor cells were obtained from fetal livers of staged embryos from timed mated Gata2 −77+/− or C57BL/6J mice. All animal protocols were approved by the University of Wisconsin–Madison Institutional Animal Care and Use Committee in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC International) regulations.
Primary cell culture
Fetal livers were harvested on ice in PBS containing 2% FBS, 10 mM glucose, and 2.5 mM EDTA. Cells were dissociated and passed through a single-cell strainer. Cells expressing lineage markers were removed using biotin-conjugated antibodies CD3e, CD11b, CD19, CD45R (B220), GR-1, Ter119, CD71, and MojoSort Streptavidin Nanobeads, all purchased from BioLegend. The remaining Lin− cells were cultured for up to 3 d in Gibco IMDM (Life Technologies) containing 20% FBS, 4% stem cell factor (SCF)–conditioned media, 4% IL-3–conditioned media, and 1% penicillin-streptomycin (Gemini Bio). IFN responsiveness was assayed by treating Lin− cells with either IFNγ, IFNα, IFNβ, or PBS with 0.1% BSA (vehicle) at the time of culture for 24 h. Alternatively, unfractionated E14.5 fetal liver cells were plated in M3434 methylcellulose media at 20,000 cells per 35-mm dish with 20 ng/ml IFNγ or an equal volume of PBS, 0.1% BSA. After 7–8 d, cells were recovered for Giemsa staining. For JAK1/2 inhibition, ruxolitinib was added to the Lin− cells at 0.25 or 1 µM and cultured for 48 h, at which point RNA was isolated with TRIzol reagent (Thermo Fisher Scientific). Cells were cultured in a humidified 5% CO2 incubator at 37°C.
For generation of ER-HoxB8-immortalized (hi) progenitors, fetal liver Lin− cells were immortalized by retroviral expression of estrogen-regulated HoxB8 as described previously (Wang et al., 2006). Cells were cultured in OPTI-MEM supplemented with 10% FBS, 1% penicillin-streptomycin, 1% SCF-conditioned medium, 30 mM 2-mercaptoethanol, 1 µM β-estradiol, and 500 µg/ml G418. For transient expression of IRF8, hi−77+/+ cells were resuspended in 100 µl of Nucleofector Solution R and transfected with 20 µg of IRF8 expression vector or control vector using the G-016 program of Nucleofector II (Lonza). Cells were harvested for RNA extraction 72 h after transfection.
Quantitative RT-PCR
Total RNA was purified from cells with TRIzol reagent and treated with DNase I (Thermo Fisher Scientific) for 15 min at room temperature. Following heat inactivation for 10 min at 65°C, RNA was incubated with 125 ng of a 5:1 mixture of oligo(dT) primers and random hexamer at 68°C for 10 min. RNA/primers were incubated with Moloney murine leukemia virus reverse transcription (Thermo Fisher Scientific), 10 mM dithiothreitol (Thermo Fisher Scientific), RNAsin (Promega), and 0.5 mM deoxynucleoside triphosphates (New England Biolabs) at 42°C for 1 h and then heat inactivated at 98°C for 5 min. Quantitative gene expression analyses was performed by real-time PCR using Power SYBR Green Master Mix (Applied Biosystems) and run on a ViiA 7 Real-Time PCR System (Applied Biosystems).
Immunohistochemistry
Biopsies were fixed in buffered formalin, decalcified, paraffin embedded, cut into 4-μm sections, and stained with H&E. Paraffin-embedded biopsy sections (4 μm) were stained for CD68 and for CD14 in a Ventana Benchmark Ultra automated staining instrument (Ventana Medical Systems) according to the manufacturer’s protocols. Images were captured on an Olympus BX41 microscope equipped with an Olympus DP72 camera using Olympus cellSens Entry software.
Flow cytometry and cell sorting
Flow cytometric analysis of bone marrow aspirates was performed using a FACSCanto II analyzer (BD Biosciences) equipped with three lasers and eight fluorescence detectors. Antibodies used to identify monocytic populations were CD14 APCH7 (clone M7P9) and CD64 PE (clone 10.1). Cells were stained with the above-mentioned antibodies in appropriate dilutions for 15 min. RBCs were lysed with BD FACS lysing solution, and cells were washed with PBS containing 1% albumin. Cells were fixed in a 1% paraformaldehyde solution, and 105 events were acquired using FACSDiva software (BD Biosciences). The list mode files were analyzed with FCS Express (DeNovo Software).
Proteomic and single-cell transcriptomic analyses were performed on a CMP/GMP pool (Lin−Sca−c-Kit+CD34+) sorted from E14.5 fetal livers using a FACSAria II cell sorter (BD Biosciences). Lineage markers were stained with FITC-conjugated B220, CD3, CD4, CD5, CD8, CD19, IgM, Il7Ra, AA4.1, and TER-119 antibodies. Other surface proteins were detected with PE-conjugated FcγR, eFluor 660–conjugated CD34, peridinin chlorophyll (PerCP)-Cy5.5–conjugated Sca1, and PE-Cy7–conjugated c-Kit. After staining, cells were washed with PBS, 2% FBS, 10 mM glucose, and 2.5 mM EDTA, then resuspended in the same buffer containing DAPI and passed through 25-µm cell strainers to obtain single-cell suspensions for sorting. For proteomic analysis, sorted cell pellets were frozen in a dry ice/ethanol bath and stored at −80°C until the time of processing. For single-cell analysis, cells were sorted in PBS with 10% FBS, adjusted to 1,000 cells/µl, and cell viability was measured with a Countess II automated cell counter (Thermo Fisher Scientific) before processing. Quantitation of Lin−Sca1−cKit+CD34+FcgRHiLy6C+Flt3−CD115Hi monocyte progenitors and Lin−Sca1−cKit+CD34+FcgRHiLy6C+Flt3−CD115Lo granulocyte progenitors was performed using the LSR Fortessa flow cytometer (BD Biosciences). Antibodies are listed in Table S3.
Proteomics
Cells sorted by flow cytometry were frozen for storage at −80°C. Cell pellets were resuspended in 20–30 µl of lysis buffer (100 mM Tris, 8 M urea, 10 mM tris(2-carboxyethyl)phosphine, 40 mM 2-chloracetamide) and sonicated in a Qsonica Q700 sonicator at an amplitude of 35 and 4°C for 20 s on/10 s off with a total processing time of 10 min. The contents of two or three tubes were then combined on the basis of the number of cells counted during the sorting to obtain enough material for proteomic analysis (5–6e5 cells). The NanoDrop OneC Microvolume UV-Vis spectrophotometer (Thermo Fisher Scientific) at 280 nm was used to determine protein concentrations of the samples. Lysates were diluted with 50 mM Tris to a final urea concentration of ∼1.5 M before the addition of LysC in a 1:50 ratio (enzyme/protein; FUJIFILM Wako Chemicals) and overnight digestion at room temperature, followed by additional digestion with trypsin (1:50 ratio of enzyme/protein; Promega) for 3 h, acidification with 10% trifluoroacetic acid, desalting over 10-mg StrataX solid-phase extraction columns (Phenomenex), and lyophilization to dryness in a SpeedVac (Thermo Fisher Scientific). Peptides were resuspended in 0.2% liquid chromatography–mass spectrometry (LC-MS) grade formic acid (Pierce; Thermo Fisher Scientific), and the resultant peptide concentrations were determined using the NanoDrop spectrophotometer.
A 1260 Infinity II High Pressure Liquid Chromatography (HPLC) system with an Analytical-Scale Fraction Collector (Agilent) was used to separate ∼35 µg of peptides across the XBridge Peptide Ethylene Bridged Hybrid C18 Column, 130 Å, 3.5 µm, 4.6-mm × 150-mm column (Waters) at a flow rate of 0.8 ml/min over a 25-min gradient into 16 fractions. For high pH reverse-phase fractionation, mobile phase A consisted of 10 mM ammonium formate (Sigma-Aldrich) in HPLC MS-grade water (Thermo Fisher Scientific), buffered to pH 10.0 with ammonium hydroxide (Sigma-Aldrich), and mobile phase B contained 10 mM ammonium formate at pH 10.0 in 80% HPLC-MS–grade methanol (Thermo Fisher Scientific). Fractions were collected into conical bottomed 96–deep-well plates (Analytical Sales & Services) and concatenated by hand into eight final fractions. The plates were lyophilized to dryness in a SpeedVac, and peptides were resuspended in 0.2% formic acid for analysis by LC with tandem MS (LC–MS/MS) analysis.
For nanoscale LC–MS/MS, capillary columns were fabricated in-house. Self-pack PicoFrit 75–360-µm inner–outer diameter bare-fused silica capillary columns with 10-µm electrospray emitter tips (New Objective) were packed using an in-house–built ultrahigh-pressure column packing station (Shishkova et al., 2018) with 1.7-µm, 130-Å pore size Ethylene Bridged Hybrid C18 particles (Waters) to a final length of ∼40 cm and installed on a Dionex Ultimate 3000 nano-HPLC system (Thermo Fisher Scientific). Mobile phase buffer A was composed of 0.2% formic acid in water; mobile phase B was composed of 0.2% formic acid in 70% HPLC-MS grade acetonitrile (Thermo Fisher Scientific). Peptides (∼1 µg) from each fraction were loaded onto a column, which was kept at 50°C inside an in-house–made heater and separated at a flow rate of 300 nl/min over a 120-min gradient, including column wash and reequilibration time. Peptide ions were analyzed on a quadrupole ion trap hybrid Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). During data-dependent acquisition (whole-proteome analysis) Orbitrap survey scans were performed at a resolving power of 240,000 at 200 m/z with an automatic gain control target of 1.5 × 106 ions and maximum injection time of 50 ms. The instrument was operated in the top-speed mode with 1-s cycle times using an advanced precursor determination algorithm (Hebert et al., 2018) for monoisotopic precursor selection. Precursors were isolated using a quadrupole with an isolation window of 0.7 Th. MS-MS scans were performed in the ion trap using the rapid scan rate on precursors with two to four charge states using higher-energy collisional dissociation fragmentation with normalized collision energy of 25 and dynamic exclusion of 20 s. The ion trap MS/MS ion count target was set to 3 × 104 with a maximum injection time of 18 ms and fixed m/z range of 200–1,200. For parallel reaction monitoring analysis (Peterson et al., 2012), Orbitrap MS2 scans of targeted peptides were performed over a 200–2,000 m/z scan range at a resolving power of 60,000 at 200 m/z with an automatic gain control target of 1.5 × 106 ions and maximum injection time of 425–750 ms, depending on the number of peptides targeted simultaneously. Precursor ions were isolated using a quadrupole with 1.6 daltons isolation window and fragmented using higher-energy collisional dissociation with normalized collisional energy of 25.
The whole-proteome data were processed using the MaxQuant quantitative software package (version 1.6.1.0) and searched against the UniProt Mus musculus database (downloaded on June 18, 2018), containing protein isoforms. If not specified, default MaxQuant settings were used. Label-free quantitation (LFQ) was performed using an LFQ minimum ratio count of 1 and no MS/MS requirement for LFQ comparisons. Carbamidomethylation of cysteine residues was included as a fixed modification; oxidation of methionine and acetylation of protein N-termini were set as variable modifications. Match between runs was performed using default settings. Ion trap mobility spectrometry tandem MS tolerance was set to 0.3 daltons, and first search tolerance was set to 27 ppm. Lists of quantified proteins were filtered to remove reverse identifications, potential contaminants, and proteins, which were identified only by a modification site. Missing quantitative values were imputed for proteins that were observed in at least four of seven samples by randomly drawing values from the low end of the distribution of all measured protein abundance values (Cox et al., 2014).
GATA2 rescue assay
The reduced levels of GATA2 observed in −77−/− hematopoietic progenitors were rescued by infection of E14.5 Lin− fetal liver cells with retrovirus carrying the murine Gata2 cDNA in the murine stem cell virus plasmid (pMSCV; Katsumura et al., 2018). Ecotropic retrovirus was packaged in 293T cells, and retrovirus-containing supernatants were collected 24 and 48 h after transfection. Cells were infected with infectious supernatant by spinoculation for 90 min at 1,315 ×g, followed by 3 d of culturing in IMDM containing 20% FBS, 1% penicillin-streptomycin, 4% IL-3–conditioned media, and 4% SCF-conditioned media. RNA was purified with TRIzol. Global changes in gene expression were determined by RNA-seq of four biological replicates each of −77+/+ infected with empty vector, −77−/− infected with empty vector, and −77−/− infected with the Gata2 pMSCV-PIG expression vector. RNA libraries were prepared by the University of Wisconsin Gene Expression Center and sequenced using an Illumina HiSeq 2500 sequencer. Sequencing reads were aligned by STAR (version 2.5.2b) to the mouse genome (mm10; chromosomes 1 to 19; X, Y, and M) with GENCODE comprehensive gene annotation (version M16) on the reference chromosomes only. Gene expression levels were quantified by RSEM (version 1.3.0). STAR and RSEM runnings followed the RNA-seq quantification protocol from ENCODE (https://github.com/ENCODE-DCC/long-rna-seq-pipeline/blob/master/dnanexus/quant-rsem/resources/usr/bin/lrna_rsem_quantification.sh).
scRNA-seq
scRNA-seq was performed by the University of Wisconsin Gene Expression Center using the Chromium Single Cell Gene Expression platform (10x Genomics). Single-cell suspensions of CMP/GMP pools sorted from E14.5 fetal livers of two −77+/+ and two −77−/− embryos were loaded onto the Chromium Controller to generate single-cell barcoded gel bead emulsions for preparation of cDNA libraries and sequencing. Sequences were obtained from 5,167 −77+/+ and 10,028 −77−/− cells. Mean reads per cell were as follows: −77+/+ (37,365 and 25,498) and −77−/− (11,599 and 12,368). CellRanger (Zheng et al., 2017) was used with default parameter settings for alignment of sequencing reads, quantification of unique molecular identifier (UMI) counts, and filtering of empty barcodes. Cells with unusually high or low total UMI counts, a low number of detected genes, and a high proportion of UMI counts originating from mitochondrial genes were filtered with the isOutlier function in the R package scater (version 1.10.1; McCarthy et al., 2017) with nmads = 3. Genes that did not have UMI counts >4 in more than five cells were filtered out as well. Scran (version 1.10.2; Lun et al., 2016a) was used to normalize data across cells, and PCA, t-SNE (van der Maaten and Hinton, 2008), and SPRING (Weinreb et al., 2018) were used for dimension reduction. For PCA and t-SNE, the Seurat function (version 2.3.4; Butler et al., 2018) ScaleData was used to regress out total UMI count and average mitochondrial gene expression to ensure that principal components and t-SNE components were independent of these sources of variation. PCA and t-SNE were fit using the most variable genes via Seurat’s FindVariableGenes function. For trajectory analysis, SPRING was used with default settings after subsampling cells to ensure equal numbers of −77+/+ and −77−/− cells. Clustering analysis was performed with k-means using the R package cluster. We maximized average silhouette, which quantifies how similar a cell is to its own cluster compared with other clusters to determine the numbers of clusters, using the function silhouette from the R package cluster.
Quantification and statistical analysis
Quantitative RT-PCR
Results are presented as either the mean ± SEM or as box-and-whisker plots with whiskers ranging from minimum to maximum values. Multiple independent cohorts were used in each experiment. Statistical comparisons were performed using two-tailed Student’s t tests (significance cutoff of P value <0.05), with correction of statistical overrepresentation of functions calculated using the two-stage stepup method of Benjamini, Krieger, and Yekutieli as calculated using Prism software (GraphPad Software).
Proteomics
Statistical significance of changes in protein abundance between WT and mutant samples was determined using two-tailed Student’s t test followed by the correction for multiple hypothesis testing according to the Benjamini-Hochberg method (q < 0.05).
RNA-seq for GATA2 rescue
Differentially expressed genes were detected by DESeq2 (version 1.16.1), requiring that genes have at least twofold changes and an adjusted P value <0.05. Heatmaps of gene expression levels were prepared using ComplexHeatmap (version 1.99.7). Fragments per kilobase of transcript per million mapped reads values were added by 10−4 to avoid taking logarithm on zero.
scRNA-seq
For differential expression analysis, MAST (version 1.8.2; Finak et al., 2015) was used along with the Benjamini-Hochberg false discovery rate control to adjust for multiple comparisons. Genes with an adjusted P value <0.05 were considered differentially expressed.
Data and software availability
All raw files associated with proteomic analysis were deposited in the Proteomics Identifications Database (PRIDE) archive (Vizcaíno et al., 2016) under project accession no. PXD013855. RNA-seq raw files and RSEM quantification results were deposited in the Gene Expression Omnibus database under accession no. GSE133606. scRNA-seq raw files were deposited in the Gene Expression Omnibus database under accession no. GSE134439.
Online supplemental material
Fig. S1 shows STRING analysis of differentially expressed proteins in −77−/− CMP/GMP cells. Fig. S2 shows GATA2 Western blot and hierarchical clustering related to population RNA-seq datasets. Fig. S3 shows scRNA-seq cluster number optimization and cluster-specific Gene Ontology term analysis. Fig. S4 shows a comparison of cell proliferation features of −77+/+ and −77−/− progenitors mined from scRNA-seq data. Fig. S5 shows responsiveness of Tlr9 and Irf8 to IFN treatment and evidence for regulation of Hdac11 by GATA2. Table S1 reports dysregulated expression of immune machinery proteins in −77−/− progenitors. Table S2 shows the relationship between −77-regulated proteins and mRNAs. Table S3 lists resources and reagents used in the study.
Supplementary Material
shows dysregulated expression of innate immune machinery proteins in −77−/− progenitors.
lists relationship between −77-regulated proteins and mRNAs.
lists resources and reagents.
Acknowledgments
E.H. Bresnick was supported by the National Institutes of Health (grant DK68634), the Evans MDS Foundation, Midwest Athletes Against Childhood Cancer, and the University of Wisconsin Carbone Cancer Center (grant P30CA014520). D.J. Conn was supported by a National Library of Medicine training grant (NLM 5T15LM007359) to the University of Wisconsin Computation and Informatics in Biology and Medicine Training Program. This work was also supported by the University of Wisconsin Carbone Cancer Center (support grant P30 CA014520).
Author contributions: Conceptualization: K.D. Johnson and E.H. Bresnick; Methodology: K.D. Johnson, D.J. Conn, S. Shen, E. Shishkova, K.R. Katsumura, J.J. Coon, S.G. Kraus, K.R. Calvo, E.A. Ranheim, and E.H. Bresnick; Investigation: K.D. Johnson, D.J. Conn, E. Shishkova, K.R. Katsumura, K.R. Calvo, W. Wang, and A.P. Hsu; Formal analysis: K.D. Johnson, D.J. Conn, E. Shishkova, K.R. Katsumura, P. Liu, S. Keles, and E.H. Bresnick; Writing (original draft): K.D. Johnson and E.H. Bresnick; Writing (review and editing): K.D. Johnson, D.J. Conn, E. Shishkova, K.R. Katsumura, P. Liu, J.J. Coon, S. Keles, K.R. Calvo, S.M. Holland, and E.H. Bresnick; Funding acquisition: E.H. Bresnick; Supervision: J.J. Coon, S. Keles, S.M. Holland, and E.H. Bresnick.
References
- Baldridge M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., and Newell E.W.. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37:38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bigley V., Maisuria S., Cytlak U., Jardine L., Care M.A., Green K., Gunawan M., Milne P., Dickinson R., Wiscombe S., et al. 2018. Biallelic interferon regulatory factor 8 mutation: A complex immunodeficiency syndrome with dendritic cell deficiency, monocytopenia, and immune dysregulation. J. Allergy Clin. Immunol. 141:2234–2248. 10.1016/j.jaci.2017.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann L.H., Schubert A., Vu Manh T.P., De Andres L., Desbordes S.C., Parra M., Zimmermann T., Rapino F., Rodriguez-Ubreva J., Ballestar E., et al. 2009. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 5:554–566. 10.1016/j.stem.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo K.R., Vinh D.C., Maric I., Wang W., Noel P., Stetler-Stevenson M., Arthur D.C., Raffeld M., Dutra A., Pak E., et al. 2011. Myelodysplasia in autosomal dominant and sporadic monocytopenia immunodeficiency syndrome: diagnostic features and clinical implications. Haematologica. 96:1221–1225. 10.3324/haematol.2011.041152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Sun L., Aramsangtienchai P., Spiegelman N.A., Zhang X., Huang W., Seto E., and Lin H.. 2019. HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc. Natl. Acad. Sci. USA. 116:5487–5492. 10.1073/pnas.1815365116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churpek J.E., and Bresnick E.H.. 2019. Transcription factor mutations as a cause of familial myeloid neoplasms. J. Clin. Invest. 129:476–488. 10.1172/JCI120854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M.. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13:2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino J.D., Lodish M.B., MacKay J.P., and Orkin S.H.. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell. 3:219–228. 10.1016/S1097-2765(00)80312-3 [DOI] [PubMed] [Google Scholar]
- de Bruin A.M., Libregts S.F., Valkhof M., Boon L., Touw I.P., and Nolte M.A.. 2012. IFNγ induces monopoiesis and inhibits neutrophil development during inflammation. Blood. 119:1543–1554. 10.1182/blood-2011-07-367706 [DOI] [PubMed] [Google Scholar]
- de Bruin A.M., Voermans C., and Nolte M.A.. 2014. Impact of interferon-γ on hematopoiesis. Blood. 124:2479–2486. 10.1182/blood-2014-04-568451 [DOI] [PubMed] [Google Scholar]
- de Pater E., Kaimakis P., Vink C.S., Yokomizo T., Yamada-Inagawa T., van der Linden R., Kartalaei P.S., Camper S.A., Speck N., and Dzierzak E.. 2013. Gata2 is required for HSC generation and survival. J. Exp. Med. 210:2843–2850. 10.1084/jem.20130751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R.E., Griffin H., Bigley V., Reynard L.N., Hussain R., Haniffa M., Lakey J.H., Rahman T., Wang X.N., McGovern N., et al. 2011. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 118:2656–2658. 10.1182/blood-2011-06-360313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R.E., Milne P., Jardine L., Zandi S., Swierczek S.I., McGovern N., Cookson S., Ferozepurwalla Z., Langridge A., Pagan S., et al. 2014. The evolution of cellular deficiency in GATA2 mutation. Blood. 123:863–874. 10.1182/blood-2013-07-517151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16:278 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi M., Di Stefano B., Berenguer C., de Andrés-Aguayo L., Plana-Carmona M., Mendez-Lago M., Guillaumet-Adkins A., Rodriguez-Esteban G., Gut M., Gut I.G., et al. 2019. Single cell RNA-seq identifies the origins of heterogeneity in efficient cell transdifferentiation and reprogramming. eLife. 8 e41627 10.7554/eLife.41627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47(D1):D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Browne C.P., Cunniff K., Goff S.C., and Orkin S.H.. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA. 93:12355–12358. 10.1073/pnas.93.22.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Johnson K.D., Chang Y.I., Boyer M.E., Dewey C.N., Zhang J., and Bresnick E.H.. 2013. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J. Exp. Med. 210:2833–2842. 10.1084/jem.20130733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi A., Paul F., Herzog Y., Lubling Y., Weiner A., Yofe I., Jaitin D., Cabezas-Wallscheid N., Dress R., Ginhoux F., et al. 2018. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20:836–846. 10.1038/s41556-018-0121-4 [DOI] [PubMed] [Google Scholar]
- Grass J.A., Boyer M.E., Pal S., Wu J., Weiss M.J., and Bresnick E.H.. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA. 100:8811–8816. 10.1073/pnas.1432147100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass J.A., Jing H., Kim S.-I., Martowicz M.L., Pal S., Blobel G.A., and Bresnick E.H.. 2006. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 26:7056–7067. 10.1128/MCB.01033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröschel S., Sanders M.A., Hoogenboezem R., de Wit E., Bouwman B.A.M., Erpelinck C., van der Velden V.H.J., Havermans M., Avellino R., van Lom K., et al. 2014. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 157:369–381. 10.1016/j.cell.2014.02.019 [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Haas S., Trumpp A., and Milsom M.D.. 2018. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 22:627–638. 10.1016/j.stem.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Hahn C.N., Chong C.E., Carmichael C.L., Wilkins E.J., Brautigan P.J., Li X.C., Babic M., Lin M., Carmagnac A., Lee Y.K., et al. 2011. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 43:1012–1017. 10.1038/ng.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton S., Salem S., Bustamante J., Bigley V., Boisson-Dupuis S., Azevedo J., Fortin A., Haniffa M., Ceron-Gutierrez L., Bacon C.M., et al. 2011. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 365:127–138. 10.1056/NEJMoa1100066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert A.S., Thöing C., Riley N.M., Kwiecien N.W., Shiskova E., Huguet R., Cardasis H.L., Kuehn A., Eliuk S., Zabrouskov V., et al. 2018. Improved precursor characterization for data-dependent mass spectrometry. Anal. Chem. 90:2333–2340. 10.1021/acs.analchem.7b04808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe P.S., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Moritz N., Endele M., Filipczyk A., Gambardella A., Ahmed N., et al. 2016. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 535:299–302. 10.1038/nature18320 [DOI] [PubMed] [Google Scholar]
- Hsu A.P., Sampaio E.P., Khan J., Calvo K.R., Lemieux J.E., Patel S.Y., Frucht D.M., Vinh D.C., Auth R.D., Freeman A.F., et al. 2011. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 118:2653–2655. 10.1182/blood-2011-05-356352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A.P., Johnson K.D., Falcone E.L., Sanalkumar R., Sanchez L., Hickstein D.D., Cuellar-Rodriguez J., Lemieux J.E., Zerbe C.S., Bresnick E.H., et al. 2013. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 121:3830–3837–S7. 10.1182/blood-2012-08-452763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., and Ivashkiv L.B.. 2009. Cross-regulation of signaling pathways by interferon-γ: implications for immune responses and autoimmune diseases. Immunity. 31:539–550. 10.1016/j.immuni.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.D., Hsu A.P., Ryu M.J., Wang J., Gao X., Boyer M.E., Liu Y., Lee Y., Calvo K.R., Keles S., et al. 2012. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J. Clin. Invest. 122:3692–3704. 10.1172/JCI61623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.D., Kong G., Gao X., Chang Y.-I., Hewitt K.J., Sanalkumar R., Prathibha R., Ranheim E.A., Dewey C.N., Zhang J., et al. 2015. Cis-regulatory mechanisms governing stem and progenitor cell transitions. Sci. Adv. 1 e1500503 10.1126/sciadv.1500503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S., Suzuki M., Yamaoka A., Keleku-Lukwete N., Katsuoka F., Otsuki A., Kure S., Engel J.D., and Yamamoto M.. 2017. GATA2 haploinsufficiency accelerates EVI1-driven leukemogenesis. Blood. 130:908–919. 10.1182/blood-2016-12-756767 [DOI] [PubMed] [Google Scholar]
- Katsumura K.R., Yang C., Boyer M.E., Li L., and Bresnick E.H.. 2014. Molecular basis of crosstalk between oncogenic Ras and the master regulator of hematopoiesis GATA-2. EMBO Rep. 15:938–947. 10.15252/embr.201438808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura K.R., Ong I.M., DeVilbiss A.W., Sanalkumar R., and Bresnick E.H.. 2016. GATA factor-dependent positive-feedback circuit in acute myeloid leukemia cells. Cell Rep. 16:2428–2441. 10.1016/j.celrep.2016.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura K.R., and Bresnick E.H.; GATA Factor Mechanisms Group . 2017. The GATA factor revolution in hematology. Blood. 129:2092–2102. 10.1182/blood-2016-09-687871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura K.R., Mehta C., Hewitt K.J., Soukup A.A., Fraga de Andrade I., Ranheim E.A., Johnson K.D., and Bresnick E.H.. 2018. Human leukemia mutations corrupt but do not abrogate GATA-2 function. Proc. Natl. Acad. Sci. USA. 115:E10109–E10118. 10.1073/pnas.1813015115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Hernandez L.D., Galán J.E., Janeway C.A. Jr., Medzhitov R., and Flavell R.A.. 2002. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 110:191–202. 10.1016/S0092-8674(02)00827-9 [DOI] [PubMed] [Google Scholar]
- Kurotaki D., Osato N., Nishiyama A., Yamamoto M., Ban T., Sato H., Nakabayashi J., Umehara M., Miyake N., Matsumoto N., et al. 2013. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 121:1839–1849. 10.1182/blood-2012-06-437863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Esain V., Teng L., Xu J., Kwan W., Frost I.M., Yzaguirre A.D., Cai X., Cortes M., Maijenburg M.W., et al. 2014. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28:2597–2612. 10.1101/gad.253302.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K.W., Ottersbach K., van Hamburg J.P., Oziemlak A., Tsai F.Y., Orkin S.H., Ploemacher R., Hendriks R.W., and Dzierzak E.. 2004. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200:871–882. 10.1084/jem.20031556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T.L., Bach K., and Marioni J.C.. 2016a Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17:75 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T.L., McCarthy D.J., and Marioni J.C.. 2016b A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000 Res. 5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., and MacCoss M.J.. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26:966–968. 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancino A., Termanini A., Barozzi I., Ghisletti S., Ostuni R., Prosperini E., Ozato K., and Natoli G.. 2015. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29:394–408. 10.1101/gad.257592.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marecki S., Riendeau C.J., Liang M.D., and Fenton M.J.. 2001. PU.1 and multiple IFN regulatory factor proteins synergize to mediate transcriptional activation of the human IL-1 β gene. J. Immunol. 166:6829–6838. 10.4049/jimmunol.166.11.6829 [DOI] [PubMed] [Google Scholar]
- Martinon F., Burns K., and Tschopp J.. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Masters S.L., Gerlic M., Metcalf D., Preston S., Pellegrini M., O’Donnell J.A., McArthur K., Baldwin T.M., Chevrier S., Nowell C.J., et al. 2012. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity. 37:1009–1023. 10.1016/j.immuni.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Campbell K.R., Lun A.T., and Wills Q.F.. 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver S.C., Hewitt K.J., Gao X., Mehta C., Zhang J., and Bresnick E.H.. 2018. Dissecting regulatory mechanisms using mouse fetal liver-derived erythroid cells. Methods Mol. Biol. 1698:67–89. 10.1007/978-1-4939-7428-3_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McReynolds L.J., Calvo K.R., and Holland S.M.. 2018. Germline gata2 mutation and bone marrow failure. Hematol. Oncol. Clin. North Am. 32:713–728. 10.1016/j.hoc.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta C., Johnson K.D., Gao X., Ong I.M., Katsumura K.R., McIver S.C., Ranheim E.A., and Bresnick E.H.. 2017. Integrating enhancer mechanisms to establish a hierarchical blood development program. Cell Rep. 20:2966–2979. 10.1016/j.celrep.2017.08.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraro D., Gleit-Kielmanowicz M., Hauser H., and Levi B.Z.. 2002. IFN-stimulated gene 15 is synergistically activated through interactions between the myelocyte/lymphocyte-specific transcription factors, PU.1, IFN regulatory factor-8/IFN consensus sequence binding protein, and IFN regulatory factor-4: characterization of a new subtype of IFN-stimulated response element. J. Immunol. 168:6224–6231. 10.4049/jimmunol.168.12.6224 [DOI] [PubMed] [Google Scholar]
- Nerlov C., Querfurth E., Kulessa H., and Graf T.. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 95:2543–2551. 10.1182/blood.V95.8.2543 [DOI] [PubMed] [Google Scholar]
- Olsson A., Venkatasubramanian M., Chaudhri V.K., Aronow B.J., Salomonis N., Singh H., and Grimes H.L.. 2016. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 537:698–702. 10.1038/nature19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., and Zon L.I.. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644. 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P., Simpson M.A., Connell F.C., Steward C.G., Brice G., Woollard W.J., Dafou D., Kilo T., Smithson S., Lunt P., et al. 2011. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat. Genet. 43:929–931. 10.1038/ng.923 [DOI] [PubMed] [Google Scholar]
- Palii C.G., Cheng Q., Gillespie M.A., Shannon P., Mazurczyk M., Napolitani G., Price N.D., Ranish J.A., Morrissey E., Higgs D.R., et al. 2019. Single-cell proteomics reveal that quantitative changes in co-expressed lineage-specific transcription factors determine cell fate. Cell Stem Cell. 24:812–820.e5. 10.1016/j.stem.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A., et al. 2015. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 163:1663–1677. 10.1016/j.cell.2015.11.013 [DOI] [PubMed] [Google Scholar]
- Peterson A.C., Russell J.D., Bailey D.J., Westphall M.S., and Coon J.J.. 2012. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics. 11:1475–1488. 10.1074/mcp.O112.020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.F., D’Agati V., Orkin S.H., and Costantini F.. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 349:257–260. 10.1038/349257a0 [DOI] [PubMed] [Google Scholar]
- Pevny L., Lin C.S., D’Agati V., Simon M.C., Orkin S.H., and Costantini F.. 1995. Development of hematopoietic cells lacking transcription factor GATA-1. Development. 121:163–172. [DOI] [PubMed] [Google Scholar]
- Richards A.L., Merrill A.E., and Coon J.J.. 2015. Proteome sequencing goes deep. Curr. Opin. Chem. Biol. 24:11–17. 10.1016/j.cbpa.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N.P., Janzen V., Forkert R., Dombkowski D.M., Boyd A.S., Orkin S.H., Enver T., Vyas P., and Scadden D.T.. 2005. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 106:477–484. 10.1182/blood-2004-08-2989 [DOI] [PubMed] [Google Scholar]
- Ronald P.C., and Beutler B.. 2010. Plant and animal sensors of conserved microbial signatures. Science. 330:1061–1064. 10.1126/science.1189468 [DOI] [PubMed] [Google Scholar]
- Rossi L., Lin K.K., Boles N.C., Yang L., King K.Y., Jeong M., Mayle A., and Goodell M.A.. 2012. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 11:302–317. 10.1016/j.stem.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönheit J., Kuhl C., Gebhardt M.L., Klett F.F., Riemke P., Scheller M., Huang G., Naumann R., Leutz A., Stocking C., et al. 2013. PU.1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep. 3:1617–1628. 10.1016/j.celrep.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Shishkova E., Hebert A.S., Westphall M.S., and Coon J.J.. 2018. Ultra-high pressure (>30,000 psi) packing of capillary columns enhancing depth of shotgun proteomic analyses. Anal. Chem. 90:11503–11508. 10.1021/acs.analchem.8b02766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup A.A., Zheng Y., Mehta C., Wu J., Liu P., Cao M., Hofmann I., Zhou Y., Zhang J., Johnson K.D., et al. 2019. Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. J. Clin. Invest. 129:1180–1192. 10.1172/JCI122694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner M.A., Sanchez L.A., Hsu A.P., Shaw P.A., Zerbe C.S., Calvo K.R., Arthur D.C., Gu W., Gould C.M., Brewer C.C., et al. 2014. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood. 123:809–821. 10.1182/blood-2013-07-515528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., and Darnell J.E. Jr.. 2012. The JAK-STAT pathway at twenty. Immunity. 36:503–514. 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.-Y., Keller G., Kuo F.C., Weiss M., Chen J., Rosenblatt M., Alt F.W., and Orkin S.H.. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 371:221–226. 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Tsai F.-Y., and Orkin S.H.. 1997. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 89:3636–3643. 10.1182/blood.V89.10.3636 [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Fujiwara Y., Hom D.B., and Orkin S.H.. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176–1188. 10.1101/gad.12.8.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura H., Tamura T., Kong H.J., Nishiyama A., Ishii K.J., Klinman D.M., and Ozato K.. 2004. Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J. Immunol. 172:6820–6827. 10.4049/jimmunol.172.11.6820 [DOI] [PubMed] [Google Scholar]
- Tusi B.K., Wolock S.L., Weinreb C., Hwang Y., Hidalgo D., Zilionis R., Waisman A., Huh J.R., Klein A.M., and Socolovsky M.. 2018. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. 555:54–60. 10.1038/nature25741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L.J.P., and Hinton G.E.. 2008. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9:2579–2605. [Google Scholar]
- Vizcaíno J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44:11033 10.1093/nar/gkw880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J.C., DeKoter R.P., Lee H.-J., Smith E.D., Lancki D.W., Gurish M.F., Friend D.S., Stevens R.L., Anastasi J., and Singh H.. 2002. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 17:665–676. 10.1016/S1074-7613(02)00452-1 [DOI] [PubMed] [Google Scholar]
- Wang G.G., Calvo K.R., Pasillas M.P., Sykes D.B., Häcker H., and Kamps M.P.. 2006. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods. 3:287–293. 10.1038/nmeth865 [DOI] [PubMed] [Google Scholar]
- Watcham S., Kucinski I., and Gottgens B.. 2019. New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood. 133:1415–1426. 10.1182/blood-2018-08-835355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C., Wolock S., and Klein A.M.. 2018. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics. 34:1246–1248. 10.1093/bioinformatics/btx792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman I. 2016. How one thing led to another. Annu. Rev. Immunol. 34:1–30. 10.1146/annurev-immunol-032414-112003 [DOI] [PubMed] [Google Scholar]
- Woods D.M., Woan K.V., Cheng F., Sodré A.L., Wang D., Wu Y., Wang Z., Chen J., Powers J., Pinilla-Ibarz J., et al. 2017. T cells lacking HDAC11 have increased effector functions and mediate enhanced alloreactivity in a murine model. Blood. 130:146–155. 10.1182/blood-2016-08-731505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dao Thi V.L., Huang Y., Billerbeck E., Saha D., Hoffmann H.H., Wang Y., Silva L.A.V., Sarbanes S., Sun T., et al. 2018. Intrinsic immunity shapes viral resistance of stem cells. Cell. 172:423–438.e25. 10.1016/j.cell.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Suzuki M., Otsuki A., Shimizu R., Bresnick E.H., Engel J.D., and Yamamoto M.. 2014. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 25:415–427. 10.1016/j.ccr.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez A., and Goodridge H.S.. 2016. Interferon regulatory factor 8 and the regulation of neutrophil, monocyte, and dendritic cell production. Curr. Opin. Hematol. 23:11–17. 10.1097/MOH.0000000000000196 [DOI] [PubMed] [Google Scholar]
- Yáñez A., Ng M.Y., Hassanzadeh-Kiabi N., and Goodridge H.S.. 2015. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood. 125:1452–1459. 10.1182/blood-2014-09-600833 [DOI] [PubMed] [Google Scholar]
- Zhang P., Zhang X., Iwama A., Yu C., Smith K.A., Mueller B.U., Narravula S., Torbett B.E., Orkin S.H., and Tenen D.G.. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 96:2641–2648. 10.1182/blood.V96.8.2641 [DOI] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoumbos N.C., Gascon P., Djeu J.Y., and Young N.S.. 1985. Interferon is a mediator of hematopoietic suppression in aplastic anemia in vitro and possibly in vivo. Proc. Natl. Acad. Sci. USA. 82:188–192. 10.1073/pnas.82.1.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows dysregulated expression of innate immune machinery proteins in −77−/− progenitors.
lists relationship between −77-regulated proteins and mRNAs.
lists resources and reagents.
Data Availability Statement
All raw files associated with proteomic analysis were deposited in the Proteomics Identifications Database (PRIDE) archive (Vizcaíno et al., 2016) under project accession no. PXD013855. RNA-seq raw files and RSEM quantification results were deposited in the Gene Expression Omnibus database under accession no. GSE133606. scRNA-seq raw files were deposited in the Gene Expression Omnibus database under accession no. GSE134439.