Morphometric characterization of TAMs in human colorectal liver metastases captures individual populations, corresponding to single-cell clusters and associated with distinct clinical outcome and transcriptional profiles. Quantitative analysis of macrophage morphology can serve as a simple readout of functional diversity with prognostic significance.
Abstract
It has long been known that in vitro polarized macrophages differ in morphology. Stemming from a conventional immunohistology observation, we set out to test the hypothesis that morphology of tumor-associated macrophages (TAMs) in colorectal liver metastasis (CLM) represents a correlate of functional diversity with prognostic significance. Density and morphological metrics of TAMs were measured and correlated with clinicopathological variables. While density of TAMs did not correlate with survival of CLM patients, the cell area identified small (S-TAM) and large (L-TAM) macrophages that were associated with 5-yr disease-free survival rates of 27.8% and 0.2%, respectively (P < 0.0001). RNA sequencing of morphologically distinct macrophages identified LXR/RXR as the most enriched pathway in large macrophages, with up-regulation of genes involved in cholesterol metabolism, scavenger receptors, MERTK, and complement. In single-cell analysis of mononuclear phagocytes from CLM tissues, S-TAM and L-TAM signatures were differentially enriched in individual clusters. These results suggest that morphometric characterization can serve as a simple readout of TAM diversity with strong prognostic significance.
Introduction
The liver is the most common site of metastases for colorectal cancer (CRC), a major cause of mortality worldwide (Siegel et al., 2020). Surgical resection of colorectal liver metastases (CLMs), combined with systemic chemotherapy has the potential to be curative, projecting the 5-yr survival rate up to 50% and the 10-yr overall survival rate up to 35% (Garden et al., 2006; Cucchetti et al., 2015). However, CLM patients present with heterogeneous clinical outcomes and degrees of therapeutic responsiveness (Halama et al., 2011; Mlecnik et al., 2018), and only an improved definition of the distinct features associated to clinical behaviors would refine patient stratification and ameliorate therapeutic output. Immune cells and mediators populate the tumor microenvironment both at the primary and metastatic sites (Hanahan and Weinberg, 2011) and their quantitative evaluation in tumor tissues holds promise toward the important clinical endpoint of better patient profiling (Fridman et al., 2017).
Tumor-associated macrophages (TAMs), essential elements of the tumor microenvironment (Mantovani et al., 2008; Ruffell et al., 2012; Murray et al., 2014; Mantovani et al., 2017; DeNardo and Ruffell, 2019), pave the way to tissue invasion and intravasation and provide a nurturing microenvironment for metastasis, serving as a component of the cancer cell niche at distant sites (Peinado et al., 2017). Their clinical relevance has been investigated in large cohorts of cancer patients, confirming TAMs as prognostic indicators of cancer progression (Forssell et al., 2007; Steidl et al., 2010) and key determinants of the efficacy of anticancer strategies (Mantovani and Allavena, 2015; Di Caro et al., 2016; Malesci et al., 2017; Mantovani et al., 2017; Cassetta and Pollard, 2018; Cortese et al., 2019; De Palma et al., 2019; DeNardo and Ruffell, 2019). Diverse methodological approaches have been adopted, including morphological identification, evaluation of conventional macrophage-related markers (Forssell et al., 2007; Steidl et al., 2010; Di Caro et al., 2016; Malesci et al., 2017), gene expression profiling (Galon et al., 2013), single-cell RNA sequencing (scRNAseq; Lavin et al., 2017; Zhang et al., 2019; Zilionis et al., 2019). Collectively, these clinical studies have highlighted the profound intratumor heterogeneity of TAMs (Laoui et al., 2014) in terms of polarization, functions and tissue localization, raising concerns about evaluating the macrophage population as a whole. Plasticity of mononuclear phagocytes (Locati et al., 2019) can be modeled in vitro by stimulating macrophages with IFN-γ and microbial components (M1, M1-like) or IL-4 and alternative signals (M2, M2-like), resulting in extreme polarization states that correspond to distinct morphologies (Waldo et al., 2008; Ballotta et al., 2014; Rodell et al., 2018). Specifically, M1-like macrophages more frequently appear round and flattened, as opposed to M2-like cells that present with an elongated morphology (Waldo et al., 2008; Ballotta et al., 2014; Rodell et al., 2018). Similarly, macrophages acquire distinct geometries in vivo (Geissmann et al., 2010; Yona and Gordon, 2015), such as in tissues with fibrous architectures (McWhorter et al., 2015), or in chronic inflammatory lesions (Russell et al., 2009). Finally, modulation of macrophage shape by physical cues can result in alterations of their polarization state (Sussman et al., 2014; McWhorter et al., 2015), suggesting that morphological features are not merely a correlate of macrophage function but rather that macrophage shape and function are connected by a causative link.
Macrophages are essential components of liver architecture in both homeostasis and disease (Wynn et al., 2013; Krenkel and Tacke, 2017; Keirsse et al., 2018). Their functions are tightly controlled by transcription factors, including the liver X receptors (LXRs), ligand-activated nuclear receptors that control several physiological processes such as cholesterol efflux, inflammatory response, and phagocytosis (Spann et al., 2012; Spann and Glass, 2013; Hong and Tontonoz, 2014; Tall and Yvan-Charvet, 2015). These three tasks are critically executed by macrophages as part of their function as regulators of tissue homeostasis and are remarkably interconnected. Inflammatory cues and LXR-mediated handling of cholesterol are counterregulated, resulting in the anti-inflammatory polarization of macrophages upon LXR activation and in the inhibition of LXR activation during the acute-phase response (Spann et al., 2012; Spann and Glass, 2013; Hong and Tontonoz, 2014; Tall and Yvan-Charvet, 2015). Metabolism is a key determinant of macrophage function and the interplay of these cells with tumor cells, vasculature, and immunocompetent cells (Colegio et al., 2014; Wenes et al., 2016; Saha et al., 2017; Bekkering et al., 2018).
Upon inspection of TAMs in human CLMs using conventional immune histology, we appreciated their heterogeneity in terms of size and morphology. In an attempt to refine patient stratification by identification of distinct features of the tumor immune microenvironment, we tested the hypothesis that discrete TAM populations can be identified in CLMs on the basis of their morphology and be exploited as correlates of clinical outcome.
Results and discussion
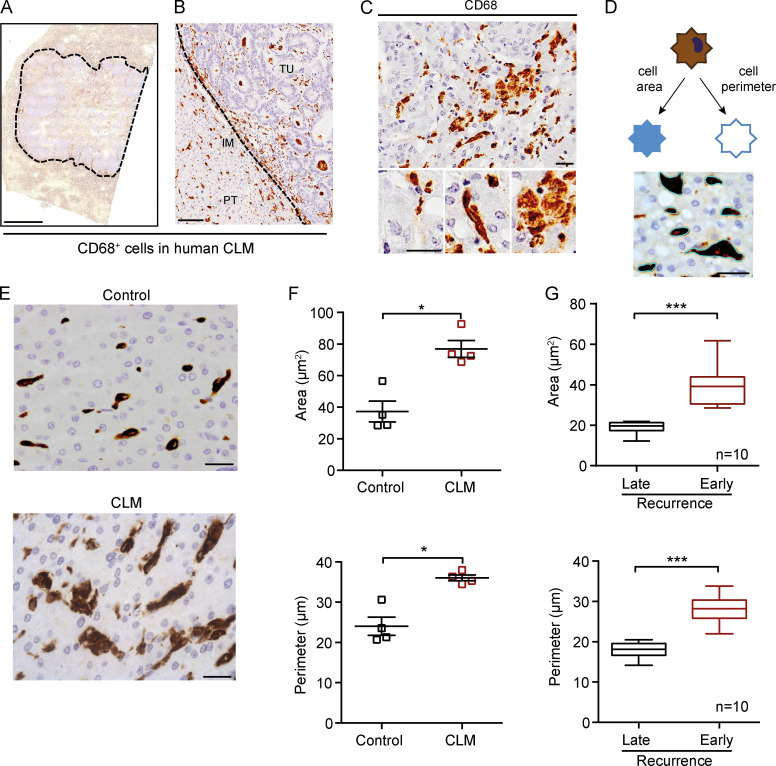
The immune landscape of CLMs has been shown to impact the outcome and therapeutic response in CLMs, with a critical contribution from CD3+ and CD8+ T cells (Halama et al., 2011; Mlecnik et al., 2018). For their plasticity and variable association to clinical outcome across cancers, we focused on macrophages, which we initially identified with CD68 staining. Human CLMs were infiltrated by CD68+ TAMs (Fig. 1 A), which variably localized both in the peritumor (PT) area, invasive margin (IM), and tumor center (Fig. 1 B). TAMs were heterogeneous in terms of size and morphology (e.g., Fig. 1 C, top), ranging from small and round to spiky and elongated to large disk-like cells (Fig. 1 C, bottom). It has long been known that in vitro polarized macrophages differ in morphology (Waldo et al., 2008; Ballotta et al., 2014; Rodell et al., 2018). Therefore, we set out to assess TAM morphological diversity in human CLMs using rigorous quantitative criteria (i.e., cell area and perimeter; Fig. 1 D) and test whether it could have a prognostic relevance. We initially probed our hypothesis by measuring macrophage shape in an exploratory set of four CLM patients and controls. Both macrophage area and perimeter were significantly higher in CLMs compared with control tissues (P = 0.029 and P = 0.028, respectively; Fig. 1, E and F). Next, we conducted an exploratory study in 20 patients selected because they were representative of opposite clinical outcomes. Larger macrophages, operationally defined large TAMs (L-TAMs), were more frequently found in CLM patients with worse prognosis compared with patients with good prognosis (early disease recurrence, within 24 mo after surgery, or late disease recurrence, after 24 mo from surgery, respectively; n = 10 each group; P < 0.001; Fig. 1 G), suggesting that TAM morphology is a critical feature associated to distinct clinical outcomes in CLM patients.
Figure 1.
Morphological assessment of macrophages in human CLMs. (A) Representative whole slide immunohistochemistry of CD68+ cells in a CLM specimen. Dotted line indicates the tumor lesion. (B) CD68+ macrophages infiltrate both the PT area, IM, and tumor (TU) of CLM tissues. (C) Morphological features of macrophages in CLM. Three exemplificative types of macrophages in the same region are shown: small and round shaped, spiky and elongated, and big pancake-like. (D) Representative scheme of morphological features of macrophages analyzed (area and perimeter). Picture depicts the image analysis procedure. (E) Representative pictures of CD68+ macrophages in a control liver (symptomatic giant liver hemangioma, top) and a CLM specimen (bottom). (F) Both macrophage area and perimeter are significantly higher in CLM specimens compared with controls (i.e., patients who had undergone surgery for symptomatic giant liver hemangiomas). Represented are mean ± SEM of three pictures from each specimen (n = 4 specimens each group; *, P = 0.029 [area] and P = 0.028 [perimeter] by Mann-Whitney test). (G) Quantitation of macrophage area and perimeter in CLM specimens from patients with late recurrence (DFS >24 mo) or early recurrence (DFS <24 mo). Box plots give median, lower, and upper quartile values by the box and minimum and maximum values by the whiskers from three pictures for each specimen (n = 10 specimens each group; ***, P < 0.001 by unpaired t test). Scale bars: 2 mm (A), 100 µm (B), and 50 µm (C–E).
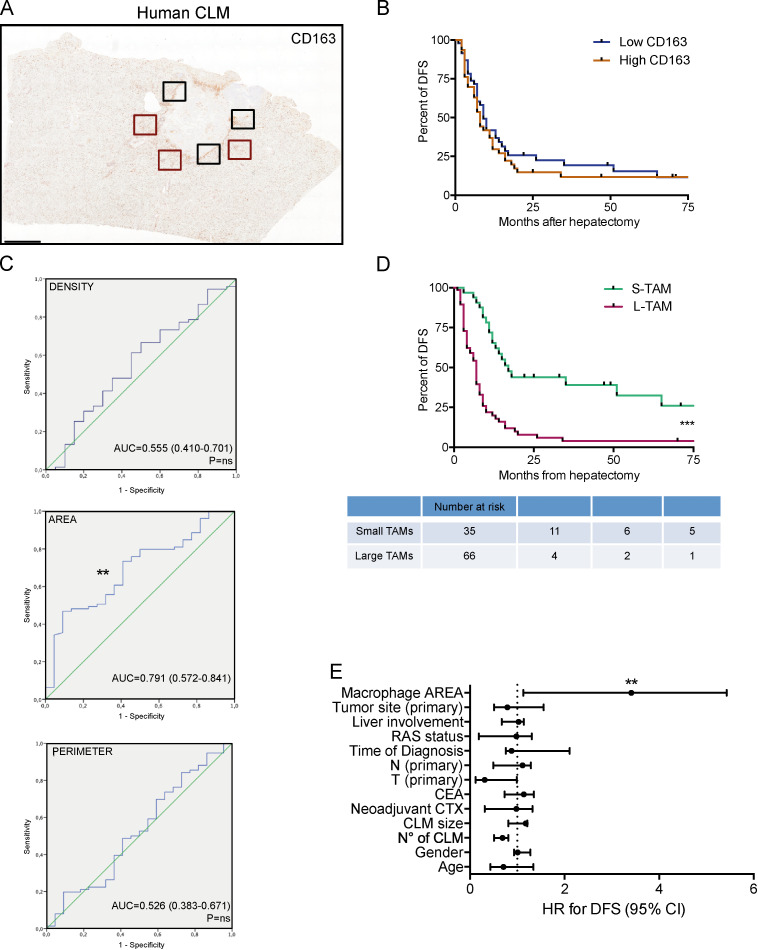
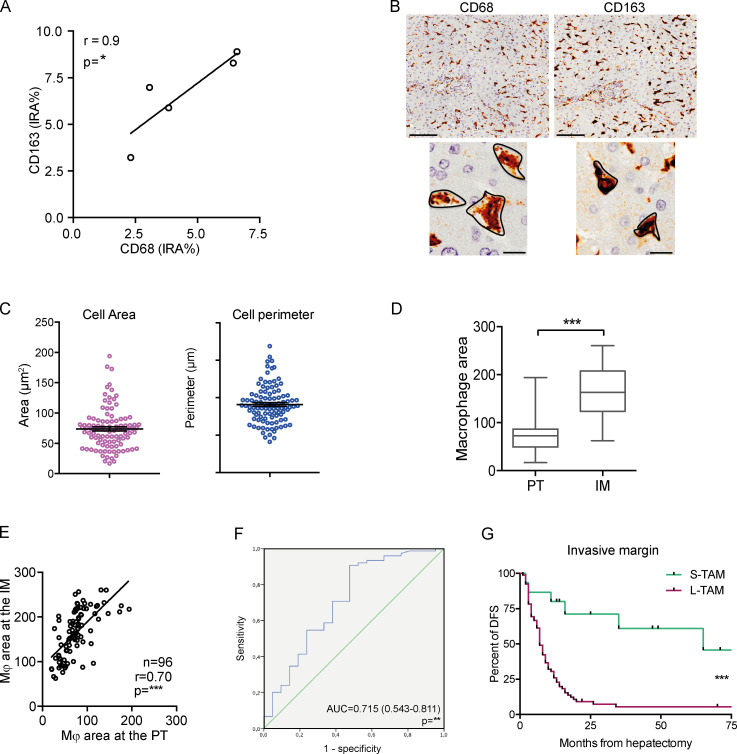
We then quantified macrophage area and perimeter in the PT area of CLM sections (Fig. 2 A) in a large validation set of 101 CLM patients and concomitantly assessed their density (percentage of immune reactive area), which is a common parameter used to test the clinical relevance of immune cell populations. The validation set (Table S1) comprised patients with severe and heterogeneous tumor burden (73% synchronous presentation; median number of metastases, 4 [range, 1–31]; median size, 4 cm [range, 0.5–18 cm]), highly representative of the clinical and pathological presentations of liver metastases. Density of CD68+ macrophages correlated with density of CD163+ macrophages (n = 5, r = 0.9, P = 0.083; Fig. S1 A), an M2-like macrophage marker commonly used in immunohistochemical analyses (Forssell et al., 2007; Steidl et al., 2010; Di Caro et al., 2016; Malesci et al., 2017). We thus opted to stain with anti-CD163, because this molecule is more uniformly distributed on the cell surface (and not intracellularly as is CD68), allowing for a better appreciation of morphometric indexes (Fig. S1 B). Moreover, in other tumors, non-macrophage expression of CD68 has been reported (Ruffell et al., 2012). Macrophage area ranged from 16.85 μm2 to 193.88 μm2, and perimeter ranged from 16.43 μm to 67.59 μm (Fig. S1 C). The density of CD163+ macrophages was not associated to differences in survival time (Fig. 2 B), while comparison of the areas under the receiver operating characteristic (ROC) curves for density, area, and perimeter revealed that only the macrophage area was statistically associated with disease recurrence (Fig. 2 C). We then defined small TAMs (S-TAMs) and L-TAMs by using the best cutoff value extrapolated from the ROC curve (area = 58.55 μm2; sensitivity = 0.79; specificity = 0.44) and found that S-TAM and L-TAM were associated with significantly different 5-yr disease-free survival (DFS) rates of 27.8% and 0.2%, respectively (P < 0.0001; Fig. 2 D). Patients with more aggressive disease had TAMs with average area above cutoff (median DFS, 16 mo [range, 10.8–21.1] and 6.5 mo [range, 5.1–7.8] in patients with S-TAMs and L-TAMs, respectively; Fig. 2 D). Notably, at multivariate analysis, only TAM area was found to be independently statistically associated with DFS (hazard ratio [HR] for recurrence = 3.41; 95% confidence interval [CI] = 1.13–5.43; P = 0.001; Fig. 2 E and Table 1). Similar assessment of macrophage area was performed at the IM. Macrophage area at the IM was in general higher compared with area of macrophages in the PT region (cutoff value extrapolated from the ROC: area = 97 μm2), but the values significantly correlated (Fig. S1, D–F). Notably, macrophage area at the IM retained a significant association with shorter DFS (P = 0.0008 by Mantel–Cox; Fig. S1 G), confirming a strong correlation between area of TAMs and worse prognosis. It should be pointed out, however, that assessment of TAM morphology in this region could be distorted by alterations in tissue architecture typical of tumor areas.
Figure 2.
Macrophage morphology is a prognostic factor in human CLMs. (A) Representative whole-slide immunohistochemistry of CD163+ cells in a CLM specimen. Area and perimeter of macrophages were quantitated in three non-contiguous areas of the PT region (red line) and IM (black line) of curatively resected metastases from 101 metastatic CRC patients. Scale bar: 2 mm. (B) Kaplan-Meier curve of CD163+ macrophages in 95 CLM specimens. Represented are mean ± SEM of three pictures from each specimen (P = not significant [ns] by log-rank Mantel–Cox test). (C) ROC curves for density, area, and perimeter of CD163+ macrophages to predict disease recurrence in CLM patients. P = ns (density); **, P = 0.006 (area); P = ns (perimeter). AUC, area under the curve. (D) Kaplan-Meier curve of macrophage area in 101 CLM specimens (S-TAM = average area below ROC cutoff value; L-TAM = average area above ROC cutoff value; represented are mean ± SEM of three pictures from each specimen; ***, P < 0.0001 by log-rank Mantel–Cox test). (E) Forest plot showing the results of multivariate regression analysis for DFS in 101 CLM patients. The x axis represents the HR for recurrence with the reference line (dashed), HRs (circles), and 95% CI (whiskers). ***, P < 0.001 by multiple regression analysis. Liver involvement: bilateral versus unilateral. Time of diagnosis: synchronous versus metachronous. N (node) and T (tumor) refer to the primary tumor. CEA, carcinoembryonic antigen. CTX, chemotherapy.
Figure S1.
Measurement of macrophage morphological metrics. (A) Density (percentage of immunoreactive area [IRA%]) of CD68+ macrophages correlates with density of CD163+ macrophages (n = 5, r = 0.9, P = 0.083 by Spearman analysis). (B) Expression of CD68 and CD163 on liver macrophages from one representative CLM specimen. CD163 (right) is more uniformly distributed on the cell surface and not intracellularly as CD68 (left). Scale bars: 100 µm (top panels); 50 µm (bottom panels). (C) Distribution of macrophage morphometric indexes (area and perimeter) in 101 patients. (D–G) Prognostic value of macrophage area at the IM in 96 CLM patients. Distribution of macrophage area (D) and correlation between area of macrophages in PT and IM regions (E) are depicted. ***, P < 0.001 by Mann-Whitney (D) and ***, P < 0.001 by Spearman correlation analysis (E). (F) ROC curve for area to predict disease recurrence for 96 CLM patients. (G) Kaplan-Meier curve of macrophage area in the IM region in 96 CLM specimens (S-TAM = average area below median value; L-TAM = average area above median value; ***, P = 0.0008 by log-rank Mantel–Cox test).
Table 1. Multivariate analysis of prognostic factors for DFS.
| Factor | Cox model | ||
|---|---|---|---|
| HR | 95% CI | P value | |
| Age, per increase of 1 yr | 0.71 | 0.43–1.34 | 0.127 |
| Sex, men vs. women | 1.01 | 0.93–1.28 | 0.414 |
| CLM number, per increase of 1 unit | 0.71 | 0.51–0.81 | 0.312 |
| CLM size, per increase of 1 cm | 1.18 | 0.81–1.21 | 0.063 |
| Preoperative chemotherapy, yes vs. no | 0.98 | 0.31–1.32 | 0.849 |
| CEA level, elevated vs. normal | 1.14 | 0.73–1.35 | 0.073 |
| CA19-9 level, elevated vs. normal | 1.31 | 1.58–2.61 | 0.061 |
| T of the primary tumor, T3-4 vs. T1-T2 | 0.31 | 0.12–0.99 | 0.789 |
| N of the primary tumor, positive vs. negative | 1.01 | 0.49–1.29 | 0.071 |
| Synchronous vs. metachronous presentation | 0.88 | 0.76–2.11 | 0.061 |
| Grading of primary tumor, G1-2 vs. G3-4 | 1.19 | 0.18–1.81 | 0.097 |
| RAS status, mutated vs. wild-type | 0.98 | 0.19–1.31 | 0.739 |
| Liver involvement, bilateral vs. unilateral | 1.01 | 0.67–1.14 | 0.076 |
| Site of the primary tumor, right vs. left | 0.79 | 0.51–1.56 | 0.364 |
| TAMs area, L-TAM vs. S-TAM | 3.41 | 1.13–5.43 | 0.001 |
CA 19-9, cancer antigen 19-9; CEA, carcinoembryonic antigen; T, tumor, N, node; RAS, RAS gene.
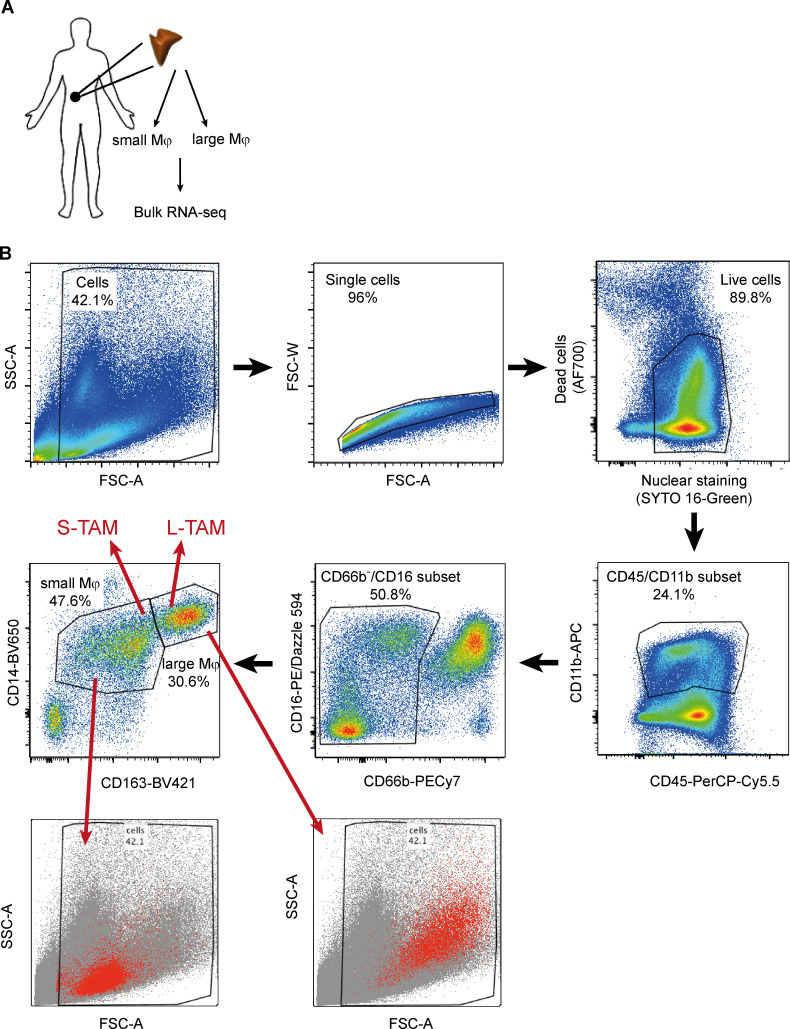
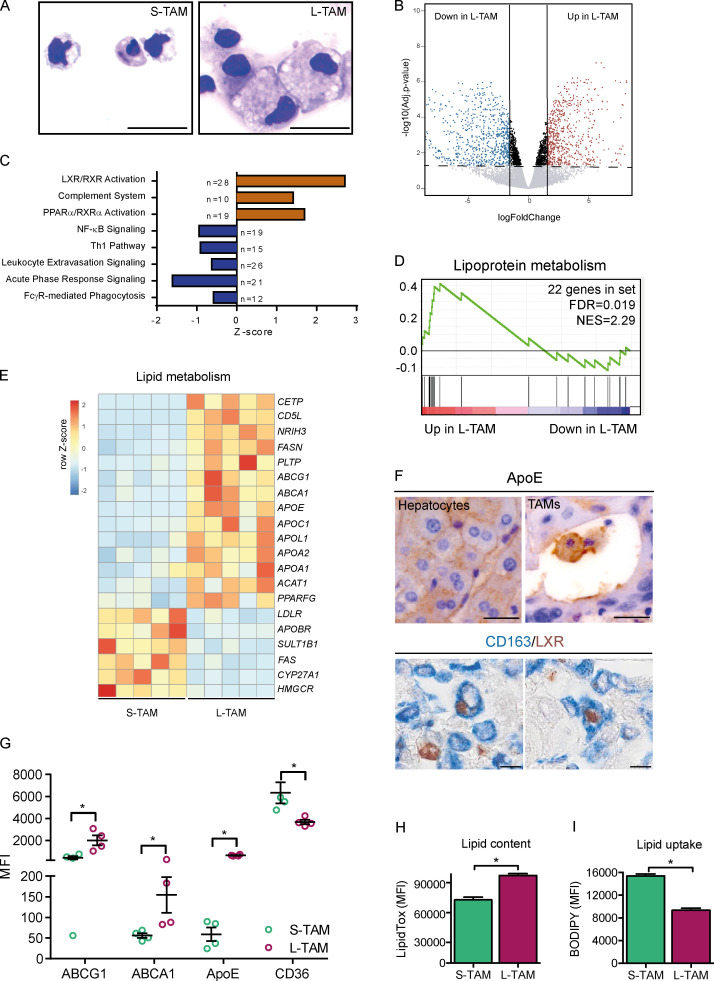
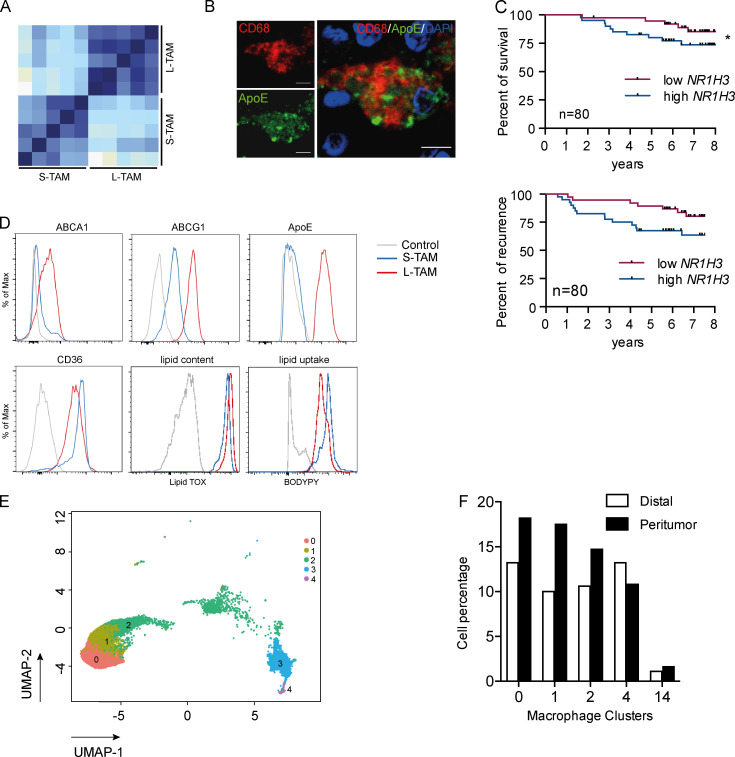
To get insights into the molecular programs linking macrophage shape to their function, we performed RNA sequencing on small and large macrophages from five CLM patients (Fig. S2 A). Cytometric isolation of macrophages based on their size (Fig. S2 B) allowed us to appreciate that, besides having a larger cell area, L-TAMs displayed a higher intracellular complexity, frequently presenting intracellular vacuoles, which could signify activation of phagolysosomes (A-Gonzalez et al., 2017) or lipid accumulations typically seen in foamy cells (Fig. 3 A). L-TAMs from five different patients clustered better between them rather than with small macrophages from the same patient (Fig. S3 A), suggesting that morphology identifies distinct macrophage clusters. 1,172 genes were found significantly differentially expressed in L-TAMs versus S-TAMs (false discovery rate [FDR] < 0.05; log2 fold change > |1.5|). (Fig. 3 B). Pathway analysis identified a number of pathways related to lipid metabolism, imputable to activation of the LXR pathway, which was the most up-regulated one (FDR = 3.34 × 10−11) in L-TAMs (Fig. 3 C). Notably, inflammation-related pathways (e.g., leukocyte extravasation, acute phase response, and NF-κB signaling) were mostly down-regulated (Fig. 3 C). Gene enrichment analysis (GSEA) confirmed lipoprotein metabolism as a significantly enriched biological process (Fig. 3 D). In homeostatic conditions, LXR activation in tissue macrophages critically controls the balance among phagocytosis, cholesterol efflux, and inflammatory response, ensuing in the anti-inflammatory polarization of macrophages upon LXR activation and in the inhibition of LXR activation during the acute phase response (Spann et al., 2012; Spann and Glass, 2013; Hong and Tontonoz, 2014; Tall and Yvan-Charvet, 2015). Notably, in publicly available datasets of breast cancer (GSE1456; Pawitan et al., 2005), a tumor in which macrophages are associated with worse prognosis (Gentles et al., 2015; Cassetta and Pollard, 2018), the LXR gene (NR1H3) significantly correlated with shorter survival (Fig. S3 C).
Figure S2.
Experimental workflow to sort small and large macrophages from CLM tissues. (A) Schematic overview of the experimental approach to sequence large and small macrophages from CLM specimens. (B) Gating strategy to sort macrophages from five CLM specimens. Small macrophages (S-TAMs) were sorted as alive CD45+/CD11b+/CD66b−/CD14dim/CD163dim/FSClo and large macrophages (L-TAMs) as CD45+/CD11b+/CD66b−/CD14dim/CD163hi/FSChi cells. Bottom panels confirm physical parameters of the small and large populations sorted. FSC-A, forward scatter area; FSC-W, forward scatter width; SSC-A, side scatter area.
Figure 3.
Morphology as a correlate of lipid metabolism in human CLMs. (A) Representative S-TAMs and L-TAMs sorted from one CLM specimen, cytospun, and stained with Diff-Quick. Scale bar: 20 µm. (B) Volcano plot of the expression profile of L-TAMs and S-TAMs sorted from five CLM specimens. Horizontal dashed line shows FDR (−log10 adjusted P value) 0.05; vertical continuous lines show log2 fold change (logFC) between −1.5 and 1.5. (C) Pathways enriched in L-TAMs versus S-TAMs from five CLM specimens. Z-score is shown on the x axis. For each pathway, the number of genes modulated is reported. (D) GSEA results showing lipoprotein metabolism as a significantly enriched biological process in L-TAMs compared to S-TAMs from five CLM specimens. The green curve represents the enrichment score, showing the measure to which the genes are overrepresented at the top or bottom of a ranked list of genes. Vertical black bars indicate the position in the ranked list of each gene, belonging to the gene set. Genes positioned in the red and blue sides are up-regulated and down-regulated, respectively, in large macrophages compared with small ones. Number of genes in set (genes = 22; FDR = 0.019) and normalized enrichment score (NES = 2.29) are shown. (E) Heatmap representing selected differentially expressed genes (−1.5 < LogFC > 1.5; FDR < 0.05) between L-TAMs and S-TAMs from five CLM specimens. Genes related to cholesterol and lipid metabolism are shown. (F) Representative pictures of immunohistochemical staining of macrophages expressing ApoE (top) and LXR (bottom) in CLM specimens. ApoE is expressed both in hepatocytes (left) and macrophages (right). Scale bars: 50 µm (top panels); 20 µm (bottom panels). (G) Flow cytometry analysis of cholesterol transporters ABCA1, ABCG1, ApoE, and CD36 in L-TAMs and S-TAMs isolated from four CLM specimens. Graphs represent mean ± SEM of four CLM samples analyzed in different experiments. *, P < 0.05 by Mann-Whitney test. (H and I) Lipid content, assessed as MFI of LipidTOX and lipid uptake, obtained as MFI of BODIPY. Bars represent mean + SEM of four different samples. *, P < 0.05 by Mann-Whitney test.
Figure S3.
Profile of small and large macrophages. (A) Correlation matrix of the whole dataset of genes in small and large macrophages from five CLM patients. Heatmap displays Pearson correlation coefficients according to the color code (light blue for low correlation, dark blue for high correlation). (B) Immunofluorescence on a CLM specimen showing a macrophage (CD68 in red) rich in ApoE protein (in green). Scale bars: 20 µm. (C) Kaplan-Meier survival curves of breast cancer patients from public datasets (GSE1456), stratified by first and last quartiles of NR1H3 expression score (LXR gene). Curves show disease-specific survival (top) and DFS (bottom). P = 0.09 (top panel) and *, P = 0.043 by Gehan-Breslow-Wilcoxon test. (D) Representative FACS plots of lipid transporters, ApoE, CD36, LipidTOX (lipid content), and BODIPY (lipid uptake) in S-TAMs and L-TAMs. (E) UMAP projection of myeloid cells from PT (n = 3) and distal (n = 3) human livers. Only macrophage clusters (c0, c1, c2, c3, and c4) are shown (n = 13,768 cells). Each dot represents an individual cell. (F) Bar graph showing the relative abundance of each macrophage cluster respect to the total dataset in distal and PT regions.
Consistent with the up-regulation of the LXR pathway, L-TAMs in human CLM presented up-regulation of the LXR gene (NR1H3) and LXR-downstream genes involved in cholesterol transport (ABCA1, ABCG1, and CETP), extracellular lipid acceptors (APOE, APOA1, and APOA2), and enzymes involved in lipid metabolism (CD5L and FASN), with concomitant down-regulation of genes regulating lipid uptake and biosynthesis (LDLR, CYP27A1, and HMGCR; Fig. 3 E), a profile suggesting a specific alteration of reverse cholesterol transport in L-TAMs. ApoE and LXR were strongly expressed in macrophages in CLM tissues (Fig. 3 F and Fig. S3 B), confirming that L-TAMs have features of foamy cells. L-TAMs isolated from CLM tissues expressed significantly higher amounts of ApoE and of the membrane cholesterol transporters ABCA1 and ABCG1 compared with S-TAM (Fig. 3 G and Fig. S3 D), while the expression of CD36, the receptor that mediates macrophage scavenging of oxidized lipoproteins, was sharply decreased (Fig. 3 G and Fig. S3 D), suggesting an impairment of this essential macrophage task. Finally, the altered lipid profile was mirrored by the increased lipid content (Fig. 3 H and Fig. S3 D) and decreased lipid uptake by L-TAMs compared with S-TAMs (Fig. 3 I and Fig. S3 D), confirming a defective execution of cholesterol handling.
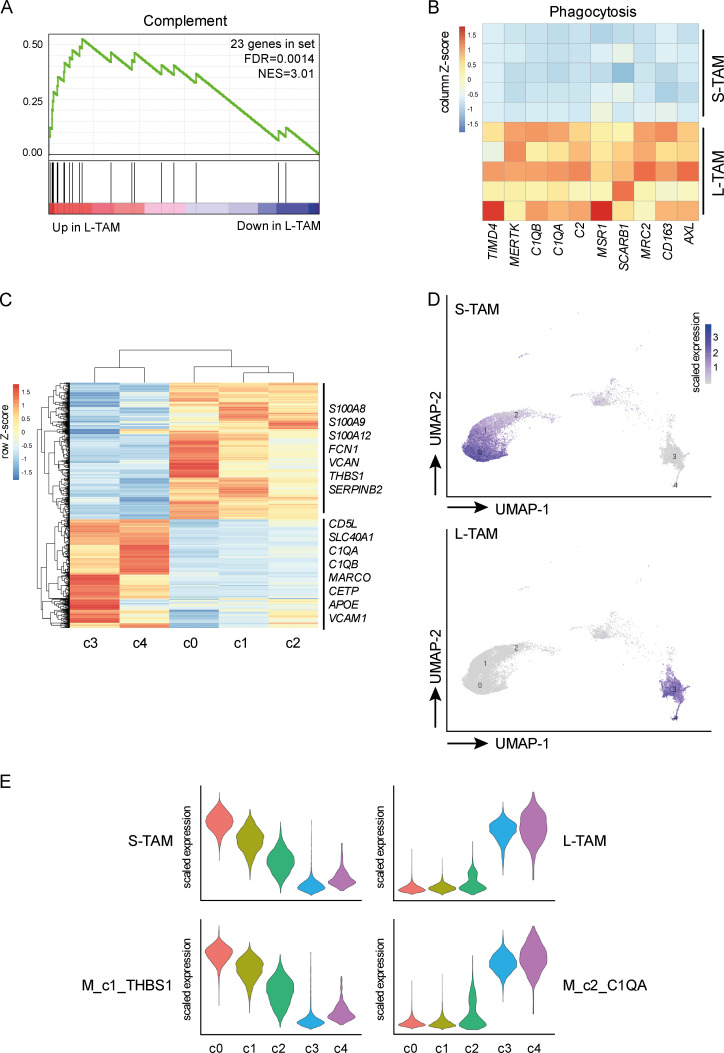
Evidence suggests that lipid metabolism and cholesterol in particular, is a key regulator of macrophage function and is associated with tumor promotion and suppression of T cell responses (Pencheva et al., 2014; Tavazoie et al., 2018; Domínguez-Andrés et al., 2019; Goossens et al., 2019; Gruosso et al., 2019). Alongside cholesterol metabolism, L-TAMs presented up-regulation of phagocytic and scavenger receptors, including MERTK, MSR1, MRC2, and members of the complement family (C1QA, C1QB, and C2; Fig. 4, A and B). These pathways have been linked to tumor promotion (Neyen et al., 2013). Engulfment of apoptotic material by MerTK favors an anti-inflammatory immunosuppressive microenvironment (Graham et al., 2014), and evidence suggests that at least one member of the scavenger receptor family is a driver of the tumor promoting function of TAMs (Stanford et al., 2014). Finally, complement has recently emerged as an important orchestrator of the recruitment and tumor-promoting functions of myeloid cells in preclinical models and selected human tumors (Bonavita et al., 2015; Medler et al., 2018; Reis et al., 2018; Roumenina et al., 2019). Notably, in scRNAseq analyses of CD45+ immune cells in hepatocellular carcinoma (HCC), a cluster of macrophages expressing C1QA was selectively associated to worse prognosis (Zhang et al., 2019).
Figure 4.
Morphology identifies individual populations of macrophages in CLM. (A and B) GSEA (Reactome) results showing complement as a significantly enriched biological process in L-TAMs compared with S-TAMs from five CLM specimens. Number of genes in complement gene set (genes = 23; FDR = 0.0014) and normalized enrichment score (NES = 3.01) are shown (A). Heatmaps showing up-regulation of phagocytosis (MERTK, MSR1, C1QA, and C1QB) in L-TAM samples. Columns represent signature genes. Rows represent individual samples. Gene expression values are reported as normalized trimmed mean of M values (B). (C–E) scRNAseq of three CLM specimens. Heatmap showing differential transcriptional profiles of five macrophage clusters; c0-c1-c2 were compared with c3-c4. Differentially expressed genes are shown (n = 1,282; adjusted P value < 0.05). Selected relevant genes are reported. Columns represent macrophage clusters, and rows represent differentially expressed genes (C). UMAP projections showing unsupervised Seurat-guided clustering of macrophage clusters colored by the expression score of L-TAM and S-TAM signatures (D). Violin plots showing expression values of selected genes in S-TAM and L-TAM from CLM specimens compared with clusters from HCC (E).
The distinct profile of L-TAM compared with S-TAM suggests that morphology identifies individual populations of macrophages in CLMs. We tested this hypothesis by performing scRNAseq of mononuclear phagocytes from CLM tissues. This is the first time a single-cell analysis of myeloid cells is performed on human metastases. Out of 18 clusters identified, including various monocyte, macrophage, and dendritic cell populations (and other less abundant cell types), 5 clusters could be convincingly annotated as macrophages (Fig. S3, E and F). In particular, cluster 0 (c0), c1, and c2 were enriched in expression of inflammatory genes (S100A8, S100A9, and SERPINB2), while c3 and c4 presented strong expression of genes of the complement family, scavenger receptors, and cholesterol-related genes (C1QA, C1QB, MARCO, CD5L, APOE, and CETP; n = 1,282 genes; adjusted P value < 0.05; Fig. 4 C). Of note, the S-TAM signature (including S100A8, S100A9, S100A12, FCN1, VCAN, THBS1, and SERPINB2) and L-TAM signature (CD5L, SLC40A1, C1QA, C1QB, MARCO, CETP, APOE, and VCAM1), obtained from their bulk sequencing profile, were differentially enriched in these individual clusters, specifically S-TAMs in c0, c1, and c2 and L-TAMs in c3 and c4 (Fig. 4, D and E), confirming that morphology of TAMs captures their diversity. Comparing our single-cell macrophage clusters to published scRNAseq data of immune cells from HCC patients (Zhang et al., 2019), we found an enrichment pattern strongly resembling that of S-TAMs and L-TAMs in two clusters enriched in tumors and with distinct phenotypes (namely myeloid derived suppressor cell–like Mφ_c1_THBS1 and TAM-like Mφ_c2_C1QA; Fig. 4 E). The L-TAM signature was highly superimposable to that of C1QA+ TAM-like macrophages, in turn also similar to TAMs previously found in lung cancer (Lavin et al., 2017). Notably, only the gene signature of C1QA+ TAM-like macrophages was associated with poor prognosis in The Cancer Genome Atlas analysis (Zhang et al., 2019).
Thus, L-TAM are endowed with orientation of lipid metabolism, receptor repertoire, and complement components which are likely to underlie their strong prognostic significance. Dissection of their relative importance may pave the way to translating a strong prognostic indicator in a metastatic setting, CLM, into a therapeutic target. For the time being, the results reported here suggest that accurate quantitative morphometric characterization of TAMs can serve as an easily quantifiable correlate of functional diversity with strong prognostic significance. Once validated in multi-institutional cohorts, this tool could pave the way to better patient stratification and improvement of therapeutic output.
Materials and methods
Patients and study design
The cohort study included 101 patients aged >18 yr with histologically proven CLMs that underwent hepatectomy at the Humanitas Clinical and Research Center–Istituto di Ricovero e Cura a Carattere Scientifico between 2005 and 2017. The study protocol was submitted to the international clinical trial registry (ClinicalTrials.gov registration number NCT03888638) and was designed to identify macrophage-related morphological features associated to distinct clinical outcomes. Written informed consent was obtained from each patient included in the study. The study protocol was in accordance with the ethical guidelines established in the 1975 Declaration of Helsinki and compliant to the procedures of the local ethical committee of the Humanitas Clinical and Research Center. Patient demographic, clinical, surgical, and histopathological data (detailed in Table S1) from the institutional intranet were assembled in a clinical retrospective database for following analyses. Only patients with partial response to preoperative therapy (for details, see Table S1) or stable disease were included in the study. Patients with the following criteria were excluded from the study: progressive disease, combination of hepatectomy with radiofrequency or microwave ablation, and nonradical hepatectomy. The preoperative workup consisted of total-body contrast-enhanced computed tomography and liver-specific magnetic resonance imaging, performed maximum 30 d before surgery. Patient postoperative follow up was performed every 3 mo and included serum oncological markers, abdominal ultrasonography, computed tomography, or magnetic resonance imaging. Control patients were patients that had undergone surgery for symptomatic giant liver hemangiomas.
Immunohistochemistry
2-μm-thick consecutive tissue sections were prepared from formalin-fixed and paraffin-embedded tissues, provided by the Pathology Department of the Humanitas Clinical and Research Center, and processed for immunohistochemistry. Briefly, after deparaffinization and rehydration, antigen retrieval was performed by heat treatment using EDTA buffer (Dako; 0.25 mM, pH 8) in water bath at 98°C for 20 min. Endogenous peroxidases were blocked by incubation with Peroxidase-Blocking Solution for 15 min at room temperature, followed by incubation for 20 min with Background Sniper (Biocare Medical) to block nonspecific binding. The sections were then incubated with primary antibodies anti-human CD68 (Dako; KP-1 clone, diluted 1:1,000), CD163 (Leica Biosystems, 10D6 clone, diluted 1:200) for 1 h at room temperature, followed by incubation with the detection system EnVision+System HRP-labeled anti-mouse (Dako). Diaminobenzidine tetrahydrochloride (Biocare Medical) was used as chromogen. Nuclei were lightly counterstained with a freshly made hematoxylin solution (Dako). LXR staining was performed with anti-human LXRα antibody (LSBio; polyclonal, diluted 1:100), together with anti-CD163, followed by incubation with the detection system Mach2 Double Stain 1 (Biocare Medical). Ferangi Blue (Biocare Medical) and 3,3′-Diaminobenzidine were used as chromogens.
Image analysis
To obtain the density of CD163+ cells, tissue slides were digitized after staining procedure using a computer-aided slide scanner (Olympus VS120 DotSlide). An expert pathologist blinded to clinical data selected three non-contiguous, non-overlapping microscopic areas of CD163-stained slides in the PT area at 20× magnification. Since most CLM patients receive neoadjuvant systemic chemotherapy leading to tumor necrosis to variable extents, we considered macrophages in the PT area to avoid necrotic areas often found in the tumor core region. The PT region was defined by the pathologist, based on a manual definition of the tumor and normal parenchyma. An image analysis software (Image Pro Premiere) was used to automatically determine the percentage of immune reactive area of the digitized images. The mean value, obtained from the three different microscopic areas, was calculated for each patient and used for subsequent analyses.
Analysis of macrophage morphology was performed with MATLAB (MathWorks) software. Three non-contiguous, non-overlapping microscopic areas in the PT area, including CD163+ cells, were captured at 63× magnification using an LMD7 inverted microscope (Leica Biosystems). For each digitized image, at least seven randomly selected macrophages were analyzed. For each cell, area and perimeter were calculated by manually tracing cell outlines on digitized images. Similarly, macrophage morphology at the IM was analyzed in 96 CLM patients of the same cohort. 5 (out of 101) slides had to be discarded due to absence of IM or other tissue alterations that would have compromised the accuracy of the analysis.
Immunofluorescence
Immunofluorescence was performed on 2-μm-thick formalin-fixed paraffin-embedded CLM sections using the Opal Kit (Perkin Elmer) following the manufacturer’s recommendations. The following primary antibodies were used: anti-human CD68 (Dako; clone KP.1, diluted 1:1,000) and anti-ApoE (Abcam; clone EPR19392, diluted 1:1,500). Representative images were acquired using an SP8II confocal microscope (Leica).
Cell sorting of small and large macrophages
Macrophages were FACS sorted from surgically resected CLM tissues from five patients. Single-cell suspensions were obtained by manually mincing tissue into small fragments and incubating the fragmented tissues for 1 h at 37°C in HBSS (Euroclone) containing 1 mg/ml Type IV Collagenase (Sigma-Aldrich), 2% FBS (Sigma-Aldrich), 50 μg/ml DNase I (Sigma-Aldrich), and 10 mM Hepes (Lonza). The resulting cell suspension was filtered through a 70-μm cell strainer and erythrocytes were lysed with ACK Lysing Buffer (Lonza). Cells were then incubated with a blocking solution containing 1% human serum in saline solution and stained with the following fluorophore-conjugated primary antibodies: anti-CD45 (BD Biosciences; clone HI30), anti-CD11b (BD Biosciences; clone ICRF44), anti-CD16 (BioLegend; clone 3G8), anti-CD14 (BD Biosciences; clone M5E2), anti-CD66b (BioLegend; clone G10F5), and anti-CD163 (BD Biosciences; clone GHI/61). Fixable Viability Stain 700 fluorescent dye (BD Biosciences) was used to perform dead cell exclusion. SYTO 16 Green Fluorescent Nucleic Acid Stain (ThermoFisher) was used to identify nucleated cells. Large and small macrophages were FACS sorted on a FACSAria III (BD Biosciences) following the gating strategy shown in Fig. S2.
Determination of membrane markers, lipid content, and lipid uptake by flow cytometry
Single-cell suspensions from surgically resected CLM tissues from four patients were obtained as described in the previous paragraph. To assess expression of lipid transporters (ABCA1 and ABCG1), ApoE, and CD36, the following antibodies were included in the cocktail: anti-ABCA1 (Invitrogen; rabbit polyclonal), anti-ABCG1 (Novus Biologicals; rabbit polyclonal), anti-ApoE (BioLegend; clone E6D7), and anti-CD36 (BioLegend; clone 5–271). Detection of ABCG1 and ApoE was performed by intracellular staining with BD Cytofix/Cytoperm Solution (BD Biosciences). Cells were acquired on a FACSAria III (BD Biosciences) following the gating strategy shown in Fig. S2.
For lipid staining, cells were stained with the same cocktail used for cell sorting and then incubated with HCS LipidTOX Deep Red Neutral Lipid Stain (ThermoFisher; dilution 1:200) in PBS for 30 min at room temperature. Cells were immediately washed with PBS and acquired by flow cytometer. Lipid content of L-TAMs and S-TAMs was evaluated as mean fluorescence intensity (MFI) of the dye.
To test lipid uptake, cells were incubated with 0.8 µM of the fluorescent dye BODIPY 558/568 C12 (ThermoFisher) in PBS containing 0.1% BSA (fatty acid–free) for 1 min at 37°C. Cells were immediately washed with cold PBS containing 0.2% BSA and acquired by flow cytometer. Lipid uptake of L-TAMs and S-TAMs was evaluated as MFI of the dye.
RNA extraction and 3′-mRNA sequencing
Total RNA was extracted from large and small macrophages using the Single Cell RNA Purification Kit (Norgen Biotek), following the manufacturer’s recommendations. Total RNA extracted from sorted macrophages were subjected to poly(A) mRNA sequencing (Table S2). Libraries were constructed by SMARTer-Stranded Total RNA Kit (Clontech), according to manufacturer’s instructions. Sequencing was performed with the NextSeq 500 (Illumina). All libraries were sequenced in paired-end mode (75-bp length).
3′-mRNA sequencing analysis
Raw reads were preprocessed for adapter trimming and quality check was assessed using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were aligned to the reference genome (Ensembl Homo sapiens release GRC38) using the STAR algorithm (Dobin et al., 2013). Differential expression analysis was performed using the Generalized Linear Model approach implemented in the R/Bioconductor edgeR (Robinson et al., 2010) package (R version 3.5; edgeR version 3.24.3). Sample correlation was estimated through Pearson correlation analysis. Results of differential analysis are provided in Data S1. The resulting gene lists were analyzed through the use of Ingenuity Pathways Analysis. Differentially expressed genes (large macrophages vs. small macrophages) were subjected to GSEA using the Reactome database. Significantly enriched pathways are available in Data S2.
Public gene expression data analysis
As breast cancer dataset, we used a publicly available microarray dataset (Gene Expression Omnibus accession no. GSE1456), including a total of 159 tumor samples (Pawitan et al., 2005). Expression profiles were downloaded as normalized data. Quartile distribution was considered for correlation with prognosis. The association between expression score of NR1H3 gene and clinical outcome was evaluated by the PRECOG system (PREdiction of Clinical Outcome from Genomic profiles; https://precog.stanford.edu; Gentles et al., 2015). Survival correlation analysis was performed by Prism, applying the Gehan-Breslow-Wilcoxon test.
scRNAseq
Mononuclear myeloid cells sorted from distal and PT region of three CLM specimens were subjected to single-cell RNA analysis. Single-cell suspensions were prepared by tissue mincing and enzyme digestion. FACS-sorted cells were resuspended in 0.5 ml PBS 1X plus 0.04% BSA and washed once by centrifugation at 450 rcf for 7 min. Cells were then resuspended in 50 μl and counted with an automatic cell counter (ThermoFisher; Countess II). Approximately 10,000 cells of each sample were loaded into one channel of the Chromium Chip B using the Single Cell reagent kit v3 (10X Genomics) for gel bead emulsion generation into the Chromium system. Following capture and lysis, cDNA was synthesized and amplified for 14 cycles following the manufacturer’s protocol. 50 ng of the amplified cDNA was then used for each sample to construct Illumina sequencing libraries. Sequencing was performed on the NextSeq550 Illumina sequencing platform following 10X Genomics' instructions for read generation, reaching at least 35,000 reads as mean reads per cell.
scRNAseq analysis
Raw sequencing data in the format of BCL files were converted in FASTQ files and aligned to the human reference genome GRCh38, taking advantage of the Cell Ranger Pipeline version 3.0.1 provided by 10X Genomics. After a quality check, we obtained a total of 12,181 and 12,568 cells for distal and PT tissue, respectively. Filtered gene expression matrices from Cell Ranger were used as input for clustering analysis by Seurat R package (version 3.1.1; R version 3.6.1). We first processed each individual dataset separately, considering the thresholds of 200, 20,000, and 0.2 for number of genes, number of unique molecular identifiers, and mitochondrial content, respectively. For each dataset, we selected the 2,000 most variable genes. Subsequently, we used the FindIntegrationAnchors function to combine the datasets together, choosing 2,000 anchor genes for integration. After integration, we ran principal component analysis and used the first 67 principal components to perform Louvain clustering and uniform manifold approximation and projection (UMAP) embedding. Finally, we obtained a total of 18 clusters (resolution level = 0.6).
The identified macrophage clusters (n = 5) were subsetted from the integrated dataset resulting in a total of 13,768 cells. Differential expression analyses were performed through the FindMarkers function, applying the Wilcoxon test by default.
Statistical analysis
Statistical computations were performed using the software IBM-SPSS and the software GraphPad Prism 7 (GraphPad Software). For image analyses, the mean value was calculated from three images for each patient. Correlations between percentage of immunoreactive area values of CD163 and CD68 were estimated by non-parametric Spearman rank correlation coefficient test and linear regression analysis. The difference between macrophage morphometric variables in CLM and control specimens was estimated by non-parametric Mann-Whitney test. The difference between macrophage morphometric variables in CLM specimens from patients with good and bad prognosis was estimated by unpaired t test. All datasets were tested for normal distribution before analyses by the D’Agostino and Pearson method. For each test, only two-sided P values < 0.05 were considered statistically significant. The categorical variables were reported as a number and percentage, while continuous variables were reported as the median and range or as mean and SD. Only patients with complete data were considered. ROC curves and associated area under the curves were generated for the relevant TAMs metrics to estimate the discriminatory ability for detecting patient survival. Univariate analysis was performed to examine association among different covariates and patient outcome. Covariates that showed a tendency of association with patient outcome were inserted into a multivariate model based on Cox regression analysis in order to investigate independent predictors of survival. All time-to-event endpoints were summarized using the Kaplan–Meier method. Differences in these endpoints between groups were examined using a log-rank test.
Data and materials availability
The bulk RNA-sequencing dataset has been deposited in the Gene Expression Omnibus under the accession no. GSE131353. All data presented in this article are available from the corresponding authors upon reasonable request.
Online supplemental material
Fig. S1 shows the comparison between CD68 and CD163 macrophage expression by immunohistochemistry in human CLMs, provides details on the macrophage morphological metrics, and shows the prognostic role of macrophage area at the IM. Fig. S2 shows the gating strategy to sort small and large macrophages from human CLM specimens. Fig. S3 provides additional information on the lipid and receptor profile of S-TAMs and L-TAMs, shows the relevance of LXR in human datasets, and presents the clusters of macrophages in human CLMs and their relative frequency. Table S1 provides the patients’ baseline characteristics. Table S2 contains the sequencing template. Data S1 shows results of differential analysis. Data S2 shows significantly enriched pathways.
Supplementary Material
shows the patients’ baseline characteristics.
contains the sequencing template.
shows results of differential analysis.
shows significantly enriched pathways.
Acknowledgments
We thank Dr. Matteo Cimino, Dr. Angela Palmisano, Dr. Luca Viganò, Dr. Daniele Del Fabbro, and Dr. Fabio Procopio (Humanitas Clinical and Research Center–Istituto di Ricovero e Cura a Carattere Scientifico, Rozzano, Italy), for their contribution in providing tissue samples. We thank Professor Mauro Perretti (The William Harvey Research Institute) for helpful discussion and advice on lipid profiling and analysis.
This work was supported by Associazione Italiana per la Ricerca sul Cancro (grant AIRC 5x1000 IG-21147 to A. Mantovani and grant IG2016-18443 to F. Marchesi). The funding agencies had no role in the design of the study or collection and analysis of data.
Author contributions: Study design: M. Donadon, A. Mantovani, and F. Marchesi. Patient and biomaterial recruitment (tissue specimens and clinical data): M. Donadon, G. Torzilli, and A. Lleo. Patient and biomaterial recruitment (tissue slides): L. Di Tommaso and M. Roncalli. Data acquisition, analysis, and interpretation: M. Donadon, N. Cortese, C. Soldani, B. Franceschini, M. Barbagallo, A. Rigamonti, R. Carriero, G. Maggi, and F. Marchesi. Development of methodology (tissue digestion, sequencing, and bioinformatics): N. Cortese, M. Barbagallo, R. Carriero, P. Kunderfranco, J. Cibella, and C. Peano. Development of methodology (flow cytometry and cell sorting): A. Anselmo and F.S. Colombo. Development of methodology (image analysis): C. Soldani, B. Franceschini, G. Maggi, L. Di Tommaso, and F. Marchesi. Statistical analysis: M. Donadon and R. Carriero. Manuscript writing: M. Donadon, N. Cortese, A. Mantovani, and F. Marchesi. Study supervision: M. Donadon, G. Torzilli, A. Mantovani, and F. Marchesi. Original observations: A. Anselmo and C. Soldani. All authors critically reviewed the manuscript.
References
- A-Gonzalez N., Quintana J.A., García-Silva S., Mazariegos M., González de la Aleja A., Nicolás-Ávila J.A., Walter W., Adrover J.M., Crainiciuc G., Kuchroo V.K., et al. 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214:1281–1296. 10.1084/jem.20161375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballotta V., Driessen-Mol A., Bouten C.V., and Baaijens F.P.. 2014. Strain-dependent modulation of macrophage polarization within scaffolds. Biomaterials. 35:4919–4928. 10.1016/j.biomaterials.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Bekkering S., Arts R.J.W., Novakovic B., Kourtzelis I., van der Heijden C.D.C.C., Li Y., Popa C.D., Ter Horst R., van Tuijl J., Netea-Maier R.T., et al. 2018. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell. 172:135–146.e9. 10.1016/j.cell.2017.11.025 [DOI] [PubMed] [Google Scholar]
- Bonavita E., Gentile S., Rubino M., Maina V., Papait R., Kunderfranco P., Greco C., Feruglio F., Molgora M., Laface I., et al. 2015. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 160:700–714. 10.1016/j.cell.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Cassetta L., and Pollard J.W.. 2018. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17:887–904. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M., et al. 2014. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 513:559–563. 10.1038/nature13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese N., Donadon M., Rigamonti A., and Marchesi F.. 2019. Macrophages at the crossroads of anticancer strategies. Front. Biosci. 24:1271–1283. 10.2741/4779 [DOI] [PubMed] [Google Scholar]
- Cucchetti A., Ferrero A., Cescon M., Donadon M., Russolillo N., Ercolani G., Stacchini G., Mazzotti F., Torzilli G., and Pinna A.D.. 2015. Cure model survival analysis after hepatic resection for colorectal liver metastases. Ann. Surg. Oncol. 22:1908–1914. 10.1245/s10434-014-4234-0 [DOI] [PubMed] [Google Scholar]
- De Palma M., Nassiri S., and Cianciaruso C.. 2019. Macrophage interference on chemotherapy. Nat. Cell Biol. 21:411–412. 10.1038/s41556-019-0303-8 [DOI] [PubMed] [Google Scholar]
- DeNardo D.G., and Ruffell B.. 2019. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19:369–382. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Caro G., Cortese N., Castino G.F., Grizzi F., Gavazzi F., Ridolfi C., Capretti G., Mineri R., Todoric J., Zerbi A., et al. 2016. Dual prognostic significance of tumour-associated macrophages in human pancreatic adenocarcinoma treated or untreated with chemotherapy. Gut. 65:1710–1720. 10.1136/gutjnl-2015-309193 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Andrés J., Novakovic B., Li Y., Scicluna B.P., Gresnigt M.S., Arts R.J.W., Oosting M., Moorlag S.J.C.F.M., Groh L.A., Zwaag J., et al. 2019. The Itaconate Pathway Is a Central Regulatory Node Linking Innate Immune Tolerance and Trained Immunity. Cell Metab. 29:211–220.e5. 10.1016/j.cmet.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Forssell J., Oberg A., Henriksson M.L., Stenling R., Jung A., and Palmqvist R.. 2007. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin. Cancer Res. 13:1472–1479. 10.1158/1078-0432.CCR-06-2073 [DOI] [PubMed] [Google Scholar]
- Fridman W.H., Zitvogel L., Sautès-Fridman C., and Kroemer G.. 2017. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14:717–734. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- Galon J., Angell H.K., Bedognetti D., and Marincola F.M.. 2013. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 39:11–26. 10.1016/j.immuni.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Garden O.J., Rees M., Poston G.J., Mirza D., Saunders M., Ledermann J., Primrose J.N., and Parks R.W.. 2006. Guidelines for resection of colorectal cancer liver metastases. Gut. 55(Suppl 3):iii1-iii8. 10.1136/gut.2006.098053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Gordon S., Hume D.A., Mowat A.M., and Randolph G.J.. 2010. Unravelling mononuclear phagocyte heterogeneity. Nat. Rev. Immunol. 10:453–460. 10.1038/nri2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., et al. 2015. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21:938–945. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens P., Rodriguez-Vita J., Etzerodt A., Masse M., Rastoin O., Gouirand V., Ulas T., Papantonopoulou O., Van Eck M., Auphan-Anezin N., et al. 2019. Membrane Cholesterol Efflux Drives Tumor-Associated Macrophage Reprogramming and Tumor Progression. Cell Metab. 29:1376–1389.e4. 10.1016/j.cmet.2019.02.016 [DOI] [PubMed] [Google Scholar]
- Graham D.K., DeRyckere D., Davies K.D., and Earp H.S.. 2014. The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer. 14:769–785. 10.1038/nrc3847 [DOI] [PubMed] [Google Scholar]
- Gruosso T., Gigoux M., Manem V.S.K., Bertos N., Zuo D., Perlitch I., Saleh S.M.I., Zhao H., Souleimanova M., Johnson R.M., et al. 2019. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Invest. 129:1785–1800. 10.1172/JCI96313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halama N., Michel S., Kloor M., Zoernig I., Benner A., Spille A., Pommerencke T., von Knebel D.M., Folprecht G., Luber B., et al. 2011. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 71:5670–5677. 10.1158/0008-5472.CAN-11-0268 [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hong C., and Tontonoz P.. 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug Discov. 13:433–444. 10.1038/nrd4280 [DOI] [PubMed] [Google Scholar]
- Keirsse J., Van Damme H., Geeraerts X., Beschin A., Raes G., and Van Ginderachter J.A.. 2018. The role of hepatic macrophages in liver metastasis. Cell. Immunol. 330:202–215. 10.1016/j.cellimm.2018.03.010 [DOI] [PubMed] [Google Scholar]
- Krenkel O., and Tacke F.. 2017. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17:306–321. 10.1038/nri.2017.11 [DOI] [PubMed] [Google Scholar]
- Laoui D., Van Overmeire E., Di Conza G., Aldeni C., Keirsse J., Morias Y., Movahedi K., Houbracken I., Schouppe E., Elkrim Y., et al. 2014. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res. 74:24–30. 10.1158/0008-5472.CAN-13-1196 [DOI] [PubMed] [Google Scholar]
- Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., et al. 2017. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 169:750–765.e17. 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locati M., Curtale G., and Mantovani A.. 2019. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 10.1146/annurev-pathmechdis-012418-012718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malesci A., Bianchi P., Celesti G., Basso G., Marchesi F., Grizzi F., Di Caro G., Cavalleri T., Rimassa L., Palmqvist R., et al. 2017. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. OncoImmunology. 6 e1342918 10.1080/2162402X.2017.1342918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., and Allavena P.. 2015. The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 212:435–445. 10.1084/jem.20150295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., and Balkwill F.. 2008. Cancer-related inflammation. Nature. 454:436–444. 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Mantovani A., Marchesi F., Malesci A., Laghi L., and Allavena P.. 2017. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14:399–416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter F.Y., Davis C.T., and Liu W.F.. 2015. Physical and mechanical regulation of macrophage phenotype and function. Cell. Mol. Life Sci. 72:1303–1316. 10.1007/s00018-014-1796-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medler T.R., Murugan D., Horton W., Kumar S., Cotechini T., Forsyth A.M., Leyshock P., Leitenberger J.J., Kulesz-Martin M., Margolin A.A., et al. 2018. Complement C5a Fosters Squamous Carcinogenesis and Limits T Cell Response to Chemotherapy. Cancer Cell. 34:561–578.e6. 10.1016/j.ccell.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B., Van den Eynde M., Bindea G., Church S.E., Vasaturo A., Fredriksen T., Lafontaine L., Haicheur N., Marliot F., Debetancourt D., et al. 2018. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J. Natl. Cancer Inst. 110:97–108. 10.1093/jnci/djx123 [DOI] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41:14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyen C., Plüddemann A., Mukhopadhyay S., Maniati E., Bossard M., Gordon S., and Hagemann T.. 2013. Macrophage scavenger receptor a promotes tumor progression in murine models of ovarian and pancreatic cancer. J. Immunol. 190:3798–3805. 10.4049/jimmunol.1203194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan Y., Bjöhle J., Amler L., Borg A.L., Egyhazi S., Hall P., Han X., Holmberg L., Huang F., Klaar S., et al. 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 7:R953–R964. 10.1186/bcr1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Zhang H., Matei I.R., Costa-Silva B., Hoshino A., Rodrigues G., Psaila B., Kaplan R.N., Bromberg J.F., Kang Y., et al. 2017. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 17:302–317. 10.1038/nrc.2017.6 [DOI] [PubMed] [Google Scholar]
- Pencheva N., Buss C.G., Posada J., Merghoub T., and Tavazoie S.F.. 2014. Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell. 156:986–1001. 10.1016/j.cell.2014.01.038 [DOI] [PubMed] [Google Scholar]
- Reis E.S., Mastellos D.C., Ricklin D., Mantovani A., and Lambris J.D.. 2018. Complement in cancer: untangling an intricate relationship. Nat. Rev. Immunol. 18:5–18. 10.1038/nri.2017.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodell C.B., Arlauckas S.P., Cuccarese M.F., Garris C.S., Li R., Ahmed M.S., Kohler R.H., Pittet M.J., and Weissleder R.. 2018. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng. 2:578–588. 10.1038/s41551-018-0236-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumenina L.T., Daugan M.V., Noé R., Petitprez F., Vano Y.A., Sanchez-Salas R., Becht E., Meilleroux J., Clec’h B.L., Giraldo N.A., et al. 2019. Tumor Cells Hijack Macrophage-Produced Complement C1q to Promote Tumor Growth. Cancer Immunol. Res. 7:1091–1105. 10.1158/2326-6066.CIR-18-0891 [DOI] [PubMed] [Google Scholar]
- Ruffell B., Au A., Rugo H.S., Esserman L.J., Hwang E.S., and Coussens L.M.. 2012. Leukocyte composition of human breast cancer. Proc. Natl. Acad. Sci. USA. 109:2796–2801. 10.1073/pnas.1104303108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.G., Cardona P.J., Kim M.J., Allain S., and Altare F.. 2009. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10:943–948. 10.1038/ni.1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Shalova I.N., and Biswas S.K.. 2017. Metabolic regulation of macrophage phenotype and function. Immunol. Rev. 280:102–111. 10.1111/imr.12603 [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., and Jemal A.. 2020. Cancer statistics, 2020. CA Cancer J. Clin. 70:7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- Spann N.J., and Glass C.K.. 2013. Sterols and oxysterols in immune cell function. Nat. Immunol. 14:893–900. 10.1038/ni.2681 [DOI] [PubMed] [Google Scholar]
- Spann N.J., Garmire L.X., McDonald J.G., Myers D.S., Milne S.B., Shibata N., Reichart D., Fox J.N., Shaked I., Heudobler D., et al. 2012. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 151:138–152. 10.1016/j.cell.2012.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford J.C., Young C., Hicks D., Owens P., Williams A., Vaught D.B., Morrison M.M., Lim J., Williams M., Brantley-Sieders D.M., et al. 2014. Efferocytosis produces a prometastatic landscape during postpartum mammary gland involution. J. Clin. Invest. 124:4737–4752. 10.1172/JCI76375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl C., Lee T., Shah S.P., Farinha P., Han G., Nayar T., Delaney A., Jones S.J., Iqbal J., Weisenburger D.D., et al. 2010. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 362:875–885. 10.1056/NEJMoa0905680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman E.M., Halpin M.C., Muster J., Moon R.T., and Ratner B.D.. 2014. Porous implants modulate healing and induce shifts in local macrophage polarization in the foreign body reaction. Ann. Biomed. Eng. 42:1508–1516. 10.1007/s10439-013-0933-0 [DOI] [PubMed] [Google Scholar]
- Tall A.R., and Yvan-Charvet L.. 2015. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 15:104–116. 10.1038/nri3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M.F., Pollack I., Tanqueco R., Ostendorf B.N., Reis B.S., Gonsalves F.C., Kurth I., Andreu-Agullo C., Derbyshire M.L., Posada J., et al. 2018. LXR/ApoE Activation Restricts Innate Immune Suppression in Cancer. Cell. 172:825–840.e18. 10.1016/j.cell.2017.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo S.W., Li Y., Buono C., Zhao B., Billings E.M., Chang J., and Kruth H.S.. 2008. Heterogeneity of human macrophages in culture and in atherosclerotic plaques. Am. J. Pathol. 172:1112–1126. 10.2353/ajpath.2008.070513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenes M., Shang M., Di Matteo M., Goveia J., Martín-Pérez R., Serneels J., Prenen H., Ghesquière B., Carmeliet P., and Mazzone M.. 2016. Macrophage Metabolism Controls Tumor Blood Vessel Morphogenesis and Metastasis. Cell Metab. 24:701–715. 10.1016/j.cmet.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Wynn T.A., Chawla A., and Pollard J.W.. 2013. Macrophage biology in development, homeostasis and disease. Nature. 496:445–455. 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., and Gordon S.. 2015. From the Reticuloendothelial to Mononuclear Phagocyte System - The Unaccounted Years. Front. Immunol. 6:328 10.3389/fimmu.2015.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., et al. 2019. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 179:829–845.e20. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M., et al. 2019. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 50:1317–1334.e10. 10.1016/j.immuni.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the patients’ baseline characteristics.
contains the sequencing template.
shows results of differential analysis.
shows significantly enriched pathways.