Macrophage morphology could be used to stratify metastasis risk for CRC patients.
Abstract
Using macrophage morphology in human colorectal cancer liver metastasis, Donadon et al. in this issue of JEM (https://doi.org/10.1084/jem.20191847) provide a window into lipid metabolism and foamy macrophages, which accrue in numerous pathological states and here are shown to have clinical application.
Large, lipid-laden “foamy” macrophages are frequently observed in cancer, atherosclerosis, and infectious diseases such as tuberculosis (TB; Molgora et al., 2020; Neyrolles, 2014; Remmerie and Scott, 2018). While potentially linked to increased phagocytosis, for example of lipids and cholesterol in atherosclerosis, the formation and function of foamy macrophages remain a mystery. Through an unbiased assessment of macrophage morphology in human colorectal cancer (CRC) liver metastasis, Donadon et al. (2020) provide an unexpected window into understanding macrophage lipid metabolism, concurrent with a new and simple way of stratifying metastasis risk for CRC patients. Donadon et al. (2020) found a striking correlation between the presence of L-TAMs (large tumor-associated macrophages) in CRC metastasis and poor outcomes; the difference compared with the numbers of S-TAMs (small TAMs) in the same anatomical confine was sufficiently significant so that it could be readily reduced to a simplified and automated pathology screening tool.

Insights from Peter J. Murray.
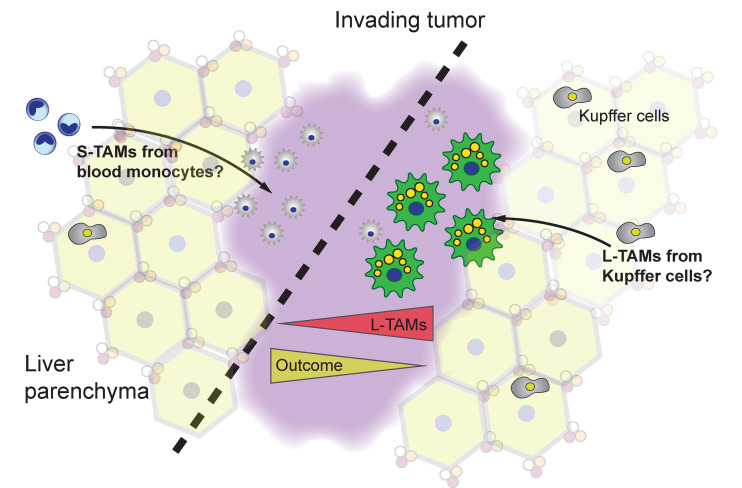
Metastatic seeding of the liver from colon carcinomas is an important driver of morbidity and mortality from colon cancer. Within the liver, metastatic lesions are readily distinguished from the normal liver architecture and are infiltrated with macrophages within the tumor bed and at the invasive margins (Oliveira et al., 2019). Donadon et al. (2020) stained CRC liver sections for CD68 or CD163 (commonly used markers for macrophages, especially in tissues). As they note, an initiating idea for the study was to try to distinguish different macrophage activation states in and around the tumor. However, CD68 or CD163 stained two types of macrophages they named S-TAM and L-TAM that were readily distinguished by size differential, allowing quantification by morphometric methods (see figure). (In this regard, the regular histological structure of the normal liver is an advantage to spot both metastatic margins and macrophages.) Based on their corresponding RNA sequencing and single-cell RNA sequencing data, we can safely assume that L-TAMs are a type of Kupffer cell (KC), or locally activated KC. L-TAMs express CD5L, MARCO, and VCAM, components of a hallmark gene expression profile of KCs observed in other granular studies of the human liver (MacParland et al., 2018; Ramachandran et al., 2019). L-TAMs were enriched with many mRNAs associated with lipid metabolism, consistent with their foamy appearance. By contrast, S-TAMs have a distinct mRNA profile characterized by inflammatory gene expression. Perhaps S-TAMs derive from the blood monocyte pool and locally differentiate under the influence of the inflammatory tumor environment. However, at this point, the exact relationships between the different macrophages in the liver bearing metastatic lesions can only be inferred by correlation of gene expression to known patterns.
S-TAMs and L-TAMs populate metastatic CRC in the liver. The quantity of L-TAMs in and around the invading tumor dictate, in part, survival outcomes from metastatic disease in the liver. The origins of the S- and L-TAMs can only be inferred from correlation to gene expression patterns. Depicted is a hypothetical organization where blood monocytes seed the S-TAM pool and L-TAMs arise from local KCs. However, further experiments are necessary to establish the precise ontogeny of the different macrophages.
Donadon et al. (2020) then analyzed the association between L-TAMs and S-TAMs at the peritumor regions of 101 CRC patients and their clinical outcomes. First, they found that neither macrophage density nor expression of CD163 had any clear link to key clinical parameters and outcomes; this is somewhat surprising, as macrophage number is often correlated with poor outcomes in cancer (Cassetta and Pollard, 2018), which may be indicative of the liver microenvironment in CRC. Second, using morphometric techniques, they found a remarkable relationship between L-TAMs and survival after hepatectomy: the disease-free survival of patients with a high L-TAM area was 0.2% compared with patients with higher S-TAMs of 27.8%. From these findings, a clear clinical implication is that macrophage morphology can stratify CRC patients, which could be facilitated by the simplicity of the approach: simple histopathology and unbiased morphometric scoring could speed patient analysis. Although Donadon et al. (2020) used a relatively limited sample number, their results are so clear-cut that expanding to a larger and blinded test group should be an imperative. However, the key scientific question that emerged concerns the underlying reasons for the association between L-TAMs and cancer. Why are lipid-laden macrophages linked to cancer progression? Clearly, much new biology needs to be learned about the specific macrophage activation and metabolic states in specific tissue environs in and around cancer.
What about the biology of the foamy macrophages and their relationship to disease? As noted above, lipid-laden macrophages are frequently observed in different pathological states. In TB, they correlate with caseating necrosis and may be a “safe harbor” for bacterial growth because they could supply a reservoir of triglycerides for bacterial growth (Neyrolles, 2014). However, the fact that foamy macrophages are common to diverse pathologies argues that they arise because of specific metabolic adaptations. There are two obvious pathways that could influence the formation of foamy macrophages. First, they could arise as a consequence of a relative increase in phagocytic activity. It is easy to imagine that considerable cell death and debris accrue in TB and inside the necrotic regions of solid tumors or in atherosclerotic lesions. The continual consumption of cell material would certainly require ways of breaking down biological polymers, macromolecules, and of course lipids from membranes. A second non–mutually exclusive possibility concerns the bioenergetic requirements of macrophages under hypoxia. In hypoxia, oxygen is obviously limited, and, therefore, electron disposal along the electron transport chain (ETC) of mitochondria is also limited. Similarly, activated microbial and interferon-activated murine macrophages generate free radicals such as nitric oxide (NO) that poisons components of the ETC such as aconitase, disabling mitochondrial function (Everts et al., 2012; Palmieri et al., 2020). (Human macrophages do not make NO at anywhere close to the amount of murine macrophages, but still generate free radicals; thus, the same principle may apply.) Under inflammation, hypoxia, or both, glycolysis will be the main way of generating energy. The “glycolytic switch” to Warburg-type metabolism occurs during M1-biased macrophage activation and is associated with increased triglyceride synthesis, which is essential for full inflammatory function (Castoldi et al., 2020). These correlations form a greater puzzle about foamy macrophages. However, since lipid-laden macrophages are common to so many disease states involving hypoxia (e.g., TB granulomas, the relative hypoxia of the liver, especially in cancer), one plausible pathway to account for foamy macrophage formation is that hypoxia or disabled mitochondria force the cell to dispose of electrons from other catabolic pathways through the biosynthesis of lipids, which are then stored until they can be metabolized by oxidation through a functioning ETC.
From studying CRC metastasis pathology, Donadon et al. (2020) therefore provided two new insights. The first is practical: can liver histology with automated image analysis and one stain be used to stratify risk in CRC? Second, if foamy macrophages (in this case, L-TAMs) accumulate and contribute to pathology, a clear implication from their negative correlation with disease-free survival, does the metabolic adaptation of macrophages to the foamy phenotype give us clues about weak points in which they could be eliminated in disease? For example, should lipid droplet formation be an obligate requirement for survival under hypoxia, a drug that disrupted lipid biosynthesis or storage may force the death of these specific macrophages. Given the tremendous research activity on the field of immune cell metabolism, lipid homeostasis is likely to come to the forefront given its clear link to diverse diseases.
Acknowledgments
Research in the Murray laboratory is supported by the Max-Planck-Gesellschaft and Deutsche Forschungsgemeinschaft Forschergruppe (grant FOR 2599).
References
- Cassetta, L., and Pollard J.W.. 2018. Nat. Rev. Drug Discov. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- Castoldi, A., et al. 2020. Nat. Commun. 10.1038/s41467-020-17881-3 [DOI] [Google Scholar]
- Donadon, M., et al. 2020. J. Exp. Med. 10.1084/jem.20191847 [DOI] [Google Scholar]
- Everts, B., et al. 2012. Blood. 10.1182/blood-2012-03-419747 [DOI] [Google Scholar]
- MacParland, S.A., et al. 2018. Nat. Commun. 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molgora, M., et al. 2020. Cell. 10.1016/j.cell.2020.07.013 [DOI] [Google Scholar]
- Neyrolles, O. 2014. Infect. Immun. 10.1128/IAI.01512-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, R.C., et al. 2019. J. Oncol. 10.1155/2019/6280347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, E.M., et al. 2020. Nat. Commun. 10.1038/s41467-020-14433-7 [DOI] [Google Scholar]
- Ramachandran, P., et al. 2019. Nature. 10.1038/s41586-019-1631-3 [DOI] [Google Scholar]
- Remmerie, A., and Scott C.L.. 2018. Cell. Immunol. 10.1016/j.cellimm.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]