Abstract
We report the identification of three cyclic peptide ligands of K-Ras(G12D) using an integrated in vitro translation–mRNA display selection platform. These cyclic peptides show preferential binding to the GTP-bound state of K-Ras(G12D) over the GDP-bound state and block Ras-Raf interaction. A co-crystal structure of peptide KD2 with K-Ras(G12D)·GppNHp reveals that this peptide binds in the Switch II groove region with concomitant opening of the Switch II loop and a 40° rotation of the α2 helix, and that a threonine residue (Thr10) on KD2 has direct access to the mutant aspartate (Asp12) on K-Ras. Replacing this threonine with non-natural amino acids afforded peptides with improved potency at inhibiting the interaction between Raf1-RBD and K-Ras(G12D) but not wildtype K-Ras. The union of G12D over wildtype selectivity and GTP state/GDP state selectivity is particularly desirable, considering that oncogenic K-Ras(G12D) exists predominantly in the GTP state in cancer cells, and wildtype K-Ras signaling is important for the maintenance of healthy cells.
Short abstract
We report the discovery of GTP-state-selective cyclic peptide ligands of K-Ras(G12D), a prevalent oncogenic driver, using an integrated in vitro translation−mRNA display selection platform.
Introduction
Missense mutations of the RAS genes (KRAS, HRAS, and NRAS) occur frequently in human cancer and drive oncogenic transformation.1 Among these, KRAS G12D is the most prevalent point mutation associated with poor clinical outcome. The G12D mutation impairs both intrinsic and GTPase-accelerating protein (GAP)-mediated GTP hydrolysis and liberates K-Ras protein from functional control by GTPase activity.2,3 As a result, K-Ras(G12D) is enriched in its GTP-bound, signaling-competent state, given the near 10-fold higher concentration of GTP than GDP inside the cell.4
Though Ras proteins were historically considered “undruggable” due to their picomolar affinity for guanine nucleotides and the lack of deep accessible pockets, recent efforts have fueled the discovery of both small-molecule and macromolecule direct binders of Ras.5,6,15−23,7−14 Work from our laboratory identified a class of compounds that bind to Ras in a highly dynamic pocket near the Switch II region (SIIP) and leverage the nucleophilicity of the acquired cysteine in K-Ras(G12C) to covalently capture K-Ras in its inactive, GDP-bound state.11 Further chemical optimization has yielded ligands with potent K-Ras(G12C)-dependent antitumor effects,24−26 and four compounds (AMG510,27,28 MRTX849,29 JNJ-74699157,30 and LY349944631) have entered clinical trials in patients with K-Ras(G12C)-mutant tumors.32 This strategy is uniquely suited for the G12C mutant because intrinsic GTPase activity is not affected by the G12C mutation,3 allowing the conversion from GTP-bound state to the compound-accessible GDP-bound state at a clinically relevant rate. Efforts to develop K-Ras(G12C) inhibitors capable of binding to the GTP state have so far been unsuccessful. The SIIP is occluded in the GTP state of K-Ras, effectively blocking the drug-binding pocket in this protein state.
The loss of intrinsic GTPase activity in K-Ras(G12D) presents an additional challenge—in cells with the G12D mutation, only a minor fraction of the K-Ras protein will be GDP-bound, and conversion from the GTP-bound state to the GDP-bound state is extremely slow. To target K-Ras(G12D), we envision that a GTP-state-selective K-Ras ligand will be advantageous because (1) it will bind to the predominant K-Ras population in G12D-mutant cells, and (2) it will be less likely to affect healthy cells, which express wildtype K-Ras with a significant GDP-bound population. Previous work from our lab as well as others has demonstrated that targeting the GTP state of Ras is feasible. Gentile et al. discovered Switch II groove (SIIG) ligands that can recognize both GDP and GTP states of Ras, albeit with a preference for the GDP state.33 Sakamoto et al. reported K-Ras(G12D)-targeting cyclic peptides generated from a T7 phage display library that are weakly selective for the GDP state and inhibit Sos-mediated nucleotide exchange.16,34,35 Wu et al. identified artificial cyclic peptides that block K-Ras(G12V)/Raf1-RBD interaction from a bead-display library but did not study the nucleotide state preference.36 To the best of our knowledge, GTP-state-selective ligands have not been documented. Here we present the discovery of GTP-state-selective and mutant-selective cyclic peptide ligands of K-Ras(G12D) using the Random non-standard Peptides Integrated Discovery (RaPID) platform,37 an in vitro translation–mRNA display technology encoding >1012 macrocyclic peptides.
Results and Discussion
To target the GTP state of K-Ras(G12D), we first considered a potential challenge: a ligand identified from a binding rather than a functional screen may not antagonize K-Ras oncogenic function. For example, an allosteric ligand that stabilizes the active conformation K-Ras(G12D)·GTP, but still allows effector binding, will render the protein constitutively active. To minimize this possibility, we took advantage of the two conformational states of Ras·GTP with distinct properties: State 1 (effector binding incompetent) and State 2 (effector binding competent).38 While most Ras proteins exist in an equilibrium of these two interconverting states, a mutation of Thr35 to Ser locks the protein in State 1.39 We therefore elected to use the double mutant K-Ras(G12D/T35S) for our initial selection, with the GppNHp-loaded protein as the selection target, and the GDP-loaded protein as the counter-target (Figure 1a).
Figure 1.
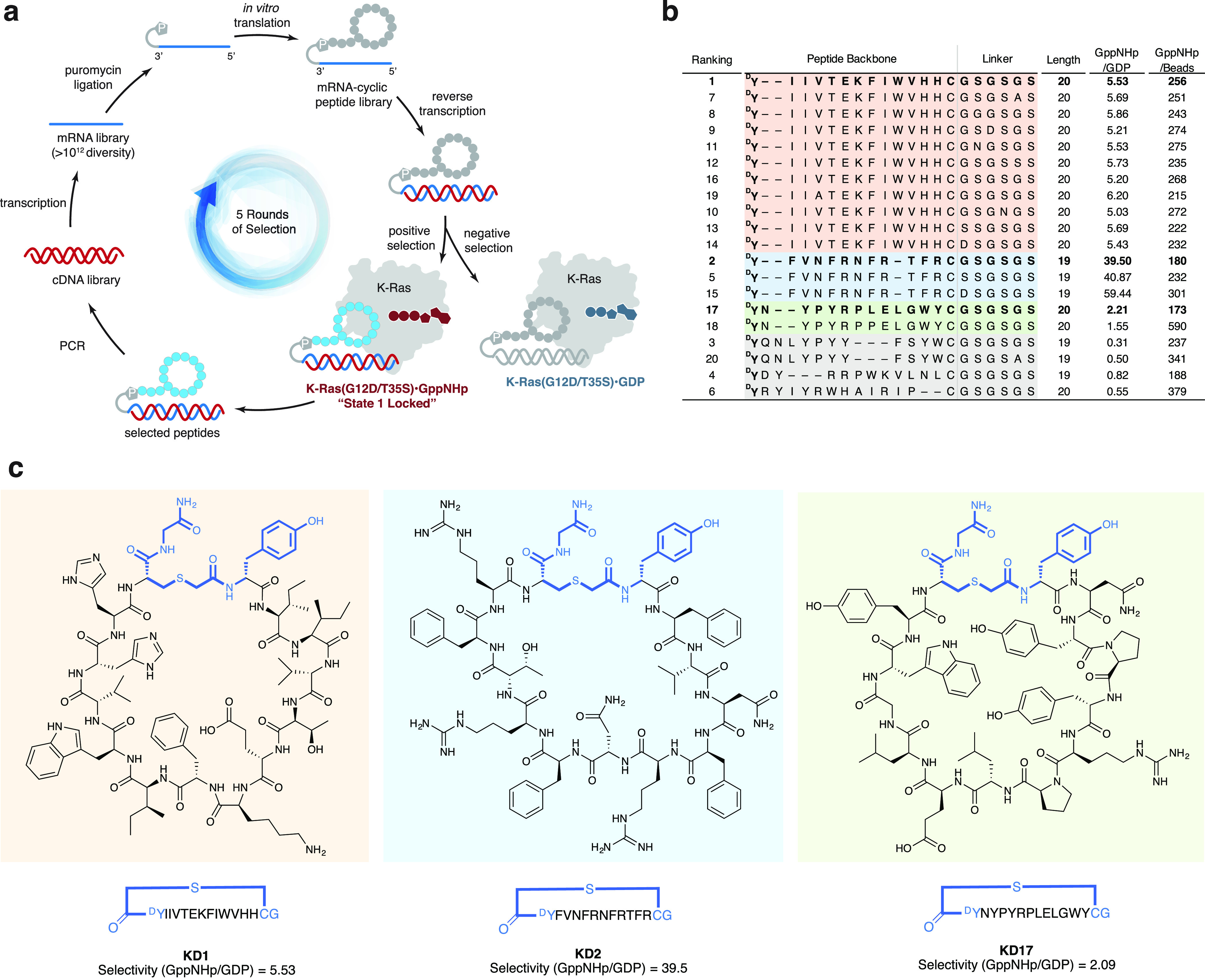
Selection of cyclic peptides that selectively bind to State 1 of GTP-bound K-Ras(G12D) from the Random non-standard Peptide Integrated Discovery (RaPID) mRNA display library. (a) Cyclic peptide selection was performed for a total of five rounds using K-Ras(G12D/T35S)·GppNHp as the positive selection target and K-Ras(G12D/T35S)·GDP as the negative selection target. (b) Top 20 hits clustered by sequence alignment. Peptides are ranked based on the number of unique NGS reads in the output library (not shown, see Supporting Information for detailed methods). Colored shades group peptides in the same cluster; gray shades represent peptides that have higher affinity for the GDP state, which were not further studied. One peptide from each cluster (bold typeface) was chosen for further characterization. (c) Structures of three distinct peptides identified from the selection. Blue color indicates the constant regions of the cyclic peptide backbone, including sulfide bridge, the starting amino acid chloroacetyl-d-tyrosine, and the ending amino acids cysteine and glycine.
Starting from a high-diversity cDNA library (>1012 encoded compounds), we performed five rounds of selection and examined the output cDNA libraries by next generation sequencing (NGS). Each round of selection included in vitro transcription, puromycin ligation, N-chloroacetyl-d-tyrosine-initiated in vitro translation with in situ cyclization, reverse transcription, negative and positive selection with target protein, K-Ras(G12D), immobilized on streptavidin beads, and PCR amplification of the selected cDNA libraries. At the end of the fifth round of selection, we subjected the output library to an additional round of selection for quantitative comparison of the binders to empty beads, immobilized K-Ras(G12D)·GDP, or immobilized K-Ras(G12D)·GppNHp using PCR and NGS. This allowed for calculation of the GTP/GDP selectivity index (to evaluate GTP state selectivity) and the GTP/beads selectivity index (to assess the level of non-specific binding). The top 20 binders (ranked by the number of NGS reads after the fifth round) contained 16 cyclic peptides with GTP/GDP selectivity index of >1 (Figure 1b). These 16 peptides clustered into three distinct sequence families, and members in each cluster displayed surprisingly high sequence homology. We chose one representative member from each cluster (namely, KD1, KD2, and KD17) and chemically synthesized linker-free cyclic peptides in multi-milligram quantities for further study.
Interestingly, when we performed a separate GTP-state positive selection using empty beads in lieu of GDP-loaded protein for negative selection, we obtained predominantly GDP-state-selective binders (Figure S1), despite the fact that the positive selection target was the same GppNHp-loaded protein.
With these three GTP-state-selective cyclic peptide ligands, we assessed their impact on effector binding to K-Ras(G12D). We used a time-resolved fluorescence energy transfer (TR-FRET) assay that allows the quantitation of the Ras·Raf complex formation (Figure 2a). All three peptides inhibited the interaction between K-Ras(G12D)·GppNHp and Raf1-RBD at micromolar concentrations. Meanwhile, all three peptides were less potent at inhibiting the interaction of wildtype K-Ras·GppNHp with Raf1-RBD, with different levels of selectivity. Surprisingly, KD2 did not exhibit an inhibitory effect against wildtype K-Ras at the highest solubility-permitting concentration. We reasoned that the observed selectivity could come from two sources: (1) K-Ras(G12D) is known to have a weaker affinity for Raf RBD than wildtype K-Ras (Kd = 270 ± 46 and 56 ± 6 nM, respectively),3 and (2) the cyclic peptide may exhibit preferential recognition for the aspartate-12 residue.
Figure 2.
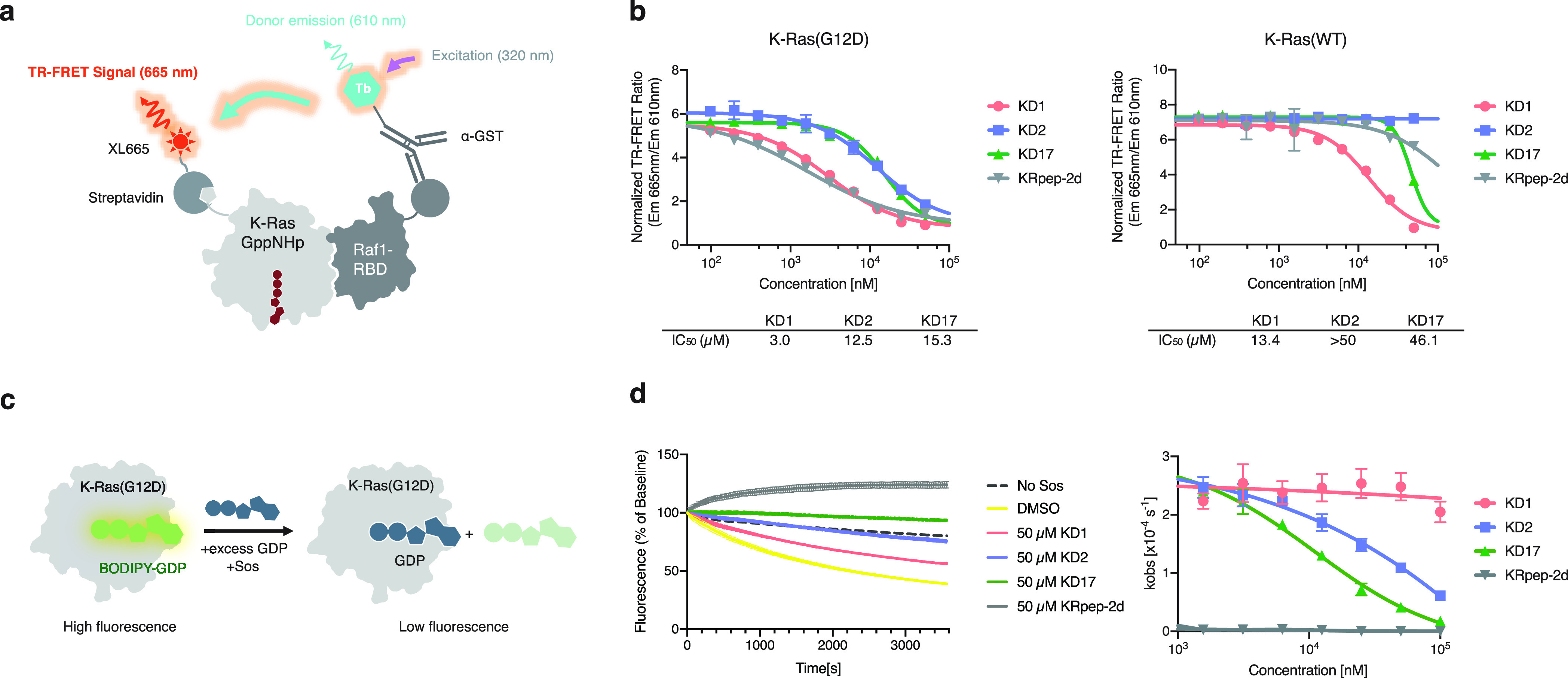
Cyclic peptides block the interaction of K-Ras(G12D) and effector proteins. (a) Illustration of a biochemical assay that detects Ras-Raf interaction by time-resolved fluorescence energy transfer (TR-FRET). (b) Cyclic peptides block Ras-Raf interaction with selectivity for the G12D mutant over wildtype K-Ras. (c) Illustration of a biochemical assay that monitors Sos-mediated nucleotide exchange using a fluorescent-GDP analog. (d) KD2 and KD17, but not KD1, inhibit Sos-mediated nucleotide exchange of K-Ras(G12D).
We next determined whether these cyclic peptides affect Sos-mediated nucleotide exchange, which takes GDP-bound Ras as the substrate. Inhibition of Sos-mediated nucleotide exchange required high concentrations of KD2 and KD17, and even near the solubility limit of 50 μM, KD1 only had a small effect on the rate of nucleotide exchange. By contrast, KRpep2d (a cyclic peptide ligand of K-Ras reported by Takeda16) is a highly potent inhibitor of nucleotide exchange, with an IC50 below the lower assay limit of the current assay format. The weaker potency of KD1 and KD2 at inhibiting Sos-mediated nucleotide exchange than at inhibiting Ras·Raf interaction was consistent with their preference for the GTP state of K-Ras (Figure 1b).
Dose-dependent binding of KD2 and KD17 to surface-immobilized or solution-phase K-Ras(G12D)·GppNHp could be readily detected using biolayer interferometry (BLI), surface plasmon resonance (SPR), and isothermal titration calorimetry (ITC) (Figure S2). None of these three peptides stabilized K-Ras(G12D) against thermal denaturation either in their GDP-bound or GppNHp-bound state (ΔTm < 1.0 °C, Figure S3). By contrast, KRpep2d increased the melting temperature of K-Ras(G12D)·GDP by 6.0 °C, but had little stabilization effect on K-Ras(G12D)·GppNHp.
To probe the structural perturbation of K-Ras(G12D) by these three cyclic peptides, we performed 1H–15N heteronuclear single quantum coherence (HSQC) experiments with 100 μM 15N-labeled K-Ras(G12D)·GppNHp in the presence of 400 μM cyclic peptide. The low solubility of KD1 in aqueous buffer (∼50 μM) precluded the acquisition of the HSQC spectrum for this peptide. Both KD2 and KD17 caused large perturbations of nearly all peaks in the HSQC spectrum of K-Ras(G12D)·GppNHp, with additional new peaks that are not present in the spectrum of unliganded K-Ras(G12D)·GppNHp (Figure S4). Unfortunately, titration of the ligand did not allow us to trace the chemical shift change of each peak (instead, we observed two distinct populations). This result is consistent with a tightly binding ligand with a slow dissociation rate. Among the easily identifiable peaks, G77 showed a +0.10 ppm 1H chemical shift change upon the binding of either KD2 or KD17.
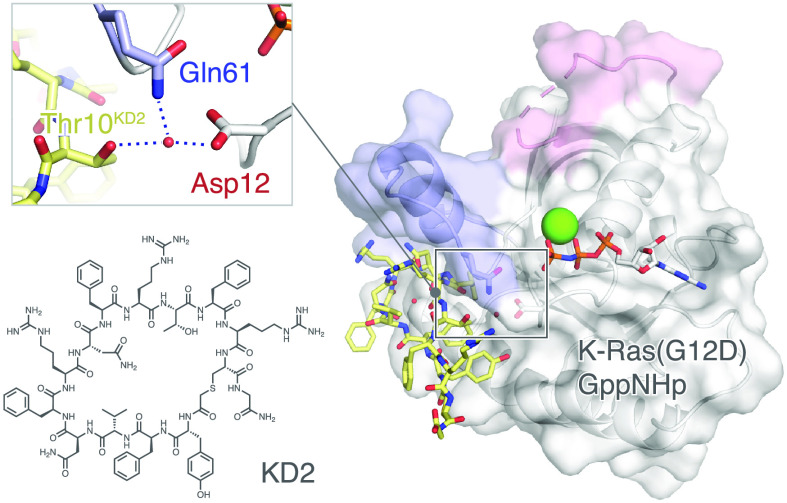
Owing to its pronounced GTP/GDP state selectivity (as revealed by NGS reads in the selection) and G12D/wildtype selectivity (as revealed by the Ras·Raf interaction assay), we focused our additional efforts on KD2. We successful obtained co-crystals of KD2 and K-Ras(G12D)·GppNHp in a construct where all cysteines have been mutated to serine or leucine and which we previously found to have improved crystallization properties. The crystal structure was determined by molecular replacement and refined to 1.6 Å (Table S1). The overall complex structure is shown in Figure 3a, with the mutant aspartate residue (Asp12) highlighted in cyan, and KD2 and GppNHp shown in stick models. KD2 binds in the Switch II groove region, below the α2 helix and the Switch II loop, the same pocket as previously reported for covalent ligands targeting the engineered cysteine in H-Ras(M72C)·GppNHp.33 Strikingly, this pocket is not observable in any published structures of the non-liganded K-Ras in the GTP state. For example, examination of the structure of K-Ras(G12D)·GppNHp (PDB: 5USJ) reveals a closed surface between Switch II and the α3 helix. However, KD2 appears to have expanded the pocket by inducing a large shift of the α2 helix and the Switch II loop (Figure 3d, Figure S5, Movie S1). The conformation adopted by these regions differs from that observed in either GDP-state or GTP-state K-Ras(G12D) (Figure 3d). We also observed concomitant changes of the Switch I conformation upon KD2 binding. However, the lack of strong electron density for several residues and possible crystal packing biases precluded further conclusions from being made (Figure S5c).
Figure 3.
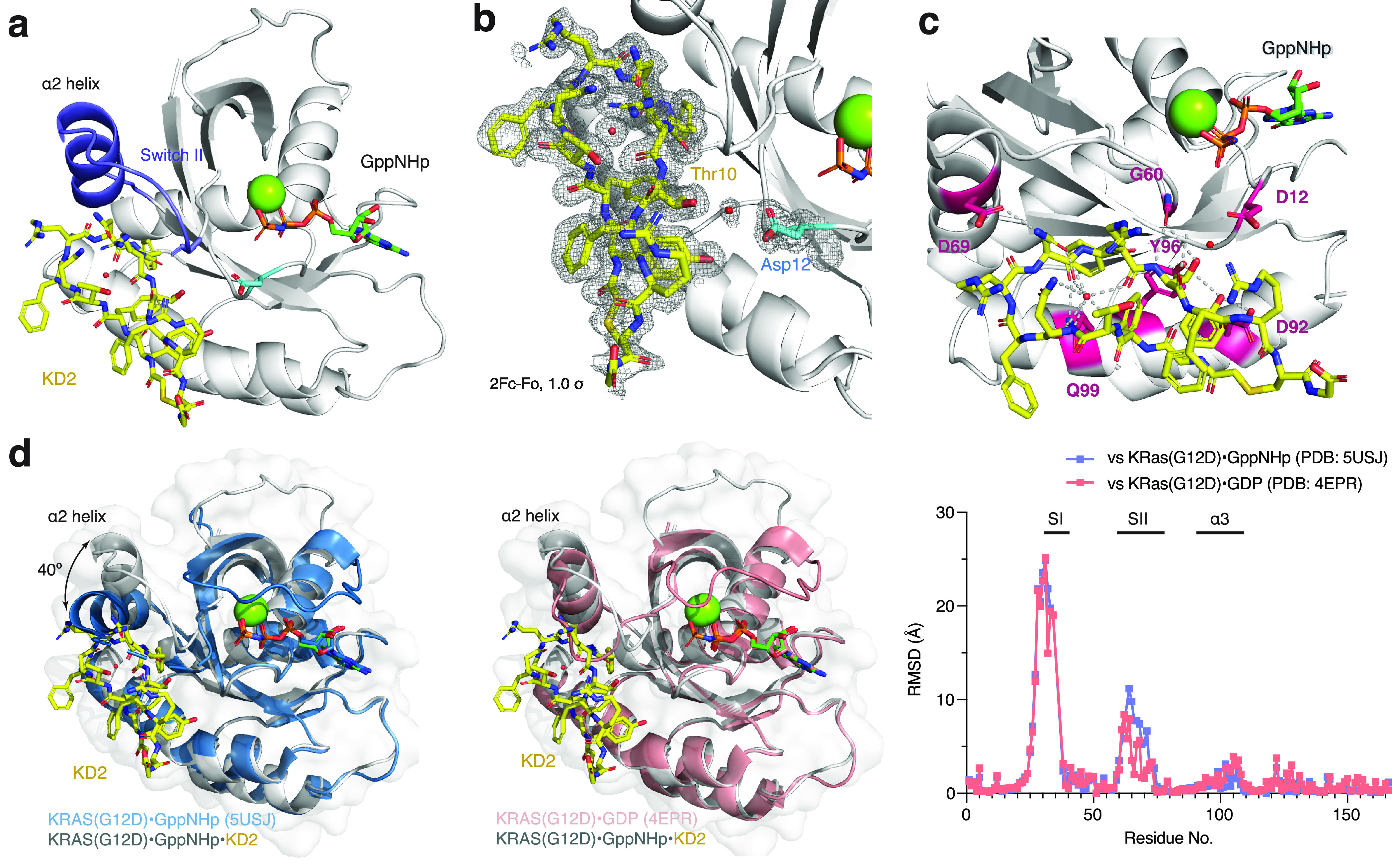
Crystal structure of KD2 bound to K-Ras(G12D)·GppNHp. (a) KD2 binds in the Switch II groove of K-Ras(G12D)·GppNHp. (b) 2Fo – Fc map showing the electron density of KD2, Asp12, Gln61, and relevant water molecules, contoured at 1.0σ. (c) KD2 forms an intricate hydrogen bond network intramolecularly and intermolecularly with K-Ras. (d) Comparison of K-Ras(G12D)·GppNHp·KD2 structure with unliganded K-Ras(G12D)·GppNHp (PDB: 5USJ) and K-Ras(G12D)·GDP (PDB: 4EPR). Root-mean-square deviations (RMSDs) were calculated on a residue-by-residue basis for each pairwise comparison. SI, Switch I; SII, Switch II.
KD2 forms an intricate hydrogen bond network both within the macrocycle and with residues on K-Ras(G12D). One intriguing observation was an ordered water molecule in the center of the macrocycle, forming hydrogen bonds with both side-chain and backbone elements of cyclic KD2. We surmise that this water molecule may be critical to maintaining the conformation of the macrocycle. KD2 interacts with K-Ras(G12D) through residues on various domains (Figure 3c), including G60 (Switch II), D69 (α2 helix), D92 (α3 helix), Y96 (α3 helix), and Q99 (α3 helix). Particularly remarkable is that the mutant aspartate residue (Asp12) is directly accessible from Thr10 on the cyclic peptide. We observed a low-occupancy water molecule bridging Thr10 (KD2), Gln61 (K-Ras), and Asp12 (K-Ras) in the crystal structure (Figure 3b). To the best of our knowledge, this is the first ligand-bound crystal structure of K-Ras(G12D) in its GTP state where the ligand makes direct contact with the mutant residue at position 12.
We next sought to improve the affinity and mutant/wildtype selectivity of KD2 to K-Ras(G12D) through structure-guided chemical modification. Asking whether we could enhance side-chain interactions between the cyclic peptide and Asp12 of K-Ras, we synthesized a set of KD2 analogs varying at the Thr10 position (Figure 4a,b). These included the His, Lys, and Arg mutants, as well as a few non-proteogenic amino acids such as l-1,2-diaminopropanoic acid (Dap), l-citrulline (Cit), and l-β-azidoalanine (Aza). During cyclic peptide synthesis, an unexpected side reaction led the conversion of the azidoalanine residue into 4-methylpiperidinylalanine (labeled Aza-X). We do not have a full mechanistic understanding of this transformation, but it likely occurred during a Fmoc deprotection step where 40% 4-methylpiperidine was used. (We hypothesize that this happened via base-mediated β-elimination of azide followed by Michael addition of 4-methylpiperidine onto the resulting dehydroalanine; as a result, the stereochemistry of Aza-X is uncertain.)
Figure 4.
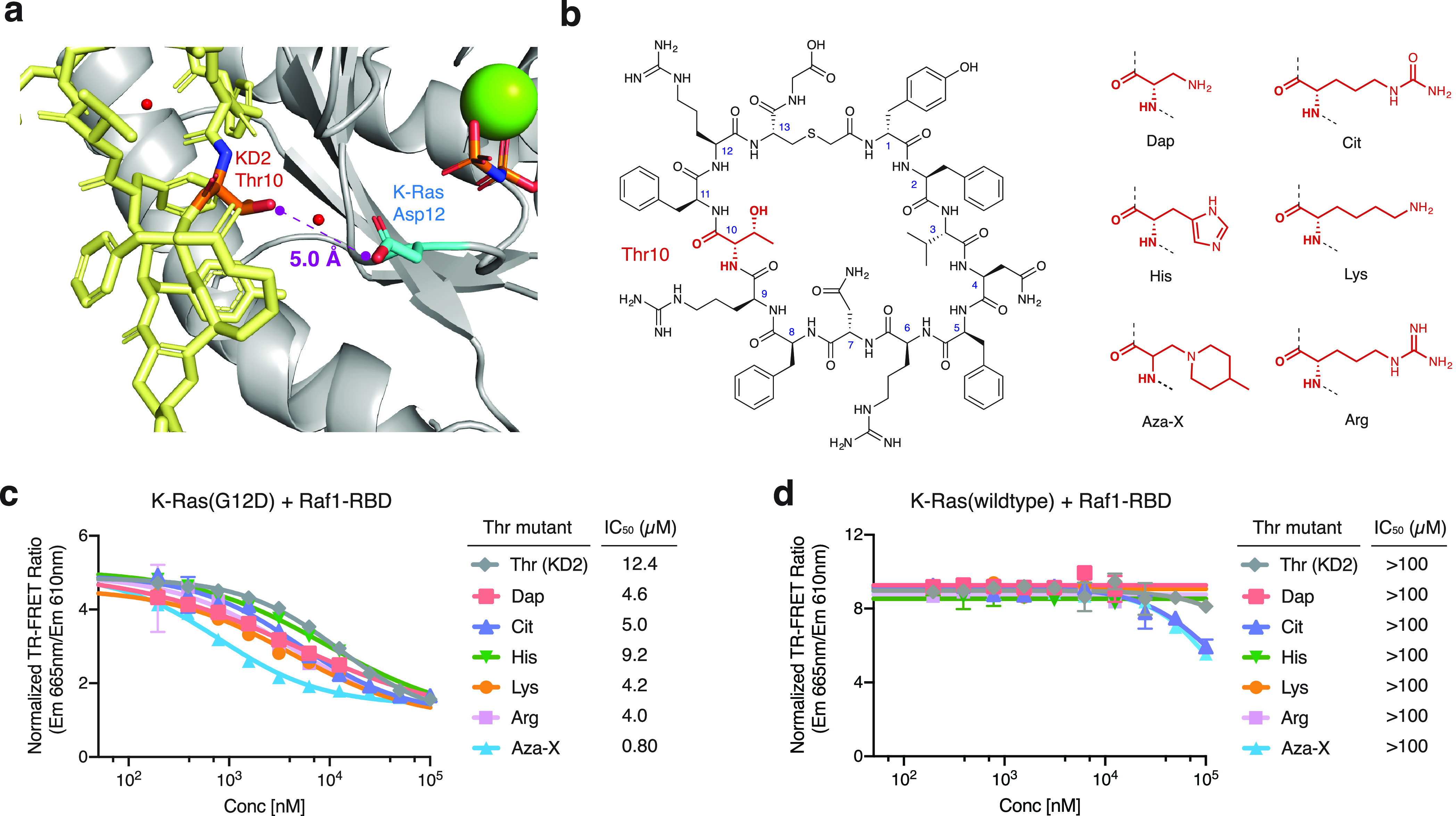
Substitution of Thr10 in KD2 improves the potency for blocking Ras-Raf interaction. (a) Thr10 in KD2 is in proximity with Asp12 of K-Ras(G12D). (b) Structure of KD2 with Thr10 highlighted in red. (c) Thr10 mutants of KD2 are more potent inhibitors of Ras-Raf interaction. (d) Thr10 mutants of KD2 do not inhibit wildtype K-Ras-Raf interaction.
We tested these KD2 analogs in the TR-FRET-based Ras·Raf interaction assay (Figure 4c,d). The IC50 values for the Lys and Arg mutants were found to be about 3-fold lower than for KD2 (however, the solubility of these two mutants was significantly lower than that of the parent peptide KD2, preventing their testing at concentrations greater than 11.1 μM). Most strikingly, the byproduct (Aza-X) from the failed attempt to incorporate azidoalanine was the most potent among all compounds tested, with an IC50 of 0.80 μM, a >10-fold improvement compared to KD2. Meanwhile, these analogs appeared to have maintained the G12D-mutant selectivity: none of these KD2 mutants inhibited the interaction between Raf1-RBD and wildtype K-Ras·GppNHp by more than >50% at the highest concentrations tested (11.1 μM for the Lys, Arg, His, and Dap mutants due to limited solubility, 100 μM for all others).
We also considered rigidification of the cyclic peptide scaffold through the formation of a transannular bridge, a structural feature that has been found in both natural and synthetic peptides to improve their activity.40−42 To achieve this, we first identified two amino acids in KD2, Val3 and Arg9, that do not participate in intramolecular interactions or binding interaction with K-Ras (Figure 5a). By replacing these two amino acids with two cysteines or a combination of azidohomoalanine and propargylglycine, we synthesized two bicyclic variants of KD2 (Figure 5b). These peptides were more potent inhibitors of the interaction between K-Ras(G12D) and Raf1-RBD (Figure 5c). Although these two bicyclic peptides also inhibited the interaction between wildtype K-Ras and Raf1-RBD, the IC50 values were more than 20-fold higher than those for K-Ras(G12D).
Figure 5.
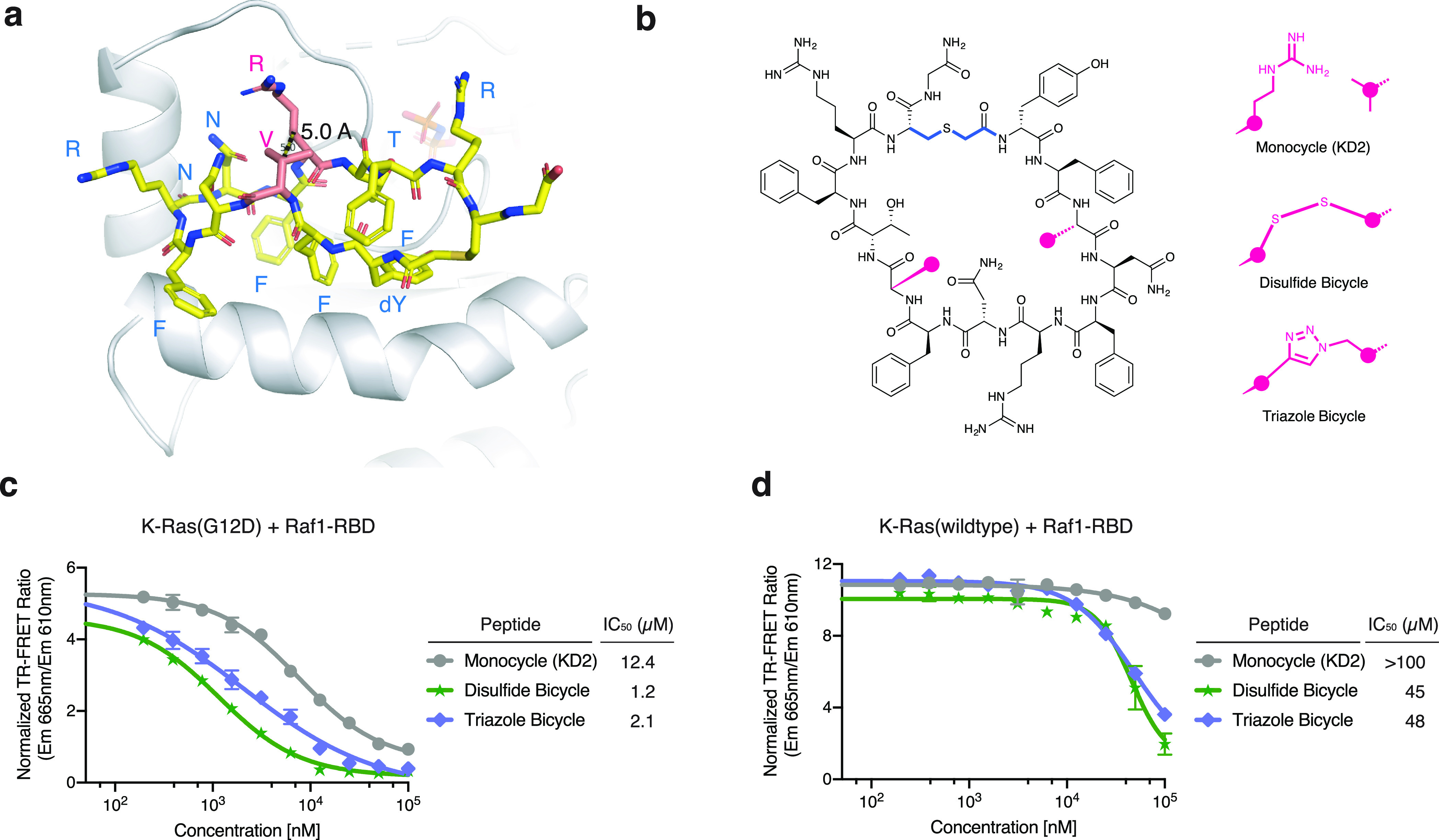
Bicyclic variants of KD2 exhibit improved potency for block Ras-Raf interaction. (a) Val3 and Arg9 on KD2 are solvent exposed and participate in neither interaction with K-Ras nor intramolecular interactions. (b) Structures of bicyclic variants of KD2. (c) Bicyclic variants of KD2 are more potent inhibitors of K-Ras(G12D)-Raf interaction. (d) Bicyclic variants of KD2 show improved inhibition of K-Ras(wildtype)-Raf interaction at high concentrations.
While we cannot extract a comprehensive structure–activity relationship from this relatively small compound set, it is conceivable that both the potency and the mutant selectivity of these peptides can benefit from further structural optimization with two distinct approaches—Thr10 modification and scaffold rigidification. Importantly, the data here indicates that substitution of Thr10 is well tolerated and may be tailored to target the GTP state of other G12 mutants of K-Ras.
Conclusion
Employing a high-throughput selection platform (the RaPID system), we have identified three distinct cyclic peptide scaffolds that preferentially bind to K-Ras(G12D) in its GTP state from an initial library of 1012 members. These cyclic peptides inhibit the interaction between K-Ras(G12D) and Raf1-RBD but are less effective for wildtype K-Ras protein. X-ray crystallography showed that one of these peptides binds to K-Ras in the Switch II groove region previously discovered in a fragment screen.33 Structure-guided chemical diversification allowed rapid optimization of one initial hit into a compound with sub-micromolar potency at inhibiting Ras-Raf interaction.
One remaining challenge is that these cyclic peptides do not readily enter cells, impeding their utility in a cellular setting (Figures S6–S8). We tested the cellular activity of these peptides in phospho-signaling (Figure S6a,c) and cell viability (Figure S6b,c) but observed no evidence of on-pathway effects. We next assessed cell permeability in multiple formats—passive artificial membrane permeability assay (PAMPA) (Figure S7a), Caco-2 permeability assay (Figure S7a), chloroalkane penetration assay43 (Figure S7b–d), and by using a fluorescein-conjugated derivative of KD2 (Figure S7e–g). These results suggested low cell permeability as a potential reason for the lack of detectable cellular activity, although other factors might have also contributed, and further studies are merited to understand the activity of these cyclic peptides (see Supporting Information for an additional discussion on cellular activity). Cellular permeability of cyclic peptides is a complex problem under active research.44−49 With guidance from abundant empirical rules and contemporary computational modeling, we are optimistic that this will be a surmountable problem. While these cyclic peptides in their current forms are unsuitable as cellular leads, they have demonstrated that selective targeting of the GTP state of mutant K-Ras is achievable and may serve as useful starting points or molecular probes for future inhibitor development.
Our study has overturned the current understanding of the Switch II pocket (SIIP) first revealed by the discovery of covalent ligands for K-Ras (G12C) and now extended by many more analogs which have advanced to the clinic. The K-Ras (G12C) ligands do not bind to the GTP state of K-Ras(G12C) and only bind in its GDP state, when Switch II of K-Ras is open, exposing the SIIP. This understanding has dominated clinical trials for K-Ras (G12C) ligands because of the inability to identify ligands for the GTP state of K-Ras (G12C). Previous studies have also revealed the Switch II groove (SIIG), a shallow surface adjacent to SIIP but accessible in both GDP and GTP states. Ligands that bind to SIIG have so far shown preference for the GDP state. This study set out to identify what we initially hypothesized would have to be a distinct pocket on K-Ras (G12D) in the GTP state since previous studies appeared to suggest the SIIP was inaccessible. The co-crystal structure of KD2 bound to K-Ras(G12D)·GppNHp was surprising because KD2 occupies both SIIP and SIIG. This unified drug pocket allowed ligands to simultaneously achieve selectivity for the GTP state over the GDP state and for the G12D mutant over wildtype K-Ras. This result suggests a previously unappreciated dynamic aspect of the Switch II loop of K-Ras (G12D) in the GTP state exposing the SIIP for drug access. This is a promising therapeutic approach for many oncogenic K-Ras mutants that are devoid of GTPase activity and enriched in their GTP-bound state.
Acknowledgments
We would like to thank Xi Liu, Keelan Guiley, and the staff at A.L.S. beamline 8.2.1 for help with X-ray data collection and processing. We would also like to thank the former and current co-workers in the Suga lab for their valuable input and effort in maintenance of the RAPID platform. Z.Z. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2281-17). Q.H. is also a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2229-15). K.M.S. would like to acknowledge The Waxman Foundation, The Mark Foundation (SU2C), and NIH 1R01CA244550. This work is supported by Japan Agency for Medical Research and Development (AMED), Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) under JP20am0101090 to H.S.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c00514.
Accession Codes
The atomic coordinates of the K-Ras(G12D)·GppNHp·KD2 crystal data have been deposited in the Protein Data Bank, www.pdb.org (PDB ID: 6WGN).
Author Contributions
§ Z.Z. and R.G. contributed equally to this work.
The authors declare the following competing financial interest(s): Kevan Shokat has consulting agreements for the following companies involving cash and/or stock compensation: Black Diamond Therapeutics, BridGene Biosciences, Denali Therapeutics, Dice Molecules, eFFECTOR Therapeutics, Erasca, Genentech/Roche, Janssen Pharmaceuticals, Kumquat Biosciences, Kura Oncology, Merck, Mitokinin, Petra Pharma, Qulab Inc. Revolution Medicines, Type6 Therapeutics, Venthera, Wellspring Biosciences (Araxes Pharma).
Supplementary Material
References
- Prior I. A.; Lewis P. D.; Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res. 2012, 72 (10), 2457–2467. 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem J. M. L.; Shokat K. M. Direct Small-Molecule Inhibitors of KRAS: From Structural Insights to Mechanism-Based Design. Nat. Rev. Drug Discovery 2016, 15 (11), 771–785. 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- Hunter J. C.; Manandhar A.; Carrasco M. A.; Gurbani D.; Gondi S.; Westover K. D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015, 13 (9), 1325–1335. 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- Traut T. W. Physiological Concentrations of Purines and Pyrimidines. Mol. Cell. Biochem. 1994, 140 (1), 1–22. 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Taveras A. G.; Remiszewski S. W.; Doll R. J.; Cesarz D.; Huang E. C.; Kirschmeier P.; Pramanik B. N.; Snow M. E.; Wang Y. S.; Del Rosario J. D.; Vibulbhan B.; Bauer B. B.; Brown J. E.; Carr D.; Catino J.; Evans C. A.; Girijavallabhan V.; Heimark L.; James L.; Liberles S.; Nash C.; Perkins L.; Senior M. M.; Tsarbopoulos A.; Ganguly A. K.; Aust R.; Brown E.; Delisle D.; Fuhrman S.; Hendrickson T.; Kissinger C.; Love R.; Sisson W.; Villafranca E.; Webber S. E. Ras Oncoprotein Inhibitors: The Discovery of Potent, Ras Nucleotide Exchange Inhibitors and the Structural Determination of a Drug-Protein Complex. Bioorg. Med. Chem. 1997, 5 (1), 125–133. 10.1016/S0968-0896(96)00202-7. [DOI] [PubMed] [Google Scholar]
- Patgiri A.; Yadav K. K; Arora P. S; Bar-Sagi D. An Orthosteric Inhibitor of the RAS-SOS Interaction. Nat. Chem. Biol. 2011, 7, 585–587. 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hons M.; Niebel B.; Karnowski N.; Weiche B.; Famulok M. Pan-Selective Aptamers for the Family of Small GTPases. ChemBioChem 2012, 13 (10), 1433–1437. 10.1002/cbic.201200164. [DOI] [PubMed] [Google Scholar]
- Maurer T.; Garrenton L. S.; Oh A.; Pitts K.; Anderson D. J.; Skelton N. J.; Fauber B. P.; Pan B.; Malek S.; Stokoe D.; Ludlam M. J. C.; Bowman K. K.; Wu J.; Giannetti A. M.; Starovasnik M. A.; Mellman I.; Jackson P. K.; Rudolph J.; Wang W.; Fang G. Small-Molecule Ligands Bind to a Distinct Pocket in Ras and Inhibit SOS-Mediated Nucleotide Exchange Activity. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (14), 5299–5304. 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.; Burke J. P.; Phan J.; Burns M. C.; Olejniczak E. T.; Waterson A. G.; Lee T.; Rossanese O. W.; Fesik S. W. Discovery of Small Molecules That Bind to K-Ras and Inhibit Sos-Mediated Activation. Angew. Chem., Int. Ed. 2012, 51 (25), 6140–6143. 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima F.; Yoshikawa Y.; Ye M.; Araki M.; Matsumoto S.; Liao J.; Hu L.; Sugimoto T.; Ijiri Y.; Takeda A.; Nishiyama Y.; Sato C.; Muraoka S.; Tamura A.; Osoda T.; Tsuda K. -i.; Miyakawa T.; Fukunishi H.; Shimada J.; Kumasaka T.; Yamamoto M.; Kataoka T. In Silico Discovery of Small-Molecule Ras Inhibitors That Display Antitumor Activity by Blocking the Ras-Effector Interaction. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (20), 8182–8187. 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem J. M.; Peters U.; Sos M. L.; Wells J. A.; Shokat K. M. K-Ras(G12C) Inhibitors Allosterically Control GTP Affinity and Effector Interactions. Nature 2013, 503 (7477), 548–551. 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. M.; Westover K. D.; Ficarro S. B.; Harrison R. A.; Choi H. G.; Pacold M. E.; Carrasco M.; Hunter J.; Kim N. D.; Xie T.; Sim T.; Jänne P. A.; Meyerson M.; Marto J. A.; Engen J. R.; Gray N. S. Therapeutic Targeting of Oncogenic K-Ras by a Covalent Catalytic Site Inhibitor. Angew. Chem., Int. Ed. 2014, 53 (1), 199–204. 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. C.; Gurbani D.; Ficarro S. B.; Carrasco M. A.; Lim S. M.; Choi H. G.; Xie T.; Marto J. A.; Chen Z.; Gray N. S.; Westover K. D. In Situ Selectivity Profiling and Crystal Structure of SML-8–73–1, an Active Site Inhibitor of Oncogenic K-Ras G12C. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (24), 8895–8900. 10.1073/pnas.1404639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh T. B.; Upadhyaya P.; Qian Z.; Pei D. Discovery of a Direct Ras Inhibitor by Screening a Combinatorial Library of Cell-Permeable Bicyclic Peptides. ACS Comb. Sci. 2016, 18 (1), 75–85. 10.1021/acscombsci.5b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R.; Koide A.; Zhou Y.; Eguchi R. R.; Sha F.; Gajwani P.; Santana D.; Gupta A.; Jacobs M.; Herrero-Garcia E.; Cobbert J.; Lavoie H.; Smith M.; Rajakulendran T.; Dowdell E.; Okur M. N.; Dementieva I.; Sicheri F.; Therrien M.; Hancock J. F.; Ikura M.; Koide S.; O’Bryan J. P. Inhibition of RAS Function through Targeting an Allosteric Regulatory Site. Nat. Chem. Biol. 2017, 13 (1), 62–68. 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.; Kamada Y.; Sameshima T.; Yaguchi M.; Niida A.; Sasaki S.; Miwa M.; Ohkubo S.; Sakamoto J.; Kamaura M.; Cho N.; Tani A. K-Ras(G12D)-Selective Inhibitory Peptides Generated by Random Peptide T7 Phage Display Technology. Biochem. Biophys. Res. Commun. 2017, 484 (3), 605–611. 10.1016/j.bbrc.2017.01.147. [DOI] [PubMed] [Google Scholar]
- Martín-Gago P.; Fansa E. K.; Klein C. H.; Murarka S.; Janning P.; Schürmann M.; Metz M.; Ismail S.; Schultz-Fademrecht C.; Baumann M.; Bastiaens P. I. H.; Wittinghofer A.; Waldmann H. A PDE6δ-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity That Prevents Efficient Inhibitor Release by Arl2. Angew. Chem., Int. Ed. 2017, 56 (9), 2423–2428. 10.1002/anie.201610957. [DOI] [PubMed] [Google Scholar]
- Guillard S.; Kolasinska-Zwierz P.; Debreczeni J.; Breed J.; Zhang J.; Bery N.; Marwood R.; Tart J.; Overman R.; Stocki P.; Mistry B.; Phillips C.; Rabbitts T.; Jackson R.; Minter R. Structural and Functional Characterization of a DARPin Which Inhibits Ras Nucleotide Exchange. Nat. Commun. 2017, 8 (1), 16111. 10.1038/ncomms16111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch M. E.; Kaplan A.; Chambers J. M.; Stokes M. E.; Bos P. H.; Zask A.; Zhang Y.; Sanchez-Martin M.; Badgley M. A.; Huang C. S.; Tran T. H.; Akkiraju H.; Brown L. M.; Nandakumar R.; Cremers S.; Yang W. S.; Tong L.; Olive K. P.; Ferrando A.; Stockwell B. R. Multivalent Small-Molecule Pan-RAS Inhibitors. Cell 2017, 168 (5), 878–889. 10.1016/j.cell.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee J. H.; Shim S. Y.; Lee S. J.; Swanson P. K.; Jiang S. Y.; Durney M. A.; Verdine G. L. Exceptionally High-Affinity Ras Binders That Remodel Its Effector Domain. J. Biol. Chem. 2018, 293 (9), 3265–3280. 10.1074/jbc.M117.816348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H.; Zhang Y.; Bos P. H.; Chambers J. M.; Dupont M. M.; Stockwell B. R. K-RasG12D Has a Potential Allosteric Small Molecule Binding Site. Biochemistry 2019, 58 (21), 2542–2554. 10.1021/acs.biochem.8b01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D.; Gmachl M.; Mantoulidis A.; Martin L. J.; Zoephel A.; Mayer M.; Gollner A.; Covini D.; Fischer S.; Gerstberger T.; Gmaschitz T.; Goodwin C.; Greb P.; Häring D.; Hela W.; Hoffmann J.; Karolyi-Oezguer J.; Knesl P.; Kornigg S.; Koegl M.; Kousek R.; Lamarre L.; Moser F.; Munico-Martinez S.; Peinsipp C.; Phan J.; Rinnenthal J.; Sai J.; Salamon C.; Scherbantin Y.; Schipany K.; Schnitzer R.; Schrenk A.; Sharps B.; Siszler G.; Sun Q.; Waterson A.; Wolkerstorfer B.; Zeeb M.; Pearson M.; Fesik S. W.; McConnell D. B. Drugging an Undruggable Pocket on KRAS. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (32), 15823–15829. 10.1073/pnas.1904529116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. H.; Alexander P.; Dharmaiah S.; Agamasu C.; Nissley D. V.; McCormick F.; Esposito D.; Simanshu D. K.; Stephen A. G.; Balius T. E. The Small Molecule BI-2852 Induces a Nonfunctional Dimer of KRAS. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (7), 3363–3364. 10.1073/pnas.1918164117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli M. P.; Janes M. R.; Li L. S.; Hansen R.; Peters U.; Kessler L. V.; Chen Y.; Kucharski J. M.; Feng J.; Ely T.; Chen J. H.; Firdaus S. J.; Babbar A.; Ren P.; Liu Y. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discovery 2016, 6 (3), 316–329. 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- Janes M. R.; Zhang J.; Li L. S.; Hansen R.; Peters U.; Guo X.; Chen Y.; Babbar A.; Firdaus S. J.; Darjania L.; Feng J.; Chen J. H.; Li S.; Li S.; Long Y. O.; Thach C.; Liu Y.; Zarieh A.; Ely T.; Kucharski J. M.; Kessler L. V.; Wu T.; Yu K.; Wang Y.; Yao Y.; Deng X.; Zarrinkar P. P.; Brehmer D.; Dhanak D.; Lorenzi M. V.; Hu-Lowe D.; Patricelli M. P.; Ren P.; Liu Y. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172 (3), 578–589. 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Hansen R.; Peters U.; Babbar A.; Chen Y.; Feng J.; Janes M. R.; Li L. S.; Ren P.; Liu Y.; Zarrinkar P. P. The Reactivity-Driven Biochemical Mechanism of Covalent KRAS G12C Inhibitors. Nat. Struct. Mol. Biol. 2018, 25 (6), 454–462. 10.1038/s41594-018-0061-5. [DOI] [PubMed] [Google Scholar]
- Fakih M.; O’Neil B.; Price T. J.; Falchook G. S.; Desai J.; Kuo J.; Govindan R.; Rasmussen E.; Morrow P. K. H.; Ngang J.; Henary H. A.; Hong D. S. Phase 1 Study Evaluating the Safety, Tolerability, Pharmacokinetics (PK), and Efficacy of AMG 510, a Novel Small Molecule KRASG12C Inhibitor, in Advanced Solid Tumors. J. Clin. Oncol. 2019, 37 (15_suppl), 3003. 10.1200/JCO.2019.37.15_suppl.3003. [DOI] [Google Scholar]
- Canon J.; Rex K.; Saiki A. Y.; Mohr C.; Cooke K.; Bagal D.; Gaida K.; Holt T.; Knutson C. G.; Koppada N.; Lanman B. A.; Werner J.; Rapaport A. S.; San Miguel T.; Ortiz R.; Osgood T.; Sun J. R.; Zhu X.; McCarter J. D.; Volak L. P.; Houk B. E.; Fakih M. G.; O’Neil B. H.; Price T. J.; Falchook G. S.; Desai J.; Kuo J.; Govindan R.; Hong D. S.; Ouyang W.; Henary H.; Arvedson T.; Cee V. J.; Lipford J. R. The Clinical KRAS(G12C) Inhibitor AMG 510 Drives Anti-Tumour Immunity. Nature 2019, 575 (7781), 217–223. 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- Papadopoulos K. P.; Ou S.-H. I.; Johnson M. L.; Christensen J.; Velastegui K.; Potvin D.; Faltaos D.; Chao R. C. A Phase I/II Multiple Expansion Cohort Trial of MRTX849 in Patients with Advanced Solid Tumors with KRAS G12C Mutation. J. Clin. Oncol. 2019, 37 (15), TPS3161–TPS3161. 10.1200/JCO.2019.37.15_suppl.TPS3161. [DOI] [Google Scholar]
- Janssen Research & Development, LLC . First-in-Human Study of JNJ-74699157 in Participants With Tumors Harboring the KRAS G12C Mutation, https://clinicaltrials.gov/ct2/show/NCT04006301, accessed July 27, 2019.
- Eli Lilly and Company . A Study of LY3499446 in Participants With Advanced Solid Tumors With KRAS G12C Mutation, https://clinicaltrials.gov/ct2/show/NCT04165031, accessed July 27, 2019.
- McCormick F. Sticking It to KRAS: Covalent Inhibitors Enter the Clinic. Cancer Cell 2020, 37 (1), 3–4. 10.1016/j.ccell.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile D. R.; Rathinaswamy M. K.; Jenkins M. L.; Moss S. M.; Siempelkamp B. D.; Renslo A. R.; Burke J. E.; Shokat K. M. Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States. Cell Chem. Biol. 2017, 24 (12), 1455–1466. 10.1016/j.chembiol.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogabe S.; Kamada Y.; Miwa M.; Niida A.; Sameshima T.; Kamaura M.; Yonemori K.; Sasaki S.; Sakamoto J.; Sakamoto K. Crystal Structure of a Human K-Ras G12D Mutant in Complex with GDP and the Cyclic Inhibitory Peptide KRpep-2d. ACS Med. Chem. Lett. 2017, 8 (7), 732–736. 10.1021/acsmedchemlett.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida A.; Sasaki S.; Yonemori K.; Sameshima T.; Yaguchi M.; Asami T.; Sakamoto K.; Kamaura M. Investigation of the Structural Requirements of K-Ras(G12D) Selective Inhibitory Peptide KRpep-2d Using Alanine Scans and Cysteine Bridging. Bioorg. Med. Chem. Lett. 2017, 27 (12), 2757–2761. 10.1016/j.bmcl.2017.04.063. [DOI] [PubMed] [Google Scholar]
- Wu X.; Upadhyaya P.; Villalona-Calero M. A.; Briesewitz R.; Pei D. Inhibition of Ras-Effector Interactions by Cyclic Peptides. MedChemComm 2013, 4 (2), 378–382. 10.1039/C2MD20329D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T.; Suga H. A RaPID Way to Discover Nonstandard Macrocyclic Peptide Modulators of Drug Targets. Chem. Commun. 2017, 53, 1931–1940. 10.1039/C6CC06951G. [DOI] [PubMed] [Google Scholar]
- Geyer M.; Schweins T.; Herrmann C.; Prisner T.; Wittinghofer A.; Kalbitzer H. R. Conformational Transitions in P21(Ras) and in Its Complexes with the Effector Protein Raf-RBD and the GTPase Activating Protein GAP. Biochemistry 1996, 35 (32), 10308–10320. 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- Spoerner M.; Herrmann C.; Vetter I. R.; Kalbitzer H. R.; Wittinghofer A. Dynamic Properties of the Ras Switch I Region and Its Importance for Binding to Effectors. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (9), 4944–4949. 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y.; Morimoto J.; Murakami H.; Suga H. Ribosomal Synthesis of Bicyclic Peptides via Two Orthogonal Inter-Side-Chain Reactions. J. Am. Chem. Soc. 2008, 130 (23), 7232–7234. 10.1021/ja800953c. [DOI] [PubMed] [Google Scholar]
- Rhodes C. A.; Pei D. Bicyclic Peptides as Next-Generation Therapeutics. Chem. - Eur. J. 2017, 23 (52), 12690–12703. 10.1002/chem.201702117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thombare V. J.; Hutton C. A. Bridged Bicyclic Peptides: Structure and Function. Pept. Sci. 2018, 110 (3), e24057. 10.1002/pep2.24057. [DOI] [Google Scholar]
- Peraro L.; Deprey K. L.; Moser M. K.; Zou Z.; Ball H. L.; Levine B.; Kritzer J. A. Cell Penetration Profiling Using the Chloroalkane Penetration Assay. J. Am. Chem. Soc. 2018, 140 (36), 11360–11369. 10.1021/jacs.8b06144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty P. G.; Sahni A.; Pei D. Understanding Cell Penetration of Cyclic Peptides. Chem. Rev. 2019, 119 (17), 10241–10287. 10.1021/acs.chemrev.9b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M.; Porter N.; Orekoya O.; Hebert J. R.; Adams S. A.; Bennett C. L.; Steck S. E. Beyond Cyclosporine A: Conformation-Dependent Passive Membrane Permeabilities of Cyclic Peptide Natural Products Christopher. Breast Cancer Res. Treat. 2016, 155 (1), 3–12. 10.1007/s10549-015-3663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockus A. T.; Lexa K. W.; Pye C. R.; Kalgutkar A. S.; Gardner J. W.; Hund K. C. R.; Hewitt W. M.; Schwochert J. A.; Glassey E.; Price D. A.; Mathiowetz A. M.; Liras S.; Jacobson M. P.; Lokey R. S. Probing the Physicochemical Boundaries of Cell Permeability and Oral Bioavailability in Lipophilic Macrocycles Inspired by Natural Products. J. Med. Chem. 2015, 58 (11), 4581–4589. 10.1021/acs.jmedchem.5b00128. [DOI] [PubMed] [Google Scholar]
- Schwochert J.; Turner R.; Thang M.; Berkeley R. F.; Ponkey A. R.; Rodriguez K. M.; Leung S. S. F.; Khunte B.; Goetz G.; Limberakis C.; Kalgutkar A. S.; Eng H.; Shapiro M. J.; Mathiowetz A. M.; Price D. A.; Liras S.; Jacobson M. P.; Lokey R. S. Peptide to Peptoid Substitutions Increase Cell Permeability in Cyclic Hexapeptides. Org. Lett. 2015, 17 (12), 2928–2931. 10.1021/acs.orglett.5b01162. [DOI] [PubMed] [Google Scholar]
- Ahlbach C. L.; Lexa K. W.; Bockus A. T.; Chen V.; Crews P.; Jacobson M. P.; Lokey R. S. Beyond Cyclosporine A: Conformation-Dependent Passive Membrane Permeabilities of Cyclic Peptide Natural Products. Future Med. Chem. 2015, 7 (16), 2121–2130. 10.4155/fmc.15.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitty A.; Zhong M.; Viarengo L.; Beglov D.; Hall D. R.; Vajda S. Quantifying the Chameleonic Properties of Macrocycles and Other High-Molecular-Weight Drugs. Drug Discovery Today 2016, 21 (5), 712–717. 10.1016/j.drudis.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.