Abstract
At present, there is no cure or vaccine for the devastating new highly infectious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has affected people globally. Herein, we identified potent phytocompounds from two antiviral plants Momordica charantia L. and Azadirachta indica used locally for the treatment of viral and parasitic infections. Structure-based virtual screening and molecular dynamics (MD) simulation have been employed to study their inhibitory potential against the main protease (Mpro) SARS-CoV-2. A total of 86 compounds from M. charantia L. and A. indica were identified. The top six phytocompounds; momordicine, deacetylnimninene, margolonone, momordiciode F2, nimbandiol, 17-hydroxyazadiradione were examined and when compared with three FDA reference drugs (remdesivir, hydroxychloroquine and ribavirin). The top six ranked compounds and FDA drugs were then subjected to MD simulation and pharmacokinetic studies. These phytocompounds showed strong and stable interactions with the active site amino acid residues of SARS-CoV-2 Mpro similar to the reference compound. Results obtained from this study showed that momordicine and momordiciode F2 exhibited good inhibition potential (best MMGBA-binding energies; −41.1 and −43.4 kcal/mol) against the Mpro of SARS-CoV-2 when compared with FDA reference anti-viral drugs (Ribavirin, remdesivir and hydroxychloroquine). Per-residue analysis, root mean square deviation and solvent-accessible surface area revealed that compounds interacted with key amino acid residues at the active site of the enzyme and showed good system stability. The results obtained in this study show that these phytocompounds could emerge as promising therapeutic inhibitors for the Mpro of SARS-CoV-2. However, urgent trials should be conducted to validate this outcome.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, natural phytocompounds, MMGBA, molecular dynamics simulations
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel strain of the coronavirus which is considered a global public health emergency (WHO, 2020; Read et al., 2020). It was first discovered in December 2019 in the city of Wuhan, Hubei province, China, and has since spread across the globe affecting over 36 million people with over 1.1 million deaths as of 10 October 2020. In humans, SARS-CoV-2 is believed to cause mild respiratory infections, such as those observed in the common cold to severe pulmonary disease with an extreme case of death (Khan et al., 2020; Lu et al., 2020). Studies suggest that the SARS-CoV-2 can be transmitted through infected droplet (respiratory secretions) and close person-to-person contact (Khan et al., 2020) where it binds to the primary target cells such as enterocytes and pneumocytes, thereby establishing a cycle of infection and replication (Wu et al., 2020).
Coronaviruses are a large family of enveloped, positive-sense, single stranded-RNA viruses with nucleocapsid belonging to the family of Coronaviridae and order Nidovirales (Zhou et al., 2020). They are classified into four main subgroups: alpha, beta, gamma and delta depending on their genomic structure (Fehr & Perlman, 2015). So far, seven strains of the human CoVs, which include 229E, NL63, OC43, HKU1, Middle East respiratory syndrome (MERS)-CoV, SARS-CoV and 2019-novel coronavirus, which are responsible for mild respiratory tract infection associated (both lower and upper respiratory tract) (Walls et al., 2020). The seventh and highly contagious “COVID-19” is taxonomically positioned in the genus Betacoronavirus and exhibits 89.1% and 60% nucleotide sequence similarity with SARS and MERS coronaviruses, respectively (Wu et al., 2020). Coronaviruses main protease (Mpro) also known as 3-chymotrypsin-like protease (3CLpro) is an important protein that is required for proteolytic maturation of the coronavirus (Wu, et al.). The main protease is considered the “Achilles hill” of coronaviruses (Fehr & Perlman, 2015) inhibiting the activity of this enzyme could block viral replication. Moreover, the Mpro is highly conserved across coronaviruses thus, the main proteases are considered an attractive target for the development of effective antiviral drugs in structure-based drug design for the treatment of SARS-CoV-2 and other coronaviruses (García-Fernández et al., 2016; Krishnan & Anirudhan, 2002; Zhang et al., 2020).
Currently, there is no known cure or vaccine for the COVID-19. A rapid search is on-going for the development of vaccines and new antiviral drugs for the effective treatment of the highly transmissible SARS-CoV-2. Many researchers are attempting to find new inhibitors for the SARS-CoV-2 main protease Mpro both from synthetic and naturally active compounds (Elfiky, 2020; Khan, et al., 2020). Several drugs have been repurposed and used as a frontline therapy for SARS-CoV-2 and relief of symptoms for patients infected with COVID 19, and these drugs include hydroxychloroquine anti-malaria and anti-HIV drugs. On the 1 May 2020, the FDA reference drug an emergency use authorization of remdesivir for the treatment of hospitalized patients with COVID-19 (FDA, 2020). Yet the clinical safety and efficacy against COVID-19 remain to be fully established.
Several crystal structures of SARS-CoV-2 3C-like proteinase have been solved and made are available on protein databank. However, only one complex structure of SARS-CoV-2 3C-like proteinase (PDB: 6W63) was bound to a broad spectrum non-covalent covalent inhibitor (X77) in Data Bank (https://www.rcsb.org). It has been observed that most of the current research effort focus on finding potential SARS-CoV-2 Mpro covalent inhibitors while the possibilities to identify non-covalent inhibitors remain less investigated. For that reason, we decided to screen new natural phytocompounds against the 6W63 crystal structure to find potential non-covalent pharmacophores against SARS-CoV-2 3C-like protease. Previous studies have shown that covalent and non-covalent inhibitors have been designed and discovered to inhibit 3C proteinases and therefore to treat related diseases. These inhibitors can be used as guidance to design drugs for SARS-CoV-2 (Zhavoronkov et al. 2020). Active compounds have been obtained by covalent bonding with the cysteine at the catalytic site. However, side effects and toxicity can often be caused by covalent bonding ( Zhavoronkov et al. 2020). Thus, prevents their use for clinic therapy. Hence, for SARS-CoV-2 treatment, it is more attractive to design and discover noncovalent inhibitors. Virtual screen by molecular docking of chemical databases is one of the most powerful approaches to discover non-covalent inhibitors. We report herein the screening of potential Phyto non-covalent inhibitors against SARS-CoV-2 3C-like proteinase.
Medicinal plants provide an excellent source for antiviral drugs due to fewer side-effects, low cost, bioavailability and easy availability along with less potential to cause resistance (Mukherjee et al., 2007). Hence, the utilization of herbal drugs in viral infections is an important future therapeutic strategy. Medicinal plants contain numerous phytochemicals responsible to exhibit the antiviral effect. Two commonly used anti-viral herbs: Momordica charantia L. (Cucurbitaceae) and Azadirachta indica A. Juss. (Meliaceae) are potent medicinal plants used in traditional medicine in Nigeria and tropical countries for the treatment of many ailments including inflammatory, anti-diabetic, parasitic and viral infections. Traditionally, these plants have been reported to exhibit potent antiviral activities by inhibiting several viruses including herpes simplex virus, hepatitis B virus, smallpox virus and human immunodeficiency virus (Liu et al., 2009; Mohammad et al., 2016; Pongthanapisith et al., 2013). Considering the wide potent antiviral activity of these plants, it is, however, possible that potent non-inhibitors could emerge from these plants against SARS-CoV-2 main protease Mpro. In this study, virtual screening of potent medicinal bioactive compounds from plants of M. charantia L. and A. indica as potential inhibitors against SARS-CoV-2 main protease Mpro was carried out. Based on the docking scores, six compounds we ranked and preceded for molecular dynamics (MD) simulations and pharmacokinetic studies.
Materials and methods
Virtual screening and molecular docking
Three-dimensional X-ray crystallographic structure of SARS-CoV-2 Mpro was downloaded from the RCSB Protein Data Bank (PDB). The SARS-CoV Mpro bound to a noncovalent inhibitor with PDB ID: 6W63 (resolution 2.1 Å) (Zhavoronkov et al., 2020) was used for this study. Before the virtual screening, the protein structures were prepared using the ‘Protein Preparation Wizard’ workflow in the Schrodinger suite. This involved the addition of hydrogen atoms to the protein, assignment of bond orders, and deletion of unnecessary water molecules. The key water molecules interacting with the active site residues of the enzyme were retained. Sidechains were added, disulphide bonds were formed, missing atoms were added, and the partial charges were assigned. Energy minimization was done using OPLS_2005 (Optimized Potentials for Liquid Simulations) force field. As the downloaded protein was co-crystallized with X77(N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide) a non-covalent inhibitor, the ligand-binding site was used to define the active site of the protein. Receptor grid generation workflow was used to define a grid (box) around the ligand, to keep all the functional residues in the grid (Sastry et al., 2013). The residue used for preparations is Cys145.
Molecular dynamic simulation
Structural preparation
2-Dimensional structures of 86 compounds from M. charantia and A indica were identified and retrieved from the PubMed literature as test ligand molecules, two antiviral drugs and 1 anti-malarial drug were retrieved from PubChem database as a reference. Ligprep, pre-processing of the ligands was done, which includes the formation of tautomers and ionization states at pH 7.0 ± 2.0 using Epik, hydrogen atoms were added, charged groups were neutralized and geometry of the ligands was optimized. Virtual screening of compounds was performed in three stages involving a high throughput virtual screening (HTVS), standard precision (SP) and extra precision (XP) using the using glide tool (grid-based ligand docking with energetics) algorithm implemented in Schrodinger (Friesner et al., 2006). Stage one and two, HTVS and SP employ the self-scoring function, whereas XP reduces the intermediate conformations and thoroughness of the torsional refinement and sampling. A total of 86 compounds and three reference FDA drugs was docked against each target protein with the co-ligands X77 and α-ketoamide was used as a positive control. Next, top scorers were set forth for SP docking, and the output of SP docking was put forward in XP docking with positive control. All the results were analysed in XP visualizer. Compounds with the highest binding energies against SARS-CoV-2 Mpro was selected for further MD and in silico pharmacokinetic studies.
Production step
The six top screened Phyto-ligands (of the 86 compounds) with high binding energy, three FDA reference drugs and the apo-enzyme (APO) of the target was subjected to MD simulation.
The graphical processing unit (GPU) version of AMBER 18 software package was used to perform the molecular dynamic (MD) simulations of all the systems. Optimization of the systems was carried out using ANTECHAMBER (Wang et al., 2001) and LEap module of Amber 18 to ensure all the appropriate parameters were available for MD simulations. The protein parameters were assigned using FF14SB (Perez et al., 2015) version of the Amber force field. The LEap module of Amber 18 was applied for adding the missing hydrogen atoms and counter ion to neutralize the systems. The systems were suspended with an orthorhombic water box of TIP3P (Jorgensen et al., 1983) to restrain the protein within 10 Å of the box edge. Long-range electrostatics were treated with the particle-mesh Ewald method (Kholmurodov et al., 2000) implemented in AMBER 18 with direct space and a 12°A van der Waals cut-off while periodic boundary conditions were adopted. Before the commencement of MD, initial and final minimization, gradual heating and the equilibration steps were performed. The final MD production was performed as previously reported (Emmanuel et al., 2019b; Ibeji, 2020). The CPPTRAJ and PTRAJ modules (Roe & Cheatham, 2013) of the Amber18 suite was used to carry out post-analysis such as Radius of Gyration, root mean square deviation (RMSD), root mean square fluctuation (RMSF) and solvent-accessible surface area (SASA). Structural visualization and plots were done using UCSF Chimera software package molecular modelling tool and Origin data analysis software version 6 (Seifert, 2014) (http://www.originlab.com).
Thermodynamic calculations
MM/GBSA a key tool used in predicting macromolecular stability and protein–ligand binding affinity (Genheden & Ryde, 2015; Zhou & Madura, 2004; Zhou et al., 2009) were applied. It gives an idea of the binding mechanism which equally includes the contribution of enthalpy, entropy to the molecular recognition and ligand–protein association (Genheden & Ryde, 2015). The binding free energy was averaged over 1000 snapshots which were extracted from the 100 ns trajectory as described by the set of equation.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where Egas is the gas-phase energy; Eint is the internal energy; Eele stands for the Coulomb and Evdw are the van der Waals energies. Egas is obtained from the FF14SB force field terms. The solvation free energy, Gsol, is split into polar and non-polar solvation state of contribution. Here the polar solvation, GGB, contribution is calculated by solving the GB equation while the non-polar solvation contribution, GSA is estimated from the SASA calculated with the water probe radius of 1.4 Å. T and S are the temperatures and the solute entropy, respectively. To examine the individual amino acid contribution to the total binding free energy between the natural products and main protease, the per-residue free energy decomposition analysis was computed at the atomic level using the MM/GBSA method in AMBER 18.
Cheminformatic evaluation of pharmacological properties
A comparative evaluation of the pharmacological properties of the natural phytocompounds and FDA reference drugs was performed. This was done by predicting their individual pharmacokinetic (ADMET) properties with online and offline tools. The bioactivity of the various compounds was estimated using selected and offline tools. The prediction tools employed include Molsoft program (http://molsoft.com/mprop/), Molinspiration Cheminformatics (Husain et al., 2016), ProTox webserver (Banerjee et al., 2018) and the OSIRIS DataWarrior property explorer (López-López et al., 2019; Sander et al., 2015). More than one prediction tools were used for precise validation and reproducibility of comparative analysis. The pharmacokinetic (ADMET) properties of the studied compounds were evaluated using Molsoft online program, this is to investigate if they adhered to Lipinski’s rule of five (RO5). An additional step was equally taken to validate the obtained pharmacokinetic properties by employing data warrior property explorer (Sander et al., 2015), this program helps to predict ligand efficiency (LE) (Abad-Zapatero, 2007; Hopkins et al., 2014), lipophilic ligand efficiency (LLE) and lipophilicity-correlated ligand efficiency (LELP) (Hopkins et al., 2014; Johnson et al., 2018). Toxicity of compounds reveals their safety state for consumption. Thus, in silico prediction of oral toxicity gives a faster route of obtaining information about doses that could be toxic in animals.
Results and discussion
Virtual screening and molecular docking
In the present study, we screened 86 compounds from M. charantia L. and A. indica as potential inhibitors against SARS-CoV-2 Mpro (PDB ID: 6W63) using structure-based drug design. This approach is based on computationally fitting compounds into the active site of the target protein, followed by a ranking of these compounds based on their low binding energies and interaction with the residues of the binding pocket. The docking scores of the compounds were when compared with a standard Mpro inhibitor, α-ketoamide 13b and X77. The α-ketoamide 13b is a broad-spectrum inhibitor with an IC50 value of 0.67 ± 0.18 μM against purified recombinant SARS-CoV-2 Mpro (Zhang et al., 2020). A comprehensive list of the 86 docked phytocompounds can be found in the supplementary file (Supplementary material Table S1). Thus, among the 86 phytocompounds and 3 antiviral drugs screened (Supplementary material Table S1), 6 phyto ligands exhibited appreciable binding energies against the SARS-CoV-2 Mpro and strong interactions within the binding pocket. The top six hit phyto-compounds were momordicine, deacetylnimninene, margolonone, momordiciode F2, nimbandiol, 17-hydroxyazadiradione with binding energies of 6.6, 6.1,−5.5, −5.2, −4.8 and −4.0 kcal/mol, respectively. Also, the FDA reference anti-viral drug remdesivir and ribavirin showed binding energies of −8.4 and −6.2 kcal/mol, respectively (Table 1). In comparison, standard Mpro inhibitor; α-ketoamide 13b and X77 displayed binding energies of −8.5 and −9.1 kcal/mol, respectively. The results show that all the lead compounds were tightly bound into the substrate-binding pocket, through a good number of hydrogen bonds as well as hydrophobic interactions (Figure S1). The glide 2D interaction suit was used to reveal the interacting residue information. In particular, the best phyto ligand “momordicine” (M. charantia) was seen to interact with the target enzyme with a binding energy of −6.6 kcal/mol. Momordicine formed five hydrophobic interactions with Leu 27, Met 49, Cys145, Met165, Leu167, Pro168 and established two hydrogen bonds with Glu 166 and Asn 142 (Figure S1(a)). The second lead phytocompound i.e. Deacetylnimninene (A. indica) showed two hydrogen bonds with residues Cys 44 and Glu 166 and eight hydrophobic interactions with Cys 44, Met 49, Pro 52, Tyr 54, Cys145, Met 165, Leu 167 and Pro 168 (Figure S1(b)). Furthermore, it was observed that the FDA antiviral drug; remdesivir exhibited the highest binding affinity among all tested compounds with a binding affinity of −8.4 kcal/mol (Table 1). This displayed three hydrogen bond interactions with Cys145, Gln166 and Thr 190 residues and eight hydrophobic interaction with key residues (Cys 44, Met 49, Tyr 54, Pro 52, Phe 140, Leu141, Cys145, Met 165, Leu 167, Pro 168, Val 186, Ala 191) (Figure S1(g)). Notably, the top lead phyto ligand and redemsivir were able to establish hydrogen bond and hydrophobic interactions with residues crucial for the inhibition of SARS-CoV-2 replication similar to phyto ligands and the standard Mpro inhibitor, alpha- ketoamide and X77 (Figure S1(h,j)). The SARS-CoV-2Mpro substrate-binding site mainly consists of a cysteine–histidine dyad (His41 and Cys145) which controls the catalytic activity of SARS CoV-2 Mpro. Also, our share similar binding interaction patterns with a previous study on virtual screening of novel non-covalent inhibitors against SARS-CoV Mpro (Liu et al., 2005). Inhibition by these compounds reflects the possible inhibitory tendencies against the COVID-19. SARS-CoV-2 Mpro with other papain-like proteases is important for processing the polyproteins into various non-structural proteins by cleaving at specific sites that are translated from the viral RNA. Several key amino acids in the active site of Mpro enzyme have been documented to be Leu, Gln, Ser, Ala, Gly along with the Cys-His dyad which marks the cleavage site. Based on the results obtained we further subjected the 6 phyto ligands and the antiviral drugs remdesivir and ribavirin to MD simulation to establish the mode of interaction with Mpro protease.
Table 1.
Interaction of the SARS-CoV-2 main protease with phyto-compounds and FDA anti-viral drugs.
| Compounds name | Source | PubChem CID | ABB | Docking score (kcal/mol) | Key residue interaction |
|---|---|---|---|---|---|
| Momordicine | M. charantia L. | 14807332 | MDC | –6.6a | H-bond: Asn 142; Glu 166 |
| Deacetylnimninene | A. indica | 102004582 | DCN | –6.1a | H-bond: Cys 44; Glu 166 |
| Margolonone | M. charantia L. | 189726 | MGN | –5.5 | H-bond: His 163, Glu 166, Ser 144, Gly 143 |
| Momordiciode F2 | M. charantia L. | 44445567 | MDF | –5.2 | H-bond: Pro 168, Glu 166 |
| Nimbandiol | A. indica | 157277 | NBD | –4.8 | H-bond: Glu 166, Asn 142, Gln 198, Pi-pi: His 41 |
| 17-Hydroxyazadiradione | A. indica | 52951892 | HXD | −4.0 | H-bond: Glu 166 |
| Ribavirin | 37542 | RBV | –6.2a | H-bond: His 163; Phe 140 | |
| Remdesivir | 121304016 | RMS | –8.4a | H-bond: Glu 166; Cys 145; Thr 190 | |
| Hydroxychloroquine | 178396 | HCQ | –3.4 | H-bond: Asn 142; Cys 44 Pi-cation: His 44 |
|
| Alpha-ketoamide | 10129151 | AK | –8.5 | H-bond: Cys 44, Hie 41, Asn 142, His 163, Glu 166 | |
| X77 | 145998279 | X77 | –9.6 | H-bond: Cys 44, Hie 41, Asn 142, Hie 163, Glu 166, Gly 143 |
ABB, abbreviations.
The lowest binding energy for respective ligand with the target protein.
Figure 1.
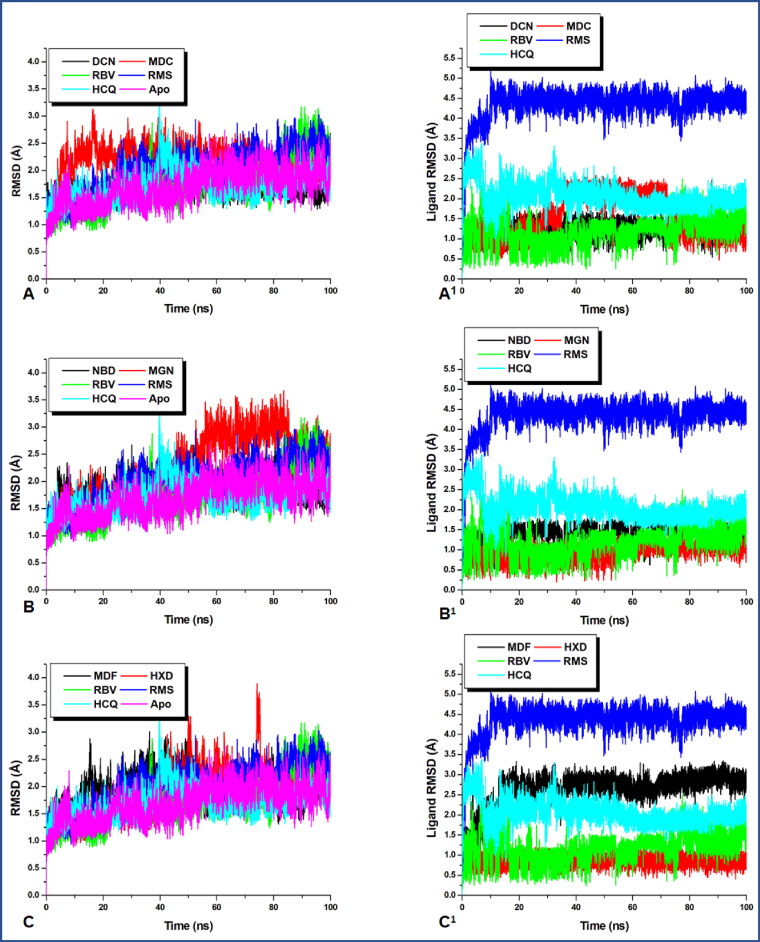
A graphical representation of the evolving conformational trend at the time of the MD simulation. Although the three reference drugs were present in all the plots, the first codes in all the figure legend represent the investigated compounds. (A–C) The RMSD of the entire protein structure whereas (A1–C1) is that of the ligand, which reveals their respective momentary re-orientations during the MD run.
Exhaustive evaluation of protein systems conformational dynamics
Previous reports have suggested main protease (Mpro, 3CLpro) as a very attractive target for coronaviruses due to the crucial role it plays in processing the translated polyproteins of the viral RNA (Zhang et al., 2020). To verify the dynamics of the protein–ligand complex a 100 ns MD simulation was run, to reveal the conformational trends which occurred in the protein’s backbone to the initial conformations. From Figure 1, all the studied systems showed a favourable stability state during the 100 ns of the MD simulation after converging at around 8 ns. From the average RMSD of all the systems in Table 2, the apo-enzyme exhibited almost the lowest average RMSD when compared with the ligand-bound systems. However, exceptions exist in the case of hydroxychloroquine with almost a close mean RMSD (1.711 Å) value to that of the apo (1.713 Å) and deacetylnimbinene with lower RMSD of 1.667 Å. The average RMSD observed in all the systems revealed a tractable structural trend in the natural products and the reported FDA reference drugs. The slightly lower average RMSD of the apo corroborate with the reported structural behaviour of proteases (Munsamy et al., 2018). The observed collective marginal structural deviation could be an indication of the characteristic mechanistic inhibitory mode of Mpro. Previous reports suggested that proteases exhibit consistent open/close and twisting dynamic motion mechanisms at the flap, flexible domain and hinge regions which promotes ligand binding (Munsamy et al., 2018). It is therefore suspected that this reported structural motion is transmitted to the entire protein structure which resulted in a higher conformational deviation of the complexed systems relative to the apo. Hence the observed higher structural deviation of the ligand-bound systems relative to the apo is an indication of inactivity of the viral Mpro due to ligand binding. Although high RMSD is an indication of atomic deviation, which suggest structural instability, based on the reported structural/conformational behaviour of proteases, the high RMSD of the bound systems relative to the apo does not suggest structural instability but rather a functional display in line with the reported structural behaviour of proteases. This indicates viral mechanism of viral protein inactivity.
Table 2.
An estimated value of the average parameter used to decode structural stability.
| Compound names | Average values |
|||||
|---|---|---|---|---|---|---|
| RMSD (Å) | RMSF (Å) | RoG (Å) | SASA (Å2) | Ligand RMSD (Å) | Active site RMSF (Å) | |
| DCN | 1.667 | 0.908 | 22.105 | 14072.19 | 1.228 | 12.794 |
| MDC | 2.210 | 0.983 | 22.045 | 13915.73 | 1.516 | 6.922 |
| NBD | 1.862 | 0.903 | 22.108 | 14146.30 | 1.389 | 6.389 |
| MGN | 2.239 | 1.361 | 22.028 | 14044.94 | 0.992 | 9.756 |
| MDF | 2.006 | 1.002 | 22.056 | 13841.17 | 2.655 | 9.970 |
| HXD | 1.965 | 1.343 | 22.366 | 14223.53 | 0.881 | 9.792 |
| RBV | 2.061 | 1.171 | 22.055 | 14122.35 | 1.331 | 9.118 |
| RMS | 2.135 | 1.210 | 22.189 | 14297.35 | 4.397 | 8.077 |
| HCQ | 1.711 | 1.051 | 22.189 | 14190.15 | 2.079 | 12.778 |
| APO | 1.713 | 1.040 | 21.931 | 14159.60 | N/A | 9.075 |
From the plots, momordicine stabilized immediately after attending convergence and maintained this stabilized trajectory till the end of the simulation whereas margolonone fluctuated from 50–85 ns within an RMSD between 2.5 and 3.5 Å and eventually reassumed stability till the end of the simulation whereas the RMSD of the other systems remained below 3 Å.
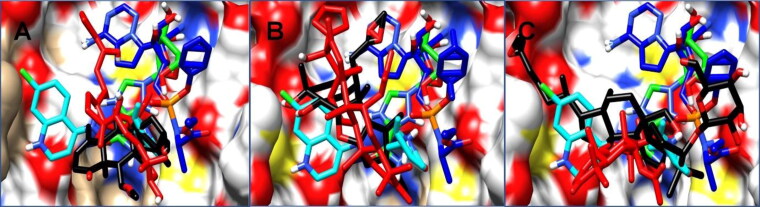
However, the lower average RMSD value of deacetylnimbinene and hydroxychloroquine when compared with the apo and observed little conformational changes in momordicine and margolonone were not of much different and does not indicate that ligand binding causes serious conformational changes. The structural dynamics of the ligands to the Mpro at the active site was further investigated by tracking the ligand confirmation and/or orientations during the MD simulation. The ligand RMSD indicate the stability of the ligands concerning the protein and its active site residues. As it is anticipated that binding pocket architecture could influence the mobility, orientation and interaction of drug molecule with protein residues. From Figure 1(A1–C1) remdesivir exhibited serious orientational/positional deviation to the other ligands examined, however, all the natural products exhibited lower conformational deviation relative to the reference drugs with ribavirin maintaining a very similar conformational trajectory with all the natural products. The plot revealed that momordicine fluctuated from 35 to 72 ns before stabilizing again. On the other hand, momordicoide F2 elicited higher structural deviation to the starting conformation while exhibiting higher structural deviation relative to the other natural products and the reference drugs except for remdesivir. A large conformational change was exhibited by remdesivir at the beginning of the simulation before attaining and maintaining stability; this initial wide conformational change of remdesivir could be a result of the orientational adjustment to properly fit into the binding pocket. To further gain insight into the conformational trends which resulted to the high conformation deviation of remdesivir in the ligand RMSD plot, a visual analysis/representation of the average conformational orientation of all the ligand at the active site of Mpro was done. As shown in Figure 2, remdesivir (blue) drifted towards the edge of the pocket and does not have contact with virtually all the ligands in the pocket except ribavirin. However, a look at the pocket, revealed that the rest of the ligands have a common contact point(s). The nature of motion exhibited by these ligands could have a relationship with the most favourable optimal orientation needed for most suitable interaction with active site residues. At the long run remdesivir has the highest average RMSD followed by momordicoide F2 and finally hydroxychloroquine this is an indication of close conformational/orientational behaviour of the natural product with that of the reference drugs as both ligand types did not have much difference in conformational trend. This minor difference in the mean RMSD values of all the simulated systems indicates that the binding of the ligand does not severely perturb the global structure of the protease.
Figure 2.
A visual representation of the average PDB orientation of the various ligands at the end of the MD simulation. All ligands were colour coded to match with their respective plots in Figure 1(A1–C1). (A) The ligands represented in systems plotted in Figure 1(A1), (B) Figure 1(B1) and (C) Figure 1(C1).
Systematic estimation of whole protein and active site residues flexibility
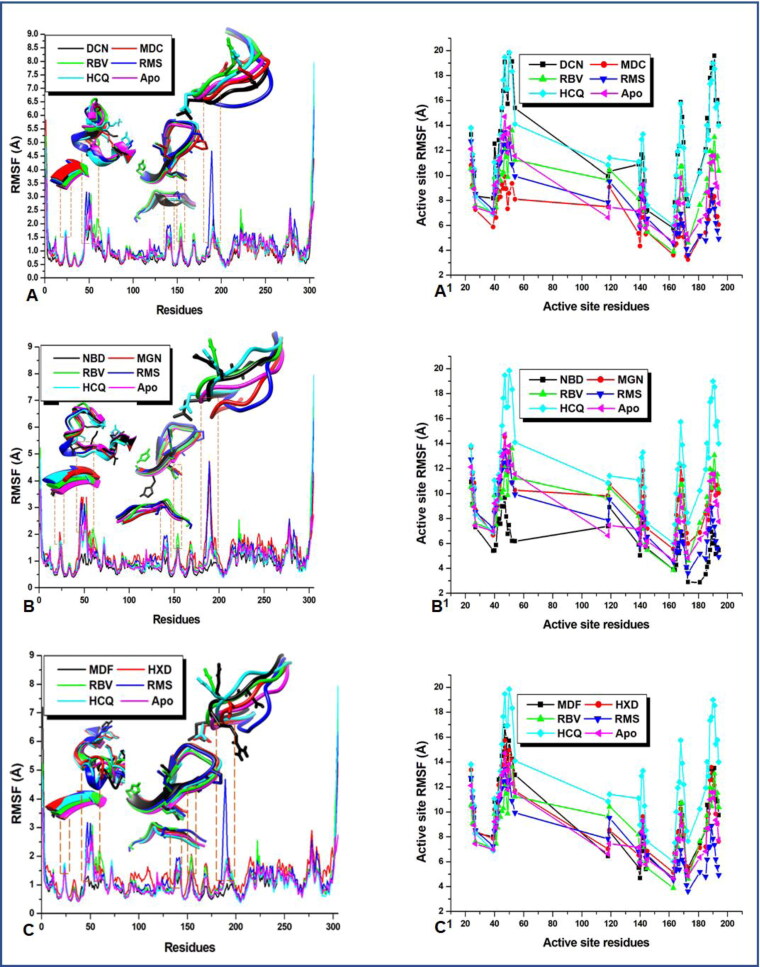
To gain insight into the nature and feature of the fluctuations displayed by the backbone atoms of the proteins, the C-α RMSF was investigated. RMSF plots reveal the precise conformational transition within the residues that make up the proteins secondary structure during MD simulation. It further shows the nature of residual fluctuation and motion to ligand binding and unbinding. A higher value of RMSF indicates an increase in flexibility while a lower value correlates with a decline in flexibility. Figure 3(A–C) shows that the residues in all the systems maintained a relatively similar pattern but varied levels of fluctuations.
Figure 3.
A plot of per residue fluctuation; revealing regions of interest in the active sites that contributes to ligand stabilization and high-affinity binding. (A–C) The proteins whole structure with unique regions of outstanding interactions whereas (A1–C1) gives clues as to the nature/degree of active site residue fluctuations.
As indicated in Table 2, all the systems had a near approximate value of average RMSF with deacetylnimbinene, momordicine and nimbandiol exhibiting the lowest residual fluctuation. However, regions of interest can be noticed in the plots, these are areas where the residues exhibited peak fluctuations. The nature/degree of residual drift from their mean positions in these interesting regions has been indicated with 3D images in the plots in Figure 3 for clarity. These are residues in the active site the play crucial role in system stabilization. They are residues that are actively involved in protein–ligand interactions(Ibeji, 2020; Olotu et al., 2019). These residues with residue numbers 25–27, 39–54, 139–145, 163–173, 185–194 exhibited wider and unusual fluctuations relative to others. To gain further insight into the degree and distinctiveness of these fluctuations in these regions, we performed an active site analysis of the RMSF of these residues and the results are presented in Figure 3(A1–C1).
A close look at the average active site RMSF, Table 2 indicates that the phytocompounds have similar average active site RMSF values to those of the reference drugs. It was also observed that higher active site RMSF of the entire ligand systems when compared with apo corroborates with initially mentioned open/close and twisting mechanism of ligand inactivation of the viral protease. However, Nimbandiol and momordicine had a very low average active site RMSF, this could be due to nonoptimal orientation for maximum interaction with the active site residues. Remdesivir equally exhibited lower mean active site RMSF, though not too far from that of the apo. This lower mobility in the active site RMSF of these compounds could be suggestive of the need for ligand optimization. This further broadened the understanding of the mobility of the crucial residues in the active site which is the location of the flap and hinge regions.
Impact of conformation compactness on structural mobility
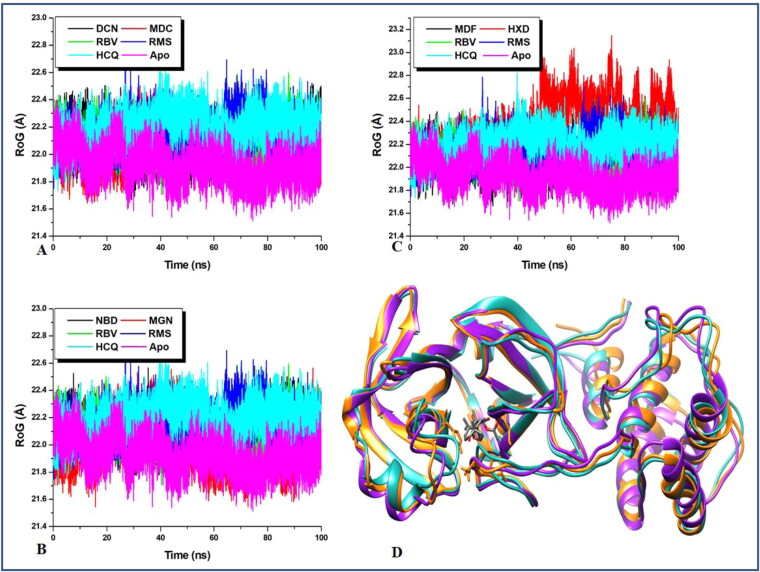
RoG measures the degree of compactness of the backbone carbon atoms of a protein and accounts for the level of mobility of a protein structure which is an indication of stability in the protein backbone structure. The conformational shift/alteration initially suggested to be the mechanism of activity of proteases in the RMSD plots is validated by the results obtained in the RoG plot. RoG gives an idea of type/nature structural deviation exhibited by the RMSD plot. The average RoG values from the plot in Figure 4 revealed that the apo has an RoG value that is slightly the lowest (21.931 Å) relative to the other systems. This low RoG value which suggests the reduction in mobility validates the mechanism of ligand binding at the active site of protease and the entire proteases. As the previous report has suggested the ligand-binding mechanism to be via open/close and twisting motion which induces a high level of conformational deviation in protein 3D structure.
Figure 4.
A comparative investigation of Protein 3D structural evolution during the MD simulation. The RoG trajectory reveals the evolutionary pattern of 3D structural packing and unpacking during the 100 ns of the MD simulation.
Elevated average RoG indicates a decrease in compact structural packing which suggests increased mobility (Emmanuel et al., 2019a) and a stable state favourable for ligand binding to proteases. The nature of the trajectories exhibited by the RoG plot of the ligand-bound relative to the apo is suggestive of the mechanism of ligand inactivation of proteases. Hence, inhibiting the activity of this enzyme will inhibit viral replication and transcription. Figure 4(D) shows a superimposed 3 D images of the Mpro starting structure, that of 17-hydroxyazadiradione complex with the highest mean RoG value (22.36575 Å) and that of the apo at the end of the simulation. The image reveals a comparative minor level of the structural deviation which could be suggestive of the level of compact packing of the protein.
Simulation induced re-orientation and intra-migration of the hydrophobic and hydrophilic residues
Residual mobility which induces side-chain re-orientation was further studied by examining the intermittent burial and exposure of the hydrophobic and hydrophilic residues during the time duration of the MD simulation. Protein folding and unfolding could correlate with the re-orientation of the side chains from the hydrophilic phase to the hydrophobic phase. Hence structural transformation of the surface residues towards the hydrophobic core could be indicative of protein unfolding whereas the inward transition of these surface residues suggests protein folding. This parameter was employed to examine protein activity and inactivity as indicated in Figure 5.
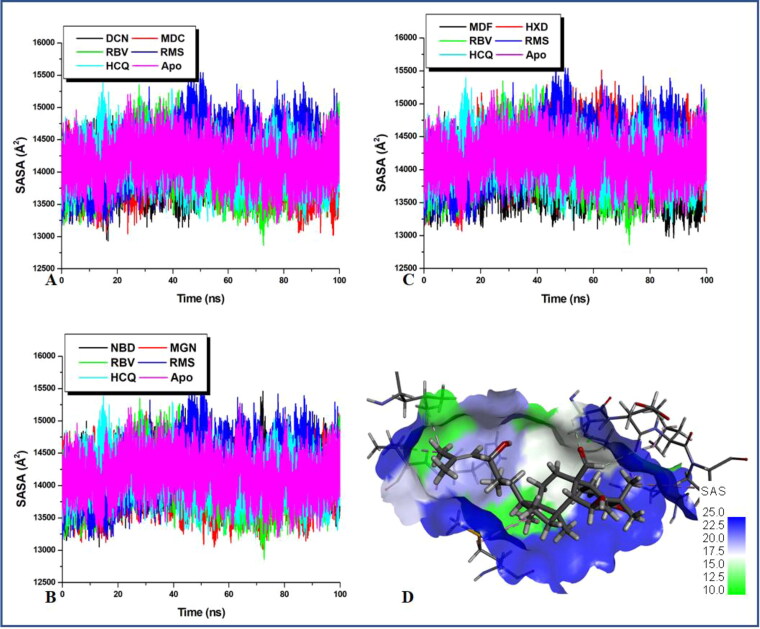
Figure 5.
Degree of the solvent of exposure of the active site residues goes hand in hand to determine protein–ligand interaction. The plot indicates the level/degree of surface residue exposure and burial which is an indirect indicator of catalytic activity.
Previous reports have revealed that high SASA values connote decrease in the exposure of the buried hydrophobic residues which indicates the reduction in systems stability while expulsion of the buried hydrophobic residues from the hydrophobic core indicates an increase in system stability (Emmanuel et al., 2019a). From average SASA values in Table 1 which were obtained in Figure 5, it can be observed that all the natural products have slightly lower average SASA values to the apo except for 17-hydroxyazadiranone with slightly higher average SASA values than the apo. Two of the reference drugs; remdesivir and hydroxychloroquine individually produced the highest respective average SASA values relative to the natural products except for 17-hydroxyazadiranone thus indicating that the natural products were more stable than the drugs. Therefore, this lower SASA values in the natural products systems relative to the apo and the two reference drugs systems are indicative of an increase in hydrophobicity induced by ligand binding. The results obtained here corroborate with the observations made from the RMSD, RMSF and RoG plots. This structural unfolding due to the exposure of the hydrophobic residues as observed from the lower average SASA values correlates with the initially mentioned open/close and twisting mechanism of the protein activity and inactivity. Conformation transition from the folded state to the unfolded state could indicate the inhibitory mechanism of ligand-bound protease. Figure 4(D) is a 3D representation of an MDC-bound Mpro (with almost lowest average SASA value); showing the solvent assessable surface (SAS). It reveals that the SAS (hydrophilic) occupy the smaller area around the ligand at the active site than the non-SAS (hydrophobic). This trend at the active site is believed to be pass on to the entire protein residue.
Disclosure of the possible inhibitory potency selected compounds via MM/GBSA calculation
Previous studies have suggested that inhibition of the activity of proteases equally inhibit the viral replication and transcription and henceforth viral survival and disease progression (Ibeji, 2020; Olotu et al., 2019). This gives an idea of the interaction that is likely to be harnessed to design novel and more potent anti-viral inhibitors that will block further viral protein replication. We went further to investigate the energetic inhibitory potency of the selected natural product in comparison with reference drugs. MM/GBSA has recently grown as a very popular computational tool for estimation of the binding propensity of chemical compounds to protein. The results obtained in Table 3 revealed thermodynamic interaction of ligands to protein and hence suggests the capacity of the ligand to block the replication and transcription of the viral protein which indirectly indicate the inhibitory prowess of the compound. Our finding as indicated in Table 3 shows that all the selected reference drugs and the natural product have favourable binding free energy, efficient enough for good binding interaction. As outlined in the table, the three FDA reference drugs gave binding free energy (ΔGbind) that are close to the values obtained for most of the natural products except for momordicine and momordiciode F2 with very high binding −41.363 and −43.413 kcal/mol, respectively. This higher binding energy exhibited by these two compounds could be due to the similarity in their structural features. And this large binding energy of these two compounds is suggestive of the need for further studies to identify the unique moieties which are responsible for high energetic interaction that produced this high-affinity binding.
Table 3.
MM/PBSA-binding energy profile of the studied compounds to Mpro.
| Compounds names | Energy components (kcal/mol) |
||||
|---|---|---|---|---|---|
| ΔEvdW | ΔEele | ΔEgas | ΔEsol | ΔGbind | |
| DCN | –36.7338 | –9.8173 | –46.5511 | 18.2133 | –28.3378 |
| MDC | –46.0194 | –15.5171 | –61.5365 | 20.1702 | –41.3663 |
| NBD | –39.6483 | –21.7271 | 23.9521 | 23.9521 | –37.4233 |
| MGN | –30.4557 | 25.1539 | 25.1539 | –45.9030 | –20.7491 |
| MDF | –52.6626 | –20.0494 | –72.7120 | 29.2992 | –43.4129 |
| HXD | –37.2274 | –6.2504 | –43.4778 | 16.5647 | –26.9131 |
| RBV | –25.0251 | –27.7128 | –52.7379 | 30.8470 | –21.8909 |
| RMS | –41.4271 | –26.4729 | –67.9001 | 34.9582 | –32.9419 |
| HCQ | –37.5839 | –250.1627 | –287.7465 | 252.7917 | –34.9548 |
ΔEele, electrostatic energy; ΔEvdW, van der Waals energy; ΔGbind, total binding energy; ΔGsol, solvation energy; ΔGgas, gas phase energy.
Some literature has highlighted crucial residues that play a vital role in the binding interaction of compounds with Mpro with inhibitors, some examples are the catalytic residues His41 and Cys145 (Zhang et al., 2020). The decomposition of the total energy into electrostatic and van der Waals (Figures 6–8) has equally enabled us to obtain vital information about more residues that contributed to the energetic interaction and stabilization of the studied compounds to the main protease.
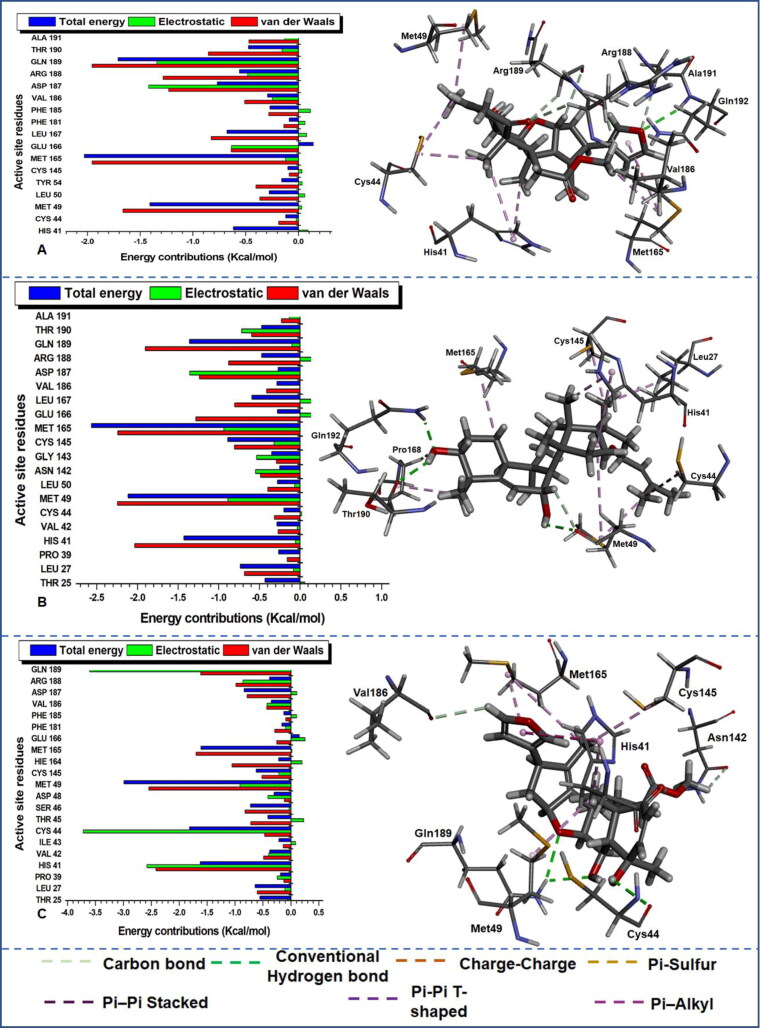
Figure 6.
Per-residue energy decomposition of active site residues and their corresponding energy contributions towards the binding and stability of (A) DCN, (B) MDC and (C) NBD. Left: Interacting residues are indicated with distinct colourations. Right: Occurring inter-molecular interactions in the active site and compounds-functional moieties.
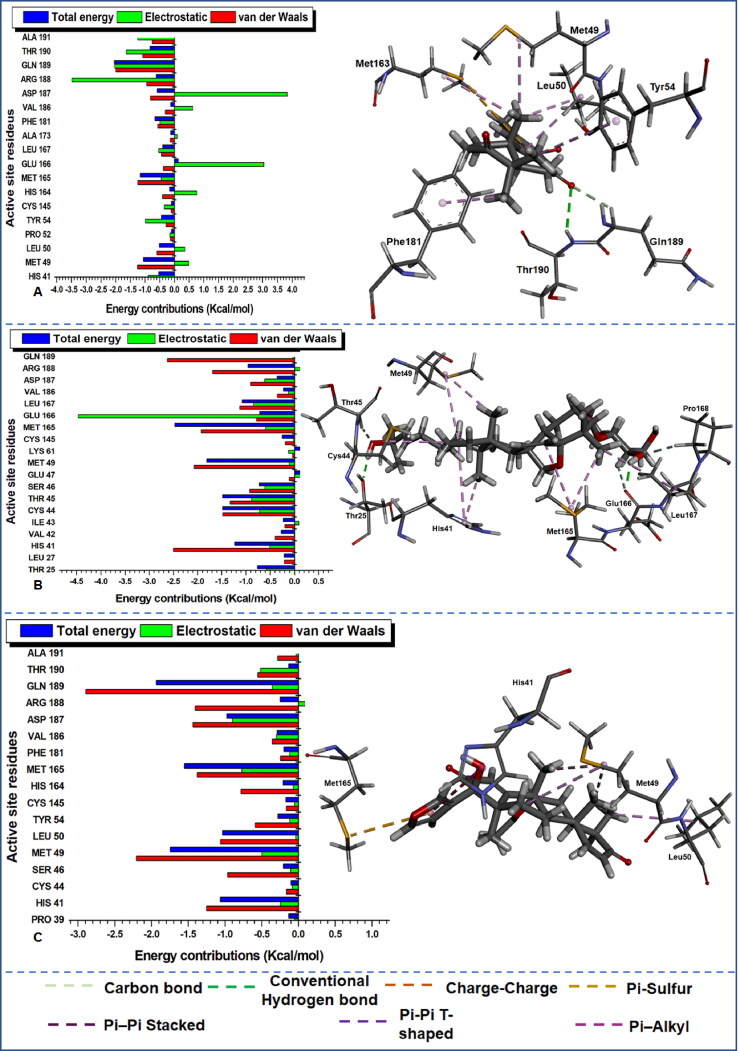
Figure 7.
Per-residue energy decomposition of active site residues and their corresponding energy contributions towards the binding and stability of (A) MGN, (B) MDF and (C) HXD. Left: Interacting residues are indicated with distinct colourations. Right: Occurring inter-molecular interactions in the active site and compounds-functional moieties.
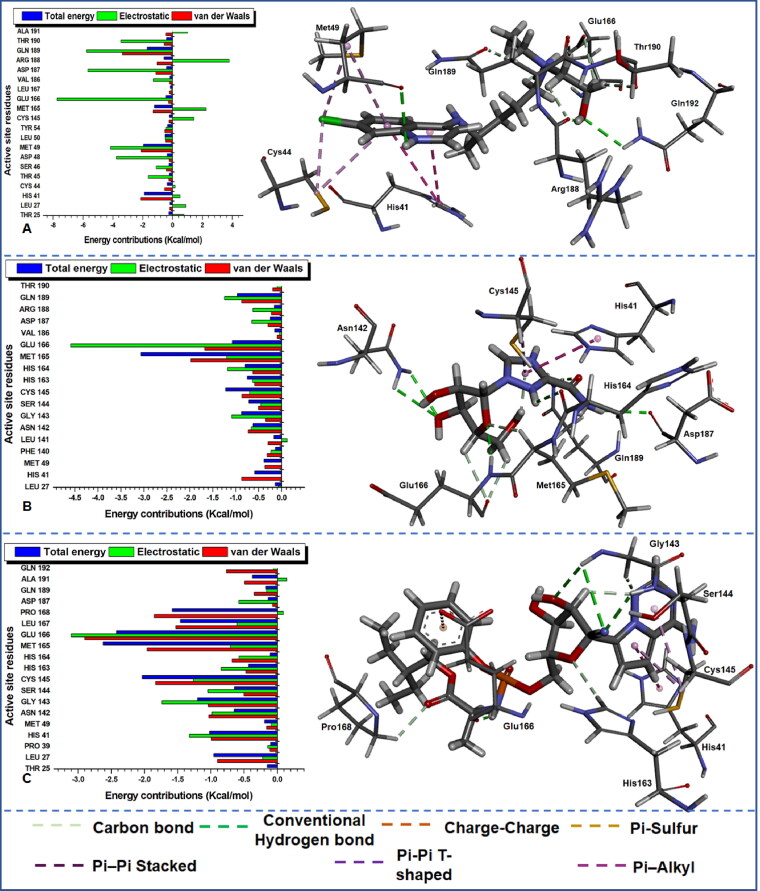
Figure 8.
Per-residue energy decomposition of active site residues and their corresponding energy contributions towards the binding and stability of (A) HCQ, (B) RBV and (C) RMS. Left: Interacting residues are indicated with distinct colourations. Right: Occurring inter-molecular interactions in the active site and compounds-functional moieties.
The estimation was essential to identify crucial active site residues and their respective energy contributions towards favourable inhibitory activities of various compounds and stabilization of the protein. From the plot of the per-residue energy contribution prominent residues with most outstanding total energy (with a threshold from 0.5 kcal/mol) drawn from van der Waals, electrostatic, Polar Solvation and Non-Polar Solvation are indicated in Table 4 with their corresponding energy values. The consistent reoccurrence of some residues in addition to the earlier mentioned catalytic residue His41 and Cys145 is suggestive of the crucial role they play in the inhibitory activity of different compounds to Mpro proteases; this will equally provide insight into the design of compounds that distinctively target these important residues. The dominance of electrostatic and van der Waals in the total energy outcome cannot equally be over-emphasized as the plot also depict the level of the contribution of the individual residues to these energy types.
Table 4.
An overview of crucial active site residues with strong energy contributions.
| Compound names | Energy contributions (kcal/mol) |
|||
|---|---|---|---|---|
| Residues | Energy value | Residues | Energy value | |
| DCN | Met165 | –2.029 | Leu167 | –0.677 |
| Gln189 | –1.711 | His41 | –0.615 | |
| Met49 | –1.408 | Arg188 | –0.558 | |
| Asp187 | –0.767 | |||
| MDC | Met165 | –2.562 | Cys145 | –0.89 |
| His41 | –1.428 | Leu167 | –0.592 | |
| Gln189 | –1.36 | Asp187 | –0.834 | |
| NBD | Met49 | –2.989 | Ser46 | –0.721 |
| Cys44 | –1.815 | Cys145 | –0.619 | |
| His41 | –1.62 | Leu27 | –0.64 | |
| Met165 | –1.608 | |||
| MGN | Gln189 | –2.045 | Arg188 | –0.615 |
| Met49 | –1.052 | Asp187 | –0.589 | |
| Met165 | –1.165 | Thr25 | –0.55 | |
| Thr190 | –0.831 | His41 | –0.533 | |
| Ala191 | –0.679 | Leu50 | –0.511 | |
| Phe181 | –0.664 | |||
| MDF | Met165 | –2.469 | Gln189 | –1.068 |
| Met49 | –1.81 | Arg188 | –0.956 | |
| Cys44 | –1.483 | Thr25 | –0.759 | |
| Thr45 | –1.48 | Ser46 | –0.723 | |
| His41 | –1.233 | Glu166 | –0.716 | |
| Leu167 | –1.077 | |||
| HXD | Gln189 | –1.936 | His41 | –1.066 |
| Met49 | –1.746 | Leu50 | –1.032 | |
| Met165 | –1.553 | Asp187 | –0.976 | |
| RBV | Met165 | –3.063 | His163 | –0.747 |
| Cys145 | –1.215 | His164 | –0.793 | |
| Glu166 | –1.068 | Ser144 | –0.712 | |
| Gln189 | –0.963 | Asn142 | –0.616 | |
| Gly143 | –0.871 | His41 | –0.587 | |
| RMS | Met165 | –2.622 | Gly143 | –1.2 |
| Glu166 | –2.422 | Leu50 | –1.032 | |
| Cys145 | –2.033 | Asn142 | –0.648 | |
| Pro168 | –1.582 | Ser144 | –0.645 | |
| Leu167 | –1.46 | |||
| HCQ | Met49 | –1.948 | Leu27 | –0.958 |
| His41 | –1.888 | Arg188 | –0.569 | |
| Gln189 | –1.705 | |||
| Met165 | –1.215 | |||
Note: The residues were colour-coded such that the once that were not absent in more than five systems were coloured. The colour coding is such that residues that were absent in five systems have five of the ID uncoloured, while the once that were absent in four had four of its ID uncoloured, the same happened for three and so on. The once that were absent in more than five systems were not coloured. The catalytic residue (Cys145) was equally double coloured to reveal where it occurred. The second catalytic residue (His41) occurred consistently.
To further unveil the interaction dynamics in protein–ligand complexes, we proceeded to visually analyse the nature and/or type of interactions that produced the strong high-affinity binding in the various systems. Different bond types indicated in the right of Figures 6–8 were noticed between the active site residues of Mpro and various ligands due to diverse constituent functional groups in the ligands that interacted concurrently.
This enabled us to identify crucial residues that played an active role in the binding interaction between the ligands and the protein. From the visual analysis, the dominant interactions include conventional hydrogen bonds, carbon bonds, charge-charge, Pi-Alky, Pi-Sulphur, Pi-Pi T-shaped and Pi-Pi stacked bonds. From the residue interactions, it can be observed that a “consistent” conventional hydrogen bond, carbon bond and Pi-Alkyl interactions were common in all the systems apart from 17-hydroxyazadiradione which did not interact with Mpro via conventional hydrogen bond.
Visual inspection in Figure 6(A) revealed that a conventional (NH–O) hydrogen bond occurred between Ala191 of Mpro and oxygen atom of Deacetylnimbinene. In another case (Figure 6(B)), a conventional (NH–O) hydrogen bond was again observed between Gln192 of Mpro and oxygen atom of momordicine whereas an oxygen and sulphur atoms in met49 of Mpro equally established another conventional (OH–O) hydrogen bond with an OH group in momordicine. In Figure 6(C), nimbandiol equally had two conventional (NH–O, OH–O) hydrogen bond; NH group in Met49 interacted with an oxygen atom in nimbandiol, while the OH group in nimbandiol interacted with an oxygen atom in Cys44. In Figure 7(A), only one conventional (NH–O) existed between oxygen atom of margolonone and NH group of main proteases. In Figure 7(B), two different OH groups in momordicoide F2 maintained conventional (OH–O) hydrogen bonds with one oxygen atom in Glu166 and Cys44.
Among the reference drugs (Figure 8), hydroxychloroquine has two conventional (NH–O) hydrogen bonds; one from the NH group in Gln192 interacting with the oxygen atom on hydroxychloroquine while another NH group from hydroxychloroquine interacted with an oxygen atom in Met49 (Figure 8(A)). Ribavirin and remdesivir have three conventional hydrogen bonds each; this is suggestive of a stronger high-affinity binding. In ribavirin (Figure 8(B)), three conventional (NH–O) hydrogen bonds occurred between this compound and main protease; NH from Asn142 and Glu166 interacted with different oxygen atoms in ribavirin, while NH in ribavirin interacted with an oxygen atom in the main protease. Finally, in Figure 8(C), NH–O and NH–N conventional interactions were observed between remdesivir and main protease. Here NH from Gly143 and Ser144 interacted with different oxygen atoms in remdesivir while another NH group in remdesivir interacted with an oxygen atom in Glu166. Visual observation indicates that conventional hydrogen bond did not occur in 17-hydroxyazadiradione, but trends from the occurrence of this strong high affinity short distant bonding interactions suggest that it is required for the stabilization of the protein–ligand complexes. Hence further structural optimization is needed to improve the binding dynamics of 17-hydroxyazadiradione to Mpro active site. The types of reoccurring bonding interaction are indicative of variations in ligand positioning and proximity to active site residues of the proteins. Table 4 gives a brief description of the residues that played an active role in the binding interaction of this compounds to Mpro and will pave way for the design of novel inhibitors that will specifically target this enzyme. From the table, Met165 consistently interacted with all the inhibitors and could be very vital residue for the design of novel inhibitors
In silico evaluation of biological activities, toxicities and pharmacokinetic properties
The pharmacological profiles of the natural products were when compared with the minimum and maximum acceptable range and those of reference drugs by predicting their pharmacokinetic (ADMET) properties as indicated in Tables 5 and 6. It is worth mentioning that the LD50 as a parameter is very vital in dictating variable toxicities of compounds that are taken via oral routes (Lipinski, 2004). Hence elevated LD50 depicts an increase in toxicity whereas a decline in LD50 correlated with a decrease in toxicity(B).
Table 5.
In silico predicted toxicity properties of the investigated compounds.
| Compounds names | Predicted pharmacokinetic properties |
||
|---|---|---|---|
| LD50 (mg/kg) | Hepatoxicity | Cytotoxicity | |
| DCN | 600 | 0.72 | 0.75 |
| MDC | 186 | 0.65 | 0.82 |
| NBD | 600 | 0.67 | 0.77 |
| MGN | 570 | 0.68 | 0.83 |
| MDF | 1190 | 0.89 | 0.52 |
| HXD | 1000 | 0.76 | 0.80 |
| RBV | 2700 | 0.52 | 0.72 |
| RMS | 1000 | 0.56 | 0.55 |
| HCQ | 1240 | 0.94 | 0.72 |
Table 6.
Comparative evaluation of physicochemical properties.
| Compounds names | Pharmacological properties |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MW (Da) | Log P | Log S (mol/L) | TPSA (A2) | HBA | HBD | Rotatable bonds (kcal/mol/heavy atom) | LE | LLE | LELP | |
| DCN | 440.22 | 3.19 | –3.23 | 85.98 | 6 | 1 | 4 | 0.0425 | –2.469 | 81.418 |
| MDC | 472.36 | 5.60 | –5.50 | 77.75 | 4 | 3 | 5 | – | – | – |
| NBD | 456.21 | 2.25 | –2.27 | 106.2 | 7 | 2 | 4 | 0.285 | 1.337 | 15.597 |
| MGN | 314.15 | 4.13 | –4.26 | 71.44 | 4 | 1 | 1 | 0.421 | 0.624 | 13.956 |
| MDF | 618.41 | 4.93 | –4.58 | 128.84 | 8 | 5 | 7 | 0.209 | –2.539 | 92.3 |
| HXD | 466.24 | 3.63 | –3.64 | 93.81 | 6 | 1 | 3 | 0.279 | –2.520 | 73.722 |
| RBV | 244.08 | –2.08 | –0.84 | 143.73 | 7 | 5 | 3 | 0.604 | 7.375 | –8.261 |
| RMS | 602.23 | 2.44 | –2.68 | 203.5 | 12 | 5 | 14 | 0.231 | 0.611 | 10.186 |
| HCQ | 335.18 | 3.69 | –3.98 | 48.38 | 3 | 2 | 9 | 0.429 | 0.666 | 13.786 |
| Acceptable threshold | ˂500 Da | ˂5 | 0 to −6 | ≤140 | ≤10 | ≤5 | ˂10 | >∼0.3 | LLE > ∼5 | −10–10 |
We performed the toxicity class and labelling study with ProTox webserver; it uses the globally harmonized system for characterizing the toxicity class and labelling of chemicals (Banerjee et al., 2018). As estimated and indicated in Table 5. Ribavirin an FDA reference drug has the highest LD50 which could be suggestive of high oral toxicity. However, ribavirin is reported with good bioactivity, bioavailability and no reported case of toxicity. The observation made from ribavirin is indicative of the promising attributes of the studied compounds as possessing the good quality to pass toxicity test. The estimation of cytotoxicity of the compounds with ProTox revealed a hepatotoxicity and cytotoxicity values similar and close to those of the reference drugs thus revealing their respective safety state at consumption. This hepatotoxicity and cytotoxicity values corroborates with the findings in LD50.
A good drug candidate is expected to have an MW threshold ˂500 Da (g/mol) based on Lipinski's RO5 (Lipinski, 2004; Lipinski et al., 1997; Omran & Rauch, 2014; Veber et al., 2002). Several studies have revealed a link between the molecular weight (MW) of drugs and its toxicity, wherein the higher the MW of a drug, the higher the toxicity whereas compounds with lower MW produces reduced toxicity (Chapman et al., 2013). Therefore lower molecular weight is better (Omran & Rauch, 2014) because high MW in drugs decreases the concentration of the compounds at the intestinal epithelium surface and thus diminish. From Table 6, all the compounds obeyed Lipinski's RO5 of MW threshold ˂500 Da (g/mol), but, surprisingly, momordicoside F2 had a MW more than 500 Da and remdesivir which is an already approved drug that has no reported case of toxicity equally had an MW more than 500 Da. This could be suggesting a need for optimization
The Log P gives an idea of the hydrophobicity of chemical entities, it is universally defined as the negative of the logarithm of the partition coefficient between n-octanol and water (Coctanol/Cwater) (Chapman et al., 2013). Hence an increase in Log P is an indication of a decline in aqueous solubility, which results in a reduction in absorption. Previous reports have revealed the chemical entities with Log P range of −0.4 to 5.6 tend to exhibit a high level of absorption while values greater than 5.6 and quite lower than −0.4 possess low hydrophilicity, poor permeability and absorption (Llorach-Pares et al., 2017; Remko, 2009; Remko et al., 2011). As presented in Table 6, all the natural products and the approved drugs exhibited an acceptable level of solubility which will enhance absorption and distribution. Although momordicine has a value that is slightly higher than the acceptable threshold (Sander et al., 2015), the value is within the range that is suggested to tend towards being well absorbed. This indicates that these studied compounds did not violate Lipinski’s rule of 6 (Log P < 5). We took another step to use Log S parameter to further evaluate the aqueous solubility of the studied compounds. The estimation is very crucial because it determines the oral bioavailability of drugs in line with membrane permeability (Bennion et al., 2017). Studies available on about 95% of existing drugs has revealed the acceptable threshold for Log S to be between 0 to −6. A look at Table 6, Section on Log S indicates that all the studied compounds were within the suggested acceptable range for Log S, this finding goes further corroborate the deductions made from the evaluation of Log P. An important observation to be noted is the value of Log P and Log S of momordicine. Though it is said to be within the range of chemical entities that are with the range described to tend to exhibit a high level of absorption. However, it is slightly above the acceptable range for Log P and very close to the limit for the acceptable range for Log S. This is there suggestive of the need for structural optimization to be done on this compound.
The topological polar surface area (TPSA) parameter totals the polar atoms at the surface which are primarily oxygen and nitrogen in addition to the hydrogen atoms that are attached to them. This parameter predicts cell permeation ability of chemical compounds; it is therefore put that the lower the TPSA value the better (Ertl et al., 2000; Prasanna & Doerksen, 2009). The metrics describes the size and volume of compounds which is an indication of physiological transport across the tight junction of the lipid bilayer membrane. These lipid bilayer membrane transport routes include the GIT and the blood–brain barrier (Shityakov et al., 2013). Therefore, the increase in TPSA is suggested to diminish the transportation capability of drugs which in turn affect their biological activities (Daga et al., 2018). The table revealed that all the selected natural products adhered to the acceptable threshold TPSA value of ≤140Å2. One is left to wonder why two of the approved drugs ribavirin and remdesivir with TPSA values of 143.73 and 203.57, respectively, had values quite above that of the threshold and are still able to permeate cells and give reasonable efficacy. We suspect that there could have been other structural modification which was implemented in this compound; however, this observation is outside the scope of this present study. The results that we have obtained however suggests that the studied compounds possessed better physiological transport qualities than the approved drugs. Hydrogen bonding is connected with constituent oxygen and nitrogen moieties and largely related with TPSA which is an indirect indication of polarity and the capacity of hydrogen bonding(Di & Kerns, 2006; Prasanna & Doerksen, 2009). The descriptors for hydrogen bonding are several hydrogen bond donors (HBD) and acceptors (HBA) present in a molecule. These parameters have been extensively employed in the evaluation of drug-likeness of compounds. Lipinski's RO5 states that for a drug to be orally active, it must have an HBD count of ≤5 and HBA of ≤10 (Doak et al., 2014). As predicted from the table, all the natural products and the approved drugs pass the test for oral bioactivity, however, remdesivir with HBD of 12 once again left an impression for structural optimization.
The drug-likeness of the compounds under investigation was further assessed using LE, LLE and LELP. These metrics have previously been employed to optimize ligands and equally identify ligands with improved binding efficiency for physiological targets (Abad-Zapatero, 2007; Hopkins et al., 2014; Johnson et al., 2018). The suggested acceptable range for potential drug candidate for each of the parameters are LE > ∼0.3 kcal/mol/heavy atom, LLE > ∼5 while optimal drug LELP value ranges between −10 and 10 (Abad-Zapatero, 2007; Hopkins et al., 2014; Johnson et al., 2018). Most of the compounds were within the indicated range for some of the investigated parameters, however, a few exceptions exist which is suggestive of further structural optimization. As indicated, deacetylnimbinene fell short of all the studied parameters, this a clear indication of a need for critical structural optimization. On an additional note, nimbandiol has an LE value that is close to the acceptable >∼0.3, but the LLE and LELP values are somehow out of the required range thus suggesting the need for structural improvement. Momordicoside and remdesivir exhibited LE value that is slightly lower than the acceptable range. In the case of LLE and LELP, only ribavirin had LLE and LELP values within the suggested range. But of least importance is lack of metrics for momordicine coupled with the shortcomings on some of the required qualities, this, therefore, raises a need for additional structural optimization to be carried to improve the drug-likeness properties of these compounds.
Conclusion
Coronaviruses main protease (Mpro) is an essential protein critically required for proteolytic maturation of the coronavirus. In this study, 86 compounds and 3 FDA drugs were screened from M. charantia L. and Azadirachta indica for possible therapy against SARS-CoV-2 Mpro using structure-based drug design and MD approach. Analysis from the Root mean square fluctuations revealed that the higher active site RMSF of the phytocompounds when compared with apo Mpro corroborates with the reported open/close and twisting mechanism of ligand inactivation of the viral protease. Furthermore, two of the Phyto ligands momordicine and momordiciode F2, gave binding free energy that is higher when compared with FDA drugs (Ribavirin, remdesivir and hydroxychloroquine). Analysis of the per-residue additional showed that studied compounds interact with these key amino acid residues in the active site of the main protease, suggesting that these phytocompounds could emerge as ideal candidate’s inhibitors against SARS-CoV-2 Mpro and another virus protease. Pharmacokinetics suggests that the phytocompounds possessed better physiological transport qualities when compared with the reference drugs. Our results present urgent attention since there are no drugs obtainable against SARS-CoV-2. These findings may provide insight for developing new treatments for SARS-CoV-2.
Supplementary Material
Acknowledgements
C.U.I. and E.A.I. are thankful to CHPC (http://www.chpc.ac.za) for operational and infrastructural support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abad-Zapatero, C. (2007). Ligand efficiency indices for effective drug discovery. Expert Opinion on Drug Discovery, 2(4), 469–488. 10.1517/17460441.2.4.469 [DOI] [PubMed] [Google Scholar]
- Banerjee, P., Eckert, A. O., Schrey, A. K., & Preissner, R. (2018). ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Research, 46(W1), W257–W263. 10.1093/nar/gky318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennion, B. J., Be, N. A., McNerney, M. W., Lao, V., Carlson, E. M., Valdez, C. A., Malfatti, M. A., Enright, H. A., Nguyen, T. H., Lightstone, F. C., & Carpenter, T. S. (2017). Predicting a drug's membrane permeability: A computational model validated with in vitro permeability assay data. The Journal of Physical Chemistry. B, 121(20), 5228–5237. 10.1021/acs.jpcb.7b02914 [DOI] [PubMed] [Google Scholar]
- Chapman, K. L., Holzgrefe, H., Black, L. E., Brown, M., Chellman, G., Copeman, C., Couch, J., Creton, S., Gehen, S., Hoberman, A., Kinter, L. B., Madden, S., Mattis, C., Stemple, H. A., & Wilson, S. (2013). Pharmaceutical toxicology: Designing studies to reduce animal use, while maximizing human translation. Regulatory Toxicology and Pharmacology, 66(1), 88–103. 10.1016/j.yrtph.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Daga, P. R., Bolger, M. B., Haworth, I. S., Clark, R. D., & Martin, E. J. (2018). Physiologically based pharmacokinetic modeling in lead optimization. 2. Rational bioavailability design by global sensitivity analysis to identify properties affecting bioavailability. Molecular Pharmaceutics, 15(3), 831–839. 10.1021/acs.molpharmaceut.7b00973 [DOI] [PubMed] [Google Scholar]
- Di, L., & Kerns, E. H. (2006). Biological assay challenges from compound solubility: Strategies for bioassay optimization. Drug Discovery Today, 11(9-10), 446–451. 10.1016/j.drudis.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Doak, B. C., Over, B., Giordanetto, F., & Kihlberg, J. (2014). Oral druggable space beyond the rule of 5: Insights from drugs and clinical candidates. Chemistry & Biology, 21(9), 1115–1142. 10.1016/j.chembiol.2014.08.013 [DOI] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592. 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel, I. A., Olotu, F., Agoni, C., & Soliman, M. E. (2019. a). Broadening the horizon: Integrative pharmacophore-based and cheminformatics screening of novel chemical modulators of mitochondria ATP synthase towards interventive Alzheimer's disease therapy. Medical Hypotheses, 130, 109277. 10.1016/j.mehy.2019.109277 [DOI] [PubMed] [Google Scholar]
- Emmanuel, I. A., Olotu, F. A., Agoni, C., & Soliman, M. E. (2019. b). Deciphering the 'Elixir of Life': dynamic perspectives into the allosteric modulation of mitochondrial ATP synthase by J147, a novel drug in the treatment of Alzheimer's disease. Chemistry & Biodiversity, 16(6), e1900085. 10.1002/cbdv.201900085 [DOI] [PubMed] [Google Scholar]
- Ertl, P., Rohde, B., & Selzer, P. (2000). Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. Journal of Medicinal Chemistry, 43(20), 3714–3717. 10.1021/jm000942e [DOI] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis coronaviruses (pp. 1–23). Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- García-Fernández, R., Ziegelmüller, P., González, L., Mansur, M., Machado, Y., Redecke, L., Hahn, U., Betzel, C., & de Los Ángeles Chávez, M. (2016). Two variants of the major serine protease inhibitor from the sea anemone Stichodactyla helianthus, expressed in Pichia pastoris. Protein Expression and Purification, 123, 42–50. 10.1016/j.pep.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, A. L., Keserü, G. M., Leeson, P. D., Rees, D. C., & Reynolds, C. H. (2014). The role of ligand efficiency metrics in drug discovery. Nature Reviews Drug Discovery, 13(2), 105–121. 10.1038/nrd4163 [DOI] [PubMed] [Google Scholar]
- Husain, A., Ahmad, A., Khan, S. A., Asif, M., Bhutani, R., & Al-Abbasi, F. A. (2016). Synthesis, molecular properties, toxicity and biological evaluation of some new substituted imidazolidine derivatives in search of potent anti-inflammatory agents. Saudi Pharmaceutical Journal, 24(1), 104–114. 10.1016/j.jsps.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeji, C. U. (2020). Molecular dynamics and DFT study on the structure and dynamics of N-terminal domain HIV-1 capsid inhibitors. Molecular Simulation, 46(1), 62–70. 10.1080/08927022.2019.1674850 [DOI] [Google Scholar]
- Johnson, T. W., Gallego, R. A., & Edwards, M. P. (2018). Lipophilic efficiency as an important metric in drug design. Journal of Medicinal Chemistry, 61(15), 6401–6420. 10.1021/acs.jmedchem.8b00077 [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kholmurodov, K., Smith, W., Yasuoka, K., Darden, T., & Ebisuzaki, T. (2000). A smooth‐particle mesh Ewald method for DL_POLY molecular dynamics simulation package on the Fujitsu VPP700. Journal of Computational Chemistry, 21(13), 1187–1191. [DOI] [Google Scholar]
- Krishnan, K. A., & Anirudhan, T. (2002). Uptake of heavy metals in batch systems by sulfurized steam activated carbon prepared from sugarcane bagasse pith. Industrial & Engineering Chemistry Research, 41(20), 5085–5093. [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today Technologies, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. (1997). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 23(1-3), 3–25. 10.1016/S0169-409X(96)00423-1 [DOI] [PubMed] [Google Scholar]
- Liu, J.-Q., Chen, J.-C., Wang, C.-F., & Qiu, M.-H. (2009). New cucurbitane triterpenoids and steroidal glycoside from Momordica charantia. Molecules, 14(12), 4804–4813. 10.3390/molecules14124804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorach-Pares, L., Nonell-Canals, A., Sanchez-Martinez, M., & Avila, C. (2017). Computer-aided drug design applied to marine drug discovery: Meridianins as Alzheimer’s disease therapeutic agents. Marine Drugs, 15(12), 366. 10.3390/md15120366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-López, E., Naveja, J. J., & Medina-Franco, J. L. (2019). DataWarrior: An evaluation of the open-source drug discovery tool. Expert Opinion on Drug Discovery, 14(4), 335–341. 10.1080/17460441.2019.1581170 [DOI] [PubMed] [Google Scholar]
- Lu, H., Stratton, C. W., & Tang, Y. W. (2020) Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. Journal of Medical Virology, 92, 401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad, A.-N., Syed, S., Andreas, D. (2016). Medicine 4.0. Proceedings of the Ninth International Conference on Mobile Computing and Ubiquitous Networking. [Google Scholar]
- Mukherjee, P. K., Kumar, V., & Houghton, P. J. (2007). Screening of Indian medicinal plants for acetylcholinesterase inhibitory activity. Phytotherapy Research. An International Journal Devoted to Pharmacological and Toxicological Evaluation of Natural Product Derivatives, 21(12), 1142–1145. [DOI] [PubMed] [Google Scholar]
- Munsamy, G., Ramharack, P., & Soliman, M. E. (2018). Egress and invasion machinery of malaria: An in-depth look into the structural and functional features of the flap dynamics of plasmepsin IX and X. RSC Advances, 8(39), 21829–21840. 10.1039/C8RA04360D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olotu, F. A., Agoni, C., Adeniji, E., Abdullahi, M., & Soliman, M. E. (2019). Probing gallate-mediated selectivity and high-affinity binding of epigallocatechin gallate: A way-forward in the design of selective inhibitors for anti-apoptotic bcl-2 proteins. Applied Biochemistry and Biotechnology, 187(3), 1061–1080. 10.1007/s12010-018-2863-7 [DOI] [PubMed] [Google Scholar]
- Omran, Z., & Rauch, C. (2014). Acid-mediated Lipinski’s second rule: Application to drug design and targeting in cancer. European Biophysics Journal, 43(4-5), 199–206. 10.1007/s00249-014-0953-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, A., MacCallum, J. L., Brini, E., Simmerling, C., & Dill, K. A. (2015). Grid-based backbone correction to the ff12SB protein force field for implicit-solvent simulations. Journal of Chemical Theory and Computation, 11(10), 4770–4779. 10.1021/acs.jctc.5b00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongthanapisith, V., Ikuta, K., Puthavathana, P., & Leelamanit, W. (2013). Antiviral protein of Momordica charantia L. inhibits different subtypes of Influenza A. Evidence-Based Complementary and Alternative Medicine, 2013, 1–6. 10.1155/2013/729081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna, S., & Doerksen, R. (2009). Topological polar surface area: A useful descriptor in 2D-QSAR. Current Medicinal Chemistry, 16(1), 21–41. 10.2174/092986709787002817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, J. M., Bridgen, J. R., Cummings, D. A., Ho, A., & Jewell, C. P. (2020). Novel coronavirus 2019-nCoV: Early estimation of epidemiological parameters and epidemic predictions. MedRxiv. 10.1101/2020.01.23.20018549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remko, M. (2009). Molecular structure, lipophilicity, solubility, absorption, and polar surface area of novel anticoagulant agents. Journal of Molecular Structure: Theochem, 916(1-3), 76–85. 10.1016/j.theochem.2009.09.011 [DOI] [Google Scholar]
- Remko, M., Boháč, A., & Kováčiková, L. (2011). Molecular structure, pKa, lipophilicity, solubility, absorption, polar surface area, and blood brain barrier penetration of some antiangiogenic agents. Structural Chemistry, 22(3), 635–648. 10.1007/s11224-011-9741-z [DOI] [Google Scholar]
- Roe, D. R., & Cheatham, T. E. (2013). Cheatham 3rd TE. 2013. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Sander, T., Freyss, J., von Korff, M., & Rufener, C. (2015). DataWarrior: An open-source program for chemistry aware data visualization and analysis. Journal of Chemical Information and Modeling, 55(2), 460–473. 10.1021/ci500588j [DOI] [PubMed] [Google Scholar]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Seifert, E. (2014). OriginPro 9.1: Scientific data analysis and graphing software: Software review. ACS Publications. [DOI] [PubMed] [Google Scholar]
- Shityakov, S., Neuhaus, W., Dandekar, T., & Förster, C. (2013). Analysing molecular polar surface descriptors to predict blood-brain barrier permeation. International Journal of Computational Biology and Drug Design, 6(1/2), 146–156. 10.1504/IJCBDD.2013.052195 [DOI] [PubMed] [Google Scholar]
- Veber, D. F., Johnson, S. R., Cheng, H.-Y., Smith, B. R., Ward, K. W., & Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. Journal of Medicinal Chemistry, 45(12), 2615–2623. 10.1021/jm020017n [DOI] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2001). Antechamber: An accessory software package for molecular mechanical calculations. Journal of the American Chemical Society., 222, U403. [Google Scholar]
- Wu, F., Zhao, S., & Yu, B. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov, A., Zagribelnyy, B., Zhebrak, A., Aladinskiy, V., Terentiev, V., Vanhaelen, Q., Bezrukov, D. S., Polykovskiy, D., Shayakhmetov, R., Filimonov, A., Bishop, M., McCloskey, S., Leija, E., Bright, D., Funakawa, K., Lin, Y.-C., Huang, S.-H., Liao, H.-J., Aliper, A., & Ivanenkov, Y. (2020). Potential non-covalent SARS-CoV-2 3C-like protease inhibitors designed using generative deep learning approaches and reviewed by human medicinal chemist in virtual reality. ChemRxiv.
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., & Madura, J. D. (2004). Relative free energy of binding and binding mode calculations of HIV‐1 RT inhibitors based on dock‐MM‐PB/GS. Proteins: Structure, Function, and Bioinformatics, 57(3), 493–503. 10.1002/prot.20223 [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Wang, Y., & Bryant, S. H. (2009). Computational analysis of the cathepsin B inhibitors activities through LR-MMPBSA binding affinity calculation based on docked complex . Journal of Computational Chemistry, 30(14), 2165–2175. 10.1002/jcc.21214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.