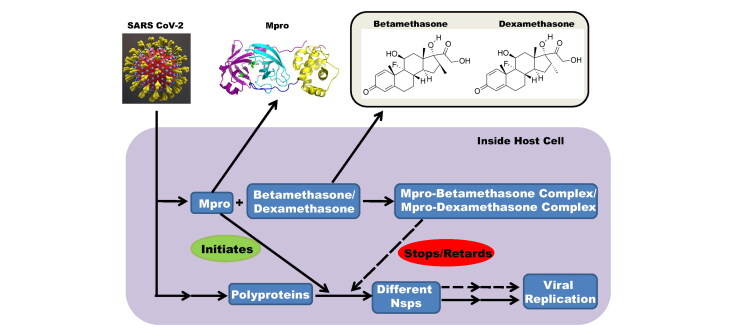
Abstract
The outbreak of COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS CoV-2), represents a pandemic threat to global public health. To date, ∼530,000 people died of this disease worldwide. Presently, researchers/clinicians are adopting the drug repurposing strategy to combat this disease. It has also been observed that some repurposed anti-viral drugs may serve as potent inhibitors of SARS CoV-2 Mpro, a key component of viral replication. Apart from these anti-viral drugs, recently dexamethasone (an important corticosteroid) is effectively used to treat COVID-19 patients. However, the mechanism behind the mode of its action is not so clear. Additionally, the effect of other well-known corticosteroids to control this disease by inhibiting the proteolytic activity of Mpro is ambiguous. In this study, we have adopted computational approaches to understand these aspects. Six well-known corticosteroids (cortisone, hydrocortisone, prednisolone, methylprednisolone, betamethasone and dexamethasone) and two repurposed drugs (darunavir and lopinavir) against COVID-19 were subjected for molecular docking studies. Two of them (betamethasone and dexamethasone) were selected by comparing their binding affinities with selected repurposed drugs toward Mpro. Betamethasone and dexamethasone interacted with both the catalytic residues of Mpro (His41 and Cys145). Molecular dynamics studies further revealed that these two Mpro-corticosteroid complexes are more stable, experience less conformational fluctuations and more compact than Mpro-darunavir/lopinavir complexes. These findings were additionally validated by MM-GBSA analysis. This study provides corroboration for execution of anti-COVID-19 activity of dexamethasone. Our study also emphasizes on the use of another important corticosteroid (betamethasone) as potential therapeutic agent for COVID-19 treatment.
Keywords: COVID-19, SARS CoV-2 main protease, docking and molecular dynamics simulation, corticosteroids, dexamethasone
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
A novel coronavirus disease identified as COVID-19 has been declared as a global pandemic by the World Health Organization (WHO) (Cucinotta & Vanelli, 2020). Till now, there have been more than 11.3 million reported cases and more than 530,000 deaths around the globe (https://www.worldometers.info/coronavirus/). The epicenter of the disease was China, but gradually it spread worldwide and infected people from 215 countries (Zhu et al., 2020). The United States remains the most affected country in terms of the total number of cases, followed by Brazil, Russia and India. COVID-19 is a highly contagious disease and can be easily transmitted through nasal droplets and close contacts (Cascella et al., 2020). Similar to other coronavirus infections, the most common symptoms of COVID-19 are high fever, cough, shortness of breath and fatigue (Chen et al., 2020; Guan et al., 2020). Some cases of this disease are life-threatening, thus it possesses a great threat to global health and safety.
The etiological agent of COVID-19 is severe acute respiratory syndrome corona virus-2 (SARS CoV-2). It belongs to the order Nidovirales and genus β-coronavirus (Zheng, 2020). It is a non-segmented, enveloped, positive-sense, single-stranded RNA virus with the genome size of ∼29.9 kb (Wu et al., 2020; Zhu et al., 2020). This large RNA genome of SARS CoV-2 is comprised of ∼30,000 nucleotides that encode two overlapping polyproteins required for viral replication and transcription (Grum-Tokars et al., 2008; Marra et al., 2003; Thiel et al., 2003). The functional proteins are released by extensive proteolysis of the polyproteins by two proteases namely the papain-like protease (PLpro) and the 3C-like protease (3CLpro) (Grum-Tokars et al., 2008; Marra et al., 2003; Thiel et al., 2003). 3CLpro is also named as the main protease (Mpro) as it executes the maximum number of proteolytic cleavages (11 sites) within the polyprotein (Harcourt et al., 2004; Thiel et al., 2003). Mpro is reported to be a cysteine protease with a molecular weight of 33.8 kDa (Blanchard et al., 2004; Dai et al., 2020; Jin et al., 2020; Osman et al., 2020). Each protomer of the homodimeric SARS CoV-2 Mpro protein have three domains – domain I (amino acid residues 8–101), domain II (amino acid residues 102–184) and domain III (amino acid residues 201–303) (Jin et al., 2020). Domains II and III are connected via a long loop (amino acid residues 185–200). Among them, domains I and II are consist of β-barrels while, domain III has predominantly α-helix structure (Jin et al., 2020). The catalytic site/active site/substrate-binding site comprising of His-Cys dyad i.e. cysteine (Cys145) and histidine (His41) amino acid moieties, are located at the cleft of domains I and II (Blanchard et al., 2004; Dai et al., 2020; Jin et al., 2020; Osman et al., 2020). Cysteine145 serves as a common nucleophile and plays a vital role in the proteolytic functioning of Mpro (Anand et al., 2003; Chou et al., 2003; Hsu et al., 2005). As Mpro plays a vital role in polyprotein processing and virus maturation, it is considered to be an important drug target (Anand et al., 2003; Yan et al., 2003). Besides, the absence of similar protease in humans makes it an automatic choice of drug target for designing antiviral drugs against SARS CoV-2 (Kim et al., 2016). Recently Yang and colleagues have reported the structure of Mpro, co-crystallized with an inhibitor N3 by X-ray crystallography (Jin et al., 2020). They have also identified six Mpro inhibitors with IC50 values of enzyme inhibition in the range of 0.67–21.4 μM by using a fluorescence resonance energy transfer (FRET) based high throughput enzyme activity assay. Thus, they have revealed, both the structures of substrate-binding site of Mpro as well as structure-based approach for designing potential drug molecules against COVID-19. Based on these, natural products and phytochemicals have been studied to find an effective inhibitor of Mpro (Bhardwaj et al., 2020; Das et al., 2020; Ghosh et al., 2020a, 2020b; Gorla et al., 2020; Gurung et al., 2020; Joshi et al., 2020a, 2020b; Mazzini et al., 2020).
Another approach that has been opted for the treatment of COVID-19 is drug repurposing (Baby et al., 2020; Beck et al., 2020; Bharadwaj et al., 2020; Hage-Melim et al., 2020; Hakmi et al., 2020; Jimenez-Alberto et al., 2020; Kandeel & Al-Nazawi, 2020; Kumar et al., 2020). Drug repurposing is commonly employed to identify potential drugs against different diseases. It has been achieved massive attention for the ability to reuse drugs that are already used for the treatment of various diseases, besides the specific diseases for which those were originally developed. Many drugs have multiple targets and several diseases share a common overlapping molecular and biochemical pathways (Hodos et al., 2016). In such cases, reusing drugs for more than one purpose and finding their new uses can significantly reduce the cost, time and risk of the drug development process (Xue et al., 2018). This concept of drug repurposing has been used for well-known diseases successfully for many years. For example, favipiravir, an approved influenza virus drug, and sofosbuvir, a hepatitis C virus drug have a strong repurposing potential against Ebola and Zika viruses (Mercorelli et al., 2018). Different drugs such as nelfinavir, lopinavir, oseltamivir, atazanavir and ritonavir have been used to cure MERS and SARS (Dobson et al., 2015; Lv et al., 2015). Similarly, several drug repurposing options are being considered and are under investigation to control the COVID-19, as there is an urgent requirement for an effective drug or combination of drugs to combat this disease. Several efforts were executed to design novel inhibitors or employ drug repurposing approach to identify anti-COVID-19 drugs, which can act as promising inhibitors against coronavirus protease (Baby et al., 2020; Beck et al., 2020; Bharadwaj et al., 2020; Hage-Melim et al., 2020; Hakmi et al., 2020; Jimenez-Alberto et al., 2020; Kandeel & Al-Nazawi, 2020; Kumar et al., 2020).
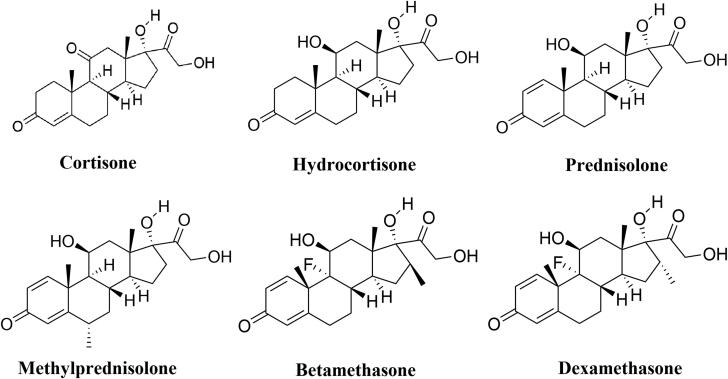
Recently, a large-scale trial conducted by a group of Oxford University revealed that dexamethasone, a repurposed drug has shown to reduce the mortality rate of COVID-19 patients (Horby et al., 2020). They also found that dexamethasone has reduced the deaths by about one-third, in patients receiving invasive mechanical ventilation while by about one-fifth, in patients receiving oxygen without invasive mechanical ventilation. But how dexamethasone abated the death of COVID-19 patients is far from clear. In fact, dexamethasone is widely used for the treatment of various inflammatory diseases, which include severe allergies, some types of nausea and vomiting, arthritis, swelling of the brain and spinal cord, severe asthma and for breathing difficulties in newborn babies (Henzi et al., 2000; Tang et al., 2014; Yoder et al., 1991). Dexamethasone is also reported to be used for the treatment of various viral infections (Clement et al., 2011; Turner et al., 2014; Yim et al., 2006). Dexamethasone belongs to a class of drugs known as corticosteroids. Other well-known members of this corticosteroids class are cortisone, hydrocortisone, prednisolone, methylprednisolone and betamethasone. All these corticosteroids are broadly used to treat a range of conditions related to inflammation (Barnes, 2006). These corticosteroids are also employed for effective treatment related to various viral diseases (Gagyor et al., 2019; Sullivan et al., 2016; Thaera et al., 2010). Cortisone and hydrocortisone are reported to inhibit the multiplication of influenza A & influenza B virus and herpes simplex virus (Kilbourne, 1957; Wheeler et al., 1961). Prednisolone is known to reduce the viral upper respiratory tract infections in children (Abeyagunawardena & Trompeter, 2008). Methylprednisolone is also used in case of various viral diseases such as H1N1 infection, H7N9 infection, influenza B, cytomegalovirus infection, etc. (Confalonieri et al., 2010; Emery et al., 2000; Liu et al., 2019; Spoto et al., 2019; Wright & O'Driscoll, 2005). The clinical efficacy of betamethasone is described against human T-cell lymphotropic virus type I (HTLV-I)-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (Alberti et al., 2011). Dexamethasone and other corticosteroid drugs are previously used for the treatment of other coronavirus diseases such as SARS and MERS (Arabi et al., 2018; Yu et al., 2004). But whether these corticosteroid drugs can inhibit the propagation of SARS CoV-2 is not clear still now. Furthermore, whether these drugs can bind to the substrate-binding site/catalytic site of the main protease is unexplained. In this current study, we have undertaken a thorough attempt to find out the potency of above discussed six corticosteroid drugs (cortisone, hydrocortisone, prednisolone, methylprednisolone, betamethasone and dexamethasone) (Figure 1) to bind and inhibit the catalytic activity of Mpro using in silico docking studies, molecular dynamics simulations and MM-GBSA analysis. This study reports betamethasone and dexamethasone as more effective inhibitors of SARS CoV-2 Mpro than previously recommended repurposed drugs (darunavir and lopinavir).
Figure 1.
Chemical structure of corticosteroid drugs. The two-dimensional structure of six corticosteroid drugs with their respective names.
2. Materials and methods
2.1. Preparation of the ligands
The structures of six corticosteroid drugs (cortisone, hydrocortisone, prednisolone, methylprednisolone, betamethasone and dexamethasone) were downloaded from the PubChem crystal database server in SDF format (https://pubchem.ncbi.nlm.nih.gov). Using the PyMol, all the SDF files were converted to PDB format after that each of the corticosteroid drug structures was optimized with B3LYP/6-31G* basis set by using Gaussian09 software (Frisch et al., 2009). Autodock Tools were then used for the generation pdbqt files.
2.2. Preparation of Mpro
The crystal structure of the SARS CoV-2 Mpro was taken from the RCSB Protein Data Bank (http://www.rcsb.org) (PDB ID: 6LU7) (Jin et al., 2020). After correcting the improper bonds, missing hydrogens, side-chain anomalies, etc. (if any), the structure file was inserted into AutoDock Tools and pdbqt file format of Mpro was obtained (Morris et al., 2008, 2009).
2.3. Molecular docking
AutoDockVina was used to perform docking studies to get docked posses with binding energies of Mpro with two anti-HIV drugs and six corticosteroid drugs (Morris et al., 2008, 2009). As per the position of active site region, the grid box was assigned with a 10.0 Å radius throughout the initial inhibitor, where the ligands can easily be fitted and which covers the entire active site pocket. To obtain different docked conformations, the same grid box size and other parameters were used for docking studies of two anti-HIV drugs along with all the six corticosteroid drugs. We have chosen the conformations with the lowest root mean square deviation (RMSD) values along with the highest Vina score for Mpro and six corticosteroid drug complexes. The output from AutoDock Vina was rendered with DS visualizer software (Biovia, 2017).
2.4. Molecular dynamics simulation
The molecular dynamics (MD) simulations were performed using the GROningen MAchine for Chemical Simulations GROMACS 2019 (Abraham et al., 2015). The GROMOS9653a6 force field and SPC water model were used for all the MD simulations (Oostenbrink et al., 2004). The ligand topologies were obtained from the PRODRG server (Schuttelkopf & van Aalten, 2004). All bond lengths of protein and anti-HIV drugs/corticosteroid drugs were constrained using the LINCS algorithm, while water molecules were restrained by the SETTLE algorithm (Hess et al., 1997; Miyamoto & Kollman, 1992). The system was accommodated in a cubic box and solvated by the SPC water model. Each system was energy-minimized using the steepest descent algorithm and equilibrated to achieve the appropriate volume. The leapfrog algorithm with time step 2 fs was used and at every five steps, the neighbor list was updated. The Particle Mesh Ewald method is used to treat the Long-range electrostatics with cut off 1.2 nm and with a Fourier grid spacing of 1.2 nm (Essmann et al., 1995). Periodic boundary conditions were applied in all three directions. Equilibration of the systems was carried out in two main stages. First, the system was allowed to heat gradually to 300 K in the NVT ensemble using the Berendsenbarostat method for 10 ns (Berendsen et al., 1984). Then the NPT ensemble using the Parrinello-Rahman method was employed for 10 ns by positional restraining of the complexes (unligated Mpro, Mpro-darunavir, Mpro-lopinavir and Mpro-corticosteroid complexes), slowly allowing the solvent molecules to relax around it (Parrinello & Rahman, 1981). The equilibrated systems were then subjected to unrestrained production MD simulations of 100 ns each, maintaining target pressure (1 bar) and temperature (300 K). The root mean square deviation (RMSD), the total number of hydrogen bonds, root mean square fluctuation (RMSF), the radius of gyration (Rg), solvent accessible surface area (SASA) for each system were calculated from the MD trajectories (Ghosh et al., 2020a, 2020b).
2.5. MM-GBSA
MM-GBSA is a popular method to calculate binding energy, which uses energy properties of free ligand, free receptor and receptor-ligand complex for binding affinity calculation. The prime module of the Schrodinger suite (Schrödinger Release 2020-1: Prime, Schrödinger, LLC, New York, NY, 2020) was used for all MM-GBSA calculations (Ghosh et al., 2020a). We have used the MM-GBSA method to calculate the relative binding free energies of anti-HIV as well as corticosteroid drugs to Mpro. We have calculated the MM-GBSA from the entire trajectory of the MD simulation run for each Mpro-complex system. For that, we have taken the entire 100 ns trajectory of the MD simulation and extracted the coordinate file at every 5 ns interval. From these 20 points, we have calculated the average MM-GBSA value. The free energy of binding can be calculated as ΔGbind=ΔH−TΔS.
ΔH = ΔEelec+ΔEvdW+ΔGpolar+ΔGnon-polar, where Eelec and EvdW are the electrostatic and van der Waal’s contributions, and Gploar and Gnon-polar are the polar and non-polar solvation terms, respectively. As similar types of ligands bind to the receptor, the entropic contribution is neglected here. Therefore, our calculated values referred to as relative binding free energies (ΔGbind).
3. Result and discussion
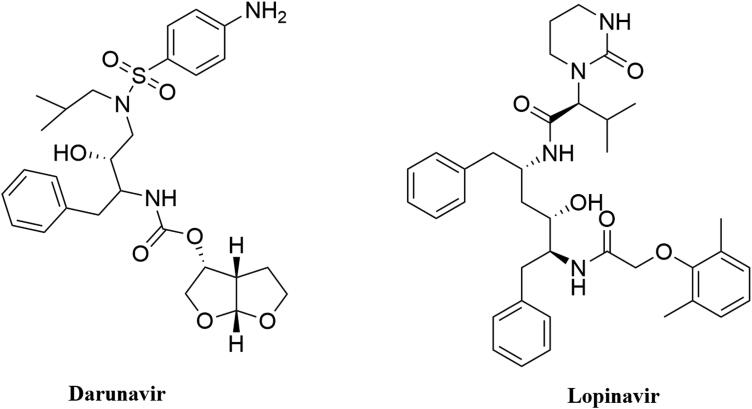
The crystal structure of Mpro in complexation with a Michael inhibitor N3 (PDB ID 6LU7) and inhibitor alpha-ketoamide (PDB ID 6Y2E) provide valuable information about the structure of Mpro (Jin et al., 2020; Zhang et al., 2020). We have discussed the structural elements of Mpro in detail in Section 1. These two crystal structures also laid the basis toward the structure-based drug design against Mpro. Many small molecules are being proposed as effective SARS CoV-2 Mpro inhibitor (Bhardwaj et al., 2020; Das et al., 2020; Ghosh et al., 2020a, 2020b; Gorla et al., 2020; Gurung et al., 2020; Joshi et al., 2020a, 2020b; Mazzini et al., 2020 ). Several anti-HIV drugs (darunavir, lopinavir, atazanavir, etc.) also exhibit excellent binding affinity toward the active site of Mpro (Beck et al., 2020). In recent past, two well-known anti-HIV drugs (darunavir and lopinavir) (structure of them mentioned in Figure 2) have been selected by many investigators as standard substrates for comparing the binding affinity and/or binding modes of various small molecules with that of ‘Mpro-darunavir/lopinavir complex’ (Bhardwaj et al., 2020; Ghosh et al., 2020b). Therefore, we have also decided to take these two anti-HIV drugs as standard Mpro inhibitors for this study.
Figure 2.
Chemical structure of anti-HIV drugs. The two-dimensional structure of two anti-HIV drugs (darunavir and lopinavir).
3.1. Molecular docking studies
Six well-known corticosteroids and two repurposed drugs against Mpro (darunavir and lopinavir) were docked to assess the corticosteroid(s) exhibiting the higher or comparable binding energy to that of ‘Mpro-darunavir/lopinavir interaction’. The binding energy of darunavir and lopinavir toward Mpro was −7.4 and −7.3 kcal/mol, respectively (Table 1). We also estimated the binding energy of six corticosteroids (cortisone, hydrocortisone, prednisolone, methylprednisolone, betamethasone and dexamethasone) toward Mpro using molecular docking studies. It was found that the binding energy of cortisone, hydrocortisone, prednisolone and methylprednisolone was in the range of −5.1 to −5.5 kcal/mol which was much lower in compared to the standards, darunavir and lopinavir (Table 1). On the contrary, the other two corticosteroids (betamethasone and dexamethasone) exhibited higher binding affinity (−7.8 to −7.9 kcal/mol) toward Mpro than that of darunavir and lopinavir (Table 1). To validate these findings, we also performed blind docking. Amongst 200 different docked conformations, majority of the interactions of dexamethasone (180 docked conformations), betamethasone (176 docked conformations), darunavir (178 docked conformations) and lopinavir (177 docked conformations) were within the active/catalytic site of Mpro. The binding energy (lowest) of darunavir, lopinavir, dexamethasone and betamethasone toward Mpro was −7.33, 7.21, −7.85 and −7.72 kcal/mol, respectively. The binding energy values from blind docking were in close agreement with those obtained from the molecular docking using a grid box around the active site of Mpro. As betamethasone and dexamethasone had higher binding affinity than darunavir and lopinavir, we decided to proceed further with these two corticosteroids.
Table 1.
The binding energy of different anti-HIV drugs and corticosteroids with Mpro.
| Drug | Binding energy (kcal/mol) |
|---|---|
| Darunavir | −7.4 |
| Lopinavir | −7.3 |
| Cortisone | −5.3 |
| Prednisolone | −5.1 |
| Hydrocortisone | −5.4 |
| Methylprednisolone | −5.5 |
| Betamethasone | −7.8 |
| Dexamethasone | −7.9 |
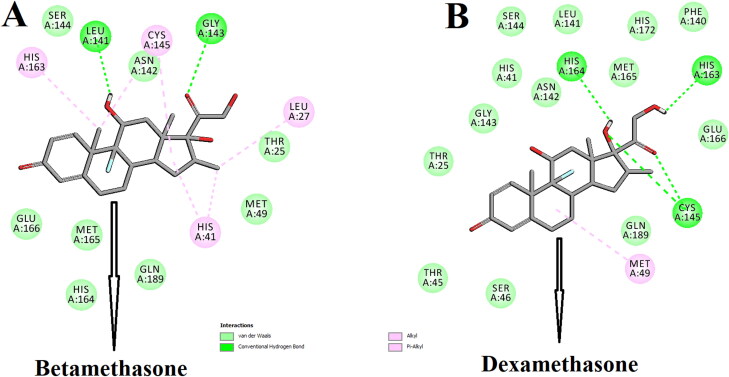
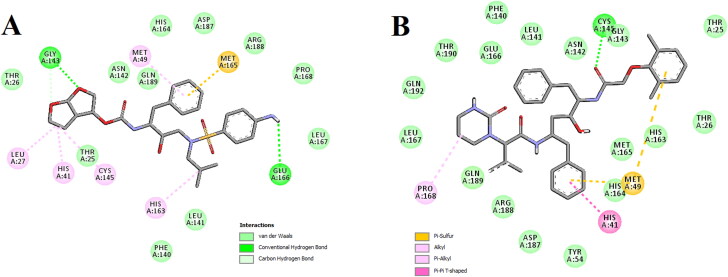
The amino acid residues within the active site of Mpro which were interacting with these two selected corticosteroids were carefully examined with the aid of discovery studio visualizer. It was evidenced that betamethasone and dexamethasone efficiently interacted with different amino acid residues of domain I and II of Mpro (Figure 3 and Table 2).
Figure 3.
Molecular docking of corticosteroids with Mpro. The docked conformation of the Mpro-betamethasone complex (A) and Mpro-dexamethasone complex (B) depicting the possible interactions with various amino acids of Mpro. Both the corticosteroid drug molecules interact with many amino acid residues including His41 and Cys145 of Mpro.
Table 2.
Hydrogen bond interactions of anti-HIV drugs and corticosteroids with the active site of SARS CoV-2 Mpro.
| Drugs interact with Mpro | Number of H-bonds | Amino acids of Mpro involved in H-bonding | Hydrogen bond distance (Å) |
|---|---|---|---|
| Betamethasone | 2 | Leu141 | 2.3 |
| Gly143 | 2.4 | ||
| Dexamethasone | 4 | Cys145 | 2.4 |
| Cys145 | 2.5 | ||
| His163 | 2.4 | ||
| His164 | 2.4 | ||
| Darunavir | 2 | Gly143 | 2.3 |
| Glu166 | 2.4 | ||
| Lopinavir | 1 | Cys145 | 2.3 |
When betamethasone was docked into the active site of Mpro, two hydrogen bond interactions [Leu141 (2.3 Å) and Gly143 (2.4 Å)] and eight van der Waals interactions (Thr25, Met49, Asn142, Ser144, His164, Met165, Glu166 and Gln189) were evidenced (Figure 3A and Table 2). Beside these, multiple alkyl (Leu27 and Cys145) and π-alkyl (His41 and His163) were evidenced in the Mpro-betamethasone complex (Figure 3A and Table 2). Interestingly, more number of H-bond interactions was observed when dexamethasone was docked to the active site of Mpro. Dexamethasone formed a single hydrogen bond with His163 and His164 as well as formed double hydrogen bonds with Cys145 of Mpro (Figure 3B and Table 2). Mpro-dexamethasone complex was further stabilized by 1 alkyl (Met49) and 13 van der Waals interactions (Thr25, His41, Thr45, Ser46, Phe140, Leu141, Asn142, Gly143, Ser144, Met165, Glu166, His172 and Gln189) (Figure 3B). Even, two repurposed drugs (darunavir and lopinavir) also interacted with several critical residues within the active site of Mpro (Figure 4 and Table 2). Darunavir interacted with Mpro via two hydrogen bonds [Gly143 (2.3 Å) and Glu166 (2.4 Å)], one π-sulfur bond (Met165) and multiple alkyl/π-alkyl bonds (Leu27, His41, Met49, Cys145 and His163) (Figure 4A and Table 2). It also formed many van der Waals interactions with different amino acid residues of Mpro (Figure 4A). Lopinavir formed only one hydrogen bond with Cys145 and several other non-covalent bonds with various important amino acid residues (such as Thr26, His41, Met49, Phe140, Glu166, Leu167, etc) within the active site of Mpro (Figure 4B and Table 2). Altogether, molecular docking studies clearly revealed that selected two corticosteroids (betamethasone and dexamethasone) interacted with both the key residues (His41 and Cys145) of Mpro via hydrogen bonding and/or other non-covalent forces (Figure 3 and Table 2). Even, in blind docking, the docked conformation having the lowest binding energy depicted that dexamethasone and betamethasone have non-covalent interaction (other than H-bond interaction) with these two important amino acid residues of Mpro (Supplementary Figure S1). In fact, there are many reports available in the literature where investigators noticed the formation of non-covalent bonds (other than H-bond) including alkyl bond(s) between compounds and residues of catalytic site (His41 and Cys145) of the Mpro (Bhardwaj et al., 2020; Das et al., 2020; Gurung et al., 2020; Islam et al., 2020; Joshi et al., 2020; Odhar et al., 2020). We believe that the availability and/or accessibility of Cys145 as well as His41 of Mpro may be ceased down due to the formation of alkyl bond(s) between compounds and these two residues of Mpro and such alterations possibly can inhibit its (Mpro) catalytic activity. Moreover, their binding affinity toward Mpro was more than the binding affinity of darunavir/lopinavir to Mpro (Table 1). Thus, it can be concluded that betamethasone and dexamethasone may possibly inhibit the proteolytic activity of Mpro and may potentially be repurposed to treat patients with COVID-19. We also computed the inhibition constant (ki) so as to get the idea about the inhibition potency of dexamethasone and betamethasone toward Mpro (Gurung et al., 2020). Inhibition constant (ki) was estimated using the following equation:
| (1) |
where ΔG is the binding energy in kcal/mol, R is the universal gas constant (1.987 cal K−1 mol−1) and T is the temperature (298 K).
Figure 4.
Molecular docking of anti-HIV drugs with Mpro. Interactions of various amino acids of Mpro with darunavir (A) and lopinavir (B) are presented with the best docking pose.
For the docked structure (having lowest energy) of Mpro-dexamethasone complex as well as Mpro-betamethasone complex, the in silico determined ki value at 298 K was 1.6 × 10−6 M and 1.9 × 10−6 M, respectively. These computed ki values are far lower than their toxicity dose range [LD50 for dexamethasone >3000 mg/kg and LD50 for betamethasone = 4096 mg/kg] which further validates the strong candidature of these two corticosteroids as target drugs to bind Mpro.
These two Mpro-corticosteroid complexes were further subjected to molecular dynamics simulations as well as binding free energy computations to assess the stability of these complexes.
3.2. Molecular dynamics simulation studies
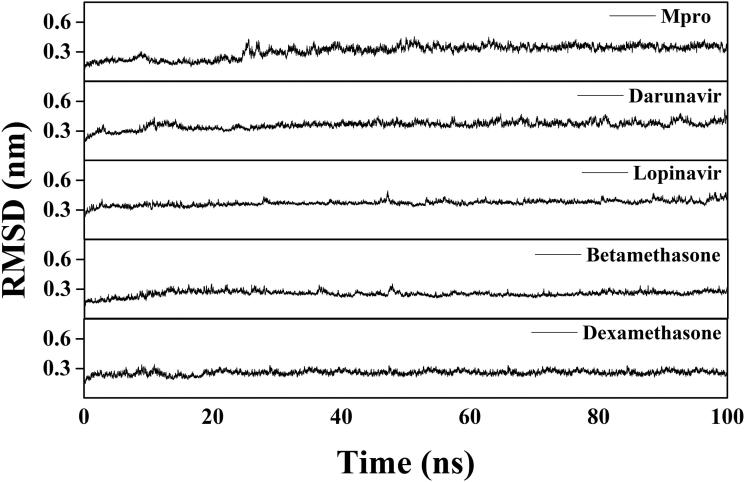
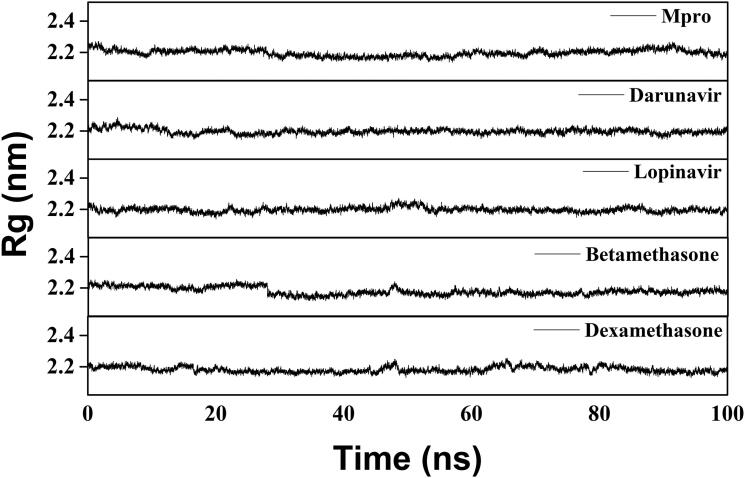
Several structural properties like overall complex stability (RMSD), conformational fluctuations (RMSF), structural compactness (Rg) and solvent accessibility (SASA) were investigated by MD simulations. We performed production run for 100 ns using GROMOS9653a6 force field of Mpro alone/Mpro (unligated) and Mpro complexed with two anti-HIV drugs, as well as betamethasone and dexamethasone. The information about the structural stability of the protein-ligand complexes could be analyzed by RMSD. We estimated the RMSD of alpha-carbon atoms of all these systems (Figure 5). The RMSD of Mpro (unligated) maintained a constant value (∼0.21–0.22 nm) from 2 to 17 ns. Thereafter, the RMSD value gradually increased till 25 ns and reached ∼0.35 nm. Then, the value was slightly decreased and persisted at ∼0.31 nm from 65 ns till the end of the MD run. The RMSD values for both Mpro-darunavir and Mpro-lopinavir complexes were found to remain almost constant (∼0.36–0.37 nm) from 10 to 100 ns with some marginal fluctuations (Figure 5). On the other hand, the magnitude of RMSD corresponding to Mpro-betamethasone and Mpro-dexamethasone complexes attained an equilibrium value after 20 ns (∼0.25–0.26 nm) and remained almost the same throughout the 100 ns simulation.
Figure 5.
RMSD plots of Mpro (unligated), Mpro-darunavir, Mpro-lopinavir and two Mpro-corticosteroid complexes. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of RMSD.
The average RMSD values for Mpro (unligated), Mpro-darunavir complex and Mpro-lopinavir complex were found to be ∼0.31 nm, ∼0.36 nm and ∼0.37 nm, respectively, which are in agreement with previously reported values (Table 3) (Bhardwaj et al., 2020). Whereas the RMSD values of Mpro-betamethasone and Mpro-dexamethasone complexes were ∼0.25 nm and ∼0.26 nm (Table 3), respectively, which were quite lower than Mpro (unligated), Mpro-darunavir and Mpro-lopinavir complexes. These results suggested that Mpro-betamethasone and Mpro-dexamethasone complexes were relatively more stable than that of Mpro-darunavir/Mpro-lopinavir complexes.
Table 3.
Average values of the RMSD, RMSF, Rg, SASA and the total number of intermolecular hydrogen bonds formed during MD simulation for different systems.
| System | RMSD (nm) | RMSF (nm) | Rg (nm) | SASA (nm2) | Total number of H-bonds formed |
|---|---|---|---|---|---|
| Mpro (unligated) | 0.309 | 0.1937 | 2.195 | 151.4483 | 547 |
| Mpro-darunavir | 0.361 | 0.1952 | 2.197 | 151.1540 | 550 |
| Mpro-lopinavir | 0.371 | 0.1948 | 2.196 | 151.2825 | 551 |
| Mpro- betamethasone | 0.254 | 0.1661 | 2.179 | 151.2791 | 556 |
| Mpro-dexamethasone | 0.258 | 0.1652 | 2.182 | 151.2388 | 555 |
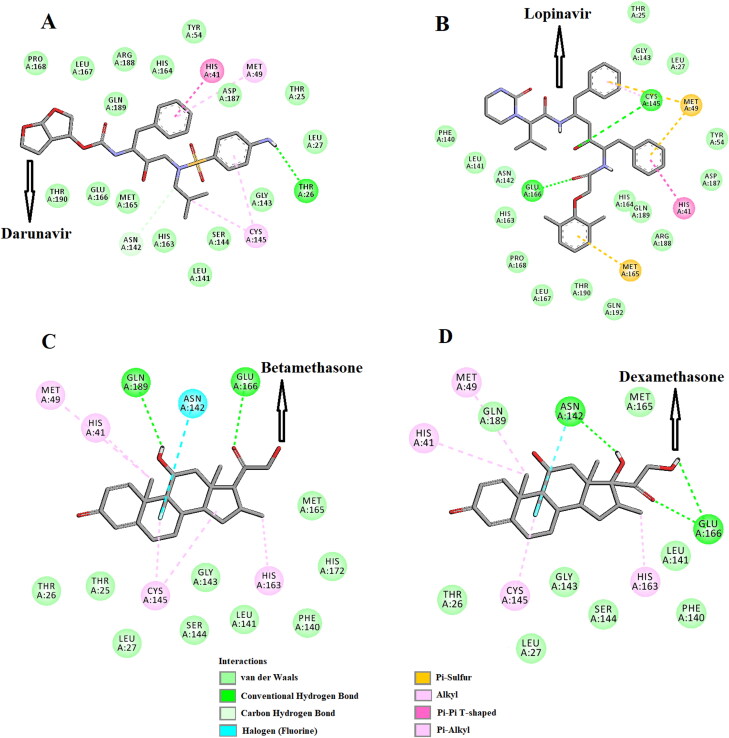
The conformational stability of these two Mpro-corticosteroids complexes was further analyzed by estimating the total number of intermolecular hydrogen bonds formed during the entire simulation as determined previously (Ghosh et al., 2020a, 2020b; Islam et al., 2020) (Table 3). The average number of intermolecular hydrogen bonds in the Mpro (unligated) system was 547, whereas, for Mpro-darunavir and Mpro-lopinavir complexes, the values were 550 and 551, respectively. In the case of Mpro-betamethasone and Mpro-dexamethasone complexes, a higher number of intermolecular hydrogen bonds (556 and 555, respectively) were found (Table 3). We also evaluated the existence of H-bonds between Mpro and these four compounds after the completion of MD run. In Mpro-darunavir and Mpro-lopinavir complexes, we observed one and two hydrogen bond interactions, respectively (Figure 6A,B and Table 4). On the other hand, after MD run, we evidenced three and two hydrogen bond interactions in Mpro-dexamethasone complex and Mpro-betamethasone complex, respectively (Figure 6C,D and Table 4). It is noteworthy to mention here that the interaction between these two corticosteroids and many important amino acid residues (including His41 and Cys145) of Mpro is retained even after MD run (Figure 6C,D and Table 4). Altogether, these results further suggested that these two Mpro-corticosteroids complexes (Mpro-betamethasone and Mpro-dexamethasone) are more stable than those of two selected Mpro-anti-HIV drug complexes.
Figure 6.
Identification of binding residues/modes within Mpro-darunavir, Mpro-lopinavir, Mpro-betamethasone and Mpro-dexamethasone complexes after MD run. The docked conformation of the Mpro-darunavir complex (A), Mpro-lopinavir complex (B), Mpro-betamethasone complex (C) and Mpro-dexamethasone complex (D) depicting the possible interactions with various amino acids of Mpro after 100 ns MD run. All these drugs are shown to interact with His41 and Cys145 of Mpro.
Table 4.
Different interactions of Mpro-darunavir, Mpro-lopinavir, Mpro-betamethasone and Mpro-dexamethasone complexes after 100 ns MD simulation.
| Drug | Hydrogen bond interaction | van der Waals interaction | C–H bond interaction | Alkyl interaction | π-alkyl interaction | π-π interaction | π-sulfur interaction | Halogen bond interaction |
|---|---|---|---|---|---|---|---|---|
| Darunavir | Thr26 (2.4 Å) | Thr25, Leu27, Tyr54, Leu141, Gly143, Ser144, His163, His164, Met165, Glu166, Leu167, Pro168, Asp187, Arg188, Gln189, Thr190 | Asn142 | Met49, Cys145 | His41 | |||
| Lopinavir | Cys145 (2.5 Å), Glu166 (2.3 Å) | Thr25, Leu27, Tyr54, Phe140, Leu141, Asn142, Gly143, His163, His164, Leu167, Pro168, Asp187, Arg188, Gln189, Thr190, Gln192 | His41 | Met49, Met165 | ||||
| Betamethasone | Glu166 (2.4 Å), Gln189 (2.4 Å) | Thr25, Thr26, Leu27, Phe140, Leu141, Gly143, Ser144, Met165, His172 | Met49, Cys145 | His41, His163 | Asn142 | |||
| Dexamethasone | Asn142 (2.3 Å), Glu166 (2.3 Å), Glu166 (2.4 Å) | Thr26, Leu27, Phe140, Leu141, Gly143, Ser144, Met165, Gln189 | Met49, Cys145 | His41, His163 | Asn142 |
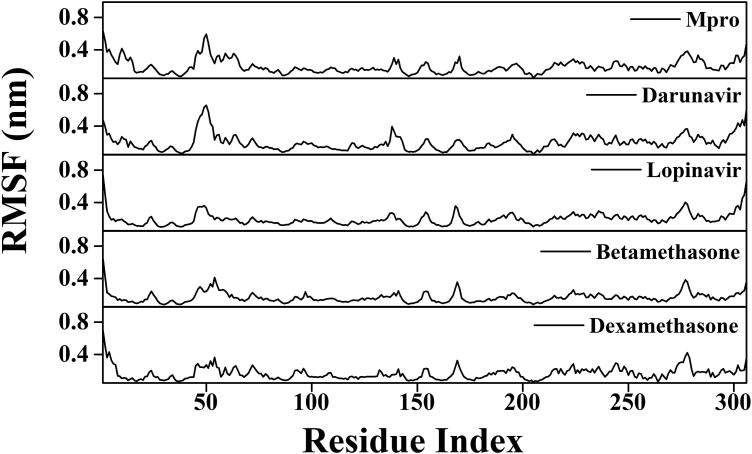
To find out the conformational flexibility of Mpro (unligated) and other Mpro-complexes, we have calculated the RMSF of alpha-carbon atoms for all systems (Figure 7). It was quite evident from the RMSF profiles that all systems experience more conformational fluctuations in domain III. In case of Mpro (unligated) system, we additionally observed higher fluctuations (upto ∼0.6 nm) in certain portion of domain I (residues 45–60). In fact, most of the amino acid residues within the domain I and II of this system had RMSF fluctuation below 0.3 nm. The average RMSF value for Mpro (unligated) system was ∼0.194 nm (Table 3). The Mpro-darunavir and Mpro-lopinavir system experienced more or less similar conformational fluctuations to that of Mpro (unligated) system (Figure 7). In fact, the fluctuations for the residues 45–60, were reduced upon the binding of lopinavir to Mpro (up to 0.35 nm). For both Mpro-darunavir and Mpro-lopinavir complexes, the average RMSF value was ∼0.195 nm (Table 3). Upon analyzing all the RMSF profiles, it was clearly observed that Mpro-betamethasone and Mpro-dexamethasone complexes showed lower fluctuations (especially in domain I and II) as compared to the Mpro (unligated) and Mpro-darunavir/lopinavir complexes (Figure 7). The average RMSF values of Mpro-betamethasone and Mpro-dexamethasone complexes were ∼0.166 nm and ∼0.165 nm, respectively (Table 3). Most importantly, the fluctuations of many key residues of the binding region of Mpro were ceased down after binding to betamethasone and dexamethasone. These findings suggested that the overall conformational fluctuations of Mpro-betamethasone and Mpro-dexamethasone complexes are relatively less than that of Mpro-darunavir/Mpro-lopinavir complex.
Figure 7.
RMSF profiles of Mpro (unligated), Mpro-anti-HIV drugs, Mpro-betamethasone and Mpro-dexamethasone complexes. The RMSF values of Mpro (unligated) and Mpro-anti-HIV drug complexes as well as Mpro-betamethasone and Mpro-dexamethasone complexes were plotted against the amino acid residues of Mpro.
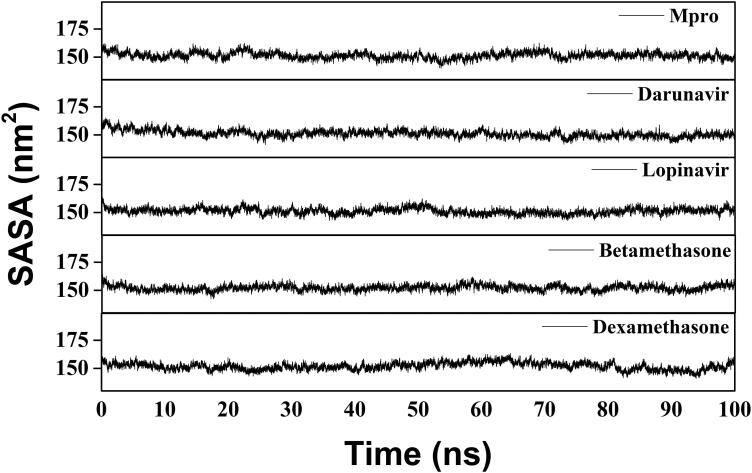
We have also estimated the Rg value to assess the compactness of all the complexes (Figure 8 and Table 3). The average Rg value for Mpro (unligated) and the other two complexes (Mpro-darunavir and Mpro-lopinavir) was almost identical (∼2.20 nm). We observed a slightly less average Rg value for Mpro-betamethasone (2.179 nm) and Mpro-dexamethasone (2.182 nm) complexes (Table 3). These results suggested that our Mpro-betamethasone and Mpro-dexamethasone complexes were slightly more compact as compared to the two Mpro-anti-HIV drug complexes. SASA values were also calculated to assess the extent of expansion of protein volume in each system (Figure 9 and Table 3). The average SASA values of Mpro-darunavir complex (151.154 nm2), Mpro-lopinavir complex (∼151.283 nm2), Mpro-betamethasone (151.279 nm2) and Mpro-dexamethasone complexes (∼151.239 nm2) were found to be in the similar range with Mpro (unligated) (∼151.448 nm2) (Table 3). Thus, it can be suggested that almost a similar degree of expansion of the Mpro occur upon interaction with two anti-HIV drugs as well as betamethasone and dexamethasone.
Figure 8.
Estimation of Rg values of Mpro (unligated), two Mpro-anti-HIV drugs and two Mpro-corticosteroid complexes. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of Rg.
Figure 9.
SASA value analysis of Mpro (unligated), Mpro-anti-HIV drugs, Mpro-betamethasone and Mpro-dexamethasone complexes. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of SASA.
3.3. MM-GBSA analysis
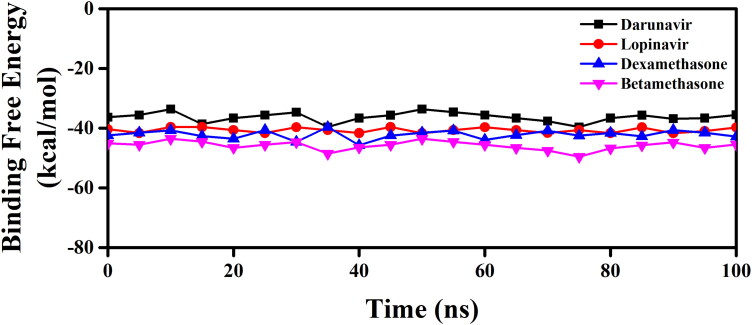
We have taken the configurations from 100 ns trajectory corresponding to every 5 ns time interval and determined the binding free energy of Mpro-betamethasone, Mpro-dexamethasone as well as Mpro-anti-HIV drugs by using MM-GBSA method. The determined binding free energy values throughout the trajectory for each system were presented in Figure 10.
Figure 10.
MM-GBSA binding free energy profiles of two Mpro-anti-HIV drug complexes, Mpro-dexamethasone and Mpro-betamethasone complexes. The binding free energy values of two Mpro-anti-HIV drug complexes as well as Mpro-dexamethasone and Mpro-betamethasone complexes were represented throughout the entire 100 ns simulation trajectory.
The resultant/average binding free energy value computed from the whole trajectory was listed in Table 5. The binding free energy values of Mpro-darunavir and Mpro-lopinavir complexes were found to be −35.60 kcal/mol and −40.66 kcal/mol, respectively, whereas for Mpro-betamethasone and Mpro-dexamethasone complexes were −45.85 kcal/mol and −42.76 kcal/mol, respectively (Table 5).
Table 5.
MM-GBSA values of different Mpro-drug complexes.
| System | Binding free energy (kcal/mol) |
|---|---|
| Mpro-darunavir | −35.60 ± 1.62 |
| Mpro-lopinavir | −40.66 ± 0.85 |
| Mpro-betamethasone | −45.85 ± 1.48 |
| Mpro-dexamethasone | −42.76 ± 1.46 |
It is quite evident from these MM-GBSA values that two corticosteroid drugs (betamethasone and dexamethasone) interact with Mpro with higher binding free energy than that of darunavir/lopinavir. The higher MM-GBSA values (ΔGbind) in the case of Mpro-betamethasone and Mpro-dexamethasone mostly contributed by the coulombic and hydrophobic interactions.
4. Conclusion
This study is aimed to test the inhibition potency of six well-known corticosteroids against SARS CoV-2 Mpro using a computational approach. Molecular docking studies were conducted to compare their binding affinities (toward Mpro) with two repurposed drugs (darunavir and lopinavir) against COVID-19. Only two of them (betamethasone and dexamethasone) had higher AutoDock Vina energy values than the darunavir and lopinavir. These two corticosteroids interacted with both the key catalytic residues (His41 and Cys145) of Mpro. The RMSD and RMSF profiles corresponding to Mpro-betamethasone and Mpro-dexamethasone complexes clearly suggested that they (complexes) are highly stable and experience less conformational fluctuations. The Rg and SASA analysis revealed that Mpro-betamethasone and Mpro-dexamethasone complexes are slightly more compact and somewhat less expanded. The existence of a higher number of intermolecular hydrogen bonds in the complexes with two corticosteroids (betamethasone and dexamethasone) was evidenced than in Mpro-darunavir/lopinavir complex suggesting greater stability of these corticosteroids in the binding pockets of Mpro. Apart from this non-structural protein, other ones (such asPLpro/Nsp3, Nsp9, Nsp16, etc.) could also be potent targets for dexamethasone as well as for betamethasone. Such possibilities should be examined thoroughly. Nevertheless, we can say that possessing Mpro inhibitory activity possibly makes dexamethasone effective for COVID-19 treatment. Our findings further revealed that betamethasone also possesses the potential to inhibit the proteolytic activity of Mpro. Thus, betamethasone may be used for the COVID-19 treatment. But detailed experimental studies are necessary before repurposing betamethasone against COVID-19.
Supplementary Material
Acknowledgements
RG acknowledges IIT Bhubaneswar for providing fellowship. The authors thank IIT Delhi HPC facility for computational resources.
Glossary
Abbreviations
- COVID-19
corona virus disease 2019
- SARS CoV-2
severe acute respiratory syndrome corona virus-2
- Mpro
main protease
- MD
molecular dynamics
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- Rg
radius of gyration
- SASA
solvent accessible surface area.
Disclosure statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Abeyagunawardena, A. S., & Trompeter, R. S. (2008). Increasing the dose of prednisolone during viral infections reduces the risk of relapse in nephrotic syndrome: A randomised controlled trial. Archives of Disease in Childhood, 93(3), 226–228. 10.1136/adc.2007.116079 [DOI] [PubMed] [Google Scholar]
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Alberti, C., Cartier, L., Valenzuela, M. A., Puente, J., Tanaka, Y., & Ramirez, E. (2011). Molecular and clinical effects of betamethasone in human T-cell lymphotropic virus type-I-associated myelopathy/tropical spastic paraparesis patients. Journal of Medical Virology, 83(9), 1641–1649. 10.1002/jmv.22131 [DOI] [PubMed] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Arabi, Y. M., Mandourah, Y., Al-Hameed, F., Sindi, A. A., Almekhlafi, G. A., Hussein, M. A., Jose, J., Pinto, R., Al-Omari, A., Kharaba, A., Almotairi, A., Al Khatib, K., Alraddadi, B., Shalhoub, S., Abdulmomen, A., Qushmaq, I., Mady, A., Solaiman, O., Al-Aithan, A. M., Fowler, R. A., … Saudi Critical Care Trial, G (2018). Corticosteroid therapy for critically Ill patients with middle east respiratory syndrome. American Journal of Respiratory and Critical Care Medicine, 197(6), 757–767. 10.1164/rccm.201706-1172OC [DOI] [PubMed] [Google Scholar]
- Baby, K., Maity, S., Mehta, C. H., Suresh, A., Nayak, U. Y., & Nayak, Y. (2020). Targeting SARS-CoV-2 main protease: A computational drug repurposing study. Archives of Medical Research. 10.1016/j.arcmed.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, P. J. (2006). How corticosteroids control inflammation: Quintiles Prize Lecture 2005. British Journal of Pharmacology, 148(3), 245–254. 10.1038/sj.bjp.0706736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, B. R., Shin, B., Choi, Y., Park, S., & Kang, K. (2020). Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Computational and Structural Biotechnology Journal., 18, 784–790. 10.1016/j.csbj.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., Gunsteren, W. F. v., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bharadwaj, S., Lee, K. E., Dwivedi, V. D., & Kang, S. G. (2020). Computational insights into tetracyclines as inhibitors against SARS-CoV-2 Mpro via combinatorial molecular simulation calculations. Life Sciences, 257, 118080. 10.1016/j.lfs.2020.118080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia, D. S. (2017). Discovery studio modeling environment [Release 4]. San Diego: Dassault Systemes. [Google Scholar]
- Blanchard, J. E., Elowe, N. H., Huitema, C., Fortin, P. D., Cechetto, J. D., Eltis, L. D., & Brown, E. D. (2004). High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chemistry & Biology, 11(10), 1445–1453. 10.1016/j.chembiol.2004.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. C., Di Napoli, R. (2020). Features, evaluation and treatment coronavirus (COVID-19) (StatPearls). Bethesda, MD: National Center for Biotechnology Information, U.S. National Library of Medicine. http://www.ncbi.nlm.nih.gov/pubmed/32150360 [Google Scholar]
- Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., Qiu, Y., Wang, J., Liu, Y., Wei, Y., Xia, J., Yu, T., Zhang, X., & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet (London, England), 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, K. C., Wei, D. Q., & Zhong, W. Z. (2003). Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochemical and Biophysical Research Communications, 308(1), 148–151. 10.1016/S0006-291X(03)01342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, C., Capriotti, J. A., Kumar, M., Hobden, J. A., Foster, T. P., Bhattacharjee, P. S., Thompson, H. W., Mahmud, R., Liang, B., & Hill, J. M. (2011). Clinical and antiviral efficacy of an ophthalmic formulation of dexamethasone povidone-iodine in a rabbit model of adenoviral keratoconjunctivitis. Investigative Ophthalmology & Visual Science, 52(1), 339–344. 10.1167/iovs.10-5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri, M., Cifaldi, R., Dreas, L., Viviani, M., Biolo, M., & Gabrielli, M. (2010). Methylprednisolone infusion for life-threatening H1N1-virus infection. Therapeutic Advances in Respiratory Disease, 4(4), 233–237. 10.1177/1753465810376951 [DOI] [PubMed] [Google Scholar]
- Cucinotta, D., & Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Bio-Medica: Atenei Parmensis, 91(1), 157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W., Zhang, B., Jiang, X. M., Su, H., Li, J., Zhao, Y., Xie, X., Jin, Z., Peng, J., Liu, F., Li, C., Li, Y., Bai, F., Wang, H., Cheng, X., Cen, X., Hu, S., Yang, X., Wang, J., … Liu, H. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (New York, N.Y.), 368(6497), 1331–1335. 10.1126/science.abb4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, J., Whitley, R. J., Pocock, S., & Monto, A. S. (2015). Oseltamivir treatment for influenza in adults: A meta-analysis of randomised controlled trials. The Lancet, 385(9979), 1729–1737. 10.1016/S0140-6736(14)62449-1 [DOI] [PubMed] [Google Scholar]
- Emery, V. C., Cope, A. V., Sabin, C. A., Burroughs, A. K., Rolles, K., Lazzarotto, T., Landini, M. P., Brojanac, S., Wise, J., & Maine, G. T. (2000). Relationship between IgM antibody to human cytomegalovirus, virus load, donor and recipient serostatus, and administration of methylprednisolone as risk factors for cytomegalovirus disease after liver transplantation. The Journal of Infectious Diseases, 182(6), 1610–1615. 10.1086/317636 [DOI] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Frisch, M., Clemente, F., Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., & Zhe, G.. 2009. Gaussian 09, Revision A. 01. Wallingford CT: Gaussian, Inc.
- Gagyor, I., Madhok, V. B., Daly, F., & Sullivan, F. (2019). Antiviral treatment for Bell's palsy (idiopathic facial paralysis). Cochrane Database of Systematic Reviews, 9 (9), CD001869. 10.1002/14651858.CD001869.pub9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020. a). Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – An in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1779818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020. b). Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1802347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorla, U. S., Rao, G. K., Kulandaivelu, U. S., Alavala, R. R., & Panda, S. P. (2020). Lead finding from selected flavonoids with antiviral (SARS-CoV-2) potentials against COVID-19: An in-silico evaluation. Combinatorial Chemistry & High Throughput Screening. 10.2174/1386207323999200818162706 [DOI] [PubMed] [Google Scholar]
- Grum-Tokars, V., Ratia, K., Begaye, A., Baker, S. C., & Mesecar, A. D. (2008). Evaluating the 3C-like protease activity of SARS-Coronavirus: Recommendations for standardized assays for drug discovery. Virus Research, 133(1), 63–73. 10.1016/j.virusres.2007.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., Liu, L., Shan, H., Lei, C. L., Hui, D. S. C., Du, B., Li, L. J., Zeng, G., Yuen, K. Y., Chen, R. C., Tang, C. L., Wang, T., Chen, P. Y., Xiang, J., … China Medical Treatment Expert Group For, C, China Medical Treatment Expert Group for Covid-19 (2020). Clinical characteristics of coronavirus disease 2019 in China. The New England Journal of Medicine, 382(18), 1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung, A. B., Ali, M. A., Lee, J., Farah, M. A., & Al-Anazi, K. M. (2020). Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sciences, 255, 117831. 10.1016/j.lfs.2020.117831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Melim, L., Federico, L. B., de Oliveira, N. K. S., Francisco, V. C. C., Correia, L. C., de Lima, H. B., Gomes, S. Q., Barcelos, M. P., Francischini, I. A. G., & da Silva, C. (2020). Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sciences, 256, 117963. 10.1016/j.lfs.2020.117963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakmi, M., Bouricha, E. M., Kandoussi, I., Harti, J. E., & Ibrahimi, A. (2020). Repurposing of known anti-virals as potential inhibitors for SARS-CoV-2 main protease using molecular docking analysis. Bioinformation, 16(4), 301–306. 10.6026/97320630016301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt, B. H., Jukneliene, D., Kanjanahaluethai, A., Bechill, J., Severson, K. M., Smith, C. M., Rota, P. A., & Baker, S. C. (2004). Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. Journal of Virology, 78(24), 13600–13612. 10.1128/JVI.78.24.13600-13612.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzi, I., Walder, B., & Tramer, M. R. (2000). Dexamethasone for the prevention of postoperative nausea and vomiting: A quantitative systematic review. Anesthesia and Analgesia, 90(1), 186–194. 10.1097/00000539-200001000-00038 [DOI] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Hodos, R. A., Kidd, B. A., Shameer, K., Readhead, B. P., & Dudley, J. T. (2016). In silico methods for drug repurposing and pharmacology. Wiley Interdisciplinary Reviews. Systems Biology and Medicine, 8(3), 186–210. 10.1002/wsbm.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby, P., Lim, W. S., Emberson, J., Mafham, M., Bell, J., Linsell, L., Staplin, N., Brightling, C., Ustianowski, A., Elmahi, E., Prudon, B., Green, C., Felton, T., Chadwick, D., Rege, K., Fegan, C., Chappell, L. C., Faust, S. N., Jaki, T., … Landray, M. J. (2020). Effect of Dexamethasone in Hospitalized Patients with COVID-19: Preliminary Report. medRxiv, 10.1101/2020.06.22.20137273 [DOI]
- Hsu, M. F., Kuo, C. J., Chang, K. T., Chang, H. C., Chou, C. C., Ko, T. P., Shr, H. L., Chang, G. G., Wang, A. H., & Liang, P. H. (2005). Mechanism of the maturation process of SARS-CoV 3CL protease. The Journal of Biological Chemistry, 280(35), 31257–31266. 10.1074/jbc.M502577200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, M. R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Alberto, A., Ribas-Aparicio, R. M., Aparicio-Ozores, G., & Castelan-Vega, J. A. (2020). Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Computational Biology and Chemistry, 88, 107325. 10.1016/j.compbiolchem.2020.107325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., Bhatt, V., & Chandra, S. (2020. a). In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. European Review for Medical and Pharmacological Sciences, 24(8), 4529–4536. 10.26355/eurrev_202004_21036 [DOI] [PubMed] [Google Scholar]
- Joshi, T., Sharma, P., Joshi, T., Pundir, H., Mathpal, S., & Chandra, S. (2020. b). Structure-based screening of novel lichen compounds against SARS Coronavirus main protease (Mpro) as potentials inhibitors of COVID-19. Mol Divers, 10.1007/s11030-020-10118-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627. 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne, E. D. (1957). The influence of cortisone on experimental viral infection. IV. Negation of interference as the mechanism by which cortisone induces increased virus yields. Journal of Experimental Medicine, 106(6), 863–881. 10.1084/jem.106.6.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Liu, H., Galasiti Kankanamalage, A. C., Weerasekara, S., Hua, D. H., Groutas, W. C., Chang, K. O., & Pedersen, N. C. (2016). Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathogens, 12(3), e1005531. 10.1371/journal.ppat.1005531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Y., Singh, H., & Patel, C. N. (2020). In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. Journal of Infection and Public Health, 13(9), 1210–1223. 10.1016/j.jiph.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Li, J., Chen, M., & Su, J. (2019). Glucocorticoid treatment of suspected organizing pneumonia after H7N9 infection: A case report. Medicine, 98(34), e16839. 10.1097/MD.0000000000016839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, Z., Chu, Y., & Wang, Y. (2015). HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV/AIDS (Auckland, N.Z.), 7, 95–104. 10.2147/HIV.S79956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., Cloutier, A., Coughlin, S. M., Freeman, D., Girn, N., Griffith, O. L., Leach, S. R., Mayo, M., McDonald, H., Montgomery, S. B., … Roper, R. L. (2003). The Genome sequence of the SARS-associated coronavirus. Science (New York, N.Y.), 300(5624), 1399–1404. 10.1126/science.1085953 [DOI] [PubMed] [Google Scholar]
- Mazzini, S., Musso, L., Dallavalle, S., & Artali, R. (2020). Putative SARS-CoV-2 M(pro) inhibitors from an in-house library of natural and nature-inspired products: A virtual screening and molecular docking study. Molecules, 25(16), 3745. 10.3390/molecules25163745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercorelli, B., Palu, G., & Loregian, A. (2018). Drug repurposing for viral infectious diseases: How far are we? Trends in Microbiology, 26(10), 865–876. 10.1016/j.tim.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., & Kollman, P. A. (1992). Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of Computational Chemistry, 13(8), 952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., & Olson, A. J. (2008). Using AutoDock for ligand-receptor docking. Current Protocols in Bioinformatics, 24(1), 8.14.1–8.14.40. 10.1002/0471250953.bi0814s24 [DOI] [PubMed] [Google Scholar]
- Odhar, H. A., Ahjel, S. W., Albeer, A., Hashim, A. F., Rayshan, A. M., & Humadi, S. S. (2020). Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation, 16(3), 236–244. 10.6026/97320630016236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink, C., Villa, A., Mark, A. E., & van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. Journal of Computational Chemistry, 25(13), 1656–1676. 10.1002/jcc.20090 [DOI] [PubMed] [Google Scholar]
- Osman, E. E. A., Toogood, P. L., & Neamati, N. (2020). COVID-19: Living through another pandemic. ACS Infectious Diseases, 6(7), 1548-1552. 10.1021/acsinfecdis.0c00224 [DOI] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Schuttelkopf, A. W., & van Aalten, D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Spoto, S., Valeriani, E., Locorriere, L., Anguissola, G. B., Pantano, A. L., Terracciani, F., Riva, E., Ciccozzi, M., Costantino, S., & Angeletti, S. (2019). Influenza B virus infection complicated by life-threatening pericarditis: A unique case-report and literature review. BMC Infectious Diseases, 19(1), 40. 10.1186/s12879-018-3606-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, F., Daly, F., & Gagyor, I. (2016). Antiviral agents added to corticosteroids for early treatment of adults with acute idiopathic facial nerve paralysis (Bell Palsy). JAMA, 316(8), 874–875. 10.1001/jama.2016.10160 [DOI] [PubMed] [Google Scholar]
- Tang, E., Chen, Y., & Luo, Y. (2014). Dexamethasone for the prevention of acute mountain sickness: Systematic review and meta-analysis. International Journal of Cardiology, 173(2), 133–138. 10.1016/j.ijcard.2014.03.019 [DOI] [PubMed] [Google Scholar]
- Thaera, G. M., Wellik, K. E., Barrs, D. M., Dunckley, E. D., Wingerchuk, D. M., & Demaerschalk, B. M. (2010). Are corticosteroid and antiviral treatments effective for bell palsy? A critically appraised topic. The Neurologist, 16(2), 138–140. 10.1097/NRL.0b013e3181d35775 [DOI] [PubMed] [Google Scholar]
- Thiel, V., Ivanov, K. A., Putics, Á., Hertzig, T., Schelle, B., Bayer, S., Weißbrich, B., Snijder, E. J., Rabenau, H., Doerr, H. W., Gorbalenya, A. E., & Ziebuhr, J. (2003). Mechanisms and enzymes involved in SARS coronavirus genome expression. The Journal of General Virology, 84(Pt 9), 2305–2315. 10.1099/vir.0.19424-0 [DOI] [PubMed] [Google Scholar]
- Turner, M. T., Nayak, S., Kuhn, M., & Roehm, P. C. (2014). The effects of dexamethasone and acyclovir on a cell culture model of delayed facial palsy. Otology & Neurotology: Official Publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology, 35(4), 712–718. 10.1097/MAO.0000000000000231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, C. E., Harvie, E. J., Jr., & Canby, C. M. (1961). The effect of hydrocortisone on the production of herpes simplex virus in tissue culture. The Journal of Investigative Dermatology, 36, 89–97. http://www.ncbi.nlm.nih.gov/pubmed/13784752 10.1038/jid.1961.18 [DOI] [PubMed] [Google Scholar]
- Wright, J. J., & O'Driscoll, G. (2005). Treatment of parainfluenza virus 3 pneumonia in a cardiac transplant recipient with intravenous ribavirin and methylprednisolone. The Journal of Heart and Lung Transplantation: The Official Publication of the International Society for Heart Transplantation, 24(3), 343–346. 10.1016/j.healun.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., Hu, Y., Tao, Z. W., Tian, J. H., Pei, Y. Y., Yuan, M. L., Zhang, Y. L., Dai, F. H., Liu, Y., Wang, Q. M., Zheng, J. J., Xu, L., Holmes, E. C., & Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, H., Li, J., Xie, H., & Wang, Y. (2018). Review of drug repositioning approaches and resources. International Journal of Biological Sciences, 14(10), 1232–1244. 10.7150/ijbs.24612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Velikanov, M., Flook, P., Zheng, W., Szalma, S., & Kahn, S. (2003). Assessment of putative protein targets derived from the SARS genome. FEBS Letters, 554(3), 257–263. 10.1016/S0014-5793(03)01115-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim, E. K., Lee, M. J., Lee, K. H., Um, S. J., & Park, J. S. (2006). Antiproliferative and antiviral mechanisms of ursolic acid and dexamethasone in cervical carcinoma cell lines. International Journal of Gynecological Cancer: Official Journal of the International Gynecological Cancer Society, 16(6), 2023–2031. 10.1111/j.1525-1438.2006.00726.x [DOI] [PubMed] [Google Scholar]
- Yoder, M. C., Jr., Chua, R., & Tepper, R. (1991). Effect of dexamethasone on pulmonary inflammation and pulmonary function of ventilator-dependent infants with bronchopulmonary dysplasia. The American Review of Respiratory Disease, 143(5 Pt 1), 1044–1048. 10.1164/ajrccm/143.5_Pt_1.1044 [DOI] [PubMed] [Google Scholar]
- Yu, W. C., Hui, D. S., & Chan-Yeung, M. (2004). Antiviral agents and corticosteroids in the treatment of severe acute respiratory syndrome (SARS). Thorax, 59(8), 643–645. 10.1136/thx.2003.017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science, 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. (2020). SARS-CoV-2: An emerging coronavirus that causes a global threat. International Journal of Biological Sciences, 16(10), 1678–1685. 10.7150/ijbs.45053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., Tan, W., China Novel Coronavirus, I., Research, T,… China Novel Coronavirus Investigating and Research Team (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.