Abstract
Methods that allow labeling and tracking of proteins have been instrumental for understanding their function. Traditional methods for labeling proteins include fusion to fluorescent proteins or self-labeling chemical tagging systems such as SNAP-tag or Halo-tag. These latter approaches allow bright fluorophores or other chemical moieties to be attached to a protein of interest though a small fusion tag. In this work, we sought to improve the versatility of self-labeling chemical-tagging systems by regulating their activity with light. We used light-inducible dimerizers to reconstitute a split SNAP-tag (modified human O6-alkylguanine-DNA-alkyltransferase, hAGT) protein, allowing tight light-dependent control of chemical labeling. In addition, we generated a small split SNAP-tag fragment that can efficiently self-assemble with its complement fragment, allowing high labeling efficacy with a small tag. We envision these tools will extend the versatility and utility of the SNAP-tag chemical system for protein labeling applications.
INTRODUCTION
Cellular proteins are often highly dynamic, rapidly changing their location and functionality over space and time. Methods that allow direct tracking and labeling of proteins, such as fusion to fluorescent proteins (FPs), have been instrumental in elucidating protein function and dynamics. Standard FPs can be used for tracking localization, while further monitoring of protein dynamics can be achieved using photoactivatable or photoswitchable FPs, which can be converted into an alternate fluorescent state upon photoexcitation.1,2 Despite their high utility, FPs do have limitations. Many FPs are derived from oligomeric proteins and thus can oligomerize or aggregate, disrupting the localization and/or function of the target protein.3,4 Monomeric versions of FPs are available that exhibit reduced self-association, but these have mutations that can also reduce photostability or brightness.3 FPs can alter function of attached proteins,5,6 and are subject to photobleaching.7 For studying dynamic events, photoswitched FPs can be used in imaging studies, but cannot be biochemically distinguished from their non-photoexcited forms, precluding use with affinity columns or other biochemical preparations.
An alternate strategy increasingly used for labeling involves fusion to a protein tag that reacts with a chemical substrate, allowing conjugation of a chemical label. A number of such systems allowing direct coupling to proteins have been developed, including SNAP-tag, CLIP-tag, and Halo-tag technologies.8–10 Since fluorescent labels are not limited to genetically-encoded proteins, brighter, more photostable organic fluorophores can be used. In addition to fluorescent tags, proteins can be labeled with a variety of other chemical moieties, such as biotin or pharmacological compounds,11,12 enabling versatile functional studies or biochemical analysis. Because the chemical substrate is added to cells at a specific time, chemical labeling approaches can be controlled temporally with high resolution. While chemical tagging systems have fine temporal resolution, such methods have limited spatial resolution. Exceptions to these are methods using photocaged or photocleavable compounds such as photocaged dyes,13,14 compounds in which the labeling functional group is photocaged (eg, benzylguanine for SNAP-tag),15 or compounds with photocleavable linkers such as MeNV-HaXS (which links SNAP-tag and Halo-tag).16
To extend the versatility of chemical tagging methods to enable both temporal and spatial control over protein labeling without use of photocaged compounds, we explored whether we could regulate the SNAP-tag enzyme with light. SNAP-tag consists of an engineered variant of the 20 kDa human DNA repair enzyme O6-alkylguanine-DNA alkyltransferase (hAGT).8 The modified hAGT (ie, ‘SNAP-tag’) reacts specifically with O6-benzylguanine (BG) derivatives, forming a permanent covalent bond at Cys145.8 A variety of BG-conjugated chemicals, including fluorophores and biotin,12 are available, with applications that include protein trafficking studies,17 protein function analysis,18 protein half-life,19 and superresolution microscopy.20,21 SNAP-tag and the related Halo-tag10 technology have also been used to tether chemical antagonists to the cell surface or other anchored locations, where they modulate the function of nearby proteins.11,22–24 The ability to spatially control SNAP-tag activity would allow users to direct chemical labels to specific cells in a tissue, or even subcellular regions of a cell. Because the SNAP-tag label is permanent once conjugated, proteins tagged with photostable dyes could be tracked over long periods of time.
To regulate SNAP-tag with light, we took two approaches. First, we split the protein into N- and C-terminal fragments, which were then fused to light-induced dimerizers to enable light-induced reconstitution of protein activity. In a separate approach, we tested whether tethering a light-sensitive domain (AsLOV2) to the fragments could affect their self-association in a light-dependent manner. While each of these strategies alone showed modest light regulation, a combined approach was most successful. In the course of this work we also developed two new versions of split SNAP-tag, one of which consists of a small (30 residue) peptide fragment that robustly self-assembles with its complementary fragment. We expect these new tagging reagents will be useful for diverse protein labeling applications including protein trafficking studies, modulating protein function at user-specified locations, and reporting intersectional gene expression.
RESULTS AND DISCUSSION
Reconstituting split SNAP(N) and SNAP(C) fragments with light-induced dimerizers.
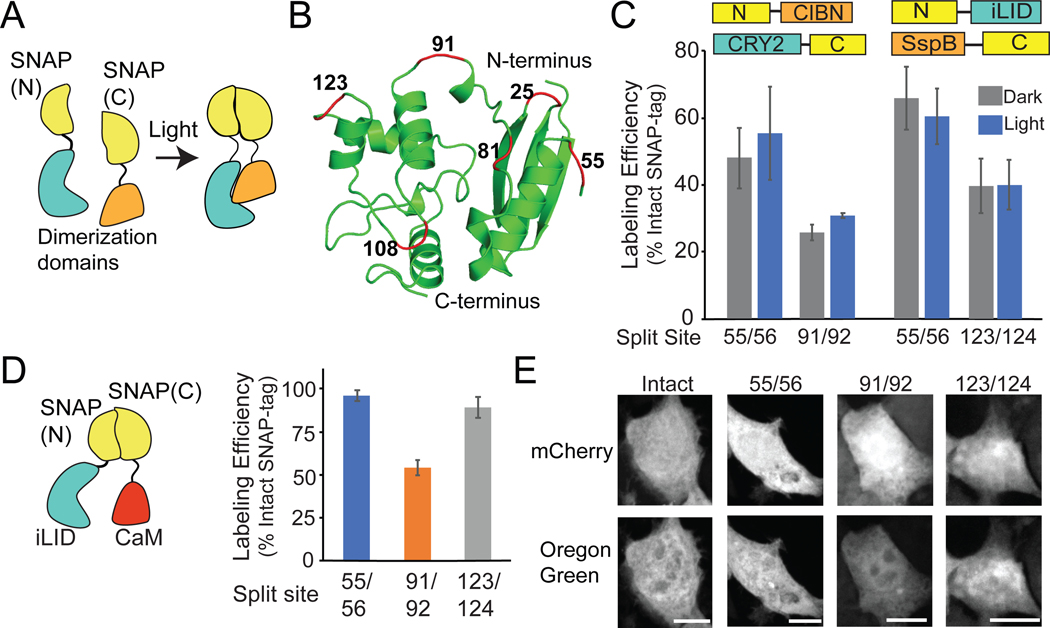
To regulate SNAP-tag activity with light, we tested an approach that was previously successful in our hands: dimerization-dependent split protein complementation.25,26 In this approach, a protein is split into two fragments that are fused to light-inducible dimerization domains. The split site is chosen such that in the dark, when dimerization is not induced, the attached fragments cannot reassemble on their own. Induction of dimerization with light brings the fragments in proximity and allows activity (Figure 1A). To identify an optimal site to split SNAP-tag, we tested six solvent-exposed surface loops, at residues 25/26, 55/56, 81/82, 91/92, 108/109, and 123/124 (Figure 1B). Four of the sites (81/82, 91/92, 108/109, 123/124) were at or near locations where SNAP-tag was previously split and reconstituted using chemical dimerizers;27,28 however in these previous studies the degree of background (uninduced) assembly of SNAP-tag fragments had not been clearly assessed.
Figure 1.
SNAP-tag activity can be reconstituted from split fragments. A) Schematic illustrating the reconstitution of SNAP(N) and SNAP(C) using dimerization domains. B) Location of tested split sites (red) within SNAP-tag (PDB:3KZY). C) Quantification of SNAP-tag activity (using BG-Oregon Green) with CRY2/CIBN or iLID/SspB photodimerizers. Labeling efficiency is expressed as a percent of intact SNAP-tag labeling. D, E) Quantification and representative images of BG-Oregon Green labeling with SNAP 55/56, 91/92, and 123/124 fused to non-interacting proteins iLID and CaM. Cells were labeled for 1 hr. Graphs represent average and error (S.E.M.) of 3 biological replicates, with over 30 cells quantified for each condition. Scale bars, 10 μm.
As our goal was to reconstitute SNAP-tag in a light dependent manner, we fused N-terminal and C-terminal fragments of SNAP-tag (SNAP(N) and SNAP(C), respectively) to Arabidopsis cryptochrome 2 (CRY2) and its binding partner CIBN (a truncated version of Arabidopsis CIB1), which dimerize in the presence of blue light.25 In parallel, we tested the same split sites with an alternate dimerizer, the iLID-SspB system. The iLID system is an engineered photodimerizer system derived from Avena sativa phototropin1 that is engineered to interact with E. coli SspB in blue light.29,30 We expressed the SNAP(N) and SNAP(C) fusion proteins in HEK293T cells and tested labeling using cell-permeable BG-conjugated Oregon Green. Of six split sites initially tested, three (55/56, 91/92, and 123/124) showed labeling activity (Supplementary Table 1), but limited or no light/dark differences (Figure 1C; Supplementary Figure 1).
We chose SNAP-tag split at residues 55/56, which showed slight light/dark differences with CRY2/CIBN (SNAP(N)-CIBN + CRY2-SNAP(C)) and the highest labeling efficiency with both CRY2/CIBN and iLID/SspB dimerizer systems, to explore for further testing. As a control, we verified that the SNAP(C) fragment (residues 56–182) is not enzymatically active on its own (Supplementary Figure 2). Because prior studies using dimerizers to reconstitute split protein fragments have shown that the N- or C-terminal orientation can greatly affect reconstitution,26 we tested other attachment orientations (Supplementary Figure 3), but these resulted in lower labeling activity. We focused on the iLID/SspB system (SNAP(N)-iLID + SspB-SNAP(C)), as it uses a smaller protein (13.1 kDa for SspB compared with 69.3 kDa for CRY2) fused to the chemically modified SNAP(C) fragment.
Disrupting self-assembly of split SNAP-tag fragments.
Optimal control of split protein reconstitution requires that the split protein fragments maintain moderate affinity such that they can assemble when brought together by dimerizers, but their affinity should be low enough such that they do not self-assemble on their own. The high dark background activity observed with both iLID/SspB and CRY2/CIBN systems suggested the fragments were of sufficiently high affinity to direct self-assembly independent of the dimerizers. To test this, we fused the 55/56 split SNAP-tag fragments to proteins that do not interact, generating SNAP(N)-iLID and Calmodulin-SNAP(C) (CaM-SNAP(C)). Use of these constructs resulted in nearly as efficient labeling (~93%) as observed with an intact SNAP-tag control (SNAP 1–182) (Figure 1D, 1E), indicating the 1–55 and 56–182 domains efficiently self-assemble. Using the same iLID/CaM non-interacting pair, SNAP-tag split at 91/92 and 123/124 also showed self-assembly (54% and 89% labeling compared with intact SNAP-tag, respectively) (Figure 1D, 1E).
The results showing substantial self-assembly of SNAP-tag split at 91/92 and 123/124 stand in contrast to several previous studies, which suggested that SNAP-tag split at these sites could be used to detect protein-protein interactions.27,28 SNAP(N)(1–91) was previously reported to assemble with SNAP(C)(92–182) only when fused to interacting proteins (for example, Fos and Jun). However, the control experiments used in these studies only tested interaction of the SNAP(1–91) and SNAP(92–182) fragments expressed on their own, rather than as a fusion with non-interacting proteins. In our hands, we found that both SNAP(1–91) and SNAP(92–182) are unstable when expressed on their own (Supplementary Figure 4) but are stably expressed in fusions with other proteins such as mCherry or calmodulin. When expressed as a fusion, the 91/92 split fragments show robust labeling with non-interacting proteins (Figure 1D, 1E). Our results suggest that use of these fragments to detect protein-protein interactions may yield false positive interaction results, although expressing at much lower levels may help to mitigate self-assembly.
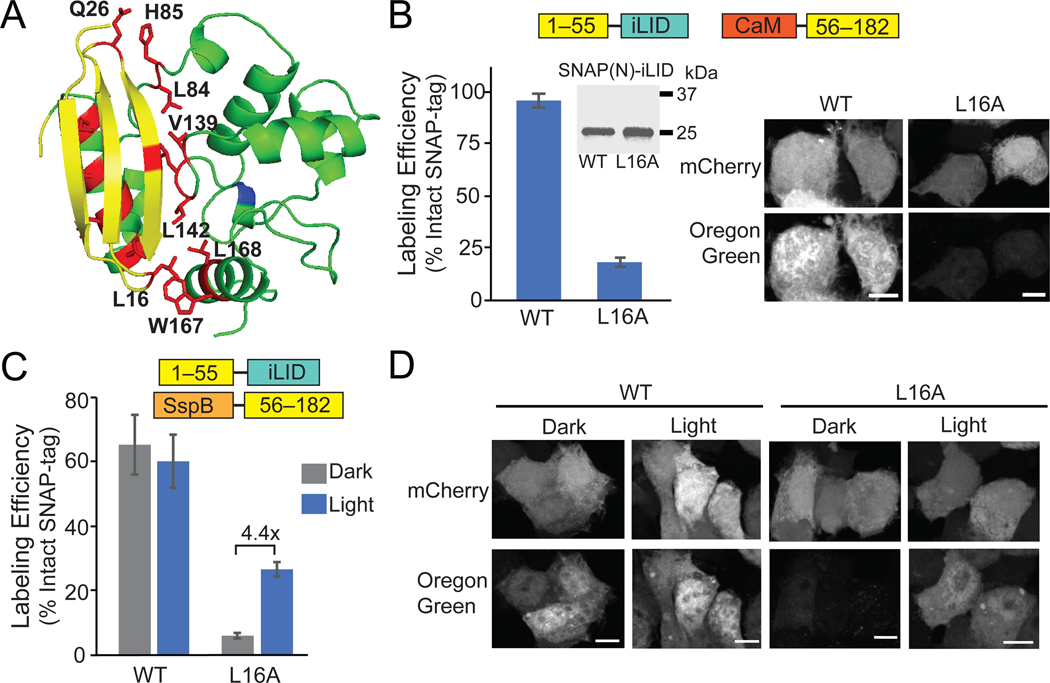
Based on previous studies with self-assembling split proteins,26 we hypothesized that we could identify point mutations along the SNAP(1–55)/(56–182) fragment interface that would reduce affinity sufficiently to disrupt self-assembly, but allow for the recovery of activity with dimerization. Analysis of the structure (PDB:3KZY) identified 15 polar and hydrophobic amino acid residues along the SNAP(N)/(C) interface, which we targeted for mutation to alanine (Figure 2A). Ten of these mutants showed reduced self-assembly using the mismatched dimerizer pair SNAP(N)-iLID and CaM-SNAP(C) (Supplementary Table 2).
Figure 2.
A L16A mutation disrupts SNAP 1–55/56–182 self-assembly and allows light-dependent activity. A) Residues (red) along SNAP(N) and SNAP(C) interface targeted for mutation. B) Quantification and representative images of BG-Oregon Green labeling using Calmodulin-SNAP(C) and SNAP(N)-iLID (wt or L16A). Inset shows immunoblot of HA-SNAP(N)-iLID (wt or L16A). C, D) Quantification (C) and images (D) of labeling of SspBmicro-SNAP(C) with SNAP(N)-iLID (wt or L16A), kept in dark or treated with light (1 s pulse 461 nm, every 30s for 2 hr). Graphs show average and error (S.E.M.) of 3 biological replicates. For each biological replicate, over 30 cells were quantified for each condition. Scale bars, 10 μm.
We focused on SNAP(N)(L16A)-iLID, which expressed equivalently to wild-type SNAP(N)-iLID but showed significantly reduced labeling when paired with CaM-SNAP(C) (from 94% to 23%) (Figure 2B). Leu16 forms a hydrophobic pocket along the N and C interface with five other hydrophobic residues (F70, I141, V164, W167, and L168) (Supplementary Figure 5). We hypothesize that mutating Leu16 to Ala destabilizes the hydrophobic pocket and reduces self-assembly and enzymatic activity. When we tested activity of the iLID/SspB-fused SNAP-tag fragments with the L16A mutation in SNAP(N), we observed significantly reduced dark background activity (6% labeling compared to 66% with wild-type SNAP(N)) (Figure 2C, 2D). Labeling in light was also reduced, but only about half compared with wild-type (26.5% with L16A compared to 60% with wt SNAP(N)).
Sterically blocking the SNAP-tag fragment interface with AsLOV2.
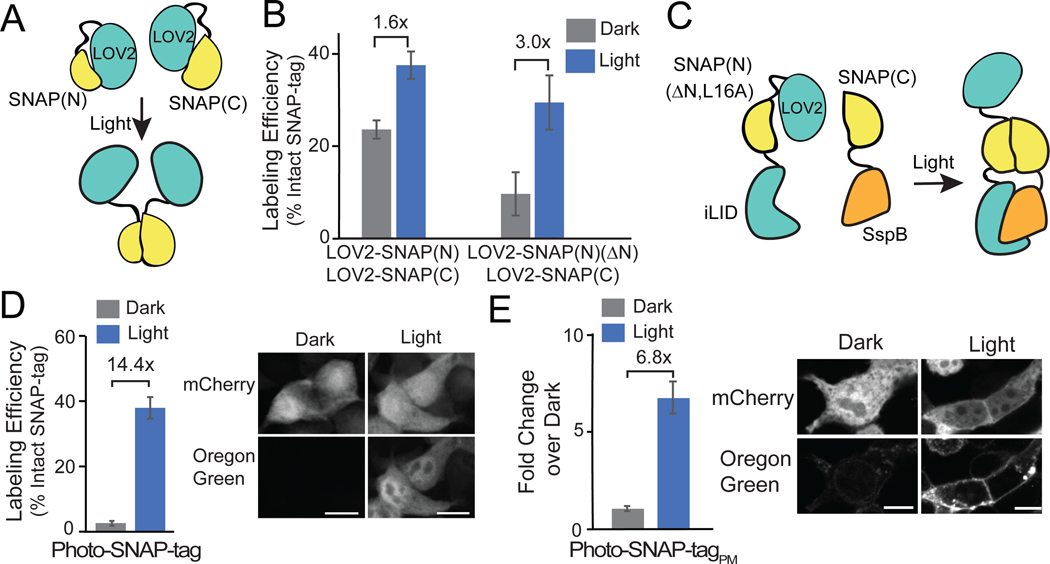
Our initial strategy using photodimerizers to reconstitute split SNAP-tag fragments showed promise, but light-induced labeling activity was still much lower that than seen with intact SNAP-tag (~25% as shown in Figure 2C). One inherent limitation of the split protein complementation approach is that the dynamic range of the system is limited by the dimerizer binding affinities in light and dark. As an alternative approach to generate a more robust labeling system, we tested a different photocontrol strategy: steric blocking of split wild-type fragment self-assembly using the AsLOV2-Jα photosensory domain. In the dark, a Jα-helix at the C-terminus is tightly bound to the core of the AsLOV2 protein, which can sterically prevent fused proteins or peptides from interacting with binding partners. With light, dissociation of the Jα-helix from the AsLOV2 core creates a configuration that can be more permissive for interaction.31 This conformational change has been used to regulate binding of peptides to their partners,29,32,33 as well as to regulate assembly of split intein protein fragments in a light-dependent manner.34
To test a steric-blocking approach to regulate fragment self-assembly, we attached AsLOV2-Jα at the N-terminus of SNAP(N)(1–55) and SNAP(C)(56–182) fragments (Figure 3A). The SNAP-tag fragments were attached to the Jα-helix at residue 542, a location previously successful.35 Cells treated with light for 2 hrs showed a modest light/dark difference of 1.6-fold (24% of intact protein labeling in dark; 38% in light) but high background, indicating inefficient steric blocking in the dark (Figure 3B). To improve the system, we truncated the first four residues of SNAP(N), which appear unstructured (PDB:3KZY), to bring SNAP(N) in closer proximity to the AsLOV2-Jα helix for more efficient blocking of activity. This version, LOV2-SNAP(N)(ΔN), showed reduced dark activity, but the light/dark fold change was still fairly low (10% labeling in dark; 30% in light) (Figure 3B).
Figure 3.
Steric blocking the N-terminus of SNAP(N) using AsLOV2 improves light regulation. A) Schematic of strategy to control interaction of SNAP(N) and SNAP(C) with AsLOV2. B) Quantification of labeling, comparing initial AsLOV2-fused version (left), and improved version (right, SNAP(N)(ΔN)), for cells kept in dark or light-treated (1s 461 nm pulse every 30s for 2h). Graph shows average and error (S.E.M.) of 3 biological replicates for graph at left, average and range for 2 for graph at right C) Strategy combining AsLOV2 steric blocking and dimerization. D) Quantification and images of Photo-SNAP-tag labeling in the cytosol in cells expressing LOV2-SNAP(N)(ΔN,L16A)-iLID and SspBmicro-SNAP(C). Graph shows average and error (S.E.M) of 3 biological replicates. E) Light/dark fold change and images of SNAP-Tag labeling (not normalized to intact SNAP-tag) in cells expressing mCh-LOV2-SNAP(N)(ΔN,L16A)-iLID(V416I) and plasma-membrane localized SspBmicro-SNAP(C)-CAAX. Graph reports average and error (S.E.M) of one experiment. 30 cells were quantified for each condition in all graphs. All scale bars, 10 μm.
Combined strategy for achieving light control combining steric blocking and dimerization.
While the dimerization and AsLOV2 steric blocking strategies each resulted in light-dark differences, on their own these differences were fairly modest (between 3 and 4.4-fold change in light vs dark). We hypothesized that a combined strategy, using AsLOV2-Jα and the L16A point mutation to prevent undesired self-assembly of the fragments in dark, then inducing fragment assembly in light through dimerization, could be effective. We fused LOV2-SNAP(N)(ΔN,L16A) to iLID and tested light-induced dimerization and reconstitution of activity with SspBmicro-SNAP(C) (Figure 3C). This combined approach, which we named ‘Photo-SNAP-tag,’ gave the best results so far. When used in the cytosol, we observed high activity in light (38% labeling) and minimal labeling in dark (2.5%), a 14.4-fold change (Figure 3D). A version anchored at the plasma membrane also showed low dark activity and good labeling in light (6.8-fold increase in light vs dark) (Figure 3E). We tested if we could improve labeling further by altering the ratio of expressed SNAP(N) and SNAP(C) fragments or generating a bicistronic vector using a P2A cleavable peptide to equalize split fragment expression (Supplementary Figure 6). The changes had minor effects on dark background, but overall did not improve light/dark dynamic range of labeling activity.
While generating ‘Photo-SNAP-tag,’ we also explored other design strategies to improve dynamic range. As previous studies have shown that different truncations of AsLOV2-Jα can have different effects on activity of fused proteins,32,33,35 we tested truncations at sites other than Ala542, finding low dark background using a truncation at Asp540. Although light-induced activity was also low, when combined with a higher affinity SspB-iLID pair (SspBwt-iLID) and wild-type SNAP-tag (rather than L16A), we could achieve similar labeling as with Photo-SNAP-tag (~7% labeling in dark; ~45% in light) (Supplementary Figure 7). We refer to this version as ‘Photo-SNAP-tagV2’.
Spatial control over SNAP-tag labeling.
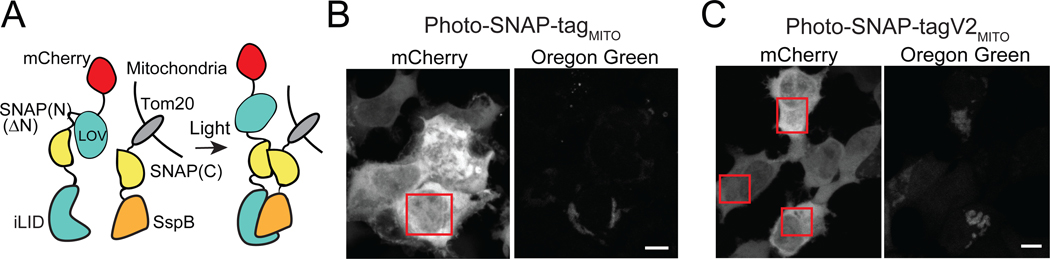
A main advantage of light is the capacity for spatial control, allowing users to steer light to cells to regulate activity in select locations. To examine spatial control of protein labeling, we generated mitochondrial-anchored versions of Photo-SNAP-tag, both original and V2 (‘Photo-SNAP-tagMITO’ and ‘Photo-SNAP-tagV2MITO’) (Figure 4A). We fused the SNAP(C) component (SspBmicro-SNAP(C) or SspBwt-SNAP(C) with V2) to Tom20, which localizes to the outer mitochondria membrane, and added a mCherry tag to the SNAP(N) fragment, to allow tracking of recruitment kinetics. Using the V2 version and global illumination, we observed robust recruitment of up to 35% of soluble SNAP(N) to mitochondria within 0.5–1 min after after light addition, resulting in a 11-fold light/dark difference in BG-Oregon Green labeling (Supplementary Figure 8).
Figure 4.
Spatial control of Photo-SNAP-tag at the mitochondria. A) Schematic of constructs. B,C) HEK293T cells were incubated with 1 μM BG-Oregon Green for 20 min (Photo-SNAP-tagMITO) (B) or 30 min (Photo-SNAP-tagV2MITO) (C). The boxed regions were focally stimulated with 488 nm light. Scale bars, 10 μm.
To test spatial control of labeling, we added 1 μM BG-Oregon Green to cells expressing each version of Photo-SNAP-tagMITO, then used a digital mirror device to steer blue light (488 nm, 150 ms pulse every 30 s (original system) or every 3 min (V2) to select cells. With both systems, cells exposed to blue light showed mitochondrial labeling specific to the illuminated areas after 20–30 min of light treatment (Figure 4C). We could observe initial labeling after ~10 min (not shown), but optimal labeling required >20 min. Minimal labeling was observed in unilluminated neighboring cells. These results indicate that Photo-SNAP-tag can be used to label proteins at user-defined sites in cells or tissues.
Split SNAP-tag as an intersectional reporter.
While our focus in these studies was on a method to spatially control SNAP-tag, our results showing strong self-assembly of wild-type SNAP(1–55) and SNAP(56–182) fragments (Figure 1D) led us to investigate other applications. We tested whether wild-type SNAP(N) and SNAP(C) fragment could act as intersectional reporters marking cells expressing both of two different genes. Such an application would be useful when construct size is an issue, such as when working with viral vectors that have size limitations. In addition, use of a single conjugated fluorophore to mark the expression of two plasmids would free up a fluorescent channel for other purposes.
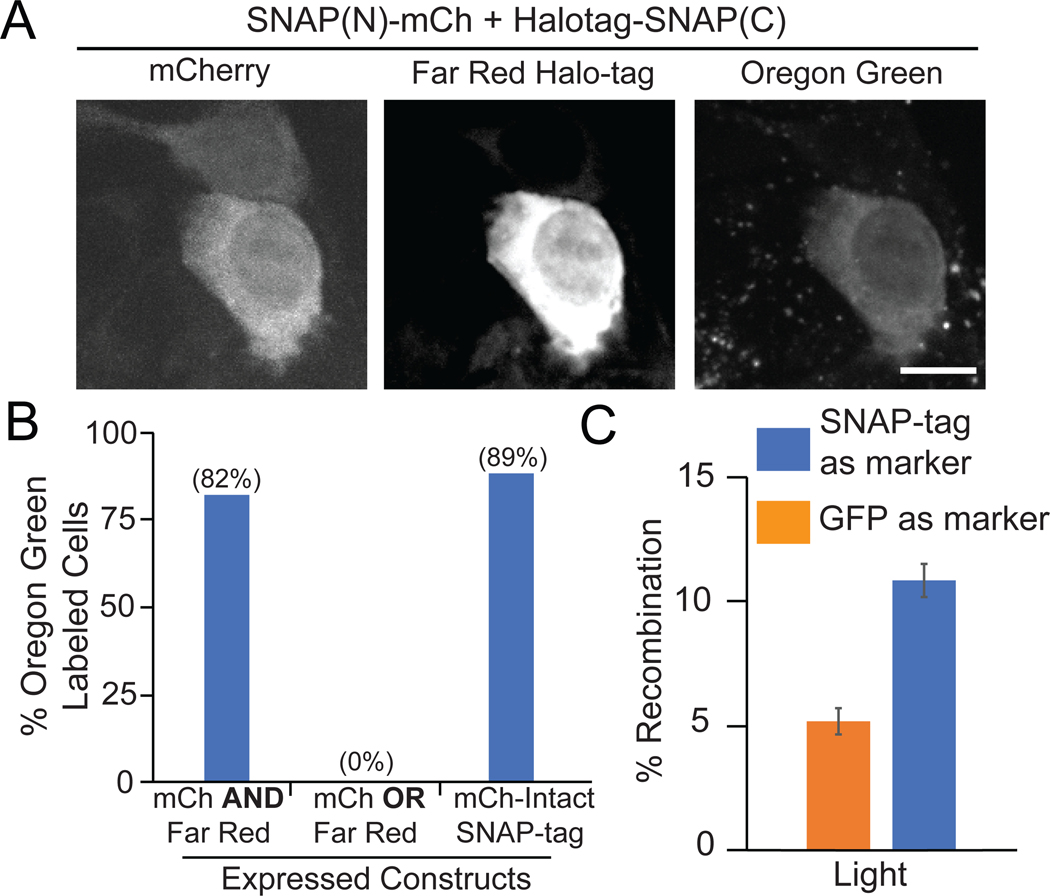
To evaluate use of split SNAP-tag as an intersectional reporter, we attached SNAP(N) to mCherry and SNAP(C) to Halo-tag,10 allowing independent monitoring of expression of each plasmid construct. 82% of cells expressing both constructs, indicated by co-expression of mCherry and a far-red Halo-tag JF646 dye, were labeled with BG-Oregon Green. In cases where mCh/far-red signals did not overlap with Oregon Green, the mCh or far-red expression level was very low. In comparison, a mCherry-Intact SNAP-tag control showed 89% of cells expressing mCherry labeled with Oregon Green, while cells expressing either mCherry or Halo-tag alone were not Oregon Green labeled (Figure 5A and 5B).
Figure 5.
Split SNAP(N)/(C) as intersectional reporter. A) Representative images of cells expressing mCherry and Halo-tag, or only mCherry, with BG-Oregon Green. Scale bars, 10 μm. B) Quantification of BG-Oregon Green labeling of cells expressing both mCherry and Halo-tag, only one construct, or a mCh-fused intact SNAP-tag control. Data represents a single experiment, 21–45 cells quantified each condition. C) Use of split SNAP-tag to quantify efficiency of a two-component split photoactivable Cre system. Cells were transfected with SNAP(N)-iLID-IRES-CIB1-Cre(C)/SNAP(C)-IRES-CRY2(L348F)-Cre(N) or CIB1-Cre(C)/EGFP-IRES-CRY2(L348F)-Cre(N), along with a floxed dsRed Cre reporter. Recombination efficiency was calculated using the split SNAP-tag, or a single transfection marker (EGFP-IRES upstream of the Cre recombinase fragment). Graph shows average and range of two biological replicates.
In a further test, we examined use of split SNAP-tag to mark cells expressing both components of a two plasmid photoactivatable split Cre system.36 SNAP(N)-iLID or SNAP(C) was inserted upstream of an IRES element, with each half of the photoactivatable Cre recombinase expressed downstream of the IRES (ie, SNAP(N)-iLID-IRES-CIB1-Cre(C) and SNAP(C)-IRES-CRY2(L348F)-Cre(N)). In this way, we could use the SNAP-tag label as a proxy to quantify the total number of transfected cells expressing both Cre fragments. When cells were transfected sparsely with a dsRed Cre reporter, use of the split SNAP-tag resulted in more accurate quantification of Cre efficacy compared with a single transfection reporter (EGFP-IRES reporter on one of the two Cre plasmids) that would be expected to underestimate activity (Figure 5C).
A 30-residue peptide tag for SNAP labeling applications.
We explored whether we could reduce the size of the SNAP(1–55) fragment to generate a smaller peptide tag that would self-assemble with SNAP(C), similar to approaches taken with EGFP (self-assembling GFP1–10/11 system)37 and S. pyogenes FbaB (eg., SpyTag/SpyCatcher).38 A smaller tag could be used to label proteins that do not tolerate larger fusions. For example, yeast Pma1 is incorrectly trafficked when fused to GFP at the C-terminus, but can be properly localized if first fused to a small SpyTag peptide.39 After trafficking, induction of GFP-SpyCatcher binds SpyTag and allows labeling of the correctly localized Pma1.
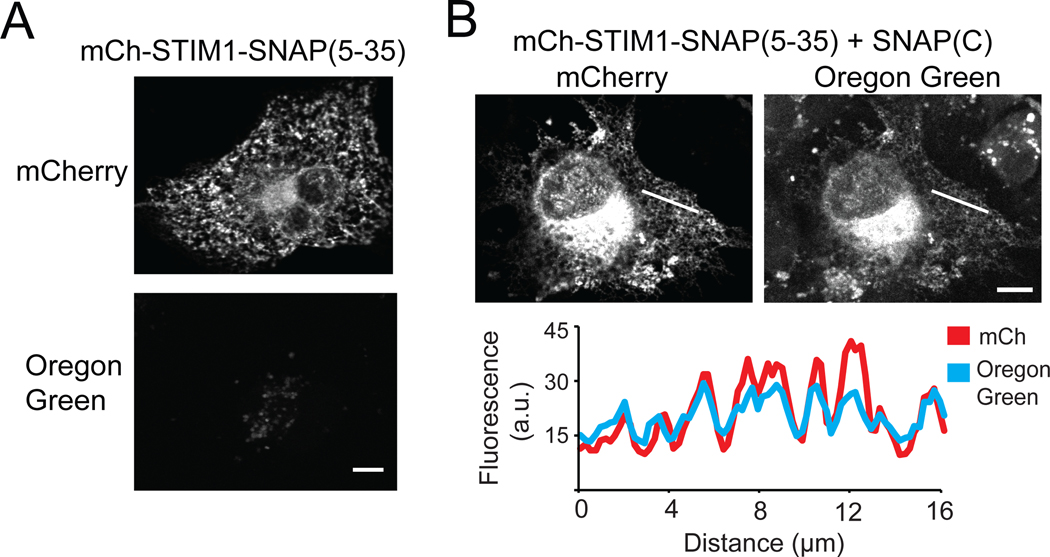
To identify a minimal SNAP fragment, we truncated residues 1–4 of SNAP(1–55), which we previously found dispensable (Figure 3B), as well as residues 36–54, which form an unstructured linker (PDB:3KZY) that we hypothesized could be removed without affecting function. We fused the SNAP(5–35) peptide at the C-terminus of the endoplasmic reticulum (ER) transmembrane protein STIM140 (with mCherry at the N-terminus) and examined localization in COS-7 cells. mCh-STIM1-SNAP(5–35) expressed well and was localized at ER (Figure 6A). When coexpressed with SNAP(C), we observed Oregon Green labeling that colocalized with mCherry at the ER (Figure 6B). These results show that residues 36–55 of SNAP-tag are dispensable for activity, and that SNAP(5–35) can reconstitute with its complementary SNAP(C) fragment to label proteins of interest.
Figure 6.
Using SNAP(5–35)/SNAP(C) to label ER-localized STIM1. A) COS-7 cell expressing mCh-STIM1-SNAP(5–35) showing mCherry localized to the ER and no Oregon Green labeling. B) Cell expressing mCh-STIM1-SNAP(5–35) and soluble SNAP(C) showing mCherry and Oregon Green labeling at the ER. Fluorescence intensity profiles at the white line are shown below. Scale bars, 10 μm.
Summary and Future Directions.
In these studies, we tested two different optogenetic strategies for regulating activity of the split SNAP-tag enzyme: a light-induced dimerization strategy and a strategy using steric blocking of self-assembling fragments. On their own, each strategy showed a light-dependent difference, but was only modestly effective. A combined approach was ultimately most successful, showing tighter light regulation and greater dynamic range than either system alone. We named this system Photo-SNAP-tag and demonstrated use of light to label mitochondria with an Oregon Green fluorophore at user-specified locations.
We envision that Photo-SNAP-tag could be used in similar ways as photoactivatable fluorescent proteins, but should provide additional benefits, including the ability to conjugate to a broader range of dyes, as well as to chemicals other than dyes, and the ability to permanently label a protein. This method is compatible with live cell imaging, thus light can be steered to individual cells or cell regions to selectively label targets. In this manner, proteins or organelles in specific subcellular locations could be labeled then tracked, or select cells in a tissue could be labeled (for example using BG-biotin) then biochemically separated. Such reagents would be useful for studying protein trafficking in large polarized cell types, such as neurons. Photo-SNAP-tag could also be used to increase the subcellular concentration of a drug to modulate the function of a protein or an organelle at a user-defined location, similar to strategies that have been taken with intact SNAP-tag, which for example has been used to target a kinase inhibitor to a specific subcellular location.11 Photo-SNAP-tag could expand on the utility of these existing approaches by providing spatial control. For example, light could be used to tag a subset of mitochondria with a drug that impacts mitochondrial function.
In addition to work developing Photo-SNAP-tag, we also identified split versions of SNAP-tag that undergo self-assembly. We demonstrate use as intersectional reporters to mark the expression of two plasmids. A split GFP 1–10/11 system has been used for intersectional applications, as well as other applications such as tagging endogenous proteins via CRISPR/Cas9, visualizing contact sites between different organelles, and sensing protein aggregation.41–44 Split FPs have also been used to map neuronal connections at synapses (eg., GFP Reconstitution Across Synaptic Partners or GRASP).45 Self-assembling SNAP-tag could be used in similar ways, but offers the versatility of chemical labeling, providing access to other fluorophores that are brighter, more stable, and are far-red shifted, activated by light that can penetrate more deeply in tissue.
We identified a minimal 30 residue peptide, SNAP(5–35), that self-assembles with its complementary SNAP(56–182) fragment and can be used to tag and label proteins at different subcellular locations. We expect this peptide can be used with proteins of interest that do not tolerate fusion to larger fluorescent proteins, or for tagging endogenous proteins using CRISPR methods, which are more efficient with smaller peptide tags.42
While we believe the Photo-SNAP-tag approach shows high potential, we acknowledge several limitations. One limitation is that the degree of reassembly is dependent on the level of expression of the two fragments. Because the SNAP-tag enzyme also serves as substrate, the degree of labeling is also dependent on expression of the C-terminal SNAP-tag fragment. For other photoactivatable split enzymes such as photoactivatable Cre recombinase,36 where a single enzyme can act on multiple substrates, only a small amount of enzyme may be required for high activity. In such cases, background activity can be reduced by reducing enzyme expression level, without impacting activity in the induced state. In contrast, because the Photo-SNAP-tag system acts to enzymatically modify itself, reducing expression of the enzyme also reduces the amount of substrate available for labeling. Because of these issues, it was challenging to maintain a high degree of labeling in light with minimal dark background. Even with these limitations, by altering the site of fusion with AsLOV2 and tuning the dimerizer and split fragment binding affinities, we were able to optimize two different Photo-SNAP-tag systems (original and V2, Figures 3D and Supplementary Figure 7A). While Photo-SNAP-tagV2 performed comparably and was not improved over the original version, these results highlight the different engineering modalities that could be further tuned to enhance function in future work.
A second limitation of Photo-SNAP-tag is the slow kinetics of labeling activity, as it required tens of minutes of light application to achieve visible cellular labeling (20 min in Figure 4B). The minutes-long time required to achieve labeling limits the spatial resolution of the approach. Our results showing rapid recruitment of a cytosolic SNAP(N) fragment to SNAP(C) anchored at mitochondria suggest that the slow kinetics of the process is due to a later step after the initial light-triggered heterodimerization, either due to slow self-assembly of the split fragments or poor enzymatic activity, requiring minutes to accumulate sufficient levels of tagged product for visualization. Another engineered optogenetic system, the FLARE system that uses TEV protease activity to mark light and calcium flux in neurons, has a similar limitation: its temporal resolution is limited by the cleavage kinetics of the enzyme.46 However, a recent study used yeast-based directed evolution to improve the temporal kinetics of FLARE.47 We envision a similar approach could be taken to evolve intact SNAP-tag and/or Photo-SNAP-tag to improve the labeling kinetics, allowing for greater subcellular resolution in future studies.
MATERIALS AND METHODS
Cloning and Mutagenesis.
Full sequences of the constructs used in this manuscript and the primers used in cloning are provided in Supplementary Table S3. Additional cloning details are provided in the Supplementary Information.
Cell Culture Experiments.
HEK293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C with 5% CO2. To quantify SNAP-tag labeling, HEK293T cells were seeded onto coverslips in 24-well plates and transfected with 500 ng of plasmid DNA using standard calcium phosphate transfection methods. For plasma membrane labeling experiments, 80 ng each of p1700 (mCh-LOV2-SNAP(N)(ΔN,L16A)-iLID(V416I)) and p1720 (SspBmicro-SNAP(C)-CAAX) were transfected into HEK293T. For the mitochondrial global cell labeling, 300 ng of p1726 (mCh-LOV2(540)-SNAP(N) (ΔN)-iLID(wt)) and 300 ng of p1725 (TOM20-SspBwt-SNAP(C)) were transfected into HEK293T. For ER localization experiments, COS-7 cells were seeded onto coverslips in 24-well plates and transfected with 200 ng of the specified plasmid DNA using Lipofectamine 2000 (ThermoFisher) following the manufacturer’s protocol.
Cells were wrapped in aluminum foil after transfection and kept in the dark until the following day. For the iLID-SspB systems, 1 s light pulses every 30 s were delivered (unless otherwise noted); for CRY2-CIBN systems, 2 s pulses were delivered every 3 min (461 nm delivered from custom-built LED arrays, 7–18 mW/cm2). Dark samples were kept in the dark the entire experiment. Light samples were pretreated with light for 1 hr before adding SNAP Oregon Green (New England Biolabs), and light treatment was continued during the 1 hr labeling reaction. Cells were labeled with 5 μM SNAP Oregon Green for 30 min, washed 3x with media, left for 30 min, then washed a final time. After the labeling reaction, coverslips were washed in 1x phosphate buffered saline (PBS), fixed in 4% paraformaldehyde, and mounted on a glass slide. Fixed cells were imaged on an Andor Dragonfly 301 spinning disc imaging system with Olympus IX73 base. Images were taken using a 60x UplanSApo 1.35 NA oil objective and collected on a 1024 × 1024 pixel Andor iXon EM-CCD camera. Data was acquired using Fusion or iQ3 (Andor).
Immunoblotting.
For immunoblotting, cells were harvested 24 hrs after transfection, washed with 1x PBS, and lysed in 2x Laemmli sample buffer with boiling. Proteins were separated by gel electrophoresis on a SDS-PAGE gel and transferred to nitrocellulose membranes followed by probing with primary (anti-HA, BioLegend #901501) and secondary (donkey anti-mouse IR-Dye 800CW, LiCOR 926–32212) antibodies. An Odyssey FC Imager (Li-COR) was used to visualize labeled immunoblots.
Image Analysis.
All images from fixed slides were quantified using ImageJ. For all images, Z-stacks were compiled into a maximal projection. To quantify SNAP-tag labeling, individual cells were manually selected and average fluorescence of the mCherry and Oregon Green channels were quantified. The background fluorescence was determined by measuring the fluorescence of each channel in untransfected cells, then subtracted from the mCherry and Oregon Green average fluorescence. For each cell, the Oregon Green labeling was normalized to the mCherry fluorescence in the same cell and multiplied by 100. The mCherry normalized Oregon Green labeling was then normalized to the Oregon Green labeling of cells expressing an intact SNAP-tag positive control within each experiment. Within each experiment, at least 30 cells were quantified per experimental condition. Reported data shows the average and error (S.E.M.) of three biological replicates performed independently, or in indicated graphs, average and range of two biological replicates.
Live Cell Imaging and Labeling.
HEK293T cells were seeded onto a 35 mm glass bottom culture dish and transfected with 150ng of p1701 (mCh-LOV2-SNAP(N)(ΔN,L16A)-iLID(wt)) or p1726 (mCh-LOV2(540)-SNAP(N)(ΔN)-iLID(V416I)) and 250ng p1717 (TOM20-SspBmicro-SNAP(C)) or p1725 (TOM20-SspBwt-SNAP(C)) for mitochondrial labeling experiments using calcium phosphate methods. The V416I mutation in iLID in p1700 and p1726 was included to lengthen the photocycle half-life from 30 s to 3 min, to enable treatment with less frequent light pulses.32 Cells were incubated in dark, wrapped in foil, with all manipulations performed under a red safe light. 12 to 24 hrs after transfection, cells were moved to Hanks Balanced Salt Solution (1.26 mM CaCl2, 0.41 mM MgSO4, .49 mM MgCl2, 5.33 mM KCL, 138 mM NaCl, 0.44 mM KH2PO4, 0.34 mM Na2HPO4, 4.17 mM NaHCO3, 5.56 mM D-glucose, and 20 mM HEPES) for live cell imaging. 1 μM BG-Oregon Green was also added. Live cell imaging was performed at 34°C on the Andor Dragonfly confocal system. For mitochondrial labeling experiments, regions of interest were locally stimulated with 488 nm light using a Mosaic digital mirror device (Andor), 150 ms every 30 s for 20 min for Photo-SNAP-tag, and 150 ms every 3 min for 30 min for Photo-SNAP-tagV2. Cells were washed 5x with imaging buffer (maintaining the position on the microscope), rested for 30 min, washed then imaged.
Cre recombinase experiments.
HEK293T cells were seeded on coverslips in 24-well plates and 250 ng of p1704 and 500 ng of p1714 were transfected using Lipofectamine 2000. The following day, cells were labeled with SNAP-Oregon Green and 3 μM Halotag JF646 dye, then fixed and imaged. To determine Cre recombinase activity, cells were seeded in 96-well plates and transfected with 125 ng of each construct (p1705 and p1706, or p1134 and p970) and a floxed DsRed Cre reporter48 (Addgene 13769), using Lipofectamine 2000. Cells were kept in the dark for 24 hrs then treated with a 4 s pulse of blue light. 24 hrs after light treatment cells were imaged to quantify reporter fluorescence. Quantification of Cre recombinase activity was carried out in live cells using a Leica DM IL LED Fluo microscope with L5 ET and TX2 ET filters. Images were taken with an iPhone7 (Apple) equipped with an iDu microscope adapter (iDu Optics) and visualized using ImageJ. The percent of transfected cells (determined by either SNAP-Oregon Green labeling or EGFP expression) showing DsRed reporter activity was quantified from images by manual count and is indicated in graphs as ‘% Cre recombination’.
Supplementary Material
ACKNOWLEDGEMENT
We thank B. Kuhlman for providing the iLID constructs used in this study and C. Cepko for providing the plasmid pCALNL-DsRed (Addgene, #13769).
FUNDING INFORMATION
This research was supported by grants (GM100225, GM136367) from the National Institutes of Health to C.L.T.
Footnotes
SUPPORTING INFORMATION
The Supporting Information is available free of charge via the Internet and includes:
Supplemental Figures 1 to 8; Supplemental Tables 1 and 2; Supplementary Note 1 (PDF) Supplemental Table 3 with details on all plasmids (Excel file)
REFERENCES
- (1).Patterson GH, and Lippincott-Schwartz J. (2002) A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877. [DOI] [PubMed] [Google Scholar]
- (2).Adam V, Berardozzi R, Byrdin M, and Bourgeois D. (2014) Phototransformable fluorescent proteins: Future challenges. Curr. Opin. Chem. Biol. 20, 92–102. [DOI] [PubMed] [Google Scholar]
- (3).Cranfill PJ, Sell BR, Baird MA, Allen JR, Lavagnino Z, De Gruiter HM, Kremers GJ, Davidson MW, Ustione A, and Piston DW (2016) Quantitative assessment of fluorescent proteins. Nat. Methods 13, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Suzuki T, Arai S, Takeuchi M, Sakurai C, Ebana H, Higashi T, Hashimoto H, Hatsuzawa K, and Wada I. (2012) Development of cysteine-free fluorescent proteins for the oxidative environment. PLoS One 7, e37551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, and Lippincott-Schwartz J. (2003) Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zacharias DA, Violin JD, Newton AC, and Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916. [DOI] [PubMed] [Google Scholar]
- (7).Nienhaus K, and Nienhaus GU (2016) Photoswitchable Fluorescent Proteins: Do Not Always Look on the Bright Side. ACS Nano 10, 9104–9108. [DOI] [PubMed] [Google Scholar]
- (8).Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, and Johnsson K. (2003) A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89. [DOI] [PubMed] [Google Scholar]
- (9).Gautier A, Juillerat A, Heinis C, Corrêa IR, Kindermann M, Beaufils F, and Johnsson K. (2008) An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Chem. Biol. 15, 128–136. [DOI] [PubMed] [Google Scholar]
- (10).Los GV, Encell LP, Mcdougall MG, Hartzell DD, Karassina N, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, and Zhu J. (2008) HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 3, 373–382. [DOI] [PubMed] [Google Scholar]
- (11).Bucko PJ, Lombard CK, Rathbun L, Garcia I, Bhat A, Wordeman L, Smith FD, Maly DJ, Hehnly H, and Scott JD (2019) Subcellular drug targeting illuminates local kinase action. Elife 8, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Xue L, Karpenko IA, Hiblot J, and Johnsson K. (2015) Imaging and manipulating proteins in live cells through covalent labeling. Nat. Chem. Biol. 11, 917–923. [DOI] [PubMed] [Google Scholar]
- (13).Maurel D, Banala S, Laroche T, and Johnsson K. (2010) Photoactivatable and photoconvertible fluorescent probes for protein labeling. ACS Chem. Biol. 5, 507–516. [DOI] [PubMed] [Google Scholar]
- (14).Banala S, Maurel D, Manley S, and Johnsson K. (2012) A caged, localizable rhodamine derivative for superresolution microscopy. ACS Chem. Biol. 7, 289–293. [DOI] [PubMed] [Google Scholar]
- (15).Banala S, Arnold A, and Johnsson K. (2008) Caged substrates for protein labeling and immobilization. ChemBioChem 9, 38–41. [DOI] [PubMed] [Google Scholar]
- (16).Zimmermann M, Cal R, Janett E, Hoffmann V, Bochet CG, Constable E, Beaufils F, and Wymann MP (2014) Cell-permeant and photocleavable chemical inducer of dimerization. Angew. Chemie - Int. Ed. 53, 4717–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Roed SN, Wismann P, Underwood CR, Kulahin N, Iversen H, Cappelen KA, Schäffer L, Lehtonen J, Hecksher-Soerensen J, Secher A, Mathiesen JM, Bräuner-Osborne H, Whistler JL, Knudsen SM, and Waldhoer M. (2014) Real-time trafficking and signaling of the glucagon-like peptide-1 receptor. Mol. Cell. Endocrinol. 382, 938–949. [DOI] [PubMed] [Google Scholar]
- (18).Foraker AB, Camus SM, Evans TM, Majeed SR, Chen CY, Taner SB, Corrêa IR, Doxsey SJ, and Brodsky FM (2012) Clathrin promotes centrosome integrity in early mitosis through stabilization of centrosomal ch-TOG. J. Cell Biol. 198, 591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Bojkowska K, Santoni De Sio F, Barde I, Offner S, Verp S, Heinis C, Johnsson K, and Trono D. (2011) Measuring in vivo protein half-life. Chem. Biol. 18, 805–815. [DOI] [PubMed] [Google Scholar]
- (20).Klein T, Löschberger A, Proppert S, Wolter S, Van De Linde S, and Sauer M. (2011) Live-cell dSTORM with SNAP-tag fusion proteins. Nat. Methods 8, 7–9. [DOI] [PubMed] [Google Scholar]
- (21).Pellett PA, Sun X, Gould TJ, Rothman JE, Xu M-Q, Corrêa IR, and Bewersdorf J. (2011) Two-color STED microscopy in living cells. Biomed. Opt. Express 2, 2364–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hill ZB, Gayani B, Perera K, and Maly DJ (2009) A chemical genetic method for generating bivalent inhibitors of protein kinases. J. Am. Chem. Soc. 131, 6686–6688. [DOI] [PubMed] [Google Scholar]
- (23).Broichhagen J, Damijonaitis A, Levitz J, Sokol KR, Leippe P, Konrad D, Isacoff EY, and Trauner D. (2015) Orthogonal optical control of a G protein-coupled receptor with a SNAP-tethered photochromic ligand. ACS Cent. Sci. 1, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shields BC, Kahuno E, Kim C, Apostolides PF, Brown J, Lindo S, Mensh BD, Dudman JT, Lavis LD, and Tadross MR (2017) Deconstructing behavioral neuropharmacology with cellular specificity. Science 356. [DOI] [PubMed] [Google Scholar]
- (25).Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, and Tucker CL (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Liu Q, Sinnen BL, Boxer EE, Schneider MW, Grybko MJ, Buchta WC, Gibson ES, Wysoczynski CL, Ford CP, Gottschalk A, Aoto J, Tucker CL, and Kennedy MJ (2019) A Photoactivatable Botulinum Neurotoxin for Inducible Control of Neurotransmission. Neuron 101, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mie M, Naoki T, Uchida K, and Kobatake E. (2012) Development of a split SNAP-tag protein complementation assay for visualization of protein-protein interactions in living cells. Analyst 137, 4760–4765. [DOI] [PubMed] [Google Scholar]
- (28).Mie M, Naoki T, and Kobatake E. (2016) Development of a split SNAP-CLIP double labeling system for tracking proteins following dissociation from protein-protein complexes in living cells. Anal. Chem. 88, 8166–8171. [DOI] [PubMed] [Google Scholar]
- (29).Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, and Kuhlman B. (2015) Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. 112, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zimmerman SP, Hallett RA, Bourke AM, Bear JE, Kennedy MJ, and Kuhlman B. (2016) Tuning the Binding Affinities and Reversion Kinetics of a Light Inducible Dimer Allows Control of Transmembrane Protein Localization. Biochemistry 55, 5264–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Harper SM, Neil LC, and Gardner KH (2003) Structural Basis of a Phototropin Light Switch. Science 301, 1541–1545. [DOI] [PubMed] [Google Scholar]
- (32).Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, and Glotzer M. (2012) TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, and Hahn KM (2009) A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wong S, Mosabbir AA, and Truong K. (2015) An engineered split intein for photoactivated protein trans-splicing. PLoS One 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Spiltoir JI, Strickland D, Glotzer M, and Tucker CL (2016) Optical Control of Peroxisomal Trafficking. ACS Synth. Biol. 5, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, and Tucker CL (2016) Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 12, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kamiyama D, Sekine S, Barsi-Rhyne B, Hu J, Chen B, Gilbert LA, Ishikawa H, Leonetti MD, Marshall WF, Weissman JS, and Huang B. (2016) Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, and Howarth M. (2012) Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hinrichsen M, Lenz M, Edwards JM, Miller OK, Mochrie SGJ, Swain PS, Schwarz-Linek U, and Regan L. (2017) A new method for post-translationally labeling proteins in live cells for fluorescence imaging and tracking. Protein Eng. Des. Sel. 30, 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Veliçelebi G, and Stauderman KA (2005) STIM1, an essential and conserved component of store-operated Ca 2+ channel function. J. Cell Biol. 169, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cabantous S, Terwilliger TC, and Waldo GS (2005) Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107. [DOI] [PubMed] [Google Scholar]
- (42).Leonetti MD, Sekine S, Kamiyama D, Weissman JS, and Huang B. (2016) A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl. Acad. Sci. 113, E3501–E3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Romei MG, and Boxer SG (2019) Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annu. Rev. Biophys. 48, 19–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kakimoto Y, Tashiro S, Kojima R, Morozumi Y, and Endo T. (2018) Visualizing multiple inter-organelle contact sites using the organelle- targeted split-GFP system. Sci. Rep. 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Feinberg EH, VanHoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008) GFP Reconstitution Across Synaptic Partners (GRASP) Defines Cell Contacts and Synapses in Living Nervous Systems. Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- (46).Wang W, Wildes CP, Pattarabanjird T, Sanchez MI, Glober GF, Matthews GA, Tye KM, and Ting AY (2017) A light-and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol. 35, 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Sanchez MI, and Ting AY (2020) Directed evolution improves the catalytic efficiency of TEV protease. Nat. Methods 17, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Matsuda T, and Cepko CL (2007) Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. 104, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.