Abstract
Adult T-cell leukemia/lymphoma (ATL) is an aggressive peripheral T-cell malignancy with a markedly poor prognosis. The low prevalence of ATL among human T-cell leukemia virus type-1 (HTLV-1) carriers and the long latency period before ATL onset suggest that additional genetic lesions are required for ATL leukemogenesis. Recently, a large-scale genetic analysis clarified the entire picture of genetic alterations, identified a number of novel driver genes, and delineated their characteristics. Frequent alterations are observed in the molecules belonging to T-cell receptor/NF-κB signaling and other T-cell-related pathways. A notable feature of the ATL genome is the predominance of gain-of-function alterations, including activating mutations in PLCG1, PRKCB, and CARD11. As many as one-fourth of all ATL cases harbor structural variations disrupting the 3′-untranslated region of the PD-L1 gene, leading to immune evasion of tumor cells. The frequency and pattern of these somatic alterations differ among clinical subtypes. Aggressive subtypes are associated with an increased burden of genetic alterations, and higher frequencies of TP53 and IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions than indolent subtypes. In contrast, STAT3 mutations are more characteristic of indolent ATL. Furthermore, these subtypes are further classified into molecularly distinct subsets with a different prognosis by genetic alterations. We present an overview of the current understanding of somatic alterations in ATL, with specific focus on their utility in clinical settings. Furthermore, we highlight their genetic features by exploring their similarities and differences among peripheral T-cell lymphomas.
Keywords: : Adult T-cell leukemia/lymphoma, genetic alterations, T-cell receptor signaling, immune evasion, clinical application
INTRODUCTION
Adult T-cell leukemia/lymphoma (ATL) is an aggressive peripheral T-cell malignancy with a markedly poor prognosis.1-3 This neoplasm is caused by human T-cell leukemia virus type-1 (HTLV-1) retroviral infection and is often reported in HTLV-1 endemic regions such as southwestern Japan, the Caribbean Basin, Latin America, Australia, Melanesia, the Middle East, Romania, and Central Africa. Only a small proportion of HTLV-1-infected individuals (6-7% of males and 2-3% of females) progress to ATL, typically more than 40-60 years after HTLV-1 infection.1-3 HTLV-1-coded gene products, such as Tax and HBZ, have been widely examined and demonstrated to play an essential role in ATL development and/or maintenance.4 However, the low prevalence of ATL among HTLV-1 carriers and the long latency period before ATL onset suggest the necessity of additional genetic alterations for ATL leukemogenesis.5
DISTINCT FEATURES OF GENETIC ALTERATIONS IN ATL
A large-scale genetic analysis clarified the overall picture of somatic alterations, identified a number of novel drivers, and characterized their unique genetic features6 (Table 1). This study demonstrated the strong enrichment of driver lesions in the T-cell receptor (TCR)/NF-κB signaling pathway, in which mutations in the proximal mediators of TCR signaling (PLCG1, VAV1, RHOA, and FYN) and downstream components belonging to the NF-κB pathway (PRKCB and CARD11), in addition to gene fusions involving a co-stimulatory and co-inhibitory molecule (CD28, CTLA4, and ICOS), are frequently observed.6 Of note, frequently altered genes, such as PLCG1, PRKCB, and CARD11, are predominantly targeted by activating mutations. Moreover, negative regulators of this pathway, including CSNK1A1, TNFAIP3, and CBLB, are commonly affected by loss-of-function mutations and focal deletions. Together, somatic alterations in the TCR/NF-κB pathway are noted in more than 90% of ATL cases, which is consistent with almost all ATL cells exhibiting constitutive activation of the NF-κB pathway.7 Indeed, the HTLV-1 protein Tax induces the nuclear localization and subsequent activation of NF-κB, which was proposed to play an important role in the transformation of T-cells by HTLV-1.4,8,9 However, HTLV-1 sense-strand genes, including Tax, are downregulated or even genetically silenced in most ATL cases.6 Several recent studies reported that Tax is expressed in short, self-terminating bursts in a minor fraction of ATL cells, leading to persistent infection and leukemogenesis.10,11 Therefore, ATL cells may exploit these host and viral mechanisms to activate the NF-κB pathway required for their maintenance and progression. Furthermore, other T-cell-related signaling pathways, including the JAK/STAT (STAT3) and NOTCH (NOTCH1) pathways, are frequently altered in ATL.
Table 1.
| Pathway | Gene | Altered frequency (WES) | Association with clinical subtype | Association with prognosis | Other alterations |
|---|---|---|---|---|---|
| TCR/NF-κB pathway | PLCG1 | 33% | |||
| PRKCB | 25% | Frequent in aggressive ATL | Worse in aggressive ATL | ||
| CARD11 | 21% | Intragenic deletions in 8% | |||
| VAV1 | 14% | ||||
| CD28 | 14% | Fusions in 7% | |||
| RHOA | 9% | ||||
| FYN | 1% | ||||
| CSNK1A1 | 6% | ||||
| TNFAIP3 | 6% | ||||
| CBLB | 5% | ||||
| Chemokine receptor | CCR4 | 31% | |||
| CCR7 | 11% | ||||
| GPR183 | 7% | Frequent in aggressive ATL | |||
| Tumor suppressor | TP53 | 25% | Frequent in aggressive ATL | ||
| CDKN2A | 22% | Worse in indolent ATL | |||
| TP73 | 1% | Intragenic deletions in 10% | |||
| Transcriptional regulation | IRF4 | 16% | Frequent in aggressive ATL | Worse in indolent ATL | |
| IRF2BP2 | 10% | Frequent in aggressive ATL | |||
| GATA3 | 14% | ||||
| PRDM1 | 12% | Frequent in aggressive ATL | |||
| IKZF2 | 4% | Intragenic deletions in 35% | |||
| Other signaling | NOTCH1 | 10% | |||
| STAT3 | 23% | Frequent in indolent ATL | |||
| Immune evasion | PD-L1 | 10% | Frequent in aggressive ATL | Worse in both ATL | 3′-UTR truncation in 27% |
| PDCD1 | 12% | ||||
| CD58 | 20% | Frequent in aggressive ATL | |||
| B2M | 4% | Frequent in aggressive ATL | |||
| HLA-A | 10% | ||||
| HLA-B | 7% | ||||
| FAS | 10% | ||||
| Epigenetic regulation | TET2 | 11% | Frequent in aggressive ATL | ||
| DNMT3A | 2% | ||||
| IDH2 | 2% | ||||
| EP300 | 5% |
Frequencies and clinical implications of somatic alterations in ATL (n = 81)6 analyzed by whole-exome sequencing (WES). Other alterations include somatic alterations detected by whole-genome or RNA sequencing.
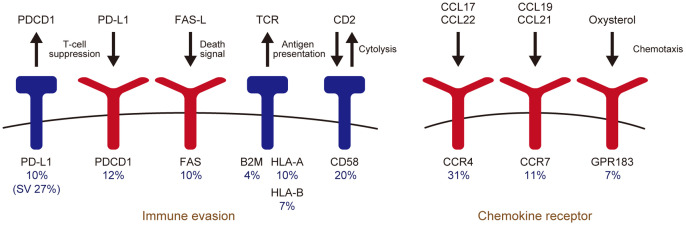
There are several lines of evidence that acquired immune response to HTLV-1 limits the proviral load, which is a major determinant for the risk of ATL development, suggesting the relevance of immune evasion in ATL pathogenesis.12 Indeed, immunogenic viral genes, especially Tax, are almost not expressed in most ATL cases.6 Moreover, ATL cells commonly acquire genetic lesions in immune-related molecules (Figure 1). In particular, as many as one-fourth of ATL cases harbor structural variations (SV) causing disruption of the PD-L1 3′-untranslated region (UTR).13 PD-L1 is an immune checkpoint molecule expressed by a variety of tumor cells, which binds to PD-1 receptor on effector T-cells, enabling tumor escape from immune surveillance.14 As a consequence of SV, such as deletion, inversion, tandem duplication, and translocation, 3′-UTR disruption causes PD-L1 overexpression, resulting in immune escape and tumor growth.13 Loss-of-function alterations of MHC class 1 (HLA-A, HLA-B, and B2M) and other immune-associated molecules (CD58 and FAS) are also present in more than half of all ATL cases.6 In addition to genetic alterations, DNA hypermethylation frequently affects the CpG islands of MHC class 1 genes, leading to reduced expression. Together, genetic and epigenetic abnormalities targeting antigen presentation machinery are present in approximately 90% of cases, demonstrating the importance of immune evasion in ATL.
Fig. 1.
Genetic alterations associated with immune evasion and chemotaxis. Altered frequencies by whole-exome sequencing (n = 81)6 are shown.
Another frequently altered pathway in ATL is chemokine receptors (CCR) mediating T-cell migration such as CCR4, CCR7, and GPR1836,15 (Figure 1). C-terminal truncating mutations of CCR4 and CCR7 induce upregulation of surface receptor expression, and augment ligand-mediated chemotaxis and downstream PI3K-AKT signaling, suggesting a gain-of-function property of these mutations.6,15 In contrast, nonsense and frameshift mutations and focal deletions frequently affect GPR183, encoding Epstein–Barr virus-induced gene 2 (EBI2), which is involved in positioning and cell fate determination of activated CD4+ T-cells.6,16 Mutations in epigenetic and histone modifying genes, such as TET2 and EP300, are also observed in ATL.6,17 These mutations were reported to be more prevalent in North American patients than in Japanese ATL patients, suggesting a possible difference in mutational profile among different ethnic cohorts.17
In addition to PD-L1 SV, ATL cells also harbor several unique SV such as intragenic deletions and/or inversions of IKZF2, TP73, and CARD11.6 Intragenic deletions and inversions commonly involve exons 5 and 6 of IKZF2 (or HELIOS), encoding an essential transcription factor in T-cell function, which accounts for approximately one-third of ATL cases. These intragenic deletions create aberrantly spliced transcripts, which were reported to act in a dominant-negative manner against IKZF1 (or IKAROS) and IKZF2.18 Another target of interest of intragenic deletion is TP73, a TP53 homolog. Found in approximately 10% of ATL cases, intragenic deletions exclusively affect exons 2 and 3 of the TP73 gene. These deletions lead to mutant p73 lacking the trans-activation domain, which was reported to function in a dominant-negative manner against p53 and p73.19 In contrast, small intragenic deletions targeting the inhibitory domain of CARD11 are recurrently observed in ATL. Similar to mutations, these intragenic deletions are thought to disrupt the interaction between the coiled-coil and inhibitory domains, resulting in constitutive activation of CARD11 and downstream NF-κB activation.20 Taken together, ATL exhibits a distinct profile of genetic lesions, which largely converge on the TCR/NF-κB pathway and other T-cell-related molecules. In addition, a unique constellation of somatic alterations, such as PD-L1 3′-UTR truncation and IKZF2 intragenic deletions, characterize the ATL genome, which may underlie its distinct pathological and clinical manifestations.
DIFFERENCES AND SIMILARITIES OF GENETIC ALTERATIONS AMONG PERIPHERAL T-CELL LYMPHOMAS
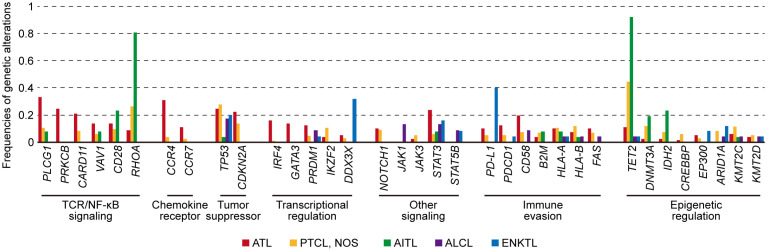
Peripheral T-cell lymphoma (PTCL) is a heterogeneous group of T-cell malignancies with different clinical, biological, and molecular features.21 In addition to ATL, this category includes PTCL, not otherwise specified (PTCL, NOS), angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), and extranodal natural killer (NK)/T-cell lymphoma, nasal type (ENKTL). The genetic landscapes of these PTCL subtypes demonstrate extensive heterogeneity within and across subtypes, in addition to several common themes through similar altered molecules and pathways, and overlapping mechanisms of lymphomagenesis (Figure 2).
Fig. 2.
Comparison of frequencies of genetic alterations among ATL (n = 81),6 PTCL, NOS (n = 133),27 AITL (n = 26),27 ALCL (n = 23),29 and ENKTL (n = 25)31 analyzed by whole-exome or targeted sequencing.
PTCL, NOS is a diagnosis of exclusion, being the most common subtype of PTCL. A subset of PTCL, NOS was reported to exhibit a T-follicular helper (TFH) cell phenotype, such as PD-1, CD10, CXCL13, BCL6, and ICOS expression, and to share pathological characteristics with AITL.21 In addition, these cases exhibit a similar pattern of genetic alterations to AITL such as TET2, RHOA, DNMT3A, and IDH2 mutations.22-25 In particular, the RHOA G17V mutation is highly specific to these two PTCL subtypes, and induces TFH lineage specification and the development of T-cell lymphomas, in cooperation with TET2 loss in vivo.26 Based on these findings, in the revised World Health Organization (WHO) classification of hematological malignancies, this subset of PTCL, NOS is classified as PTCL with a TFH cell phenotype as a provisional entity.21 On the other hand, recent genetic studies identified a new molecular subtype characterized by TP53 and/or CDKN2A mutations and deletions in non-TFH PTCL, NOS.27,28 This subtype exhibits distinct genetic features associated with widespread genomic instability, which preferentially affects molecules involved in immune escape such as HLA-A and HLA-B.27 In addition to these subtype-defining alterations, recurrent mutations are found in other epigenetic regulators (KMT2C and ARID1A) and TCR signaling molecules (PLCG1 and CD28).
ALCL is characterized by the proliferation of predominantly large lymphoid cells with strong CD30 expression and classified into two groups based on the expression of ALK.21 ALK-positive ALCL usually has a favorable outcome and has a defining translocation involving the ALK gene, such as NPM1-ALK, leading to STAT3 constitutive activation. In contrast, ALK-negative ALCL has recurrent activating mutations in JAK1 and/or STAT3, and translocations involving tyrosine kinases genes other than ALK.29 These genetic events also activate STAT3, which can explain the biological and pathological similarities between ALK-positive and ALK-negative ALCL.
ENKTL is a predominantly extranodal lymphoma of NK-cell or T-cell lineage, characterized by vascular damage and destruction, prominent necrosis, cytotoxic phenotype, and strong association with Epstein-Barr virus.21 The most common genetic alterations in ENKTL are PD-L1-involving structural variations, mainly consisting of 3′-UTR truncations, which are found in as many as 40% of cases.30 Recurrent mutations are also frequently located in the RNA helicase DDX3X, tumor suppressors (TP53 and MGA), and JAK/STAT pathway molecules (STAT3 and STAT5B).31
ATL shares many altered molecules and pathways with other PTCL subtypes. For example, most PTCL subtypes have a high prevalence of TP53 and STAT3 mutations. In addition, immune-related molecules and epigenetic regulators are altered in more than one subtype, with a range of frequencies. Although the TCR/NF-κB pathway is commonly affected in many PTCL subtypes, the altered frequency is highest in ATL, which is consistent with gene expression profiling demonstrating strong enhancement of this pathway in ATL.32 In this pathway, ATL harbors frequent alterations in PLCG1, CARD11, PRKCB, and IRF4, among which, the latter two are characteristic of ATL. On the other hand, RHOA G17V mutation is more characteristic of TFH-related lymphomas. Moreover, there are other genetic lesions with a high specificity for ATL such as those in chemokine receptors (CCR4 and CCR7). Therefore, although many somatic lesions are shared across PTCL subtypes, ATL exhibits a specific pattern of genetic alterations, which may characterize the distinct pathogenesis of this neoplasm.
ASSOCIATION OF GENETIC ALTERATIONS WITH DISEASE PHENOTYPE AND CLINICAL OUTCOME IN ATL
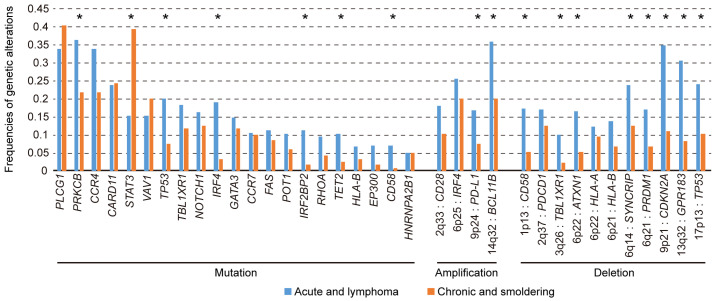
Based on clinical manifestations, ATL is classified into four subtypes: acute, lymphoma, chronic, and smoldering.1-3 Acute and lymphoma ATL are highly aggressive diseases with a markedly poor prognosis and usually require intensive treatment, including allogeneic hematopoietic stem cell transplantation (HSCT), which is the only potentially curative therapy for this neoplasm. The chronic and smoldering subtypes generally exhibit a slowly progressive clinical course, but many cases eventually progress to more aggressive disease. As many genetic alterations are shared across aggressive and indolent diseases, these subtypes are closely related from a genetic perspective. However, there are still substantial differences among them.33 Aggressive subtypes exhibit an increased burden of mutations and copy number alterations, suggesting that accumulated genetic alterations drive ATL progression. In particular, TP53 and IRF4 mutations, PD-L1 amplification, and CDKN2A deletion are more common in aggressive subtypes, suggesting that these alterations can lead to acute transformation of indolent ATL (Figure 3). Among these, the most significantly enriched is IRF4 mutation, preferentially targeting the DNA-binding domain, such as K59, L70, and S114, which suggests a gain-of-function nature. Indeed, K59R mutation was reported to increase nuclear localization and transcriptional activation, resulting in the expansion of T-cells in vivo.34
Fig. 3.
Comparison of frequencies of genetic alterations between aggressive (acute and lymphoma) and indolent (chronic and smoldering) ATL analyzed by targeted sequencing (n = 414) and SNP array (n = 463).33
On the other hand, only STAT3 mutation is more frequent in indolent ATL, affecting 39% of indolent cases compared with 15% of aggressive cases,33 suggesting that STAT3 mutation characterizes indolent behavior. The clinical course until the development of aggressive ATL is diverse; some patients acutely develop aggressive diseases from an asymptomatic status, whereas indolent disease transforms into acute disease after many years in others. The presence of STAT3 mutation in aggressive ATL suggests a transformation from indolent ATL. Almost all STAT3 mutations affect the SH2 domain, forming prominent hotspots at Y640 and D661, which were demonstrated to induce increased STAT3 transcriptional activity.6,33,35 A high frequency of STAT3 mutation is also observed in other mature T- or NK-cell neoplasms, such as large granular lymphocytic leukemia and chronic lymphoproliferative disorders of NK cells, implicating gain-of-function STAT3 mutation in the slowly progressive clonal expansion of T- and NK cells.36,37 As all ATL cases harboring STAT3 mutations were reported to exhibit distinct genetic features, such as rare TP53 mutations, further investigation of this entity is warranted.6
It is well known that several clinical factors are associated with the clinical outcome of ATL.1-3 For example, a prognostic index for acute and lymphoma subtypes (ATL-PI; based on stage, performance status, age, albumin, and soluble IL-2 receptor level) and Japan Clinical Oncology Group prognostic index (JCOG-PI; based on calcium level and performance status) can predict patient survival in aggressive ATL.38,39 Moreover, albumin, blood urea nitrogen (BUN), and lactate dehydrogenase (LDH) levels are adverse prognostic factors for indolent ATL.1-3 In addition to these clinical factors, genetic alterations were recently reported to influence the prognosis of ATL.33 First, higher numbers of somatic mutations and copy number alterations are significantly correlated with a poorer prognosis of ATL patients. Second, among these somatic alterations, PRKCB mutations and PD-L1 amplification are independent poor prognostic indicators in aggressive ATL, even after adjusting for clinical factors. In indolent ATL, IRF4 mutation, PD-L1 amplification, and CDKN2A deletion independently predict shorter survival, suggesting that genetic aberrations characterizing aggressive ATL are associated with a poorer prognosis in indolent ATL. Taken together, these observations suggest that ATL clinical subtypes can be divided into genetically different subsets that have a different prognosis. Genetic profiling by next-generation sequencing can improve prognostic stratification and provide a better therapeutic strategy such as early intervention by systemic chemotherapy and/or allogeneic HSCT for indolent ATL.
GENETIC ALTERATIONS AS THERAPEUTIC TARGETS FOR ATL
In addition to the prognosis, the associations of genetic alterations with the response to a specific targeted therapy have been reported. CCR4 mutation was found to predict a better response to the anti-CCR4 antibody mogamulizumab, suggesting the possibility of this mutation as a biomarker.40 In addition, patients with Hodgkin lymphoma harboring PD-L1 genetic alteration exhibit superior outcomes following PD-1 blockade therapy, although this alteration is associated with shorter survival after conventional chemotherapy.41,42 Thus, PD-L1 amplification and 3′-UTR truncation may function as predictive biomarkers for immune checkpoint blockade therapy in ATL. On the other hand, ATL has frequent focal deletions in PDCD1, encoding PD-1, which is a master gene suppressing oncogenic T-cell signaling.6,43 Indeed, a small prospective study found that patients with ATL had rapid disease progression and increased viral replication after treatment using the anti-PD-1 antibody nivolumab, although it is not clear whether these patients had PD-L1 genetic alterations.44 Therefore, further studies carefully examining the efficacy of immune checkpoint therapy and its associations with somatic alterations involving PDCD1 and PD-L1 are needed for ATL. In addition, inhibitors of MALT1, which forms signalosome complexes with CARD11 and BCL10, were previously reported to suppress NF-κB and JAK/STAT signaling, and induce growth retardation in an activated B-cell-like subtype of diffuse large B-cell lymphoma.45,46 The predominance of gain-of-function mutations in the TCR/NF-κB signaling pathway provides theoretical rationale for exploiting these agents against ATL. Therefore, molecular profiling can not only improve prognostic stratification, but also help in predicting the efficacy of molecularly targeted agents.
CONCLUSION
Next-generation sequencing technologies have delineated the entire landscape of genetic alterations and identified many driver genes in ATL. The ATL genome is characterized by frequent alterations in molecules associated with TCR/NF-κB signaling, immune evasion, and other T-cell-related pathways. In addition, these somatic alterations differ in frequency among subtypes and further characterize molecularly distinct subsets with different prognoses within the subtype. These observations provide useful information for exploiting these alterations as predictive biomarkers and novel drug targets in ATL. Taken together, the genetic findings provide novel insights into the molecular basis of ATL, which can be used for further therapeutic and diagnostic development to improve the management of this disease.
ACKNOWLEDGMENTS
This study was supported by a Grant-in-Aid from the Japan Agency for Medical Research and Development [Research on Development of New Drugs (19ak0101064)] and a Grant-in-Aid for Scientific Research on Innovative Areas (KAKENHI 18H04907).
Footnotes
CONFLICT OF INTEREST
K.K. owns stock in Asahi Genomics. The remaining author declares no relevant competing financial interests.
REFERENCES
- 1.Tsukasaki K, Hermine O, Bazarbachi A, et al. Definition, prognostic factors, treatment, and response criteria of adult T-cell leukemia-lymphoma: a proposal from an international consensus meeting. J Clin Oncol. 2009; 27: 453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishitsuka K, Tamura K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol. 2014; 15: e517-e526. [DOI] [PubMed] [Google Scholar]
- 3.Cook LB, Fuji S, Hermine O, et al. Revised Adult T-Cell Leukemia-Lymphoma International Consensus Meeting Report. J Clin Oncol. 2019; 37: 677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007; 7: 270-280. [DOI] [PubMed] [Google Scholar]
- 5.Kogure Y, Kataoka K. Genetic alterations in adult T-cell leukemia/lymphoma. Cancer Sci. 2017; 108: 1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015; 47: 1304-1315. [DOI] [PubMed] [Google Scholar]
- 7.Mori N, Fujii M, Ikeda S, et al. Constitutive activation of NF-kappaB in primary adult T-cell leukemia cells. Blood. 1999; 93: 2360-2368. [PubMed] [Google Scholar]
- 8.Ruben S, Poteat H, Tan TH, et al. Cellular transcription factors and regulation of IL-2 receptor gene expression by HTLV-I tax gene product. Science. 1988; 241: 89-92. [DOI] [PubMed] [Google Scholar]
- 9.Ballard DW, Böhnlein E, Lowenthal JW, et al. HTLV-I tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science. 1988; 241: 1652-1655. [DOI] [PubMed] [Google Scholar]
- 10.Mahgoub M, Yasunaga J, Iwami S, et al. Sporadic on/off switching of HTLV-1 Tax expression is crucial to maintain the whole population of virus-induced leukemic cells. Proc Natl Acad Sci USA. 2018; 115: E1269-E1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura M, Dey S, Ramanayake S, et al. Kinetics of HTLV-1 reactivation from latency quantified by single-molecule RNA FISH and stochastic modelling. PLoS Pathog. 2019; 15: e1008164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bangham CRM, Osame M. Cellular immune response to HTLV-1. Oncogene. 2005; 24: 6035-6046. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016; 534: 402-406. [DOI] [PubMed] [Google Scholar]
- 14.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016; 16: 275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa M, Schmitz R, Xiao W, et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J Exp Med. 2014; 211: 2497-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Lu E, Yi T, Cyster JG. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature. 2016; 533: 110-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah UA, Chung EY, Giricz O, et al. North American ATLL has a distinct mutational and transcriptional profile and responds to epigenetic therapies. Blood. 2018; 132: 1507-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asanuma S, Yamagishi M, Kawanami K, et al. Adult T-cell leukemia cells are characterized by abnormalities of Helios expression that promote T cell growth. Cancer Sci. 2013; 104: 1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002; 2: 605-615. [DOI] [PubMed] [Google Scholar]
- 20.Sommer K, Guo B, Pomerantz JL, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005; 23: 561-574. [DOI] [PubMed] [Google Scholar]
- 21.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lypm, IARC Press. 2017. [Google Scholar]
- 22.Cairns RA, Iqbal J, Lemonnier F, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012; 119: 1901-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012; 366: 95-96. [DOI] [PubMed] [Google Scholar]
- 24.Palomero T, Couronné L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014; 46: 166-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014; 46: 171-175. [DOI] [PubMed] [Google Scholar]
- 26.Cortes JR, Ambesi-Impiombato A, Couronné L, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018; 33 : 259-273 e7. [DOI] [PMC free article] [PubMed]
- 27.Watatani Y, Sato Y, Miyoshi H, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019; 33: 2867-2883. [DOI] [PubMed] [Google Scholar]
- 28.Heavican TB, Bouska A, Yu J, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019; 133: 1664-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crescenzo R, Abate F, Lasorsa E, et al. European T-Cell Lymphoma Study Group, T-Cell Project: Prospective Collection of Data in Patients with Peripheral T-Cell Lymphoma and the AIRC 5xMille Consortium “Genetics-Driven Targeted Management of Lymphoid Malignancies”. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015; 27: 516-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kataoka K, Miyoshi H, Sakata S, et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia. 2019; 33: 1687-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L, Gu ZH, Yan ZX, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015; 47: 1061-1066. [DOI] [PubMed] [Google Scholar]
- 32.Iqbal J, Weisenburger DD, Greiner TC, et al. International Peripheral T-Cell Lymphoma Project. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010; 115: 1026-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kataoka K, Iwanaga M, Yasunaga J, et al. Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood. 2018; 131: 215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherian MA, Olson S, Sundaramoorthi H, et al. An activating mutation of interferon regulatory factor 4 (IRF4) in adult T-cell leukemia. J Biol Chem. 2018; 293: 6844-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellon M, Lu L, Nicot C. Constitutive activation of Pim1 kinase is a therapeutic target for adult T-cell leukemia. Blood. 2016; 127: 2439-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012; 120: 3048-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koskela HLM, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012; 366: 1905-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsuya H, Yamanaka T, Ishitsuka K, et al. Prognostic index for acute- and lymphoma-type adult T-cell leukemia/lymphoma. J Clin Oncol. 2012; 30: 1635-1640. [DOI] [PubMed] [Google Scholar]
- 39.Fukushima T, Nomura S, Shimoyama M, et al. Japan Clinical Oncology Group (JCOG) prognostic index and characterization of long-term survivors of aggressive adult T-cell leukaemia-lymphoma (JCOG0902A). Br J Haematol. 2014; 166: 739-748. [DOI] [PubMed] [Google Scholar]
- 40.Sakamoto Y, Ishida T, Masaki A, et al. CCR4 mutations associated with superior outcome of adult T-cell leukemia/lymphoma under mogamulizumab treatment. Blood. 2018; 132: 758-761. [DOI] [PubMed] [Google Scholar]
- 41.Roemer MGM, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol. 2016; 34: 2690-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roemer MGM, Redd RA, Cader FZ, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol. 2018; 36: 942-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wartewig T, Kurgyis Z, Keppler S, et al. PD-1 is a haploinsufficient suppressor of T cell lymphomagenesis. Nature. 2017; 552: 121-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratner L, Waldmann TA, Janakiram M, Brammer JE. Rapid progression of adult T-cell leukemia-lymphoma after PD-1 inhibitor therapy. N Engl J Med. 2018; 378: 1947-1948. [DOI] [PubMed] [Google Scholar]
- 45.Fontán L, Qiao Q, Hatcher JM, et al. Specific covalent inhibition of MALT1 paracaspase suppresses B cell lymphoma growth. J Clin Invest. 2018; 128: 4397-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferch U, Kloo B, Gewies A, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell–like diffuse large B cell lymphoma cells. J Exp Med. 2009; 206: 2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]