Abstract
MYC is a transcriptional factor that regulates growth and proliferation through cell cycle pathways. MYC alterations, in particular MYC rearrangements, are important in assessing the prognosis of aggressive B-cell lymphoma. In this study, we focused on the impact of nine major cell cycle genes for MYC-driven aggressive mature B-cell lymphoma and analyzed the mutational status using targeted next generation sequencing. Our 40 cases of aggressive mature B-cell lymphomas included 5 Burkitt lymphomas, 17 high-grade B-cell lymphomas and 18 diffuse large B-cell lymphomas with MYC breaks in 100%, 88% and 11%, respectively. Our data allowed a molecular classification into four categories partially independent from the histopathological diagnosis but correlating with the Ki-67 labelling index: (I) harboring TP53 and CDKN2A mutations, being highly proliferative, (II) with MYC rearrangement associated with MYC and/or ID3 mutations, being highly proliferative, (III) with MYC rearrangement combined with additional molecular changes, being highly proliferative, and (IV) with a diverse pattern of molecular alterations, being less proliferative. Taken together, we found that mutations of TP53, CDKN2A, MYC and ID3 are associated with highly proliferative B-cell lymphomas that could profit from novel therapeutic strategies.
Keywords: Cell cycle, Diffuse large B-cell lymphoma, High-grade B-cell lymphoma, MYC
INTRODUCTION
MYC is a transcriptional factor and proto-oncoprotein that mediates apoptosis, differentiation and proliferation, deregulation of its gene is detected in 50-70% of all human malignancies.1,2 In particular, MYC rearrangement is often strongly correlated with a poor prognosis in a variety of malignant hematological tumors and is known not only as a hallmark of Burkitt lymphoma (BL), but also as a prognostic factor in other aggressive mature B-cell lymphomas such as diffuse large B-cell lymphoma (DLBCL) and high-grade B-cell lymphoma (HBL).3,4 HBL was established as a new disease entity in the current 2017 World Health Organization (WHO) classification of Haematopoietic and Lymphoid Tissue, and partly replaced the provisional entity of B-cell lymphoma, unclassifiable with features intermediate between DLBCL and BL (BCL-U) of the previous classification.5 The HBL category is further divided into two subtypes: (1) HBL associated with MYC and BCL2 and/or BCL6 rearrangements [HBL-double hit (DH)/triple hit (TH)], and (2) HBL not otherwise specified (NOS). HBL-DH/TH harbors chromosomal rearrangements of MYC at the 8q24 locus and BCL2 at the 18q21 locus, and/or BCL6 at the 3q27 locus. HBL with MYC/BCL2 rearrangements accounts for most cases of this entity and has a more dismal prognosis than typical DLBCL.6,7 On the other hand, HBL NOS does not harbor double rearrangements, but approximately 20-35% of these have isolated MYC rearrangement with or without copy number alteration or amplification of the BCL2 region. Some cases harbor BCL2 rearrangement with MYC copy number alteration or amplification.8-10 However, the precise definition of HBL NOS is currently controversial and not well defined. Additionally, the differentiation from DLBCL with MYC abnormalities or BL and HBL NOS is impeded due to limited knowledge of the biological background, allowing merely insufficient morphological or immunophenotypical categorization. Therefore, a more reliable, easily applicable and accurate method to classify these categories is required. Next generation sequencing (NGS) of BL has elucidated recurrent somatic mutations in CCND3, ID3, TCF3 and TP53, which add to the malignant potential of BL by activating the cell cycle pathway.11-13 Momose et al. reported that recurrent somatic mutations of CCND3, ID3, TCF3 and MYC were detected not only in BL, but also in the other MYC-driven aggressive mature B-cell lymphomas, like HBL and DLBCL with MYC rearrangement.14 Furthermore, inactivating mutations of tumor suppressor genes (e.g. BTG1, BTG2, CDKN2A, and TP53) mediating cell cycle arrest were detected in MYC-driven aggressive mature B-cell lymphomas.9,15-18 Recently, many alterations of cell cycle- or apoptosis-genes were detected in approximately 40% of HBL-DH/TH cases.19 In this context, it is conceivable that the molecular change in these specific cell cycle genes affects many MYC-driven aggressive mature B-cell lymphomas. Moreover, the mutational status may allow a sub-classification into molecular subgroups independent from the histopathological diagnosis.
In this study, we focused on the mutational status of cell cycle genes (BTG1, BTG2, CCND3, CDKN2A, ID3, MAX, MYC, TCF3 and TP53) to gain deeper insights into the molecular mechanisms of MYC-driven aggressive mature B-cell lymphoma.
MATERIALS AND METHODS
Case selection
We selected 168 cases of aggressive mature B-cell lymphoma diagnosed as BL (n=12), HBL-DH/TH (n=17), HBL NOS (n=8) and DLBCL NOS (n=131) according to the criteria of the current WHO classification between 2016 and 2018 diagnosed at the Institute of Pathology at the Charité - Universitätsmedizin Berlin. These cases were reviewed by two pathologists (I.A and T.Y). Next, we selected 68 of 168 cases that fulfilled our criteria for MYC-driven aggressive mature B-cell lymphoma, which were analyzed for not only MYC expression (≥40%), but also the Ki-67 labeling index (LI) (≥80%). As we used the Ki-67 LI for case selection, Ki-67 was both a useful prognostic marker and cell cycle-related cellular marker for cell proliferation in the cases of aggressive lymphoma. Recent studies reported that overexpression of Ki-67 was a predictor of a dismal prognosis in patients treated using rituximab.20-22 Moreover, MYC expression was independent of Ki-67 and mature B-cell lymphomas harboring high MYC expression did not always have high proliferation rates.23 Our criteria of MYC-driven aggressive mature B-cell lymphoma referenced the previous report, which compared the expression of MYC with that of Ki-67.16,23 Finally, we excluded 28 cases, in particular CD5-positive DLBCL, DLBCL with coexistent or transformed follicular lymphoma (FL), human immune deficiency virus-related lymphoma, primary central nervous system lymphoma, and cases mostly exhibiting necrotic tissue or insufficient material for our analysis (Supplementary figure 1).
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Commission of the Charité-Universitätsmedizin (approval number: EA4/129/15).
Immunohistochemistry (IHC)
The following antibodies were used for IHC: anti-CD3 (clone: LN10, dilution: 1:100, Novocastra, Leica Biosystems, Germany), anti-CD10 (clone: 56C6, dilution: 1:25, Novocastra, Leica Biosystems), anti-CD20 (clone: L26, dilution: 1:100, DAKO, Agilent-Dako, USA), anti-BCL2 (clone: 124, dilution: 1:25, Agilent-Dako), anti-BCL6 (clone: PG-B6p, dilution: 1:25, Agilent-Dako), anti-IRF4 (clone: MUM1p, dilution: 1:25, Agilent-Dako), anti-MYC (clone: Y69, dilution: 1:300, Epitomics, USA) and anti-Ki-67 (clone: MIB-1, dilution: 1:200, Agilent-Dako). Immunohistochemical staining was performed using BOND-MAX (Leica, Germany) according to the manufacturer’s instructions. Protein expression was assessed independently by two pathologists (I.A and T.Y) and was categorized into two groups according to the proportion of expressing cells in several areas of the specimens by visual examination. The cut-off value for positive expression of each antibody was set as follows: ≥90% of lymphoma cells stained for CD20, ≥30% of lymphoma cells stained for CD10, BCL2, BCL6 and IRF4, and ≥40% of lymphoma cells stained for MYC. The definition of double expressor (DE) lymphoma followed the WHO criteria for MYC expression (≥40%) and BCL2 expression (≥50%). The percentage of lymphoma cells with Ki-67 nuclear immunostaining was defined as Ki-67 LI. We separated our cases into 3 groups according to the expression rate of MYC as follows: M1: 40-59%, M2: 60-79% and M3: 80-100%. A further subdivision was made according to Ki-67 LI in the following manner: less proliferative K1: 80-94% and highly proliferative K2: 95-100% because a Ki-67 LI over 95% is one of the criteria for highly aggressive lymphoma such as BL.24
Fluorescence in situ hybridization (FISH) analysis
The interphase FISH analyses were carried out to detect translocations of MYC (Vysis LSI MYC-dual color break-apart rearrangement probe, Abbott, Germany), BCL2 (dual color break-apart probe, DAKO) and BCL6 (Vysis LSI BCL6-dual color break-apart rearrangement probe, Abbott). The cut-off levels for positivity were 18%, 7% and 11% for MYC, BCL2 and BCL6 in the routine diagnostic setting, respectively. The total counted number of target cells was 50 to 100 cells for the detection of breaks, and all cases were independently evaluated by two or more investigators. Initially, we selected 168 cases consisting of 46 cases (27%) of MYC rearrangement, 39 cases (23%) of BCL2 rearrangement, 43 cases (26%) of BCL6 rearrangement and 68 cases (40%) without rearrangements (Supplementary figure 1).
Next generation sequencing (NGS) DNA preparation
DNA was extracted from formalin-fixed and paraffin-embedded (FFPE) tissue sections using the Maxwell 16 FFPE plus LEV DNA purification kit (Promega, Germany) according to the manufacturer’s protocol, followed by fluorescence-based quantification using Qubit dsDNA HS assay kit (ThermoFisher Scientific, USA).
Design of B-cell lymphoma gene panel
The B-cell lymphoma specific gene panel spans a genomic region of 6.72 kb covering proportions of the coding regions of nine genes relevant for cell cycle regulation (BTG1, BTG2, CCND3, CDKN2A, ID3, MAX, MYC, TCF3 and TP53) and was custom-designed using the Ion Ampliseq Designer (Version v7.0.9, Thermo Fisher Scientific) (Supplementary table 1).
Library preparation, sequencing and data analysis
Amplification of the selected gene regions by multiplex polymerase chain reaction (PCR) was performed with 40 ng of DNA input for each primer pool. (Supplementary table 2). Subsequently, adaptors were ligated following the instructions of NEXTflex DNA Sequencing Kit for Ion Torrent, Manual V15.12 (Bioo Scientific, USA). Finally, library quality was analyzed by microfluidic electrophoresis using Fragment Analyzer (Agilent Technologies, USA) and quantified using Ion Library TaqMan Quantitation Kit (Thermo Fisher Scientific). Libraries were sequenced on the Ion S5XL system (Thermo Fisher Scientific). Fastq files were analyzed using the CLC Genomics Workbench, version 5.0.1 (Qiagen, Hilden, Germany) after alignment, mapping to the hg19 human reference genome and corresponding target regions as determined in the custom panel bed file. The quality of base scoring and minimum depth of coverage met the criteria of Q20, which is equal to an error rate of 1% and a minimum of > 250 reads.25 The cut-off value of allele frequency for mutation detection was set to 5% for tissue sample analysis according to our results of the limit of detection (LoD).25
Determining the limit of detection (LoD)
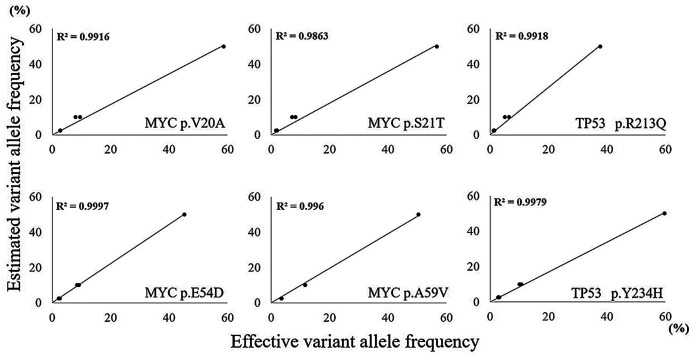
In order to determine the LoD of our NGS assay, DNA from BL cell line Raji with known mutations (MYC, NP_002458: p.V20A, p.S21T, p.E54D, p.A59V; TP53, NP_000537: p.R213Q, p.Y234H) was serially diluted to 5%, 20% or undiluted (100%) with un-mutated tonsil reference DNA, and analyzed in duplicate by NGS as described above.13
Prediction analysis of single nucleotide variants
To investigate the impact of the DNA variants detected in the histological samples on protein structure/function, we used the web-based tools MutationAssessor (cBio@MSKCC, release 3),26 Functional Analysis through Hidden Markov Model (FATHMM, version 2.3)27 and Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines (InterVar, version 0.1.7).28
RESULTS
Immunohistochemistry and FISH
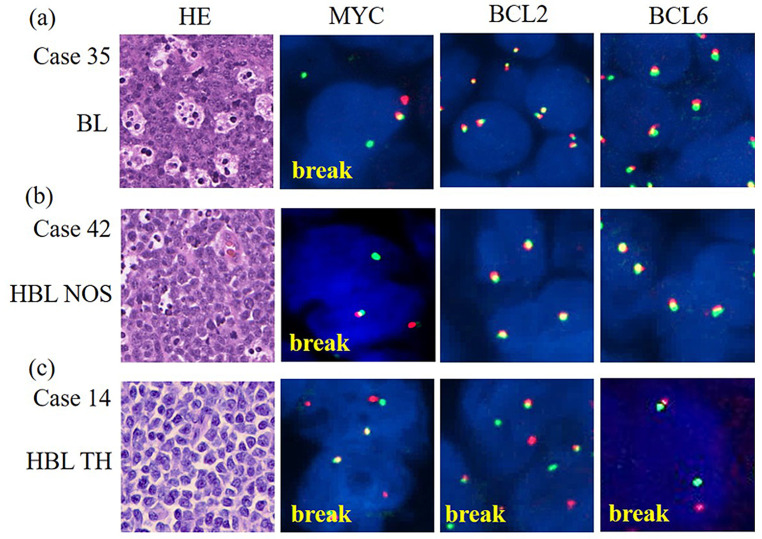
The majority of cases (28 out of 40; 70%) displayed a germinal center B-cell lymphoma immunophenotype (GCB) according to the Hans algorithm,29 whereby 12 cases (30%) were assigned to non-germinal center B-cells (non-GCB). All HBL cases with the exception of one (16 out of 17; 94%) were also of the GCB type. Thirty-one (89%) of 35 cases of DLBCL or blastoid morphology (not including BL) demonstrated co-expression of MYC and BCL2 (DE lymphoma), and were divided into subgroups according to their percentage of MYC or Ki-67-expressing tumor cells. All cases of the BL subgroup belonged to M3/K2, whereas a decreasing percentage of HBL cases (71% M3, 88% K2) and DLBCL cases (61% M3, 56% K2) was assigned to these two subgroups (Table 1 and Supplementary figure 2). The detection of chromosomal breaks was performed by FISH for MYC, BCL2 and BCL6 genes. MYC breaks were detectable in all 5 cases of BL, 15 out of 17 cases (88%) of HBL and 2 out of 18 cases (11%) of DLBCL. Correlation with MYC expression demonstrated the presence of MYC breaks in 50% (1/2) of subgroup M1, 30% (3/10) of subgroup M2 and 64% (18/28) of subgroup M3. BCL6 breaks were detectable in 7 cases of DLBCL and in 3 cases of HBL; BCL2 breaks were found in 8 cases of HBL and 3 cases of DLBCL. Seventeen HBL cases were classified as HBL-DH, 9 cases as HBL-TH and 8 cases as HBL NOS using a combined evaluation of FISH, and morphological and immunohistochemical findings (Figure 1).
Table 1. FISH and IHC results.
| BL | HBL-DH/TH | HBS NOS | DLBCL | Total | ||
|---|---|---|---|---|---|---|
| Case number | 5 | 9 | 8 | 18 | 40 | |
| Hans algorithm | GCB/non-GCB | 5/0 | 9/0 | 7/1 | 7/11 | 28/12 |
| FISH | ||||||
| Rearrangement | ||||||
| MYC | 5 (100%) | 9 (100%) | 6 (75%) | 2 (11%) | 21 (53%) | |
| BCL2 | 0 (0%) | 8 (89%) | 0 (0%) | 3 (17%) | 11 (28%) | |
| BCL6 | 0 (0%) | 2 (22%) | 1 (13%) | 7 (39%) | 10 (25%) | |
| IHC | ||||||
| MYC | M1 (40-59%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (10%) | 2 (5%) |
| M2 (60-79%) | 0 (0%) | 3 (33%) | 2 (25%) | 5 (28%) | 10 (25%) | |
| M3 (80-100%) | 5 (100%) | 6 (67%) | 6 (75%) | 11 (61%) | 28 (70%) | |
| BCL2 | 0 (0%) | 9 (100%) | 5 (65%) | 17 (94%) | 31 (78%) | |
| BCL6 | 5 (100%) | 8 (89%) | 7 (88%) | 17 (94%) | 37 (93%) | |
| CD10 | 5 (100%) | 8 (89%) | 6 (75%) | 7 (39%) | 26 (65%) | |
| IRF4 | 5 (100%) | 2 (22%) | 4 (50%) | 16 (89%) | 27 (68%) | |
| Ki-67 | K1 (80-94%) | 0 (0%) | 2 (22%) | 0 (100%) | 8 (44%) | 10 (25%) |
| K2 (95-100%) | 5 (100%) | 7 (78%) | 8 (100%) | 10 (56%) | 30 (75%) | |
| DE lymphoma | Positive | 0 (0%) | 9 (100%) | 5 (63%) | 17 (94%) | 31 (78%) |
FISH: Fluorescence in situ hybridization, IHC: Immunohistochemistry, BL: Burkitt lymphoma, HBL-DH/TH: high-grade B-cell lymphoma double hit/triple hit, HBL NOS: high-grade B-cell lymphoma not otherwise specified, DLBCL: diffuse large B-cell lymphoma, GCB: germinal center B-cell lymphoma immunophenotype, DE lymphoma: double expressor lymphoma
Fig. 1.
Representative FISH results
(a) Case 35 (BL) MYC (+)/BCL2 (-)/BCL6 (-), (b) Case 42 (HBL NOS) MYC (+)/BCL2 (-)/BCL6 (-), (c) Case14 (HBL-TH) MYC (+)/BCL2 (+)/BCL6 (+). FISH: Fluorescence in situ hybridization, BL: Burkitt lymphoma, HBL NOS: high-grade B-cell lymphoma, not otherwise specified, HBL TH: high-grade B-cell lymphoma triple hit.
Targeted next generation sequencing (NGS)
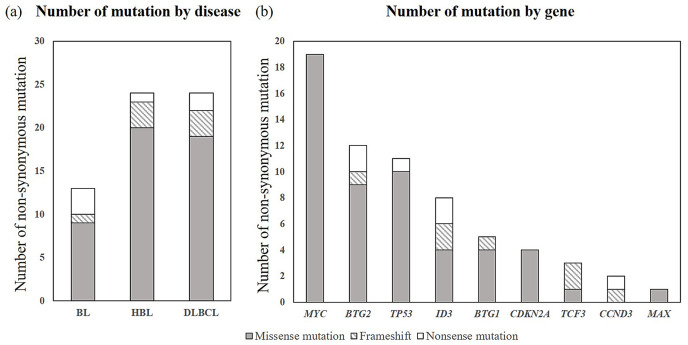
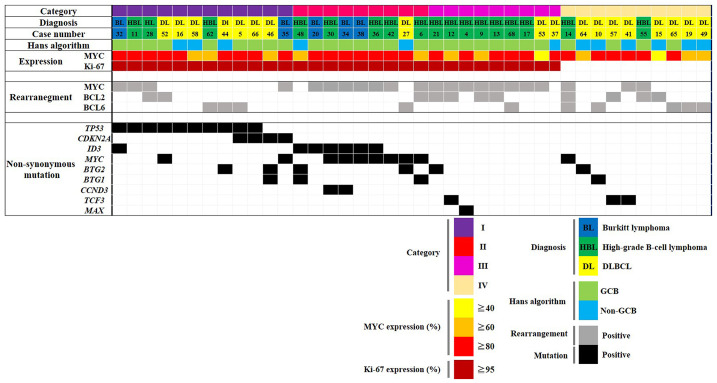
In order to determine the sensitivity of mutation detection and uniformity of the amplicon distribution, we employed serial dilutions of DNA of BL cell line Raji extracted from non-diseased human tonsils. The resulting allelic frequencies of mutations, as exemplified by the MYC and TP53 genes, demonstrated high correlation with the calculated dilution of cell line DNA ranging from 100% to 5% (Figure 2 and Supplementary table 3). Non-synonymous mutations determined by NGS of clinical samples consisted of 57 missense mutations, 7 frameshifts and 6 nonsense mutations (Figure 3, Supplementary table 4). All analyzed cell cycle regulating genes were mutated at varying frequencies: MYC (number of variants/number of samples: 19/10), TP53 (11/10), ID3 (8/7), BTG2 (11/5), BTG1 (5/4), CDKN2A (4/4), TCF3 (3/3), CCND3 (2/2) and MAX (1/1), respectively. Five variants were excluded because of non-cancerous change and low impact defined on all predictive analyses. According to our results, almost all cases were able to be classified molecularly into four categories (Figure 4). Category I (12/40 cases) is characterized by the dominance of TP53 and CDKN2A mutations, which were exclusively found in this subgroup. The majority of cases belonged to DLBCL (7 cases), and the remaining cases belonged to HBL (3 cases) and BL (2 cases). For example, case 46 (DLBCL) had three mutations in CDKN2A (p.A148T), BTG1 (p.L37M) and BTG2 (p.L10F), and no chromosomal rearrangements. BTG1 mutation (p.L37M, MSKCC impact: medium, FATHMM impact: oncogenic/non-cancer associated) located in the potential phosphorylation site commonly found in aggressive B-cell lymphoma has a significant function to accelerate the cell cycle.30 The in silico analysis predicted a functional impact of the CDKN2A mutation p.A148T (MSKCC impact: medium, FATHMM impact: neutral/cancer-associated) (Table 2). Category II (9/40 cases) was dominated by mutations in ID3 and MYC, and samples harbored MYC translocations in the majority of cases (7 cases). With the exception of one case, all others belonged to HBL (5 cases) or BL (3 cases). Almost all cases of category III (9 cases) carried MYC breaks, and 6 cases displayed additional BCL2 (5 cases) or BCL6 (1 case) breaks. Only three of the investigated genes (BTG2, TCF3 and MAX) were associated with mutations. Category IV (10 cases) is composed of mainly DLBCL cases; only two cases belonged to the HBL group. Based on the molecular status, this category is heterogeneous with few scattered mutations and a predominance of BCL6 breaks. Categories I, II and III are characterized by a very high proliferation rate (Ki-67 LI: ≥95%) and categories I and II are dominated by a high load of mutations in genes involved in proliferation control. Interestingly, category III had only few mutations in proliferation-associated genes despite a high proportion of HBL-DH/TH cases. Taken together, our approach allows a molecular categorization of MYC-driven aggressive mature B-cell lymphoma into four groups, which is partially independent from pathological diagnosis.
Fig. 2.
Determining the limit of detection
Sequencing results of serial dilutions of mutated Burkitt lymphoma cell line Raji with unmutated tonsil DNA. A high correlation of expected and effective variant allele frequencies was noted, even at tumor cell dilutions down to 5%.
Fig. 3.
Summary of non-synonymous variants detected by the B-cell lymphoma specific NGS panel
(a) Splitting the number of non-synonymous variants by disease revealed the highest amount for HBL and DLCBL; (b) Number of non-synonymous detected variants by gene; Most alteration types were missense mutations (81%), with MYC as the most frequently altered gene (23 variants). NGS: next generation sequencing, HBL: high-grade B-cell lymphoma, DLBCL: diffuse large B-cell lymphoma.
Fig. 4.
Molecular classification of MYC-driven aggressive mature B-cell lymphoma based on IHC, FISH and NGS
The block color represents the type of rearrangement and mutational status lined by expression of MYC and Ki-67. Categories I and II are included in the aggressive type (K2 group) and Category III (K1 group) is a less aggressive type than the others. IHC: immunohistochemistry, FISH: fluorescence in situ hybridization, NGS: next generation sequencing, DLBCL: diffuse large B-cell lymphoma.
Table 2. TP53 and CDKN2A mutations.
| N | Diag. | Gene | Mut. | Changed | DBD | Func. | MSKCC impact | FATHMM Impact |
|
|---|---|---|---|---|---|---|---|---|---|
| DNA | amino acid | ||||||||
| 5 | DL | TP53 | N | c.586C>T | p.R196 | + | LOF | NA | Onco./ Cancer |
| 11 | HBS NOS | TP53 | M | c.738G>A | p.M246I | + | LOF | Med. | Onco./ Cancer |
| 16 | DL | TP53 | M | c.818G>A | p.R273H | + | LOF | Med. | Onco./ Cancer |
| 28 | HBL DH |
TP53 | M | c.417G>T c.700T>G | p.K139N p.Y234D | + | LOF LOF | Med. | Onco./ Cancer |
| 32 | BL | TP53 | M | c.843C>A | p.D281E | + | LOF | Med. | Onco./ Cancer |
| 44 | DL | TP53 | M | c.413C>T | p.A138V | + | LOF | Med. | Onco./ Cancer |
| 52 | DL | TP53 | M | c.817C>G | p.R273G | + | LOF | Med. | Onco./ Cancer |
| 58 | DL | TP53 | M | c.701A>G | p.Y234C | + | LOF | Med. | Onco./ Cancer |
| 62 | HBL NOS |
TP53 | M | c.847C>T | p.R283C | + | LOF | Low | Onco./ Cancer |
| 66 | DL | TP53 | M | c.542G>C | p.R181P | + | LOF | Med. | Onco./ Cancer |
| 5 | DL | CDKN2A | M | c.442G>A | p.A148T | - | LOF? | Med. | Neut./ Cancer |
| 35 | BL | CDKN2A | M | c.442G>A | p.A148T | - | LOF? | Med. | Neut./ Cancer |
| 46 | DL | CDKN2A | M | c.442G>A | p.A148T | - | LOF? | Med. | Neut./ Cancer |
| 66 | DL | CDKN2A | M | c.442G>A | p.A148T | - | LOF? | Med. | Neut./ Cancer |
N: case number, Diag: diagnosis, DL: diffuse large B-cell lymphoma, HBL NOS: high-grade B-cell lymphoma not otherwise specified, HBL DH: high-grade B-cell lymphoma double hit, BL: Burkitt lymphoma, Mut: mutational type, N: nonsense mutation, M: missense mutation, DBD: DNA-binding region, Func: function, LOF: loss of function, NA: not available, Med: medium impact, Onco: oncogenic impact, Neut: neutral impact, Cancer: cancer-associated variant
DISCUSSION
In this study, we examined the mutational status of nine cell cycle genes of MYC-driven aggressive mature B-cell lymphoma, and identified four molecular categories according to the combined results of targeted NGS and FISH that correlated with proliferative activity but only partially with the histopathological diagnosis. Of note, the highly proliferative types (Categories I and II) demonstrated a high mutational frequency in the analyzed cell cycle-regulating genes. Category I is mainly based on the mutations of TP53 and CDKN2A (Table 2). The missense and nonsense mutations of these tumor suppressor genes are known to be driver mutations and prognostic factors in numerous tumors, and directly affect cell cycle progression.31-34 In particular, the DNA-binding site of TP53, which ranges from exon 4 to 9, is very important for p53 activity and mutations in this region lead to not only loss-of-function due to their ability to act as dominant-negative function of wild type p53, but also to a gain-of-function independent of the effects on wild type p53.35 In our cases, the missense mutations of this region matched those described in previous reports.36,37 Moreover, the cases with point mutations in the DNA binding site of TP53 had a significantly poorer prognosis than those with mutations in the non-DNA binding domain.38 This is in concordance with our results, as Category I (highly proliferative) consists of cases with mutations in the TP53 DNA-binding domain. The other frequent molecular alterations in Category I are alterations of CDKN2A that encode two cycle-dependent kinase inhibitors, p16INK4a and p14AR, which directly negatively regulate the cell cycle like p53.39 Deletion of this gene is a prognostic factor in many tumors40 and missense mutations of this gene were detected previously in some DLBCL cases.41 The common CDKN2A polymorphism p.A148T found in 4 cases was previously described in several cancer types such as colon-, lung-, breast-, bladder-cancer and melanoma.40,42 This variant and amino acid change located in exon 2 of CDKN2A and in the open reading frame of p16INK4a are not directly involved in binding to cyclin kinases, CDK4 or CDK6. Thus, this variant (p.A148T) may not have direct effects on the inhibitor activity, but the functional impact is unclear. However, previous functional studies suggested that this alteration (p.A148T) can reduce its ability to inhibit the cell cycle in both melanoma and acute lymphoblastic leukemia cell lines.43,44 Debniak et al. reported that the variant p.A148T may affect the function of p16INK4a through the CDKN2A promoter.45,46 Our in silico analysis demonstrated that this alteration induced cancer-associated ability (MSKCC impact; medium, FATHMM impact; cancer-associated). Taken together, these mutations of TP53 and CDKN2A may affect the progression, development and survival of MYC-driven aggressive mature B-cell lymphoma.
The highly proliferative cases of Category II are characterized by MYC rearrangement with MYC and/or ID3 mutation, and consisted of 5 HBL cases (63%) and 3 BL cases (37%). Most of the Category II samples had two or more mutations in all nine cell-cycle associated genes. Both HBL with MYC mono-rearrangement and BL cases of this category are similar in terms of the mutational status (MYC, ID3 and CCND3). These BL-specific mutations may give the aggressive malignant ability not only for BL, but also for HBL and DLBCL as previously assumed.14,47 For example, case 30 (HBL NOS) displayed not only BL-like morphological features, but also a BL-like mutational status of MYC (p.E54D, p.S107R, p.G152A), ID3 (p.V67I, p.E68fs) and CCND3 (p.L292fs), with MYC rearrangement and, in addition, a strong expression of BCL2 (Figure 5). In particular, both the transactivation domain (aa position: 12-155) in exon 2 of MYC and loop-helix-loop (HLH) domain (aa position: 43-80) in exon 1 of ID3 commonly mutated in BL (known as a hot spot region) and may be important for the pathogenesis of these tumors.12,13,30 Almost all cases of Category III had MYC rearrangements – comparable to Category II - but additional chromosomal rearrangements are found frequently. The high proliferative activity of this category is likely supported by the chromosomal rearrangements and modulated by other molecular changes. Evrard et al. recently reported that CREBBP mutations - most frequently mutated in HBL-DH/TH cases - lead not only to accelerated BCL6 activity, but were also able to suppress p53 function.19 For this reason, Category III may be influenced by the other alterations of cell cycle pathway. Taken together, Categories I, II and III had increased proliferative activity, which is associated with characteristic molecular alterations. Lastly, 15 (83%) of 18 DLBCL cases were classified into either Category I (7 cases) or IV (8 cases). Seven cases of 8 DLBCL cases in Category IV had at least one rearrangement and half had additional alterations (MYC, BTG1, BTG2 and TCF3) of cell cycle genes. This suggested that TP53 and CDKN2A alterations separate Category I (highly proliferative type) from Category IV (less proliferative type) in MYC-driven DLBCL cases.
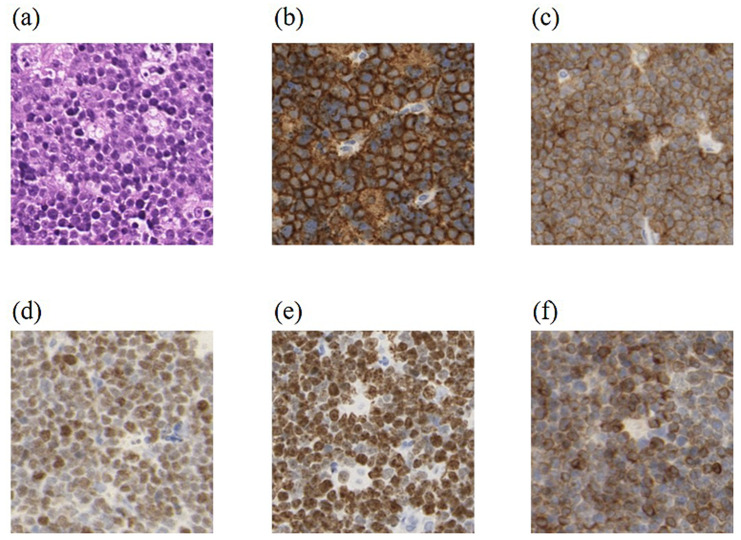
Fig. 5.
Histopathological findings in HBL NOS
Case 30 (HBL, NOS: Category II) exhibited (a) morphology (H&E stain) and expression of (b) CD20, (c) CD10, (d) MYC, and (e) Ki-67 similar to BL, but (f) with the additional expression of BCL2. HBL NOS: high-grade B-cell lymphoma not otherwise specified.
In conclusion, our targeted NGS approach using a gene panel for cell cycle related genes in conjunction with chromosomal rearrangements was able to identify four molecular categories of aggressive B-cell lymphomas. Category I based on TP53 and CDKN2A mutations, Category II based on MYC rearrangement with MYC and/or ID3 mutation, Category III based on MYC rearrangement with one additional molecular alteration, and Category IV based on one rearrangement of BCL2 or BCL6 without TP53, CDKN2A, ID3 or MYC mutations. Thus, our study demonstrated, despite the limited number of cases and lack of clinical information, that alterations in cell cycles genes provide added value for a more precise sub-classification of highly aggressive B-cell lymphoma. Consequently, Categories I, II and III having characteristic molecular features and the highest Ki-67 LI should be further evaluated for novel treatment strategies.
Supplemental Materials and Methods
ACKNOWLEDGMENTS
We thank the molecular pathology team at the Charité, especially Anke Sommerfeld, Erika Berg, Edda von der Wall, Hedwig Lammert and Arndt Boshof, for their technical assistance.
Footnotes
FUNDING
This study was not supported by any commercial foundation.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Ellrott K, Bailey MH, Saksena G, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018; 6 : 271-281. e7. [DOI] [PMC free article] [PubMed]
- 2.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999; 18: 3004-3016. [DOI] [PubMed] [Google Scholar]
- 3.Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009; 114: 3533-3537. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Weiss VL, Wang XJ, et al. High-grade B-cell lymphoma with MYC rearrangement and without BCL2 and BCL6 rearrangements is associated with high P53 expression and a poor prognosis. Am J Surg Pathol. 2016; 40: 253-261. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016; 127: 2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkozy C, Traverse-Glehen A, Coiffier B. Double-hit and double-protein-expression lymphomas: aggressive and refractory lymphomas. Lancet Oncol. 2015; 16: e555-e567. [DOI] [PubMed] [Google Scholar]
- 7.Zeng D, Desai A, Yan F, et al. Challenges and opportunities for high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangement (double-hit lymphoma). Am J Clin Oncol. 2019; 42: 304-316. [DOI] [PubMed] [Google Scholar]
- 8.Lin P, Dickason TJ, Fayad LE, et al. Prognostic value of MYC rearrangement in cases of B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma. Cancer. 2012; 118: 1566-1573. [DOI] [PubMed] [Google Scholar]
- 9.Perry AM, Crockett D, Dave BJ, et al. B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and burkitt lymphoma: study of 39 cases. Br J Haematol. 2013; 162: 40-49. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Seegmiller AC, Lin P, et al. B-cell lymphomas with concurrent MYC and BCL2 abnormalities other than translocations behave similarly to MYC/BCL2 double-hit lymphomas. Mod Pathol. 2015; 28: 208-217. [DOI] [PubMed] [Google Scholar]
- 11.Campo E. New pathogenic mechanisms in Burkitt lymphoma. Nat Genet. 2012; 44: 1288-1289. [DOI] [PubMed] [Google Scholar]
- 12.Richter J, Schlesner M, Hoffmann S, et al. ICGC MMML-Seq Project Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012; 44: 1316-1320. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012; 490: 116-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momose S, Weißbach S, Pischimarov J, et al. The diagnostic gray zone between Burkitt lymphoma and diffuse large B-cell lymphoma is also a gray zone of the mutational spectrum. Leukemia. 2015; 29: 1789-1791. [DOI] [PubMed] [Google Scholar]
- 15.Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012; 30: 3452-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kluk MJ, Chapuy B, Sinha P, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS One. 2012; 7: e33813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu-Monette ZY, Deng Q, Manyam GC, et al. Clinical and biologic significance of MYC genetic mutations in de novo diffuse large B-cell lymphoma. Clin Cancer Res. 2016; 22: 3593-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zenz T, Kreuz M, Fuge M, et al. German High-Grade Non-Hodgkin Lymphoma Study Group (DSHNHL) TP53 mutation and survival in aggressive B cell lymphoma. Int J Cancer. 2017; 141: 1381-1388. [DOI] [PubMed] [Google Scholar]
- 19.Evrard SM, Péricart S, Grand D, et al. Targeted next generation sequencing reveals high mutation frequency of CREBBP, BCL2 and KMT2D in high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements. Haematologica. 2019; 104: e154-e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szczuraszek K, Mazur G, Jeleń M, et al. Prognostic significance of Ki-67 antigen expression in non-Hodgkin’s lymphomas. Anticancer Res. 2008; 28: 1113-1118. [PubMed] [Google Scholar]
- 21.Yoon DH, Choi DR, Ahn HJ, et al. Ki-67 expression as a prognostic factor in diffuse large B-cell lymphoma patients treated with rituximab plus CHOP. Eur J Haematol. 2010; 85: 149-157. [DOI] [PubMed] [Google Scholar]
- 22.Li ZM, Huang JJ, Xia Y, et al. High Ki-67 expression in diffuse large B-cell lymphoma patients with non-germinal center subtype indicates limited survival benefit from R-CHOP therapy. Eur J Haematol. 2012; 88: 510-517. [DOI] [PubMed] [Google Scholar]
- 23.Lynnhtun K, Renthawa J, Varikatt W. Detection of MYC rearrangement in high grade B cell lymphomas: correlation of MYC immunohistochemistry and FISH analysis. Pathology. 2014; 46: 211-215. [DOI] [PubMed] [Google Scholar]
- 24.Broyde A, Boycov O, Strenov Y, et al. Role and prognostic significance of the Ki-67 index in non-Hodgkin’s lymphoma. Am J Hematol. 2009; 84: 338-343. [DOI] [PubMed] [Google Scholar]
- 25.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: A joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017; 19: 341-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011; 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013; 34: 57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP Guidelines. Am J Hum Genet. 2017; 100: 267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004; 103: 275-282. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Grubor V, Love CL, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2013; 110: 1398-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012; 26: 1268-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017; 170: 1062-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilischkis R, Sarcevic B, Kennedy C, Warlters A, Sutherland RL. Cancer-associated mis-sense and deletion mutations impair p16INK4 CDK inhibitory activity. Int J Cancer. 1996; 66: 249-254. [DOI] [PubMed] [Google Scholar]
- 34.Monti S, Chapuy B, Takeyama K, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012; 22: 359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018; 25: 154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young KH, Leroy K, Møller MB, et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood. 2008; 112: 3088-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peroja P, Pedersen M, Mantere T, et al. Mutation of TP53, translocation analysis and immunohistochemical expression of MYC, BCL-2 and BCL-6 in patients with DLBCL treated with R-CHOP. Sci Rep. 2018; 8: 14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young KH, Weisenburger DD, Dave BJ, et al. Mutations in the DNA-binding codons of TP53, which are associated with decreased expression of TRAILreceptor-2, predict for poor survival in diffuse large B-cell lymphoma. Blood. 2007; 110: 4396-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine. 2016; 8: 30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borkowska E, Jędrzejczyk A, Kruk A, et al. Significance of CDKN2A gene A148T variant in patients with bladder cancer. Cent European J Urol. 2011; 64: 168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017; 171 : 481-494. e15. [DOI] [PMC free article] [PubMed]
- 42.Dębniak T, Scott RJ, Huzarski T, et al. CDKN2A common variant and multi-organ cancer risk—a population-based study. Int J Cancer. 2006; 118: 3180-3182. [DOI] [PubMed] [Google Scholar]
- 43.Xu H, Zhang H, Yang W, et al. Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nat Commun. 2015; 6: 7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker GJ, Gabrielli BG, Castellano M, Hayward NK. Functional reassessment of P16 variants using a transfection-based assay. Int J Cancer. 1999; 82: 305-312. [DOI] [PubMed] [Google Scholar]
- 45.Debniak T, Górski B, Huzarski T, et al. A common variant of CDKN2A (p16) predisposes to breast cancer. J Med Genet. 2005; 42: 763-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riese U, Dahse R, Fiedler W, et al. Tumor suppressor gene p16 (CDKN2A) mutation status and promoter inactivation in head and neck cancer. Int J Mol Med. 1999; 4: 61-65. [DOI] [PubMed] [Google Scholar]
- 47.Bouska A, Bi C, Lone W, et al. Adult high-grade B-cell lymphoma with Burkitt lymphoma signature: genomic features and potential therapeutic targets. Blood. 2017; 130: 1819-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.