Abstract
Despite the continued global spread of the current COVID-19 pandemic, the nonavailability of a vaccine or targeted drug against this disease is still prevailing. The most established mechanism of viral entry into the body is considered to be via angiotensin-converting enzyme 2 (ACE2) acting as a receptor for viral spike protein thereby facilitating its entry in the cell. However, ACE2 is also involved in providing the protection from severe pathological changes. This article provides a computational and bioinformatics-based analysis of ACE2 with an objective of providing further insight into the earnest efforts to determine its true position in COVID-19 pathology. The results of this study show that ACE2 has strikingly low expression in healthy human lung tissue and was absent from the list of differentially expressed genes. However, when transcription factors were analyzed, we found a significant upregulation of FOS and downregulation of FOXO4 and FOXP2. Moreover, the miRNA prediction analysis revealed that miR-1246, whose upregulation has been experimentally established to be a cause of acute respiratory distress syndrome (ARDS), was found to be targeting the coding DNA sequence (CDS) of ACE2. This study presents a wide range of potentially important transcription factors as well as miRNA targets associated with ACE2 which can be potentially used for drug designing amid this challenging pandemic situation.
Communicated by Ramaswamy H. Sarma
Keywords: Angiotensin-converting enzyme -2, Acute respiratory distress syndrome Predicted ACE2 structure, miRNA-1246, FOXO4, FOS, MiR-212-5p
1. Introduction
Emerging and re-emerging of viral outbreaks has had been a significant threat to human health since ancient times (Gao 2018). In the Wuhan city of China, a cluster of pneumonia cases was reported in humans during the end of 2019 (Wang, Horby, et al. 2020), and the disease was referred to as CVOID-19 (coronavirus disease 2019). This disease presented several symptoms such as cough, fever, occasional diarrhea and pneumonia (Guan, Ni et al. 2020; Holshue, DeBolt et al. 2020; Huang, Wang et al. 2020). Consequently, the whole-genome sequencing depicted the causative agent to be a member of the coronavirus family and named as 2019-nCoV by World Health Organization (WHO) initially (Wu, Zhao et al. 2020; Zhou, Yang et al. 2020; Zhu, Zhang et al. 2020). However, the virus was officially designated as SARS-CoV-2 by International Committee on Taxonomy of Viruses later on (Gorbalenya, Baker et al. 2020).
Coronaviruses (CoVs) are enveloped viruses containing positive-sense single-stranded RNA as their genetic material (Lai 2007; Lu and Liu 2012). The phylogenetic analyses of its genome indicated SARS-CoV-2 as a member of the genus Betacoronavirus, which also includes SARS-CoV, SARSr-CoV, MERS-CoV and many other viruses as well reported to have been isolated from humans and other animal species (Li, Li et al. 2005; Lu, Zhao et al. 2020; Wu, Zhao et al. 2020, Zhou, Yang et al. 2020; Zhu, Zhang et al. 2020). Although SARS-CoV-2 shares more than 93.1% sequence similarity of spike (S) gene with of BatCoV RaTG13, the similarity percentage with SARS-CoV and other SARSr-CoVs has been exhibited to be lower than 80% (Zhou, Yang et al. 2020).
The initiation of a vital infection takes place after binding of virus proteins with cellular receptors present on host cell surface and subsequent entry of the virus inside the host cell. This receptor recognition is a significant checkpoint in determining the host range and cross-species viral infection. In this regard, angiotensin converting enzyme 2 (ACE2) has been recognized as a cellular receptor for SARS-CoV-2 spike protein (Zhou, Yang et al. 2020). Initially, ACE2 was identified as an exopeptidase being expressed in vascular endothelial cells of heart and kidney where it catalyzes the conversion of angiotensins into its various forms (Donoghue, Hsieh et al. 2000; Ferrario, Trask et al. 2005). Later on, this enzyme became well known as a receptor for SARS-CoV binding (Kuba, Imai et al. 2005). Therefore, the use of ACE2 by both SARS-CoV-2 and SARS-CoV is an important classifier of both viruses in the same subgenus. On the other hand, although ACE2 is expressed in most of the vertebrates but all ACE2 forms cannot be utilized as receptors by SARS-CoV-2. For example, recent investigations have indicated that SARS-CoV-2 can infect Chinese horseshoe bats, swine and civet but cannot use mouse ACE2 receptor for its entry thereby presenting specificity to some species and not others (Zhou, Yang et al. 2020).
In coronaviruses, the process of virus entry is mediated by envelope-embedded surface-located spike (S) glycoprotein (Lu, Wang et al. 2015). Generally, this trimeric S protein is cleaved by host proteases into S1 and S2 subunits where S1 subunits are released in the transition to the post fusion conformation (Simmons, Reeves et al. 2004; Belouzard, Chu et al. 2009; Simmons, Zmora et al. 2013; Song, Gui et al. 2018). The S1 bears the receptor binding domain (RBD), which binds directly to peptidase domain (PD) of ACE2 receptor (Li, Li et al. 2005) while S2 is involved in the membrane fusion. When S1 binds to ACE2, another cleavage site is exposed on S2 which gets cleaved from host protease, a critical process for viral infection (Simmons, Gosalia et al. 2005; Belouzard, Chu et al. 2009; Millet and Whittaker 2015). The computational modeling (Lu, Zhao et al. 2020; Xu, Chen et al. 2020) and other experiments of viral infection using HeLa cells (Zhou, Yang et al. 2020) have provided the evidences of ACE2 as receptor for SARS-CoV-2. Cryo-EM experiments have indicated that SARS-CoV-2 exhibited 10 time more affinity for ACE2 compared to SARS-CoV, which is also consistent with higher SARS-CoV-2 infection efficiency (Wrapp, Wang et al. 2020).
Paradoxically, although ACE2 provides the entry point for some coronaviruses into the body, this enzyme has been found to play a protective role in several pathophysiological processes too. These include alleviating pathological changes in acute lung injury and acute respiratory distress syndrome (Imai, Kuba et al. 2005; Kuba, Imai et al. 2006; Hamming, Cooper et al. 2007), protection against diabetes, hypertension, cardiovascular disease and organ damage as well, (Cheng, Wang et al. 2020) by regulation of the renin-angiotensin system (RAS). The RAS maintains the blood pressure homeostasis in addition to fluids and salts balance which is crucial for the pathological and physiological regulation of several organs including kidney, heart and lungs (Patel, Rauf et al. 2017). In this scenario, ACE2 primarily functions by negatively regulating the RAS (Hamming, Cooper et al. 2007; Kuba, Imai et al. 2010) and via degrading and converting the Angiotensin II (potent in vasoconstriction, pro-fibrosis and pro-inflammation) to Angiotensin 1-7 (vasodialatic, apoptotic and proliferative) (Hamming, Cooper et al. 2007). Moreover, ACE2 also exerts local regulatory effects for pathological changes in different organs such as kidneys, heart and lungs e.g. it regulates the absorption of amino acids in kidney and gut by modulating the expression of amino acid transporters (Kuba, Imai et al. 2010).
Consequently, ACE2 serves as a double‐edged sword playing dual roles of a demon as well as an angel. It not only opens the doors for SARS-CoV-2 as a receptor but also helps protects the body from severe pathological changes (Cheng, Wang et al. 2020; Xiao, Sakagami et al. 2020; Yan, Xiao et al. 2020). In the current COVID-19 outbreak, ACE2 is a topic of pronounced interest and several studies are being carried out to assess the interaction of SARS-CoV-2 with ACE2, the receptor.
In this perspective, this study was designed to investigate human ACE2 via its computational and bioinformatics characterization so as to demonstrate its functionality and biological roles ultimately leading not only opening insights to the use of its associated molecules for potential therapeutic interventions but also to get clarified whether it is rightfully getting the attention of the scientific community or other targets/pathways must also be explored in order to hasten the most important cause of the present time – finding cure to COVID-19.
2. Methodology
2.1. Homology and phylogenetic analysis
The sequence of human ACE2 gene was obtained from NCBI database (https://www.ncbi.nlm.nih.gov/) and its genome localization was assessed. The phylogenetic analysis describes the useful information related the gene variations among different species. Human ACE2 peptide sequence was retrieved from Uniprot database (https://www.uniprot.org/) along with other species which include Mus musculus (MOUSE), Rattus norvegicus (RAT), Paguma larvata (PAGLA), Felis catus (FELCA), Bos Taurus (BOVIN), Pongo abelii (PONAB), Equus caballus (HORSE), Pan troglodytes (PANTR), Mustela putorius (MUSPF), Canis lupus familiaris (CANLF), Gallus gallus (CHICK), Ornithorhynchus anatinus (ORNAN), Anolis carolinensis (ANOCA), Nomascus leucogenys (NOMLE), Danio rerio (DANRE), Ictidomys tridecemlineatus (ICTTR), Chlorocebus sabaeus (CHLSB), Macaca fascicularis (MACFA), Myotis lucifugus (MYOLU), Cavia porcellus (CAVPO), Pan paniscus (PANPA), Papio Anubis (PAPAN), Ovis aries (SHEEP), Loxodonta Africana (LOXAF), Rhinopithecus roxellana (RHIRO), Cercocebus atys (CERAT), Macaca nemestrina (MACNE), Mandrillus leucophaeus (MANLE), Heterocephalus glaber (HETGA), Capra hircus (CAPHI), Tarsius syrichta (TARSY), Pelodiscus sinensis (PELSI), Xenopus tropicalis (XENTR), Propithecus coquereli (PROCO), Ursus maritimus (URSMA), Macaca mulatta (MACMU), Ficedula albicollis (FICAL), Otolemur garnettii (OTOGA), Saimiri boliviensis boliviensis (SAIBB), Meleagris gallopavo (MELGA), Mesocricetus auratus (MESAU), Cebus capucinus imitator (CEBCA), Ailuropoda melanoleuca (AILME), Dipodomys ordii (DIPOR), Bos indicus x Bos Taurus (BOBOX), Vombatus ursinus (VOMUR), Aotus nancymaae (AOTNA), Vulpes Vulpes (VULVU), Physeter macrocephalus (PHYMC), Ursus americanus (URSAM), Anas platyrhynchos platyrhynchos (ANAPP), Rhinopithecus bieti (RHIBE), Rousettus leschenaultia (ROULE), Rhinolophus sinicus (CHIR), R. macrotis (RHIMR), Procyon lotor (PROLO), Sus scrofa domesticus (PIG), Nothobranchius furzeri (NOTFU). Multiple sequence alignment was performed by Clustal Omega tool to generate homology/phylogenetic (Neighbour-joining) tree with default settings for the determination of evolutionary trend for ACE2 (McWilliam, Li et al. 2013).
2.2. Physiochemical characterization
ProtParam tool of ExPASy (https://web.expasy.org/protparam/) was applied to assess the physicochemical characterization and functional/structural motifs for human ACE2 peptide sequence (Garg, Avashthi et al. 2016). Signal peptide prediction of ACE2 was performed with the application of SignalP-4.1 tool (http://www.cbs.dtu.dk/services/SignalP-4.1/) (Emanuelsson, Brunak et al. 2007). Protscale server (https://web.expasy.org/protscale/) calculated further physicochemical properties such as buried residues (%), polarity, average flexibility, hydrophilicity, relative mutability and bulkiness for human ACE2 (Wilkins, Gasteiger et al. 1999).
2.3. Single nucleotide polymorphism (SNP) analysis
Association of diseases with genetic polymorphism was evaluated by BioMuta v4.0 database (https://hive.biochemistry.gwu.edu/biomuta). This database provides the information of single-nucleotide variation (SNV) and association with disease specially if related to oncogenesis (Dingerdissen, Torcivia-Rodriguez et al. 2018). The non-synonymous single nucleotide polymorphisms (nsSNPs) were retrieved from NCBI SNPs database (dbSNP) (http://www.ncbi.nlm.nih.gov/snp), selected and analyzed with the application of prediction tools such as SIFT, PolyPhen-2, PROVEAN and PANTHER to predict human ACE2 gene mutations.
SIFT (https://sift.bii.a-star.edu.sg/) predicts the impact of substitutions of amino acid on protein functionality and phenotype. This program has attained the standard to assess and characterize the missense variations. The principle of this tool is to align the homologous sequences in large numbers and assign the scores to each residue varying from in value 0 to 1. The threshold score of nsSNPs was set as ≤ 0.05 (Sim, Kumar et al. 2012). The mutations selected by SIFT were further analyzed by other programs. PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) also predicts the substitution impact of amino acid on protein structure and function on the basis of structural and phylogenetic information (Kumar, Henikoff et al. 2009). PROVEAN (http://provean.jcvi.org/index.php) is another tool to predict the effect of amino acid change on protein biological function. This is helpful to filter the sequence variants for the identification of indels or nonsynonymous variants which are predicted as functionally important (Choi and Chan 2015). PANTHER (http://www.pantherdb.org/tools/) estimates the chances of nonsynonymous coding SNP for its impact on protein function. It estimates the time length (millions of years) for an amino acid in which it has been preserved in lineage resulting in the specific protein. Longer the preservation time, more the chances of a functional impact (Tang and Thomas 2016).
2.4. Sub cellular localization
Subcellular Localization or the prediction of protein’s residence in a cell plays a crucial role in determining the comparative protein expression levels in various cell types, which can ultimately help us in exploring the functionality of protein. For ACE2, this prediction was performed using online protein subcellular localization prediction servers namely CELLO (Yu, Cheng et al. 2014), TMHMM (Krogh, Larsson et al. 2001), HMMTOP (Tusnády and Simon 2001) and UniProt (Bairoch and Apweiler 2000).
2.5. Methylation sites assessment
Regulation of a gene expression is the central key to maintain homeostasis and this regulation is primarily controlled by DNA methylation. DNA methylation patterns can provide a deep insight into tissue–specific gene transcription. The methylation sites for ACE2 were determined using an online methylation site prediction tool known as MethyCancer (http://methycancer.psych.ac.cn/), a part of Cancer Epigenomics project in China (He, Chang et al. 2008). ACE2 was scanned for possible CpG islands using the default settings.
2.6. Post translational modifications’ assessment
Protein biosynthesis is followed by enzymatic or covalent post-translational modifications. Prediction of glycosylation, phosphorylation and mannosylation sites of ACE2 was done via online tools NetCGlyc1.0 (Julenius 2007), NetCorona 1.0 (Kiemer, Lund et al. 2004), NetOGlyc 1.0 (Steentoft, Vakhrushev et al. 2013), NetGlycate1.0 (Johansen, Kiemer et al. 2006), NetPhos 3.1 (Blom, Sicheritz-Pontén et al. 2004), NetNGlyc 1.0 (Gupta, Jung et al. 2004) and ProP 1.0 (Duckert, Brunak et al. 2004) provided by Center for Biological Sequence Analysis CBS (http://www.cbs.dtu.dk/services/).
2.7. Functional networks of protein associations
The coherence between various seemingly unconnected biological activities of an individual are governed by protein-protein interactions. This network of protein interactions plays a central role in explaining how one dysfunctional protein can contribute to heterogeneous symptomatology of a single disease. The protein-protein interactions and orthologue associations for ACE2 were predicted using Search Tool for Retrieval of Interacting Genes and Proteins codenamed STRING (Szklarczyk, Morris et al. 2017) (https://string-db.org/).
2.8. Protein structure building
Three dimensional structure of ACE2 was predicted using Swiss Model online (https://swissmodel.expasy.org/) (Guex and Peitsch 1997). First, a template search was performed on Swiss model to select the appropriate template for ACE2 structure prediction. Template with high sequence identity and in range GMQE and QSQE score was finalized for ACE2 protein modeling. The Protein-protein Interaction (PPI) fingerprint was also evaluated for this template. The predicted ACE2 model was further evaluated via Ramachandran plot using Swiss Model Structure Assessment feature. The Local Quality Estimate and QMEAN score for ACE2 predicted model was also checked.
2.9. Tfs of ACE2
Among the regulatory elements for ACE2, the predicted transcription factors for the promoter/enhancer of ACE2 were extracted from the GeneCards database selecting from the queries showing the highest scores (GeneCards 2020). After the identification of putative transcription factors, these were arranged in a tabular form with respect to the organ in which they are being most abundantly expressed and this information was extracted using NCBI gene database (Fagerberg, Hallström et al. 2014). Meanwhile, the graph of the expression profiles of ACE2 in different tissues of human body was also fetched from NCBI database (Fagerberg, Hallström et al. 2014) to have them as the base reference of its expression under normal conditions.
2.10. Identification of miRNAs targeting ACE2
miRNAs are small non-coding RNA molecules which play an important function of orchestrating gene regulation post transcriptionally. To determine the putative miRNAs targeting ACE2 transcript, two databases were used: miRWalk (Sticht, De La Torre et al. 2018) (to predict all miRNAs targeting the 3′ UTR, CDS, and 5′ UTR (Supplementary Figure S1) and miRDB (Chen and Wang 2020) to further sort these miRNAs on the basis of scoring, similar prediction and cross referencing. miRWalk is an open source online tool which utilizes random-forest-based approach for miRNA prediction. It is also integrated with other miRNA databases in its platform to increase validity of the prediction. The miRDB function integrated in the miRWalk was used to further sort the miRNAs of ACE2.
2.11. Analysis of RNA sequencing data of lung biopsy samples
2.11.1. Rna-seq data retrieval
The RNA sequencing data from lungs of two SARS-CoV-2 patients and two healthy controls was taken from GEO database (Blanco-Melo, Nilsson-Payant et al. 2020). The raw read counts of the human data were already available in the .tsv format which was downloaded and used for further analysis of the required samples in R Studio (3.6.3) (R Development Core Team 2010).
2.11.2. Differential expression analysis
The raw read counts were processed for further analysis of the differentially expressed genes between the healthy lung biopsies and SARS-CoV-2 patients’ lung biopsies using different packages of R Bioconductor. The required packages included limma, edgeR, gplots, org.Hs.eg.db, RcolorBrewer and Glimma (Warnes, Bolker et al. 2005; McCarthy, Chen et al. 2012; Ritchie, Phipson et al. 2015; Su, Law et al. 2017).
The analysis of the in vivo samples started from the filtration of the raw counts to remove genes with low expression values. Each group of this study contains 2 biological replicates, therefore counts per million (CPM) threshold was set to filter those genes above 0.5 in both samples. By converting values to CPM, the normalization of sequencing depths of samples was also done. This filtered data was then stored in DGEList object which was then processed further for quality control checks and modifications before getting the plots of differentially expressed genes.
2.12. Analysis of cellular pathways regulated by transcription factors
The transcription factors selected form the list of differentially expressed genes were then further investigated to see which cellular pathways they are involved in and whether any of them matches with the predicted transcription factors of ACE2 or not. This was done using the ‘Pathway from PubChem’ function from NCBI gene database for all the significantly differentially expressed TFs (NCBI Gene, NCBI Gene, NCBI Gene).
3. Results
3.1. Sequence retrieval and phylogenetic analysis
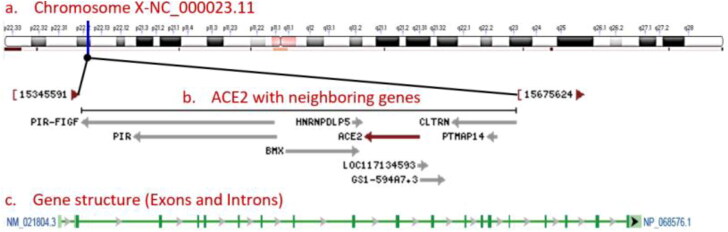
The human ACE2 gene was found to be located on p arm of X chromosome (Xp22.2) assuredly between 15561033 and 15602158 position. The gene spans on 41,126 bp, contains 19 exons and encodes 805 amino acids. The chromosome localization and exons have been displayed in Figure 1.
Figure 1.
Human ACE2 gene location, structure and domains. (a) ACE2 Chromosomal localization on p arm of X chromosome (Xp22.2) highlighted in blue; (b) ACE2 gene (red) flanked by LOC117134593 and GS1-594A7.3 genes (grey); and (c) ACE2 gene contains 19 exons (green bars) and 16 introns (grey arrows).
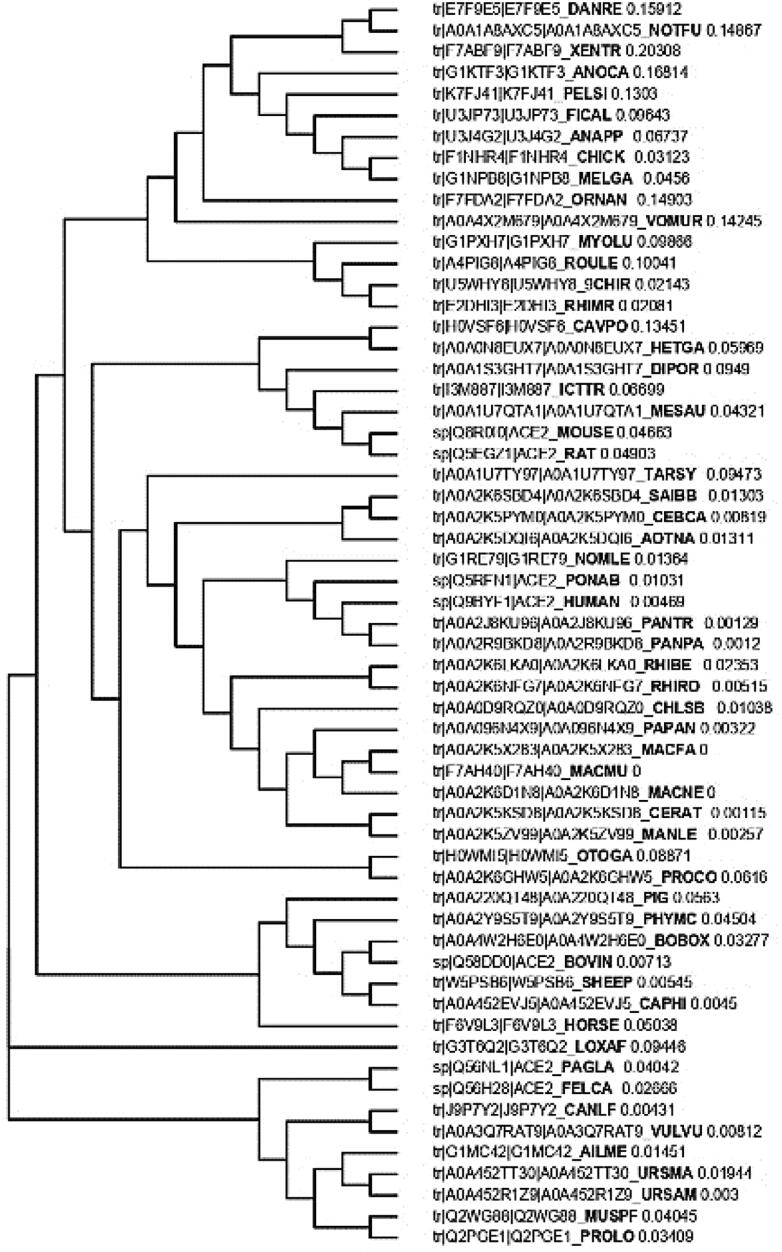
The human ACE2 (Q9BYF1) peptide sequence along with other species were subjected to Clustal Omega for multiple alignment to create the homology/phylogenetic (Neighbor-joining) tree (Figure 2). Human ACE2 protein was clustered along with Pan troglodytes (PANTR), P. paniscus (PANPA), Pongo abelii (PONAB) and Nomascus leucogenys (NOMLE), where first three members were from same family, Hominidae while Nomascus leucogenys belonged to Hylobatidae family which showed the distinct relatedness based on peptide in this cluster. The multiple peptide sequence alignment has been provided in Supplementary Data.
Figure 2.
Phylogenetic (Neighbor-joining) tree showing relationship among different animals based on multiple sequence alignment of ACE2 peptide.
3.2. Physicochemical characterization
Physicochemical characterization was evaluated using Expasy-ProtParam and Protscale server. Human ACE2 has molecular weight of 92463.04 with formula C4170H6358N1092O1222S35 having total 12877 atoms. The theoretical pI was found to be 5.36 while total number of negatively charged (Asp + Glu):and positively charged (Arg + Lys) residues were 99 and 78, respectively. The estimated half-life in mammalian reticulocytes (in vitro) was 30 h while in yeast (in vivo) it was >20 h and in Escherichia coli (in vivo) was >10 h. The protein was stable with instability index (II) of 40.09. Aliphatic index was found as 80.55 while GRAVY as −0.375. The Protscale properties has been displayed in Table 1. The least buried residue was 479 aa with the value of 1.322 while maximum value (11.222) was shown by 755 aa. Lowest polarity was shown by 775 aa with 0.101 value and the highest polarity was 769 with value of 39.468. The average flexibility values varied between 0.381 (372aa) and 0.497 (217aa). The values of hydrophobicity were estimated using Hopp and Woods method with minimum and maximum values of −1.756 (745aa) and 2.3 (770aa) respectively. The values of relative mutability indicated the residue of 60aa with highest mutability (111.222) while the least mutable position was 51.111 (264). The bulkiness ranged from 11.472 (399aa) to 19.523 (742aa) while accessibility was estimated as 3.622 (746aa) and 8.344 (773aa).
Table 1.
Different physicochemical parameters for ACE2 analyzed by ProtParam and Protscale tools.
| Lowest |
Highest |
Lowest |
Highest |
Lowest |
Highest |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buried residues % | Polarity/Zimmerman | Average flexibility | |||||||||||
| Position | Score | Position | Score | Position | Score | Position | Score | Position | Score | Position | Score | ||
| 479 | 1.322 | 755 | 11.222 | 755 | 0.101 | 769 | 39.468 | 372 | 0.381 | 217 | 0.497 | ||
| 478 | 1.333 | 756 | 11.211 | 756 | 0.116 | 770 | 39.453 | 460 | 0.388 | 790 | 0.491 | ||
| 477 | 1.522 | 751 | 10.956 | 757 | 0.116 | 771 | 39.453 | 387 | 0.387 | 768 | 0.493 | ||
| 474 | 1.611 | 752 | 10.956 | 758 | 0.14 | 772 | 34.117 | 458 | 0.387 | 736 | 0.492 | ||
| 476 | 1.644 | 11 | 10.744 | 7 | 1.033 | 479 | 33.992 | 240 | 0.386 | 767 | 0.492 | ||
| Hphob./Hopp and Woods | Relative mutability | Bulkiness | |||||||||||
| Position | Score | Position | Score | Position | Score | Position | Score | Position | Score | Position | Score | ||
| 745 | −1.756 | 770 | 2.300 | 264 | 51.111 | 60 | 111.222 | 399 | 11.472 | 742 | 19.523 | ||
| 746 | −1.700 | 771 | 2.300 | 462 | 52.333 | 61 | 111.000 | 217 | 11.602 | 756 | 19.476 | ||
| 758 | −1.578 | 769 | 2.156 | 591 | 52.889 | 55 | 107.778 | 398 | 11.619 | 757 | 19.457 | ||
| 742 | −1.556 | 772 | 2.000 | 271 | 53.333 | 57 | 107.333 | 397 | 11.698 | 745 | 19.309 | ||
| 743 | −1.556 | 768 | 1.822 | 592 | 54.556 | 443 | 107.333 | 549 | 11.796 | 758 | 19.279 | ||
| Accessible residues (%) | |||||||||||||
| Position | Score | Position | Score | ||||||||||
| 746 | 3.622 | 773 | 8.344 | ||||||||||
| 377 | 3.778 | 772 | 8.1 | ||||||||||
| 745 | 3.822 | 110 | 7.944 | ||||||||||
| 459 | 3.844 | 770 | 7.911 | ||||||||||
| 558 | 3.911 | 771 | 7.911 | ||||||||||
3.3. Prediction of ACE2 signal peptide and structural domains
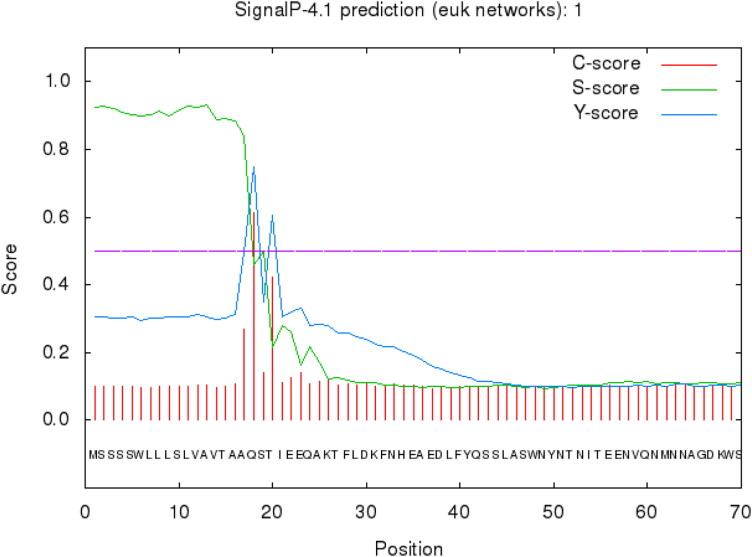
SignalP-4.1 predicted a cleavage site between the positions of 17 and 18 as depicted in Figure 3. For ACE2, C-, S-, and Y-scores were computed. Amino acid 18 showed the maximum C- and Y-scores. The maximum S-score was obtained for position 13 all greater than standard value of 0.5.
Figure 3.
Signal peptide detection for ACE2 protein from peptide sequence. SignalP 4.1 analysis predicted signal peptide cleavage site between 17 and 18 aa.
3.4. Ace2 single nucleotide polymorphism (SNP)
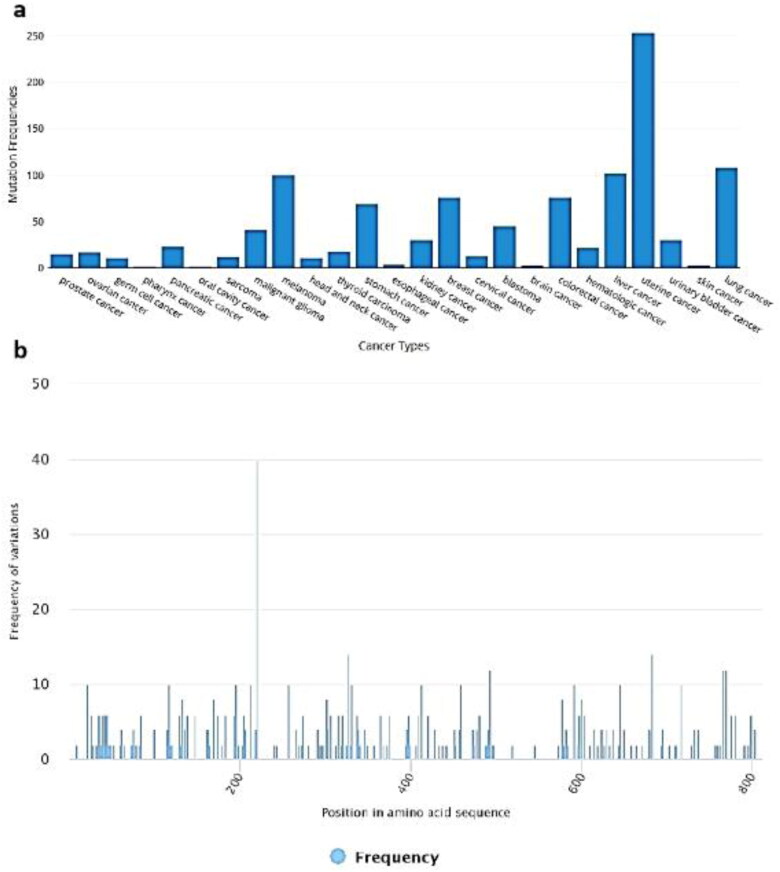
The BioMuta displayed the detailed overview of ACE2 single nucleotide polymorphism with frequency and the type of cancer. The SNP observed for gene were: Uterine cancer: 253, Lung cancer: 108, Liver cancer: 102, Melanoma 100, Colorectal cancer, 76 and Breast cancer 76 (Figure 4(a)). Three frequent SNPs observed were at the positions of 219 aa in 40 patients, 325 aa in 14 patients and 683aa in 14 patients (Figure 4(b)).
Figure 4.
SNP analysis of ACE2 in cancer patients. (a) SNPs observed for ACE2 gene in response different cancer types; and (b) Position-wise frequency of SNP.
The nsSNPs after retrieval from dbSNP tool of NCBI, the SIFT tool selected and analyzed fifty (50) nsSNPs which were predicted as deleterious as described in Supplementary Table S1. These deleterious nsSNPs were further evaluated by PolyPhen-2, PROVEAN and PANTHER software for the prediction of damaging SNPs (Supplementary Tables S1 and S2). PolyPhen-2 predicted 33, PROVEAN 23 and 28 nsSNPs as damaging as the result of their analysis. In Panther analysis, the preservation time (millions of years) depicts the duration for a position in a given protein for which it has been preserved from back to reconstructed direct ancestors. Greater the time of preservation for a position, higher the chances of deleterious effect (Thomas, Kejariwal et al. 2003).
3.5. Sub cellular localization
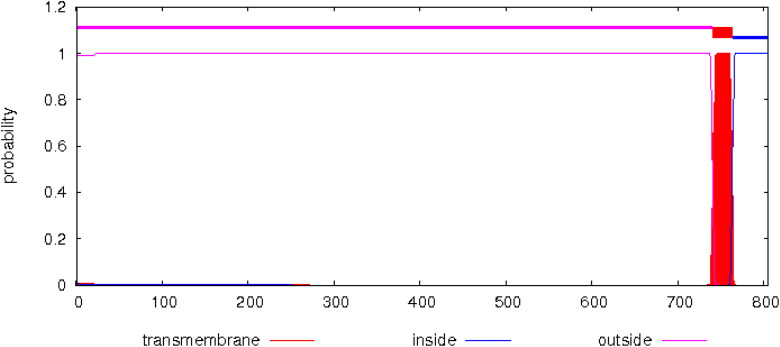
Results of CELLO and UniProt showed that ACE2 has high concentrations in plasma membrane and secreted in extracellular spaces. Apart from these subcellular locations, cytoplasm and cell membrane of neurons also have a detectable amount of ACE2. HMMTOP and TMHMM revealed the presence of a single transmembrane helix for ACE2, the expected number of amino acids in this transmembrane helix was predicted to be 23.23853, with N-terminus towards outside (Figure 5).
Figure 5.
TMHMM predicted posterior probabilities for Human ACE2.
3.6. Methylation sites assessment
The ACE2 gene sequence analysis using MethyCancer revealed absence of any methylation site, moreover the MethyCancer CpG island prediction server failed to predict any methylation site for ACE2.
3.7. Post translational modifications’ assessment
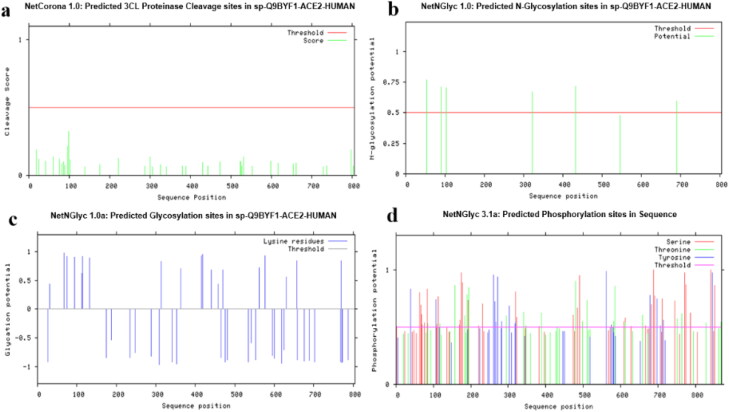
Numerous post-translational modifications were predicted by CBS server for ACE2. The threshold value was set at 0.5 (default value) and residues with value greater than this threshold would indicate high probability. NetCGlyc revealed the absence of c-mannosylation sites for ACE2. No 3CL cleavage sites were predicted for ACE2 by NetCorona (Figure 6a). ProP 1.0 server predicted a signal peptide cleavage site between 17 and 18 amino acids, but no propeptide cleavage site was predicted. NetGlycate 1.0 server predicted 19 glycosylation sites for ACE2 (Figure 6b). NetNGlyc 1.0 showed presence of 6 N-linked glycosylation sites (Figure 6c), whereas no O-glycosylation site was predicted for ACE2 by NetOGlyc 4.0. NetPhos3.1 predicted a total of 111 phosphorylation sites for ACE2. Highest number of phosphorylation sites were identified for Serine (58), followed by threonine which had 30 phosphorylation sites and lastly tyrosine having only 23 sites (Figure 6d). Different type of kinases with their total number of phosphorylation sites are shown in Table 2.
Figure 6.
Human ACE2 Post-Translational Modifications predicted by CBS servers. (a) Predicted 3CL Proteinase cleavage sites by NetCorona 1.0. (b) 19 glycosylation sites predicted by NetGlycate 1.0. (c) 6-N glycosylation sites predicted by NetNGlyc 1.0. (d) Predicted phosphorylation sites by NetPhos 3.1.
Table 2.
Kinases with their total number of phosphorylation sites.
| Kinases | Phosphorylation sites |
|---|---|
| PKC | 19 |
| PKA | 11 |
| CKII | 11 |
| cdc2 | 10 |
| INSR | 6 |
| CKI | 5 |
| DNAPK | 4 |
| P38MAPK | 3 |
| EGFR | 1 |
| cdk5 | 1 |
| PKG | 1 |
| SRC | 1 |
| Unspecific | 38 |
3.8. Predicted ACE2 protein structure and assessment of model stability
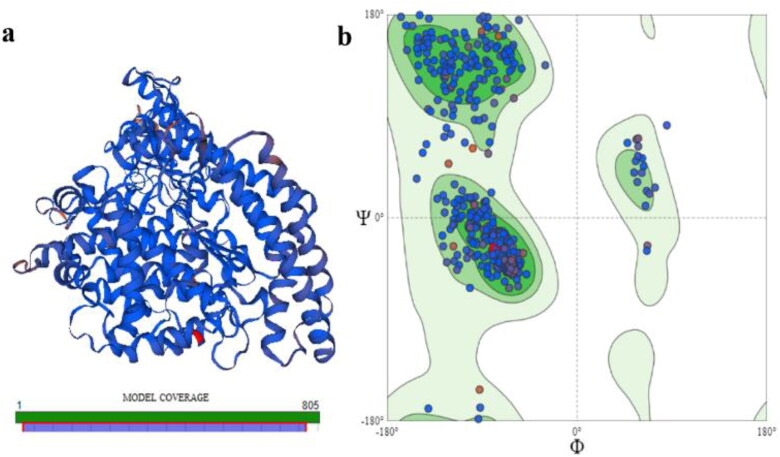
Template search on Swiss Model yielded a total of eight templates with variable sequence identity to our target protein ACE 2. Based on sequence identity, GMQE and QSQE scores, Template 6m18.1B (Title: Angiotensin converting enzyme 2) was finalized as template for model construction. This template showed 100% sequence identity with ACE2, with a perfect GMQE score at 0.99 and QSQE score at 0.85. PPI fingerprint for this template was also within the predicted range for homology modeling. The predicted model of ACE2 showed in range quality estimate and QMEAN value. ACE2 protein consists of 805 aa however our model coverage range was up to 768 aa. Due to unavailability of any suitable template for the last 32 aa segments of ACE2, no structure could be predicted for this protein segment (Figure 7a). The predicted ACE2 model was further evaluated via Ramachandran plot using Swiss Model Structure Assessment feature (Figure 7b). The MolProbity score and Clash score was 0.88 and 0.25, respectively. Ramachandran favored regions were 95.84% with outliers at 0.27%.
Figure 7.
Predicted Protein Structure for Human ACE2. (a) 3-D structure and model coverage; and (b) Ramachandran plot for stability assessment.
3.9. Ace2 protein-protein association pathways and orthologue clustering
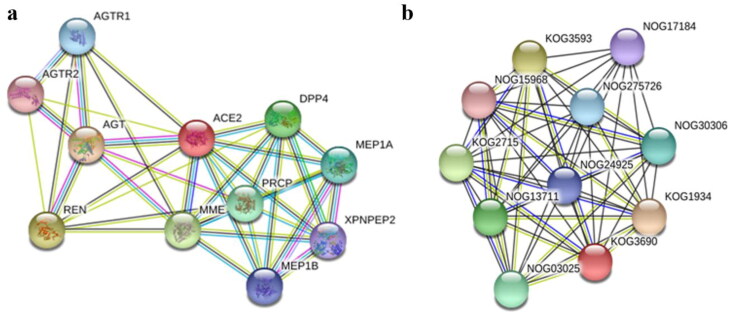
A multiple node interaction map was constructed using STRING database. The results showed interaction of ACE2 with ten other proteins. The analysis of this protein-protein interaction network revealed 11 nodes, 37 edges and average node degree of 6.73. The PPI enrichment value and average local clustering co-efficient for this interactive mesh of proteins was 0.844 and 1.29e-09 respectively (Figure 8a). Ortholog analysis of ACE2 revealed that it belonged to KOG3690 orthologous group of M2-family peptidases (Figure 8b). The primary molecular functions of ACE2 were exopeptidase/endopeptidase activity (GO:0008238, GO:0004175), amino peptidase activity (GO:0004177), metallopeptidase activity (GO:0008237) and peptidase activity on peptides of L-amino acids (GO:0070011). ACE2 plays a primary role in a number of biological processes: modulation of systematic arterial blood pressure via renin-angiotensin system (GO:0003073), adjustment of blood volume (GO:0002016), production of aldosterone (GO:0002018), metabolism of reactive oxygen species (GO:2000377), maintaining balance of hormone level (GO:0010817), controlling diameter of blood vessel via renin-angiotensin system (GO:0002034), maturation of angiotensin (GO:0002003), involved in renin-angiotensin system of brain (GO:0002035) and much more. Cellular pathways in which ACE2 play a central role include Secretion of renin (hsa04924), Protein digestion and assimilation (hsa04974), Adrenergic signaling in cardiac tissues (hsa04261), Receptor-ligand interaction in neuronal pathways (hsa04080) and Renin-angiotensin system (hsa04614).
Figure 8.
Protein-protein Interaction and Ortholog clustering of Human ACE2 (a) Protein-Protein Interaction network analysis via STRING revealed 11 nodes, 37 edges and average node degree of 6.73. The PPI enrichment value was 1.29e-09 whereas average local clustering co-efficient was 0.844. (b) Protein family analysis placed ACE2 in KOG3690 orthologous group of M2-family peptidases.
3.10. Mirnas targeting ACE2
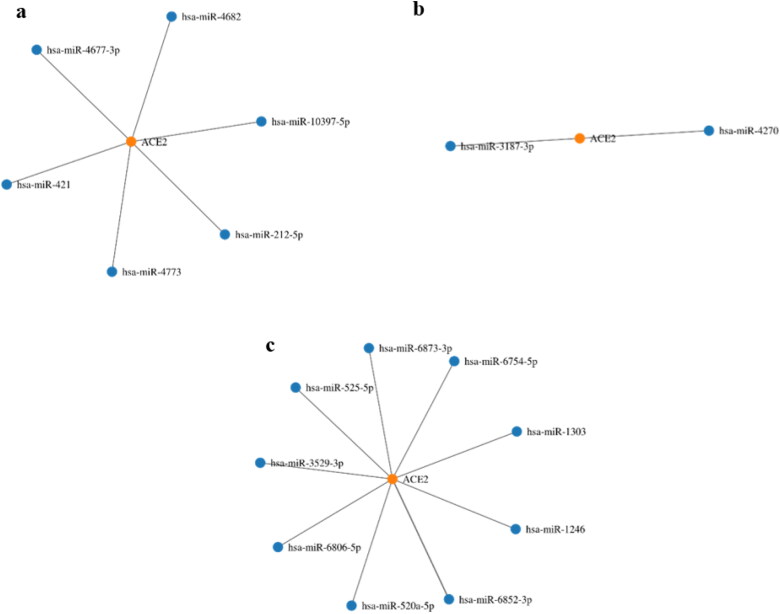
The predicted miRNAs for ACE2 are shown in the Figure 9. When the literature survey was done to validate these predicted miRNAs against the experimental data, only miR-1246 was found to have experimental validation as its upregulation is associated with acute respiratory distress syndrome (ARDS) (Fang, Gao et al. 2017).
Figure 9.
Node graphs of ACE2 specific miRNAs predicted by miRWalk and also cross matched and filtered with miRDB (Sticht, De La Torre et al. 2018). (a) miRNAs targeting 3’ UTR; (b) miRNAs targeting 5’ UTR; and (c) miRNAs targeting CDS (coding DNA sequence).
3.11. Tfs of ACE2
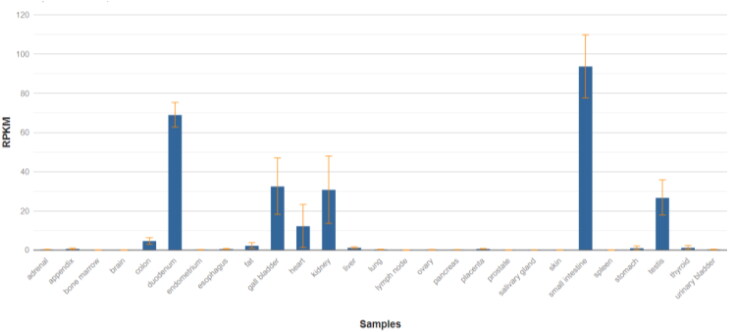
Since the binding of viral spike protein with ACE2 has been proposed to be a mechanism for viral entry into lungs, the first sensible step was to check the expression of this gene in lungs along with other tissues to get a clearer picture. The graph for mRNA expression of ACE2 for different tissues of adult human being is shown in the Figure 10 (Fagerberg, Hallström et al. 2014). Furthermore, the RPKM values ACE2 for healthy tissues (of all the organs reported to have been affected by COVID-19 infection) are also presented in Table 3. Quite surprisingly, the normal expression of ACE2 in lung tissue is very low as compared to other tissues.
Figure 10.
Expression level of ACE2 in different tissues of adult human (Fagerberg, Hallström et al. 2014). Graph courtesy from NCBI database (NCBI Gene 2020).
Table 3.
Expression of ACE2 in different normal human tissues, arranged in a descending order (Fagerberg, Hallström et al. 2014).
| Tissue type | RPKM values for ACE2 expression |
|---|---|
| Small intestine | 93.724 ± 16.1 |
| Duodenum | 69.059 ± 6.292 |
| Kidneys | 30.81 ± 17.148 |
| Heart | 12.309 ± 10.958 |
| Stomach | 1.177 ± 0.937 |
| Lungs | 0.162* |
| Brain | NS |
| Colon | NS |
RPKM value of a single lung sample via Illumina bodyMap2 transcriptome.
NS – non-significant.
The potential transcription factors for ACE2 were identified using GeneCards database (GeneCards 2020) resulting in 38 TFs selected on the basis of highest scoring. The GeneHancer score for these 38 transcription factors was 1.2 while gene association score was 500.7 with the transcription start site (TSS) location was +2.4 kb. These TFs were then further sorted on the basis of their ubiquitous expression in specific tissues of the body (Table 4) (Fagerberg, Hallström et al. 2014). This resulted in the identification of FOXP1 (forkhead box protein P1) and RARA (Retinoic acid receptor alpha) as the TFs having abundant expression in the lungs.
Table 4.
Tissue specific expression of transcription factors acting on promoter/enhancer region of ACE2 gene.
| Tissue type | Putative Transcription Factors regulating expression of ACE2 arranged with respect to their high expression in the corresponding tissue (38 TFs) | Transcription factors with no available transcriptomic data |
|---|---|---|
| Adrenal gland | – | CEBPB, USF1, JUN, JUND, CEBPA |
| Appendix | – | |
| Bone marrow | ZNF384, EP300, | |
| Colon | NCOR1, NR2F6, | |
| Duodenum | HNF4A, | |
| Endometrium | SMARCE1, | |
| Esophagus | – | |
| Fat | BCL6, | |
| Gall bladder | MAFF | |
| Heart | – | |
| Kidney | – | |
| Liver | FOXA3, | |
| Lung | FOXP1, RARA, | |
| Lymph node | TCF7, MIXL1, | |
| Ovary | – | |
| Pancreas | – | |
| Placenta | TEAD3, ARID3A, SOX13, | |
| Prostate | FOXA1, | |
| Salivary gland | – | |
| Skin | GATA3, RXRA, SP1, | |
| Small intestine | – | |
| Spleen | HMG29A, | |
| Stomach | FOXA2, | |
| Testis | SAP130, HOMEZ, KDM1A, RFX1, SOX5, GATAD2A, MNT, | |
| Thyroid | SMAD4, ZNF614, CEBPG, RXRB, | |
| Urinary bladder | – |
3.12. Analysis of RNA-seq data of lung biopsy samples
The identification of lung specific ACE2 transcription factors further urged us to analyze the in vivo RNA seq data of COVID-19 infected lungs to find if any of these TFs might be differentially expressing. The results of the RNA-seq analysis are described below.
3.12.1. Quality control, composition bias normalization and hierarchical clustering
The distribution of the samples’ data and library sizes was checked with the help of different plots. The library sizes of each sample were barplotted to find the major discrepancies between the samples. The barplot of library sizes demonstrated that the data is not normally distributed (Supplementary Figure S2 (a)). Further examination of the raw counts was done via plotting the log2 counts per million to correct for different library sizes. This resulted in similar distributions of log intensities except for the COVID-19 lung 1 sample which appeared far below the median line (Supplementary Figure S2 (b)). Furthermore, the determination of the greatest source of variation in this data and visualization of the principle components analysis was done via multidimensional scaling plot (MDS plot) (Supplementary Figure S2 (c)). The MDS plot shows a great deal of variation between the lung biopsy samples from COVID-19 patients.
TMM normalization was performed for each library to eliminate composition bias and mean difference plots were generated side by side for each sample to see the difference (Supplementary Figure S3).
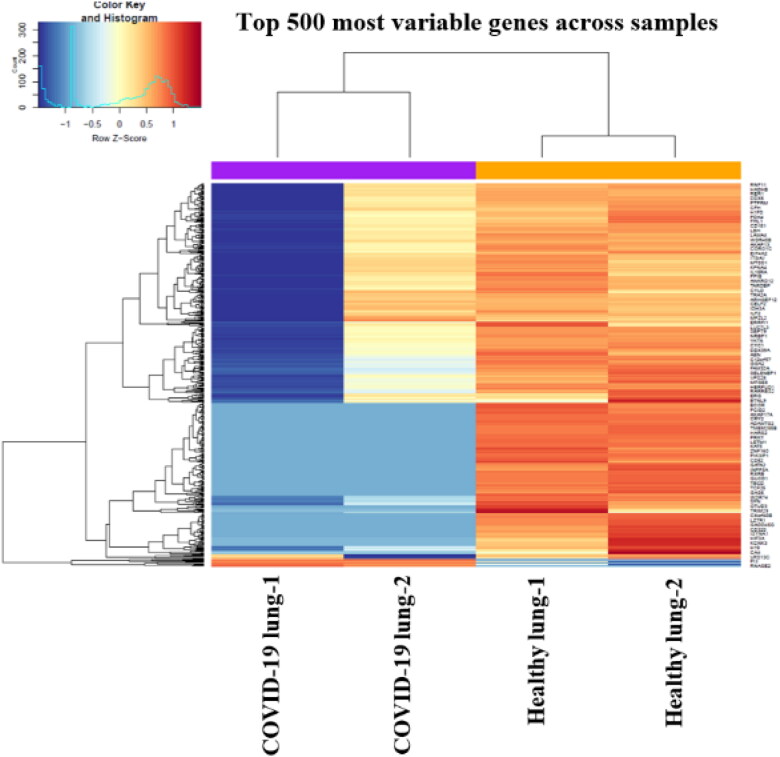
For further examining the relationship between the samples of this study, hierarchical clustering was done and heatmap was plotted for the top 500 variable genes (Figure 11) using heatmap.2 function of the gplots package.
Figure 11.
Heatmap for 500 most variable genes between healthy and COVID-19 lung biopsy samples. Purple – COVID-19 lung biopsy samples, orange – healthy lung samples.
3.12.2. Limma-Voom analysis of differential expression
The voom function of the limma package transformed the read counts into logCPMs and then a linear model was built on the voom-transformed data. The design matrix was built and used to plot the mean variance trend. Moreover, the boxplots were generated again as a comparison between before and after TMM normalization which showed that the variance in the density distributions between libraries is now reduced much and all the samples are now arranged near the median line (Supplementary Figure S4).
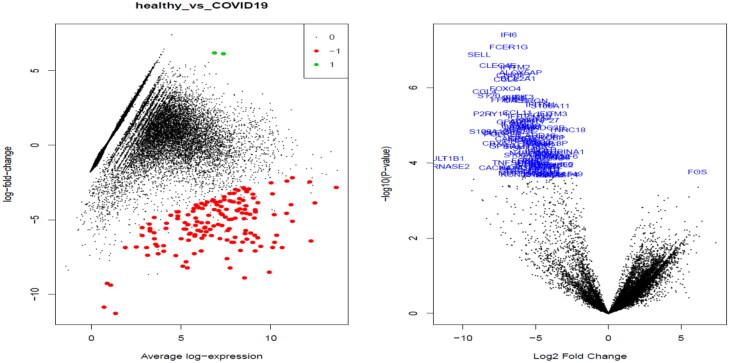
The differentially expressed genes were tested using voom transformed data and contrast matrix was made between COVID19 patients and healthy controls (supplementary excel file). After testing for differential expression, MDplots and volcano plots were generated to see the variance of the genes being significantly expressed differentially (Figure 12).
Figure 12.
Plotting to view the differentially expressed genes. Left – MDplot; and right – volcano plot.
3.12.3. Sorting transcription factors
The differentially expressed genes were further sorted to find any transcription factors being expressed. As a result, FOXO4, FOXP2 were found to be downregulated whereas FOS was found to be upregulated as a result of COVID-19 infection (Table 5). Although these transcription factors are not the same as identified (Table 4), however the only similarity we got was that the TFs from Forkhead box protein family were common among the two.
Table 5.
Description of transcription factors from the down regulated genes.
| SYMBOL | GENENAME | Average expression | Class | Family |
|---|---|---|---|---|
| FOXO4 | Forkhead Box O4 | 9.134273 | Fork head / winged helix factors | Forkhead box (FOX) factors |
| FOXP2 | Forkhead Box P2 | 5.165177 | Fork head / winged helix factors | Forkhead box (FOX) factors |
| FOS | FOS proto-oncogene, AP-1 transcription factor subunit | 7.36853 | Basic leucine zipper factors (bZIP) | FOS-related factors |
3.13. Analysis of cellular pathways regulated by transcription factors
All the three TFs sorted from the expression analysis were further investigated to see which cellular pathways they might regulate and to get a picture of how their downregulation might be affecting cell survival thereby determining their probabilistic contribution in cell death or proliferation of lung cells as a result of COVID-19 infection. Of the three transcription factors, no cellular pathways were found to be existing for FOXP2, whereas 1 and 3 relevant pathways were found associated with FOXO4 and FOS respectively (Supplementary Figures S5-S8) (Slenter, Kutmon et al. 2018).
FOXO4 was found to be present in pathways associated with cellular signaling of glioblastoma. Present as a downstream molecular component of the AKT signaling pathway, it is an upstream effector of JNK pathway via intermediacy of some other molecules and found to be involved in G1/S phase of cell cycle progression (Supplementary Figure S5) (Slenter, Kutmon et al. 2018).
The three cellular pathways of which FOS was found to be an integral component are: Toll-like receptor (TLR) signaling pathway, FOS Corticotropin-releasing hormone signaling pathway and estrogen signaling pathway (Slenter, Kutmon et al. 2018). In the TLR signaling pathway, FOS is a downstream mediator which leads to complement and coagulation cascades whereas its role in estrogen signaling pathway is of the regulation of the expression anti-apoptotic and proliferative genes (Supplementary Figure S6 and S8). However, looking at the FOS Corticotropin-releasing hormone signaling pathway, the definite role of FOS is still not clear despite its presence as a downstream effector of this pathway (Supplementary Figure S7).
4. Discussion
Coronaviruses have caused massive pandemics twice in the last two decades, SARS in 2002, and the Middle East respiratory syndrome (MERS) in 2012 (Xu, Zhong et al. 2020). Novel coronavirus SARS-CoV-2 was identified in Wuhan, China in December 2019. This viral outbreak has been declared as a pandemic by WHO and has 8,018,963 active cases with 436,138 deaths till June 15, 2020, worldwide. Among these cases, 144,478 cases are of Pakistan with 2,729 deaths (https://www.worldometers.info/coronavirus/).
Infection routes and treatment of COVID-19 have not been understood so far. However, SARS-CoV-2/COVID-19 is reported to share the same receptor, Angiotensin-converting enzyme 2 (ACE2), with SARS-CoV. ACE2 acts as a receptor for viral spike protein thereby facilitating its entry in the cell. Thus, the study of COVID-19 host cell receptor ACE2 can be useful for the prevention and treatment of the COVID-19 (Zhao, Zhao et al. 2020). In this study, we have analyzed ACE2 protein using in silico approaches for acquiring a better understanding of its pathology and its potential as a target therapeutic target.
The Human ACE2 sequence was subjected to structural analysis. ACE2 protein has one transmembrane helix with 23.23853 amino acids and its N-terminal to be outside the membrane. While gene sequence analysis of ACE2, it was observed that there is no methylation site for ACE2, and the server also failed to predict any CpG site. Eight templates were found for ACE2 by using homology modeling with high sequence similarity. One of those templates was selected based on sequence similarity and structure was predicted. ACE2 protein consists of 801 amino acids and the predicted structure was of 768 amino acids. For the last few amino acids, no appropriate template was found so those specific regions were not synthesized. Then the model was subjected to Ramachandran plot analysis for assessment. Due to the use of a good template, the quality of the predicted structure was found satisfactory. Functional association and ortholog clustering of ACE2 was done, the interaction of ACE2 with other proteins was observed in the functional association. In ortholog analysis, it was observed that ACE2 belongs to the orthologous group of M2-family peptidases. ProtParam computations predicted the protein to be stable with precise aliphatic (Guruprasad, Reddy et al. 1990) and stability indexes (Ikai 1980). Prominent kinases with the total number of their respective phosphorylation sites were predicted with 38 non-specific kinases. BioMuta analysis for the ACE2 displayed three frequent SNPs at the positions of 219 aa in 40 patients, 325 aa in 14 patients and 683aa in 14 patients, and types of cancer. Damaging SNPs were predicted by using PolyPhen-2 and PROVEAN software. In literature, correlation of radical expression of ACE2 and Ang II with metastasis and diagnosis of endometrial cancer has been reported by Watanabe et al., 2003; Shibata et al., 2005; Delforce et al., 2017 (Jing, Run-Qian et al. 2020)
Different miRNAs were predicted for ACE2 and only miR-1246 was found to have experimental validation due to the association with Acute Respiratory Distress Syndrome. MiR-1246 was found to repress ACE2 expression by binding to its 3′-UTR (Liu, Du et al. 2017). It is possible that this miRNA might also be getting upregulated in the respiratory distress caused by COVID-19. However, no data is available in this regard and it would be useful to study the expression of miR-1246 as a result of COVID-19 infection in the wet lab along with other predicted miRNAs of this study. Parallel studies of other predicted miRNAs showed involvement in different diseases. ACE2 specific MiR-212-5p suppresses the epithelial-mesenchymal transition and metastasis by downregulating Prrx2 and can act as prognostic marker for breast cancer (Lv, Yang et al. 2017). MiR-212-5p is also involved in chronic obstructive pulmonary disease (COPD), where it exerts a protective effect in COPD by promoting cell proliferation and down-regulating the expression of the related genes and proteins (Jia, Chang et al. 2018).
Different transcription factors of ACE2 were selected based on the highest scoring and their expression in different organs was also checked. FOXP1 (forkhead box protein P1) and RARA (Retinoic acid receptor alpha) were the predicted TFs with the highest expression in the lungs. Three transcription factors (TFs) including Signal transducer and activator of transcription 4 (STAT4), Estrogen related receptor α (ESRRA), and Signal transducer and activator of transcription 3 (STAT3) which express in different organs other than lungs were sorted as potential targets of ACE2 in a study. These TFs might act as possible therapeutic agents against the COVID-19 infection by regulation of the transcriptional activation of ACE2 (Yang, Zhao et al. 2020). In this study, we have also focused on those TFs which are highly expressed in the lungs and tried to validate that data with RNA sequence data. Significant upregulation of FOS and downregulation of FOXO4 and FOXP2 was found and upregulation FOS was found because of the COVID-19 infection. This is the first study that elaborates the potential targets related to ACE2.
5. Conclusion and future perspective
Transcription factors and miRNAs both constitute a huge class of biological molecules actively playing their roles in health and disease. Their implication in various human diseases and also their presence in a tissue-specific manner makes them a useful target for therapeutic interventions under both acute and chronic conditions. The same applies to our findings of this study concerning the development of new treatments for COVID-19. We propose that inhibitors and activators of the up and downregulated molecules, respectively, could present valuable new targets for this currently incurable pandemic disease.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contributions
Abeedha Tu-Allah Khan designed the study and worked on prediction of miRNAs, prediction of TFs, RNA seq analysis and analysis of cellular pathways. Zumama Khalid, Muhammad Abrar Yousaf and Hafsa Zahid contributed in Phylogenetic analysis, physiochemical characterization, protein structure building and SNPs, methylation, PTM and functional network analysis. Abdul Rauf Shakoori supervised and guided the whole study.
References
- Bairoch, A., & Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research, 28(1), 45–48. 10.1093/nar/28.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard, S., Chu, V. C., & Whittaker, G. R. (2009). Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences of the United States of America, 106(14), 5871–5876. 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo, D., Nilsson-Payant, B. E., Liu, W.-C., Møller, R., Panis, M., Sachs, D., Albrecht, R. A., & tenOever, B. R. (2020). SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv: 2020.2003.2024.004655.
- Blom, N., Sicheritz-Pontén, T., Gupta, R., Gammeltoft, S., & Brunak, S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics, 4(6), 1633–1649. 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- Chen, Y., & Wang, X. (2020). miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Research, 48(D1), D127–D131. 10.1093/nar/gkz757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Wang, Y., & Wang, G. Q. (2020). Organ‐protective effect of angiotensin‐converting enzyme 2 and its effect on the prognosis of COVID‐19. Journal of Medical Virology, 92(7), 726–730. 10.1002/jmv.25785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y., & Chan, A. P. (2015). PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England), 31(16), 2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingerdissen, H. M., Torcivia-Rodriguez, J., Hu, Y., Chang, T.-C., Mazumder, R., & Kahsay, R. (2018). BioMuta and BioXpress: Mutation and expression knowledgebases for cancer biomarker discovery. Nucleic Acids Research, 46(D1), D1128–D1136. 10.1093/nar/gkx907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue, M., Hsieh, F., Baronas, E., Godbout, K., Gosselin, M., Stagliano, N., Donovan, M., Woolf, B., Robison, K., Jeyaseelan, R., Breitbart, R. E., & Acton, S. (2000). A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circulation Research, 87(5), E1–e9. 10.1161/01.res.87.5.e1 [DOI] [PubMed] [Google Scholar]
- Duckert, P., Brunak, S., & Blom, N. (2004). Prediction of proprotein convertase cleavage sites. Protein Engineering, Design & Selection : Peds, 17(1), 107–112. 10.1093/protein/gzh013 [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Brunak, S., von Heijne, G., & Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols, 2(4), 953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Fagerberg, L., Hallström, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., Habuka, M., Tahmasebpoor, S., Danielsson, A., Edlund, K., Asplund, A., Sjöstedt, E., Lundberg, E., Szigyarto, C. A.-K., Skogs, M., Takanen, J. O., Berling, H., Tegel, H., Mulder, J., Nilsson, P., Schwenk, J. M., Lindskog, C., Danielsson, F., Mardinoglu, A., Sivertsson, A., von Feilitzen, K., Forsberg, M., Zwahlen, M., Olsson, I., Navani, S., Huss, M., Nielsen, J., Ponten, F., & Uhlén, M. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics : Mcp, 13(2), 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., Gao, F., Hao, J., & Liu, Z. (2017). microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. American Journal of Translational Research, 9(3), 1287–1296. [PMC free article] [PubMed] [Google Scholar]
- Ferrario, C. M., Trask, A. J., & Jessup, J. A. (2005). Advances in biochemical and functional roles of angiotensin-converting enzyme 2 and angiotensin-(1-7) in regulation of cardiovascular function. American Journal of Physiology. Heart and Circulatory Physiology, 289(6), H2281–H2290. 10.1152/ajpheart.00618.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, G. F. (2018). From “A"IV to "Z"IKV: Attacks from emerging and re-emerging pathogens. Cell, 172(6), 1157–1159. 10.1016/j.cell.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, V. K., Avashthi, H., Tiwari, A., Jain, P. A., Ramkete, P. W., Kayastha, A. M., & Singh, V. K. (2016). MFPPI - Multi FASTA ProtParam Interface. Bioinformation, 12(2), 74–77. 10.6026/97320630012074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeneCards, T. H. G. D. (2020). ACE2 Gene. Retrieved 11 June, 2020, from https://www.genecards.org/cgi-bin/carddisp.pl?gene=ACE2.
- Gorbalenya, A., Baker, S., Baric, R., de Groot, R., Drosten, C., Gulyaeva, A., Haagmans, B., Lauber, C., Leontovich, A., & Neuman, B. (2020). The species severe acute respiratory syndrome related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, W.-J., Ni, Z.-Y., Hu, Y., Liang, W.-H., Ou, C.-Q., He, J.-X., Liu, L., Shan, H., Lei, C.-L., Hui, D. S. C., Du, B., Li, L.-J., Zeng, G., Yuen, K.-Y., Chen, R.-C., Tang, C.-L., Wang, T., Chen, P.-Y., Xiang, J., … China Medical Treatment Expert Group for Covid-19 (2020). Clinical characteristics of coronavirus disease 2019 in China. The New England Journal of Medicine, 382(18), 1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18(15), 2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Gupta, R., Jung, E., & Brunak, S. (2004). Prediction of N-glycosylation sites in human proteins. 46, 203–206. [Google Scholar]
- Guruprasad, K., Reddy, B. B., & Pandit, M. W. (1990). Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering, 4(2), 155–161. 10.1093/protein/4.2.155 [DOI] [PubMed] [Google Scholar]
- Hamming, I., Cooper, M. E., Haagmans, B. L., Hooper, N. M., Korstanje, R., Osterhaus, A. D., Timens, W., Turner, A., Navis, G., & van Goor, H. (2007). The emerging role of ACE2 in physiology and disease. The Journal of Pathology, 212(1), 1–11. 10.1002/path.2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Chang, S., Zhang, J., Zhao, Q., Xiang, H., Kusonmano, K., Yang, L., Sun, Z. S., Yang, H., & Wang, J. (2008). MethyCancer: The database of human DNA methylation and cancer. Nucleic Acids Research, 36(Database issue), D836–841. (Database issue): 10.1093/nar/gkm730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., Spitters, C., Ericson, K., Wilkerson, S., Tural, A., Diaz, G., Cohn, A., Fox, L., Patel, A., Gerber, S. I., Kim, L., Tong, S., Lu, X., & Lindstrom, S. (2020). First case of 2019 novel coronavirus in the United States. The New England Journal of Medicine, 382(10), 929–936. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., & Liu, M. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikai, A. (1980). Thermostability and aliphatic index of globular proteins. The Journal of Biochemistry, 88(6), 1895–1898. [PubMed] [Google Scholar]
- Imai, Y., Kuba, K., Rao, S., Huan, Y., Guo, F., Guan, B., Yang, P., Sarao, R., Wada, T., Leong-Poi, H., Crackower, M. A., Fukamizu, A., Hui, C.-C., Hein, L., Uhlig, S., Slutsky, A. S., Jiang, C., & Penninger, J. M. (2005). Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature, 436(7047), 112–116. 10.1038/nature03712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Q., Chang, J., Hong, Q., Zhang, J-j., Zhou, H., & Chen, F-h. (2018). MiR-212-5p exerts a protective effect in chronic obstructive pulmonary disease. Discovery Medicine, 26(144), 173–183. [PubMed] [Google Scholar]
- Jing, Y., Run-Qian, L., Hao-Ran, W., Hao-Ran, C., Ya-Bin, L., Yang, G., & Fei, C. (2020). Potential influence of COVID-19/ACE2 on the female reproductive system. Molecular Human Reproduction, 26(6), 367–373. 10.1093/molehr/gaaa030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, M. B., Kiemer, L., & Brunak, S. (2006). Analysis and prediction of mammalian protein glycation. Glycobiology, 16(9), 844–853. 10.1093/glycob/cwl009 [DOI] [PubMed] [Google Scholar]
- Julenius, K. (2007). NetCGlyc 1.0: Prediction of mammalian C-mannosylation sites. Glycobiology, 17(8), 868–876. 10.1093/glycob/cwm050 [DOI] [PubMed] [Google Scholar]
- Kiemer, L., Lund, O., Brunak, S., & Blom, N. (2004). Coronavirus 3CLpro proteinase cleavage sites: Possible relevance to SARS virus pathology. BMC Bioinformatics, 5, 72–72. 10.1186/1471-2105-5-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., & Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology, 305(3), 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kuba, K., Imai, Y., Ohto-Nakanishi, T., & Penninger, J. M. (2010). Trilogy of ACE2: A peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacology & Therapeutics, 128(1), 119–128. 10.1016/j.pharmthera.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., Huan, Y., Yang, P., Zhang, Y., Deng, W., Bao, L., Zhang, B., Liu, G., Wang, Z., Chappell, M., Liu, Y., Zheng, D., Leibbrandt, A., & Wada, T. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nature Medicine, 11(8), 875–879. 10.1038/nm1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba, K., Imai, Y., Rao, S., Jiang, C., & Penninger, J. M. (2006). Lessons from SARS: Control of acute lung failure by the SARS receptor ACE2. Journal of Molecular Medicine (Berlin, Germany)), 84(10), 814–820. 10.1007/s00109-006-0094-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P., Henikoff, S., & Ng, P. C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols, 4(7), 1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Lai, M. (2007). Coronaviridae. Fields Virology, I, 1305–1318. [Google Scholar]
- Li, F., Li, W., Farzan, M., & Harrison, S. C. (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.).), 309(5742), 1864–1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- Liu, Q., Du, J., Yu, X., Xu, J., Huang, F., Li, X., Zhang, C., Li, X., Chang, J., Shang, D., Zhao, Y., Tian, M., Lu, H., Xu, J., Li, C., Zhu, H., Jin, N., & Jiang, C. (2017). miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discovery, 3(1), 1–17. 10.1038/celldisc.2017.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G., & Liu, D. (2012). SARS-like virus in the Middle East: A truly bat-related coronavirus causing human diseases. Protein & Cell, 3(11), 803–805. 10.1007/s13238-012-2811-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G., Wang, Q., & Gao, G. F. (2015). Bat-to-human: Spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends in Microbiology, 23(8), 468–478. 10.1016/j.tim.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., & Zhao, L. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, Z.-D., Yang, D.-X., Liu, X.-P., Jin, L.-Y., Wang, X.-G., Yang, Z.-C., Liu, D., Zhao, J.-J., Kong, B., Li, F.-N., & Wang, H.-B. (2017). MiR-212-5p Suppresses the epithelial-mesenchymal transition in triple-negative breast cancer by targeting Prrx2. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology, 44(5), 1785–1795. 10.1159/000485785 [DOI] [PubMed] [Google Scholar]
- McCarthy, D. J., Chen, Y., & Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research, 40(10), 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., Cowley, A. P., & Lopez, R. (2013). Analysis tool web services from the EMBL-EBI. Nucleic Acids Research, 41(Web Server issue), W597–W600. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet, J. K., & Whittaker, G. R. (2015). Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Research, 202, 120–134. 10.1016/j.virusres.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Gene . (2020, 7th June). FOS Fos proto-oncogene, AP-1 transcription factor subunit [Homo sapiens (human)]. https://www.ncbi.nlm.nih.gov/gene/2353.
- NCBI Gene . (2020, 1st June). FOXO4 forkhead box O4 [Homo sapiens (human)]. https://www.ncbi.nlm.nih.gov/gene/4303.
- NCBI Gene . (2020, 7th June). FOXP2 forkhead box P2 [Homo sapiens (human)]. https://www.ncbi.nlm.nih.gov/gene/93986.
- NCBI Gene . (2020). ACE2 angiotensin I converting enzyme 2 [Homo sapiens (human)]. Retrieved June 9th, 2020. https://www.ncbi.nlm.nih.gov/gene/59272.
- Patel, S., Rauf, A., Khan, H., & Abu-Izneid, T. (2017). Renin-angiotensin-aldosterone (RAAS): The ubiquitous system for homeostasis and pathologies. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 94, 317–325. 10.1016/j.biopha.2017.07.091 [DOI] [PubMed] [Google Scholar]
- R Development Core Team . (2010). R: A language and environment for statistical computing, R Foundation for Statistical Computing. Austria.. [Google Scholar]
- Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., & Smyth, G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research, 43(7), e47–e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, N. L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., & Ng, P. C. (2012). SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Research, 40(Web Server issue), W452–457. (Web Server issue): 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, G., Gosalia, D. N., Rennekamp, A. J., Reeves, J. D., Diamond, S. L., & Bates, P. (2005). Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proceedings of the National Academy of Sciences of the United States of America, 102(33), 11876–11881. 10.1073/pnas.0505577102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, G., Reeves, J. D., Rennekamp, A. J., Amberg, S. M., Piefer, A. J., & Bates, P. (2004). Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proceedings of the National Academy of Sciences of the United States of America, 101(12), 4240–4245. 10.1073/pnas.0306446101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, G., Zmora, P., Gierer, S., Heurich, A., & Pöhlmann, S. (2013). Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Research, 100(3), 605–614. 10.1016/j.antiviral.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenter, D. N., Kutmon, M., Hanspers, K., Riutta, A., Windsor, J., Nunes, N., Mélius, J., Cirillo, E., Coort, S. L., Digles, D., Ehrhart, F., Giesbertz, P., Kalafati, M., Martens, M., Miller, R., Nishida, K., Rieswijk, L., Waagmeester, A., Eijssen, L. M. T., Evelo, C. T., Pico, A. R., & Willighagen, E. L. (2018). WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Research, 46(D1), D661–D667. 10.1093/nar/gkx1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W., Gui, M., Wang, X., & Xiang, Y. (2018). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathogens, 14(8), e1007236 10.1371/journal.ppat.1007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft, C., Vakhrushev, S. Y., Joshi, H. J., Kong, Y., Vester-Christensen, M. B., Schjoldager, K. T., Lavrsen, K., Dabelsteen, S., Pedersen, N. B., Marcos-Silva, L., Gupta, R., Bennett, E. P., Mandel, U., Brunak, S., Wandall, H. H., Levery, S. B., & Clausen, H. (2013). Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. The EMBO Journal, 32(10), 1478–1488. 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticht, C., De La Torre, C., Parveen, A., & Gretz, N. (2018). miRWalk: An online resource for prediction of microRNA binding sites. PloS One, 13(10), e0206239. 10.1371/journal.pone.0206239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, S., Law, C. W., Ah-Cann, C., Asselin-Labat, M. L., Blewitt, M. E., & Ritchie, M. E. (2017). Glimma: Interactive graphics for gene expression analysis. Bioinformatics (Oxford, England)), 33(13), 2050–2052. 10.1093/bioinformatics/btx094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., Santos, A., Doncheva, N. T., Roth, A., Bork, P., Jensen, L. J., & von Mering, C. (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research, 45(D1), D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H., & Thomas, P. D. (2016). PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics (Oxford, England)), 32(14), 2230–2232. 10.1093/bioinformatics/btw222 [DOI] [PubMed] [Google Scholar]
- Thomas, P. D., Kejariwal, A., Campbell, M. J., Mi, H., Diemer, K., Guo, N., Ladunga, I., Ulitsky-Lazareva, B., Muruganujan, A., Rabkin, S., Vandergriff, J. A., & Doremieux, O. (2003). PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Research, 31(1), 334–341. 10.1093/nar/gkg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády, G. E., & Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics (Oxford, England)), 17(9), 849–850. 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- Wang, C., Horby, P. W., Hayden, F. G., & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes, G., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., Lumley, T., Mächler, M., Magnusson, A., & Möller, S. (2005). gplots: Various R programming tools for plotting data.
- Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., & Hochstrasser, D. F. (1999). Protein identification and analysis tools in the ExPASy server. Methods in Molecular Biology (Clifton, N.J.), 112, 531–552. [DOI] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.).), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L., Sakagami, H., & Miwa, N. (2020). ACE2: The key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: Demon or angel. Viruses, 12(5), 491. 10.3390/v12050491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Zhong, L., Deng, J., Peng, J., Dan, H., Zeng, X., Li, T., & Chen, Q. (2020). High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. International Journal of Oral Science, 12(1), 1–5. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., Zhong, W., & Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China. Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, T., Xiao, R., & Lin, G. (2020). Angiotensin-converting enzyme 2 in severe acute respiratory syndrome coronavirus and SARS-CoV-2: A double-edged sword? FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 34(5), 6017–6026. 10.1096/fj.202000782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Zhao, H., Li, Y., & Zhang, P. (2020). Angiotensin converting enzyme 2 in coronavirus disease (COVID-19): Evidence from bioinformatics analysis.
- Yu, C. S., Cheng, C. W., Su, W. C., Chang, K. C., Huang, S. W., Hwang, J. K., & Lu, C. H. (2014). CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PloS One, 9(6), e99368 10.1371/journal.pone.0099368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Zhao, Z., Wang, Y., Zhou, Y., Ma, Y., & Zuo, W. (2020). Single-cell RNA expression profiling of ACE2, thereceptor of SARS-CoV-2. Biorxiv. [DOI] [PMC free article] [PubMed]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., & Wang, X. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.