Abstract
Outbreaks of avian orthoreovirus (ARV) infection with primary symptoms of arthritis/tenosynovitis syndrome have been occurring more frequently in broiler flocks in China in recent years. This study aimed to investigate the genetic characteristics of ARV field strains in broiler flocks exhibiting arthritis/tenosynovitis syndrome from 9 cities in Shandong province during 2015 to 2017. A total of 64 synovial and tendon samples were obtained from broilers with significant arthritis/tenosynovitis syndrome, and 21 ARV field strains were obtained. Phylogenetic analysis of the σC nt/aa sequences revealed that only 4 isolates were clustered in genotype I, including vaccine strains S1133, 1733, and most of the ARV field strains identified previously in China. Eleven and 6 ARV field isolates were identified in genotypes II and V, sharing 70.9 to 76.0% and 53.0 to 55.2% nt identities with the vaccine strains, respectively. Previous studies in China have not reported these 2 serotypes of field strains, and prevalence of these ARV variants may be increasing in Chinese broiler flocks. Results of this study suggest that large-scale investigation of epidemic ARV should be conducted to explore the genetic diversity of ARV field isolates in China.
Key words: avian orthoreovirus, arthritis/tenosynovitis, phylogenetic analysis, σC protein, genetic diversity
Introduction
Avian orthoreoviruses (ARV), which belong to the Orthoreovirus genus in the Reoviridae family, are icosahedral, nonenveloped double-stranded RNA viruses (Spandidos and Graham, 1976). Avian orthoreovirus can infect various avian species, including chickens, turkeys, pheasants, ducks, geese, and other domestic poultry (Yun et al., 2012, Tang and Lu, 2016, Farkas et al., 2018). Based on their electrophoretic mobility, the 10 genomic segments are classified into 3 size classes: 3 L-class segments (L1, L2, and L3), 3 M-class segments (M1, M2, and M3), and 4 S-class segments (S1, S2, S3, and S4), encoding at least 8 structural proteins (λA, λB, λC, μA, μB, σA, σB, and σC) and 4 non-structural proteins (μNS, P10, P17, and σNS) (Benavente and Martinez-Costas, 2007, Day, 2009). The outer capsid consists of 3 more variable proteins (μB, σB, and σC). σC, a minor component of the outer capsid of the virion, is encoded by the S1 gene and is the viral cell attachment protein that elicits ARV-specific neutralizing antibodies (Benavente and Martinez-Costas, 2007, Huang et al., 2011). Depending on the genetic characterization of σC protein, ARV isolates are divided into multiple genotypes (Kant et al., 2003, Day, 2009, Goldenberg et al., 2010). Recent studies also have shown that all ARV isolates are divided into 4 to 6 clusters (Dandar et al., 2013, Lu et al., 2015, Farkas et al., 2016, Tang et al., 2016, Noh et al., 2018).
Subclinical symptoms have been observed in many ARV-infected chickens. Clinical manifestations associated with ARV in young broilers, turkeys, and pheasants include arthritis/tenosynovitis syndrome, runting–stunting syndrome, hepatitis, myocarditis, enteritis, osteoporosis, and respiratory disturbance (Jones, 2000, Van de Zande and Kuhn, 2007, Shivaprasad et al., 2009). The disease is highly contagious in birds, and viral arthritis/tenosynovitis syndrome is the most serious disorder caused by ARV. The affected broilers are reported to exhibit arthroncus, lameness, feed conversion ratio, depression, growth retardation, a high elimination rate, and occasional high mortality rate. Avian orthoreovirus infection can also result in immunosuppression, causing the birds to become more susceptible to other pathogens, which often results in co-infection or secondary infection (Hoerr, 2010). Low-pathogenic ARV isolates also may exist in apparently healthy poultry flocks without causing any visible symptoms. Avian orthoreoviruses have diverse evolutionary mechanisms that make the mutation trend of ARV difficult to predict. Additionally, the current commercial ARV vaccines have not provided sufficient protection against the emerging ARV variants, resulting in more frequent outbreaks of ARV infection. Consequently, outbreaks of viral arthritis/tenosynovitis have significantly affected the poultry industry, leading to extensive economic losses worldwide.
In recent years, many ARV variants (genotype/serotype) have emerged in broiler flocks worldwide, and arthritis/tenosynovitis syndrome is the most predominant characteristic observed, including serious acute arthritis/tenosynovitis syndrome in domestic avian flocks in North China. Many infectious organisms such as E. coli, Staphylococcus, synovial species, ARV, and other agents can cause symptoms of arthritis/tenosynovitis syndrome (Thorp et al., 1993, Lockaby et al., 1998). Trauma also may result in such symptoms. In the present study, we investigated the vaccine strains and ARV field strains in 6 broiler flocks with arthritis/tenosynovitis syndrome, aiming to describe a further phylogenetic analysis of field ARV isolates based on σC and to provide evidence that these strains are clustered into distinct genotypes within classical ARV genotype I.
Materials and methods
Sampling and Virus Isolation
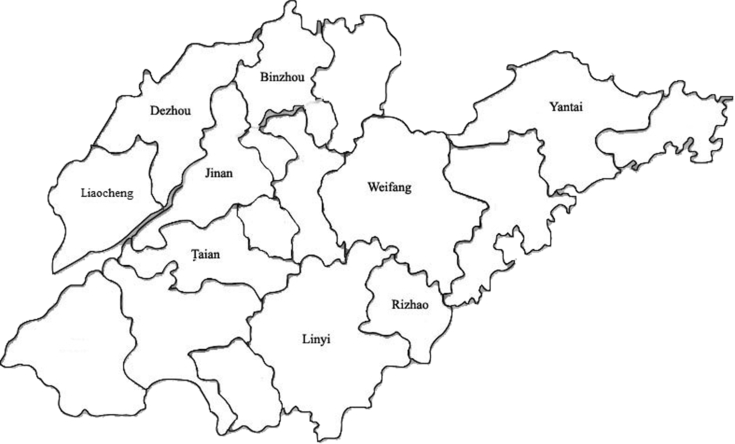
Since 2014, arthritis/tenosynovitis syndrome became prevalent in domestic broiler flocks in Shandong province. All these flocks were housed in floor pens of enclosed coops. All diseased broilers were subjected to Poultry disease laboratory of Shandong Agricultural University and Qufu Normal University under low temperature transportation (4 to 8°C) within 12 h. Fresh synovial fluid and tendon samples were collected from separated broiler/breeder flocks in 9 cities during 2015 to 2017 in Shandong province, China (Figure 1), specifically selecting broilers with signs of acute arthritis/tenosynovitis syndrome that were suspected to have ARV-infection. Total RNA was extracted from tissue samples (tendons and joints, stored at −20°C) using Trizol reagent (Invitrogen, Karlsruhe, Germany). Thirty-seven samples were found to be ARV-positive using one-step real-time RT-PCR (Ct value≤28), as previously described (Tang and Lu, 2016), and were prepared for virus isolation. Tissues were thawed and homogenized in phosphate buffer solution. This suspension was frozen and thawed 3 times and centrifuged briefly. Supernatants from homogenates were filtered and then inoculated into LMH monolayer cells (chicken hepatocellular carcinoma cell line). The LMH monolayers were grown in medium DMEM/F12 (Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (Gibco; Thermo Scientific, Waltham, MA), 100 U/ml penicillin–streptomycin and 0.025 mg/ml gentamicin and inoculated with the supernatant. The inoculated cell cultures were passaged blindly 3 times after 4 to 5 D. Cell cultures were frozen and thawed 3 times after cytopathic effects were observed in 70%. They were then stored at −70°C.
Figure 1.
Regions of ARV field samples from broiler flocks in Shandong Province, PR. China. The 9 cities affected have been marked. All field strains were isolated and confirmed by one-step qRT-PCR. ARV, avian orthoreovirus.
RNA Extraction and RT-PCR
All the virus stocks were examined using RT-PCR to verify the presence of ARV and other avian viral pathogens (e.g., Avian influenza virus, Newcastle virus, infectious bronchitis virus, infectious bursal disease virus, reticuloendothelial virus, avian leukosis virus, and chicken infectious anemia virus). Briefly, the total RNA was extracted from the samples using Trizol reagent (Invitrogen) according to the manufacturer's instructions and treated with DNase I. cDNA was synthesized with random primers (6mer) (Takara, Dalian, China) from total RNA according to the manufacturer's instructions. PCR for σC gene amplification was carried out using 100 to 200 ng cDNA, forward primer (P1: 5′- AGTATTTGTGAGTACGATTG-3′) and reverse primer (P4: 5′- GGCGCCACACCTTAGGT-3′) (Kant, et al., 2003).
Cloning and Sequencing
The amplified products of 1,088 bp sizes were purified, cloned into pMD18-T vector, and transformed into DH5α E.coli competent cells. The positive clones were sequenced with Sanger technology by Sangon Biological Engineering Technology & Service Co., (Shanghai, China). The raw sequence fragments were assembled using the Lasergene SeqMan program (version7.0; DNAStar, Inc., Madison, WI).
Phylogenetic Analysis
Partial σC gene and deduced amino acid (aa) sequences of field strains and 18 reference ARV sequences retrieved from GenBank underwent comparative analysis. Nucleotide and amino acid sequences were aligned using the ClustalW methods in the Megalin program included in the Lasergene 7.0 software program (DNASTAR Inc.). Phylogenetic trees of the aligned nucleotide and amino acid sequences were constructed using the neighbor-joining method associated with the MEGA 5 software with 1,000 bootstrap replicates, using pairwise distances (Tamura, et al., 2011). The GenBank accession numbers for ARV field isolates are listed in Table 1, and reference strains are listed in Supplementary Table S1.
Table 1.
Description of ARV field isolates involved in this study.
| ARV field strains | Origin | GenBank accession No |
|---|---|---|
| Reo/Broiler/YTMP/160427 | Yaitai | MK189464 |
| Reo/Bredder/DZSH/150309 | Dezhou | MK189482 |
| Reo/Broiler/LCSX/150411 | Liaocheng | MK189475 |
| Reo/Broiler/TADY/150416 | Tai'an | MK189473 |
| Reo/Breeder/BZHM/150814 | Binzhou | MK189483 |
| Reo/Breeder/JNSH/150826 | Jinan | MK189481 |
| Reo/Broiler/YTPL/150516 | Yantai | MK189463 |
| Reo/Broiler/DZPY/151016a | Dezhou | MK189479 |
| Reo/Broiler/DZPY/151016b | Dezhou | MK189478 |
| Reo/Broiler/BZHM/151026a | Binzhou | MK189477 |
| Reo/Broiler/BZHM/151026b | Binzhou | MK189476 |
| Reo/Broiler/YTLY/161020a | Yaitai | MK189470 |
| Reo/Broiler/YTLY/161020b | Yaitai | MK189469 |
| Reo/Broiler/YTLY/161024a | Yaitai | MK189468 |
| Reo/Broiler/YTLY/161024b | Yaitai | MK189469 |
| Reo/Broiler/LCYG/160916 | Liaocheng | MK189466 |
| Reo/Broiler/YTLY/161021 | Yaitai | MK189465 |
| Reo/Broiler/WFZH/160416 | Weifang | MK189472 |
| Reo/Breeder/YTHY/160422 | Yaitai | MK189480 |
| Reo/Broiler/HZJY/170308 | Heze | MK189474 |
| Reo/Breeder/YTHY/170108 | Yaitai | MK189471 |
All ARV field samples were collected from boilers which parent breeders were conventional immunization (s1133, 1733, 2177, or 2311 strains).
Abbreviation: ARV, avian orthoreovirus.
Results
Virus Isolation and Propagation
Twenty-one ARV field strains were isolated. The virus fluids were generated from chicken embryos and LMH monolayer cells. Giant or “bloom-like” cytopathic effects were observed in ARV-inoculated LMH cell cultures. All 21 ARV isolates were confirmed as ARV-positive using one-step real-time RT-PCR. There were no other avian viruses in ARV-positive viral stocks acquired from fresh synovial fluid and tendon samples.
Sequence Analyses
Sequence analyses of field isolates revealed that nucleotide identities of the σC gene ranged between 52.6 and 100% between the isolates themselves and 52.5 to 99.8% between the isolates and reference strains. The identities of nucleotide sequences were below 76.7% between 13 field isolates and reference strains. Similar nucleotide sequence identities were not found within the σC genes for specific regions and dates.
Genetic Diversity of ARV Isolates
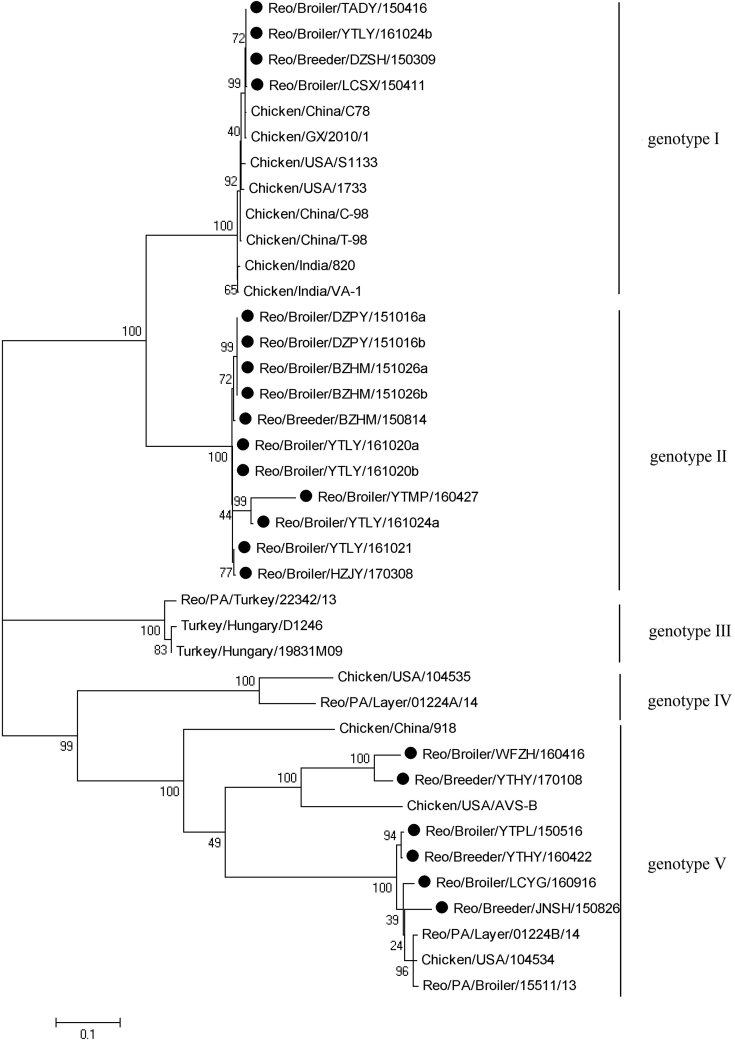
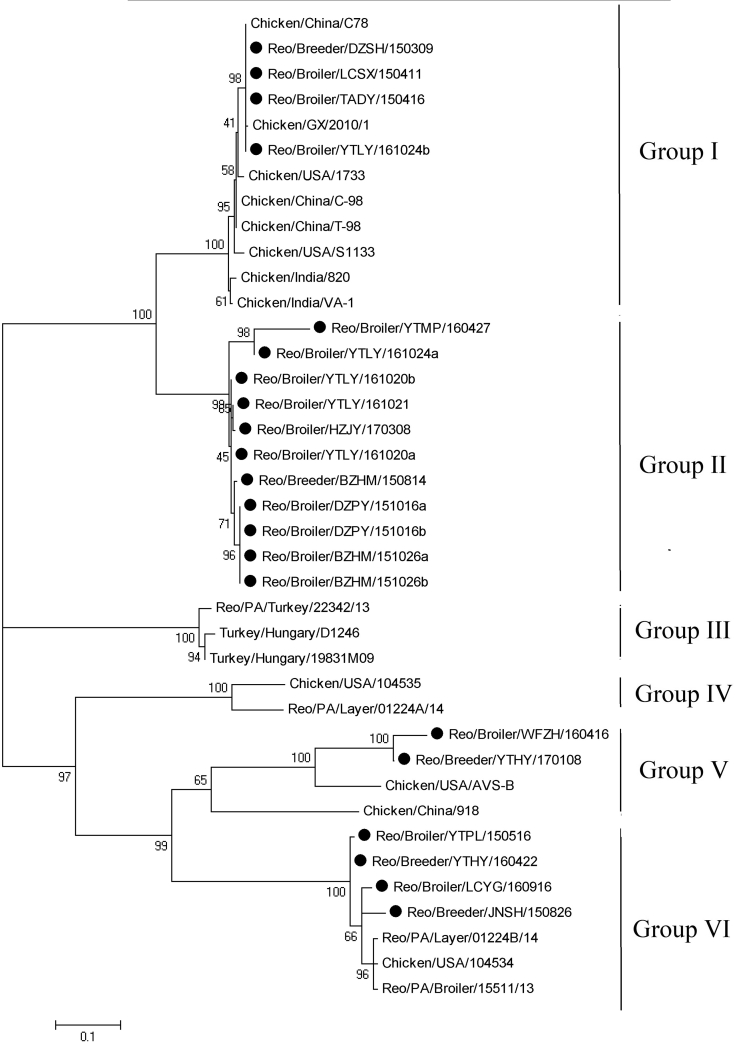
Phylogenetic analysis of the σC gene demonstrated that ARV strains could be divided into 5 genotypes (Figure 2). Four isolates were clustered together with ARV strains S1133, 1733, C78, T-98, GX2010, 820, and VA-1 in genotype I. Eleven field isolates were clustered into a distinct genotype that shared a higher rate of similarities with ARV strains in cluster I. The remaining 6 field isolates were clustered into the same genotype, which contained ARV isolates PA01224b, PA15511, PA05682, and AVS-B. The phylogenetic tree of σC proteins was constructed among those ARV strains, revealing that all ARV isolates were classified into 6 groups (Figure 3). Groups I to IV were similar to those of the phylogenetic tree of the σC genes. However, ARV isolates WFZH160416 and YTHY170106 were clustered into group V and shared high sequence similarities with AVS-B and 918. YTBY150516, YTHY160422, LCYG160916, and JNSH150826 were clustered into group VI, which shared low sequence similarities with other ARV isolates. Field isolates from different geographic regions may be clustered into the same group and isolates from adjacent regions may be clustered into different groups. No obvious relationship was seen between ARV isolate groups and the original regions from which they were acquired.
Figure 2.
Phylogenetic relationship between ARV field isolates(•) and ARV reference strains based on nucleotides sequences of σC gene. Abbreviation: ARV, avian orthoreovirus.
Figure 3.
Phylogenetic relationship between ARV field isolates(•) and ARV reference strains based on amino acid sequences of σC protein. Abbreviation: ARV, avian orthoreovirus.
Comparison of Amino Acid Sequences of σC Protein Among ARV
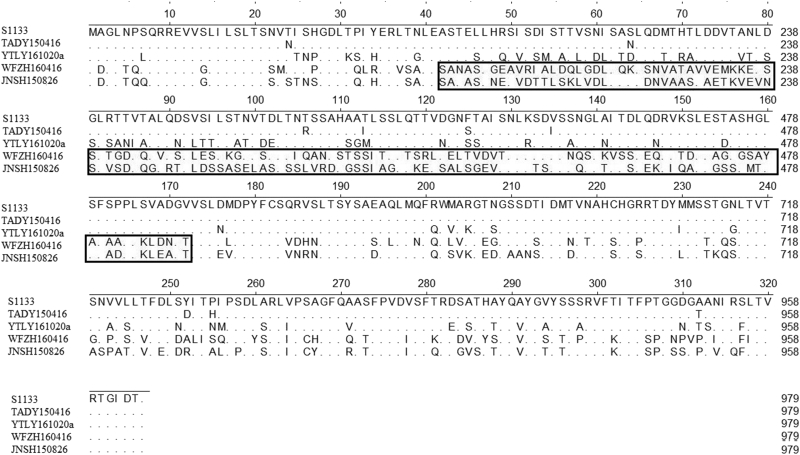
Because all field isolates were clustered into 4 groups based on the phylogenetic tree of σC proteins, 4 representative isolates were selected for comparison of σC aa sequences with vaccine strains S1133 (Figure 4). Alignment of the 5 avian reovirus σC sequences revealed that the amino terminal residues 1–41 (23 of 41 residues identified) and 174-326 (82 of 153 residues identified) showed a high degree of conservation, whereas the residues 42-173 had fewer identical residues (19 of 132 residues identified). Further, secondary structures of σC protein were predicted using the online software SWISS-MODEL. Amino acids 155–159 link the α-helical coiled-coil to the triple β-spiral. Compared with S1133, TADY150416, and YTLY161020a, a zinc ion–containing link was deleted in the hinge region of σC protein of JNSH160826 and WFZH160416.
Figure 4.
A multiple amino acid sequence alignment of ARV S1133 strain and 4 field isolates. Abbreviation: ARV, avian orthoreovirus.
Discussion
Avian orthoreovirus is ubiquitous among domestic poultry flocks worldwide and causes a range of symptoms, including tenosynovitis/arthritis syndrome, malabsorption syndrome, runting-stunting syndrome, and immunosuppression in broiler and layer flocks (Jones, 2000). Given the increased observation of teno-synovitis/arthritis syndrome in broiler flocks in China in recent years, the present study initially obtained 64 joint samples from diseased broilers, of which 37 samples were found to be ARV positive using one-step real-time RT-PCR. Phylogenetic analysis of σC revealed the separation of the 37 ARV isolates into 5 different clusters. Finally, 21 ARV isolates were acquired for epidemiologic analysis. Most ARV field isolates (17/21) were classified within genotypes other than genotype I, including the commercial vaccine viruses S1133, 1733. The 6 ARV field isolates in genotype V shared the lowest nucleotide sequence similarity (53.0 to 55.2% nt identities) with the ARV vaccine strains. The 11 field isolates clustered in genotype II shared 70.9 to 76.0% nt identities with the ARV vaccine strains. The aa similarity of σC protein among these ARV isolates is similar to that of the nucleotide sequence analysis. Based on the analysis of nt and aa sequences of all field isolates, results of the present study indicate that the current commercial ARV vaccines are not clustered in the predominant genotypes in North China and were not able to provide sufficient cross-protection for poultry against the prevalent field strains. Avian orthoreovirus adhesion fiber is formed by the protein σC, which elicits ARV-specific neutralizing antibodies and is responsible for extracellular host cell attachment.
In our study, 21 ARV isolates were finally acquired for epidemiological analysis. σC phylogenetic analysis display the separation of 39 ARV isolates into 5 different clusters. Most ARV field isolates (17/21) could be classified in other genotypes other than genotype I, which includes the commercial vaccine viruses S1133, 1733. The 6 ARV field isolates in genotype V shared lowest nucleotide sequence similarity (53.0 to 55.2% identity) with the ARV vaccine strains. The 11 field isolates clustering in genotype II shared 70.9 to 76.0% nt identities with the ARV vaccine strains. The aa similarity of σC protein among these ARV isolates is similar to the nucleotide sequence analysis. Based on the nt and aa sequences analysis of all field isolates, it indicated that the current commercial ARV vaccines are not clustered in the predominant genotypes in North China and could not provide sufficient cross-protection for poultry against the prevalent field strains.
Several studies have shown that emerging and novel ARV variants have become prevalent in different regions of the world, especially North America, during the past 5 y. The nt BLAST of 4 field isolates (YTPY150516, YTHY160422, LCYG160916, and JNSH150826) revealed that these ARV variants share high nt identities (>94%) with North America field isolates (except for CL013184 from Chile), including those that have become more prevalent in the US and Canada over the last few years. These isolates were first isolated in China as emerging ARV strains or variants. Two ARV field isolates (WFZH160416 and YTHY1701018) shared high nt similarities with the 4 isolates and also shared low nt similarities with vaccine strains. All 6 poultry flocks sampled in the present study were scattered in 4 geographically separated cities (Liaocheng, Jinan, Weifang, and Yantai) in Shandong Province. The presence of ARV variants found both east and west of Shandong indicates that the ARV variants may be a potential threat to the poultry industry.
Different from the above 6 ARV isolates, 11 ARV field isolates identified in this study were on the same main branch (not the same secondary branch) as the vaccine strains based on the nt/aa phylogenetic tree of σC. The nt BLAST of the 11 ARV isolates revealed that they share high nt identities (>95%) with strains identified in Hungary, France, and Brazil. Initially, 4599-V-04A was isolated from broiler flocks in Hungary (2004), and field isolates were also acquired from flocks in France and Brazil during the past 5 y (Troxler et al., 2013, Farkas et al., 2016). However, field strains in genotype II shared high similarity with LN160607-1, which was initially reported in China (Zhang, et al., 2019). In the present study, the ARV isolates identified in broiler flocks from Shandong Province were of the dominant genotypes. Comparison of aa sequences revealed multiple single or double aa mutations between YTLY161020a and S1133. Previous studies reported that genotype I (including vaccine strains) was prevalent in North China during the past few years (Wen et al., 2016, Zhong et al., 2016). It is possible that long-term application of ARV vaccines could induce aa mutations in field strains, resulting in an antigenic shift under immune-stress.
Based on comparison of aa sequences of σC protein, the 6 antigenic peptides (170–179aa, 218–232aa, 235–239aa, 257–265aa, 267–281aa, and 289–295aa) predicted from 5 ARV strains demonstrated multiple mutations between the different cluster groups and the S1133 strain, which was similar to those reported in a previous study. Neutralizing antibody elicited by ARV vaccine 2,177 was not able to cross-neutralize the currently reported isolates in other clusters. However, in the present study, 17 ARV field isolates may have been able to offset the protection of commercial ARV vaccines (S1133, 1733) in broilers. Since 2014, outbreaks of ARV-associated disease have become more frequent in Shandong and adjacent regions. The emerging ARV variants appear to be major agents in increasing the prevalence of the disease in China and possibly other geographic regions as well.
In conclusion, analysis of synovial fluid and tendon samples from vaccinated poultry flocks in China identified 21 ARV field strains, of which only 4 were genotype Group I. Seventeen field isolates were clustered in genotypes other than genotype Group I, distinct from vaccine stain, and might evade neutralizing antibody elicited by commercial vaccine strains. Results of the present study suggest that full length genome sequencing and bioinformatics analysis should be carried out to explore the evolution and genetic diversity of ARV. The development of efficient, effective ARV vaccines is urgently needed to prevent and control the disease in domestic flocks.
Nucleotide sequence accession numbers
Sequence data have been deposited in the GenBank database under accession number MK189463- MK189483.
Acknowledgments
The authors acknowledge TopEdit LLC for linguistic editing and proofreading during the preparation of this manuscript.
This study was supported by National Key Research and Development Program (2018YFD0500106-3), the Taishan industry leader talent project of Shandong Province (LJNY201610), the Natural Science Foundation of China (31702233), and the Funds of Shandong "Double Top" Program.
Conflict of interest: None.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi.org/10.1016/j.psj.2019.11.067.
Contributor Information
Hao Chen, Email: zqqch401@163.com.
Yi Tang, Email: tyck288@163.com.
Youxiang Diao, Email: yxdiao@126.com.
Supplementary data
References
- Benavente J., Martinez-Costas J. Avian reovirus: structure and biology. Virus Research. 2007;123:105–119. doi: 10.1016/j.virusres.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Dandar E., Balint A., Kecskemeti S., Szentpali-Gavaller K., Kisfali P., Melegh B., Farkas S.L., Banyai K. Detection and characterization of a divergent avian reovirus strain from a broiler chicken with central nervous system disease. Arch. Virology. 2013;158:2583–2588. doi: 10.1007/s00705-013-1739-y. [DOI] [PubMed] [Google Scholar]
- Day J.M. The diversity of the orthoreoviruses: molecular taxonomy and phylogentic divides. Infect. Genetics Evolution : Journal Molecular Epidemiology Evolutionary Genetics Infectious Diseases. 2009;9:390–400. doi: 10.1016/j.meegid.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Farkas S.L., Marton S., Dandar E., Kugler R., Gal B., Jakab F., Balint A., Kecskemeti S., Banyai K. Lineage diversification, homo- and heterologous reassortment and recombination shape the evolution of chicken orthoreoviruses. Scientific Reports. 2016;6:36960. doi: 10.1038/srep36960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas S.L., Varga-Kugler R., Marton S., Lengyel G., Palya V., Banyai K. Genomic sequence and phylogenetic analyses of two novel orthoreovirus strains isolated from Pekin ducks in 2014 in Germany. Virus Research. 2018;257:57–62. doi: 10.1016/j.virusres.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Goldenberg D., Pasmanik-Chor M., Pirak M., Kass N., Lublin A., Yeheskel A., Heller D., Pitcovski J. Genetic and antigenic characterization of sigma C protein from avian reovirus. Avian Pathology : Journal W.V.P.A. 2010;39:189–199. doi: 10.1080/03079457.2010.480969. [DOI] [PubMed] [Google Scholar]
- Hoerr F.J. Clinical aspects of immunosuppression in poultry. Avian Diseases. 2010;54:2–15. doi: 10.1637/8909-043009-Review.1. [DOI] [PubMed] [Google Scholar]
- Huang W.R., Wang Y.C., Chi P.I., Wang L., Wang C.Y., Lin C.H., Liu H.J. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and Src signaling pathways as well as microtubules and small GTPase Rab5 protein. J. Biological Chemistry. 2011;286:30780–30794. doi: 10.1074/jbc.M111.257154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.C. Avian reovirus infections. Rev. Sci. Tech. 2000;19:614–625. doi: 10.20506/rst.19.2.1237. [DOI] [PubMed] [Google Scholar]
- Kant A., Balk F., Born L., van Roozelaar D., Heijmans J., Gielkens A., ter Huurne A. Classification of Dutch and German avian reoviruses by sequencing the sigma C protein. Vet. Research. 2003;34:203–212. doi: 10.1051/vetres:2002067. [DOI] [PubMed] [Google Scholar]
- Lockaby S.B., Hoerr F.J., Lauerman L.H., Kleven S.H. Pathogenicity of Mycoplasma synoviae in broiler chickens. Vet. Pathology. 1998;35:178–190. doi: 10.1177/030098589803500303. [DOI] [PubMed] [Google Scholar]
- Lu H., Tang Y., Dunn P.A., Wallner-Pendleton E.A., Lin L., Knoll E.A. Isolation and molecular characterization of newly emerging avian reovirus variants and novel strains in Pennsylvania, USA, 2011-2014. Scientific Reports. 2015;5:14727. doi: 10.1038/srep14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J.Y., Lee D.H., Lim T.H., Lee J.H., Day J.M., Song C.S. Isolation and genomic characterization of a novel avian orthoreovirus strain in Korea, 2014. Arch. Virology. 2018;163:1307–1316. doi: 10.1007/s00705-017-3667-8. [DOI] [PubMed] [Google Scholar]
- Shivaprasad H.L., Franca M., Woolcock P.R., Nordhausen R., Day J.M., Pantin-Jackwood M. Myocarditis associated with reovirus in Turkey poults. Avian Diseases. 2009;53:523–532. doi: 10.1637/8916-050309-Reg.1. [DOI] [PubMed] [Google Scholar]
- Spandidos D.A., Graham A.F. Physical and chemical characterization of an avian reovirus. J. Virology. 1976;19:968–976. doi: 10.1128/jvi.19.3.968-976.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biology Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Lin L., Sebastian A., Lu H. Detection and characterization of two co-infection variant strains of avian orthoreovirus (ARV) in young layer chickens using next-generation sequencing (NGS) Scientific Reports. 2016;6:24519. doi: 10.1038/srep24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Lu H. Whole genome alignment based one-step real-time RT-PCR for universal detection of avian orthoreoviruses of chicken, pheasant and Turkey origins. Infect. Genetics Evolution : Journal Molecular Epidemiology Evolutionary Genetics Infectious Diseases. 2016;39:120–126. doi: 10.1016/j.meegid.2016.01.018. [DOI] [PubMed] [Google Scholar]
- Thorp B.H., Whitehead C.C., Dick L., Bradbury J.M., Jones R.C., Wood A. Proximal femoral degeneration in growing broiler fowl. Avian pathology. journal W.V.P.A. 1993;22:325–342. doi: 10.1080/03079459308418924. [DOI] [PubMed] [Google Scholar]
- Troxler S., Rigomier P., Bilic I., Liebhart D., Prokofieva I., Robineau B., Hess M. Identification of a new reovirus causing substantial losses in broiler production in France, despite routine vaccination of breeders. Vet. Record. 2013;172:556. doi: 10.1136/vr.101262. [DOI] [PubMed] [Google Scholar]
- Van de Zande S., Kuhn E.M. Central nervous system signs in chickens caused by a new avian reovirus strain: a pathogenesis study. Vet. Microbiology. 2007;120:42–49. doi: 10.1016/j.vetmic.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Wen C., Zhong Q., Zhang J.D., Lu J.S., Zhang L.X., Yuan X.M., Gan M.H., Cai X.P., Zhang G.Z. Sequence and phylogenetic analysis of chicken reoviruses in China. J. Integr. Agric. 2016;15:1846–1855. [Google Scholar]
- Yun T., Ye W., Ni Z., Chen L., Yu B., Hua J., Zhang Y., Zhang C. Complete genomic sequence of goose-origin reovirus from China. J. Virology. 2012;86:10257. doi: 10.1128/JVI.01692-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lei X., Ma L., Wu J., Bao E. Genetic and pathogenic characteristics of newly emerging avian reovirus from infected chickens with clinical arthritis in China. Poult. Science. 2019;98:5321–5329. doi: 10.3382/ps/pez319. [DOI] [PubMed] [Google Scholar]
- Zhong L., Gao L., Liu Y., Li K., Wang M., Qi X., Gao Y., Wang X. Genetic and pathogenic characterisation of 11 avian reovirus isolates from northern China suggests continued evolution of virulence. Scientific Reports. 2016;6:35271. doi: 10.1038/srep35271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.