Abstract
The genetic relatedness and antimicrobial susceptibility profiles of Salmonella isolated from poultry and their environment were determined. One broiler breeder flock (BBF1) and 2 broiler flocks (BF1 and BF2) were reared over a 1.75-year period on the same poultry research farm. Hatching eggs were obtained from BBF1 to produce BF1 chicks, while BF2 chicks were progeny of a separate, unsampled broiler breeder flock. BF1 and BF2 were reared in the same housing facilities but 6 mo apart. Salmonella isolates were collected via litter sock sampling (BF1), cecal excision (BF1 and BF2), or cloacal swabs (BBF1). Serotyping identified Salmonella enterica subsp. enterica serovar Altona (SA) in BBF1 and S. enterica subsp. enterica serovar Senftenberg (SS) in BF1 and BF2. Genotypic fingerprinting was achieved with Rep-PCR using the (GTG)5 primer and revealed sequence homology among Senftenberg isolates from BF1 and BF2. For each isolate, the minimum inhibitory concentration was determined for 27 antimicrobial agents using Sensititre plates with formularies specific to antimicrobials used in poultry production or those used to control gram negative pathogens. Isolates from the 3 flocks were resistant to clindamycin, erythromycin, novobiocin, penicillin, and tylosin tartrate and demonstrated intermediate resistance to azithromycin, florfenicol, and spectinomycin. These data demonstrated that serovar Altona and Senftenberg were harbored by poultry, the latter appeared to persist in broiler flocks, and both serotypes shared similar patterns of antimicrobial susceptibility in an integrated research operation. In the case of multiple Salmonella isolates, combining genotypic fingerprinting methods with serotyping of representative isolates would reduce the number of samples required for serotyping and more clearly identify relatedness of isolates. These methods facilitate effective surveillance in poultry production systems, thus allowing for implementation of precise Salmonella control measures.
Key words: Salmonella, antimicrobial resistance, broiler, microbiology, genetic characterization
Introduction
Salmonella prevalence among live poultry and their housing environment is a concern due to zoonosis and food safety risks (Scallan et al., 2011). The pathogen has become difficult to eradicate because of its resistance to antimicrobials and environmental persistence (Kalily et al., 2017, Liljebjelke et al., 2017). Antimicrobials have not typically been used to treat Salmonella-positive poultry because their use has been reduced globally and they are generally not efficacious for this application (United States Department of AgricultureFood Safety Inspection Service, 2009, Prescott, 2019). Nevertheless, Salmonella harbored by poultry possess antimicrobial resistance genes (Zhu et al., 2017) that can be transferred to other bacteria through horizontal gene transfer (von Wintersdorff et al., 2016). These resistant strains can then persist in contaminated meat products intended for human consumption (Chuanchuen and Padungtod, 2009, Abd-Elghany et al., 2015, Antunes et al., 2016). Thus, reduction and eradication of Salmonella from live poultry and associated products continue to be a top priority for producers and regulatory agencies alike.
Salmonella are spread by horizontal and vertical transmission and frequently detected in integrated poultry production systems (Liljebjelke et al., 2005, Kim et al., 2007). Thus, attempts to reduce prevalence and eradicate the pathogen must be systematic. Thorough and efficient surveillance is a critical aspect of any Salmonella control program, and rapid diagnostic tools combined with molecular typing methods facilitate this process (Stepan et al., 2011). This study aimed to improve the efficiency of surveillance methods by using genotypic fingerprinting to identify the relatedness of isolates. The PCR technique was more time- and cost-effective than pulse field gel electrophoresis and decreased the number of samples that required serotyping. The present study characterized the relatedness and antimicrobial susceptibility of Salmonella isolated from poultry over a 1.75-year period. The research-scale farm mirrored commercial conditions, allowing for surveillance of broiler breeder, hatchery, broiler, and feed milling facilities on the same premises.
Materials and methods
Broiler and Broiler Breeder Husbandry
The animal trials were conducted in accordance with the principles and specific guidelines of the Guide for the Care and Use of Agricultural Animals in Research and Teaching (FASS, 2010) and approved by the North Carolina State University Institutional Animal Care and Use Committee. Broiler flocks 1 (BF1) and 2 (BF2) were reared in a curtain-sided, fan-ventilated, 96 litter floor pen house 6 mo apart with routine cleaning and disinfection between the flocks. This consisted of pressure washing removable interior components, allowing sufficient UV exposure and drying, replacing used litter with fresh pine shavings, and treating the entire interior with a pyrethrin-based fogging insecticide. Two broiler breeder flocks (Ross 708 hens by Yield Plus Male roosters) housed on the same premises produced hatching eggs for the broiler flocks. Both were reared sex-separately in a black out, fan-ventilated housing until 21 wk of age at which point photostimulation, intermixing, and transfer to 2/3 slat and 1/3 litter floor laying facilities occurred. BF1 were progeny broilers of the sampled broiler breeder flock 1 (BBF1), while the parent flock of BF2 was not sampled. These flocks were reared for the independent research studies referenced in the following sections and were not intentionally exposed to or challenged with Salmonella at any point.
Sample Collection and Salmonella Serotyping
The following samples were collected from BF1 as described by Walker et al. (2018): feed samples from each experimental diet at the feed mill, chick paper, and eggshells in the hatchery at day of hatch, a pre-enriched sock applied to litter floor pens at 15 D, and individual ceca at 44 D. Samples were suspended in 1% buffered peptone water (BPW) and maintained on ice until further enrichment and incubation occurred. This entailed pulverizing samples with a rubber mallet, adding additional BPW so that a 1-part sample, 9 parts BPW solution was achieved, and then mechanically homogenizing the samples for 1 min. The enriched samples were then incubated for 24 h at 37°C before detection with methods described in the following sections. One month later, the parent BBF1 flock was sampled by collecting cloacal swabs of all roosters and hens in the flock. The 16-pen laying facilities housed 8 roosters and 60 hens per pen. Swabs were collected from each bird and pooled into groups of 4 and 15 for roosters and hens, respectively, resulting in n = 6 samples per pen. The swabs were suspended in BPW, maintained on ice before enrichment with additional BPW and incubated as described previously before Salmonella detection. Ceca of BF2 were sampled at 19 and 48 D of age as described by Caraway et al. (2019) and processed as described previously. After initial enrichment for all samples, Salmonella spp. identification was achieved with an enzyme-linked fluorescence assay automated instrument (VIDAS 30 Multi-parametric Immunoassay Instrument, BioMérieux, Inc., Marcy-l’Étoile, France) and confirmed by culture with Rapid Salmonella Agar (Bio-Rad #3563961; Hercules, CA) and XLT-4 agar (Oxoid Product #CM1061) as described by Walker et al. (2018). Serotyping of isolates was conducted by the United States Department of Agriculture National Veterinary Services Laboratories (Ames, IA).
PCR Genotypic Characterization
Colony PCR of the isolates was conducting using the (GTG)5 fingerprinting technique initially described by Versalovic et al. (1994) with some modifications. A single colony from an overnight trypticase soy agar culture was suspended in 100 μL of molecular-grade water, which served as the template suspension. PCR reaction mixtures were prepared by combining 12.5-μl master mix (Qiagen #201443), primer (GTG GTG GTG GTG GTG) at 0.8-μM final concentration, 1.5 μL of the template suspension and molecular-grade water to achieve a final volume of 25 μL. A 2-log DNA ladder (New England Biolabs #N3200S) was used for visualization and band normalization in the downstream analysis. A Bio-Rad (Hercules, CA) thermocycler was used for the PCR reaction with the following parameters: a single initial denaturation at 95°C for 4 min followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 1 min, and extension at 65°C for 1 min. A single final extension at 65°C for 10 min completed the PCR reaction. PCR products were size separated in a 1.5% agarose gel with incorporated ethidium bromide in tris/borate/EDTA (TBE) buffer and visualized with a gel imager (Bio-Rad #170-8,195).
Antimicrobial Susceptibility Phenotyping
Broth microdilution methods were used to determine the minimum inhibitory concentration (MIC) of 27 antimicrobial agents using Sensititre plates according to the manufacturer's protocol (TREK Diagnostic Systems; Oakwood Village, OH). For each isolate, the MICs were determined for defined drug groups based on unique 96-well plate formularies. Specifically, plates containing variable concentrations of antimicrobials used in poultry production (Thermo Fisher #AVIAN1F plate) as well as antimicrobials used against gram negative pathogens monitored by the National Antimicrobial Resistance Monitoring System (Thermo Fisher #CMV3AGNF plate) were used. Isolates were designated as resistant, intermediately resistant, or susceptible based on available breakpoint data published by the Clinical Lab Standards Institute (2010) and National Antimicrobial Resistance Monitoring System (CDC, 2014).
Genotypic Analysis
Genotypic relatedness among isolates was determined using PCR band patterns generated by the (GTG)5 protocol. Bionumerics software version 7.5 (Applied Maths, Sint-Martens-Latem, Belgium) was used for band analysis. Bands were normalized with interspersed lanes of DNA ladder before generation of a dendrogram. Similarity coefficients were band-based and determined with optimization and tolerance of 1.5% each. The unweighted-pair group method using average linkages method with arithmetic mean was used for cluster analysis. A threshold of 95% similarity was adapted from a study comparing Escherichia coli that had been genotyped with a similar PCR method (Bonacorsi et al., 2009) and was used to determine the clonal relationship among isolates.
Results and discussion
Salmonella isolates were obtained from live birds and their environment on a vertically integrated research farm over a 1.75-year period. Two Salmonella serovars, Salmonella enterica subsp. enterica serovar Altona (SA), and S. enterica subsp. enterica serovar Senfentenberg (SS) occurred naturally and were isolated from one broiler breeder flock and 2 broiler flocks, respectively. Salmonella were isolated from litter, ceca, and cloacal swab samples and were prevalent in 19, 9, and 15% of samples collected from BBF1, BF1, and BF2, respectively (Table 1). Isolates from BF1 and BBF1 (n = 2) and BF2 (n = 4) were further characterized to determine their genotypic relatedness.
Table 1.
Summary of samples collected for Salmonella testing, Salmonella prevalence, and serotyping results from 3 flocks included in this study: a broiler breeder flock (BBF1), broiler flock 1 (BF1), and broiler flock 2 (BF2).
| Flock | Samples collected |
Salmonella prevalence1 |
Serovar detected2 | Reference |
|---|---|---|---|---|
| Number (%) | ||||
| BBF1 | Cloacal Swabs3 | 18/96 (19) | Altona | This study |
| BF1 | Litter, Ceca4 | 18/192 (9) | Senftenberg | Walker et al., 2018 |
| BF2 | Ceca5 | 26/170 (15) | Senftenberg | Caraway et al., 2019 |
Positive samples were determined by an automated enzyme-linked fluorescence assay instrument (VIDAS 30 Multi-parametric Immunoassay Instrument, BioMérieux, Inc., Marcy-l’Étoile, France) and confirmed with culture methods.
Serotyping was conducted by the United States Department of Agriculture National Veterinary Services Laboratories (Ames, IA).
The BBF1 housing facility contained 16 pens of 8 roosters and 60 hens each. Pools of 4 and 15 swabs were collected from males and females, respectively, from each pen at a flock age of 63 wk.
Litter was sampled at 15 D, and ceca were sampled from broilers of each treatment at flock ages of 44 and 55 D.
Ceca were sampled from broilers of each treatment at flock ages of 19 and 48 D.
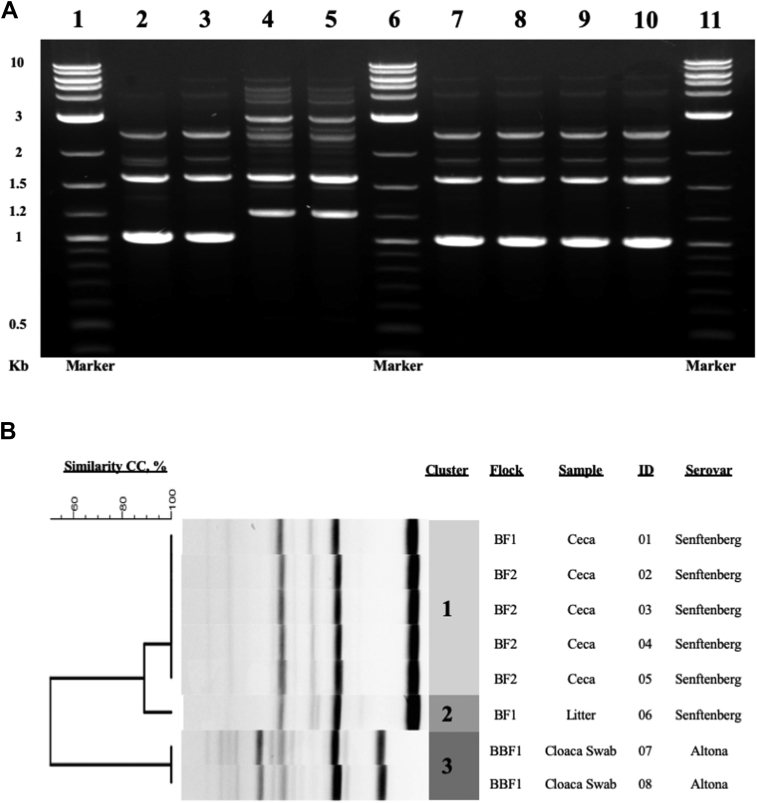
SS was isolated from bird ceca and litter in BF1. This 96-floor pen house was then disinfected, and the litter changed before placement of a second broiler flock (BF2) 6 mo later. SS appeared to persist in the housing environment and was isolated from the ceca of birds in BF2. Positive samples were isolated from birds housed in the same 3 specific pens in both BF1 and BF2. Genotyping confirmed isolate clonality (Figure 1). While SS could have been reintroduced to BF2 by environmental vectors or external fomites, isolation of Salmonella from birds housed only in the same locations in the house supports persistence. SS has demonstrated desiccation resistance (Pedersen et al., 2008) and thermotolerance (Nguyen et al., 2017). Thus, the ability of this serovar to survive amidst harsh environmental conditions could also support its persistence between the broiler flocks described in this study.
Figure 1.
Genotyping of Salmonella isolates from broiler flock 1 (BF1), broiler flock 2 (BF2), and broiler breeder flock 1 (BBF1). (A) Agarose gel electrophoresis of (GTG)5 PCR products. Lane 1: Marker; lane 2: BF1 litter isolate; lane 3: BF1 cecal isolate; lane 4-5: BBF1 cloacal swab isolates; lane 6: Marker; lane 7-10: BF2 cecal isolates; lane 11: Marker. (B) Dendrogram with similarity coefficients (CC) determined from band analysis of (GTG)5 PCR products.
The (GTG)5 Rep-PCR coupled with banding pattern analysis was a reliable determinant of genotypic relationships among isolates. Three clusters were generated from 8 isolates representing 2 distinct Salmonella serovars (Figure 1B). Cluster 1 consisted of all cecal S. serovar Senftenberg isolates from BF1 and BF2 (Salmonella ID 01-05). These shared 100% similar fingerprint profiles, confirming isolate clonality. The litter SS isolate (Salmonella ID 06) belonged to cluster 2 and shared 90% similarity with cluster 1 isolates. The similarity difference was due to one additional band in the fingerprint profile. SA (Salmonella ID 07-08) isolated from cloacal swabs from BBF1 shared 100% genotypic similarity (cluster 3) and had a different fingerprint profile than SS. The observed Salmonella serotype-specific patterns were also reported in a previous study (Rasschaert et al., 2005). These methods allowed for detailed comparison of Salmonella genotypes beyond what could be inferred from serotyping alone. The same methodology could be applied to larger integrated operations to determine sources of infection and transmission of Salmonella between multiple facilities.
We initially suspected vertical transmission of Salmonella upon isolating the bacteria from the parent breeder flock BBF1 shortly after detecting it in BF1 progeny broilers. A different serovar, SA, was verified by serotyping and confirmed to have a genotype that was distinct from SS by PCR fingerprinting (Figure 1B). In addition, SA was not detected in hatch residue or among progeny BF1 at any point. Contrary to our initial hypothesis, these data did not support vertical transmission of this Salmonella serovar. There have been reports of SA in broiler production (Marin and Lainez, 2009, Marin et al., 2011). The serovar was also linked to a hatchery during a multistate outbreak (Forshey et al., 2012) and has also been detected in table eggs (Martelli and Davies, 2012). Vertical transmission of different Salmonella serovars has been extensively reported among integrated poultry production systems and was not serovar-specific (Humphrey and Lanning, 1988, Berchieri et al., 2001, Liljebjelke et al., 2005, Oh et al., 2010, Martelli and Davies, 2012). In the present study, broilers and broiler breeders on the same premises harbored 2 different Salmonella serovars, SS and SA, respectively. The former was able to persist in broiler housing and infect broilers reared 6 mo later. While isolate serotyping provided useful insight, genotypic fingerprinting was more distinct and could more accurately confirm or refute horizontal transmission even in the case of isolation of identical serotypes.
Antimicrobial susceptibility of the isolates was determined, as the discovery of antimicrobial resistance among Salmonella harbored by food animals has serious implications for human health. Antimicrobial susceptibility phenotypes were identical among all isolates. Each was resistant to clindamycin, erythromycin, novobiocin, penicillin, and tylosin tartrate. This was expected as Salmonella are intrinsically resistant to these drugs (St. Amand et al., 2013). In addition, all isolates exhibited intermediate resistance to azithromycin (MIC = 4 μg/mL), spectinomycin (MIC = 32 μg/mL), and florfenicol (MIC = 4 μg/mL). Intermediate resistance to these drugs is noteworthy because of their use in poultry production (Hofacre et al., 2013) and in treating human salmonellosis (Sjölund-Karlsson et al., 2011). Multidrug-resistant Salmonella isolated from poultry have exhibited florfenicol resistance (Meunier et al., 2003), and these could serve as a reservoir for resistance genes in poultry production systems. Intermediate resistance indicates a drug has uncertain therapeutic effects because of its pharmacokinetic properties (Rodloff et al., 2008). As such, Salmonella that are intermediately resistant to these drugs may have significant treatment implications for humans.
Salmonella can be isolated at all stages of poultry production, posing a food safety risk. Serovars isolated in this study were responsible for human outbreaks linked to live poultry (SA; Forshey et al., 2012) and poultry food products (SS; L'Ecuyer et al., 1996). The emergence of strains resistant to antimicrobials further exacerbates the human health threat. In integrated poultry production systems, there remains a need for diagnostic tests that provide a rapid, effective, and cost-efficient means for surveillance of this foodborne pathogen. The described methodologies met these criteria for this research model production system. The (GTG)5 Rep-PCR dramatically decreased the number of Salmonella isolates to be serotyped and was not as laborious or costly as other genotyping methods, for example, pulsed field gel electrophoresis and whole genome sequencing. Antimicrobial susceptibility phenotyping using Sensititre plates allowed for simultaneous quantification of resistance to 27 drugs. Together, these methods could be used in large production systems, poultry diagnostic laboratories, and/or federal and state agencies to generate tailored Salmonella control programs. Thus, efficient surveillance with the approach described here may allow for targeted management practices that will contribute to successful reduction and elimination of the pathogen and increase the overall safety of poultry products.
Acknowledgments
The authors would like to thank Ryan Patterson and his staff who supported the research activities that made this study possible. Special thanks are given to Christina Sigmon who provided microbiology guidance during collection and preservation of the isolates. Recognition is given to Dr. Shivaramu Keelara Veerappa who assisted with data analysis. Utmost gratitude is given to Dr. John Brake, who unfortunately was unable to witness the completion of this work but whose mentorship is not forgotten.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
References
- Abd-Elghany S.M., Sallam K.I., Abd-Elkhalek A., Tamura T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 2015;143:997–1003. doi: 10.1017/S0950268814001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes P., Mourão J., Campos J., Peixe L. Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect. 2016;22:110–121. doi: 10.1016/j.cmi.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Berchieri A., Murphy C., Marston K., Barrow P. Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens: effect of bacterial and host genetic background. Avian Pathol. 2001;30:221–231. doi: 10.1080/03079450120054631. [DOI] [PubMed] [Google Scholar]
- Bonacorsi S., Bidet P., Mahjoub F., Mariani-Kurkdjian P., Ait-Ifrane S., Courroux C., Bingen E. Semi-automated rep-PCR for rapid differentiation of major clonal groups of Escherichia coli meningitis strains. Int. J. Med. Microbiol. 2009;299:402–409. doi: 10.1016/j.ijmm.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Caraway C.T., Walker G.K., Brake J. The effects of coarse corn and refined functional carbohydrates on the live performance and cecal Salmonella prevalence in coccidiosis-vaccinated broilers. Poult. Sci. 2019;98:4565–4574. doi: 10.3382/ps/pez302. [DOI] [PubMed] [Google Scholar]
- Chuanchuen R., Padungtod P. Antimicrobial resistance genes in Salmonella enterica isolates from poultry and swine in Thailand. J. Vet. Med. Sci. 2009;71:1349–1355. doi: 10.1292/jvms.001349. [DOI] [PubMed] [Google Scholar]
- Clinical Lab Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, PA: 2010. Performance standards for antimicrobial susceptibility testing; Twentieth informational supplement. [Google Scholar]
- FASS . 3rd ed. FASS, Inc.; Champaign, IL: 2010. Guide for the Care and Use of Agricultural Animals in Research and Teaching. [Google Scholar]
- Forshey T.M., Byrum B.A., Machesky K.D., Roney C.S., Gomez T.M., Mitchell J.R., Behravesh C.B., Hausman L.B., O’Connor K.A. Notes from the field: multistate outbreak of Salmonella Altona and Johannesburg infections linked to chicks and ducklings from a mail-order hatchery — United States, February–October 2011. Morb. Mortal. Wkly. Rep. 2012;61:195. [PubMed] [Google Scholar]
- Hofacre C.L., Fricke J.A., Inglis T. Antimicrobial drug use in poultry. In: Giguère S., Prescott J.F., Dowling P.M., editors. Antimicrobial Therapy in Veterinary Medicine. 5th ed. John Wiley and Sons, Inc, Hoboken, NJ; 2013. pp. 569–587. [Google Scholar]
- Humphrey T.J., Lanning D.G. The vertical transmission of Salmonellas and formic acid treatment of chicken feed: a possible strategy for control. Epidemiol. Infect. 1988;100:43–49. doi: 10.1017/s0950268800065547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalily E., Hollander A., Korin B., Cymerman I., Yaron S. Adaptation of Salmonella enterica serovar Senftenberg to linalool and its association with antibiotic resistance and environmental persistence. Appl. Environ. Microbiol. 2017;83:1–17. doi: 10.1128/AEM.03398-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A., Young J.L., Min S.K., Sang I.K., Jae K.C. Dissemination and tracking of Salmonella spp. in integrated broiler operation. J. Vet. Sci. 2007;8:155–161. doi: 10.4142/jvs.2007.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Ecuyer P.B., Diego J., Murphy D., Trovillion E., Jones M., Sahm D.F., Fraser V.J. Nosocomial outbreak of gastroenteritis due to Salmonella Senftenberg. Clin. Infect. Dis. 1996;23:734–742. doi: 10.1093/clinids/23.4.734. [DOI] [PubMed] [Google Scholar]
- Liljebjelke K. a, Hofacre C.L., Liu T., White D.G., Ayers S., Young S. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog. Dis. 2005;2:90–102. doi: 10.1089/fpd.2005.2.90. [DOI] [PubMed] [Google Scholar]
- Liljebjelke K.A., Hofacre C.L., White D.G., Ayers S., Lee M.D., Maurer J.J. Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial Poultry Farms. Front. Vet. Sci. 2017;4:1–9. doi: 10.3389/fvets.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin C., Balasch S., Vega S., Lainez M. Sources of Salmonella contamination during broiler production in Eastern Spain. Prev. Vet. Med. 2011;98:39–45. doi: 10.1016/j.prevetmed.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Marin C., Lainez M. Salmonella detection in feces during broiler rearing and after live transport to the slaughterhouse. Poult. Sci. 2009;88:1999–2005. doi: 10.3382/ps.2009-00040. [DOI] [PubMed] [Google Scholar]
- Martelli F., Davies R.H. Salmonella serovars isolated from table eggs: an overview. Food Res. Int. 2012;45:745–754. [Google Scholar]
- Meunier D., Baucheron S., Chaslus-Dancla E., Martel J.L., Cloeckaert A. Florfenicol resistance in Salmonella enterica serovar Newport mediated by a plasmid related to R55 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 2003;51:1007–1009. doi: 10.1093/jac/dkg141. [DOI] [PubMed] [Google Scholar]
- Nguyen S.V., Harhay G.P., Bono J.L., Smith T.P.L., Harhay D.M. Genome sequence of the thermotolerant foodborne pathogen Salmonella enterica Serovar Senftenberg ATCC 43845 and phylogenetic analysis of loci encoding increased protein quality control mechanisms. Appl. Environ. Microbiol. 2017;2:1–13. doi: 10.1128/mSystems.00190-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.-Y., Kang M.-S., An B.-K., Song E., Kwon J.-H., Kwon Y.-K. Occurrence of purulent arthritis broilers vertically infected with Salmonella enterica serovar Enteritidis in Korea. Poult. Sci. 2010;89:2116–2122. doi: 10.3382/ps.2010-00918. [DOI] [PubMed] [Google Scholar]
- Pedersen T.B., Olsen J.E., Bisgaard M. Persistence of Salmonella Senftenberg in poultry production environments and investigation of its resistance to desiccation. Avian Pathol. 2008;37:421–427. doi: 10.1080/03079450802216561. [DOI] [PubMed] [Google Scholar]
- Prescott J.F. Veterinary antimicrobial stewardship in North America. Aust. Vet. J. 2019;97:243–248. doi: 10.1111/avj.12811. [DOI] [PubMed] [Google Scholar]
- Rasschaert G., Houf K., Imberechts H., Grijspeerdt K., Heyndrickx M., De Zutter L. Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonella enterica isolates. J. Clin. Microbiol. 2005;43:3615–3623. doi: 10.1128/JCM.43.8.3615-3623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodloff A., Bauer T., Ewig S., Kujath P., Müller E. Susceptible, intermediate, and resistant – the intensity of antibiotic action. Dtsch. Arztebl. 2008;105:657–662. doi: 10.3238/arztebl.2008.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund-Karlsson M., Joyce K., Blickenstaff K., Ball T., Haro J., Medalla F.M., Fedorka-Cray P., Zhao S., Crump J.A., Whichard J.M. Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob. Agents Chemother. 2011;55:3985–3989. doi: 10.1128/AAC.00590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Amand J.A., Otto S.J.G., Cassis R., Annett Christianson C.B. Antimicrobial resistance of Salmonella enterica serovar Heidelberg isolated from poultry in Alberta. Avian Pathol. 2013;42:379–386. doi: 10.1080/03079457.2013.811465. [DOI] [PubMed] [Google Scholar]
- Stepan R.M., Sherwood J.S., Petermann S.R., Logue C.M. Molecular and comparative analysis of Salmonella enterica Senftenberg from humans and animals using PFGE, MLST and NARMS. BMC Microbiol. 2011;11:1–9. doi: 10.1186/1471-2180-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Agriculture, Food Safety Inspection Service Prevention, detection and control of Salmonella in poultry. Terr. Anim. Heal. Stand. Comm. Rep. 2009;6(5):1–7. [Google Scholar]
- Versalovic J., Schneider M., DeBuijn F.J., Lupski J.R. Genomic fingerprinting of bacterial using repititive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994;5:25–40. [Google Scholar]
- von Wintersdorff C.J.H., Penders J., van Niekerk J.M., Mills N.D., Majumder S., van Alphen L.B., Savelkoul P.H.M., Wolffs P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G.K., Jalukar S., Brake J. The effect of refined functional carbohydrates from enzymatically hydrolyzed yeast on the transmission of environmental Salmonella Senftenberg among broilers and proliferation in broiler housing. Poult. Sci. 2018;97:1412–1419. doi: 10.3382/ps/pex430. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Lai H., Zou L., Yin S., Wang C., Han X., Xia X., Hu K., He L., Zhou K., Chen S., Ao X., Liu S. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 2017;259:43–51. doi: 10.1016/j.ijfoodmicro.2017.07.023. [DOI] [PubMed] [Google Scholar]