Abstract
The gut microbiome is a complex ecosystem that contributes to host nutrition and health. However, our current knowledge of the relationship between ambient temperature and gut microbiota of poultry is still limited. The objective of the present study was to characterize the intestinal microbiota of ducks exposed to high ambient temperature. Sixty 60-day-old Shaoxing ducks were allocated to control and heat-treated groups. The ducks in the control group were kept at 25°C, and the ducks in the heat treatment group were raised at 30–40°C, which simulated the temperature change of day and night in summer. After 15 D, the intestinal contents of the duodenum, jejunum, and ileum were obtained from 6 ducks of each group. Genomic DNA was extracted and amplified based on the V4–V5 hypervariable region of 16S rRNA. The results showed that Firmicutes was the dominant bacterial phylum with the highest abundance in the contents of the small intestine of ducks, and the relative abundance of the phylum Firmicutes in all 3 intestinal segments was increased by high temperature. At the genus level, Lactobacillus was found to be the most dominant bacterial genus across 3 gut segments, and its abundance was increased in ducks under heat treatment. Compared with the corresponding intestine segment of control ducks, a total of 36 genera in the duodenum, 19 genera in the jejunum, and 6 genera in the ileum of heat-treated ducks were found to be significantly different in the abundance (linear discriminant analysis score >3.0, P < 0.05). Functional prediction of gut microbiota revealed that high temperature caused changes in the abundance of metabolism and transcription-related pathways. It is noteworthy that most of the altered pathways are related to metabolism. In conclusion, high temperature induced remarkable taxonomic changes in the gut microbiome of ducks, which might be related to the negative effects of high temperature in ducks. Our present study provided an important theoretical ground for high-temperature intervention.
Key words: gut microbiota, high temperature, community structure, functional pathway, Shaoxing duck
Introduction
High ambient temperature is one of the most serious environmental factors that adversely affect animal production performance and health (Nardone et al., 2010, Kamel et al., 2017), and it is responsible for heavy economic losses and has become a significant challenge in animal husbandry industry (St-Pierre et al., 2003, Nawab et al., 2018). Environmental stressors may also increase the risk of colonization with pathogens such as Salmonella and Campylobacter in the gut of animals, and some pathogens can disseminate to other animals as well as to humans, that is, it has the potential to alter coexistence in host–pathogen systems and may pose a serious threat to public health (Freestone et al., 2008, Verbrugghe et al., 2012, Lara and Rostagno, 2013, Alhenaky et al., 2017).
As an important part of the body, the gut microbiota plays vital roles in feed digestion (Yang et al., 2017, Liu et al., 2018), nutrition absorption (Krajmalnik-Brown et al., 2012, Semova et al., 2012), energy extraction (Turnbaugh et al., 2006, Heiss and Olofsson, 2018), immune response (Furusawa et al., 2013, Takiishi et al., 2017), and disease resistance (Mazmanian et al., 2008, Smith et al., 2013). Previous reports have shown that the gut microbial composition can be altered by various factors such as environmental temperature (Fontaine et al., 2018), diet (David et al., 2014), age (O'Toole and Jeffery, 2015), disease (McKenzie et al., 2017), and drugs (Modi et al., 2014). A recent report showed the fecal microbial flora of laying hens was dominated by Firmicutes, Bacteroidetes, and Proteobacteria phyla, in which the phylum Firmicutes was significantly decreased and Bacteroidetes was increased under heat conditions (Zhu et al., 2019). Alteration of the gut microbiota may have adverse effects on feed efficiency, productivity, and health of animals. Therefore, understanding the effects of stressors on microbial profiles is essential for improving gut physiological functions in host animal growth and development.
In the last decade, the use of 16S rRNA gene sequencing has substantially improved the knowledge about the composition and diversity of the gut microbiota. Modern high-throughput sequencing technology is capable of quickly shaping the structural composition of the microbiome, which is a powerful tool providing important new insights into the impacts of environmental factors on the biological and ecological roles of the gut microbiota in poultry (Sohail et al., 2015, Awad et al., 2018). However, most studies have been performed in the regulation of feed additives and nutrients in gut microflora of chickens. Few reports have been published with regard to the gut microbial ecosystem of poultry exposed to high temperature, and knowledge of the relationship between the hot environment and gut microbiota in poultry, especially in ducks, is still limited. Shaoxing ducks (Anas platyrhynchos), as a famous egg-laying duck breed, are raised in large numbers in China. To improve land utilization and feed conversion rate and reduce environmental pollution, Shaoxing ducks were normally transformed from flat culture to cage rearing at the age of 80 D. In production, we found that a certain number of ducks died early in the culture mode alteration, especially in the first 2 wk, and the high environmental temperature has greatly exacerbated this negative result. Here, we focused on the effect of high temperature on duck gut microbiome, and our previous report has shown that high temperature induced a certain degree of damage to the intestinal mucosa of ducks (Tian et al. 2019). Therefore, we hypothesized that there was some kind of connection between intestinal flora and mucosal barrier integrity. This study was carried out to evaluate the impacts of high temperature on intestinal microbiome of ducks using next-generation 16S rRNA gene deep-sequencing technologies, which may provide a theoretical basis for exploring methods to mitigate adverse reaction caused by high temperature via manipulating gut microbiota.
Materials and methods
Ethics Statement
Animals used in this study were raised and slaughtered in accordance with the national standard of Laboratory animal-Guideline for ethical review of animal welfare. All experiment procedures were approved by the Institute of Animal Husbandry and Veterinary Science, Zhejiang Academy of Agricultural Sciences (Hangzhou, China).
Animals Rearing and Sample Collection
The animal feeding experiment was conducted in the animal testing area of Zhejiang Academy of Agricultural Sciences, Hangzhou, China. Sixty healthy 60-day-old Shaoxing ducks with the same genetic background were collected and housed in a poultry cage system at a commercial production facility (Jiangsu Xiufu Technology Co. Ltd., Yancheng, Jiangsu, China). The ducks were randomly divided into 2 groups (control and heat treatment) with 3 replicates per group and 10 birds per replicate. All ducks were raised under standardized commercial rearing conditions without any added antimicrobials and kept in the temperature-controlled room for an adaptation period of 20 D under conditions of natural light and at 25°C and 65% relative humidity. Throughout the whole experimental period, feed and water were supplied ad libitum.
After the adaptation period, the feeding and management conditions remained unchanged in the control group. Ducks in the heat-treated group were maintained in a high-temperature environment, which mimicked the diurnal variation of summer temperature and ranged between 30°C and 40°C. The daily variation mode of room temperature in the heat-treated group was as follows: the temperature was increased at 5°C/h from 30°C to 40°C (10:00 am–12:00 pm), kept at 40°C for 2 h (12:00 pm–2:00 pm), and decreased at 5°C/h from 40°C to 30°C (2:00 pm–4:00 pm) and maintained at 30°C until 10:00 am the following day. After 15 consecutive days of treatment, 2 ducks from each replicate were randomly selected and euthanatized by jugular venesection. The luminal contents of the duodenum, jejunum, and ileum were sampled, transferred into liquid nitrogen immediately, and then stored at –80°C. CD, CJ, and CI were used to label the samples of duodenal, jejunal, and ileal contents from the control ducks, respectively, and HD, HJ, and HI represented the duodenal, jejunal, and ileal content samples in the heat-treated ducks, respectively.
DNA Extraction and PCR Amplification
Total bacterial genomic DNA was extracted from the luminal contents of each sample using the E.Z.N.A. Soil DNA Kit (Omega Bio-tek, Norcross, GA) as per the manufacturer's protocol. The integrity of genomic DNA was assessed by agarose gel electrophoresis.
The V4–V5 region of the bacterial 16S rRNA gene was amplified from the DNA samples by PCR using barcoded primers of 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). The PCR reaction mixtures (20 μL) contained 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mMol dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. The PCR was performed on a T100 thermal cycler (Bio-Rad, CA), and the cycling conditions were 95°C for 2 min, followed by 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min. Each reaction was carried out in 3 biological replicates. The PCR products (amplicons) were detected by 2% agarose gel electrophoresis and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA) as per the manufacturer's instructions.
Library Construction and Sequencing
The purified PCR products were quantified using Life Invitrogen Qubit2.0 (Invitrogen, CA), and every 24 amplicons with different barcodes were mixed equally. The pooled DNA products were used to construct the pair-end sequencing libraries following Illumina's genomic DNA library preparation procedure. Then, the libraries were subjected to the Illumina HiSeq 2500 platform (Illumina, San Diego, CA) as per the standard protocols, and 250-bp paired-end reads were generated. The raw sequence data generated from this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) with the accession number of SRP234307.
Bioinformatics and Statistical Analysis
Raw sequence data from 36 intestinal content samples were processed and analyzed using the Quantitative Insights Into Microbial Ecology (QIIME) software package (Schellenberg et al., 2012). The 250-bp reads were truncated at any site receiving an average quality score <20 over a 10 bp sliding window, the truncated reads shorter than 50 bp were discarded, and the reads containing more than 2 nt mismatch in primer or 1 nt mismatch in the barcode matching were removed. Then, sequences that overlap longer than 10 bp were assembled according to the overlapped sequences. Operational taxonomic units (OTU) were clustered with 97% similarity cutoff using UPARSE (http://drive5.com/uparse/), and chimeric sequences were identified and removed using UCHIME (Edgar et al., 2011). Representative sequences were picked for each OTU, and singleton OTU from the total number of OTU picked was removed. In case of the influences of sequencing depth on community diversity, the OTU was rarified to make all samples holding the same sequence number that corresponds to the sample with the least sequences, and the subsequent analyses were all based on these rarified OTU data. The OTU were further subjected to the taxonomical analysis based on the ribosomal database project classifier; then, the alpha diversity (Chao1, Observed species, Shannon, and Simpson) of each sample was analyzed using the QIIME program. The differences in microbial communities of the each small intestinal section between the control and heat-treated ducks were determined by the linear discriminant analysis (LDA) effect size (LEfSe) algorithm (Segata et al., 2011), and differences with linear discrimination analysis scores >3 and P value < 0.05 were considered statistically significant. Visualization of differences in the gut microbiota of ducks reared under the control and heat-treated conditions was performed using nonmetric multidimensional scaling (NMDS) ordinations based on weighted UniFrac distance. PICRUSt (https://picrust.github.io/picrust/) was used to conduct the functional annotation of microbial communities.
For LEfSe analysis of each intestinal microbial community, Mann-Whitney U tests were performed to detect significantly different abundances between the control and heat-treated groups, and LDA scores were used to estimate the effect degree (LDA score >3.0 and P < 0.05). The differences of alpha diversity (Chao1, Observed species, Shannon, and Simpson indices) and relative abundance of COGs and KEGG pathways between the 2 groups (control vs. heat treatment) were analyzed by the independent samples t-test using SPSS version 22.0 (SPSS Inc., Chicago, IL). Data were presented as the mean ± standard deviation. The value of P < 0.05 was considered to be statistically significant.
Results
Summary of Sequencing Data
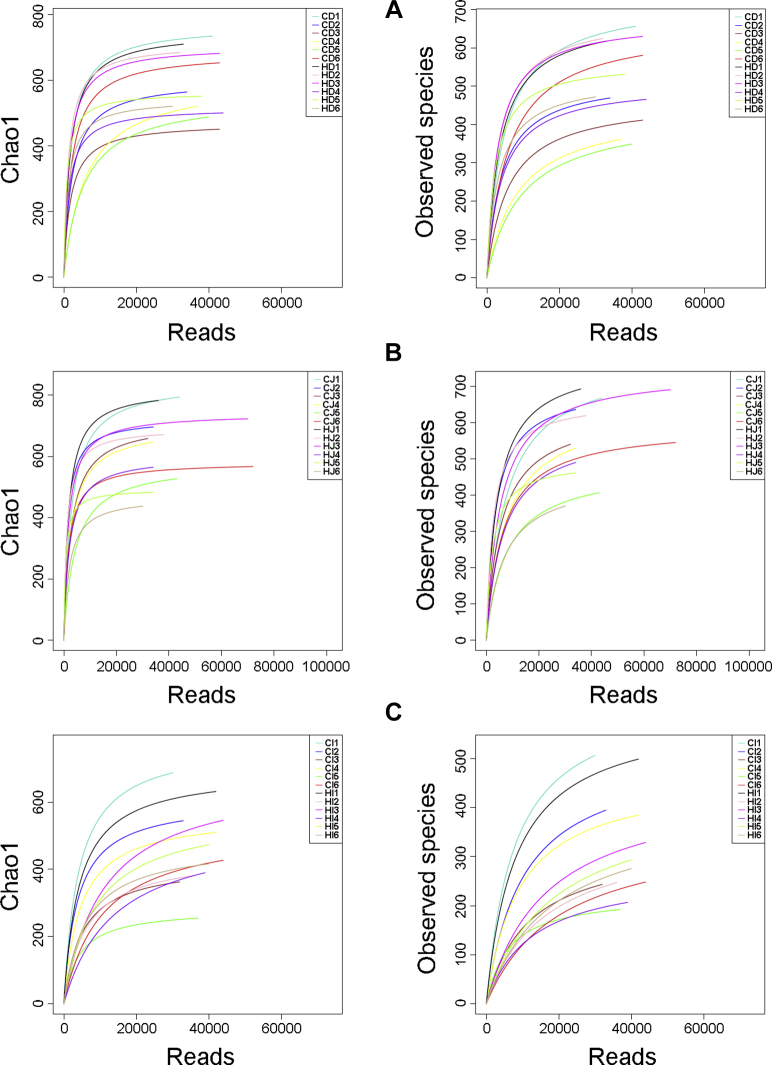
To explore the compositional changes of microbial communities in samples of small intestinal contents in ducks exposed to different ambient temperatures, the 16S rRNA gene PCR products of the V4–V5 region were sequenced. Next-generation sequencing was used for the bacterial 16S rRNA gene in each of the 36 samples. After assembling, quality filtering, and singleton removal, a total of 1,436,930 effective reads were obtained from 36 samples, with an average of 39,915 reads per sample. These sequences were clustered into OTU, and the number of OTU detected in each sample ranged from 202 to 714 (Supplementary Table 1). To avoid the influence of the sequencing depth on evaluation of microbial communities, the library size was rarefied to 30,648 reads per sample using the rarefy function in QIIME, and the dilution curves of the alpha diversity indices showed that the rarefied sequencing depth reveals microbial communities in all samples (Figure 1).
Figure 1.
Rarefaction curves of Chao1 and Observed species indices for each sample of duodenal (A), jejunal (B), and ileal (C) contents. CD, CJ, and CI represent the intestinal content samples collected from the duodenum, jejunum, and ileum of the control ducks, respectively, and HD, HJ, and HI represent the duodenal, jejunal, and ileal content samples from the heat-treated ducks, respectively.
Comparison of Microbial Community at the Phylum and Genus Levels
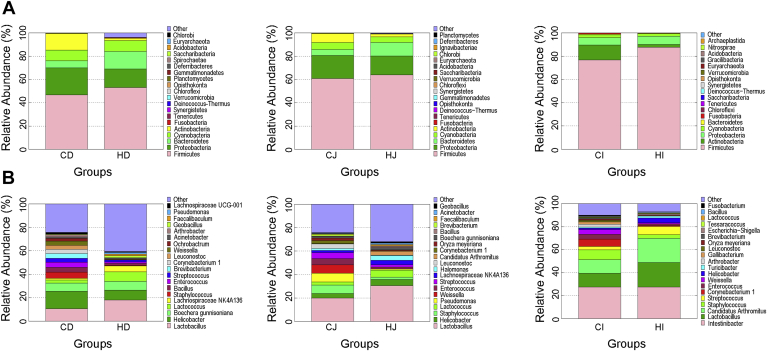
The dominant phyla and their relative abundance in the each group are shown in Figure 2A. For the samples of duodenal contents, bacteria belonging to the phylum Firmicutes were the most predominant, accounting for 46.86% in the CD group and 53.17% in the HD group, respectively, followed by Proteobacteria (23.22% in the CD and 15.79% in the HD group), Bacteroidetes (5.93% in the CD and 15.18% in the HD group), Cyanobacteria (9.35% in the CD and 9.31% in the HD group), and Actinobacteria (14.15% in the CD and 1.90% in the HD group). In the CJ and HJ groups, the predominant phyla were Firmicutes (60.90 and 64.02%, respectively), Proteobacteria (19.73 and 16.15%, respectively), Bacteroidetes (5.11 and 11.68%, respectively), Cyanobacteria (6.12 and 4.90%, respectively), and Actinobacteria (7.63 and 2.46%, respectively). In the samples of ileal contents, Firmicutes, Actinobacteria, Proteobacteria, Cyanobacteria, and Bacteroidetes were the dominant phyla. For the CI group, most of the sequences were assigned to Firmicutes (76.67%), followed by Actinobacteria (12.85%), Proteobacteria (6.34%), Cyanobacteria (2.54%), and Bacteroidetes (0.75%). In the HI group, Firmicutes also showed the overwhelming dominance, accounting for 87.78%, followed by bacteria from the phyla Proteobacteria (6.95%), Actinobacteria (2.37%), Cyanobacteria (2.19%), and Bacteroidetes (0.62%).
Figure 2.
Relative abundance of major phylum (A) and genus (B) in the duodenal, jejunal, and ileal contents of ducks in the control and high-temperature groups. CD, CJ, and CI represent the intestinal content samples collected from the duodenum, jejunum, and ileum of the control ducks, respectively, and HD, HJ, and HI represent the duodenal, jejunal, and ileal content samples from the heat-treated ducks, respectively.
At the genus level, the predominant genera and their abundance in all groups are shown in Figure 2B. The dominant bacterial genera were found across the 3 gut segments of the duck small intestines, such as Lactobacillus, Helicobacter, Lactococcus, Staphylococcus, Enterococcus, and Streptococcus. Among them, Lactobacillus was more abundant in the heat-treated intestines than in their corresponding controls, whereas the abundance of Staphylococcus and Enterococcus in the HD, HJ, and HI groups was lower than in the corresponding control groups. Helicobacter showed a lower abundance in the HD group (8.49%) than in the CD group (14.96%), but its abundance in the HJ and HI groups was 1.38 and 3.22 times higher than in the CJ and CI groups, respectively. The richness of Lactococcus in the HD and HJ groups was 3.17 and 2.17 times higher, respectively, than in the CD and CJ groups, but its abundance in the HI and CI groups was similar. In the HD and HJ groups, the abundance of Streptococcus was 1.80 and 2.58 times lower than in the corresponding controls, respectively, and its richness in the HI group was 2.44 times higher than in the CI group.
The Differences of Microbial Richness, Diversity, and Community
To evaluate the richness and diversity of gut microbiota between the control and heat-treated groups, the alpha diversity values (Chao1, Observed species, Shannon, and Simpson) were analyzed. For the duodenum, the Chao1, Observed species, Shannon, and Simpson indices were higher in the heat-treated ducks than in the controls. In the jejunum, the results of the Chao1 and Simpson analyses showed lower community diversity in the heat-treated group than in the control group, whereas the Observed species and Shannon indices of the gut microbiota of heat-treated ducks were slightly higher than those of the controls. In the ileum, the Chao1, Observed species, Shannon, and Simpson indices were lower in the heat-treated ducks than in the controls. However, there was no significantly statistical difference in alpha diversity values between the heat-treated and the control groups within each small intestinal segment (Table 1).
Table 1.
Effects of high temperature on alpha diversity of the duodenal, jejunal, and ileal microbiota in ducks.
| Index | Duodenum (n = 6) |
Jejunum (n = 6) |
Ileum (n = 6) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | Heat treatment | P value | Control | Heat treatment | P value | Control | Heat treatment | P value | |
| Chao1 | 550.44 ± 110.77 | 620.36 ± 98.96 | 0.276 | 643.18 ± 101.01 | 617.01 ± 137.12 | 0.714 | 480.91 ± 159.68 | 462.50 ± 109.16 | 0.820 |
| Observed species | 453.00 ± 120.97 | 555.50 ± 80.92 | 0.115 | 536.00 ± 95.18 | 541.67 ± 119.39 | 0.929 | 323.33 ± 128.91 | 283.67 ± 98.76 | 0.563 |
| Shannon | 4.83 ± 0.77 | 5.70 ± 0.85 | 0.094 | 4.98 ± 0.91 | 5.16 ± 1.55 | 0.814 | 3.70 ± 1.06 | 2.79 ± 0.94 | 0.148 |
| Simpson | 0.90 ± 0.05 | 0.91 ± 0.07 | 0.696 | 0.88 ± 0.08 | 0.86 ± 0.14 | 0.741 | 0.81 ± 0.13 | 0.69 ± 0.18 | 0.196 |
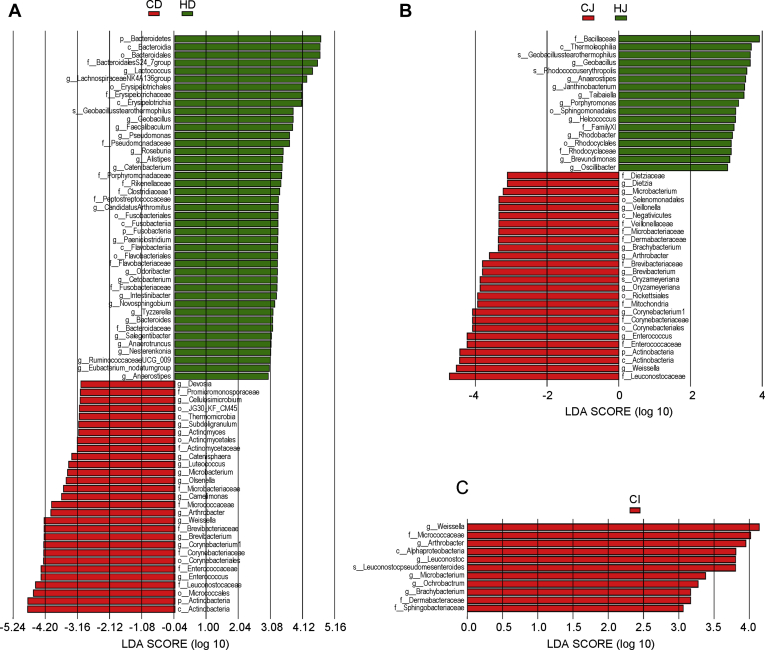
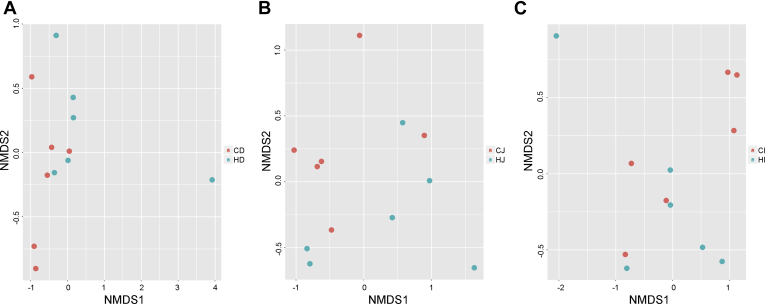
To identify the bacterial taxa that were significantly associated with high temperature, we used LEfSe to compare the gut microbiome in the same gut section of ducks reared under 2 different conditions. The results indicated that the abundance of 36 genera was significantly different in the comparison of CD and HD groups (genus level, LDA score >3.0, P < 0.05), with 14 genera (e.g., Enterococcus, Corynebacterium1, and Weissella) being enriched in the CD group and 22 genera (e.g., Lactococcus, Geobacillus, and Pseudomonas) being enriched in the HD group. The abundance of 19 genera was found to be significantly different between the CJ and HJ ducks, among which 10 genera including Weissella, Enterococcus, and Corynebacterium1 were observed to be higher in the CJ ducks, whereas the other 9 genera including Geobacillus, Anaerostipes, and Janthinobacterium were detected at higher abundance in the HJ ducks. However, only 6 genera such as Weissella, Arthrobacter, and Leuconostoc tended to be significantly different between the CI and HI groups, all of which were decreased by high temperature (Figure 3). To visualize overall similarities and differences in the gut microbial communities of ducks reared under different ambient temperature conditions, NMDS plots derived from weighted UniFrac distances were generated (Figure 4). The NMDS plot ordinations showed that the microbial communities of the duodenum, jejunum, and ileum were separately clustered in the control and heat-treated groups, but the clustering of each group was not so distinct.
Figure 3.
Differential analysis of the microbiota community between the control and high-temperature groups within each intestinal segment. The linear discrimination analysis (LDA) coupled with effect size (LEfSe) was used to identify the most differentially abundant taxa between the control and heat-treated ducks within the duodenum (A), jejunum (B), and ileum (C). The criteria of the LDA score for discriminative features was >3.0 and P < 0.05 (Mann-Whitney U tests). CD represents the duodenal contents of the control ducks. HD represents the duodenal contents of the heat-treated ducks. CJ represents the jejunal contents of the control ducks. HJ represents the jejunal contents of the heat-treated ducks. CI represents the ileal contents of the control ducks. HI represents the ileal contents of the heat-treated ducks.
Figure 4.
Weighted UniFrac distance–based nonmetric multidimensional scaling (NMDS) plots of all samples from the duodenum (A), jejunum (B), and ileum (C). Red and blue dots represent the samples collected from the control and heat-treated ducks, respectively.
Functional Changes in the Intestinal Microbiota Induced by High Temperature
Based on next-generation sequencing reads, PICRUSt was used to evaluate the functional and metabolic potentials of the duck intestinal microbiota and their possible alteration by high temperature. A total of 4,582, 4,601, and 4,511 COG protein families were identified in the samples of duodenal, jejunal, and ileal contents, respectively. In the samples of the duodenal contents, the most abundant COG families were response regulators consisting of a CheY-like receiver domain and a winged helix DNA-binding domain (COG#0745), signal transduction histidine kinase (COG#0642), glycosyltransferase (COG#0438), arabinose efflux permease (COG#2814), and transcriptional regulator (COG#1309) (Supplementary Table 2). In the jejunal microbiota, the most abundant COG families included response regulators consisting of a CheY-like receiver domain and a winged helix DNA-binding domain (COG#0745), signal transduction histidine kinase (COG#0642), transcriptional regulator (COG#0583), arabinose efflux permease (COG#2814), and glycosyltransferase (COG#0438) (Supplementary Table 3). In the ileal microbiota, the most abundant COG families were response regulators consisting of a CheY-like receiver domain and a winged helix DNA-binding domain (COG#0745), signal transduction histidine kinase (COG#0642), transcriptional regulator (COG#1309), ABC-type multidrug transport system, ATPase and permease components (COG#1132), and arabinose efflux permease (COG#2814) (Supplementary Table 4). In the 3 intestinal segments of ducks, the most abundant COG functional categories (classes) were amino acid transport and metabolism (category E), transcription (category K), and carbohydrate transport and metabolism (category G). Heat treatment seemed to have a significant effect on some functional categories of the microbiota in the duodenum and jejunum (Table 2, Table 3, Table 4). Specifically, the abundance of inorganic ion transport and metabolism (category P) and defense mechanisms (category V) in the HD group was significantly different from those in the CD group, and the abundance of 2 categories including posttranslational modification, protein turnover, chaperones (category O), and RNA processing and modification (category A) in the HJ ducks was significantly lower than in the CJ ducks.
Table 2.
COG function classes identified in the duodenal microbiome of ducks.
| COG_class | Description | CD1 | HD2 | P value |
|---|---|---|---|---|
| R | General function prediction only | 11.75 ± 0.51 | 11.58 ± 0.34 | 0.534 |
| E | Amino acid transport and metabolism | 8.36 ± 0.91 | 7.77 ± 0.38 | 0.170 |
| S | Function unknown | 8.04 ± 0.64 | 7.72 ± 0.37 | 0.320 |
| K | Transcription | 7.34 ± 1.05 | 7.57 ± 0.42 | 0.621 |
| G | Carbohydrate transport and metabolism | 7.28 ± 1.52 | 7.78 ± 0.69 | 0.486 |
| J | Translation, ribosomal structure, and biogenesis | 6.60 ± 0.68 | 6.45 ± 0.54 | 0.680 |
| M | Cell wall/membrane/envelope biogenesis | 6.16 ± 0.69 | 6.50 ± 0.14 | 0.276 |
| L | Replication, recombination, and repair | 6.05 ± 0.52 | 6.42 ± 0.24 | 0.149 |
| C | Energy production and conversion | 5.46 ± 0.24 | 5.30 ± 0.13 | 0.196 |
| T | Signal transduction mechanisms | 5.27 ± 1.17 | 5.90 ± 0.72 | 0.290 |
| P | Inorganic ion transport and metabolism | 5.27 ± 0.29 | 4.90 ± 0.14 | 0.017 |
| H | Coenzyme transport and metabolism | 4.50 ± 0.37 | 4.25 ± 0.24 | 0.192 |
| O | Posttranslational modification, protein turnover, chaperones | 3.64 ± 0.50 | 3.56 ± 0.24 | 0.743 |
| F | Nucleotide transport and metabolism | 2.95 ± 0.37 | 2.88 ± 0.28 | 0.743 |
| I | Lipid transport and metabolism | 2.90 ± 0.46 | 2.63 ± 0.12 | 0.202 |
| V | Defense mechanisms | 2.10 ± 0.32 | 2.51 ± 0.11 | 0.015 |
| U | Intracellular trafficking, secretion, and vesicular transport | 2.05 ± 0.37 | 2.10 ± 0.08 | 0.754 |
| Q | Secondary metabolites biosynthesis, transport, and catabolism | 1.59 ± 0.42 | 1.34 ± 0.12 | 0.203 |
| N | Cell motility | 1.44 ± 0.50 | 1.52 ± 0.12 | 0.691 |
| D | Cell cycle control, cell division, chromosome partitioning | 1.21 ± 0.16 | 1.28 ± 0.05 | 0.335 |
| B | Chromatin structure and dynamics | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.581 |
| A | RNA processing and modification | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.260 |
| Z | Cytoskeleton | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.812 |
| W | Extracellular structures | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.343 |
CD: duodenal content samples from the control ducks.
HD: duodenal content samples from the heat-treated ducks.
Table 3.
COG function classes identified in the jejunal microbiome of ducks.
| COG_class | Description | CJ1 | HJ2 | P value |
|---|---|---|---|---|
| R | General function prediction only | 11.72 ± 0.24 | 11.53 ± 0.26 | 0.127 |
| S | Function unknown | 8.36 ± 0.23 | 7.96 ± 0.42 | 0.269 |
| E | Amino acid transport and metabolism | 8.18 ± 0.50 | 7.93 ± 0.39 | 0.503 |
| K | Transcription | 7.71 ± 0.28 | 7.77 ± 0.40 | 0.164 |
| G | Carbohydrate transport and metabolism | 7.53 ± 0.69 | 7.99 ± 0.76 | 0.359 |
| J | Translation, ribosomal structure, and biogenesis | 6.70 ± 0.73 | 6.72 ± 0.73 | 0.818 |
| L | Replication, recombination, and repair | 6.16 ± 0.69 | 6.44 ± 0.51 | 0.299 |
| M | Cell wall/membrane/envelope biogenesis | 5.93 ± 0.28 | 6.25 ± 0.12 | 0.327 |
| C | Energy production and conversion | 5.33 ± 0.23 | 5.23 ± 0.26 | 0.243 |
| T | Signal transduction mechanisms | 5.31 ± 0.86 | 5.43 ± 0.54 | 0.979 |
| P | Inorganic ion transport and metabolism | 5.12 ± 0.18 | 4.90 ± 0.17 | 0.764 |
| H | Coenzyme transport and metabolism | 4.16 ± 0.16 | 4.00 ± 0.34 | 0.442 |
| O | Posttranslational modification, protein turnover, chaperones | 3.60 ± 0.15 | 3.48 ± 0.24 | 0.037 |
| F | Nucleotide transport and metabolism | 2.99 ± 0.38 | 3.04 ± 0.34 | 0.626 |
| I | Lipid transport and metabolism | 2.96 ± 0.35 | 2.76 ± 0.16 | 0.299 |
| V | Defense mechanisms | 2.03 ± 0.28 | 2.38 ± 0.23 | 0.053 |
| U | Intracellular trafficking, secretion, and vesicular transport | 2.03 ± 0.27 | 2.03 ± 0.09 | 0.118 |
| Q | Secondary metabolites biosynthesis, transport, and catabolism | 1.62 ± 0.28 | 1.39 ± 0.19 | 0.220 |
| N | Cell motility | 1.3 ± 0.63 | 1.44 ± 0.28 | 0.074 |
| D | Cell cycle control, cell division, chromosome partitioning | 1.21 ± 0.10 | 1.30 ± 0.11 | 0.771 |
| B | Chromatin structure and dynamics | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.945 |
| A | RNA processing and modification | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.041 |
| Z | Cytoskeleton | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.862 |
| W | Extracellular structures | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.200 |
CJ: jejunal content samples from the control ducks.
HJ: jejunal content samples from the heat-treated ducks.
Table 4.
COG function classes identified in the ileal microbiome of ducks.
| COG_class | Description | CI1 | HI2 | P value |
|---|---|---|---|---|
| R | General function prediction only | 11.57 ± 0.28 | 11.39 ± 0.20 | 0.222 |
| K | Transcription | 8.79 ± 0.84 | 8.63 ± 0.69 | 0.735 |
| E | Amino acid transport and metabolism | 8.29 ± 0.35 | 7.96 ± 0.61 | 0.270 |
| S | Function unknown | 7.90 ± 0.59 | 7.62 ± 0.42 | 0.367 |
| G | Carbohydrate transport and metabolism | 7.66 ± 0.67 | 7.21 ± 0.60 | 0.250 |
| L | Replication, recombination and repair | 6.93 ± 0.46 | 7.08 ± 0.45 | 0.568 |
| J | Translation, ribosomal structure, and biogenesis | 6.82 ± 0.30 | 7.29 ± 1.06 | 0.324 |
| M | Cell wall/membrane/envelope biogenesis | 5.46 ± 0.25 | 5.75 ± 0.26 | 0.077 |
| C | Energy production and conversion | 5.36 ± 0.09 | 5.25 ± 0.29 | 0.413 |
| T | Signal transduction mechanisms | 5.23 ± 0.83 | 5.59 ± 0.64 | 0.418 |
| P | Inorganic ion transport and metabolism | 5.02 ± 0.35 | 4.91 ± 0.37 | 0.612 |
| H | Coenzyme transport and metabolism | 3.89 ± 0.38 | 3.65 ± 0.32 | 0.255 |
| O | Posttranslational modification, protein turnover, chaperones | 3.19 ± 0.33 | 3.16 ± 0.12 | 0.827 |
| F | Nucleotide transport and metabolism | 3.00 ± 0.17 | 3.03 ± 0.27 | 0.863 |
| I | Lipid transport and metabolism | 2.71 ± 0.32 | 2.50 ± 0.11 | 0.181 |
| V | Defense mechanisms | 2.34 ± 0.37 | 2.51 ± 0.10 | 0.311 |
| U | Intracellular trafficking, secretion, and vesicular transport | 1.77 ± 0.16 | 1.93 ± 0.17 | 0.120 |
| Q | Secondary metabolites biosynthesis, transport, and catabolism | 1.39 ± 0.28 | 1.15 ± 0.12 | 0.094 |
| N | Cell motility | 1.36 ± 0.62 | 1.95 ± 0.60 | 0.126 |
| D | Cell cycle control, cell division, chromosome partitioning | 1.27 ± 0.11 | 1.41 ± 0.17 | 0.118 |
| Z | Cytoskeleton | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.568 |
| B | Chromatin structure and dynamics | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.179 |
| A | RNA processing and modification | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.106 |
| W | Extracellular structures | 0.00 ± 0.01 | 0.00 ± 0.00 | 0.437 |
CI: ileal content samples from the control ducks.
HI: ileal content samples from the heat-treated ducks.
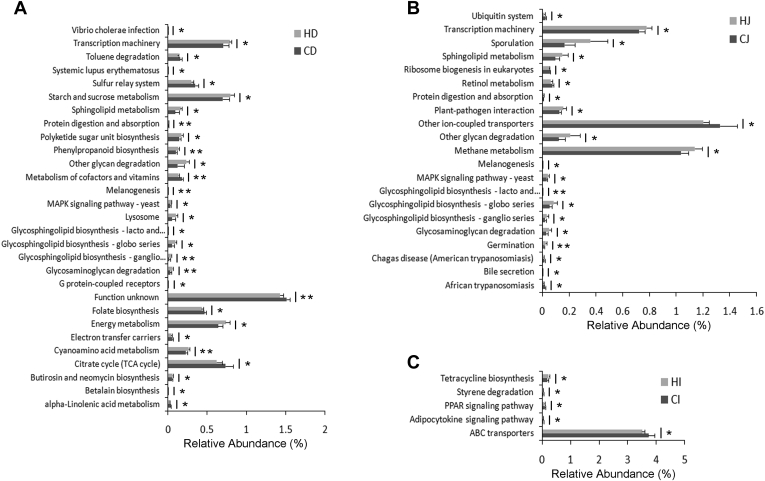
Next-generation sequencing reads were further analyzed for KEGG pathway assignment to predict the potential functions altered by heat treatment. For the duodenum, we found that the abundance of 29 KEGG pathways was significantly different between the control and heat-treated ducks (P < 0.05), among which the abundance of 8 pathways was extremely significant (P < 0.01). In the 29 KEGG pathways, 21 pathways were more abundant, and the other 8 pathways were less abundant by heat treatment. Among these pathways, starch and sucrose metabolism, transcription machinery, and energy metabolism were the most enriched functions, and the abundance of these 3 pathways were increased after heat treatment (Figure 5A). Compared with the CJ group, the abundance of 21 KEGG pathways was observed significantly different in the HJ group (P < 0.05), in which pathways related to other ion-coupled transporters, methane metabolism, and transcription machinery were the most abundant KEGG pathways. Of the 21 pathways, in the HJ group, 15 pathways were more abundant, whereas 6 pathways were less abundant than in the CJ group. And we found that heat treatment seemed to extremely significantly elevate the abundance of 2 KEGG pathways including germination and glycosphingolipid biosynthesis—lacto and neolacto series (P < 0.01) (Figure 5B). In the ileum, we identified that 5 KEGG pathways were significantly differentially abundant between the control and heat-treated ducks (P < 0.05), among which the ABC transporter pathway was the most enriched. Compared with the control ducks, the heat-treated ducks had higher abundance in the pathway of tetracycline biosynthesis but lower abundance in the pathways related to ABC transporters, adipocytokine signaling, peroxisome proliferator-activated receptor signaling, and styrene degradation (Figure 5C).
Figure 5.
Relative abundance of KEGG pathways of duodenal (A), jejunal (B), and ileal (C) microbiota in the control and heat-treated ducks. Only the KEGG pathways that were significantly affected in percentage by temperature are presented. P value was calculated based on the independent samples t-test. P < 0.05 was considered to be statistically significant (*), whereas P < 0.01 was considered to be extremely statistically significant (**). CD and HD represent the duodenal content samples from the control ducks and heat-treated ducks, respectively. CJ and HJ represent the jejunal contents of the control group and heat-treated group, respectively. CI and HI were the samples of ileal contents from the control ducks and heat-treated ducks, respectively. Abbreviations: MAPK, mitogen-actived protein kinase; PPAR, peroxisome proliferator-activated receptor.
Discussion
The gastrointestinal tract of vertebrates is colonized by a diverse microbial community which represents one of the most densely populated microbial ecosystems in the body, and there is growing evidence that the intestinal microbiota plays a key role in nutrition absorption, productive performance, and health (Flint et al., 2012, Crisol-Martínez et al., 2017). In hot seasons, poultry shows decreased pathogen resistance, which often leads to the occurrence of intestinal bacterial and viral diseases (Alhenaky et al., 2017), results in decreased productivity (Lara and Rostagno, 2013), and causes heavy economic losses in the poultry industry (Nawab et al., 2018). Previous reports have shown that the structure of chicken gut microflora is closely related to the environmental factors such as temperature (Wang et al., 2018, Zhu et al., 2019). To understand the effect of high temperature on the intestinal microbiota, we investigated the alterations of the small intestinal microbiome in ducks using high-throughput 16S rRNA gene sequencing after heat treatment.
As per the 16S rRNA gene sequencing results, we observed a complex and diverse microbial community in the 3 intestinal locations (duodenum, jejunum, and ileum) of ducks by taxonomic analysis. The results showed that Firmicutes, Proteobacteria, and Bacteroidetes were the 3 most abundant phyla in the duodenum and jejunum of ducks, and Firmicutes, Actinobacteria, and Proteobacteria were the dominant phyla in the ileum of ducks. The major phyla of the avian intestinal microbiome are Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria (Wei et al., 2013, Best et al., 2017, Xiao et al., 2017, Wang et al., 2018), which is in accordance with our findings. Compared with the duodenum and jejunum of the control ducks, the abundance of Firmicutes and Bacteroidetes was increased, but the abundance of Proteobacteria was decreased after heat treatment. In the ileum, the abundance of Firmicutes and Proteobacteria in the heat-treated group was higher than in the control group, while the abundance of Actinobacteria in the heat-treated ducks was lower than in the controls. Wang et al. (2018) demonstrated that heat stress increased the viable counts of Firmicutes and reduced the viable counts of Proteobacteria and Bacteroidetes in the small intestinal contents of broilers, which is slightly different from the results found in this study. This may be due to differences in host genetics, age, gastrointestinal tract location, diet, and environmental factors (Kurilshikov et al., 2017, Clavijo and Flórez, 2018). At the genus level, we found that the genus of Lactobacillus was the predominant bacteria across 3 intestinal locations, and the richness of the Lactobacillus bacteria was higher in the heat-treated ducks than in the controls. Previous reports have shown that Lactobacillus strains could improve intestinal microbiota, morphology, and barrier integrity and enhance performance of broiler chickens raised under heat conditions (Song et al., 2014, Faseleh Jahromi et al., 2016). Therefore, we postulate that the changes in the abundance of Lactobacillus might be associated with the adaptability of ducks subjected to prolonged heat treatment.
The alpha diversity values were used to evaluate the richness (Chao1 and Observed species) and diversity (Shannon and Simpson) of gut microbiota. High temperature increased the ileal microbial species richness (Chao1 and Observed species), but the Shannon and Simpson indices were unaltered in broilers after heat treatment (Wang et al., 2018). Similarly, we observed that the high temperature altered the Shannon and Simpson indices in the 3 intestinal locations of ducks, but there was no statistical significant difference in these changes, which indicated that high temperature might have no remarkable impact on the diversity of the gut microbiome in poultry. Although, the Chao1 and Observed species indices in each intestinal tract region were not significantly different between the controls and heat-treated ducks, the richness values of duodenal microbial community in the heat-treated group were higher than those in the control, and this trend is consistent with the previous results in broilers (Wang et al., 2018).
Furthermore, we investigated the specific taxa in duodenal, jejunal, and ileal microbiota that were significantly associated with high temperature, which allowed us to determine whether high ambient temperature could change the intestinal microbial community and functions. The present study indicated that abundance of Enterococcus decreased after the ducks were exposed to high temperature, suggesting that the abundance of Enterococcus in the gut contents was significantly affected and sensitive to high temperature, which is consistent with previous results in silkworms (Sun et al., 2017). Enterococcus is characterized by its prebiotic properties in the intestine, which has been reported to have beneficial effects on enhancing the intestinal barrier function (Pedicord et al., 2016, Huang et al., 2019). Previously, we found that high temperatures caused damage to the intestinal mucosa of ducks (Tian et al., 2019). Therefore, it can be inferred that adverse impacts on the integrity of the intestinal mucosal barrier induced by high ambient temperature may be related to the decrease in the abundance of Enterococcus. The dietary supplementation with Weissella koreensis powder improved growth performance in pigs and had a beneficial effect on the immune response during an inflammatory challenge (Wang et al., 2011). A study has revealed that Weissella species played important roles in preventing lipopolysaccharide-induced proinflammatory stress in murine macrophages and in human intestinal epithelial cells (Singh et al., 2018). The Lactococcus genus is considered a potential etiological agent in the mastitis outbreak investigation (Rodrigues et al., 2016), and Lactococcus garvieae is the causative agent of lactococcosis, a hyperacute and hemorrhagic septicemia of fish (Meyburgh et al., 2017). The reduced abundance of Lactococcus, Geobacillus, and Pseudomonas might be associated with the decrease of muscularis thickness and the height of intestinal microvillus, as well as the upregulation of relative expression of IL-1β, IL-10, and IL-17F (Miao et al., 2018). High ambient temperature causes deleterious impacts on intestinal morphology and mucosal barrier integrity in poultry (Farag and Alagawany, 2018), and this has been confirmed by our previous work (Tian et al., 2019). Thus, the variation in the abundance of Enterococcus, Weissella, Lactococcus, Geobacillus, and Pseudomonas observed in the present study may be associated with intestinal injury induced by heat treatment.
Subsequently, the differences in gut microbial community were calculated and ordinated in two-dimensional NMDS plots based on the weighted UniFrac distance. Our results showed that weak separation was found in the ordination plots of the duodenal, jejunal, and ileal microbial communities in ducks raised under different conditions. It may not be unexpected that NMDS ordination of weighted UniFrac distance did not show distinct group separation. Therefore, additional statistical tests may be more informative than ordinations for identifying temperature-associated taxa.
It has been well understood that gut bacteria play a fundamental role in maintaining the health of the host (Kåhrström et al., 2016, Buford, 2017). Functional prediction results revealed that amino acid transport and metabolism, transcription, and carbohydrate transport and metabolism were more abundant in the microbiota community from different small intestinal positions of ducks. In addition, we found that there were differences in metabolism of the small intestinal microbiome in ducks raised under different temperature conditions, which was in accordance with the previous reports on other animals (Tian et al., 2015, Zhu et al., 2019). Increased evidence suggests that environmental stressors could induce increased gut permeability allowing bacteria to cross the epithelial barrier, then activate mucosal immune response, which in turn alters the community and composition of gut microbiome (Dinan and Cryan, 2012, Brzozowski et al., 2016). It can be proposed that defense mechanisms enable animals to adapt to high ambient temperature more easily in response to environmental changes. Indeed, we found that defense mechanisms in the duodenal flora were significantly increased by heat treatment. However, our findings demonstrated that posttranslational modification, protein turnover, and chaperones were decreased by high temperature in the microflora of the 3 intestinal segments and significantly reduced in the jejunal flora, which does not agree with the report of Zhu et al. (2019) on chickens. We speculate that heat treatment time and animal species may have contributed to this difference.
The gut microbiota in ducks raised under control and high-temperature conditions was further analyzed for pathway enrichment to predict the functions and gain insights into the potential roles in management practices. Regarding the KEGG pathways with significant differences in abundance after heat treatment, the pathways related to starch and sucrose metabolism, transcription machinery, and energy metabolism were enriched in the duodenum; pathways related to other ion-coupled transporters, methane metabolism, and transcription machinery were the top 3 enriched pathways in the jejunum; and the ABC transporter pathway was enriched in the ileum. The findings of the present study showed that starch and sucrose metabolism, transcription machinery, energy metabolism, and methane metabolism were significantly enhanced after heat treatment, whereas pathways related to other ion-coupled transporters and ABC transporters significantly weakened. The enrichment of KEGG pathways including transcription machinery, starch and sucrose metabolism, and methane metabolism was found to be associated with metabolic dysfunction caused by high-fat diet in mice (Tan et al., 2018, Zheng et al., 2018). It suggests that the weight loss in animals during hot seasons may be due to changes in those pathways. Interestingly, most of the KEGG pathways with differences in abundance caused by high temperature are related to metabolism. Therefore, we expected that heat treatment would change the intestinal flora of ducks and thus affect their metabolic functions.
In summary, we provide comparative characterization of microbial communities in ducks raised under control and high-temperature conditions. Our results revealed that heat treatment caused significant taxonomic changes in the intestinal microbiome of ducks, which in turn altered the function features of intestinal flora. It is noteworthy that the major changes in these functions are associated with the metabolic pathways, which may be related to the weight loss of animals in hot seasons. However, further investigations will be required to confirm it.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 31402066), the National Waterfowl Industry Technology System of China (grant no. CARS-42-6), the basic public welfare research program of Zhejiang Province (grant no. LGN18C170003), and the Natural Science Foundation of Zhejiang Province (LQ17C170003).
Conflict of interest statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2019.12.046.
Supplementary data
References
- Alhenaky A., Abdelqader A., Abuajamieh M., Al-Fataftah A.R. The effect of heat stress on intestinal integrity and Salmonella invasion in broiler birds. J. Therm. Biol. 2017;70:9–14. doi: 10.1016/j.jtherbio.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Awad E.A., Idrus Z., Soleimani Farjam A., Bello A.U., Jahromi M.F. Growth performance, duodenal morphology and the caecal microbial population in female broiler chickens fed glycine-fortified low protein diets under heat stress conditions. Br. Poult. Sci. 2018;59:340–348. doi: 10.1080/00071668.2018.1440377. [DOI] [PubMed] [Google Scholar]
- Best A.A., Porter A.L., Fraley S.M., Fraley G.S. Characterization of gut microbiome Dynamics in developing Pekin ducks and impact of management system. Front. Microbiol. 2017;7:2125. doi: 10.3389/fmicb.2016.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski B., Mazur-Bialy A., Pajdo R., Kwiecien S., Bilski J., Zwolinska-Wcislo M., Mach T., Brzozowski T. Mechanisms by which stress affects the experimental and Clinical inflammatory Bowel disease (IBD): role of Brain-gut Axis. Curr. Neuropharmacol. 2016;14:892–900. doi: 10.2174/1570159X14666160404124127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buford T.W. (Dis) Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5:80. doi: 10.1186/s40168-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo V., Flórez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult. Sci. 2018;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisol-Martínez E., Stanley D., Geier M.S., Hughes R.J., Moore R.J. Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: linking gut microbiota and growth performance in chickens. Appl. Microbiol. Biotechnol. 2017;101:4547–4559. doi: 10.1007/s00253-017-8193-9. [DOI] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., Biddinger S.B., Dutton R.J., Turnbaugh P.J. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan T.G., Cryan J.F. Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37:1369–1378. doi: 10.1016/j.psyneuen.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M.R., Alagawany M. Physiological alterations of poultry to the high environmental temperature. J. Therm. Biol. 2018;76:101–106. doi: 10.1016/j.jtherbio.2018.07.012. [DOI] [PubMed] [Google Scholar]
- Faseleh Jahromi M., Wesam Altaher Y., Shokryazdan P., Ebrahimi R., Ebrahimi M., Idrus Z., Tufarelli V., Liang J.B. Dietary supplementation of a mixture of Lactobacillus strains enhances performance of broiler chickens raised under heat stress conditions. Int. J. Biometeorol. 2016;60:1099–1110. doi: 10.1007/s00484-015-1103-x. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Scott K.P., Louis P., Duncan S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- Fontaine S.S., Novarro A.J., Kohl K.D. Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J. Exp. Biol. 2018;221:jeb187559. doi: 10.1242/jeb.187559. [DOI] [PubMed] [Google Scholar]
- Freestone P.P., Sandrini S.M., Haigh R.D., Lyte M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 2008;16:55–64. doi: 10.1016/j.tim.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N.N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J.M., Topping D.L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Heiss C.N., Olofsson L.E. Gut microbiota-Dependent Modulation of energy metabolism. J. Innate Immun. 2018;10:163–171. doi: 10.1159/000481519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Luo L., Zhang Y., Wang Z., Xia Z. Effects of the dietary probiotic, Enterococcus faecium NCIMB11181, on the intestinal barrier and system immune Status in Escherichia coli O78-challenged broiler chickens. Probiotics Antimicrob. Proteins. 2019;11:946–956. doi: 10.1007/s12602-018-9434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kåhrström C.T., Pariente N., Weiss U. Intestinal microbiota in health and disease. Nature. 2016;535:47. doi: 10.1038/535047a. [DOI] [PubMed] [Google Scholar]
- Kamel N.N., Ahmed A.M.H., Mehaisen G.M.K., Mashaly M.M., Abass A.O. Depression of leukocyte protein synthesis, immune function and growth performance induced by high environmental temperature in broiler chickens. Int. J. Biometeorol. 2017;61:1637–1645. doi: 10.1007/s00484-017-1342-0. [DOI] [PubMed] [Google Scholar]
- Krajmalnik-Brown R., Ilhan Z.E., Kang D.W., DiBaise J.K. Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. 2012;27:201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurilshikov A., Wijmenga C., Fu J., Zhernakova A. Host genetics and gut microbiome: Challenges and Perspectives. Trends Immunol. 2017;38:633–647. doi: 10.1016/j.it.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Lara L.J., Rostagno M.H. Impact of heat stress on poultry production. Animals. 2013;3:356–369. doi: 10.3390/ani3020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Luo X., Zhao X., Zhang A., Jiang N., Yang L., Huang M., Xu L., Ding L., Li M., Guo Z., Li X., Sun J., Zhou J., Feng Y., He H., Wu H., Fu X., Meng H. Gut microbiota correlates with fiber and apparent nutrients digestion in goose. Poult. Sci. 2018;97:3899–3909. doi: 10.3382/ps/pey249. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Round J.L., Kasper D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McKenzie C., Tan J., Macia L., Mackay C.R. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev. 2017;278:277–295. doi: 10.1111/imr.12556. [DOI] [PubMed] [Google Scholar]
- Meyburgh C.M., Bragg R.R., Boucher C.E. Lactococcus garvieae: an emerging bacterial pathogen of fish. Dis. Aquat. Organ. 2017;123:67–79. doi: 10.3354/dao03083. [DOI] [PubMed] [Google Scholar]
- Miao S., Zhao C., Zhu J., Hu J., Dong X., Sun L. Dietary soybean meal affects intestinal homoeostasis by altering the microbiota, morphology and inflammatory cytokine gene expression in northern snakehead. Sci. Rep. 2018;8:113. doi: 10.1038/s41598-017-18430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S.R., Collins J.J., Relman D.A. Antibiotics and the gut microbiota. J. Clin. Invest. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone A., Ronchi B., Lacetera N., Ranieri M.S., Bernabucci U. Effects of climate changes on animal production and sustainability of livestock systems. Livest. Sci. 2010;130:57–69. [Google Scholar]
- Nawab A., Ibtisham F., Li G., Kieser B., Wu J., Liu W., Zhao Y., Nawab Y., Li K., Xiao M., An L. Heat stress in poultry production: Mitigation strategies to overcome the future Challenges facing the global poultry industry. J. Therm. Biol. 2018;78:131–139. doi: 10.1016/j.jtherbio.2018.08.010. [DOI] [PubMed] [Google Scholar]
- O'Toole P.W., Jeffery I.B. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- Pedicord V.A., Lockhart A.A.K., Rangan K.J., Craig J.W., Loschko J., Rogoz A., Hang H.C., Mucida D. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol. 2016;1:eaai7732. doi: 10.1126/sciimmunol.aai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M.X., Lima S.F., Higgins C.H., Canniatti-Brazaca S.G., Bicalho R.C. The Lactococcus genus as a potential emerging mastitis pathogen group: a report on an outbreak investigation. J. Dairy Sci. 2016;99:9864–9874. doi: 10.3168/jds.2016-11143. [DOI] [PubMed] [Google Scholar]
- Schellenberg J.J., Dumonceaux T.J., Hill J.E., Kimani J., Jaoko W., Wachihi C., Mungai J.N., Lane M., Fowke K.R., Ball T.B., Plummer F.A. Selection, phenotyping and identification of acid and hydrogen peroxide producing bacteria from vaginal samples of Canadian and East African women. PLoS One. 2012;7:e41217. doi: 10.1371/journal.pone.0041217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semova I., Carten J.D., Stombaugh J., Mackey L.C., Knight R., Farber S.A., Rawls J.F. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Bhatia R., Singh A., Singh P., Kaur R., Khare P., Purama R.K., Boparai R.K., Rishi P., Ambalam P., Bhadada S.K., Bishnoi M., Kaur J., Kondepudi K.K. Probiotic attributes and prevention of LPS-induced pro-inflammatory stress in RAW264.7 macrophages and human intestinal epithelial cell line (Caco-2) by newly isolated Weissella cibaria strains. Food Funct. 2018;9:1254–1264. doi: 10.1039/c7fo00469a. [DOI] [PubMed] [Google Scholar]
- Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., Garrett W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M.U., Hume M.E., Byrd J.A., Nisbet D.J., Shabbir M.Z., Ijaz A., Rehman H. Molecular analysis of the caecal and tracheal microbiome of heat-stressed broilers supplemented with prebiotic and probiotic. Avian Pathol. 2015;44:67–74. doi: 10.1080/03079457.2015.1004622. [DOI] [PubMed] [Google Scholar]
- Song J., Xiao K., Ke Y.L., Jiao L.F., Hu C.H., Diao Q.Y., Shi B., Zou X.T. Effect of a probiotic mixture on intestinal microflora, morphology, and barrier integrity of broilers subjected to heat stress. Poult. Sci. 2014;93:581–588. doi: 10.3382/ps.2013-03455. [DOI] [PubMed] [Google Scholar]
- St-Pierre N.R., Cobanov B., Schnitkey G. Economic losses from heat stress by US livestock industries. J. Dairy Sci. 2003;86:E52–E77. [Google Scholar]
- Sun Z., Kumar D., Cao G., Zhu L., Liu B., Zhu M., Liang Z., Kuang S., Chen F., Feng Y., Hu X., Xue R., Gong C. Effects of transient high temperature treatment on the intestinal flora of the silkworm Bombyx mori. Sci. Rep. 2017;7:3349. doi: 10.1038/s41598-017-03565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiishi T., Fenero C.I.M., Câmara N.O.S. Intestinal barrier and gut microbiota: shaping our immune responses throughout life. Tissue Barriers. 2017;5:e1373208. doi: 10.1080/21688370.2017.1373208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Caparros-Martin J.A., Matthews V.B., Koch H., O'Gara F., Croft K.D., Ward N.C. Isoquercetin and inulin synergistically modulate the gut microbiome to prevent development of the metabolic syndrome in mice fed a high fat diet. Sci. Rep. 2018;8:10100. doi: 10.1038/s41598-018-28521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Li G., Bu X., Shen J., Tao Z., Chen L., Zeng T., Du X., Lu L. Changes in morphology and miRNAs expression in small intestines of Shaoxing ducks in response to high temperature. Mol. Biol. Rep. 2019;46:3843–3856. doi: 10.1007/s11033-019-04827-2. [DOI] [PubMed] [Google Scholar]
- Tian H., Wang W., Zheng N., Cheng J., Li S., Zhang Y., Wang J. Identification of diagnostic biomarkers and metabolic pathway shifts of heat- stressed lactating dairy cows. J. Proteomics. 2015;125:17–28. doi: 10.1016/j.jprot.2015.04.014. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity- associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Verbrugghe E., Boyen F., Gaastra W., Bekhuis L., Leyman B., Van Parys A., Haesebrouck F., Pasmans F. The complex interplay between stress and bacterial infections in animals. Vet. Microbiol. 2012;155:115–127. doi: 10.1016/j.vetmic.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Wang X.J., Feng J.H., Zhang M.H., Li X.M., Ma D.D., Chang S.S. Effects of high ambient temperature on the community structure and composition of ileal microbiome of broilers. Poult. Sci. 2018;97:2153–2158. doi: 10.3382/ps/pey032. [DOI] [PubMed] [Google Scholar]
- Wang J.P., Yoo J.S., Jang H.D., Lee J.H., Cho J.H., Kim I.H. Effect of dietary fermented garlic by Weissella koreensis powder on growth performance, blood characteristics, and immune response of growing pigs challenged with Escherichia coli lipopolysaccharide. J. Anim. Sci. 2011;89:2123–2131. doi: 10.2527/jas.2010-3186. [DOI] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Xiang Y., Zhou W., Chen J., Li K., Yang H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2017;96:1387–1393. doi: 10.3382/ps/pew372. [DOI] [PubMed] [Google Scholar]
- Yang H., Huang X., Fang S., He M., Zhao Y., Wu Z., Yang M., Zhang Z., Chen C., Huang L. Unraveling the fecal microbiota and Metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 2017;8:1555. doi: 10.3389/fmicb.2017.01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Li H., Zhang X., Jiang M., Luo C., Lu Z., Xu Z., Shi J. Prebiotic Mannan- Oligosaccharides Augment the Hypoglycemic effects of Metformin in Correlation with Modulating gut microbiota. J. Agric. Food Chem. 2018;66:5821–5831. doi: 10.1021/acs.jafc.8b00829. [DOI] [PubMed] [Google Scholar]
- Zhu L., Liao R., Wu N., Zhu G., Yang C. Heat stress mediates changes in fecal microbiome and functional pathways of laying hens. Appl. Microbiol. Biotechnol. 2019;103:461–472. doi: 10.1007/s00253-018-9465-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.