Abstract
Background
Nasopharyngeal carcinoma (NPC) is primarily caused by the Epstein-Barr virus (EBV) infection in NPC endemic areas. EBNA1 is an EBV-encoded nuclear antigen, which plays a critical role in the maintenance and replication of EBV genome. However, the mechanisms of EBNA1-promoted NPC immune escape are unknown. Regulatory T (Treg) cells are among the key regulators in restraining antitumor responses. However, the mechanisms of accumulation of Treg cells in NPC have not been defined. This study attempted to identify the detailed mechanisms of EBNA1 functions as a tumor accelerator to promote NPC immune escape by enhancing chemoattraction of Treg cells.
Methods
mRNA profiles were determined by next-generation sequencing in NPC cells. In vitro and in vivo assays were performed to analyze the role of EBNA1 in regulation of recruitment of Treg cells. Colocation and coimmunoprecipitation analyzes were used to identify the SMAD3/c-JUN complex. Chromatin immunoprecipitation assay and dual luciferase reporter assays were designed to demonstrate c-JUN binding to miR-200a promoter and miR-200a targeting to CXCL12 3’Untranslated Regions. The hepatocellular carcinoma models were designed to demonstrate universality of the CXCL12-CXCR4-Treg axis in promoting immune evasion of various tumors.
Result
A novel molecular mechanism was identified that involves EBV-EBNA1-stimulated chemotactic migration of Treg cells toward NPC microenvironment by upregulation of the transforming growth factor-β1 (TGFβ1)-SMAD3-PI3K-AKT-c-JUN-CXCL12-CXCR4 axis and downregulation of miR-200a. EBV-EBNA1 promotes the chemoattraction of Treg cells by governing the protein–protein interactions of the SMAD3/c-JUN complex in a TGFβ1-dependent manner in vitro and in vivo. TGFβ1 suppresses miR-200a by enhancing the SMAD3/c-JUN complex. miR-200a negatively regulates the CXCL12 chemokine by direct targeting of the CXCL12 3’UTR region. However, CXCL12 acts as the target gene of miR-200a and as an inhibitor of miR-200a transcription, and inhibition of miR-200a by CXCL12 is mediated by c-JUN, which directly binds to the miR-200a promoter and forms a c-JUN-miR-200a-CXCL12-c-JUN feedback loop. In addition, enhanced CXCL12 efficiently attracts CXCR4-positive Treg cells to remodel an immunosuppressive microenvironment.
Conclusions
EBV-EBNA1 promotes chemotactic migration of Treg cells via the TGFβ1-SMAD3-PI3K-AKT-c-JUN-miR-200a-CXCL12-CXCR4 axis in the NPC microenvironment. These results suggest that EBV-EBNA1 may serve as a potential therapeutic target to reshape the NPC microenvironment.
Keywords: head and neck neoplasms, immunomodulation, tumor microenvironment, immune evation
Introduction
Statistical data indicate that 129 079 new nasopharyngeal carcinoma (NPC) cases were reported in 2018 worldwide, and approximately, 71% of new cases were in East and Southeast Asia.1 In the NPC-endemic areas, Epstein-Barr virus (EBV) infection is believed to be necessary for NPC development and is considered the most important aetiological factor in pathogenesis.2 Many studies demonstrated that EBV employs a wide range of immune evasion strategies to interfere with the innate and adaptive immune responses during latent infection.3 A better understanding of the immune evasion mechanisms of EBV will improve the development of novel EBV-directed therapies in NPC. However, the exact mechanisms of the process remain unknown.
EBNA1 is an EBV-encoded nuclear antigen, which plays a critical role in the maintenance and replication of EBV genome and postmitotic EBV genome segregation and is the only EBV protein expressed in all types of EBV-infected cells.4 Numerous studies have focused on the proliferation, invasion and metastasis induced by EBNA15–7 and the significance of serum EBNA1 antibody for disease diagnosis and prognosis.8–10 However, the role of EBV-EBNA1 in NPC immune escape is unknown.
Regulatory T (Treg) cells express CD4+CD25+CD127low/- cell surface markers and FOXP3+ intracellular marker11 and are regarded as a predictor of unfavorable prognosis in multiple cancers.12 Our previous report demonstrated a significant positive correlation between the density of EBV-DNA in the plasma and Treg cells in the peripheral blood of NPC patients.13 We also found that EBV-EBNA1-positive NPC cells converted naïve T cells into Treg cells by upregulation of transforming growth factor-β1 (TGFβ1) in vitro.14 However, the role and immanent mechanism of the effect of EBV-EBNA1 on the chemotactic migration of Treg cells have not been investigated. Therefore, we attempted to identify the detailed mechanisms of EBV-EBNA1-promoted chemoattraction of Treg cells, which promotes NPC immune escape.
Methods
Western blot analysis and immunofluorescence
Western blot and immunofluorescence were performed according to an established method.15
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) assay was performed to test whether c-JUN is targeted to the miR-200a promoter region using a Pierce agarose ChIP kit (Thermo Scientific, catalog: 26156) according to the manufacturer’s instructions.
Coimmunoprecipitation assay
Coimmunoprecipitation (Co-IP) assay was performed to test the protein-protein interactions (PPI) between c-JUN and SMAD3 proteins using a Pierce Classic magnetic IP/Co-IP kit (Thermo Scientific, catalog: 88804) according to the manufacturer’s instructions.
Luciferase reporter assay
Luciferase reporter assay was performed to test whether miR-200a directly binds to the CXCL12 3’UTR region and whether c-JUN directly binds to the miR-200a promoter region using a dual luciferase reporter gene assay kit (Beyotime Biotechnology, catalog: RG027) according to the manufacturer’s instructions. All dual luciferase reporter vectors contained specific wild-type target sequences or specific mutant sequences.
Facs
Single-cell suspensions were stained with monoclonal antibodies and analyzed using a BD FACScanto instrument according to an established method.16 17 Cells were resuspended in PBS and stained with CD4, CD8, CD25 and CD127 monoclonal antibodies for 15–30 min. CD4 positive, CD25 positive, CD127 negative or low expression and CD8 negative cells (CD4 +CD25+CD127-/low CD8-) were identified as Treg cells. Please refer to online supplemental table 2 for the detailed description of antibodies.
jitc-2020-001588supp001.pdf (144.5KB, pdf)
A detailed description of the methods and materials can be found in the online supplemental Information.
jitc-2020-001588supp002.pdf (193.8KB, pdf)
Results
EBV-EBNA1 promotes chemotactic migration of Treg cells by activating TGFβ1-SMAD3 signaling
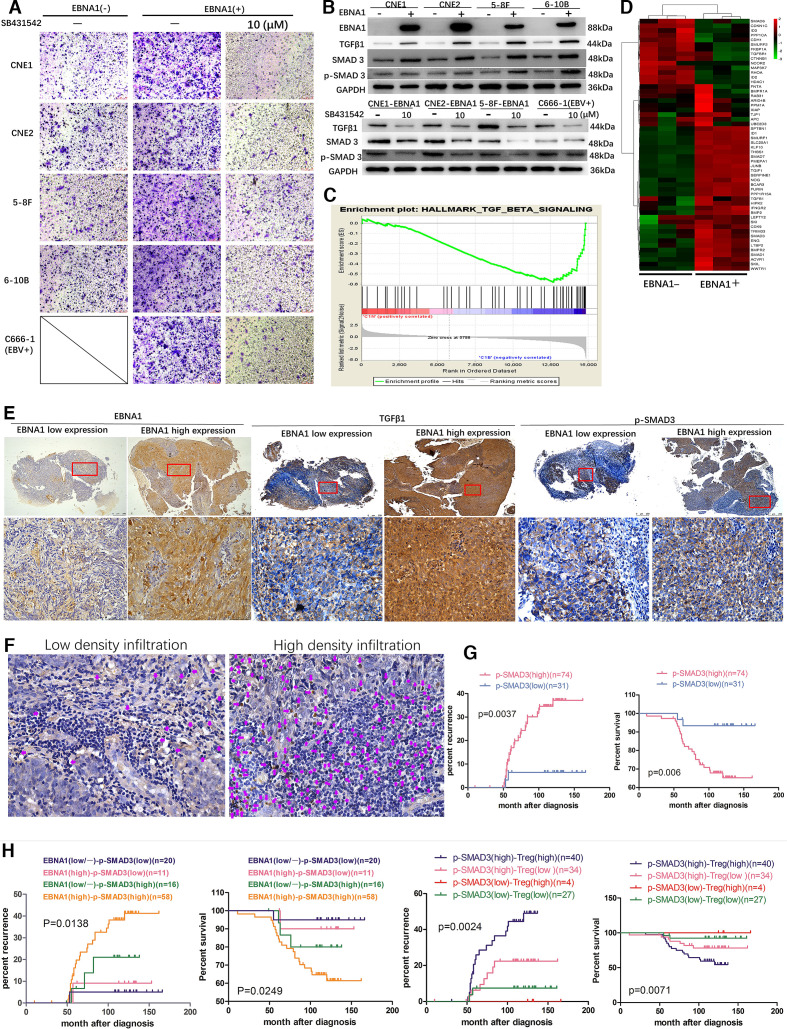
Our previous study showed a significant positive correlation between the density of EBV-DNA in the plasma and Treg cells in the peripheral blood of NPC patients.13 Moreover, EBV-EBNA1-positive NPC cells were shown to convert naïve T cells into Treg cells by upregulation of TGFβ1 in vitro.14 Hence, we speculated that TGFβ1 plays certain roles in the production of Treg cells and probably in chemotactic migration of Treg cells in EBV-EBNA1-positive NPC microenvironment. The initial results of in vitro migration assay of Treg cells and western blot analysis indicated that the culture supernatant of EBNA1-positive NPC cells promotes the tropism and migration of Treg cells compared with that in the EBNA1-negative groups; however, SB431542 (a specific inhibitor of type I TGFβ receptor kinase) blocked the expression of TGFβ1 induced by EBNA1 and reversed the migration of Treg cells driven by EBNA1 (figure 1A, B, (online supplemental figure 1A). To identify potential molecular mechanisms associated with EBV-EBNA1, TGFβ1 and migration of Treg cells, mRNA expression profile was determined by next-generation sequencing in EBNA1-negative and -positive NPC cells. Gene set enrichment analysis showed that TGFβ signaling is enriched in the EBNA1-positive group in the hallmark gene sets database (figure 1C and online supplemental figure 1B). Hierarchical clustering analysis of TGFβ signaling showed that SMAD3 was upregulated in EBNA1-positive cells (figure 1D). Since SMAD family is intracellular signal transducer and transcriptional modulator activated by the TGFβ family,18 19 the activation level of TGFβ1 signaling was measured based on the status of SMAD3. TGFβ1, SMAD3 and phospho-SMAD3 (p-SMAD3) expression levels were considerably higher in EBNA1-positive cells, which is consistent with the sequencing results (figure 1B and online supplemental figure 1C). Moreover, IHC detection in 105 tumor specimens of NPC patients indicated that p-SMAD3 expression is positively correlated with EBV-EBNA1 and TGFβ1 expression, infiltration of Treg cells, advanced clinical stage, higher risk of recurrence, metastasis and death, and shorter overall survival (OS) in NPC patients (figure 1E-G, (online supplemental figure 1D) and online supplemental tables 3 and 4). Patients with EBNA1highp-SMAD3high expression pattern or patients with p-SMAD3highTreghigh pathological feature had the worst prognosis (figure 1H). Collectively, these findings suggest that EBV-EBNA1 promotes chemotactic migration of Treg cells by activating TGFβ1-SMAD3 signaling and that enhanced TGFβ1-SMAD3 axis is an indicator of poor prognosis.
Figure 1.
EBV-EBNA1 promotes migration of Treg cells by activating TGFβ1-SMAD3 signaling. (A) SB431542 blocked the migration of Treg cells induced by EBNA1. (B) Upper: TGFβ1, SMAD3 and p- SMAD3 protein expression in EBNA1-positive and -negative NPC cell lines assayed by western blot (WB). Lower: SB431542 blocked the expression of TGFβ1, SMAD3 and p-SMAD3 induced by EBNA1. (C) TGFβ signaling is enriched in EBNA1-positive group in the hallmark gene sets database according to GSEA. (D) Differentially expressed genes in EBNA1-negative and -positive cells in TGFβ signaling according to hierarchical clustering analysis. (E) Representative EBNA1, TGFβ1 and p-SMAD3 expression in NPC patients in the EBNA1 low and high expression groups according to IHC detection. Scale bars at the bottom right: 500 μm (low magnification) and 75 μm (high magnification). (F) Representative images of tumor-infiltrating Treg cells according to FOXP3 staining. The purple arrows represent FOXP3+ Treg cells in the stroma surrounding the tumor cells. Scale bars at the bottom right: 75 μm. (G) Kaplan-Meier analysis of recurrence risk and overall survival for p-SMAD3 expression groups in 105 NPC patients. (H) Kaplan-Meier analysis of recurrence risk and overall survival for concurrent EBNA1/p-SMAD3 expression groups or concurrent p-SMAD3/Treg pathological feature groups in 105 NPC patients. EBV, Epstein-Barr virus; GSEA, Gene set enrichment analysis; TGFβ, transforming growth factor-β; Treg, regulatory T; IHC, Immunohistochemistry.
jitc-2020-001588supp003.pdf (596.6KB, pdf)
miR-200a suppresses chemotactic migration of Treg cells
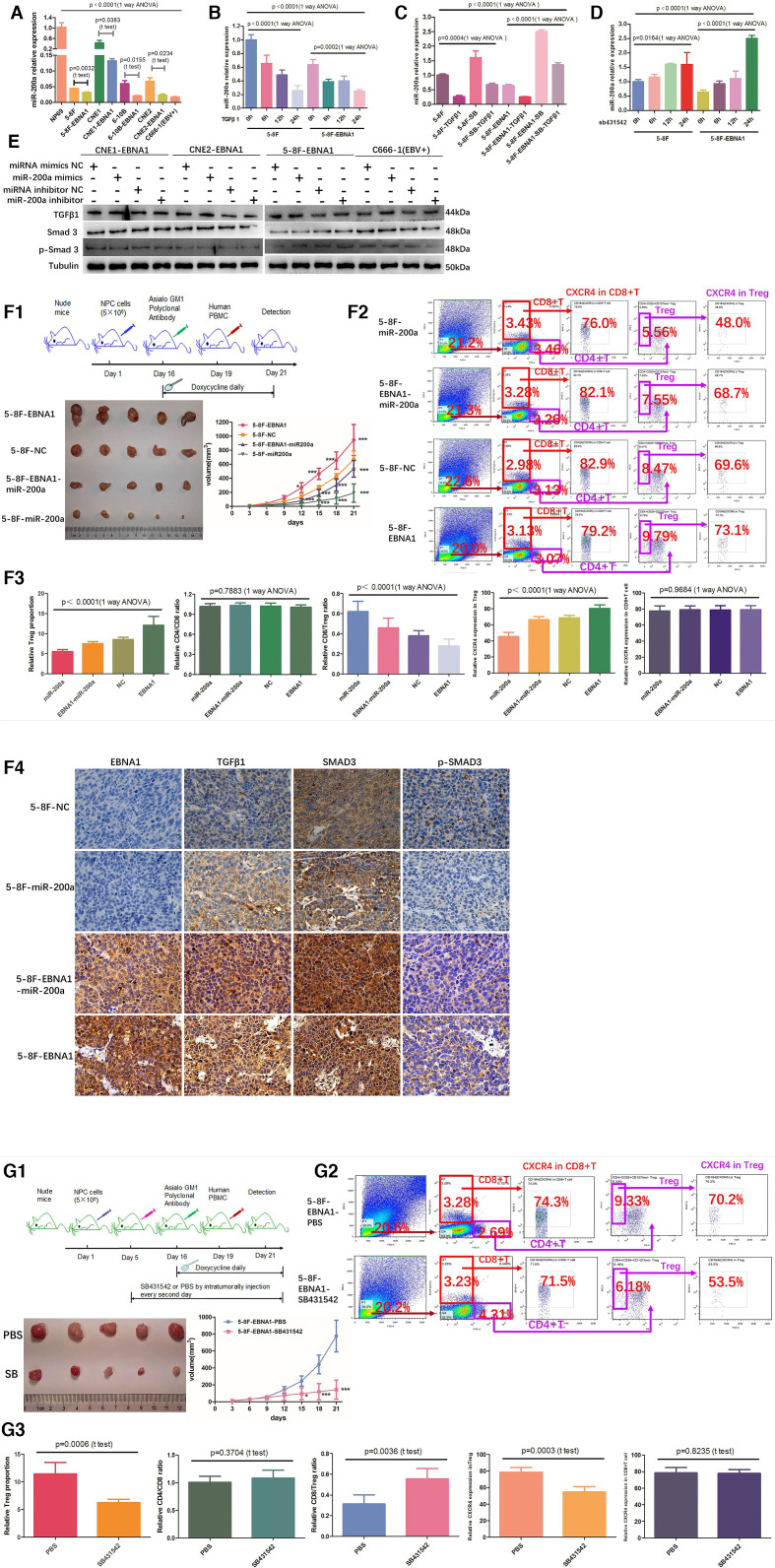
miR-200a is a known tumor suppressor in a wide variety of malignant tumors20 21 and one of the signature markers for prediction of NPC metastasis, and reduction in the expression of miR-200a at the time of diagnosis is associated with poor prognosis22–25; thus, we hypothesized that miR-200a may play a role in chemotactic migration of Treg cells. Initially, a direct effect of EBV-EBNA1-TGFβ1-SMAD3 axis on miR-200a expression was tested by qRT-PCR analysis. The expression level of miR-200a was considerably lower in EBNA1-positive NPC cells (figure 2A). Then, NPC cell lines were treated with recombinant human TGFβ1 protein. TGFβ1 induced a rapid and robust reduction in mature miR-200a in all NPC cell lines in a time-dependent manner, even though the basal level of miR-200a in EBNA1-positive cells was already very low (figure 2B, online supplemental figure 2A). However, SB431542 treatment rescued this decrease in miR-200a levels (figure 2C, online supplemental figure 2B). Then, NPC cells were treated with SB431542 at various time points; the results indicated an increase in the level of miR-200a in a time-dependent manner after the blockade of autocrine TGFβ1 signaling activity, suggesting that the suppressive effect of EBV-EBNA1 on miR-200a is mediated by autocrine action of TGFβ1 (figure 2D, online supplemental figure 2C). However, increasing the levels or blocking miR-200a had no effect on TGFβ1 and SMAD3 expression, indicating that miR-200a is a downstream factor in the TGFβ1-SMAD3 axis (figure 2E, online supplemental figure 2D).
Figure 2.
miR-200a suppresses migration of Treg cells. (A) qRT-PCR analysis of the basal expression level of miR-200a in NP69 and NPC cells with or without EBNA1 protein expression. (B) TGFβ1 suppressed miR-200a expression in a time-dependent manner. (C) SB431542 rescued the blocking effect of TGFβ1 on miR-200a and reversed a decrease in the expression levels of miR-200a induced by TGFβ1. (D) miR-200a expression was elevated in a time-dependent manner after blockade of autocrine TGFβ signaling activity by SB431542. (E) miR-200a had little effect on the TGFβ1, SMAD3 and p-SMAD3 expression according to WB. (F1) EBNA1 enhanced tumor load and accumulation of Treg cells, and miR- 200a overexpression significantly suppressed tumor load and migration of Treg cells in vivo. Upper: animal experiment flow chart. Lower: representative image and tumor growth curves of different tumor formation groups in humanized immune reconstruction mouse model (*p<0.05; ***p<0.001). (F2)(F3) The in vivo tumor-infiltrating CD4+ T cells, CD8+ T cells, Treg cells and CXCR4 (alias CD184) receptor expression levels were detected by FACS. (F4) IHC detection of EBNA1, TGFβ1, SMAD3 and p- SMAD3 expression in various NPC xenograft groups. Scale bars at the bottom right: 75 μm. (G1) Blockade of the TGFβ1-SMAD3 axis in EBNA1-positive tumor by SB431542 inhibited tumor load and migration of Treg cells in vivo. Upper: animal experimental flow diagram. Lower: representative image and tumor growth curves of various tumor formation groups in humanized immune reconstruction mouse model (*p<0.05; ***p<0.001). (G2)(G3) The in vivo tumor-infiltrating CD4+ T cells, CD8+ T cells, Treg cells and CXCR4 (alias CD184) receptor expression levels were detected by FACS. ANOVA, analysis of variance; EBV, Epstein-Barr virus; NPC, nasopharyngeal carcinoma; TGFβ1, transforming growth factor-β1; Treg, regulatory T; WB, western blot.
jitc-2020-001588supp004.pdf (909.6KB, pdf)
jitc-2020-001588supp005.pdf (920.6KB, pdf)
Furthermore, the in vitro findings were confirmed in a mouse model, indicating that tumor load and proportion of tumor-infiltrating Treg cells were significantly increased in the 5–8F-EBNA1 group and decreased in the 5–8F-miR-200a group compared with those in the 5–8F-NC group (figures 2F1–F3). However, miR-200a overexpression partly suppressed the stimulatory effect of EBNA1 on tumor burden and proportion of tumor-infiltrating of Treg cells (figures 2F1–F3). Numerous studies demonstrated that high CD8/Treg ratio is associated with better recurrence-free survival and better OS, whereas low CD8/Treg ratio is associated with tumor aggressiveness and predicted poor prognosis. In this study, the CD8/Treg ratio was considerably decreased in the 5–8F-EBNA1 group and increased in the 5–8F-miR-200a group compared with that in the 5–8F-NC group (figures 2F2–F3). However, miR-200a overexpression partly suppressed the stimulatory effect of EBNA1 on the CD8/Treg ratio (figures 2F2–F3). These results indicated that EBV-EBNA1 protein enhances chemotactic migration of Treg cells and induces Treg cell-mediated immunosuppressive microenvironment, while overexpression of miR-200a inhibits the migration of Treg cells and partially reverses the stimulatory effect of EBNA1 on the migration of Treg cells. In addition, IHC analysis of NPC xenograft showed that EBV-EBNA1 upregulated the TGFβ1-SMAD3 axis, while miR-200a overexpression had no significant effect on the expression of the upstream components of the TGFβ1-SMAD3 axis, suggesting that miR-200a is a downstream factor of the TGFβ1-SMAD3 axis (figure 2F4). Subsequently, an in vivo rescue experiment was performed to observe the effect of blockade of TGFβ1-SMAD3 axis signal transduction on migration of Treg cells. Blocking the TGFβ1-SMAD3 axis by SB431542 inhibited chemotactic migration of Treg cells and increased the CD8/Treg ratio (figures 2G1–G3). Thus, our results indicate that stimulation of the EBV-EBNA1-TGFβ1-SMAD3 axis promotes chemotactic migration of Treg cells by reducing miR-200a expression.
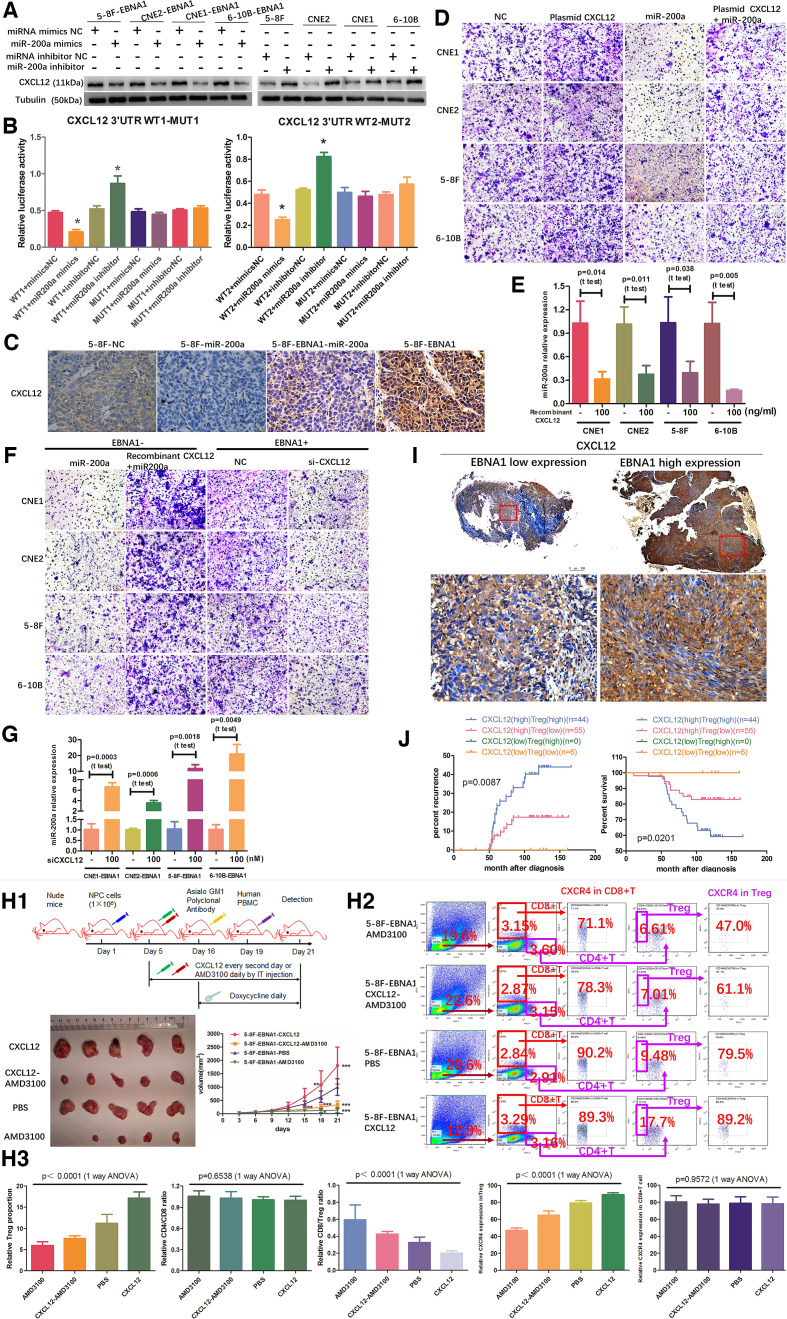
Figure 3.
miR-200a directly targets CXCL12 3’UTR region. (A) miR-200a negatively regulated Journal for immunotherapy for cancer CXCL12 protein expression according to WB. (B) Luciferase reporter assay was used to determine direct targeting of CXCL12 3’UTR by miR-200a. (C) IHC detection of CXCL12 expression in various NPC xenograft groups from online supplemental figure 3F1. Scale bars at the bottom right: 75 μm. (D) Assay of migration of Treg cells verified that overexpression of CXCL12 promotes migration of Treg cells, and miR-200a overexpression inhibited movement of Treg cells and significantly suppresses the stimulatory effect of CXCL12 on migration of Treg cells. (E) overexpression of CXCL12 suppressed the miR-200a expression in EBNA1-negative NPC cell lines. (F) overexpression of CXCL12 decreased the suppressive effect of miR-200a on migration of Treg cells, and suppression of CXCL12 by specific siRNA reduced migration of Treg cells in vitro. (G) miR-200a expression level was elevated after transfection with CXCL12-specific siRNA in EBNA1-positive NPC cell lines according to qRT-PCR. (H1) In vivo migration assay verified that overexpression of CXCL12 promotes migration of Treg cells, and suppression of CXCL12 decreases infiltration of Treg cells. Upper: animal experiment flow chart. Lower: representative image and tumor growth curves of various tumor groups in humanized immune reconstruction mouse model (*p<0.05; **p<0.01; ***p<0.001). (H2) (H3) The in vivo tumor infiltration with CD4+ T cells, CD8+ T cells and Treg cells and CXCR4 (alias CD184) receptor expression levels were detected by FACS. (I) Representative CXCL12 expression in NPC patients in the EBNA1 low and i high expression groups according to IHC. Scale bars at the bottom right: 500 μm (low magnification) and 75 μm (high magnification). (J) Kaplan-Meier analysis of recurrence risk and overall survival for concurrent CXCL12 expression and infiltration of Treg cells groups in 105 NPC patients. ANOVA, analysis of variance; NPC, nasopharyngeal carcinoma; Treg, regulatory T; WB, western blot.
miR-200a directly targets the CXCL12 3’UTR region
Bioinformatics analysis (Targetscan) predicted putative miR-200a target genes and identified CXCL12 among the top hits associated with tumor immune evasion. Two putative miR-200a-targeting sequences were identified in the 3’UTR of human CXCL12 and mouse CXCL12, suggesting that the associations between miR-200a and CXCL12 may be universally applicable in various species (online supplemental figure 3A). Overexpression of miR-200a downregulated CXCL12 protein and blocking miR-200a increased the expression of CXCL12 protein (figure 3A). Interestingly, miR-200a overexpression or suppression had no significant effect on CXCL12 transcript, implying that miR-200a inhibits CXCL12 by a posttranscriptional mechanism (online supplemental figure 3B). Moreover, the results of dual luciferase reporter assay showed that miR-200a mimics significantly suppressed luciferase reporter activity of CXCL12, and miR-200a inhibitor had the opposite effect. However, these effects on luciferase activity were abrogated by cotransfection with mutated CXCL12 3’UTR reporter (figure 3B and online supplemental figure 3C). Then, the association between miR-200a and CXCL12 was verified in an NPC xenograft model. CXCL12 expression was significantly increased in the 5–8F-EBNA1 group and decreased in the 5–8F-miR-200a group compared with that in the 5–8F-NC group (figure 3C). Moreover, miR-200a overexpression partially suppressed the stimulatory effect of EBNA1 on CXCL12 expression (figure 3C). Furthermore, migration assay of Treg cells verified that the culture supernatant of CXCL12-overexpressing NPC cells stimulated tropism and migration of Treg cells, and the culture supernatant of the miR-200a overexpression group inhibited tropism and migration of Treg cells and significantly suppressed the stimulatory effect of CXCL12 on the migration of Treg cells (figure 3D and online supplemental figure 3D).
Interestingly, treatment of NPC cells with a high dose of exogenous recombinant human CXCL12 protein suppressed miR-200a expression (figure 3E) and decreased the suppressive effect of miR-200a on the migration of Treg cells (figure 3F and online supplemental figure 3E). Conversely, miR-200a expression was elevated and tropism and migration of Treg cells was reduced after transfection with CXCL12-specific siRNA (figure 3F, G and online supplemental figure 3E). These results demonstrated that CXCL12 overexpression inhibits miR-200a transcription and blocks the suppressive effect of miR-200a on the migration of Treg cells. Collectively, our data suggest that miR-200a suppresses chemotactic migration of Treg cells by directly targeting the CXCL12 3’UTR and that CXCL12 suppresses miR-200a transcription.
To investigate the regulatory associations between CXCL12 and chemotactic migration of Treg cells, the in vitro findings were verified in an in vivo migration assay; the results indicated that overexpression of CXCL12 enhanced migration of Treg cells and decreased the CD8/Treg ratio; blockade of the CXCL12 axis with AMD3100 (a selective antagonist of CXCL12-mediated chemotaxis) suppressed the migration of Treg cells and increased the CD8/Treg ratio (figure 3H1–H3). Additionally, CXCL12 expression was positively correlated with EBV-EBNA1 expression, infiltration density of Treg cells, advanced clinical stage, high risk of recurrence, metastasis and death in NPC patients (figures 1E and 3I, (online supplemental figure 3F) and online supplemental tables 3 and 4). NPC patients with CXCL12highTreghigh pathological feature had the worst prognosis (figure 3J).
Collectively, our data indicate that EBV-EBNA1 promotes chemotactic migration of Treg cells by upregulating CXCL12 expression, and CXCL12 acts as a target gene of miR-200a and as an inhibitor of miR-200a transcription.
c-Jun directly binds to miR-200a promoter and induces a c-JUN-miR-200a-CXCL12-PI3K-AKT-c-JUN feedback loop
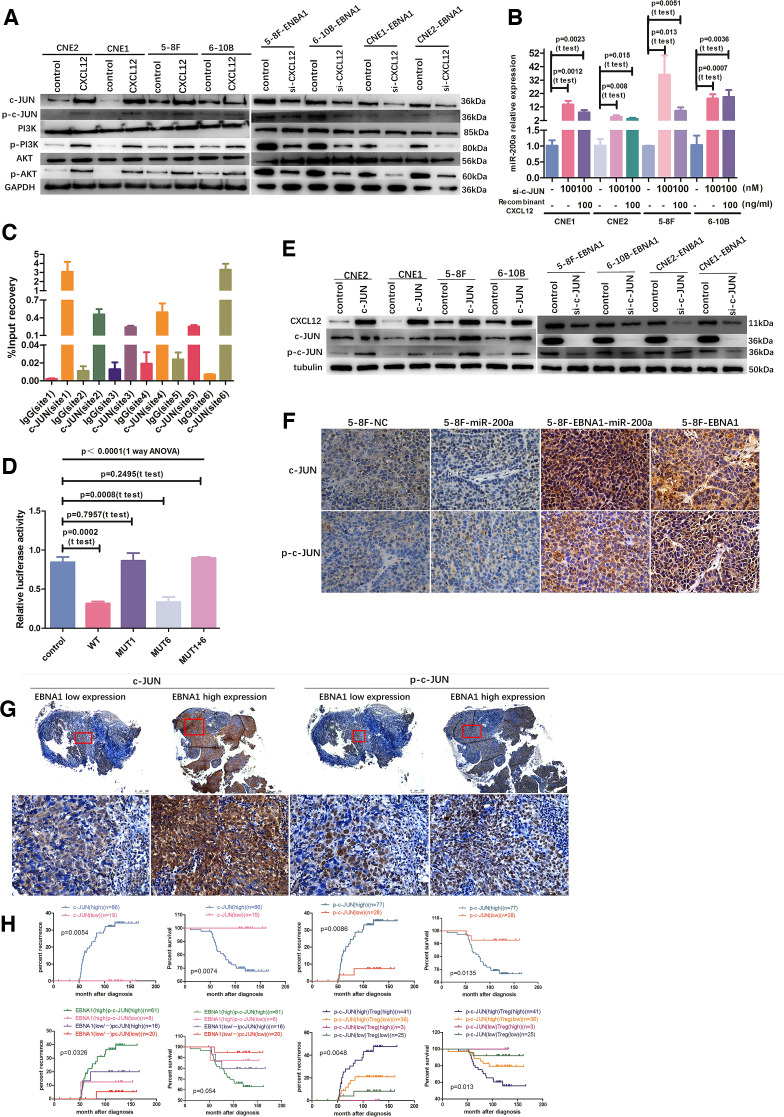
We hypothesized that CXCL12 suppresses miR-200a transcription via certain transcriptional factors, which interact with the miR-200a promoter region. Bioinformatics analysis (UCSO, PROMO, JASPAR and Genomatix) was used to predict possible transcription factors that bind upstream of the 3 kbp miR-200a promoter region. Six putative c-JUN binding sites were identified and named site1, site2, site3, site4, site5 and site6 (online supplemental figure 4A). To evaluate whether CXCL12 regulates miR-200a expression via c-JUN and the signals of the PI3K/AKT pathway upstream of c-JUN, NPC cells were treated with recombinant human CXCL12 protein or CXCL12-specific siRNA, respectively. Overexpression of CXCL12 increased p-PI3K, p-AKT, c-JUN and p-c-JUN expression and did not change the total PI3K and AKT levels; however, blockade of CXCL12 had the opposite effect, suggesting that CXCL12 is an upstream positive regulator of the PI3K-AKT-c-JUN signals (figure 4A). Moreover, c-JUN-specific siRNAs considerably increased miR-200a transcription and rescued the suppressive effect of CXCL12 on miR-200a transcription, suggesting that c-JUN is an upstream regulator of miR-200a (figure 4B). Subsequent ChIP-qPCR assay demonstrated that c-JUN protein was recruited to site1 and site6 (figure 4C). Then, miR-200a promoter dual luciferase reporter assay confirmed the exact association between c-JUN and these two sites (online supplemental figure 4B). WT and MUT6 promoter-dependent luciferase activities were significantly reduced by cotransfection with the c-JUN overexpression plasmid, and MUT1 and MUT1 +6 promoter-dependent luciferase activities did not change, implying that c-JUN suppressed miR-200a promoter-dependent transcription by directly binding at site1 (figure 4D).
Figure 4.
c-JUN directly binds to miR-200a promoter and generates a c-JUN-miR-200a-CXCL12- PI3K-AKT-c-JUN feedback loop. (A) Overexpression of CXCL12 increased PI3K-AKT-c-JUN signaling expression; however, CXCL12-specific siRNA decreased the PI3K-AKT-c-JUN signaling pathway expression. (B) The c-JUN-specific siRNAs increased miR-200a level after blockade of c-JUN and rescued the suppressive effect of CXCL12 overexpression on miR-200a transcription. (C) ChIP assay demonstrated c-JUN binding to the miR-200a promoter region at site1 and site6. (D) miR-200a promoter dual luciferase reporter assay verified that c-JUN directly binds to miR-200a promoter. (E) c-JUN positively regulates CXCL12 expression. (F) IHC detection of c-JUN and p-c-JUN expression in various NPC xenograft groups from figure 3F1. Scale bars at the bottom right: 75 μm. (G) Representative c-JUN and p-c-JUN expression in NPC patients in EBNA1 low and high expression groups according to IHC. Scale bars at the bottom right: 500 μm (low magnification) and 75 μm (high magnification). (H) Kaplan-Meier analysis of recurrence risk and overall survival for c-JUN and phospho-c-JUN expression groups, and the recurrence risk and overall survival for the concurrent EBNA1/p-c-JUN expression groups and the concurrent p-c-JUN expression and infiltration of Treg cells groups in 105 NPC patients. ANOVA, analysis of variance; ChIP, chromatin immunoprecipitation; NPC, nasopharyngeal carcinoma; Treg, regulatory T.
jitc-2020-001588supp006.pdf (648.2KB, pdf)
Interestingly, overexpression of c-JUN upregulated CXCL12 protein and blockade of c-JUN reduced CXCL12 expression, suggesting that CXCL12 is positively regulated by c-JUN and serves as a downstream factor of c-JUN (figure 4E). Moreover, IHC assay in the xenograft model confirmed that CXCL12 expression is positively associated with c-JUN. The c-JUN-CXCL12 axis was dramatically upregulated in the 5–8F-EBNA1 group and downregulated in the 5–8F-miR-200a group compared with that in the 5–8F-NC group (figures 3C and 4F). In NPC patients, c-JUN and p-c-JUN levels were positively correlated with the expression of EBNA1 and CXCL12, infiltration of Treg cells, advanced clinical stage, high risk of recurrence and metastasis, and short OS (figures 1E, 3I, 4G and H, (online supplemental figure 4C) and online supplemental tables 3 and 4). In addition, patients with the EBNA1highp-c-JUNhigh expression pattern suffered from the highest recurrence rate and had a tendency for shorter survival even if it was not statistically significant (figure 4H). Moreover, patients with p-c-JUNhighTreghigh pathological feature had the worst prognosis (figure 4H).
Thus, our study demonstrates the exact molecular mechanism of EBV-EBNA1-dependent regulation of chemotactic migration of Treg cells mediated by the c-JUN-miR-200a-CXCL12-PI3K-AKT-c-JUN feedback loop.
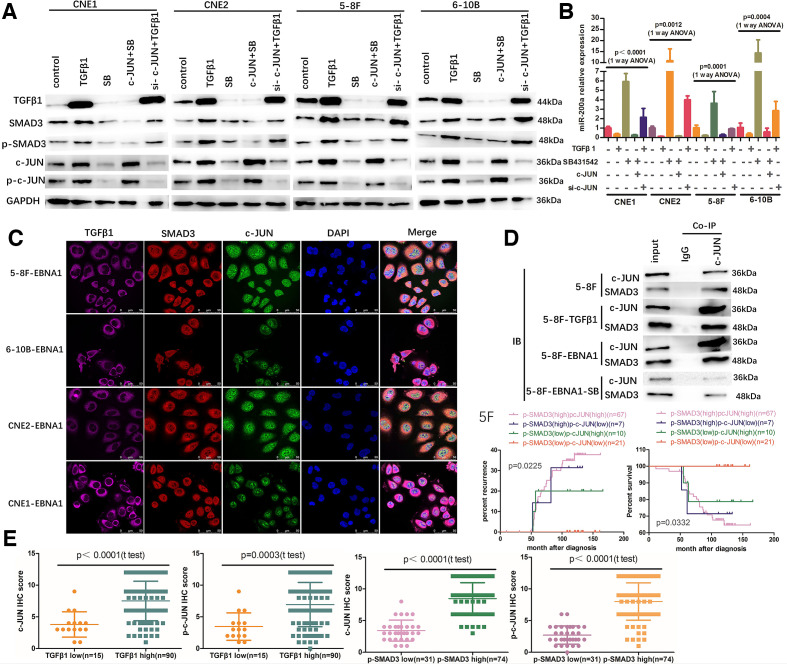
Tgfβ1 suppresses miR-200a by enhancing the formation of the Smad3/c-Jun complex
To evaluate whether TGFβ1 regulates miR200a expression through the c-JUN-dependent pathway, NPC cells were treated with recombinant human TGFβ1 protein or SB431542 in the presence or in the absence of c-JUN expression. TGFβ1 promoted the expression of c-JUN, SMAD3 and p-SMAD3 and suppressed miR200a expression and SB431542 reversed these effects (figure 5A, B). However, c-JUN overexpression by transient transfection neutralized the stimulatory effect of SB431542 on miR200a but did not neutralize the suppressive effect of SB431542 on TGFβ1, SMAD3 or p-SMAD3 (figure 5A, B). Moreover, blockade of c-JUN partially neutralized the suppressive effect of TGFβ1 on miR200a but did not neutralize the stimulatory effect of TGFβ1 on SMAD3 or p-SMAD3 (figure 5A, B). These results suggest that c-JUN is a downstream factor of the TGFβ1-SMAD3 axis and TGFβ1 suppresses miR-200a by stimulating the SMAD3-c-JUN pathway.
Figure 5.
TGFβ1 suppresses miR-200a by enhancing the formation of the SMAD3/c-JUN complex. (A) (B) WB and qRT-PCR detection verified that c-JUN is a downstream factor of the TGFβ1-SMAD3 axis; TGF-β1 suppressed miR-200a by activating the SMAD3-c-JUN pathway. (C) Quadruple-color confocal microscopy colocalization of TGFβ1, SMAD3 and c-JUN. (D) Coimmunoprecipitation (Co-IP) analysis indicated that SMAD3 and c-JUN can form a complex to mediate TGFβ1-induced transcription and that EBNA1 determines the SMAD3/c-JUN interaction in a TGF-β1-dependent manner. (E) NPC patients with high TGFβ1 or high p-SMAD3 expression had higher c-JUN and p-c-JUN expression compared to those in patients with low TGFβ1 or low p-SMAD3 expression. (F) Kaplan-Meier analysis of recurrence risk and overall survival for concurrent p-SMAD3/p-c-JUN expression groups in 105 NPC patients. ANOVA, analysis of variance; NPC, nasopharyngeal carcinoma; TGF-β1, transforming growth factor-β1; WB, western blot.
To analyze the details of the interactions of TGFβ1, SMAD3 and c-JUN, possible PPI were predicted by bioinformatics analysis (STRING, Genomatix, BIOGRID and BioPlex); the results indicated that SMAD3 and c-JUN may form a protein–protein complex at the AP-1/SMAD site. Thus, we hypothesized that TGFβ1, which is activated by EBV-EBNA1, can modulate the transcription and subsequent profound immunosuppression via the SMAD3/c-JUN complex in the microenvironment of EBV-infected NPC cells. Quadruple-color confocal microscopy colocalization analysis demonstrated that TGFβ1 protein is located in the cytoplasm, and SMAD3 and c-JUN proteins are located in the cytoplasm and nucleus in EBV-EBNA1-positive NPC cells (figure 5C). Subsequent co-IP analysis showed that SMAD3 and c-JUN interact weakly in the absence of EBV-EBNA1 and exogenous TGFβ1 stimulus (5–8F). However, the interaction between SMAD3 and c-JUN was enhanced after stimulation with exogenous recombinant human TGFβ1 protein (5–8F-TGFβ1) or exogenous recombinant active EBV-EBNA1 protein (5–8F-EBNA1) (figure 5D). Interestingly, blockade of the TGFβ1 pathway by SB431542 neutralized the stimulatory effect of EBV-EBNA1 and significantly weakened the interaction between SMAD3 and c-JUN (5–8F-EBNA1-SB) (figure 5D). These results suggest that SMAD3/c-JUN interaction is governed by the upstream EBNA1-TGFβ1 axis in EBV-EBNA1-infected NPC cells. In the clinical cohort, NPC patients with high TGFβ1 or high p-SMAD3 expression had higher c-JUN and p-c-JUN expression (figures 1E, 4G and 5E). Patients with the p-SMAD3highp-c-JUNhigh expression pattern suffered from the highest recurrence rate and the shortest OS (figure 5F).
Overall, the results indicate that TGFβ1 suppresses miR-200a by enhancing the formation of the SMAD3/c-JUN complex, and EBV-EBNA1 acts as a comprehensive regulator of cellular gene expression by governing the SMAD3/c-JUN interaction in a TGFβ1-dependent manner.
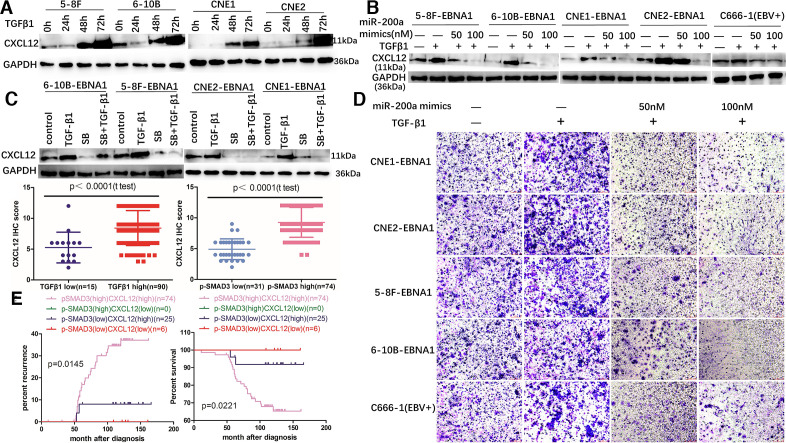
Tgfβ1 induces CXCL12 production by suppressing miR-200a
To verify whether TGFβ1 influences CXCL12 expression in a miR-200a-dependent manner, NPC cells were treated with recombinant human TGFβ1 protein for various periods of time; the results indicate that production of CXCL12 protein was elevated in a time-dependent manner (figure 6A). However, SB431542 blocked the induction effect of TGFβ1 on CXCL12 expression when NPC cells were pretreated with SB431542 in the presence or in the absence of TGFβ1 (figure 6B). These results suggest that TGFβ1 positively regulates CXCL12 expression. Moreover, various concentrations of miR-200a mimics partially neutralized the stimulatory effect of TGFβ1 on CXCL12 expression in a dose-dependent manner, suggesting that TGFβ1 regulates CXCL12 expression in a miR-200a-dependent manner (figure 6C). Interestingly, the effect of TGFβ1 or SB431542 on CXCL12 mRNA level was not significant in agreement with the finding that miR-200a inhibits CXCL12 by a posttranscriptional mechanism (online supplemental figure 5A). To detect the stimulatory effect of CXCL12 on the migration of Treg cells induced by the TGFβ1-miR-200a axis, the culture supernatant was collected after the treatment of the cells with miR-200a mimics or recombinant human TGFβ1 protein for 72 hours; exogenous and endogenous TGFβ1 proteins were depleted by incubation with TGFβ1-neutralizing antibody for 2 hours. Various concentrations of miR-200a mimics partially neutralized the stimulatory effect of TGFβ1 on CXCL12 expression and ultimately reduced the migration of Treg cells in a dose-dependent manner (figure 6C, D and online supplemental figure 5B). In NPC patients, CXCL12 level was positively correlated with TGFβ1 and p-SMAD3 expression (figures 1E, 3I and 6E). Patients with p-SMAD3highCXCL12high expression pattern suffered from the worst prognosis (figure 6F). Thus, these results suggest that the stimulatory effect of TGFβ1 on CXCL12 expression is predominantly mediated by the suppression of miR-200a, and chemotactic migration of Treg cells is controlled by the TGFβ1-SMAD3-miR-200a-CXCL12 pathway in NPC.
Figure 6.
TGF-β1 induces CXCL12 production by suppressing miR-200a. (A) TGFβ1 promoted CXCL12 expression in a time-dependent manner. (B) SB431542 blocked the induction effect of TGFβ1 on CXCL12 expression. (C, D) Various concentrations of miR-200a mimics partly neutralized the stimulatory effect of TGFβ1 on CXCL12 expression and ultimately reduced migration of Treg cells in a dose-dependent manner. (E) NPC patients with high TGFβ1 or high p-SMAD3 expression had higher CXCL12 expression compared with those in patients with low TGFβ1 or low p-SMAD3 expression. (F) Kaplan-Meier analysis of recurrence risk and overall survival for concurrent p-SMAD3/CXCL12 expression groups in NPC patients. EBV, Epstein-Barr virus; NPC, nasopharyngeal carcinoma; TGF-β1, transforming growth factor-β1; Treg, regulatory T.
jitc-2020-001588supp007.pdf (332.4KB, pdf)
CXCL12 mediates chemoattraction of Treg cells by upregulating the CXCR4 receptor of Treg cells and the CXCL12-CXCR4-Treg regulatory pathway is universal in various malignant solid tumors
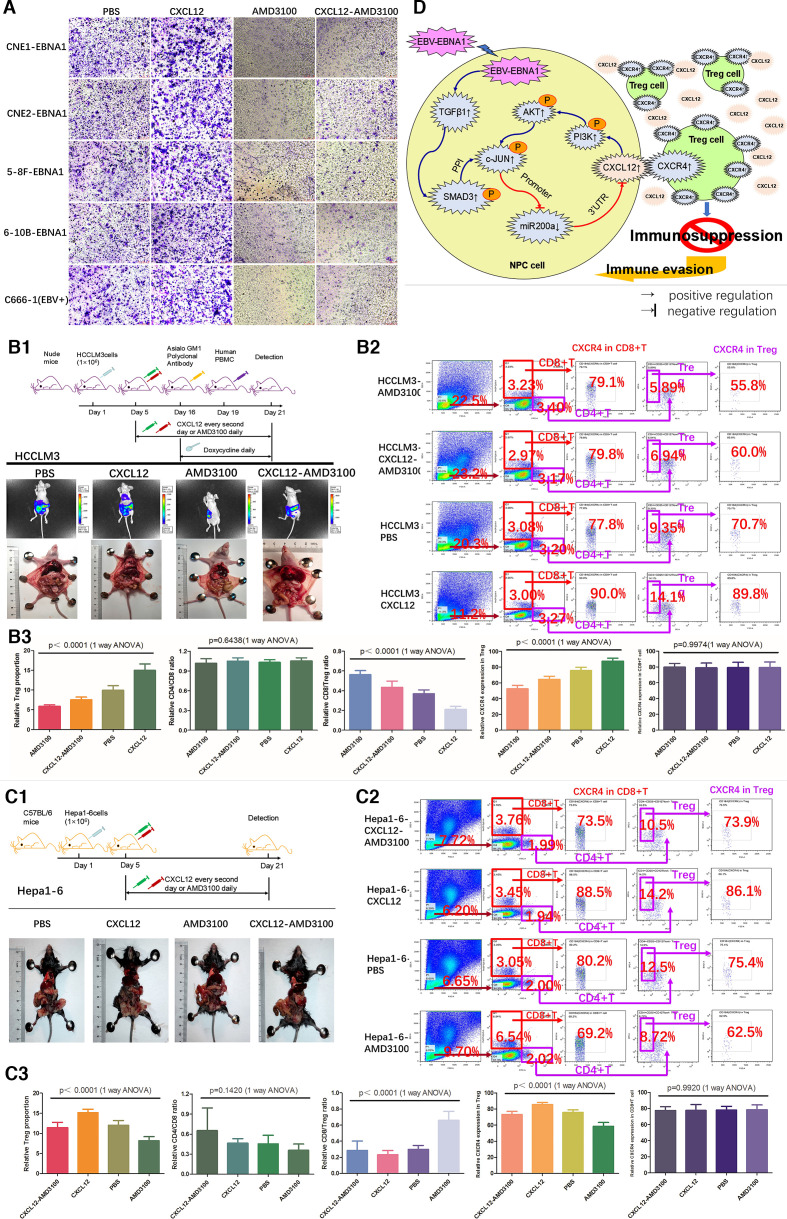
To determine the mechanism involved in CXCL12 axis-induced recruitment of Treg cells, involvement of CXCR4 receptor was investigated. The ligand-binding receptor function was blocked with AMD3100; the data indicate that the migration of Treg cells and CXCR4 expression in Treg cells were considerably suppressed by AMD3100 in vitro (figure 7A and online supplemental figure 6A) and in vivo (figure 3H1–H3) despite the stimulatory effects of CXCL12. Furthermore, similar phenomenon was detected in other animal models. The expression of CXCR4 in Treg cells was significantly increased in the 5–8F-EBNA1 group and decreased in the 5–8F-miR-200a group compared with that in the control (5–8F-NC), which explains the mechanism of EBV-EBNA1 antigen-promoted migration of Treg cells and miR-200a-suppressed migration of Treg cells (figures 2F1–F3). Blockade of the TGFβ1-SMAD3 axis by SB431542 inhibited CXCL12 expression in vitro (figure 6B) or migration of Treg cells in vivo (figures 2G1–G3) and reduced CXCR4 expression in infiltrating Treg cells (figures 2G1–G3). However, in these three animal models, the CD4/CD8 ratio and CXCR4 expression in CD8+ T cells were similar in all groups, implying that the CXCL12-CXCR4 ligand/receptor pair is unlikely to be responsible for regulation of the functional migration of CD8+ T cells in the NPC microenvironment (figures 2F1–F3, G1–G3 and 3H1–H3). Thus, the data indicate that the CXCL12-CXCR4 ligand/receptor pair is the main direct regulator of chemotactic migration of Treg cells, and the EBV-EBNA1-TGFβ1-SMAD3-miR-200a-CXCL12 axis mediates the migration of Treg cells by regulating the CXCR4 receptor of Treg cells.
Figure 7.
CXCL12 mediates recruitment of Treg cell by upregulating CXCR4 receptor of Treg cells, and the CXCL12-CXCR4-Treg regulation pathway is universally applicable to various malignant solid tumors. (A) AMD3100 suppressed migration of Treg cells induced by CXCL12 in vitro. (B1) CXCL12-CXCR4 pathway regulated migration of Treg cells in human hepatocellular carcinoma nude mouse model generated using HCCLM3 cells. Upper: animal experiment flow chart. Lower: representative images of various tumor formation groups in humanized immune reconstruction mouse model generated using intraperitoneal injection of HCCLM3 cells. (B2) (B3) The in vivo tumor-infiltrating CD4+ T cells, CD8+ T cells and Treg cells and CXCR4 (alias CD184) receptor expression levels were detected by FACS. (C1) CXCL12-CXCR4 pathway regulated migration of Treg cells in mouse hepatoma immunocompetent C57BL/6 mouse model (with fully functional T cell lineages) generated using Hepa1-6 cell. Upper: animal experiment flow diagram. Lower: representative images of various tumor formation groups in immunocompetent C57BL/6 mouse model generated using intraperitoneal injection of Hepa1-6 mouse hepatoma cells. (C2) (C3) The in vivo tumor-infiltrating CD4+ T cells, CD8+ T cells and Treg cells and CXCR4 (alias CD184) receptor expression levels were detected by FACS. (D) Schematic diagram. ANOVA, analysis of variance; EBV, Epstein-Barr virus; Treg, regulatory T.
jitc-2020-001588supp008.pdf (381.5KB, pdf)
To provide additional support for the role of CXCL12 in stimulation of the migration of Treg cells and to assess whether CXCL12-CXCR4 pathway is universal in various tumor contexts, a humanized immune reconstruction nude mouse model of human hepatocellular carcinoma was produced by intraperitoneal injection of HCCLM3 cells into immunocompromised nude mice; additionally, a well-established immunocompetent mouse model of hepatocellular carcinoma was produced by intraperitoneal injection of Hepa1-6 mouse hepatoma cells into the C57BL/6 syngeneic hosts (figure 7B1-B3, 7C1-C3). In HCCLM3 and Hepa1-6 animal models, the tumor load, proportion of tumor-infiltrating Treg cells and expression of CXCR4 in Treg cells were significantly increased in the CXCL12 treatment groups and decreased in the AMD3100 treatment groups compared with those in the control groups (figure 7B1-B3, 7C1-C3, (online supplemental figure 6B, C). Moreover, the CD8/Treg ratio was decreased in the CXCL12 treatment groups and increased in the AMD3100 treatment groups (figure 7B2-3 and 7C2-C3). Unexpectedly, the CD4/CD8 ratio was reversed due to a decrease in the proportion of CD4+ T cells and an increase in CD8+ T cells in each group in the immunocompetent C57BL/6 mouse model (figure 7C2-C3); this effect was not detected in the immunocompromised nude mouse models, which did not have fully functional T cell lineages (figures 2F2–F3, 2G2–G3, 3H2–H3 and 7B2–B3), implying that tumor caused local immune senescence in a real immunocompetent syngeneic host, which may be another synergistic mechanism for tumor immune escape. Overall, our results in NPC and hepatoma models demonstrate the universal role of the CXCL12-CXCR4 axis in chemotactic migration of Treg cells in various malignant solid tumors.
Thus, EBV-EBNA1 promotes chemoattraction of Treg cells by upregulation of the TGFβ1-SMAD3-PI3K-AKT-c-JUNCXCL12-CXCR4 axis and downregulation of miR-200a (figure 7D).
Discussion
EBV-positive NPC microenvironment is heavily infiltrated with lymphoid stroma. Latent EBV-infected NPC cells use several specific mechanisms to form and shape the tumor microenvironment to their own benefit, especially immune escape.26 Treg cells are one of the key regulators in restraining antitumor immune responses,27 and are considered to be associated with immune escape of various tumors, including human ovarian carcinoma,16 melanoma28 and hepatocellular carcinoma.17 Our previous report showed that the plasma EBV-DNA density has significant positive association with the number of Treg cells in the peripheral blood of NPC patients.13 EBNA1 is an EBV-encoded nuclear antigen which plays a critical role in EBV genome maintenance, replication and postmitotic segregation and is the only EBV protein expressed in all types of EBV-infected cells.4 We have previously demonstrated that EBV-EBNA1 promotes the production of Treg cells in a TGFβ1-dependent manner.14 However, the role and specific mechanism of EBV-EBNA1 in chemoattraction of Treg cells remains poorly understood. The present study is the first to demonstrate that EBV-EBNA1 regulates chemotactic migration of Treg cells via the TGFβ1-SMAD3-PI3K-AKT-c-JUN-miR-200a-CXCL12-CXCR4 feedback loop to create an extremely immunosuppressive microenvironment in NPC. Our results link latent EBV infection and construction of immunosuppressive microenvironment and identify EBV-EBNA1 as a therapeutic target in NPC.
Mechanistic analysis revealed that EBNA1 overexpression in NPC cells upregulates TGFβ1 expression and activates the downstream SMAD3/c-Jun protein–protein complex, which binds to the promoter region of miR-200a and represses miR-200a transcription. However, another study showed that miR-200a increases c-Jun expression by stabilizing its mRNA in proliferating cells.29 This contradiction suggests a feedback loop between miR-200a and c-Jun. In our study, CXCL12 was confirmed for the first time as a direct target of miR-200a. Importantly, in addition to the repressive effect of miR-200a on CXCL12 expression, CXCL12 overexpression can suppress miR-200a transcription through the PI3K/AKT/c-Jun pathway. These results confirm direct interaction of miR-200a, c-Jun and CXCL12, which form a c-JUN-miR-200a-CXCL12-PI3K-AKT-c-JUN feedback loop to regulate chemotactic migration of Treg cells.
Previous studies have reported that EBV-encoded latent genes can induce immune escape through several mechanisms, including downregulation of HLA expression to interfere with the antigen-presenting machinery, upregulation of the coinhibitory receptor expression to interrupt T cell activation, production of immunosuppressive cytokines and prevention of innate immune responses.30 Several recent publications focused on the modulation of the tumor microenvironment induced by EBV latent genes, especially by the well-characterized LMP1 oncogene.31 However, the role of EBNA1 in immune modification associated with escape from immune attack is incompletely understood. Our study demonstrates that EBNA1 overexpression in NPC cells induces the secretion of CXCL12 chemokine and efficiently attracts CXCR4-positive Treg cells to remodel an immunosuppressive microenvironment. This result provides a link between EBV infection and chemokine-mediated immunosuppressive microenvironment in NPC. Mrizak et al obtained similar results, which indicated that NPC-derived exosomes contain CCL20 chemokine that converts conventional CD4+ T cells into suppressive Treg cells, and recruitment of Treg cells into the tumor increases their suppressive functions.32 Interactions between EBNA1 and recruitment of Treg cells represent a newly defined mechanism of EBV-induced immune evasion in NPC.
In conclusion, our study demonstrates a previously unrecognized mechanism of EBNA1-induced chemoattraction of Treg cells mediated by the TGFβ1-SMAD3-PI3K-AKT-c-JUN-miR-200a-CXCL12-CXCR4 axis in the NPC microenvironment. Given the critical roles of Treg cells in immune escape, a more comprehensive understanding of the crosstalk between Treg cells and EBV latent infection will provide an opportunity to develop novel EBV-directed immunotherapies to reshape the tumor microenvironment.
jitc-2020-001588supp009.pdf (133.4KB, pdf)
Acknowledgments
We thank Qianbin Zhang (Cancer research institute, Southern medical university, China) for providing guidance of flow cytometry detection. Authors would to thank Jiahong Wang (Cancer research institute, Southern medical university, China) for suggestions on bioinformatics analysis. We also thank Ying Peng (Sun Yat-Sen Memorial Hospital, Sun Yat-sen University) for general support.
Footnotes
Contributors: This study was designed by SH and JL and supervised by XL. Experiments were primary conducted by SH. Patient enrolment and follow-up were done by RD. Specimen preparation was done by XL. YL, JW, LW, BZ and FW prepared NPC cells for NGS detection and conducted qRT-PCR screening according to NGS results. The administrative support was obtained from XL. The manuscript was written by SH and approved by all authors.
Funding: This study was supported by grants from National Natural Science Foundation of China (grant No. 81472535, to XL).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request. All data in our study are availability on request.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Chen Y-P, Chan ATC, Le Q-T, et al. Nasopharyngeal carcinoma. Lancet 2019;394:64–80. 10.1016/S0140-6736(19)30956-0 [DOI] [PubMed] [Google Scholar]
- 2.Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet 2016;387:1012–24. 10.1016/S0140-6736(15)00055-0 [DOI] [PubMed] [Google Scholar]
- 3.Münz C, Moormann A. Immune escape by Epstein-Barr virus associated malignancies. Semin Cancer Biol 2008;18:381–7. 10.1016/j.semcancer.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen C-L, Liu C-D, You R-I, et al. Ribosome protein L4 is essential for Epstein-Barr virus nuclear antigen 1 function. Proc Natl Acad Sci U S A 2016;113:2229–34. 10.1073/pnas.1525444113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messick TE, Smith GR, Soldan SS, et al. Structure-Based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth. Sci Transl Med 2019;11. 10.1126/scitranslmed.aau5612. [Epub ahead of print: 06 Mar 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Liu Y, Wang C, et al. Triptolide inhibits Epstein-Barr nuclear antigen 1 expression by increasing sensitivity of mitochondria apoptosis of nasopharyngeal carcinoma cells. J Exp Clin Cancer Res 2018;37:192. 10.1186/s13046-018-0865-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridgewater HE, Date KL, O'Neil JD, et al. The Epstein-Barr virus-encoded EBNA1 protein activates the bone morphogenic protein (BMP) signalling pathway to promote carcinoma cell migration. Pathogens 2020;9. 10.3390/pathogens9070594. [Epub ahead of print: 21 Jul 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X, Ji M, Cheng W, et al. Assessment of the long-term diagnostic performance of a new serological screening scheme in large-scale nasopharyngeal carcinoma screening. J Cancer 2018;9:2093–7. 10.7150/jca.23755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, Guo X, Ji M, et al. Establishment and validation of a two-step screening scheme for improved performance of serological screening of nasopharyngeal carcinoma. Cancer Med 2018;7:1458–67. 10.1002/cam4.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Li H, Sheng H, et al. A randomized controlled trial on evaluation of plasma Epstein-Barr virus biomarker for early diagnosis in patients with nasopharyngeal carcinoma. Adv Ther 2020;37:4280–90. 10.1007/s12325-020-01461-4 [DOI] [PubMed] [Google Scholar]
- 11.Wood KJ, Bushell A, Hester J. Regulatory immune cells in transplantation. Nat Rev Immunol 2012;12:417–30. 10.1038/nri3227 [DOI] [PubMed] [Google Scholar]
- 12.deLeeuw RJ, Kost SE, Kakal JA, et al. The prognostic value of Foxp3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res 2012;18:3022–9. 10.1158/1078-0432.CCR-11-3216 [DOI] [PubMed] [Google Scholar]
- 13.Bi P, Wang J, Lu J, et al. [Correlation analysis of the Epstein-Barr virus recruited regulatory T cells in nasopharyngeal carcinoma of immune escape]. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2017;52:692–7. 10.3760/cma.j.issn.1673-0860.2017.09.011 [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Luo Y, Bi P, et al. Mechanisms of Epstein-Barr virus nuclear antigen 1 favor Tregs accumulation in nasopharyngeal carcinoma. Cancer Med 2020;9:5598–608. 10.1002/cam4.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao M, Luo R, Liu Y, et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun 2016;7:11309. 10.1038/ncomms11309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004;10:942–9. 10.1038/nm1093 [DOI] [PubMed] [Google Scholar]
- 17.Yang P, Li Q-J, Feng Y, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 2012;22:291–303. 10.1016/j.ccr.2012.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebrun JJ, Takabe K, Chen Y, et al. Roles of pathway-specific and inhibitory Smads in activin receptor signaling. Mol Endocrinol 1999;13:15–23. 10.1210/mend.13.1.0218 [DOI] [PubMed] [Google Scholar]
- 19.UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47:D506–15. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinozaki A, Sakatani T, Ushiku T, et al. Downregulation of microRNA-200 in ebv-associated gastric carcinoma. Cancer Res 2010;70:4719–27. 10.1158/0008-5472.CAN-09-4620 [DOI] [PubMed] [Google Scholar]
- 21.Nishijima N, Seike M, Soeno C, et al. miR-200/ZEB axis regulates sensitivity to nintedanib in non-small cell lung cancer cells. Int J Oncol 2016;48:937–44. 10.3892/ijo.2016.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H-C, Chen G-H, Chen Y-H, et al. Microrna deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer 2009;100:1002–11. 10.1038/sj.bjc.6604948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Z, Zhang L, Li Z, et al. An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma. BMC Med Genomics 2012;5:3. 10.1186/1755-8794-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan X-X, Yi H, Qu J-Q, et al. Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues. Oncol Rep 2015;34:2585–601. 10.3892/or.2015.4237 [DOI] [PubMed] [Google Scholar]
- 25.Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun 2010;391:535–41. 10.1016/j.bbrc.2009.11.093 [DOI] [PubMed] [Google Scholar]
- 26.Tan GW, Visser L, Tan LP, et al. The microenvironment in Epstein-Barr virus-associated malignancies. Pathogens 2018;7. 10.3390/pathogens7020040. [Epub ahead of print: 13 Apr 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell DJ, Koch MA. Phenotypical and functional specialization of Foxp3+ regulatory T cells. Nat Rev Immunol 2011;11:119–30. 10.1038/nri2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viguier M, Lemaître F, Verola O, et al. Foxp3 expressing CD4+CD25(high) regulatory T cells are overrepresented in human metastatic melanoma lymph nodes and inhibit the function of infiltrating T cells. J Immunol 2004;173:1444–53. 10.4049/jimmunol.173.2.1444 [DOI] [PubMed] [Google Scholar]
- 29.Del Vecchio G, De Vito F, Saunders SJ, et al. Rna-Binding protein HuR and the members of the miR-200 family play an unconventional role in the regulation of c-Jun mRNA. RNA 2016;22:1510–21. 10.1261/rna.057588.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tangye SG, Palendira U, Edwards ESJ. Human immunity against EBV-lessons from the clinic. J Exp Med 2017;214:269–83. 10.1084/jem.20161846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshizaki T, Kondo S, Endo K, et al. Modulation of the tumor microenvironment by Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma. Cancer Sci 2018;109:272–8. 10.1111/cas.13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mrizak D, Martin N, Barjon C, et al. Effect of nasopharyngeal carcinoma-derived exosomes on human regulatory T cells. J Natl Cancer Inst 2015;107:363. 10.1093/jnci/dju363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001588supp001.pdf (144.5KB, pdf)
jitc-2020-001588supp002.pdf (193.8KB, pdf)
jitc-2020-001588supp003.pdf (596.6KB, pdf)
jitc-2020-001588supp004.pdf (909.6KB, pdf)
jitc-2020-001588supp005.pdf (920.6KB, pdf)
jitc-2020-001588supp006.pdf (648.2KB, pdf)
jitc-2020-001588supp007.pdf (332.4KB, pdf)
jitc-2020-001588supp008.pdf (381.5KB, pdf)
jitc-2020-001588supp009.pdf (133.4KB, pdf)