Abstract
Enterococcus faecalis (E. faecalis) has rapidly acquired resistance to multiple antimicrobials, and the antimicrobial resistance of E. faecalis from broiler breeders has been implicated in its vertical transmission to their offspring. The objective of this study was to investigate the antimicrobial resistance and genetic diversity of commensal E. faecalis isolated from the broiler breeder farms. Among a total of 229 E. faecalis isolates from 9 broiler breeder farms, the highest resistance rate was observed in tetracycline (78.2%), followed by doxycycline (58.1%) and erythromycin (43.7%), and the prevalence of antimicrobial resistance showed significant differences among the 9 broiler breeder farms (P < 0.05). The tetM gene (77.1%) and ermB gene (85.0%) were detected at the highest levels in 179 TE-and 100 E-resistant isolates, respectively. Twenty-four high-level gentamicin-resistant isolates carried aac(6″)Ie-aph(2″)-la gene, and 9 high-level ciprofloxacin-resistant isolates showed point mutations in both gyrA and parC genes. All high-level gentamicin-resistant or high-level ciprofloxacin-resistant isolates showed one of the two different virulence gene patterns, ace-asa1-efaA-gelE complex or ace-efaA-gelE complex. These results indicate that constant epidemiological monitoring at the breeder level is required to prevent the pyramidal transmission of antimicrobial-resistant E. faecalis.
Key words: Enterococcus faecalis, broiler breeder, poultry, antimicrobial resistance
Introduction
Enterococci are normal inhabitants of the gastrointestinal tracts of animals and humans (Cauwerts et al., 2007, Choi and Woo, 2015). However, Enterococcus faecalis (E. faecalis) and Enterococcus faecium, which are of the greatest importance to human health, could cause nosocomial bacteremia, peritonitis, surgical wound infections, and urinary tract infections (Diarra et al., 2010, Han et al., 2011). In particular, E. faecalis has rapidly acquired resistance to multiple antimicrobials in the last several decades (Kim et al., 2018) and could transfer the antimicrobial resistance genes from animals to humans through the food chain (Aarestrup et al., 2000, Cauwerts et al., 2007, Han et al., 2011).
The broiler industry is vertically integrated from breeding flocks and hatcheries to feed mills, transportation divisions, and processing plants. Dierikx et al. (2013) and Ha et al. (2018) have implicated a vertical transmission of the bacterial isolates from broiler breeding chickens to their offspring. Antimicrobial resistance of the enterococci inhabiting the intestinal tract of the parent stock can result from the use of antimicrobials and can directly reflect the dissemination in commercial chicks through hatcheries which serve as a reservoir (Osman et al., 2018).
The Korea Animal Health Products Association reported that 154 tons of antimicrobials were sold for use in the poultry industry in 2017 (APQA, 2017). In particular, antimicrobial agents such as aminoglycoside, β-lactam, and fluoroquinolone have been widely used in the broiler industry in Korea (Kim et al., 2018). Although the Korean Veterinary Antimicrobial Resistance Monitoring is an ongoing program against the zoonotic and commensal bacteria isolated from the food-producing animals including broiler chicken since 2003 (APQA, 2017), the level of antimicrobial resistance at the broiler parent stage has not yet been studied. In this study, we investigated the antimicrobial resistance and genetic diversity of commensal E. faecalis isolated from the broiler breeder farms.
Materials and methods
Sampling
Fecal samples were collected from 9 broiler breeder farms including 94 flocks from 2016 to 2018 (Table 1) in accordance with the standards set by the National Poultry Improvement Plan (USDA, 2012). Briefly, approximately 10 g of fecal droppings were sampled from each of the 15 different locations in 2 divided areas per flock. All the samples were transported to the laboratory in a cooler, individually inoculated into 100 mL of buffered peptone water (BD Biosciences, Sparks, MD) and incubated for 18 to 24 h at 37°C.
Table 1.
Distribution of Enterococcus faecalis isolated from 9 broiler breeder farms for this study.
| Farm | No. of positive flocks/no. of flocks tested (%) | No. of E. faecalis isolates1 |
|---|---|---|
| Ⅰ | 9/12 (75.0) | 28 |
| Ⅱ | 9/9 (100.0) | 25 |
| Ⅲ | 13/14 (92.9) | 31 |
| Ⅳ | 18/20 (90.0) | 50 |
| Ⅴ | 9/9 (100.0) | 23 |
| Ⅵ | 9/9 (100.0) | 24 |
| Ⅶ | 5/8 (62.5) | 16 |
| Ⅷ | 3/4 (75.0) | 9 |
| Ⅸ | 9/9 (100.0) | 23 |
| Total | 84/94 (92.4) | 229 |
If isolates from the same flock showed the same antimicrobial susceptibility patterns, only one isolate was included in this study.
Bacterial Isolation
Pre-enriched buffered peptone water was transferred to Enterococcosel broth (BD Biosciences) at a 1:10 ratio, and the broth was streaked onto Enterococcosel agar (BD Biosciences) after incubation for 18 to 24 h at 37°C. The suspected Enterococcus colonies were selected and subsequently confirmed as E. faecalis by using the polymerase chain reaction (PCR) method as previously described (Dutka-Malen et al., 1995, Kim et al., 2019). At least 3 suspicious colonies were randomly confirmed, and only one isolate was included if the isolates from the same flock showed the same antimicrobial susceptibility patterns. As a result, a total of 229 E. faecalis isolates were tested in this study (Table 1).
Antimicrobial Susceptibility Testing
All isolates were tested for resistance to 9 antimicrobial agents using the disc diffusion method on Mueller-Hinton agar (BD Biosciences), according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2013) as previously described (Kim et al., 2019). The antimicrobial discs (BD Biosciences) used were ampicillin (10 μg), chloramphenicol (30 μg), ciprofloxacin (CIP, 5 μg), doxycycline (DOX, 30 μg), erythromycin (E, 15 μg), penicillin (10 units), rifampin (5 μg), tetracycline (TE, 30 μg), and vancomycin (30 μg). Multidrug resistance (MDR) was defined as the acquired resistance to at least one agent in 3 or more antimicrobial classes.
The minimum inhibitory concentration (MIC) was determined by standard agar dilution methods on Muller-Hinton agar (BD Biosciences) and the MIC breakpoint values for high-level CIP (≥64 μg/mL) as previously described (Leavis et al., 2006). The high-level gentamicin (HLG, ≥500 μg/mL) and high-level streptomycin (HLS, ≥2,000 μg/mL) was defined by the CLSI (CLSI, 2013). The MIC breakpoint values for high-level kanamycin (HLK, ≥500 μg/mL) was determined according to European Committee on Antimicrobial Susceptibility Testing (2019). Staphylococcus aureus ATCC 25923 was used as quality control for the disk diffusion and MIC tests.
Detection of Antimicrobial Resistance and Virulence Genes
The presence of genes encoding resistance to E, including ermA, ermB, and mef genes; TE, including tetL, tetM, tetO, Int-Tn, and tndX genes; and aminoglycoside-modifying enzyme (AME), including aac(6′)-Ie–aph(2″)-Ia, aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Id, ant(3″)-Ia, and ant(6)-Ia genes, were screened by PCR, using primers and conditions as previously described (Clark et al., 1999, Vakulenko et al., 2003, Kehrenberg and Schwarz, 2006, Di Cesare et al., 2013, Choi and Woo, 2015). Furthermore, the genes encoding the virulence factors—ace, asa1, cylA, efaA, esp, gelE, and hyl—were also identified by using PCR as previously described (Choi and Woo, 2013).
Detection of IS256-Flanking Pattern for aac(6ʹ)Ie-aph(2″)-Ia
The presence of the insertion sequence IS256-flanking patterns was investigated in all the high-level gentamicin-resistant (HLGR) E. faecalis harboring aac(6ʹ)Ie-aph(2″)-Ia. PCR using 2 primer pairs as reported by Watanabe et al. (2009) was performed to determine the IS256-flanking patterns.
Screening for Mutations in gyrA and parC
The detection of gyrA and parC genes in high-level CIP-resistance (HLCR) isolates was also assessed by PCR using the primers previously described (Kwak et al., 2013). The PCR products were purified using the PCR purification kit (Qiagen, Valencia, CA) and sequenced by the automatic sequencer (Cosmogenetech, Korea). The DNA sequences were compared with those deposited in the GeneBank accession no. AF060881 and AB017811 for gyrA and parC genes, respectively (El Amin et al., 1999).
Statistical Analysis
Statistical analysis was performed using the Statistical Package for Social Science version 25 (SPSS; IBM, Korea) and conducted with the chi-square test to identify statistically significant differences. Differences were considered significant at P < 0.05.
Results
Antimicrobial Resistance Profile
The distribution of antimicrobial resistance to 12 antimicrobial agents in the 229 E. faecalis isolates from 9 broiler breeder farms is presented in Table 2. The highest resistance rate was observed to TE (78.2%), followed by DOX (58.1%), E (43.7%), rifampin (31.0%), HLS (16.2%), CIP (15.3%), HLK (14.0%), chloramphenicol (13.1%), and HLG (10.5%). The rates of antimicrobial resistance to penicillin and ampicillin were only 8.3 and 3.5%, respectively. None of the isolates were resistant to vancomycin. The prevalence of antimicrobial resistance also showed significant differences among the 9 broiler breeder farms (P < 0.05).
Table 2.
Prevalence of antimicrobial resistance of 229 Enterococcus faecalis from 9 broiler breeder farms.
| Farm (no. of isolates) | No. of antimicrobial resistance isolates (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | C | CIP | DOX | E | P | TE | RA | VA | HLC | HLG | HLK | HLS | MDR | |
| Ⅰ (28) | 0 (0.0) | 0 (0.0) | 3 (10.7) | 18 (64.3) | 6 (21.4) | 1 (3.6) | 24 (85.7) | 6 (21.4) | 0 (0.0) | 1 (3.6) | 6 (21.4) | 6 (21.4) | 8 (28.6) | 6 (21.4) |
| Ⅱ (25) | 1 (4.0) | 2 (8.0) | 5 (20.0) | 20 (80.0) | 15 (60.0) | 2 (8.0) | 23 (92.0) | 3 (12.0) | 0 (0.0) | 0 (0.0) | 1 (4.0) | 1 (4.0) | 1 (4.0) | 7 (28.0) |
| Ⅲ (31) | 0 (0.0) | 0 (0.0) | 10 (32.3) | 8 (25.8) | 7 (22.6) | 0 (0.0) | 15 (48.4) | 15 (48.4) | 0 (0.0) | 5 (16.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (9.7) |
| Ⅳ (50) | 1 (2.0) | 13 (26.0) | 7 (14.0) | 38 (76.0) | 28 (56.0) | 3 (6.0) | 45 (90.0) | 6 (12.0) | 0 (0.0) | 1 (2.0) | 12 (24.0) | 14 (28.0) | 10 (20.0) | 18 (36.0) |
| Ⅴ (23) | 0 (0.0) | 11 (47.8) | 0 (0.0) | 21 (91.3) | 17 (73.9) | 2 (8.7) | 23 (100.0) | 4 (17.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 11 (47.8) | 13 (56.5) |
| Ⅵ (24) | 0 (0.0) | 0 (0.0) | 2 (8.3) | 3 (12.5) | 9 (37.5) | 2 (8.3) | 11 (45.8) | 15 (62.5) | 0 (0.0) | 1 (4.2) | 1 (4.2) | 2 (8.3) | 0 (0.0) | 3 (12.5) |
| Ⅶ (16) | 2 (12.5) | 3 (18.8) | 4 (25.0) | 8 (50.0) | 5 (31.3) | 2 (12.5) | 12 (75.0) | 6 (37.5) | 0 (0.0) | 0 (0.0) | 3 (18.8) | 5 (31.3) | 3 (18.8) | 4 (25.0) |
| Ⅷ (9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (22.2) | 3 (33.3) | 0 (0.0) | 5 (55.6) | 7 (77.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (11.1) |
| Ⅸ (23) | 4 (17.4) | 1 (4.3) | 4 (17.4) | 15 (65.2) | 10 (43.5) | 7 (30.4) | 21 (91.3) | 9 (39.1) | 0 (0.0) | 1 (4.3) | 1 (4.3) | 4 (17.4) | 4 (17.4) | 9 (39.1) |
| Total (229)1 | 8 (3.5) | 30 (13.1) | 35 (15.3) | 133 (58.1) | 100 (43.7) | 19 (8.3) | 179 (78.2) | 71 (31.0) | 0 (0.0) | 9 (3.9) | 24 (10.5) | 32 (14.0) | 37 (16.2) | 64 (27.9) |
Abbreviations: AM, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; DOX, doxycycline; E, erythromycin; HLC, high-level ciprofloxacin; HLG, high-level gentamicin; HLK; high-level kanamycin; HLS, high-level streptomycin; MDR, multidrug-resistance; P, penicillin; RA, rifampin; TE, tetracycline; VA, vancomycin.
Differences in the prevalence of resistance between isolates from farms had chi-square P values of <0.05 for all drugs depicted except vancomycin.
Distribution of TE- and E-Resistance Genes
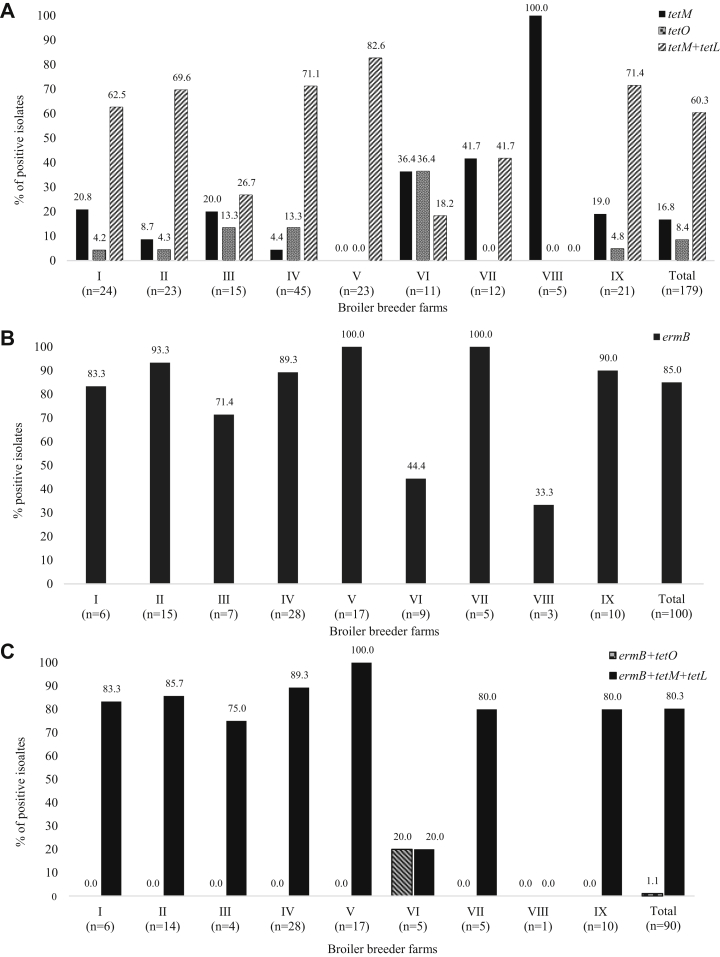
The distribution of TE- and E-resistance genes is shown in Figure 1. The tetM and tetO genes were detected in 138 (77.1%) and 15 (8.4%) among the 179 TE-resistant isolates, respectively. One hundred and eight (60.3%) isolates carried both tetM and tetL genes. The ermB gene was detected in 85 (85.0%) among the 100 E-resistant isolates. None of the E-resistant isolates had ermA and mef genes. Among the 90 both TE- and E-resistant isolates, tetM-tetL-ermB gene combinations were detected in 75 (80.3%) isolates. Only one (1.1%) isolate carried tetO-ermB gene combinations. The transposon genes, Int-Tn and tndx, were not detected in any of the isolates, and the distribution of antimicrobial resistance genes showed a significant difference among the 9 broiler breeder farms (P < 0.05).
Figure 1.
Distribution of resistance genes to tetracycline (A), erythromycin (B), and both tetracycline and erythromycin (C)-resistant Enterococcus faecalis isolated from 9 broiler breeder farms. The tetL gene for tetracycline resistance (A) and ermA and mef genes for erythromycin resistance (B) were not detected in any of the isolates. The distribution of antimicrobial resistance genes showed a significant difference among 9 broiler breeder farms (P < 0.05).
Characteristics of HLGR E. faecalis Isolates
The characteristics of 24 (10.5%) HLGR among the 229 E. faecalis isolates are shown in Table 3. The HLGR isolates were revealed in 6 broiler breeder farms, Ⅰ (n = 6, 21.4%), Ⅱ (n = 1, 4.0%), Ⅳ (n = 12, 24.0%), Ⅵ (n = 1, 4.2%), Ⅶ (n = 3, 18.8%), and Ⅸ (n = 1, 4.3%). All HLGR isolates carried the aac(6″)Ie-aph(2″)-la gene, and 2 isolates carried both aac(6″)Ie-aph(2″)-la and ant(6)-Ia genes. With respect to the distribution of IS256-flanking pattern, the pattern type C (50.0%) showed the highest prevalence, followed by type A (41.7%) and type D (8.3%). In particular, 6 (100%) isolates from farm I and 11 (91.7%) isolates from farm IV showed pattern type A and type C, respectively. Seventeen (70.8%) of the 24 HLGR E. faecalis isolates showed an MDR to 3 to 6 classes of antimicrobial agents. Two different virulence gene patterns, ace-asa1-efaA-gelE complex (54.2%) and ace-efaA-gelE complex (45.8%), were identified among the 24 HLGR E. faecalis isolates. None of the isolates carried the cyl, esp, and hyl genes.
Table 3.
Characteristics of 24 high-level gentamicin-resistant Enterococcus faecalis isolated from 9 broiler breeder farms.
| Farm | Strains | AME gene | IS256-flanking pattern | Antimicrobial resistance phenotype | Antimicrobial resistance genotype | Virulence factor | MICs (μg/mL) |
||
|---|---|---|---|---|---|---|---|---|---|
| G | K | S | |||||||
| Ⅰ | 143 | aac(6″)Ie-aph(2″)-la | A | DOX-HLG-HLK-TE | tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅰ | 225 | aac(6″)Ie-aph(2″)-la | A | DOX-E-HLG-HLK-TE | ermB, tetM, tetL | ace, efaA, gelE | 2,048 | >2,048 | <256 |
| Ⅰ | 227 | aac(6″)Ie-aph(2″)-la | A | DOX-E-HLG-HLK-TE | ermB, tetM, tetL | ace, efaA, gelE | 2,048 | >2,048 | <256 |
| Ⅰ | 228 | aac(6″)Ie-aph(2″)-la | A | DOX-E-HLG-HLK-TE | ermB, tetM, tetL | ace, efaA, gelE | 2,048 | >2,048 | <256 |
| Ⅰ | 224 | aac(6″)Ie-aph(2″)-la | A | HLG-HLK-RA | NT | ace, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅰ | 226 | aac(6″)Ie-aph(2″)-la | A | HLG-HLK-RA | NT | ace, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅱ | 167 | aac(6″)Ie-aph(2″)-la, ant(6)-Ia | D | C-DOX-E-HLG-HLK-HLS-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | 1,024 | >2,048 | >2,048 |
| Ⅳ | 103 | aac(6″)Ie-aph(2″)-la | C | AM-C-HLG-HLK-HLS | NT | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 95 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 97 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE, | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 98 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 99 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | 2,048 | 2,048 |
| Ⅳ | 101 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-P-HLG-HLK-RA-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 102 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 105 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 108 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE, | ermB, tetM, tetL | asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 112 | aac(6″)Ie-aph(2″)-la | C | C-DOX-E-HLG-HLK-HLS-TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅳ | 96 | aac(6″)Ie-aph(2″)-la | C | DOX-HLG-HLK-RA-TE | tetM, tetL | ace, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅳ | 92 | aac(6″)Ie-aph(2″)-la, ant(6)-Ia | D | C-DOX-E-HLG-HLK-HLS- TE | ermB, tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | >2,048 |
| Ⅵ | 1 | aac(6″)Ie-aph(2″)-la | A | DOX-HLG-HLK-TE | tetM, tetL | ace, asa1, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅶ | 212 | aac(6″)Ie-aph(2″)-la | A | CIP-HLG-HLK-RA | NT | ace, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅶ | 211 | aac(6″)Ie-aph(2″)-la | A | HLG-HLK-RA | NT | ace, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅶ | 210 | aac(6″)Ie-aph(2″)-la | C | HLG-HLK-RA | NT | ace, efaA, gelE | >2,048 | >2,048 | <256 |
| Ⅸ | 229 | aac(6″)Ie-aph(2″)-la | A | HLG-HLK-RA | NT | ace, efaA, gelE | >2,048 | >2,048 | <256 |
Abbreviations: AM, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; DOX, doxycycline; E, erythromycin; G, gentamicin; HLGR, high-level gentamicin; HLK; high-level kanamycin; HLS, high-level streptomycin; K, kanamycin; NT, not tested; P, penicillin; RA, rifampin; S, streptomycin; TE, tetracycline.
Characteristics of HLCR E. faecalis Isolates
The characteristics of 9 (3.9%) HLCR among the 229 E. faecalis isolates are shown in Table 4. The HLCR isolates were revealed in 5 broiler breeder farms, Ⅰ (n = 1, 3.6%), Ⅲ (n = 5, 20.0%), Ⅳ (n = 1, 3.2%), Ⅵ (n = 1, 4.2%), and Ⅸ (n = 1, 4.3%). All 9 isolates showed a point mutation at both gyrA and parC genes and MICs from 64 to 128 μg/mL to fluoroquionolones. One isolate showed resistance to 7 classes of antimicrobial agents and carried the AME gene, ant(6)-Ia gene. The 2 virulence gene patterns, ace-efaA-gelE complex (5 isolates) and ace-asa1-efaA-gelE complex (4 isolates), were only identified, but the cyl, esp, and hyl genes were not detected in any of the HLCR isolates.
Table 4.
Characteristics of 9 high-level ciprofloxacin-resistant Enterococcus faecalis isolated from 9 broiler breeder farms.
| Farm | Strains | Amino acid change |
Antimicrobial resistance phenotype | Antimicrobial resistance genotype | Virulence factor | MICs (μg/mL) |
||
|---|---|---|---|---|---|---|---|---|
| gyrA | parC | Ciprofloxacin | Enrofloxacin | |||||
| Ⅰ | 148 | S83I | S80I | HLC | NT | ace, asa1, efaA, gelE | 64 | 128 |
| Ⅲ | 63 | S83I | S80I | HLC | NT | ace, efaA, gelE | 64 | 128 |
| Ⅲ | 65 | S83I | S80I | HLC | NT | ace, asa1, efaA, gelE | 64 | 64 |
| Ⅲ | 67 | S83I | S80I | HLC | NT | ace, asa1, efaA, gelE | 64 | 64 |
| Ⅲ | 82 | S83I | S80I | HLC-E | ermB | ace, efaA, gelE | 64 | 128 |
| Ⅲ | 62 | S83I | S80I | HLC-E-RA-TE | ermB | ace, efaA, gelE | 64 | 64 |
| Ⅳ | 100 | S83Y | S80I | C-HLC-DOX-E-HLK-HLS-TE | ermB, tetM, tetL, ant(6)-Ia | ace, asa1, efaA, gelE | 64 | 64 |
| Ⅵ | 131 | S83I | S80I | HLC-E-HLK | ermB | ace, efaA, gelE | 64 | 64 |
| Ⅸ | 161 | S83I | S80I | HLC-DOX-TE | tetM, tetL | ace, efaA, gelE | 64 | 64 |
Abbreviations: C, chloramphenicol; CIP, ciprofloxacin; DOX, doxycycline; E, erythromycin; HLC, high-level ciprofloxacin; HLK; high-level kanamycin; HLS, high-level streptomycin; NT, not tested; RA, rifampin; TE, tetracycline.
Discussion
The integrated production, processing, and distribution systems of the poultry industry can vertically transfer antimicrobial resistant bacteria from the breeding chickens to their offspring (Olsen et al., 2011; Kim et al., 2012; Dierikx et al., 2013, Seo et al., 2018). Although numerous studies on antimicrobial resistance in poultry have been reported, only a few studies have investigated the breeders (Fei et al., 2018). In this study, E. faecalis from broiler breeders showed high resistance to a variety of antimicrobials and suggested a potential role as reservoirs for the transmission of resistant isolates throughout the poultry industry. In particular, the resistance rate to TE (45.8–100%), DOX (12.5–91.3%), E (21.4–73.9%), HLS (0–47.8%), CIP (0–32.3%), HLK (0–31.3%), and HLG (0–24.0%) were observed, and the significant (P < 0.05) differences in between each of the 9 broiler breeder farms were found.
The different resistance rate to antimicrobial agents might be due to differences in the amount and frequency of antimicrobial agents used for disease prevention or therapeutic purposes in each farm (Rizzotti et al., 2005). In Korea, although a variety of antimicrobial agents are required to be administered only by veterinary prescription since 2013, broiler breeder farms should also adopt guidelines for the prudent use of antimicrobial agents throughout the broiler production process to reduce the emergence and spread of resistant strains with potentially serious effects (Jeon et al., 2019).
As TEs are relatively cheap and effective against a wide variety of microorganisms, they are frequently used in poultry (Brtková et al., 2011, Choi and Woo, 2015). Macrolides are also used in both human and veterinary medicine and may be important as an alternative therapy for the treatment of enterococcal infections in human (Cauwerts et al. 2007). In this study, 189 (82.5%) among the 229 isolates showed resistance to at least TE or E. The majority of the TE-resistant E. faecalis isolates harbored the tetM gene (77.1%) or carried both tetM and tetL genes (60.3%), and E-resistant isolates harbored the ermB gene (85.0%). The tetM are cytoplasmic proteins that protect the ribosomes from the action of TE (Giovanetti et al., 2003). The tetL efflux gene encodes membrane-associated proteins that export TE from the cell (Giovanetti et al., 2003, Choi and Woo, 2015). Kim et al., (2019) reported that resistance mediated by tetM and tetL was the most frequent in the isolates from chicken meat. The ermB gene also encodes an erm methylase that modifies the 23S rRNA for resistance to macrolides, which was the most common (Poeta et al., 2005, Kim et al., 2019). This study indicates that the antimicrobial resistance and distribution of genes are similar in isolates from commercial broiler farms (Nowakiewicz et al., 2017b). Agersø et al., 2006 reported that the ermB and tetM genes can be easily transferred by conjugative transposons. In contrast to the findings of Agersø et al. (2006), E. faecalis isolates, positive for tetM and ermB genes, were negative for the Int-Tn and tndX genes already reported as major determinants of Tn916/1545 and Tn5385 families in the this study, respectively. Although the extensive use of antimicrobials in commercial broiler farms can lead to resistance, another reason could also be the vertical transmission of E. faecalis isolates through the integrated broiler operation system from broiler breeding chickens to retail chicken meat (Dierikx et al., 2013, Ha et al., 2018).
The HLGR eliminates the synergistic bactericidal effect and causes a major reduction in efficient therapeutic options (Rosvoll et al., 2012). The HLGR E. faecalis isolates were characterized by the presence of the AME gene, aac(6′)-Ie-aph(2″)-Ia, encoding a bifunctional enzyme whose activity is related to the occurrence of resistance to all clinically available aminoglycosides but not S (Vakulenko et al., 2003, Niu et al., 2016, Nowakiewicz et al., 2017b). In this study, 24 (10.5%) of the 229 E. faecalis isolates showed HLGR, which is lower than that seen in previous investigations (40.7%) in chicken meat in Korea (Han et al., 2011). However, all HLGR E. faecalis isolates carried the aac(6′)-Ie-aph(2″)-Ia gene. Because the aac(6′)-Ie-aph(2″)-Ia gene is linked to Tn5281 and is often located in plasmids for facilitating cell-to-cell dissemination (Rosvoll et al. 2012), HLGR E. faecalis from breeders could also be the transmission to commercial chickens.
The genetic diversity was related to the organization of IS256 at both ends of this gene (Kilbi et al., 2006, Watanabe et al., 2009). In this study, IS256-flanking pattern types C (50%), A (41.7%), and D (8.3%) were detected, and the prevalence of individual types was different depending on each farm. Although Watanabe et al. (2009) reported that IS256-flanking pattern type C was more prevalent than other patterns in the hospital Enterococci isolates, no relationship between the IS256-flanking patterns and resistance level to aminoglycoside was evident till now, nor the presence of genes for virulence determination. Therefore, the significance of IS256-flanking patterns for HLGR is still unknown (Kilbi et al., 2006, Watanabe et al., 2009). The ant(6′)-Ia gene, which is the most common AME gene among the HLS-resistant isolates (Udo et al., 2004), was detected in 2 (10%) of 24 HLGR isolates in this study.
Fluoroquinolone resistance is essentially mediated by the alteration of target enzymes such as DNA gyrase and topoisomerase IV encoded by the gyrA gene and parC gene, respectively (Petersen and Jensen, 2004, Lee et al., 2005, Oyamada et al., 2006). In this study, 35 (15.3%) of the 229 E. faecalis isolates showed resistance to CIP, which is lower than those reported in chicken meat (43.8%) in Korea (Kim et al., 2018). However, the prevalence of HLCR isolates (25.7%) was similar to that of chicken meat (24.9%) in Korea (Kim et al., 2018). Although the results showed a low resistance rate to fluoroquinolones in broiler breeder farms, the use of CIP has been increasing in the poultry industry in Korea (APQA, 2017). Therefore, the occurrence of HLCR isolates in broiler breeder farms might be continuously increased.
Seventeen (70.8%) of the 24 HLGR E. faecalis and 3 (33.3%) of the 9 HLCR E. faecalis isolates showed MDR. Among both these MDR isolates, the most common resistance pattern included resistance to TE, E, and aminoglycosides, which is widely used in the poultry industry in Korea (APQA, 2017). Similar resistance profiles in MDR isolates with their corresponding genotypes within and between the farms and the occurrence of the same profiles in different farms may be an indication of the ease of spread of enterococcus strains (Nowakiewicz et al., 2017a). This study also suggest that the different resistance levels and patterns in MDR isolates may be related to the use of diverse antimicrobials during poultry production on each farm (Fei et al., 2018, Li et al., 2018).
Virulence factors contribute to the pathogenesis of enterococcal infections through the mediation of adhesion, colonization, and invasion into the host tissues, modulation of the host immunity, and extracellular production of enzymes and toxins, which enhance the severity of the infection (Strateva et al., 2016). In this study, ace (collagen-binding protein), efaA (cell wall–associated protein involved in immune evasion), and gelE (gelatinase) were present in all HLCR and HLGR isolates. These results are similar to that of E. faecalis isolated from poultry in a previous study (Olsen et al., 2011). Therefore, this study indicates that E. faecalis isolates from broiler breeders could be a reservoir of genes important for the pathogenic potential of the poultry industry involving the transmission of antimicrobial related genes. This is the first study to investigate the prevalence and characteristics of antimicrobial resistant E. faecalis isolated from broiler breeders in Korea. Our findings indicate that the genotypic characteristics of E. faecalis from commercial broilers have recently emerged in broiler breeders. Therefore, constant epidemiological monitoring and studies at the breeder level are required to prevent the pyramidal transmission of antimicrobial-resistant E. faecalis.
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Agriculture, Food and Rural Affairs Research Center Support Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (716002-7).
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
References
- Aarestrup F.M., Agerso Y., Gerner-Smidt P., Madsen M., Jensen L.B. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broiler, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000;37:127–137. doi: 10.1016/s0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- Animal and Plant Quarantine Agency (APQA) Animal and Plant Quarantine Agency; Gimcheon, Republic of Korea: 2017. National Antimicrobial Resistance Monitoring Program. [Google Scholar]
- Agersø Y., Pedersen A.G., Aarestrup F.M. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J. Antimicrob. Chemother. 2006;57:832–839. doi: 10.1093/jac/dkl069. [DOI] [PubMed] [Google Scholar]
- Brtková A., Revallová M., Bujdáková H. Detection of tetracycline and macrolide resistance determinants in enterococi of animal and environmental origin using multiplex PCR. Folia. Microbiol. (Praha) 2011;56:236–240. doi: 10.1007/s12223-011-0042-0. [DOI] [PubMed] [Google Scholar]
- Cauwerts K., Decostere A., De Graef E.M., Haesebrouck F., Pasmans F. High prevalence of tetracycline resistance in Enterococcus isolates form broilers carrying the erm (B) gene. Avian Pathol. 2007;36:395–399. doi: 10.1080/03079450701589167. [DOI] [PubMed] [Google Scholar]
- Choi J.M., Woo G.J. Molecular characterization of high-level gentamicin-resistant Enterococcus faecalis from chicken meat in Korea. Int. J. Food Microbial. 2013;165:1–6. doi: 10.1016/j.ijfoodmicro.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Choi J.M., Woo G.J. Transfer of tetracycline resistance genes with aggregation substance in food-borne Enterococcus faecalis. Curr. Microbiol. 2015;70:476–484. doi: 10.1007/s00284-014-0742-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark N.C., Olsvik Ø., Swenson J.M., Spiegel C.A., Tenover F.C. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 1999;43:157–160. doi: 10.1128/aac.43.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, PA: 2013. Performance Standards for Antimicrobial Susceptibility Testing, M100-S23. [Google Scholar]
- Di Cesare A.D., Luna G.M., Vignaroli C., Pasquaroli S., Tota S., Paroncini P., Biavasco F. Aquaculture can promote the presence and spread of antibiotic-resistant enterococci in marine sediments. PLoS One. 2013;26:e62838. doi: 10.1371/journal.pone.0062838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diarra M.S., Rempel H., Champagne J., Masson L., Pritchard J., Topp E. Distribution of antimicrobial resistance and virulence genes in Enterococcus spp. and characterization of isolates from broiler chickens. Appl. Environ. Microbiol. 2010;76:8033–8043. doi: 10.1128/AEM.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierikx C.M., Goot J.V.D., Smith H.E., Kant A., Mevius D.J. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One. 2013;8:e97005. doi: 10.1371/journal.pone.0079005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka-Malen S., Evers S., Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amin N., Jalal S., Wretlind B. Alterations in gyrA and parC associated with fluoroquinolone resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 1999;43:947–949. doi: 10.1128/aac.43.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST . 2019. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters.http://www.eucast.org Version 9.0. Accessed Jan. 2019. [Google Scholar]
- Fei X., Yin K., Yin C., Hu Y., Li J., Zhou Z., Tian Y., Geng S., Chen X., Pan Z., Li Q., Jiao X. Analysis of prevalence and molecular typing reveal the spread of antimicrobial-resistant Salmonella infection across two breeder chicken farms. Poult. Sci. 2018;97:4374–4383. doi: 10.3382/ps/pey305. [DOI] [PubMed] [Google Scholar]
- Giovanetti E., Brenciani A., Lupidi R., Roberts M.C., Varaldo P. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Chemother. 2003;47:2844–2849. doi: 10.1128/AAC.47.9.2844-2849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J.S., Seo K.W., Kim Y.B., Kang M.S., Song C., Lee Y.J. Prevalence and characterization of Salmonella in two integrated broiler operations in Korea. Ir. Vet. J. 2018;71:3. doi: 10.1186/s13620-018-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Unno T., Jang J., Lim K., Lee S.-N., Ko G., Sadowsky M.J., Hur H.-G. The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. Int. J. Food Microbiol. 2011;144:387–392. doi: 10.1016/j.ijfoodmicro.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Jeon H.Y., Seo K.W., Kim Y.B., Kim D.K., Kim S.W., Lee Y.J. Characteristics of third-generation cephalosporin-resistant Salmonella from retail chicken meat produced by integrated broiler operations. Poult. Sci. 2019;98:1766–1774. doi: 10.3382/ps/pey514. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006;50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Lim T.-H., Jang J.-H., Lee D.-H., Lee B.-H., Kim B.-Y., Kwon J.-H., Choi S.-W., Noh J.-Y., Hong Y.-H., Lee S.-B., Yang S.-Y., Lee H.-J., Lee J.-B., Park S.-Y., Choi I.-S., Song C.-S. Prevalence and antimicrobial resistance of Salmonella species isolated from chicken meats produced by different integrated broiler operations in Korea. Poult. Sci. 2012;91:2370–2375. doi: 10.3382/ps.2012-02357. [DOI] [PubMed] [Google Scholar]
- Kim Y.B., Seo H.J., Seo K.W., Jeon H.Y., Kim D.K., Kim S.W., Lim S.K., Lee Y.J. Characteristics of high-level ciprofloxacin-resistant enterococcus faecalis and enterococcus faecium from retail chicken meat in Korea. J. Food Prot. 2018;81:1357–1363. doi: 10.4315/0362-028X.JFP-18-046. [DOI] [PubMed] [Google Scholar]
- Kim Y.B., Seo K.W., Jeon H.Y., Lim S.K., Sung H.W., Lee Y.J. Molecular characterization of erythromycin and tetracycline-resistant Enterococcus faecalis isolated from retail chicken meats. Poult. Sci. 2019;98:977–983. doi: 10.3382/ps/pey477. [DOI] [PubMed] [Google Scholar]
- Kilbi N., Slama K.B., Masmoudi A., Gharbi S., Ruiz-Larrea F., Fendri C., Boudabous A., Torres C. Diversity of structures carrying the aac(6')-aph(2") gene in clinical Enterococcus faecalis and Enterococcus faecium strains isolated in Tunisia. J. Chemother. 2006;18:353–359. doi: 10.1179/joc.2006.18.4.353. [DOI] [PubMed] [Google Scholar]
- Kwak Y.G., Truong-Bolduc Q.C., Kim H.B., Song K.-H., Kim E.S., Hooper D.C. Association of norB overexpression and fluoroquinolone resistance in clinical isolates of Staphylococcus aureus from Korea. J. Antimicrob. Chemother. 2013;68:2766–2772. doi: 10.1093/jac/dkt286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis H.L., Willems R.J.L., Top J., Bonten M.J.M. High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J. Clin. Microbiol. 2006;44:1059–1064. doi: 10.1128/JCM.44.3.1059-1064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K., Lee Y.S., Park Y.K., Kim B.S. Mutations in the gyrA and parC genes in ciprofloxacin-resistant clinical isolates of Acinetobacter baumannii in Korea. Microbiol. Immunol. 2005;49:647–653. doi: 10.1111/j.1348-0421.2005.tb03643.x. [DOI] [PubMed] [Google Scholar]
- Li X., Aly S.S., Su Z., Pereira R.V., Williams D.R., Rossitto P., Champagne J.D., Chase J., Nguyen T., Atwill E.R. Phenotypic antimicrobial resistance profiles of E. coli and Enterococcus from daily cattle in different management units on a central California daily. Clin. Microbiol. 2018;7:311. [Google Scholar]
- Niu H., Yu H., Hu T., Tian G., Zhang L., Guo X., Hu H., Wang Z. The prevalence of aminoglycoside-modifing enzyme and virulence genes among enterococci with high-level aminoglycoside resistance in inner Mongolia, China. Braz. J. Microbiol. 2016;47:691–696. doi: 10.1016/j.bjm.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakiewicz A., Ziółkowska G., Trosćiańczyk A., Zięba P., Gnat S. Determination of antimicrobial resistance of Enterococcus strains isolated from pigs and their genotypic characterization by method of amplification of DNA fragments surrounding rare restriction sites (ADSRRS fingerprinting) J. Med. Microbiol. 2017;66:175–183. doi: 10.1099/jmm.0.000400. [DOI] [PubMed] [Google Scholar]
- Nowakiewicz A., Ziólkowska G., Trościańczyk A., Zięba P., Gnat S. Determination of resistance and virulence genes in Enterococcus faecalis and E. faecium strains isolated from poultry and their genotypic characterization by ADSRRS-fingerprinting. Poult. Sci. 2017;96:986–996. doi: 10.3382/ps/pew365. [DOI] [PubMed] [Google Scholar]
- Olsen R.H., Schønheyder H.C., Christensen H., Bisgaaard M. Enterococcus faecalis of human and poultry origin share virulence genes supporting the zoonotic potential of E. faecalis. Zoonoses Public Health. 2011;59:256–263. doi: 10.1111/j.1863-2378.2011.01442.x. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Kappell A.D., Elhadidy M., Elmougy F., El-Ghany W.A.A., Orabi A., Mubarak A.S., Dawoud T.M., Hemeg H.A., Moussa I.M.I., Hessain A.M., Yousef H.M.Y. Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: a risk to public health and food safety. Sci. Rep. 2018;8:5859. doi: 10.1038/s41598-018-23962-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyamada Y., Ito H., Inoue M., Yamagishi J. Topoisomerase mutations and efflux are associated with fluoroquinolone resistance in Enterococcus faecalis. J. Med. Microbiol. 2006;55:1395–1401. doi: 10.1099/jmm.0.46636-0. [DOI] [PubMed] [Google Scholar]
- Petersen A., Jensen J.B. Analysis of gyrA and parC mutations in enterococci from environmental samples with reduced susceptibility to ciprofloxacin. FEMS Microbiol. Lett. 2004;231:73–76. doi: 10.1016/S0378-1097(03)00929-7. [DOI] [PubMed] [Google Scholar]
- Poeta P., Costa D., Sáenz Y., Klibi N., Ruiz-Larrea F., Rodrigues J., Torres C. Characterization of antibiotic resistance genes and virulence factors in faecal enterococci of wild animals in Portugal. J. Vet. Med. B. Infect. Dis. Vet. Public Health. 2005;52:396–402. doi: 10.1111/j.1439-0450.2005.00881.x. [DOI] [PubMed] [Google Scholar]
- Rizzotti L., Simeoni D., Cocconcelli P., Gazzola S., Dellaglio F., Torriani S. Contribution of enterococci to the spread of antibiotic resistance in the production chain of swine meat commodities. J. Food Prot. 2005;68:955–965. doi: 10.4315/0362-028x-68.5.955. [DOI] [PubMed] [Google Scholar]
- Rosvoll T.C., Lindstad B.L., Lunde T.M., Hegstad K., Aasnaes B., Hammerum A.M., Lester C.H., Simonsen G.S., Sundsfjord A., Pedersen T. Increased high-level gentamicin resistance in invasive Enterococcus faecium is associated with aac(6′)-Ie-aph(2″)-Ia-encoding transferable megaplasmids hosted by major hospital-adapted lineages. FEMS Immunol. Med. Microbiol. 2012;66:166–176. doi: 10.1111/j.1574-695X.2012.00997.x. [DOI] [PubMed] [Google Scholar]
- Seo K.W., Kim Y.B., Jeon H.Y., Lim S.K., Lee Y.J. Comparative genetic characterization of third-generation cephalosporin-resistant Escherichia coli from chicken meat produced by integrated broiler operations in South Korea. Poult. Sci. 2018;97:2871–2879. doi: 10.3382/ps/pey127. [DOI] [PubMed] [Google Scholar]
- Strateva T., Atanasova D., Savov E., Petrova G., Mitov I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016;20:127–133. doi: 10.1016/j.bjid.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo E.E., Al-Swith N., John P., Jacob L.E., Mohanakrishnan S. Characterization of high-level aminoglycoside-resistant enterococci in Kuwait hospitals. Microb. Drug Resist. 2004;10:139–145. doi: 10.1089/1076629041310037. [DOI] [PubMed] [Google Scholar]
- United States Department of Agriculture (USDA) APHIS Publication; Washington, D C: 2012. National Poultry Improvement Plan and Auxiliary Provisions. [Google Scholar]
- Vakulenko S.B., Donabedian S.M., Anatoliy M., Zervos M.J., Lerner S.A., Chow J.W., Voskresenskiy A.M. Multiplex PCR for detection of aminoglycoside resistance genes in Enterococci. Antimicrob. Agents Chemother. 2003;47:1423–1426. doi: 10.1128/AAC.47.4.1423-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Kobayashi N., Quiñones D., Nagashima S., Uehara N., Watanabe N. Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6’)-Ie-aph(2’’)-Ia or aph(2’’)-Ie in a Japanese hospital. Microb. Drug Resist. 2009;15:185–194. doi: 10.1089/mdr.2009.0917. [DOI] [PubMed] [Google Scholar]