Analysis of saliva samples obtained from 16 patients with Sjögren’s syndrome and 10 sicca controls revealed reduced methylation levels of the H19 locus imprinting control region in the patient group compared to the sicca control group. H19 RNA levels were increased in patients' peripheral blood mononuclear cells compared to the sicca controls.
Keywords: H19, methylation, saliva, Sjögren’s syndrome
Summary
Epigenetic mechanisms have been implicated in the pathogenesis of Sjögren’s syndrome (SS). Extensive alterations in DNA methylation have been described in minor salivary gland (MSG) epithelial cells and lymphocytes derived from SS patients compared to sicca controls. In an effort to identify novel potential epigenetic markers that could prove useful in diagnosis and disease monitoring, we explored whether DNA methylation differences can also be detected in saliva from SS patients compared to sicca controls. We performed DNA methylation analysis by methylation‐sensitive restriction digestion followed by quantitative real‐time polymerase chain reaction of selected genomic loci in saliva samples of 16 SS patients and 10 sicca controls with negative MSG biopsy. We identified reduced DNA methylation of the imprinting control region (ICR) of the H19 locus in SS patient saliva compared to sicca controls. Levels of saliva H19 ICR methylation were negatively correlated with C4 serum complement levels. Consistent with the reduced methylation of the ICR, H19 RNA levels were increased in SS patient peripheral blood mononuclear cells (PBMCs), while no significant change was observed in MSG H19 RNA levels compared to sicca controls. Our findings support that H19 ICR methylation could be a useful molecular epigenetic marker in monitoring patients with SS, highlighting saliva as a valuable biological sample in SS research and clinical practice. The role of H19 in SS pathogenesis remains to be addressed.
Introduction
Sjögren’s syndrome (SS) is a chronic autoimmune disease, affecting primarily the exocrine glands. In many cases the disease may also extend to epithelial structures of other organs beyond the salivary glands (kidneys, lungs, liver), and therefore it is considered as autoimmune epithelitis [1]. Approximately 15% of patients may develop extra‐epithelial manifestations in the form of vasculitis, while 5–8% of patients may develop non‐Hodgkin’s lymphoma. Similar to other autoimmune diseases, SS is characterized by a remarkable clinical heterogeneity, often challenging diagnosis and treatment options [2]. In recent years an effort has been undertaken to stratify SS patients based on the endotypes of the disease; that is, the pathogenetic mechanisms operating in different clinical phenotypes. Thus, molecular biomarkers are currently being explored as useful tools in diagnosis, prognosis, monitoring and treatment, using either targeted or high‐throughput approaches [3].
Previous studies have uncovered extensive alterations in DNA methylation in minor salivary gland (MSG) epithelial cells and peripheral blood mononuclear cells (PBMCs) of SS patients, compared to sicca controls [4]. Herein, we sought to identify whether such differences can also be detected in saliva DNA. We undertook a candidate‐based approach, including the H19 imprinting control region among the loci examined. The H19 gene encodes for a long non‐coding RNA (lncRNA) which has been implicated in autoimmunity via little‐understood mechanisms, including regulation of downstream targets [5]. The gene is normally expressed from the paternal allele, while the maternal allele is methylated and transcriptionally silent. We identify reduced DNA methylation of the imprinting control region (ICR) upstream of H19 in saliva collected from SS patients compared to sicca controls. H19 ICR methylation levels correlated with clinical parameters and its RNA levels were also found to differ in patient PBMCs, supporting a role of this long non‐coding RNA in SS pathophysiology, as well as its potential use as a biomarker.
Materials and methods
Sixteen patients with primary Sjogren’s syndrome [age = 63·2, mean ± standard deviation (s.d.) = 8·8)] who fulfilled the American–European SS classification criteria were studied [6]. Ten sicca controls with negative MSG biopsy (age = 57·2, mean ± s.d. = 13·7) were used for comparison. Sicca controls were patients who visited the physician with symptoms of persistent dry mouth with no other symptoms, clinical manifestations or obvious cause and were subjected to MSG biopsy, which was revealed as negative for SS histopathological findings. All individuals consented to participate in the study, which was also approved by the institutional ethical committee in the context of a European Union (EU)‐sponsored grant (HarmonicSS no. 731944). Whole unstimulated saliva was collected from patients and controls during morning hours, following a standard 15‐min collection protocol after a minimum of 2 h of fasting. Biopsy scoring was performed as described previously [7]. In all SS patients who participated in the study, the clinical manifestations (both glandular and extraglandular) and the laboratory data were recorded at the time of saliva collection.
DNA was extracted from saliva samples by phenol/chloroform followed by ethanol precipitation. Restriction analysis was performed using 800 ng of genomic DNA and the MspI or HpaII endonucleases at 37°C for 2 h. Following digestion, reactions were diluted and 34 ng of each sample or equal amount of undigested gDNA were used as templates in quantitative real‐time polymerase chain reaction (PCR) with specific primers. Primer sequences for genomic regions were as follows: H19 ICR forward: 5′‐GAGCCGCACCAGATCTTCAG‐3′, H19 ICR reverse: 5′‐TTGGTGGAACACACTGTGATCA‐3′; long interspersed nuclear element (LINE1) forward: 5′‐TTCCGAGTCAAAGAAAGG‐3′, LINE1 reverse: 5′‐AGGTGCGGGATATAGTCTC‐3′; histone cluster 1 H3 family member b (HIST1H3B) forward: 5′‐ CCCACACTTCTTATGCGACA‐3′, HIST1H3B reverse: 5′ CTGTGCCTGGTTGCAGATTA‐3′; and single‐stranded DNA binding. protein 1 (SSBP1)S forward: 5′‐GTACAGACGCCGTCCAGAAA‐3′, SSBP1S reverse: 5′‐ CGGTCCCTGTGCTCTGA‐3′. For expression analysis, PBMCs and MSG samples from 13 sicca controls (age = 51·1, mean ± s.d. = 3·5)]and eight SS patients (age = 56·1, mean ± s.d. = 1·767) were used. Total RNA was extracted from PBMCs and MSG tissues using the Qiagen RNeasy Micro Kit, reverse‐transcribed (PrimeScript RT reagent kit; Takara) and subjected to quantitative PCR analysis (Kapa SYBR). Primer sequences were as follows: H19 forward: 5′‐AGTGGACTTGGTGACGCTGTAT‐3′, H19 reverse: 5′‐CTCCTGAGAGCTCATTCACTCC‐3′; beta 2 microglobulin (B2M) forward: 5′‐ACACTGAATTCACCCCCAC‐3′, B2M reverse: 5′‐CATCCAATCCAAATGCGGCA‐3′. DNA immunoprecipitation was performed using an antibody specifically recognizing 5‐hydroxymethylated cytosine (Origene, AM33116PU‐N), as previously described [8]. Statistical analysis was performed using GraphPad Prism.
Results
Electrophoretic and spectrophotometric analysis, as well as preliminary experiments of PCR, restriction digestion and immunoprecipitation, confirmed that both the quality and yield of genomic DNA isolated from saliva was suitable for epigenetic studies by molecular methods. Saliva samples from 16 SS patients versus 10 sicca controls were used. The main clinical, serological and immunological characteristics of SS patients are presented in Table 1. To measure relative DNA methylation of the H19 ICR locus, genomic DNA was first digested with either HpaII endonuclease, which is inhibited by cytosine methylation, or its isoschizomer MspI, which cleaves the same sequence independently of methylation status, while an undigested aliquot of each sample was also kept. Digested and non‐digested samples were subjected to quantitative real‐time PCR analysis using specific primers for the H19 ICR. With this approach, as the primers are designed to flank the restriction site, increased DNA methylation levels will lead to reduced HpaII digestion efficiency, and thus increased levels of the amplicon in the PCR. MspI digestion followed by PCR serves as a control to exclude inhibition of the digestion by other factors and was used for normalization in the real‐time PCR. For analysis, first the levels of total (i.e. methylated and unmethylated) H19 amplicon in the MspI‐digested samples, normalized over H19 amplicon levels in their respective undigested aliquots (ΔCt), were quantitated relative to one sicca patient’s sample which was selected as control using the ΔΔCt method. This quantitation did not reveal any difference between the groups of patient and sicca samples, supporting that there was no difference in the efficiency of the restriction digestion between sicca and SS patients’ saliva. In contrast, quantitation of the levels of methylated H19 ICR, as derived by the relative amount of the H19 amplicon in the HpaII‐digested sample over its total levels in the MspI‐digested reaction, again calculated as fold‐enrichment over a sicca sample by the ΔΔCt method, revealed a statistically significant reduction of H19 ICR methylation in SS patients’ saliva compared to sicca controls (Fig. 1b). Additional control experiments examining the methylation status of a region in the Sjögren’s syndrome antigen B gene SSB P1 promoter, which is also subject to methylation, were performed [9]. Analysis of SSB P1 in the same samples using the same methodology showed no difference between patients and controls, supporting that the observed reduction in H19 ICR methylation is specific for this locus (Fig. 1c). As this method examines methylation of specific CpGs, we cannot exclude that other CpGs in the SSB P1 locus could show differences in methylation between SS and controls in saliva.
Table 1.
Clinical, laboratory and histological features of SS patients
| ID | Age | Sex | Focus score | Dry mouth | SGE | Saliva vol (ml)/15 min | Low C4 | C4 levels | Anti‐Ro | Anti‐La | RF | ANA | Cryoglobulins | Dry eyes | Arthritis | Raynaud | Palpable purpura | Sclerosing cholangitis | PBC | Interstitial renal disease | Lymphoma |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | 71 | F | 1·33 | 1 | 0 | 0·250 | 0 | 25·0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 2 | 64 | F | 1·50 | 1 | 1 | 0·115 | 1 | 19·0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Patient 3 | 66 | F | 1·00 | 1 | 0 | 0·120 | 1 | 10·0 | 1 | 0 | 0 | 1 | n.a. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 4 | 66 | F | 2·22 | 1 | 0 | 1·100 | 0 | 22·4 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Patient 5 | 72 | F | 2·00 | 1 | 0 | 1·100 | 0 | 24·1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 6 | 71 | F | 1·00 | 1 | 1 | 1·700 | 1 | 3·4 | 1 | 1 | 1 | 1 | n.a. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Patient 7 | 71 | F | 1·00 | 0 | 0 | 1·500 | 1 | 19·0 | 1 | 0 | 0 | 1 | n.a. | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Patient 8 | 53 | F | 1·35 | 1 | 1 | 0·500 | 1 | 14·0 | 0 | 0 | 0 | 0 | n.a. | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Patient 9 | 66 | F | 1·00 | 1 | 1 | 0·100 | 0 | 26·0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| Patient 10 | 52 | F | 1·71 | 1 | 1 | 0·400 | 1 | 10·0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Patient 11 | 64 | F | 2·40 | 1 | 0 | 2·500 | 1 | 15·8 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 12 | 51 | M | 1·00 | 1 | 0 | 0·500 | 0 | 20·0 | 1 | 1 | n.a. | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 13 | 57 | F | 1·56 | 1 | 0 | 0·300 | 0 | 39·0 | 1 | 1 | 1 | 1 | n.a. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 14 | 75 | F | 1·09 | 1 | 0 | 0·900 | 1 | 17·0 | 0 | 0 | 0 | 1 | n.a. | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 15 | 67 | F | 1·00 | 1 | 0 | 3·000 | 0 | 24·0 | 1 | 0 | 0 | n.a. | n.a. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Patient 16 | 46 | F | 2·33 | 1 | 0 | 1·200 | 1 | 11·0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
0 = not present/negative; 1 = present/positive; n.a. = data not available; SGE = salivary gland enlargement; RF = rheumatoid factor; ANA = anti‐nuclear antibody; PBC = primary biliary cholangitis; F = female; M = male.
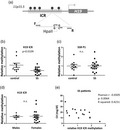
Fig. 1.
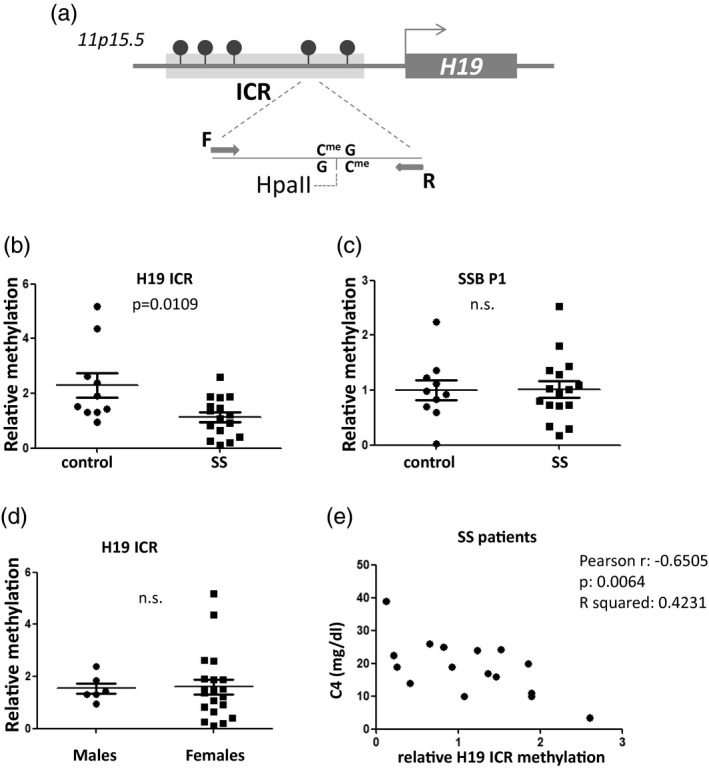
Reduced H19 imprinting control region (ICR) methylation in saliva derived from Sjögren’s syndrome (SS) patients compared to sicca controls. (a) Schematic representation of the H19 ICR genomic region analyzed by methylation‐sensitive restriction digestion and polymerase chain reaction (PCR). Arrows labelled F and R indicate forward and reverse primers used in the polymerase chain reaction. (b) Analysis using methylation‐sensitive restriction endonuclease HpaII followed by quantitative real‐time PCR. Values were normalized to MspI digestion control reactions. (c) No difference was observed in saliva Sjögren’s syndrome antigen B (SSB P1) methylation by the same method. (d) No difference was observed in H19 ICR methylation between male and female donors. (e) Significant negative correlation is observed between patient C4 levels and H19 ICR saliva methylation.
The levels of H19 ICR methylation were not significantly different between males and females (Fig. 1d). In addition, a significant negative correlation was observed between C4 levels and H19 ICR methylation (Fig. 1e). Thus, H19 ICR methylation in saliva could be a novel saliva molecular marker for SS, a finding that is worthy of validation in larger cohorts. No correlation was found between H19 ICR methylation levels and either glandular or extraglandular manifestations, probably due to the low number of SS patients.
To address if reduced ICR methylation correlates with increased H19 expression, we quantitated H19 expression in PBMC and MSG total RNA from SS patients and sicca controls by real‐time quantitative PCR using specific primers. While no significant difference was observed in MSG samples, increased H19 expression (P = 0·021) was observed in SS PBMCs compared to sicca controls, consistent with the reduced methylation observed in saliva (Fig. 2).
Fig. 2.
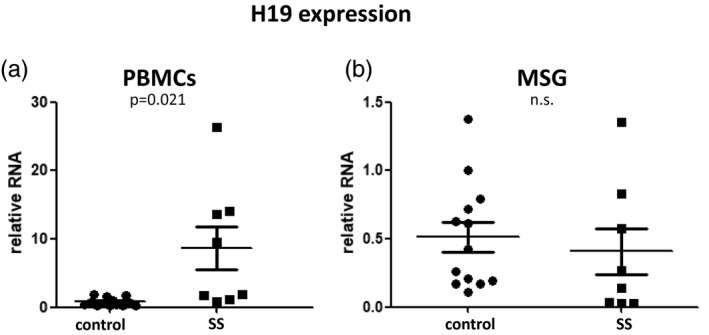
Quantitative analysis of H19 expression in peripheral blood mononuclear cells (PBMCs) and minor salivary glands (MSGs) from Sjögren’s syndrome (SS) patients and sicca controls. (a) Increased H19 expression in PBMCs derived from SS patients compared to sicca controls. (b) No significant difference was observed in MSG H19 expression between patients and sicca controls.
Finally, as 5‐methyl cytosine can be oxidized to 5‐hydroxymethyl cytosine, we addressed the levels of this modification by immunoprecipitation using a specific antibody against 5‐hydroxymethyl cytosine. Comparison of the relative enrichment of 5‐hydroxymethylation of specific loci was performed in a subset of four of the above SS and four sicca patients’ saliva DNA samples by immunoprecipitation followed by quantitative real‐time PCR. Patients were not selected based on specific clinical criteria; however, the immunoprecipitation was performed with 4 μg of sonicated DNA, and therefore samples with adequate DNA yield were selected. In addition to the H19 ICR locus, two other loci were included, the long interspersed nuclear element LINE1 locus, generally methylated and transcriptionally silent, and the HIST1H3B locus, which encodes for histone H3.1 and is generally unmethylated and transcriptionally active. Although the number of samples is too small for statistical analysis, cytosine 5‐hydroxymethylation was found slightly increased in the H19 ICR and also increased in the LINE1 locus of SS versus sicca patients’ saliva (Fig. 3). No difference was observed in the HIST1H3B locus. It should be noted that with this approach, any observed differences represent the mean outcome of possible combined changes in multiple CpGs included in the examined locus. These data complement previous reports on LINE1 methylation in SS [10]. Whether hydroxymethylation functions as an intermediate to the de‐methylation reaction or has other roles remains to be elucidated.
Fig. 3.
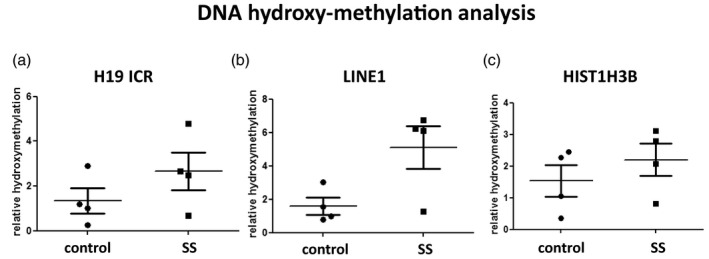
Analysis of cytosine 5‐hydroxymethylation in selected loci in saliva DNA derived from Sjögren’s syndrome (SS) patients compared to sicca controls by DNA immunoprecipitation followed by quantitative real‐time polymerase chain reaction (PCR). (a) Small increase is observed in the H19 imprinting control region (ICR). (b) Increased 5‐hydroxymethyl cytosine in the long interspersed nuclear element (LINE1) locus of SS patients versus sicca controls. (c) The histone cluster 1 H3 family member b (HIST1H3B) locus (encoding histone H3.1) was used as a control for the immunoprecipitation.
Discussion
The term ‘epigenetics’ describes modifications occurring on chromatin without altering the DNA sequence. In SS, several epigenetic changes have been described, including DNA demethylation that predominates in epithelial cells. An epigenome‐wide association study in cultured salivary gland epithelial cells (SGECs) disclosed differentially methylated CpG (DMC) in 4662 positions of 2560 genes, while 575 had two or more DMC sites in SGEC of SS patients compared to sicca controls. Interestingly, further analysis, following gene clustering, highlighted an important proportion of interferon (IFN)‐regulated genes (61%), the calcium pathway and the Wnt pathway to be involved [11]. In another study, hypomethylation of certain IFN‐regulated genes, including the MX1, IFI44L and PARP9 genes, in whole blood and CD19+ B cells, as well as the OAS2 gene in MSG biopsies, were described. Hypomethylation of IFN‐regulated genes in SS patients was associated with their increased mRNA expression in B cells [12]. The same group investigated the DNA methylation levels in certain genes associated with the IFN signature (RSAD2, IFIT1 and IFI44L) and found that they possessed a strong correlation to the RNAseq IFN score (r = 0·84, P < 0·0001), also correctly classifying patients and controls [area under the curve (AUC) = 0·96, P < 0·0001] [13].
Besides the involvement of epigenetic alterations in the effector mechanisms of the immune response, there is also strong evidence that epigenetic changes may affect the autoantigen expression. Indeed, DNA demethylation at the SSB autoantigen gene promoter P1 with a consequent overexpression at both the transcriptional and protein levels in salivary gland epithelial cells might explain the abundance of the autoantigen within the tissue lesion and, therefore, the perpetuation of the autoimmune response [9].
These findings in the literature indicate that epigenetic changes may affect the autoimmune response at all stages and at multiple levels. Prompted by the previous reports, we selected to evaluate epigenetic changes in saliva of SS patients in an attempt to explore whether we can find potential biomarkers or surrogate markers and gain further insight into the pathogenetic mechanisms. In SS, saliva is considered a valuable biological material that could reflect the biology of the inflamed tissue [14]. In a broader context, epigenetic alterations are being examined as potential biomarkers for various diseases. Along this line, we employed a simple methodology to explore DNA methylation differences in SS saliva and have identified reduced DNA methylation of the H19 ICR in saliva derived from SS patients compared to sicca controls.
As performed in the present study, methylation‐sensitive restriction analysis depicts the methylation status of a specific CpG in the restriction site. Previous studies have demonstrated that the levels of methylation of individual CpGs in the H19 ICR correlate with each other. Consistently, in various contexts where reduced methylation of the H19 ICR was observed (e.g. among patients and controls or healthy and tumour tissue), the reduction was also observed in the individual CpGs examined in that locus [15, 16, 17].
In agreement with a potential clinical significance of this epigenetic difference, we found reduced H19 ICR methylation to negatively correlate with C4 levels. Low C4 complement is considered a risk factor for lymphoma development and a measure of the biological activity of the disease [14]. The cellular source of the DNA with reduced H19 ICR methylation is unknown, but we speculate that it could derive from lymphocytes or other leucocytes present in saliva. Methylation of the H19 gene ICR has been implicated in its transcriptional silencing by interfering with the function of a downstream enhancer [5, 18]. Interestingly, H19 expression was found increased in SS patient PBMCs compared to sicca controls, while no difference in H19 expression was observed in MSG samples between the two groups. Taken together, the above data may support a protective role of the H19 up‐regulation in SS. However, two critical limitations of this study, i.e. the small number of SS patients included as well as the fact that all comparisons were made between SS and sicca rather than healthy controls, do not allow drawing more general safe conclusions without additional experimental data. This also applies to the hydroxymethylation data presented, as they were derived from only four SS and four sicca patients.
The potential role of H19 in the context of SS remains unknown. The H19 gene product belongs to the lncRNA (length > 200 nucleotides) family implicated in the regulation of gene expression by interfering with transcription and mRNA turnover [19]. Recently, it has been proposed that lncRNAs may serve as competing endogenous RNAs capable of blocking and inhibiting microRNAs via complementary base pairing [20]. Previous studies have linked this lncRNA to other inflammatory and autoimmune conditions [21, 22, 23, 24]. For instance, increased H19 levels were observed in synovial tissue from rheumatoid arthritis and knee cartilage from osteoarthritis patients compared to controls, as well as muscle biopsies from inclusion body and anti‐Jo‐1‐associated myositis patients compared to controls [21, 22, 23]. Proposed mechanisms include regulation of H19 expression by stress and inflammatory signals, while H19 in turn regulates expression of a number of downstream targets via little‐understood mechanisms. Future research, with a larger number of patients and careful phenotyping of the clinical picture, can shed light on the biological function of H19 in SS and its potential validity as a biomarker.
Disclosures
The authors have no conflicts of interest to declare.
Acknowledgements
The project was supported by the EU grant ‘HarmonicSS’ (grant agreement no: 731944). The authors would like to acknowledge Dr Efstathia Kapsogeorgou and Dr Clio Mavragani for helpful discussions.
References
- 1. Moutsopoulos HM. Sjögren’s syndrome: autoimmune epithelitis. Clin Immunol Immunopathol 1994; 72:162–5. [DOI] [PubMed] [Google Scholar]
- 2. Goules AV, Tzioufas AG. Lymphomagenesis in Sjögren’s syndrome: predictive biomarkers towards precision medicine. Autoimmun Rev 2019; 18:137–43. [DOI] [PubMed] [Google Scholar]
- 3. Tzioufas AG, Goules AV. The necessity of novel biomarkers in primary Sjögren’s syndrome. Clin Exp Rheumatol 2019; 37(Suppl 118):16–8. [PubMed] [Google Scholar]
- 4. Konsta OD, Thabet Y, Le Dantec C et al The contribution of epigenetics in Sjögren’s syndrome. Front Genet 2014; 5:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamann PD, Roux BT, Heward JA et al Transcriptional profiling identifies differential expression of long non‐coding RNAs in Jo‐1 associated and inclusion body myositis. Sci Rep 2017; 7:8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vitali C, Bombardieri S, Jonsson R et al; European Study Group on Classification Criteria for Sjögren's Syndrome . Review. European Study Group on classification criteria for Sjögren’s syndrome. Classification criteria for Sjögren’s syndrome: a revised version of the European criteria proposed by the American–European Consensus Group. Ann Rheum Dis 2002; 61:554–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fisher BA, Jonsson R, Daniels T et al Standardisation of labial salivary gland histopathology in clinical trials in primary Sjögren’s syndrome. Ann Rheum Dis 2017; 76:1161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA Immunoprecipitation (MeDIP). Methods Mol Biol 2009; 507:55–64. [DOI] [PubMed] [Google Scholar]
- 9. Konsta OD, Le Dantec C, Charras A et al Defective DNA methylation in salivary gland epithelial acini from patients with Sjogren’s syndrome is associated with SSB gene expression, anti‐SSB/LA detection, and lymphocyte infiltration. J Autoimmun 2016; 68:30–8. [DOI] [PubMed] [Google Scholar]
- 10. Mavragani CP, Sagalovskiy I, Guo Q et al Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease. Arthritis Rheumatol 2016; 68:2686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charras A, Konsta OD, Le Dantec C et al Cell‐specific epigenome‐wide DNA methylation profile in long‐term cultured minor salivary gland epithelial cells from patients with Sjogren’s syndrome. Ann Rheum Dis 2017; 76:625–8. [DOI] [PubMed] [Google Scholar]
- 12. Imgenberg‐Kreuz J, Sandling JK et al Genome‐wide DNA methylation analysis in multiple tissues in primary Sjogren’s syndrome reveals regulatory effects at interferon‐induced genes. Ann Rheum Dis 2016; 75:2029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Björk A, Richardsdotter Andersson E, Imgenberg‐Kreuz J et al Protein and DNA methylation‐based scores as surrogate markers for interferon system activation in patients with primary Sjögren’s syndrome. RMD Open 2020; 6:e000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goules AV, Tzioufas AG. Primary Sjӧgren’s syndrome: clinical phenotypes, outcome and the development of biomarkers. Autoimmun Rev 2016; 15:695–703. [DOI] [PubMed] [Google Scholar]
- 15. Mansell T, Novakovic B, Meyer B et al The effects of maternal anxiety during pregnancy on IGF2/H19 methylation in cord blood. Transl Psychiatry 2016; 6:e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peng H, Zhao P, Liu J et al Novel epigenomic biomarkers of male infertility identified by methylation patterns of CpG sites within imprinting control regions of H19 and SNRPN genes. OMICS 2018; 22:354–64. [DOI] [PubMed] [Google Scholar]
- 17. Coolen MW, Statham AL, Qu W et al Impact of the genome on the epigenome is manifested in DNA methylation patterns of imprinted regions in monozygotic and dizygotic twins. PLOS ONE 2011; 6:e25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2‐kilobase‐pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 1997; 17:4322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szabó P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal‐specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 2000; 10:607–10. [DOI] [PubMed] [Google Scholar]
- 20. Derrien T, Johnson R, Bussotti G et al The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22:1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen TB, Jensen TI, Clausen BH et al Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–8. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X, Ji S, Cai G et al H19 increases IL‐17A/IL‐23 releases via regulating VDR by Interacting with miR675‐5p/miR22‐5p in ankylosing spondylitis. Mol Ther Nucleic Acids 2019; 19:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu J, Zhang TP, Zhao YL et al Decreased H19, GAS5, and linc0597 expression and association analysis of related gene polymorphisms in rheumatoid arthritis. Biomolecules 2019; 29:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen SW, Wang PY, Liu YC et al Effect of long noncoding RNA H19 overexpression on intestinal barrier function and its potential role in the pathogenesis of ulcerative colitis. Inflamm Bowel Dis 2016; 22:2582–92. [DOI] [PubMed] [Google Scholar]


