Abstract
Several genomic methods were applied for predicting shell quality traits recorded at 4 different hen ages in a White Leghorn line. The accuracies of genomic prediction of single-step GBLUP and single-trait Bayes B were compared with predictions of breeding values based on pedigree-BLUP under single-trait or multitrait models. Breaking strength (BS) and dynamic stiffness (Kdyn) measurements were collected on 18,524 birds from 3 consecutive generations, of which 4,164 animals also had genotypes from an Affymetrix 50K panel containing 49,591 SNPs after quality control edits. All traits had low to moderate heritability, ranging from 0.17 for BS to 0.34 for Kdyn. The highest accuracies of prediction were obtained for the multitrait single-step model. The use of marker information resulted in higher prediction accuracies than pedigree-based models for almost all traits. A genome-wide association study based on a Bayes B model was conducted to detect regions explaining the largest proportion of genetic variance. Across all 8 shell quality traits analyzed, 7 regions each explaining over 2% of genetic variance and 54 regions each explaining over 1% of genetic variance were identified. The windows explaining a large proportion of genetic variance overlapped with several potential candidate genes with biological functions linked to shell formation. A multitrait repeatability model using a single-step method is recommended for genomic evaluation of shell quality in layer chickens.
Key words: eggshell quality, genomic prediction, white layers, GWAS
Introduction
The eggshell is composed of 95% calcium carbonate and 3.5% proteins and proteoglycans that constitute the shell membranes and the shell organic matrix, which is believed to play a key role in the shell formation (Hincke et al., 2012). The eggshell is the natural physical barrier which allows the egg to resist microbial contamination. It also functions to regulate the transfer of metabolic gases and water. Hence, shell quality is one of the key traits for the poultry breeding industry and is critical for maintaining egg integrity during transportation and storage. With its key role in preventing microorganism invasion, it is also crucial for food safety by preventing the spread of foodborne diseases. These issues are relevant not only for eggs used for human consumption but also for hatching eggs used to reproduce genetic stocks at the different levels of poultry multiplication stages, from pure lines to breeder stocks.
Shell quality is a complex trait depending on shell resistance to breakage, plasticity, and permeability for its entire area. Multiple factors contribute to poor eggshell quality including nutrition and the age of the hen (Hamilton et al., 1979, Roberts et al., 2013). Accordingly, measurements of shell quality were taken at 4 hen ages. Changes in hen calcium nutrition and metabolism can result, among other factors, from deterioration of the absorption capacity of the gut and inefficient mobilization of calcium body reserves, which ultimately lead to declining shell quality. Another important factor impacting eggshell quality is the influence of genetics. QTL detection studies for various shell quality traits have been performed, and chromosomes Z, 2, 4, and 8 have been most frequently reported to impact these traits (review by Roberts, 2017). From 15 QTLs detected for eggshell strength by Romé et al. (2015), 4 showed an interaction with diet, 3 with age at egg collection, and 1 QTL involved both variables.
Despite its importance for the egg production industry and recent developments in genomics, very few reports are available evaluating the genetic basis of shell strength using genomic data. We have found only 1 article (Wolc et al., 2011) which compared the accuracy of 2 genomic methods vs. pedigree for puncture score (an eggshell deformation test). Four genome-wide association studies (GWAS) using SNP chips have been reported, using experimental lines or line crosses. Liu et al. (2011) performed GWAS for eggshell weight, eggshell thickness, and eggshell strength in White Leghorn × Brown-Egg Dwarf Layers using a 60K panel; Sun et al. (2015) performed GWAS for the same traits using a 600K panel in White Leghorn × Chinese Dongxiang chickens; Romé et al. (2015) performed diet-specific GWAS for static stiffness and breaking strength (BS) using a 600K panel in an F1 white commercial cross, and Wolc et al. (2014) performed GWAS for puncture score in a brown commercial line using a 42K panel. Most of the previous studies were limited in scope to a single aspect of shell quality (1 type of measurement), a single age of birds, low density of genetic markers, low numbers of genotyped birds, or the use of artificially created mapping populations with allele combinations that may not be relevant to commercial breeding populations. Thus, there is a need for a comprehensive study including genomic prediction and GWAS of shell quality in modern layers. Better understanding of genetics of shell quality and optimization of breeding value estimation methods can enhance genetic improvement of these traits because the accuracy of breeding values is one of the key components determining the rate of genetic gain.
The objective of this study was to compare the accuracy of predictions of breeding values for shell quality traits at 4 different hen ages using pedigree BLUP models with those of single-step GBLUP using single-trait or multitrait models and with a single-trait BayesB marker-effects model. Additionally, regions explaining the largest proportion of genetic variance and candidate genes for shell quality were reported.
Material and methods
The line used for this study was a commercially relevant elite White Leghorn line that has been under selection for multiple generations for many production performance traits, including egg production, egg quality, and feed efficiency. Two lab devices were used to assess shell quality: (a) the Futura-FEST (Bröring; Lohne, Germany), which measures BS (measured in gF) and which reflects the force necessary to break the shell, and (b) the acoustic egg test (AET, De Ketelaere et al., 2000), which uses the vibrational properties of the shell around the egg's equator, by tapping repeatedly (every 90° to cover the entire perimeter) on the surface with an electromagnetic hammer and recovering the sound resonance profiles (Coucke et al., 1999). This process measures the shell integrity (described as the presence of microcracks (1) or, by default, their absence (0) in an intact shell). In intact eggs, the device also uses resonance parameters along with egg mass in an equation to calculate a quantitative value, referred to as dynamic stiffness (Kdyn), which describes the resistance of the shell over its entire surface. Because Kdyn is a quantitative trait with higher heritability than static measurements such as BS, it has been proposed as an adequate trait for selection to improve shell quality (Dunn et al., 2005, Icken et al., 2006, Arango et al., 2016). The traits used in this study (BS and Kdyn) were collected on eggs from 18,524 birds from 3 consecutive generations. Breaking strength was measured on the equator of the egg (BSe) for hens of 26 and 65 wk of age and on poles (BSp) of the egg for hens at 42 and 86 wk of life; whereas Kdyn was measured using the AET Columbus device (Octinion, Leuven, Belgium) at all 4 hen ages. Repeated measurements were taken at each age, but not all birds were evaluated for every trait at every time point. Additional details about these phenotypes can be found in Arango et al. (2016). The summary statistics of the data are presented in Table 1.
Table 1.
Summary statistics for breaking strength measured at the equator (BSe) at 26 or 65 wk of age and at poles (BSp) at 42 or 86 wk and for dynamic stiffness (Kdyn) measured at all 4 hen ages.
| Trait | Min | Max | Mean | SD | N | N BayesB | N validation | nGen |
|---|---|---|---|---|---|---|---|---|
| BSe26 wk | 599 | 6,789 | 3,246 | 734 | 46,009 | 4,685 | 1,012 | 3 |
| BSe65 wk | 855 | 5,872 | 3,010 | 725 | 4,342 | 1,448 | 635 | 3 |
| BSp42 wk | 1,100 | 6,530 | 3,432 | 742 | 13,576 | 1,022 | 1,014 | 1 |
| BSp86 wk | 1,040 | 5,740 | 2,758 | 893 | 14,681 | 2,509 | 651 | 2 |
| Kdyn26 wk | 37.5 | 357.3 | 141.3 | 14.8 | 45,096 | 4,673 | 1,006 | 3 |
| Kdyn42 wk | 37.5 | 219.6 | 152.4 | 14.5 | 20,260 | 2,989 | 1,011 | 3 |
| Kdyn65 wk | 38.1 | 206.7 | 143.3 | 16.6 | 4,209 | 1,426 | 678 | 3 |
| Kdyn86 wk | 62.1 | 357 | 132.6 | 18.1 | 17,865 | 3,485 | 608 | 3 |
Abbreviations: N, total number of phenotypes available for the analysis; N BayesB, number of phenotypes (own phenotypes + family means) used in the BayesB analysis; N validation, number of genotyped individuals with own phenotypes from the final generation used for validation; nGen, number of generations with trait records in the data.
All data used for this study were collected as part of routine data collection in the Hy-Line Int. breeding program. Birds were handled according to the company animal welfare policy that was approved by the company veterinarian.
Genotyping with a customized Affymetrix 50K panel was performed on 4,164 animals, comprising males and females that were selected to produce the subsequent generation. The panel was a subset of the SNP in the commercially available 600k Affymetrix chip (Kranis et al., 2013). The subset of SNP was selected to capture genetic variation in a specific population with some additional SNPs selected based on previous studies. The number of SNPs retained after clustering based on quality control parameters was 49,591. Of the birds with phenotypic records, 600 to 1,000 genotyped animals from the last generation were excluded from the analysis for the purpose of validation. Their nongenotyped sisters were retained in the analysis to approximate expected accuracy of estimated breeding values for males, which do not have their own performance measured for egg-related traits but do have records on female relatives.
An 8-trait animal model with the fixed effects of hatch, machine performing the measurements, and covariate of hen age, as well as random additive genetic and permanent environmental effects, as described in detail by Wolc et al. (2017), was fitted to estimate (co)-variance components using a pedigree-based relationship matrix.
y is a vector of phenotypes.
X is a known design matrix of fixed effects.
b is a vector of fixed effects of hatch of hen, machine performing the measurements and covariates of age.
Z1 is a known design matrix of random additive genetic effects.
a is a vector of random additive genetic effects.
Z2 is a known design matrix for random permanent environmental effects common to all records of a given animal.
p is a vector of permanent environmental effects to account for effects common to repeated records on the same individual.
e is a vector of random residuals.
Covariances between records for the same individual were accounted for by fitting both the genetic and permanent environmental components that were common to records on the same hen. Standard errors of genetic parameters were estimated as posterior standard deviations by POSTGIBBSF90, after 110,000 Markov chain Monte Carlo (MCMC) iterations in GIBBS2F90, discarding first 10,000 iterations as burn-in, keeping every 20th sample, with final estimates of AIREMLF90 used as priors (Misztal et al., 2015). Owing to the high computational demands and poor stability of variance components, single-step analysis was performed as BLUP (without re-estimation of variance components) for 2 groups of 4 traits: BS and Kdyn traits. The inverse of the H-matrix was created with default settings specified in the preGS program, comprising a combination of the inverse of a G matrix created according to the method proposed by VanRaden (2008) with current population allele frequencies and the inverse of the pedigree-based relationship matrix, which were pooled together with respective weights of 0.95 and 0.05. All of these analyses were performed using the BLUPF90 family of programs (Misztal et al., 2015). Additionally, a BayesB analysis was performed in GenSel (Fernando and Garrick, 2013), analyzing each trait phenotype separately and including nongenotyped animals that had genotyped parents as family means and weighting the residuals based on family size (Wolc et al., 2011). BayesB was shown to detect QTL more accurately than other genomic prediction methods used in this study (Wolc et al., 2016). In the Bayes B analysis, 95% of the markers were assumed to have no association with the analyzed traits, and pedigree-based estimates of variance components were used as priors. The MCMC chain length was 31,000 with 1,000 samples discarded as burn in. The proportion of variance explained by markers was estimated as the mean across MCMC samples of the ratio of variance of breeding values divided by phenotypic variance. To evaluate the gains from using a multitrait model, pedigree and single-step analyses were also run as single-trait models on the same data on average records within each hen age. For each model, the accuracy of estimated breeding values was estimated as the correlation of predicted merit with and phenotypes of the validation animals adjusted for fixed effects, with the resulting correlation divided by the square root of heritability. The same data were used for all methods for evaluating accuracy of prediction.
Additional data (650–2,600 individuals depending on trait) were available for an association analysis, which was performed with the Bayes B method. The genome was divided into 998 nonoverlapping one-megabase windows. The ratio of variance of genomic breeding values based on a specific window to variance of genomic breeding values based on the whole genome averaged across MCMC samples was used to measure the proportion of variance explained by that window (Wolc et al., 2012). Genes that overlapped regions explaining more than 2% of genetic variance were identified with the Ensembl BioMart webtool (http://useast.ensembl.org/biomart/) based on the Galgal4 assembly and the Ensembl Genes 85 database.
Results and discussion
Genetic Parameters
Estimates of heritability and repeatability and of correlations because of genetic and permanent environmental effects between the traits are given in Table 2. All traits had low to moderate heritability estimates, with estimates for Kdyn (0.24–0.34) higher than for BS (0.09–0.26). Similar results were obtained by Wolc et al. (2014) in an unrelated brown egg layer line, where estimates of heritability for puncture score were equal to 0.12 and 0.18 for early and late measurements, respectively. For lines representing 3 layer breeds, Arango et al. (2016) reported heritability estimates between 0.33 and 0.36 for Kdyn and between 0.14 and 0.23 for BS. In other studies using hen-average records, rather than individual observations, heritability of shell strength was estimated to be moderate to high (Dunn et al., 2005, Blanco et al., 2014), similar to our estimates of repeatability (0.33–0.58). For comparison, in a population of brown-egg dwarf layers, heritability for eggshell strength was higher than in the present study at 0.24 ± 0.08 (Zhang et al., 2005).
Table 2.
Estimates of genetic correlations (above diagonal) of permanent environmental correlations (below diagonal), of heritability (bold on the diagonal), of repeatability (diagonal), and of the proportion of variance explained by markers (last row) for breaking strength measured at the equator (BSe) at 26 or 65 wk of hen age and at poles (BSp) at hen ages of 42 or 86 wk, and for dynamic stiffness (Kdyn) measured at all 4 hen ages.
| Trait | BSe 26 wk |
BSe 65 wk |
BSp 42 wk |
BSp 86 wk |
Kdyn 26 wk |
Kdyn 42 wk |
Kdyn 65 wk |
Kdyn 86 wk |
|---|---|---|---|---|---|---|---|---|
| BSe26 wk |
0.17 ± 0.09 0.33 ± 0.00 |
0.47 ± 0.21 | 0.60 ± 0.28 | 0.11 ± 0.18 | 0.12 ± 0.17 | 0.29 ± 0.15 | 0.28 ± 0.22 | 0.26 ± 0.13 |
| BSe65 wk | 0.20 ± 0.07 |
0.20 ± 0.03 0.43 ± 0.01 |
0.76 ± 0.09 | 0.84 ± 0.11 | 0.27 ± 0.11 | 0.46 ± 0.11 | 0.70 ± 0.06 | 0.78 ± 0.08 |
| BSp42 wk | 0.20 ± 0.06 | 0.37 ± 0.10 |
0.09 ± 0.02 0.35 ± 0.01 |
0.40 ± 0.10 | 0.19 ± 0.08 | 0.41 ± 0.05 | 0.50 ± 0.06 | 0.40 ± 0.09 |
| BSp86 wk | 0.08 ± 0.04 | -0.03 ± 0.07 | 0.36 ± 0.04 |
0.26 ± 0.02 0.58 ± 0.01 |
0.14 ± 0.06 | 0.20 ± 0.07 | 0.51 ± 0.10 | 0.71 ± 0.07 |
| Kdyn26 wk | 0.98 ± 0.02 | 0.11 ± 0.07 | 0.22 ± 0.05 | -0.03 ± 0.03 |
0.34 ± 0.01 0.60 ± 0.01 |
0.83 ± 0.03 | 0.71 ± 0.06 | 0.47 ± 0.05 |
| Kdyn42 wk | 0.32 ± 0.05 | 0.46 ± 0.07 | 0.96 ± 0.02 | 0.21 ± 0.04 | 0.35 ± 0.03 |
0.33 ± 0.02 0.61 ± 0.01 |
0.93 ± 0.03 | 0.69 ± 0.05 |
| Kdyn65 wk | 0.23 ± 0.06 | 0.99 ± 0.02 | 0.50 ± 0.08 | -0.01 ± 0.10 | 0.16 ± 0.06 | 0.58 ± 0.05 |
0.31 ± 0.03 0.63 ± 0.02 |
0.88 ± 0.08 |
| Kdyn86 wk | 0.23 ± 0.07 | 0.25 ± 0.06 | 0.34 ± 0.04 | 0.94 ± 0.05 | 0.10 ± 0.03 | 0.26 ± 0.03 | 0.25 ± 0.06 |
0.24 ± 0.02 0.72 ± 0.01 |
| h2m | 0.12 | 0.15 | 0.14 | 0.14 | 0.26 | 0.25 | 0.25 | 0.15 |
As expected, estimates of genetic correlations between consecutive ages for Kdyn were high (>0.8) and eroded as the time between the measurement periods increased. A similar observation was made by Wolc et al., (2017) in both Rhode Island Red and White Leghorn lines. The BS traits had lower genetic correlations among each other (ranging from 0.11 to 0.84) than did Kdyn. The BSe measurements had slightly higher genetic correlations with each other (0.47) than the BSp measurements (0.40), despite the greater age gap between measurements (39 vs.44 wk). Estimates of genetic correlations between BSe and BSp in lines from 3 breeds ranged from 0.51 to 0.68 (Arango et al., 2016). Estimates of genetic correlations between the BS and Kdyn traits were more variable, ranging from 0.12 to 0.78. For traits were measured at the same age, this could be attributed to some extent to the covariance of permanent environmental effects between traits. Breaking strength and Kdyn measurements were only moderately correlated, which confirms that these traits measure different aspects of shell quality. The BS at poles is more critical for egg integrity after packing, but a weak BS on the equator may result in cage checks and cracks because of impacts while traveling on the conveyor systems, which transport eggs from point of lay to processing or grading stations in commercial farms. The Kdyn measures different aspect of shell integrity and was only moderately genetically correlated with BS. Similar observations had been made by Arango et al. (2016), who reported estimates of genetic correlations between BS traits and Kdyn ranging from 0.23 to 0.51.
Dynamic stiffness has been reported to be a good predictor of the incidence of shell cracks, particularly microcracks. Arango et al. (2016) estimated genetic correlations of BS and Kdyn with percentage of cracks to be around −0.3 and −0.6, respectively. These results are in agreement with Bain et al. (2006), who showed that increased Kdyn values resulted in a lower frequency of cracks during transportation.
Accuracy of Prediction
In terms of predictive ability (Table 3), for traits with lower numbers of observations, the multitrait model was advantageous over the single-trait model for both pedigree-based and single-step analyses. The highest accuracies were obtained for the multitrait single-step model. For almost all traits, any method that included marker information had higher accuracy than the corresponding model that was based solely on pedigree information. Gains in accuracy from including genomic information in breeding value estimation were smaller than reported in some previous studies (Wolc et al., 2011), but in this case, observations from contemporaries were retained in training, which creates good population structure (presence of half and full sibs) for pedigree-based predictions. BayesB, which used only genotyped animals and family means, had similar accuracies as the single-trait single step model that used all genotyped and nongenotyped individuals.
Table 3.
Accuracy of prediction of breeding values from different models (pedST—pedigree-based single trait, pedMT—pedigree-based multitrait, ssST—single-step single-trait, ssMT—single-step multitrait, BB95—Bayes B) for breaking strength measured at the equator (BSe) at 26 or 65 wk of age and at poles (BSp) at 42 or 86 wk and for dynamic stiffness (Kdyn) measured at all 4 hen ages.
| Trait | pedST | pedMT | ssST | ssMT | BB95 |
|---|---|---|---|---|---|
| BSe26 wk | 0.37 | 0.39 | 0.53 | 0.55 | 0.50 |
| BSe65 wk | 0.34 | 0.50 | 0.42 | 0.55 | 0.39 |
| BSp42 wk | 0.38 | 0.50 | 0.67 | 0.71 | 0.71 |
| BSp86 wk | 0.52 | 0.51 | 0.68 | 0.69 | 0.63 |
| Kdyn26 wk | 0.67 | 0.69 | 0.84 | 0.85 | 0.82 |
| Kdyn42 wk | 0.52 | 0.52 | 0.65 | 0.67 | 0.64 |
| Kdyn65 wk | 0.28 | 0.52 | 0.47 | 0.69 | 0.48 |
| Kdyn86 wk | 0.61 | 0.60 | 0.84 | 0.84 | 0.85 |
The method with the highest accuracy is marked in bold.
Genome Wide Association Analyses
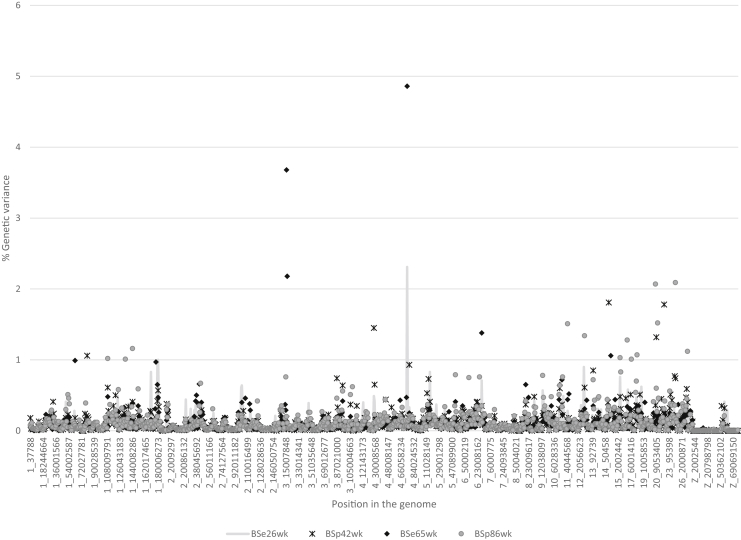
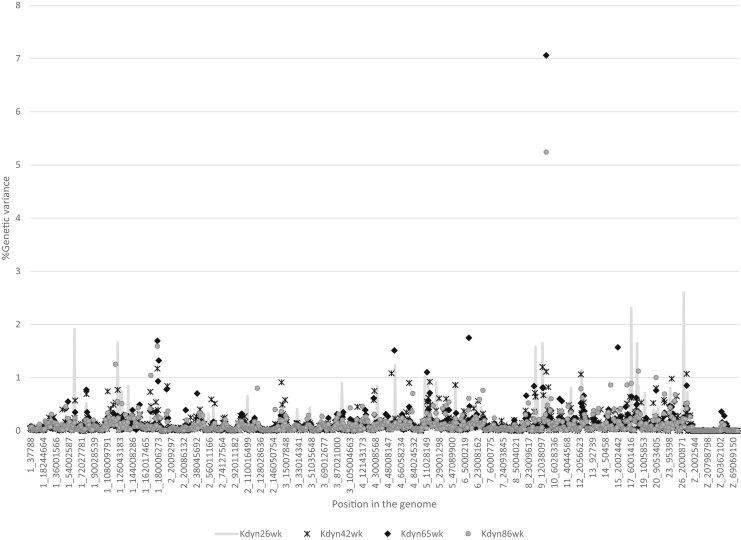
Estimates of the proportion of genetic variance explained by each 1 Mb SNP window across the genome for BSe and BSp are illustrated in Figure 1 and for Kdyn in Figure 2. The majority of regions that each explained over 2% of genetic variance (Table 4) were identified for traits measured in older hens. Several QTL regions explained a higher percentage of genetic variance at older than at younger ages. Similar results were obtained by Sun et al. (2015), who suggested the effects of some variants were age-dependent. Some studies (Abasht et al., 2009, Liu et al., 2011) suggested that variants influencing eggshell quality were characteristic for either early or late shell quality but not for both. This same trend was observed here, although some regions that explained about 1% of variance for both young and older hens were identified.
Figure 1.
Proportion of genetic variance explained by each 1 Mb SNP window across the genome for breaking strength measured at the equator (BSe) at 26 and 65 wk of age and at poles (BSp) at 42 and 86 wk.
Figure 2.
Proportion of genetic variance explained by each 1 Mb SNP window across the genome for dynamic stiffness (Kdyn) measured at 26, 42, 65 and 86 wk of age.
Table 4.
Genomic regions identified to be associated with breaking strength measured at the equator (BSe) at 26 or 65 wk of age and at poles (BSp) at 42 or 86 wk and for dynamic stiffness (Kdyn) measured at all 4 hen ages from a Bayes B analysis, along with candidate genes. Only regions explaining over 2% of genetic variance were included in the table.
| Chr | Window Mb | Trait | Variance explained [%] | P > 0 (avg) | Candidate genes (location in Mb) | QTLs from AnimalQTLdb |
|---|---|---|---|---|---|---|
| 3 | 16-17 | BSe65 wk | 3.7 | 0.997 (0.883) | SLC8A1 (15.7)—calcium ion transport, positive regulation of bone mineralization | Egg weight, Eggshell strength |
| 17-18 | BSe65 wk | 2.2 | 0.937 (0.560) | CAPN2, CAPN8 (17.0)—calcium ion binding | Bone mineral density | |
| 4 | 75-76 | BSe65 wk | 4.9 | 0.987 (0.817) | NCAPG (75.5)—actin cytoskeleton, CCKAR (72.8) and LCOLR (75.4)—effect on body weight | Egg weight, Eggshell weight |
| BSe26 wk | 2.3 | 1 (0.943) | ||||
| 9 | 19-20 | Kdyn65 wk | 7.1 | 1 (1) | RARRES1 (21.9)—coding ovocalyxin 32, an eggshell matrix protein | Egg weight |
| Kdyn86 wk | 5.2 | 1 (1) | ||||
| 17 | 7-8 | Kdyn26 wk | 2.3 | 1 (1) | GSN (8.4)—calcium ion binding, actin filament severing | Eggshell weight, Bone mineral density |
| 20 | 8-9 | BSp86 wk | 2.1 | 1 (0.94) | CDH4 (7.7)—calcium ion binding | |
| 24 | 3-4 | BSp86 wk | 2.1 | 1 (0.96) | CBL (4.2)—calcium ion binding | Eggshell weight |
| 26 | 5-6 | Kdyn26 wk | 2.6 | 1 (1) | ITPR3 (4.5)—calcium ion transport |
Of the 8 1 Mb regions reported in Table 4, 5 had been previously reported as being associated with egg weight or eggshell quality. Three regions (chr3: 17 Mb, chr20: 8 Mb, and chr26: 5 Mb) had not been previously reported as being associated with egg weight or shell quality in the QTL database as of 6/10/2019 (www.animalgenome.org). Regions with the highest percent of genetic variance explained were identified for BSe at 65 wk of age on chromosomes 3 and 4 and for Kdyn on chromosome 9. The significant region on chromosome 3 was comprised of 2 apparent regions at 16 and 17 Mb, which explained 3.9 and 2.2% of genetic variance, respectively. Two genes located in this region, CAPN2 and CAPN8, are involved in interactions with calcium ions, which is suggestive of them having a role in shell mineralization. The nearby SLC8A1 gene is involved in calcium ion transport and positive regulation of bone mineralization. A window explaining 4.9% of genetic variance for BSe at 65 wk was located on chromosome 4 (75 Mb). The same window explained 2.3% of genetic variance for the BSe at 26 wk of age. That region overlapped the NCAPG gene that was reported by Sun et al. (2015) as having both an effect on eggshell quality for young hens and for influencing eggshell weight. This gene is also associated with growth in many livestock species (see review by Takasuga, 2016), and it is hypothesized to impact eggshell quality by influencing egg weight. The other 2 genes (ITPR2, PIK3C2G) identified by Sun et al. (2015) were both located on chromosome 1 (at 64 and 67 Mb), and the closest associations in our study were for BSe at 65 wk of age (1.0% of genetic variance at 63 Mb), Kdyn at 26 wk of age (1.9% of genetic variance at 62 Mb), and Kdyn at 42 wk of age (0.6% of genetic variance at 63 Mb).
A number of proteins involved in the process of shell mineralization and antibacterial defense have been reported so far (Gautron et al., 2001, Nys et al., 2004, Hincke et al., 2012, Marie et al., 2015). Some of the eggshell matrix genes described by either Marie et al. (2015) or Dunn et al. (2009) overlapped with regions explaining a high percentage of genetic variance in the present study. For those proteins, the RARRES1 and GSN genes were located in closest proximity to the identified regions on chromosome 9 with over 2% of variance explained. The gene RARRES1 (chr 9: 21.9 Mb) encodes Ovocalyxin-32, an eggshell matrix protein found within the outer layers of the eggshell and in the cuticle. The association of SNPs within exons of Ovocalyxin-32 with eggshell quality was described in detail by Fulton et al. (2012) and in previous studies (Gautron et al., 2001, Dunn et al., 2009, Uemoto et al., 2009, Takahashi et al., 2009, Takahashi et al., 2010). Association studies revealed a significant effect of Ovocalyxin-32 on several shell-related traits, including shell color in white egg lines and line-specific significant effects on albumen height, early egg weight, puncture score, and yolk weight (Fulton et al., 2012). In the study of Dunn et al. (2012), markers located in the RARRES1 gene were associated with orientation of the calcium carbonate crystals.
The closest candidate gene to the region identified on chromosome 17 (Table 4) is GSN (chr17: 8.4 Mb), which encodes gelsolin, a calcium-binding protein and is abundant at the terminal stage of shell mineralization. The QTL regions on chromosomes 20 and 24 were located close to genes encoding calcium binding proteins, CDH4 and CBL, respectively.
The gene located in close proximity to the region on chromosome 26 (ITPR3) (Table 4) is IP3 receptor-2 and is involved in ion transfer for supplying eggshell mineral precursors in the hen's uterus (Jonchère et al., 2012). Another IP3 (ITPR2) (chr1: 67 Mb) was identified in the work of Sun et al. (2015) as a promising candidate gene involved in eggshell calcification and eggshell mechanical properties. In the study reported herein, there was no indication of a QTL in the region of this gene. Perhaps the intensive and multigenerational selection for shell quality in this commercial line has resulted in fixation of favorable alleles in this region, or the previously reported QTL was population specific. The organic matrix is believed to play a key role in shell formation (Nys et al., 2004, Hincke et al., 2012). It is hypothesized that the eggshell organic matrix plays an important regulatory role in assembly of the calcite zone with calcium carbonate. There is evidence that proteins in the shell influence the variability of calcium carbonate crystal traits, such as crystal size and orientation, which are directly related with the ultrastructure of the eggshell (Dunn et al., 2012).
Conclusions
Accuracies of predictions of breeding values for shell quality traits at different hen ages were compared using pedigree-based models and using genomic information. Use of genomic information resulted in higher accuracies than pedigree-based models for almost all traits. The highest accuracies were obtained with a multitrait single-step model. All traits had low to moderate heritability, with estimates for Kdyn being higher than those for BS. Breaking strength and Kdyn measurements were only moderately correlated, which confirms that these traits measure different aspects of shell quality. A multitrait repeatability model implemented in a single-step method is recommended for genomic evaluation of shell quality in layer chickens.
Seven regions each explaining over 2% of genetic variance and 54 regions each explaining over 1% of genetic variance were identified. Several candidate genes were identified in close proximity to those regions, some of which overlapped with previously reported QTL. Further studies are needed to determine and validate the specific roles of QTL identified for these candidate genes for eggshell strength and resilience.
Acknowledgments
This study was supported by Hy-Line International.
Acknowledgments
Conflict of interest statement: The authors did not provide any conflict of interest statement.
References
- Abasht B., Sandford E., Arango J., Settar P., Fulton J.E., O’Sullivan N.P., Hassen A., Habier D., Fernando R.L., Dekkers D.C., Lamont S.J. Extent and consistency of linkage disequilibrium and identification of DNA markers for production and egg quality traits in commercial layer chicken populations. BMC Genom. 2009;10:S2. doi: 10.1186/1471-2164-10-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango J., Wolc A., Settar P., O’Sullivan N.P. Model comparison to evaluate a shell quality bio-complex in layer hens. Poult. Sci. 2016;95:2520–2527. doi: 10.3382/ps/pew286. [DOI] [PubMed] [Google Scholar]
- Bain M.M., Dunn I.C., Wilson P.W., Joseph N., De Ketelaere B., De Baerdemaeker J., Waddington D. Probability of an egg cracking during packing can be predicted using a simple non-destructive acoustic test. Br. Poult. Sci. 2006;47:462–469. doi: 10.1080/00071660600829233. [DOI] [PubMed] [Google Scholar]
- Blanco A.E., Icken W., Ould-Ali D., Cavero D., Schmutz M. Genetic parameters of egg quality traits on different pedigree layers with special focus on dynamic stiffness. Poult. Sci. 2014;93:2457–2463. doi: 10.3382/ps.2014-04132. [DOI] [PubMed] [Google Scholar]
- Coucke P., Dewil E., Decuypere E., De Baerdemaeker J. Measuring the mechanical stiffness of an eggshell using resonant frequency analysis. Poult. Sci. 1999;40:227–232. doi: 10.1080/00071669987647. [DOI] [PubMed] [Google Scholar]
- De Ketelaere B., Coucke P., De Baerdemaeker J. Eggshell crack detection based on acoustic resonance frequency analysis. J. Agric. Engng. Res. 2000;76:157–163. [Google Scholar]
- Dunn I.C., Bain M., Edmond A., Wilson P.W., Joseph N., Solomon S., De Ketelaere B., De Baerdemaeker J., Schmutz M., Preisinger R., Waddington D. Heritability and genetic correlation of measurements derived from acoustic resonance frequency analysis; a novel method of determining eggshell quality in domestic hens. Br. Poult. Sci. 2005;46:280–286. doi: 10.1080/00071660500098574. [DOI] [PubMed] [Google Scholar]
- Dunn I.C., Joseph N.T., Bain M., Edmond A., Wilson P.W., Milona P., Nys Y., Gautron J., Schmutz M., Preisinger R., Waddington D. Polymorphisms in eggshell organic matrix genes are associated with eggshell quality measurements in pedigree Rhode Island Red hens. Anim. Genet. 2009;40:110–114. doi: 10.1111/j.1365-2052.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- Dunn I.C., Rodríguez-Navarro A.B., Mcdade K., Schmutz M., Preisinger R., Waddington D., Wilson P.W., Bain M.M. Genetic variation in eggshell crystal size and orientation is large and these traits are correlated with shell thickness and are associated with eggshell matrix protein markers. Anim. Genet. 2012;43:410–418. doi: 10.1111/j.1365-2052.2011.02280.x. [DOI] [PubMed] [Google Scholar]
- Fernando R.L., Garrick D.J. Bayesian methods applied to GWAS. In: Gondro C., van der Werf J., Hayes B., editors. Vol.1019. Springer Science+Business Media; New York, NY: 2013. pp. 237–274. (Genome-Wide Association Studies and Genomic Prediction, Methods in Molecular Biology). [Google Scholar]
- Fulton J.E., Soller M., Lund A.R., Arango J., Lipkin E. Variation in the ovocalyxin-32 gene in commercial egg-laying chickens and egg quality traits. Anim. Genet. 2012;43:102–113. doi: 10.1111/j.1365-2052.2012.02384.x. [DOI] [PubMed] [Google Scholar]
- Gautron J., Hincke M.T., Mann K., Panheleux M., Bain M., McKee M.D., Solomon S.E., Nys Y. Ovocalyxin-32, a novel chicken eggshell matrix protein. Isolation, amino acid sequencing, cloning and immunocytochemical localization. J. Bio. Chem. 2001;276:39243–39252. doi: 10.1074/jbc.M104543200. [DOI] [PubMed] [Google Scholar]
- Hamilton R.M.G., Thompson B.K., Voisey P.W. The effects of age and strain on the relationships between destructive and non-destructive measurements of eggshell strength for White Leghorn hens. Poult. Sci. 1979;58:1125–1132. [Google Scholar]
- Hincke M.T., Nys Y., Gautron J., Mann K., Rodriguez-Navarro A.B., McKee M.D. The eggshell: structure, composition and mineralization. Front. Biosci. 2012;17:1266–1280. doi: 10.2741/3985. [DOI] [PubMed] [Google Scholar]
- Icken W., Schmutz M., Preisinger R. Dynamic stiffness measurements with the “crack detector”: a new method to improve egg shell strength. Lohmann Info. 2006;41:13–19. [Google Scholar]
- Jonchère V., Brionne A., Gautron J., Nys Y. Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 2012;12:10. doi: 10.1186/1472-6793-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranis A., Gheyas A.A., Boschiero C., Turner F., Yu L., Smith S. Development of a high density 600K SNP genotyping array for chicken. BMC Genom. 2013;14:59. doi: 10.1186/1471-2164-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li D., Liu J., Chen S., Qu L., Zheng J. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PLoS One. 2011;6:1427–1430. doi: 10.1371/journal.pone.0028600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie P., Labas V., Brionne A., Harichaux G., Hennequet-Antier C., Nys Y., Gautron J. Quantitative proteomics and bioinformatic analysis provide new insight into protein function during avian eggshell biomineralization. J. Proteome. 2015;113:178–193. doi: 10.1016/j.jprot.2014.09.024. [DOI] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Lourenco D., Aguilar I., Legarra A., Vitezica Z. Manual for BLUPF90 Family of Programs. 2015. http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all2.pdf
- Nys Y., Gautron J., Garcia-Ruiz J.M., Hincke M.T. Avian eggshell mineralization: biochemical and functional characterization of matrix proteins. Comptes. Rendus. Palevol. 2004;3:549–562. [Google Scholar]
- Roberts J.R., Chousalkar K., Samiullah Egg quality and age of laying hens: implications for product safety. Anim. Prod. Sci. 2013;53:1291–1297. [Google Scholar]
- Roberts J. Burleigh Dodds Science Publishing; London, UK: 2017. Achieving Sustainable Production of Eggs. Volume 2. [Google Scholar]
- Romé H., Varenne A., Hérault F., Chapuis H., Alleno C., Dehais P., Vignal A., Burlot T., Le Roy P. GWAS analyses reveal QTL in egg layers that differ in response to diet differences. Genet. Sel. Evol. 2015;47:83. doi: 10.1186/s12711-015-0160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Qu L., Yi G., Yuan J., Duan Z., Shen M., Qu L., Xu G., Wang K., Yang N. Genome-wide association study revealed a promising region and candidate genes for eggshell quality in an F2 resource population. BMC Genom. 2015;16:565. doi: 10.1186/s12864-015-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Yang D., Sasaki O., Furukawa T., Nirasawa K. Mapping of quantitative trait loci affecting eggshell quality on chromosome 9 in an F2 intercross between two chicken lines divergently selected for eggshell strength. Anim. Genet. 2009;40:779–782. doi: 10.1111/j.1365-2052.2009.01914.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Sasaki O., Nirasawa K., Furukawa T. Association between ovocalyxin-32 gene haplotypes and eggshell quality traits in an F 2 intercross between two chicken lines. Anim. Genet. 2010;41:541–544. doi: 10.1111/j.1365-2052.2010.02034.x. [DOI] [PubMed] [Google Scholar]
- Takasuga A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016;87:159–167. doi: 10.1111/asj.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemoto Y., Suzuki C., Sato S., Sato S., Ohtake T., Sasaki O., Takahashi H., Kobayashi E. Polymorphism of the ovocalyxin-32 gene and its association with egg production traits. Poult. Sci. 2009;88:2512–2517. doi: 10.3382/ps.2009-00331. [DOI] [PubMed] [Google Scholar]
- VanRaden P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J.E., O’Sullivan N.P., Preisinger R., Habier D., Fernando R., Garrick D.J., Hill W.G., Dekkers J.C.M. Genome-wide association analysis and genetic architecture of egg weight and egg uniformity in layer chickens. Anim. Genet. 2012;43:87–96. doi: 10.1111/j.1365-2052.2012.02381.x. [DOI] [PubMed] [Google Scholar]
- Wolc A., Arango J., Jankowski T., Dunn I., Settar P., Fulton J.E., O’Sullivan N.P., Preisinger R., Fernando R.L., Garrick D.J., Dekkers J.C.M. Genome wide association study for egg production and quality in layer chickens. J. Anim. Breed. Genet. 2014;131:173–182. doi: 10.1111/jbg.12086. [DOI] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J.E., O’Sullivan N.P., Dekkers J.C.M., Fernando R., Garrick D.J. Mixture models detect large effect QTL better than GBLUP and result in more accurate and persistent predictions. J. Anim. Sci. Biotechno. 2016;7:7. doi: 10.1186/s40104-016-0066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., O’Sullivan N.P., Dekkers J.C.M. Repeatability vs. multiple-trait models to evaluate shell dynamic stiffness for layer chickens. J. Anim. Sci. 2017;95:9–15. doi: 10.2527/jas.2016.0618. [DOI] [PubMed] [Google Scholar]
- Wolc A., Stricker C., Arango J., Settar P., Fulton J.E., O’Sullivan N.P., Habier D., Fernando R., Garrick D.J., Lamont S.J., Dekkers J.C.M. Breeding value prediction for production traits in layers using pedigree and marker based methods. Genet. Sel. Evol. 2011;43:5. doi: 10.1186/1297-9686-43-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.C., Ning Z.H., Xu G.Y., Hou Z.C., Yang N. Heritabilities and genetic and phenotypic correlations of egg quality traits in brown-egg dwarf layers. Poult. Sci. 2005;84:1209–1213. doi: 10.1093/ps/84.8.1209. [DOI] [PubMed] [Google Scholar]